Abstract
Near-infrared spectroscopy (NIRS) has become a popular tool for investigating phenotypic variability in plants. We developed the Shiny NIRSpredict application to get predictions of 81 Arabidopsis thaliana phenotypic traits, including classical functional traits as well as a large variety of commonly measured chemical compounds, based from near-infrared spectroscopy values based on deep learning. It is freely accessible at the following URL: https://shiny.cefe.cnrs.fr/NirsPredict/.
NIRSpredict has three main functionalities. First, it allows users to submit their spectrum values to get the predictions of plant traits from models built with the hosted A. thaliana database. Second, users have access to the database of traits used for model calibration. Data can be filtered and extracted on user’s choice and visualized in a global context. Third, a user can submit his own dataset to extend the database and get part of the application development.
NIRSpredict provides an easy-to-use and efficient method for trait prediction and an access to a large dataset of A. thaliana trait values. In addition to covering many of functional traits it also allows to predict a large variety of commonly measured chemical compounds. As a reliable way of characterizing plant populations across geographical ranges, NIRSpredict can facilitate the adoption of phenomics in functional and evolutionary ecology.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05776-0.
Keywords: Arabidopsis thaliana, Functional traits, Genetic variability, Machine learning, Phenomics, Secondary metabolites, Trait prediction
Introduction
Plant traits are key to characterize biodiversity from a functional perspective [1, 2]. However, measuring traits that describe adaptive strategies on many individuals remains laborious. The development of near-infrared spectroscopy (NIRS) has provided a powerful tool enabling the collection of plenty of traits non-destructively [3–6], but which, until recently, required complex, sophisticated, and dataset-dependent statistical analyses.
NIRS is a non-invasive analytical technique that uses light in the near-infrared region of the electromagnetic spectrum (typically between 700 and 2500 nanometers) to analyze the chemicals composition of a sample. A sample is exposed to near-infrared light, which is differentially absorbed depending on the structure and composition of samples. In turn, the shape of the electromagnetic spectrum can be used to predict sample structure and chemical composition. Spectral information is exploited through the development of calibration models relating spectra and reference trait data, then new sample’s trait values are predicted using these models. In the 1980s and 1990s, NIRS became increasingly used as an analytical tool in a variety of fields, including food science, agriculture, pharmaceuticals, and biomedical research [5, 7–9]. The development of portable and handheld NIRS instruments in the 2000s further expanded the range of applications for NIRS. For instance, NIRS has become a popular tool for investigating phenotypic variability in plants [6, 10–12]. By analyzing the spectrum, one can quantify specific plant features, such as protein and carbohydrate concentrations, secondary metabolites, as well as physiological and morphological traits [6, 10–12]. NIRS is a particularly powerful tool because it is rapid, non-destructive, and requires minimal sample preparation. However, the negative side of the coin is that trait estimation from NIRS requires complex statistical methods that often represent a bottleneck for robust and generalizable predictions.
Until recently, statistical methods such as PCA, least squares, and support vector machines (SVM), were mostly used to exploit NIRS data and predict trait value from spectra [13–15]. However, these methods exhibit some weaknesses. First, NIRS data typically consists of numerous highly- correlated features, as wavelength range and resolution of commercial spectrometers allow for a highly multivariate signal. However, conventional methods like PLSR or PCA often imply dimension reduction, leading to a loss of information, and these methods may struggle to effectively extract the pertinent features from the spectral data [16]. Moreover, the variability associated with spectroscopic measure result in a highly noisy signal. To deal with it, PLSR usually relies on pre-treatment and removal of spectral outliers, which depends on user-guided decisions (as we lack a quantifiable method for determining a robust number of latent variables in PLSR [17]), and which can lead to loss of information, particularly when working with small datasets. The escalating array of potential pre-treatment techniques leads to the development of numerous approaches for identifying the most suitable approach to eliminate noise and linearize the signal (e.g [18–20]. , . Thus, selecting the “good” model for each dataset is highly time-consuming, and it represents a strong bottleneck for analyzing different datasets from various sources. Finally, the functional properties of a biological samples (e.g., a leaf) arise from complex non-linear relationships or threshold effects between traits, which are hardly modeled by reference methods such as PLSR [21, 22].
An alternative approach is the use of deep learning algorithms. Unlike statistical methods like PLSR, deep learning algorithms are particularly efficient in filtering input signals, which therefore requires less pre-treatment and human arbitrary intervention. For instance, Cui and Fearn [23] illustrated how the training of convolutional neural network (CNN) was able to mimic data preprocessing by continuously tuning the variables. They argued that because of the greater flexibility of convolutional layers, their algorithm was more efficient at finding the best form of preprocessing, saving time and effort compared to manual trial and error. In addition, deep learning enables to deal with noisy signals allowing for data augmentation to increase robustness (e.g., robust loss function and early stopping). Finally, deep networks proved that they can avoid dimensionality problems [24] including multiple useful techniques to deal with the overfitting risk and nonlinearity issues (e.g., batch normalization, dropout, early stopping and noise generation).The combination between NIRS and deep learning computation has been shown to be a powerful method to measure phenotypic traits including plant morphology, chemistry, and metabolism [25]. Moreover, this approach has allowed to capture a range of ecological information on plant diversity and may leads to the creation of extensive trait databases [26, 27]. To our knowledge, no study has yet been published that (1) utilizes deep-learning approaches to predict functional and metabolomic traits across multiple genotypes within a single species, and (2) develops an open-access web interface for making deep-learning-based predictions using both built-in and new models. This represents a significant advancement in the field and provides a powerful tool for the extensive community of scientists working on Arabidopsis thaliana.
Here we introduce NIRSpredict, an interactive web tool containing a database built from 5,325 unique spectra and 81 trait measurements from Arabidopsis thaliana plants grown in various conditions. This plant species was chosen because it is widely used in molecular biology and population genetics [28–30]. Plenty of natural ecotypes have been fully sequenced to examine the genetic determinism of trait variation and local adaptation (1001 Genomes Consortium [31]), . Moreover, A. thaliana exhibits a large range of functional trait variation across its geographic range [32–38]. Gathering more phenotypic information on this species, in wild and laboratory populations, is critical for the understanding of plant physiological regulation, trait diversity and local adaptation. In this context, NIRSpredict fulfills the needs for an automatic way to get predictions out of NIRS measures on A. thaliana without high knowledge in deep-learning.
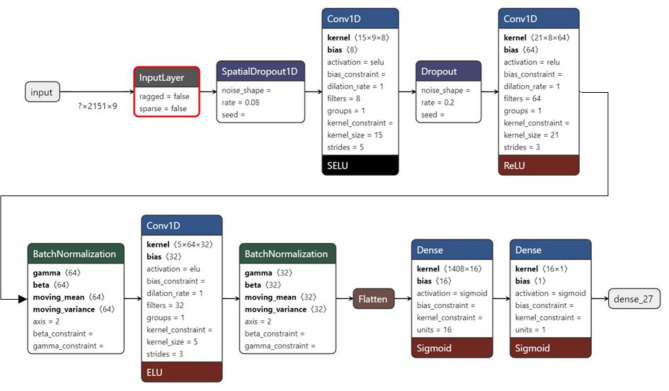
NIRSpredict (https://shiny.cefe.cnrs.fr/NirsPredict/) is a R shiny [39] application designed to make use of a large NIRS values database and predict phenotypic traits of A. thaliana by submitting NIRS spectra, using a hyperparametrized CNN approach comprising three convolutional layers followed by two layers of fully connected neural networks (see details of the CNN in Fig. 1). NIRSpredict allows users to (a) predict phenotypic traits related to leaf metabolism, physiology, and morphology using a large trait database available in A. thaliana [25]; (b) consult, visualize, and download subset of the database through filters and extend the database with submission of new datasets. NIRSpredict is not only an open access tool allowing consultation of a huge trait-linked NIRS values database, but also a modern trustful solution to make spectral predictions thanks to deep-learning algorithms (Fig. 2).
Fig. 1.
Diagram of the architecture of the neural network used by NIRSpredict. Diagram of the architecture of the convolutional neural network used to calibrate Arabidopsis thaliana near-infrared spectra prediction models. It was generated using Netron. Roeder, L. (2023, November 15). lutzroeder/netron GitHub repository. Retrieved from https://github.com/lutzroeder/netron
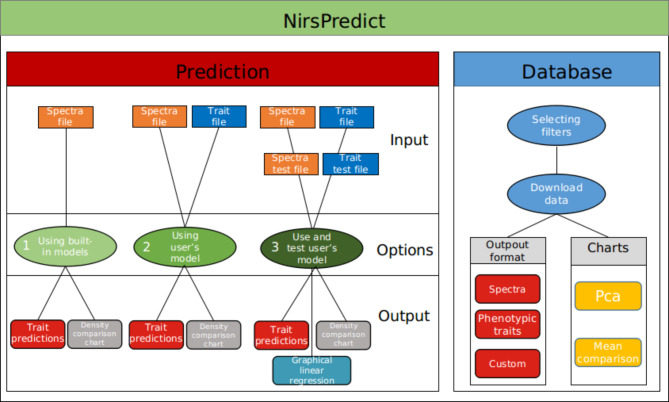
Fig. 2.
Representative diagramm of NIRSpredict features. Schema of the application showing how the application works. The available running options of the prediction features are represented with their needed input and their expected output. The database query process is represented with the available data format and the associated charts
Why and how using NIRSpredict ?
Obtain NIRS-based predictions of many phenotypic traits at different scales
The first functionality of NIRSpredict (‘Predict trait’ tab) is to submit your NIRS data set to get predictions of 81 phenotypic traits (Additional file 1), including 13 functional traits, 19 sugars, 5 hormones, 24 glucosinolates and 19 other secondary metabolites. To do so, it is possible to choose among three options according to your needs (Fig. 3a). Depending on the chosen option, one or more files can be uploaded (a valid email address must be provided to launch the job). Then, phenotypic traits of interest can be chosen among two lists, one for functional traits and one for metabolites. Once the job is complete, an email containing all the results is sent. The content of the results will depend on the selected option. It will always contain a csv file with predicted trait values, as well as a graphic representation of density comparison between users’ values and those available in the database. It can also contain graphical analysis of the predictions: a linear regression of predictions values and their residuals (Fig. 4). To predict trait values, users can choose among three options:
Fig. 3.
Screenshots showing the NIRSpredict main pages in use. (a) Predictions tab with uploaded files and fields fulfilled ; (b) Result of a query on the database matching water stress/high temperature treatment condition with graphics outputs. Mean absorption comparison between the subset and the database values. Principal component analysis of the subset spectrum values compared to the database values one
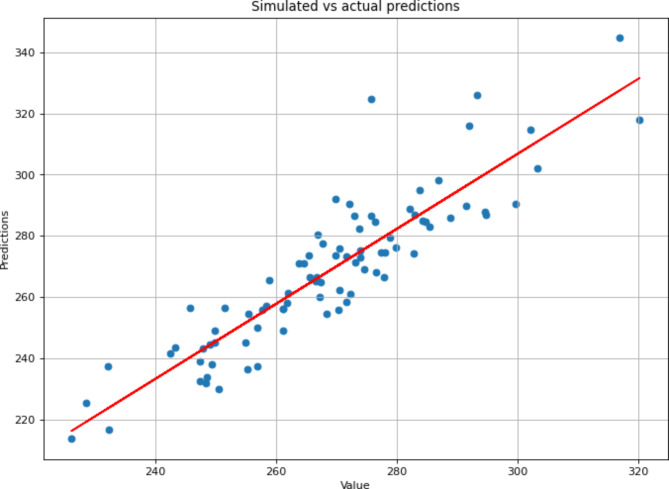
Fig. 4.
Graphical analysis associated to the predictions. Linear regression of predictions values. Predicted values have been obtained following the creation of a model and the use of a calibration and a validation dataset. The predicted values are compared to the observed values for the leaf thickness trait. The values follow the x = y pattern thus showing a correct prediction accuracy
The first option (‘Predict traits from built-in models’) allows to use trait-specific models already trained with the NIRSpredict database. In that case, only spectrum data are needed from users. A csv file must be prepared containing all spectrum data, i.e. with wavelengths from 350 to 2,500 with headers (see template in Additional file 2). Each individual is thus represented by 2,151 value of absorbance in line corresponding to a wavelength in column with a header. If the submitted file contains less than 2,151 values, the dataset will be re-sampled to match the fitted model but a minimum of 400 values is needed. A file with missing values will not be accepted since no data completion nor gap-filling method is performed in the application. The csv file can then be uploaded by clicking the ‘Browse’ button in the ‘Upload spectrum CSV file’ section.
The second option (‘Predict traits with your own model’) allows to create and use new models trained on the provided data set. As for the first option, a spectrum csv file must be supplied, and a second file containing trait values should also be prepared. This file will be split into a training and a testing dataset used for training and test the model. It is mandatory to have as many values in the spectrum file as in the trait file, and the rows need correspond to each other. It is possible to make predictions for only one trait or multiple ones since the traits file can contain from one to several columns. The file should be organized in columns whose headings correspond to the trait names. As with the first file, missing values are not accepted. The csv file can then be uploaded by clicking the ‘Browse’ button in ‘Upload traits CSV file’ section.
The third option (‘External validation’) is very similar to option 2, but it allows users to use an independent dataset (validation file) for external validation of models built on their own data (as in option 2). In this case, testing and training files must be prepared following the same rules (see above). The testing file must contain exactly the same traits in the same order as the training one. Here, we used the spectral and phenotypic data from the AraDiv public dataset [40] to test prediction robustness on independent measurements of SLA and LDMC in A. thaliana. Importantly, plants from the AraDiv dataset have been grown outside, in small pots with low-nutrient soil [40], i.e. in very different conditions than most plants used to build the NIRSpredict database. Yet, we found high prediction accuracy (predicted-versus-observed R2 = 0,89 for both SLA and LDMC, Additional file 3), which suggest that the NIRS-based predictions are robust to substantial variation in growth and measurement conditions.
Explore and extract data from the database
The second functionality of NIRSpredict (‘Explore database’ tab) allows to consult the trait database used to make predictions through NIRS and deep-learning algorithms. This database is made of the 5,325 A. thaliana individuals with spectra and 81 phenotypic trait values (Table 1). The search can be specified through several filters (Fig. 3b). Moreover a graphical analysis of the filtered samples will be printed with a mean comparison, a PCA, and finally the result of the search can be downloaded in multiple formats. The results of the search will be downloadable as a csv file, but its content depends on the output format chosen. Four options are available. First, ‘All data’ allows to get spectrum values and trait values of the individuals corresponding to the chosen filters. Second, ‘Spectrum only’ allows to get only spectrum values while the third one, ‘Functional traits only’ allows to get only trait values. Finally, (‘Additional traits’ allows to choose which traits should appear in the downloadable result file.
Table 1.
Summary of the database content. Detailed description of each experiment present in the database. Some genotypes are common between experiments and only unique ones are counted as total. Same for the traits
| Experiment name | Reference | IndOut | Condition | Treatment | Spectra | Genotypes | Traits |
|---|---|---|---|---|---|---|---|
| Exp1_Arabreed_2018 | Unpublished | Outdoor | Common garden | Control | 958 | NA | 81 |
| Herbivory | |||||||
| Water stress | |||||||
| Water stress / Herbivory | |||||||
| Exp2_Arabreed_2019 | Unpublished | Outdoor | Common garden | Control | 226 | NA | 7 |
| Herbivory | |||||||
| Water stress | |||||||
| Water stress / Herbivory | |||||||
| Exp3_Arabreed_2018 | Unpublished | Indoor | Growth chamber | Control | 702 | NA | 13 |
| Exp4_2017 | Unpublished | Indoor | Growth chamber | Control | 129 | 11 | 60 |
| Exp5_2018 | Unpublished | Indoor | Greenhouse | Control | 62 | 10 | 9 |
| Exp6_2015 | Vasseur et al. 2018 | Indoor | Greenhouse | Control | 687 | 209 | 9 |
| Herbivory | |||||||
| Exp7_2019 | Estarague et al. 2022 | Indoor | Greenhouse | Control | 1646 | 30 | 10 |
| High Temperature | |||||||
| Low Temperature | |||||||
| Water stress / High Temperature | |||||||
| Water stress / Low Temperature | |||||||
| Exp8_2017 | Sartori et al. 2022 | Indoor | Greenhouse | Control | 877 | 146 | 9 |
| Exp9_2017 | Unpublished | Outdoor | Wild | NA | 38 | NA | 8 |
| Total | 2 | 4 | 8 | 5325 | 340 | 81 |
Once the filters are established and the output format is chosen, the ‘Submit’ button should be pressed to visualize the graphical analysis of the corresponding samples (after a short loading time, a ‘Download’ button will appear at the bottom of the page to get the expected data set). A comparison between the chosen subset and the database content is visualized through (a) a representation of mean absorption and (b) a principal component analysis of spectrum values. (Fig. 3b).
Provide data to the NIRSpredict database
The third functionality of NIRSpredict (‘Become a Contributor’ tab) allows users to submit their own data set by uploading them to the administrators of NIRSpredict to be integrated into the database. As in the first functionality, a csv file must be prepared containing all spectrum data corresponding to a wavelength from 350 to 2,500 with headers. Each individual should be represented by 2,151 values of absorbance in line corresponding to a wavelength in column with a header. There is no size constraint in term of number of spectra. Once the file is ready, it can be uploaded by clicking the ‘Browse’ button in ‘Upload CSV file’ section. An email address needs to be provided so that data providers can be contacted for further information by the NIRSpredict administrators. The process is finalized when the ‘Send’ button is clicked and a confirmation message appears on the screen.
Discussion
By analyzing the NIRSpredict database and predicting traits with built-in models, we showed that NIRS accurately predicts most functional traits (Table 2). For instance, only five plant traits have a validation R² below 0.65 (Table 2). Correlations between measured and predicted values were the highest for leaf traits associated with resource-use strategies [41, 42], such as specific leaf area, leaf dry matter content and leaf nitrogen content (all R² > 0.79; Table 2). Consistently, Ecarnot and colleagues found similar prediction accuracy of NIRS for leaf traits (nitrogen content and dry mass per area) in wheat [43], using PLSR-based approaches rather than deep-learning. This suggests that NIRS-based approach can be similarly developed in different species, including crop species for which it can be a valuable tool for breeding [44, 45]. In addition, using 15 functional and metabolomic traits, it has been proven that deep learning methods outperform PLSR approach (Additional file 4). Moreover, NIRS also allows predicting a large variety of commonly measured chemical compounds. Our results show that prediction accuracy is highly variable among metabolites: validation R² ranged from 0% for the poorest predictions (e.g., glucoalysiin, gluconapin, progoitrin, see Table 2) to 90% (glucoerucin; Table 2). For sugars, the best predictions were obtained for fructose, galactose, melezitose, melibiose and raffinose (Table 2). Overall, the variation observed here in predictive power of metabolites and chemical composition of the leaves mirror the variation reported in the literature [4, 45–48]. For instance, Petit Bon and colleagues [47] found relatively high prediction accuracy for nutrient content across contrasted plant species, while Galvez-Sola and colleagues found weak predictions for many nutrients, including B, Cu, and Mn [46]. Yet, even the abundance in trace elements, such as Zn, Cu, and Mn, can be well predicted with NIRS in legume [49]. Interestingly, studies suggest that NIRS is able to predict pathogen attack in asymptomatic leaves [50, 51], which works particularly well when associated with machine learning approaches [52]. Here, we found that glucosinolates, a class of metabolites involved in plant defense against herbivores [53], showed relatively high prediction accuracy (e.g., glucoerucin and glucoraphenin with R² > 0. 75; Table 2) leading to a potential prediction of plant response to herbivore and pathogen attack at low cost. Accordingly, the jasmonic acid (JA), a hormone involved in plant response to pathogen and herbivores, was satisfactorily predicted by NIRS (R² =0.56; Table 2), although other hormones like auxin (IAA) and abscisic acid (ABA) had a weak validation R² (respectively 0.13 and 0.12; Table 2).
Table 2.
Predictions accuracy for traits and coverage
| Category | Trait name | R² | MSE | Number of calibration data | Coverage percentage |
|---|---|---|---|---|---|
| Functional traits | Leaf dry matter content (mg g-1) | 0.79 | 337.2 | 2836 | 53.3 |
| Specific leaf area (mm² mg-1) | 0.83 | 61.2 | 3399 | 63.8 | |
| Leaf nitrogen content (%) | 0.85 | 0.6 | 1958 | 36.8 | |
| Leaf thickness (µm) | 0.81 | 1162.7 | 2513 | 47.2 | |
| Leaf relative water content (%) | 0.18 | 29.9 | 1285 | 24.1 | |
| Leaf carbon content (%) | 0.45 | 3.56 | 1905 | 35.8 | |
| δ13C | 0.69 | 0.73 | 1218 | 22.9 | |
| δ15N | 0.19 | 4.23 | 1170 | 22 | |
| Plant life span (days) | 0.18 | 88.7 | 1398 | 26.3 | |
| Plant relative growth rate (mg d-1) | 0.69 | 178581.7 | 700 | 13.2 | |
| C score (%) | 0.88 | 14.7 | 2902 | 54.5 | |
| R score (%) | 0.70 | 44.38 | 2737 | 51.4 | |
| S score (%) | 0.00 | 202040307.5 | 2472 | 46.4 | |
| Sugars | Arabinose | 0.00 | 104531.2 | 105 | 1,97 |
| Cellobiose | 0.03 | 0.00 | 104 | 1,95 | |
| Fructose | 0.32 | 54.4 | 159 | 2,99 | |
| Fucose | 0.05 | 106810.3 | 111 | 2,08 | |
| Galactose | 0.39 | 0.05 | 149 | 2,8 | |
| Glucose | 0.12 | 51.5 | 160 | 3 | |
| Inositol | 0.08 | 0.20 | 116 | 2,18 | |
| Isomaltose | 0.12 | 442295.9 | 114 | 2,14 | |
| Maltose | 0.21 | 0.00 | 146 | 2,74 | |
| Mannose | 0.00 | 0.00 | 33 | 0,62 | |
| Melezitose | 0.37 | 876,203 | 158 | 2,97 | |
| Melibiose | 0.51 | 0.00 | 113 | 2,12 | |
| Palatinose | 0.01 | 0.00 | 112 | 2,1 | |
| Raffinose | 0.46 | 0.46 | 114 | 2,14 | |
| Rhamnose | 0.12 | 3646980.8 | 97 | 1,82 | |
| Ribose | 0.17 | 152967.5 | 102 | 1,92 | |
| Sucrose | 0.21 | 23.1 | 114 | 2,14 | |
| Trehalose | 0.21 | 0.00 | 108 | 2,03 | |
| Xylose | 0.11 | 0.00 | 113 | 2,12 | |
| Hormones | ABA | 0.12 | 12.4 | 142 | 2,67 |
| CMLX | 0.00 | 151220.5 | 143 | 2,69 | |
| IAA | 0.13 | 95 | 152 | 2,85 | |
| JA | 0.56 | 13135.9 | 155 | 2,91 | |
| SA | 0.00 | 39487.8 | 132 | 2,48 | |
| Glucosinolates | Butyl | 0.38 | 9.8 | 156 | 2,93 |
| Epigallocatechin | 0.39 | 18586.1 | 164 | 3,08 | |
| Epiprogoitrin | 0.17 | 30431788.3 | 162 | 3,04 | |
| Glucoalysiin | 0.00 | 44.4 | 142 | 2,67 | |
| Glucobrassicin | 0.12 | 536790.1 | 156 | 2,93 | |
| Glucoerucin | 0.90 | 0.06 | 145 | 2,72 | |
| Gluconapin | 0.00 | 16251280.3 | 160 | 3 | |
| Gluconasturtiin | 0.00 | 62.3 | 147 | 2,76 | |
| Glucoraphanin | 0.26 | 250.7 | 146 | 2,74 | |
| Glucoraphenin | 0.77 | 0.55 | 157 | 2,95 | |
| Glucosinalbin | 0.00 | 10.6 | 147 | 2,76 | |
| Hexyl | 0.01 | 182.3 | 143 | 2,69 | |
| Isobutyl | 0.00 | 154789.3 | 159 | 2,99 | |
| Negoclubrassicin Peak 1 | 0.36 | 10065.5 | 151 | 2,84 | |
| Neoglucabrassicin Peak 2 | 0.12 | 20491.1 | 147 | 2,76 | |
| Progoitrin | 0.00 | 68268.5 | 142 | 2,67 | |
| Sinigrin | 0.06 | 8346258.7 | 149 | 2,8 | |
| X3MTP | 0.33 | 4.26 | 148 | 2,78 | |
| X5MTP | 0.38 | 3.36 | 140 | 2,63 | |
| X6MSH | 0.12 | 781.9 | 150 | 2,82 | |
| X7MSH | 0.00 | 27169.3 | 153 | 2,87 | |
| X7MTH | 0.00 | 9199.5 | 153 | 2,87 | |
| X8MSO | 0.02 | 2395944.7 | 162 | 3,04 | |
| X8MTO | 0.38 | 571412.8 | 158 | 2,97 | |
| Other secondary metabolites | Apigenin rutinoside | 0.45 | 279839.3 | 158 | 2,97 |
| Caffeic acid | 0.29 | 0.71 | 162 | 3,04 | |
| Chlorogenic acid | 0.38 | 81.4 | 151 | 2,84 | |
| Citrat | 0.29 | 2695015.9 | 163 | 3,06 | |
| Cyanidin rhamnoside | 0.38 | 603090.3 | 155 | 2,91 | |
| Cyanidin sophorosid glucoside | 0.14 | 108530.9 | 155 | 2,91 | |
| Dihydro caffeoyl glucuronide | 0.74 | 154.7 | 159 | 2,99 | |
| Fumarat | 0.09 | 30909.8 | 153 | 2,87 | |
| Kaempherol glucosyl rhamnosyl glucoside | 0.10 | 189910.4 | 151 | 2,84 | |
| Kaempherol rutinoside | 0.63 | 1581260.3 | 160 | 3 | |
| Kaempherol xylosyl rhamnoside | 0.59 | 345149.6 | 156 | 2,93 | |
| Malat | 0.09 | 719645.7 | 163 | 3,06 | |
| mCoumaric acid | 0.13 | 360.7 | 150 | 2,82 | |
| pCoumaric Acid | 0.13 | 0.78 | 146 | 2,74 | |
| Pelargonidin cumaroyl diglucoside glucoside | 0.54 | 404.2 | 149 | 2,8 | |
| Pelargonidin samubioside | 0.34 | 22259.7 | 154 | 2,89 | |
| Prenyl naringenin | 0.45 | 265.6 | 160 | 3 | |
| Quercetin glucoside | 0.00 | 476.4 | 146 | 2,74 | |
| Succinat | 0.00 | 2138.9 | 163 | 3,06 |
R2: coefficient of determination; MSE : mean squared error ; The number of calibration data represents the number of spectra associated with trait values for the each traits; The coverage percentage represents the ratio between the spectra available for each trait against the total number of spectra
See (Vasseur et al. 2022) for a detailed description of each traits
The “Predict traits with built-in models” is convenient for predicting traits in A. thaliana samples grown and measured in relatively similar conditions than the samples of the present database [25]. In our external validation procedure, we obtained high prediction accuracy for leaf traits (Additional file 3), even if the plants used for external validation were grown in very contrasted conditions compared to plants of the database. However, although the database is composed of many measurements performed in various conditions [25], and although deep-learning models are rather robust to variations beyond the training dataset [54], the quality of trait prediction will inevitably fall with increasing sources of differences between hosted and provided datasets. Moreover, prediction accuracy is quite low for certain traits, particularly for metabolomic traits such as some glucosinolates and other secondary metabolites. However, instead of setting an arbitrary threshold to classify predictions as “accurate” or “meaningless,” we prefer to inform users about the variability in prediction accuracy, allowing users to determine what is acceptable based on their specific research questions and topics. In addition, it is important to note that deviation between datasets can be caused by different NIRS measurement devices (we used a LabSpec 4 spectrometer; ASD Inc., Analytik Ltd, UK), extremely stressing conditions (beyond the range of stresses in the database), specific genotypes (e.g., phenotypically altered mutants), and different sampling measurement protocols (e.g., leaf versus other organs, living tissues versus dry powder).
Hopefully, the “Predict traits with your own model” option can extend the NIRSpredict operability by giving the opportunity for the user to generate a new predictive model from fully provided data. This can be useful for analyzing datasets obtained on A. thaliana samples that may strongly deviate from those contained in the database. Moreover, this option can be used to predict traits beyond the range of genotypes and conditions hosted in the database. For instance, we used the second option of NIRSpredict to train independent models using the Aradiv dataset [40]. This external validation, with traits collected on plants grown under conditions outside of the range of the database, showed that prediction accuracy is higher when a model trained on these external data is regenerated (predicted-versus-observed R2 = 0.94 for SLA and 0.96 for LDMC, compared to 0.89 with built-in models, Additional file 3 & 5). This suggests two things: first, that the hosted database allows making predictions with high accuracy on external datasets, and second, that generating new models can be a powerful approach, even on very different, out-of-domain, NIRS data. This option is nonetheless designed for large datasets, because the prediction accuracy is linked to the size of the provided dataset. For instance, predictions made with a spectra dataset with less than one hundred individuals might not be very reliable.
Future developments can be extended to a wider range of species. We are currently working on integrating other species, notably those for which NIRS is already used in routine, such as wheat, maize, sorghum, and tomato [55, 56]. Such extension of the database will be released as soon as we have gathered enough traits and spectra in various environmental conditions. Once implemented, it will be possible to select one species from a list and choose which trait to predict among the associated ones. It would be even possible with the right calibration and validation dataset to predict values of a specific trait missing from the database through the automatic model training and building. Currently, the range of traits covered by the application is only made up of quantitative ones but with more developed algorithms it could be possible to predict qualitative traits such as the identity of a genotype. In fact, qualitative features such as survival rate or genotypes have already been predicted in A. thaliana and maize with NIRS [25, 57]. Furthermore, it would even be possible to predict environmental features and growth conditions through an automatic way leading to an improved understanding of the species environmental niche and stress response.
Overall, NIRSpredict provides an interactive tool that allows a huge saving of time and efforts by providing an automatic way of predicting plant functional traits or metabolite concentration. Using convolutional neural network led to predictions with an improved robustness than usual statistical methods, like PLSR, can provide. This approach also avoids risk of information loss induced by manual pre-treatment and arbitrary removal of outliers. Moreover, NIRSpredict compiles thousands of trait values acquired from A. thaliana plants grown in a large set of environmental conditions into a public and reliable database, thus making access to these data easier. In brief, NIRSpredict provides a reliable way of characterizing plant populations across geographical ranges and may facilitate the adoption of phenomics by functional and evolutionary ecologists.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thanks the Platform Systèmes d’Information en Écologie (SIE) of the Centre d’Écologie Fonctionnelle et Évolutive (CEFE CNRS), Montpellier, France for hosting the application and for the maintenance of the shiny server. We are grateful to the “Terrain d’expériences” platform at CEFE for supporting the conception and execution of the experiments.
Author contributions
FV, CV and DV conceived the ideas and collected the data.AV, GB, DC and LR analyzed the data.AV led the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Funding
This work was supported by INRAE, CNRS, the French Agency for Research (ANR grant ANR-17-CE02-0018-01, ‘AraBreed’), and the European Research Council (‘PHENOVIGOUR’: grant ERC-StG-2020-949843).
Data availability
Trait values are publicly available in the NIRSpredict database at https://shiny.cefe.cnrs.fr/NirsPredict/. For unpublished data, the identifier of the genotype is not accessible to users. The R code of the application is available on a GitHub repository at : https://github.com/AxelVaillant/NirsPredict. We do not have any conflict of interest.
License: CC-BY-4.0.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Violle C, Navas M-L, Vile D, Kazakou E, Fortunel C, Hummel I, et al. Let Concept Trait be Functional! Oikos. 2007;116:882–92. 10.1111/j.0030-1299.2007.15559.x. [Google Scholar]
- 2.Garnier E, Navas M-L, Grigulis K. Plant Functional Diversity Organism traits, community structure, and ecosystem properties. 2016. 10.1093/acprof:oso/9780198757368.001.0001
- 3.Foley WJ, Aragones L. Ecological applications of near infrared re¯ectance spectroscopy ± a tool for rapid, cost-effective prediction of the composition of plant and animal tissues and aspects of animal performance n.d.:13. [DOI] [PubMed]
- 4.Cozzolino D, Fassio A, Gimenez A. The use of near-infrared reflectance spectroscopy (NIRS) to predict the composition of whole maize plants. J Sci Food Agric. 2001;81:142–6. . [DOI] [Google Scholar]
- 5.Pasquini C. Near infrared spectroscopy: a mature analytical technique with new perspectives – A review. Anal Chim Acta. 2018;1026:8–36. 10.1016/j.aca.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Silva-Perez V, Molero G, Serbin SP, Condon AG, Reynolds MP, Furbank RT, et al. Hyperspectral reflectance as a tool to measure biochemical and physiological traits in wheat. J Exp Bot. 2018;69:483–96. 10.1093/jxb/erx421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepherd KD, Walsh MG. Infrared spectroscopy—enabling an evidence-based Diagnostic Surveillance Approach to Agricultural and Environmental Management in developing countries. J Near Infrared Spectrosc. 2007;15:1–19. 10.1255/jnirs.716. [Google Scholar]
- 8.Wójcicki K. Application of nir spectroscopy for whisky identification and determination the content of ethanol, 2015.
- 9.Biancolillo A, Marini F. Chemometric methods for spectroscopy-based Pharmaceutical Analysis. Front Chem 2018;6. [DOI] [PMC free article] [PubMed]
- 10.Arslan M, Xiaobo Z, Xuetao H, Elrasheid Tahir H, Shi J, Khan MR, et al. Near infrared spectroscopy coupled with chemometric algorithms for predicting chemical components in black goji berries (Lycium Ruthenicum Murr). J Near Infrared Spectrosc. 2018;26:275–86. 10.1177/0967033518795597. [Google Scholar]
- 11.Burnett AC, Serbin SP, Davidson KJ, Ely KS, Rogers A. Detection of the metabolic response to drought stress using hyperspectral reflectance. J Exp Bot. 2021;72:6474–89. 10.1093/jxb/erab255. [DOI] [PubMed] [Google Scholar]
- 12.Kothari S, Beauchamp-Rioux R, Laliberté E, Cavender-Bares J. Reflectance spectroscopy allows rapid, accurate, and non-destructive estimates of functional traits from pressed leaves 2022:2021.04.21.440856. 10.1101/2021.04.21.440856
- 13.Wold S, Martens H, Wold H. The multivariate calibration problem in chemistry solved by the PLS method. In: Kågström B, Ruhe A, editors. Matrix pencils. Volume 973. Berlin, Heidelberg: Springer Berlin Heidelberg; 1983. pp. 286–93. 10.1007/BFb0062108. [Google Scholar]
- 14.Cheng J-H, Sun D-W. Food Eng Rev. 2017;9:36–49. 10.1007/s12393-016-9147-1. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle.
- 15.Fu P, Meacham-Hensold K, Guan K, Wu J, Bernacchi C. Estimating photosynthetic traits from reflectance spectra: a synthesis of spectral indices, numerical inversion, and partial least square regression. Plant Cell Environ. 2020;43:1241–58. 10.1111/pce.13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le BT. Application of deep learning and near infrared spectroscopy in cereal analysis. Vib Spectrosc. 2020;106:103009. 10.1016/j.vibspec.2019.103009. [Google Scholar]
- 17.Mishra P, Passos D. A synergistic use of chemometrics and deep learning improved the predictive performance of near-infrared spectroscopy models for dry matter prediction in mango fruit. Chemometr Intell Lab Syst. 2021;212:104287. 10.1016/j.chemolab.2021.104287. [Google Scholar]
- 18.Li X, Zhang L, Zhang Y, Wang D, Wang X, Yu L, et al. Review of NIR spectroscopy methods for nondestructive quality analysis of oilseeds and edible oils. Trends Food Sci Technol. 2020;101:172–81. 10.1016/j.tifs.2020.05.002. [Google Scholar]
- 19.Cozzolino D. An overview of the use of infrared spectroscopy and chemometrics in authenticity and traceability of cereals. Food Res Int. 2014;60:262–5. 10.1016/j.foodres.2013.08.034. [Google Scholar]
- 20.Roggo Y, Chalus P, Maurer L, Lema-Martinez C, Edmond A, Jent N. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J Pharm Biomed Anal. 2007;44:683–700. 10.1016/j.jpba.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Cook RD, Forzani L. PLS regression algorithms in the presence of nonlinearity. Chemometr Intell Lab Syst. 2021;213:104307. 10.1016/j.chemolab.2021.104307. [Google Scholar]
- 22.Balabin RM, Safieva RZ, Lomakina EI. Comparison of linear and nonlinear calibration models based on near infrared (NIR) spectroscopy data for gasoline properties prediction. Chemometr Intell Lab Syst. 2007;88:183–8. 10.1016/j.chemolab.2007.04.006. [Google Scholar]
- 23.Cui C, Fearn T. Modern practical convolutional neural networks for multivariate regression: applications to NIR calibration. Chemom. Intell Lab Syst. 2018;182:9–20. 10.1016/j.chemolab.2018.07.008. [Google Scholar]
- 24.Poggio T, Mhaskar H, Rosasco L, Miranda B, Liao Q. Why and when can deep-but not shallow-networks avoid the curse of dimensionality: a review. Int J Autom Comput. 2017;14:503–19. 10.1007/s11633-017-1054-2. [Google Scholar]
- 25.Vasseur F, Cornet D, Beurier G, Messier J, Rouan L, Bresson J, et al. A perspective on Plant Phenomics: Coupling Deep Learning and Near-Infrared Spectroscopy. Front Plant Sci. 2022;13:836488. 10.3389/fpls.2022.836488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L, Zhang C, Taha M, Qiu Z, He Y. Determination of Leaf Water Content with a portable NIRS System based on Deep Learning and Information Fusion Analysis. Trans ASABE. 2021;64:127–35. 10.13031/trans.13989. [Google Scholar]
- 27.Ma T, Tsuchikawa S, Inagaki T. Rapid and non-destructive seed viability prediction using near-infrared hyperspectral imaging coupled with a deep learning approach. Comput Electron Agric. 2020;177:105683. 10.1016/j.compag.2020.105683. [Google Scholar]
- 28.Chan EKF, Rowe HC, Hansen BG, Kliebenstein DJ. The Complex Genetic Architecture of the Metabolome. PLoS Genet. 2010;6:e1001198. 10.1371/journal.pgen.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tohge T, Borghi M, Fernie AR. The natural variance of the Arabidopsis floral secondary metabolites. Sci Data. 2018;5:180051. 10.1038/sdata.2018.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S, Tohge T, Cuadros-Inostroza Á, Tong H, Tenenboim H, Kooke R, et al. Mapping the Arabidopsis Metabolic Landscape by untargeted metabolomics at different environmental conditions. Mol Plant. 2018;11:118–34. 10.1016/j.molp.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, et al. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–91. 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasky JR, Marais D, Mckay DL, Richards JK, Juenger JH, T. E.&, Keitt TH. Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Mol Ecol. 2012;21:5512–29. 10.1111/j.1365-294X.2012.05709.x. [DOI] [PubMed] [Google Scholar]
- 33.May R-L, Warner S, Wingler A. Classification of intra-specific variation in plant functional strategies reveals adaptation to climate. Ann Botany. 2017;119:1343–52. 10.1093/aob/mcx031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price N, Moyers BT, Lopez L, Lasky JR, Monroe JG, Mullen JL et al. Combining population genomics and fitness QTLs to identify the genetics of local adaptation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2018;115:5028–33. 10.1073/pnas.1719998115 [DOI] [PMC free article] [PubMed]
- 35.Takou M, Wieters B, Kopriva S, Coupland G, Linstädter A, De Meaux J. Linking genes with ecological strategies in Arabidopsis thaliana. J Exp Bot. 2019;70:1141–51. 10.1093/jxb/ery447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasseur F, Sartori K, Baron E, Fort F, Kazakou E, Segrestin J, et al. Climate as a driver of adaptive variations in ecological strategies in Arabidopsis thaliana. Ann Botany. 2018;122:935–45. 10.1093/aob/mcy165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sartori K, Vasseur F, Violle C, Baron E, Gerard M, Rowe N, et al. Leaf economics and slow-fast adaptation across the geographic range of Arabidopsis thaliana. Sci Rep. 2019;9:10758. 10.1038/s41598-019-46878-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Estarague A, Vasseur F, Sartori K, Bastias CC, Cornet D, Rouan L, et al. Into the range: a latitudinal gradient or a center-margins differentiation of ecological strategies in Arabidopsis thaliana? Ann Botany. 2022;129:343–56. 10.1093/aob/mcab149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang W, Cheng J, Allaire J, Sievert C, Schloerke B, Xie Y, Allen J, McPherson J, Dipert A, Borges B. (2023). shiny: Web Application Framework for R. https://shiny.posit.co/, https://github.com/rstudio/shiny
- 40.Przybylska MS, Violle C, Vile D, Scheepens JF, Lacombe B, Le Roux X, et al. AraDiv: a dataset of functional traits and leaf hyperspectral reflectance of Arabidopsis thaliana. Sci Data. 2023;10:314. 10.1038/s41597-023-02189-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich PB, Ellsworth DS, Walters MB, Vose JM, Gresham C, Volin JC, et al. Generality of Leaf Trait relationships: a test across six biomes. Ecology. 1999;80:1955–69. 10.2307/176671. [Google Scholar]
- 42.Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, et al. The worldwide leaf economics spectrum. Nature. 2004;428:821–7. 10.1038/nature02403. [DOI] [PubMed] [Google Scholar]
- 43.Ecarnot M, Compan F, Roumet P. Assessing leaf nitrogen content and leaf mass per unit area of wheat in the field throughout plant cycle with a portable spectrometer. Field Crops Res. 2013;140:44–50. 10.1016/j.fcr.2012.10.013. [Google Scholar]
- 44.Cabrera-Bosquet L, Crossa J, von Zitzewitz J, Serret MD, Araus JL. High-throughput phenotyping and genomic selection: the frontiers of crop breeding converge. J Integr Plant Biol. 2012;54:312–20. 10.1111/j.1744-7909.2012.01116.x. [DOI] [PubMed] [Google Scholar]
- 45.Cabrera-Bosquet L, Sánchez C, Rosales A, Palacios-Rojas N, Araus JL. Near-Infrared Reflectance Spectroscopy (NIRS) assessment of δ(18)O and nitrogen and ash contents for improved yield potential and drought adaptation in maize. J Agric Food Chem. 2011;59:467–74. 10.1021/jf103395z. [DOI] [PubMed] [Google Scholar]
- 46.Galvez-Sola L, García-Sánchez F, Pérez-Pérez JG, Gimeno V, Navarro JM, Moral R et al. Rapid estimation of nutritional elements on citrus leaves by near infrared reflectance spectroscopy. Front Plant Sci 2015;6. [DOI] [PMC free article] [PubMed]
- 47.Petit Bon M, Böhner H, Kaino S, Moe T, Bråthen KA. One leaf for all: Chemical traits of single leaves measured at the leaf surface using near-infrared reflectance spectroscopy. Methods Ecol Evol. 2020;11:1061–71. 10.1111/2041-210X.13432. [Google Scholar]
- 48.Prananto JA, Minasny B, Weaver T. Rapid and cost-effective nutrient content analysis of cotton leaves using near-infrared spectroscopy (NIRS). PeerJ. 2021;9:e11042. 10.7717/peerj.11042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cozzolino D, Moron A. Exploring the use of near infrared reflectance spectroscopy (NIRS) to predict trace minerals in legumes. Anim Feed Sci Technol. 2004;111:161–73. 10.1016/j.anifeedsci.2003.08.001. [Google Scholar]
- 50.Spinelli F, Noferini M, Costa G. Near Infrared spectroscopy (NIRS): perspective of fire blight detection in asymptomatic plant material. Acta Hort. 2006;704:87–90. 10.17660/ActaHortic.2006.704.9. [Google Scholar]
- 51.Gold KM, Townsend PA, Chlus A, Herrmann I, Couture JJ, Larson ER, et al. Hyperspectral measurements enable pre-symptomatic detection and differentiation of contrasting physiological effects of late blight and early blight in Potato. Remote Sens. 2020;12:286. 10.3390/rs12020286. [Google Scholar]
- 52.Fearer CJ, Conrad AO, Marra RE, Georskey C, Villari C, Slot J et al. A combined approach for early in-field detection of beech leaf disease using near-infrared spectroscopy and machine learning. Front Forests Global Change 2022;5.
- 53.Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J. Disarming the mustard oil bomb. Proc Natl Acad Sci USA. 2002;99:11223–8. 10.1073/pnas.172112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voulgaris G, Philippides A, Quadrianto N. Deep Learning Robustness to Domain Shifts During Seasonal Variations. IGARSS 2022–2022 IEEE International Geoscience and Remote Sensing Symposium, 2022, pp. 417–20. 10.1109/IGARSS46834.2022.9883940
- 55.Furbank RT, Silva-Perez V, Evans JR, Condon AG, Estavillo GM, He W, et al. Wheat physiology predictor: predicting physiological traits in wheat from hyperspectral reflectance measurements using deep learning. Plant Methods. 2021;17:108. 10.1186/s13007-021-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Abid MA, Rasheed A, Crossa J, Hearne S, Li H. DNNGP, a deep neural network-based method for genomic prediction using multi-omics data in plants. Mol Plant. 2023;16:279–93. 10.1016/j.molp.2022.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Rincent R, Charpentier J-P, Faivre-Rampant P, Paux E, Le Gouis J, Bastien C, et al. Phenomic selection is a low-cost and high-throughput method based on indirect predictions: Proof of Concept on Wheat and Poplar. G3 Genes|Genomes|Genetics. 2018;8:3961–72. 10.1534/g3.118.200760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Cheng J-H, Sun D-W. Food Eng Rev. 2017;9:36–49. 10.1007/s12393-016-9147-1. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle.
Supplementary Materials
Data Availability Statement
Trait values are publicly available in the NIRSpredict database at https://shiny.cefe.cnrs.fr/NirsPredict/. For unpublished data, the identifier of the genotype is not accessible to users. The R code of the application is available on a GitHub repository at : https://github.com/AxelVaillant/NirsPredict. We do not have any conflict of interest.
License: CC-BY-4.0.