Abstract
Introduction
Age-related hearing loss (ARHL) is a degenerative condition that involves both peripheral and central auditory system pathologies. There is a close relationship between chronic inflammation and ARHL, but there is currently a lack of in-depth exploration of this relationship, particularly regarding causality.
Methods
Using age-appropriate mice for basic experiments to examine changes in central auditory nervous system inflammation, a cohort study was conducted to select relevant clinical data and observe inflammation changes in the elderly population with ARHL. Mendelian randomization was employed to investigate the causal relationship between chronic inflammation and ARHL.
Results
Clinical results indicate that CRP levels in the ARHL group are significantly higher than those in the normal group. Chronic inflammation also occurs in the auditory centers. Mendelian randomization studies provide causal evidence that genetic chronic inflammation factors do not increase the risk of ARHL.
Discussion
This article provides reliable causal evidence of the relationship between chronic inflammation and ARHL, confirming the accumulation of inflammatory factors in the auditory center, which provides a basis for the prevention and treatment of ARHL and has a good prevention prospect. However, due to the differences of research objects, this study has limitations and needs further research.
Keywords: CRP, TNF-α, chronic inflammation, age-related hearing loss, Mendelian randomization
Introduction
Age-related hearing loss (ARHL), also known as presbycusis, is the most common sensory disorder in the elderly and a common degenerative disease in the elderly. In an aging society, ARHL will impose a significant social and welfare burden.1 Therefore, effective treatment interventions are urgently needed to improve hearing loss associated with aging. From the basic research on ARHL, it is found that ARHL is a major hearing impairment characterized by pathological changes of peripheral and central auditory system,2 and neuronal degeneration is the main sign of ARHL.3 All animal models with early onset ARHL had mild inflammation in the cochlear, suggesting that inflammation was involved in the occurrence and development of ARHL.4,5 Meanwhile, Seicol BJ further found that the activation of cochlear nucleus (CN) microglia and C1q deposition increased significantly.6 This suggests that both the immune system and the complement system are activated during ARHL development. Further studies should examine whether inflammatory activation or hyperactivation of microglia during ARHL, like peripheral inflammation, is a detrimental and complex self-program. And the activation and localization of C1q deposition and other components of the classical complement cascade in the auditory center. But it is currently unclear whether this neuroinflammatory response occurs in the auditory central cochlear nucleus.
Recent cohort studies have found a significant association between chronic inflammation and ARHL. The primary biomarkers for this association include C-reactive protein (CRP), tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and complement component 3 (C3).7,8 Elevated levels of these biomarkers in the elderly population suggest that chronic inflammation plays a role in the process of hearing loss. However, the causal relationship between chronic inflammation and ARHL remains unclear. Clarifying this causal relationship is essential, which requires not only basic and clinical experiments but also genetic causal relationship studies. We employ Mendelian randomization as our research method.
Mendelian randomization (MR) is an analytical method used to evaluate the causal relationship between observed modifiable exposures or risk factors and clinically relevant outcomes. It provides a valuable approach, especially when randomized controlled trials (RCTs) to examine causal relationships are impractical, and observational studies may offer biased associations due to confounding or reverse causation. Additionally, the extensive availability of published genetic associations makes Mendelian randomization a time-efficient and cost-effective method, contributing to its growing popularity in assessing and screening potential causal relationships.9 In this study, our primary objective is to establish a causal relationship between inflammation, particularly chronic inflammation, and ARHL, thereby providing a reliable basis for the prevention and treatment of ARHL. Our secondary objective is to demonstrate the relationship between central nervous system inflammation and ARHL through foundational experiments and clinical data.
Experimental Method
Experimental Animals
C57BL/6 mice were chosen as the experimental subjects. These mice serve as a model for ARHL and effectively simulate the progression of hearing loss with aging. This model is widely recognized and accepted internationally.
The C57BL/6 mice were divided into young group (4 weeks) and old age group (48 weeks), and 40 mice in each group. The sample size of mice was calculated in accordance with ARRIVE guidelines. Considering the rarity of cochlear nucleus of each mouse and the need for repeatability of basic experiments, 40 mice were included in each group. The body weights of the two groups of C57BL/6 mice were 19.8±0.4g and 29.8±0.3g and sensitive to sound were purchased from Hangzhou Hangse Biotechnology Co., LTD. All animals had no history of noise exposure, had never used ototoxic drugs, and had normal eardrums examined by otoscopy. All experiments were approved by the Animal Ethics Committee of Yuhuangding Hospital, Yantai, China. And follow the guidelines for the ethical Review of Laboratory Animals - Animal welfare established by the People’s Republic of China.
ABR Tests for Changes in Hearing
Detection of changes in the hearing level of mice 5 mice were randomly selected in each group to detect the hearing threshold with ABR.10 Mice were anesthetized by intraperitoneal injection of pentobarbital sodium and placed in a sound insulation shielding room. Electrodes were inserted under the skin of the head. The recording electrode was placed in the middle of the head, the reference electrode was placed behind the test ear, the earth pole was placed behind the opposite ear, and the afferent tubules were placed into the ear canal of the mice. Different frequencies of sound stimulation of 4kHz, 8kHz, 16kHz and 32kHz were given successively, and the intensity gradually decreased by 10dB, and the amplitude began to decrease by 5dB when the waveform began to smooth out. The hearing threshold of mice should be SPL when the wave crest disappeared.
Preparation of Frozen Sections of CN
Immunofluorescence was used to determine the expression of glial cell markers. According to the mouse heart perfusion method and the mouse cochlear nucleus microscope map in the previous literature, the mouse cochlear nucleus was located and frozen sections were prepared. Immunofluorescence was used to observe the relative expression of microglia surface markers (IBA1, CD16) and astrocyte markers (GFAP, C3) under fluorescence microscope in the two groups.
Frozen sections of the cochlear nucleus were washed with phosphate-buffered saline (PBS) three times, each for 5 minutes. They were then permeabilized with PBS containing 0.2% Triton X-100 for 15 minutes. After that, the sections were blocked with 2% BSA at room temperature for 1 hour. Primary antibodies IBA1, GFAP, CD16, and C3 were added and incubated overnight at 4°C. The next day, after bringing the sections back to room temperature, they were washed with PBS three times, each for 5 minutes. Corresponding fluorescent secondary antibodies were added and incubated at 37°C for 2 hours, followed by another three washes with PBS, each for 5 minutes. The nuclei were stained with DAPI, and the sections were mounted with an anti-fade mounting medium for observation under a fluorescence microscope.
Detection of Protein Expression Levels of Inflammatory Factors and Pathway Related Genes
The cochlear nucleus was peeled off under the microscope, the protein was cleaved and used to extract tissue RNA with TRIzol reagent for qPCR detection. The expression levels of inflammatory cytokines TNF-α, IL-1β and C3 were detected by qPCR.
Under sterile conditions, extract RNA or DNA samples and use reverse transcriptase to transcribe RNA into cDNA. Prepare qPCR reaction mixtures for TNF-α, IL-1β, and C3. Initiate the thermal cycling program, adjust parameters, and collect data. Finally, perform result analysis and validation.
Statistical Analysis
All data provided for statistical analysis represent ± average SEM results from at least three independent experiments. Statistical analysis was performed using t tests or using ANOVA. Statistical significance was defined as P<0. 05. The software used is GraphPadPrism8. 0. 1 and ImageJ-win64.
Screening of Clinical Data for ARHL
In order to explore the changes of inflammatory factors in ARHL patients, we included 20 patients in lesion group and 20 in control group from the hospital database, and detected the content of CRP in serum and performed statistics. The study was also approved by the Ethics committee of Yantai Yuhuangding Hospital. The sample size calculation followed the ARRIVE guidelines. We chose a small sample size with the initial aim of verifying the trend of chronic inflammation in the ARHL population, thereby providing direction for subsequent research. The screening criteria for patients with disease were as follows: 1. Age ≥60 years; 2. No history of noise exposure; 3, have hearing loss symptoms (high frequency hearing loss or full frequency hearing loss); 4, exclude external otitis, otitis media and other systemic inflammatory diseases; 5. Rule out systemic immune system diseases. The control group met the following criteria: 1. Age ≥60 years old; 2. No symptoms of hearing loss; 3. No history of noise exposure; 4. Exclude external otitis, otitis media and other systemic inflammatory diseases; 5. Rule out systemic immune system diseases. The data were summarized and analyzed by t test using GraphPadPrism8.0.1. p value <0. 05 was considered statistically significant.
Mendelian Randomization
Study Design
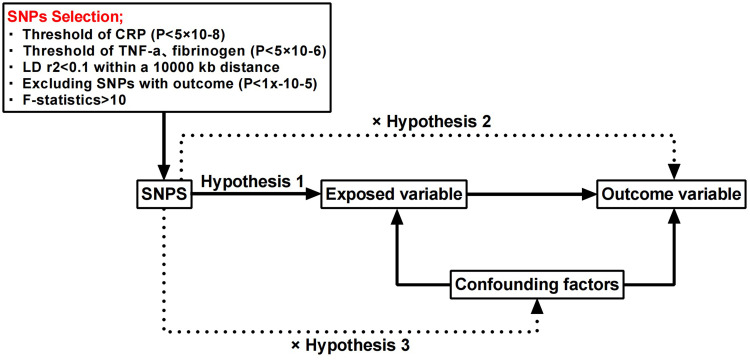
We used a two-sample MR Design to investigate the causal relationship between inflammatory markers and ARHL. A convincing MR design should be based on three key assumptions: (1) genetic variation is directly and strongly associated with exposure (markers of inflammation); (2) Instrumental variables were not affected by potential confounding factors associated with outcomes; (3) Genetic variation affects outcome only through exposure and not through other pathways (ARHL).11 The flowchart in Figure 1 Outlines this MR Approach with two samples. All summary data used in this study were publicly available, limited to European populations, and with appropriate ethical approval and informed patient consent from participants in all prior studies. This paper is published in accordance with the recommendations of the Statement on the Use of Mendelian Randomization to Enhance Reports of Observational Epidemiological Studies (STROBE-MR).12
Figure 1.
Two-sample MR Design to investigate the causal relationship between inflammatory markers and ARHL.
Instrumental Variables
SNP is a genetic instrumental variable (IVs) that acts as a proxy phenotype and is used for two-specimen MR Study. When CRP was used as an exposure, SNPs (p<5×10−8) were selected as an instrumental variable to achieve genome-wide significance. However, given the limited sample size and number of SNPS, when selecting SNPS associated with TNF-α and IL-1β, C3, we identified SNPS (p<5×10−6). In order to obtain independent SNPS, de-linkage unbalanced LD (r2<0. 01, kb=10000) was performed.13 To satisfy the third central hypothesis (that IV variants affect outcomes only through exposure), SNPS directly associated with hearing loss outcomes (p<1×10−5) were also removed in each analysis. Finally, we calculate the population F statistic: F=beta2/se.2 14 To further evaluate the strength of instrumental variables, the F statistic for each SNP was calculated, where instrumental variables with F<10 (considered as weak instrumental variables) were excluded. Summary data on expose-associated single nucleotide polymorphisms and their association with ARHL can be found in Tables 1 and 2.
Table 1.
Details of the SNPS Included in the Mendelian Randomization
| Detection Component | SNP | EA | EAF | Association with Detection Component | Association with ARHL | ||||
|---|---|---|---|---|---|---|---|---|---|
| β | SE | P | β | SE | P | ||||
| CRP | rs10240168 | G | 0.257909 | −0.028684 | 0.004343 | 4.11E-11 | 0.0117831 | 0.0261876 | 0.652746 |
| rs10512597 | C | 0.768532 | 0.036931 | 0.00489 | 4.44E-14 | 0.0210036 | 0.0270267 | 0.437075 | |
| rs1051338 | G | 0.35308 | 0.023881 | 0.003992 | 2.27E-09 | −0.00186362 | 0.0240407 | 0.93821 | |
| rs10521222 | T | 0.0258353 | −0.104411 | 0.010714 | 2.06E-22 | −0.0217164 | 0.0740045 | 0.76918 | |
| rs10778215 | A | 0.371688 | −0.033242 | 0.003583 | 1.86E-20 | −0.00355655 | 0.0238069 | 0.881245 | |
| rs10778215 | A | 0.000396085 | −0.033242 | 0.003583 | 1.86E-20 | 0.70941 | 0.696334 | 0.308308 | |
| rs10832027 | A | 0.538471 | 0.025944 | 0.003745 | 4.43E-12 | 0.0397571 | 0.0229872 | 0.0837144 | |
| rs10925027 | C | 0.611794 | −0.036035 | 0.00382 | 4.25E-21 | 0.0125527 | 0.0235715 | 0.594355 | |
| rs11108056 | G | 0.427481 | −0.027907 | 0.003708 | 5.42E-14 | −0.0245612 | 0.0231579 | 0.288872 | |
| rs12202641 | T | 0.394157 | −0.022804 | 0.003617 | 3.00E-10 | −0.0126698 | 0.0234401 | 0.588838 | |
| rs12587622 | A | 0.503851 | −0.020798 | 0.003609 | 8.52E-09 | −0.0135293 | 0.022928 | 0.555139 | |
| rs1260326 | C | 0.650802 | −0.073462 | 0.003604 | 2.72E-92 | −0.0390344 | 0.0241131 | 0.105489 | |
| rs12960928 | C | 0.195944 | 0.024 | 0.003993 | 1.91E-09 | −0.0218956 | 0.0287909 | 0.446952 | |
| rs12995480 | C | 0.837018 | 0.031261 | 0.004855 | 1.24E-10 | 0.0381527 | 0.0309407 | 0.217542 | |
| rs13233571 | T | 0.127359 | −0.056895 | 0.005476 | 2.95E-25 | 0.00558468 | 0.0342117 | 0.87033 | |
| rs13409371 | A | 0.29575 | 0.048232 | 0.003847 | 5.07E-36 | −0.0212523 | 0.0252514 | 0.399995 | |
| rs1441169 | G | 0.585671 | −0.024926 | 0.003725 | 2.27E-11 | 0.0528676 | 0.0233173 | 0.0233711 | |
| rs1490384 | T | 0.478182 | −0.024816 | 0.003545 | 2.65E-12 | 0.0312399 | 0.0229537 | 0.173515 | |
| rs1509394 | T | 0.538142 | 0.025685 | 0.004147 | 6.05E-10 | 0.0561611 | 0.0230059 | 0.01464 | |
| rs1558902 | A | 0.419083 | 0.033926 | 0.003701 | 5.20E-20 | −0.0299746 | 0.0232584 | 0.197481 | |
| rs1582763 | A | 0.281736 | −0.022107 | 0.0037 | 2.37E-09 | −0.0355807 | 0.0254022 | 0.161306 | |
| rs17658229 | C | 0.0484773 | 0.055568 | 0.009522 | 5.50E-09 | 0.00353019 | 0.0531046 | 0.946999 | |
| rs178810 | T | 0.60701 | 0.02001 | 0.003606 | 2.95E-08 | −0.00226054 | 0.0234688 | 0.923266 | |
| rs1800961 | T | 0.0452826 | −0.1115 | 0.011267 | 4.63E-23 | 0.0679937 | 0.0554232 | 0.219895 | |
| rs1805096 | A | 0.469061 | −0.104381 | 0.003614 | 2.17E-183 | 0.0395712 | 0.0230224 | 0.0856485 | |
| rs1880241 | G | 0.38889 | −0.027537 | 0.003687 | 8.41E-14 | −0.0393131 | 0.0235165 | 0.094578 | |
| rs2064009 | T | 0.527094 | 0.027111 | 0.003549 | 2.28E-14 | 0.0395771 | 0.0230364 | 0.0857926 | |
| rs2239222 | G | 0.375498 | 0.035484 | 0.003901 | 9.87E-20 | 0.0108566 | 0.023755 | 0.647652 | |
| rs2293476 | C | 0.212578 | 0.030262 | 0.004225 | 8.27E-13 | 0.0731978 | 0.0278036 | 0.00847149 | |
| rs2315008 | G | 0.740503 | 0.023467 | 0.003777 | 5.36E-10 | 0.0133725 | 0.0260421 | 0.607605 | |
| rs2352975 | C | 0.253651 | 0.024897 | 0.004026 | 6.43E-10 | −0.00577308 | 0.0265613 | 0.827936 | |
| rs2710804 | C | 0.353826 | 0.021262 | 0.003737 | 1.30E-08 | 0.0403265 | 0.023943 | 0.0921298 | |
| rs2794520 | T | 0.365927 | −0.182186 | 0.003712 | 1.00E-200 | 0.0528871 | 0.0239253 | 0.0270695 | |
| rs2836878 | A | 0.247109 | −0.042902 | 0.004079 | 7.71E-26 | −0.0834991 | 0.0269033 | 0.00191135 | |
| rs2852151 | A | 0.438545 | 0.024735 | 0.003655 | 1.36E-11 | 0.0456995 | 0.0231939 | 0.0488011 | |
| rs2891677 | T | 0.53831 | 0.019859 | 0.003511 | 1.59E-08 | 0.0152484 | 0.0229053 | 0.505593 | |
| rs3122633 | C | 0.334778 | 0.027479 | 0.00389 | 1.68E-12 | 0.0396981 | 0.0243961 | 0.103688 | |
| rs3134899 | T | 0.687515 | 0.023329 | 0.004274 | 4.93E-08 | 0.0385508 | 0.0247872 | 0.119881 | |
| rs340005 | A | 0.722085 | 0.030007 | 0.003736 | 1.01E-15 | 0.0287263 | 0.025628 | 0.262332 | |
| rs387976 | C | 0.289331 | 0.025507 | 0.003946 | 1.05E-10 | 0.0130054 | 0.0252607 | 0.606658 | |
| rs4092465 | G | 0.623961 | 0.027483 | 0.004364 | 3.11E-10 | −0.0397178 | 0.0239287 | 0.096946 | |
| rs4129267 | T | 0.298688 | −0.087519 | 0.003612 | 1.20E-129 | −0.00210177 | 0.0250229 | 0.933061 | |
| rs4246598 | A | 0.622943 | 0.022063 | 0.003547 | 5.11E-10 | −0.0350167 | 0.0236748 | 0.13912 | |
| rs4420638 | G | 0.27165 | −0.229459 | 0.006122 | 1.00E-200 | −0.0769399 | 0.0266887 | 0.00394085 | |
| rs4655802 | A | 0.592882 | −0.025012 | 0.00416 | 1.88E-09 | −0.0247645 | 0.0235855 | 0.293723 | |
| rs4656849 | G | 0.689006 | 0.057656 | 0.003723 | 4.91E-54 | 0.0294445 | 0.0248076 | 0.235263 | |
| rs469772 | T | 0.212781 | −0.031327 | 0.004542 | 5.54E-12 | 0.0407837 | 0.0280192 | 0.145513 | |
| rs4767920 | A | 0.847532 | −0.038519 | 0.0049 | 4.00E-15 | −0.0443954 | 0.0318091 | 0.162811 | |
| rs4841132 | G | 0.856831 | 0.065095 | 0.006243 | 2.00E-25 | 0.0279679 | 0.0328151 | 0.394054 | |
| rs6001193 | G | 0.357687 | −0.027809 | 0.003706 | 6.53E-14 | 0.00512404 | 0.0239273 | 0.830429 | |
| rs6601302 | G | 0.732555 | −0.030518 | 0.004478 | 9.80E-12 | 0.0335811 | 0.0262994 | 0.201646 | |
| rs6672627 | A | 0.178939 | −0.037135 | 0.005083 | 2.89E-13 | 0.00752807 | 0.0302026 | 0.803165 | |
| rs7121935 | A | 0.341699 | −0.021853 | 0.00374 | 5.28E-09 | −0.00798505 | 0.0242044 | 0.741474 | |
| rs7310409 | G | 0.585655 | 0.137075 | 0.003706 | 1.00E-200 | −0.0438609 | 0.0234579 | 0.0615149 | |
| rs9271608 | G | 0.151706 | 0.042021 | 0.004954 | 2.33E-17 | −0.0151416 | 0.0324425 | 0.6407 | |
| rs9284725 | A | 0.872477 | −0.02731 | 0.00419 | 7.34E-11 | 0.0930957 | 0.0350255 | 0.00786194 | |
| rs10240168 | G | 0.257909 | −0.028684 | 0.004343 | 4.11E-11 | 0.0117831 | 0.0261876 | 0.652746 | |
| TNF-a | rs16938466 | T | 0.050541 | 0.4278 | 0.0927781 | 4.53E-06 | 0.0318709 | 0.0521172 | 0.540853 |
| rs16984398 | T | 0.0733842 | 0.3835 | 0.0796635 | 1.71E-06 | 0.0444996 | 0.0446333 | 0.318762 | |
| rs17119113 | G | 0.128217 | 0.3826 | 0.0766273 | 7.03E-07 | 0.0187464 | 0.0341943 | 0.583532 | |
| rs9808346 | A | 0.0171235 | 0.6353 | 0.137451 | 4.31E-06 | 0.000768886 | 0.0866581 | 0.992921 | |
| IL-1β | rs6102001 | T | 0.00535703 | 0.6057 | 0.128763 | 2.91E-06 | 0.0553659 | 0.15981 | 0.729006 |
| rs7298175 | A | 0.34477 | −0.2204 | 0.047716 | 4.37E-06 | 0.00503727 | 0.0240947 | 0.834401 | |
| C3 | rs11702778 | A | 0.381989 | −0.12144 | 0.0233053 | 1.88E-07 | 0.0114173 | 0.0236387 | 0.629103 |
| rs1351769 | A | 0.450311 | −0.115696 | 0.0227924 | 3.85E-07 | −0.0478406 | 0.0231226 | 0.0385461 | |
| rs148109646 | G | 0.0198793 | 0.491122 | 0.0986872 | 6.47E-07 | −0.0117514 | 0.0830749 | 0.88751 | |
| rs16893518 | A | 0.0310123 | 0.188153 | 0.0395855 | 2.00E-06 | 0.112192 | 0.0651153 | 0.0848946 | |
| rs61946396 | T | 0.0376351 | 0.312494 | 0.0681738 | 4.57E-06 | −0.0447086 | 0.0608681 | 0.462634 | |
| rs76695941 | T | 0.0194657 | −0.28386 | 0.0579346 | 9.60E-07 | 0.120848 | 0.0827731 | 0.144294 | |
| rs77562174 | C | 0.0372661 | 0.369173 | 0.0773841 | 1.84E-06 | −0.129182 | 0.0602878 | 0.0321329 | |
| rs8066176 | G | 0.36587 | 0.126138 | 0.0258012 | 1.01E-06 | −0.0254978 | 0.0238851 | 0.285737 | |
| rs8131444 | T | 0.179535 | −0.17986 | 0.0388564 | 3.68E-06 | 0.01441 | 0.0302143 | 0.633413 | |
Table 2.
MR Analyses Showing the Associations of Genetically Inflammatory Markers with the Risk of ARHL
| Detection Component | nSNP | Method | β | SE | P |
|---|---|---|---|---|---|
| CRP | 56 | Inverse variance weighted | 0.042634578 | 0.085064154 | 0.616226865 |
| MR Egger | −0.088860608 | 0.122862405 | 0.472828632 | ||
| TNF-a | 4 | Inverse variance weighted | 0.061801496 | 0.055889156 | 0.268818777 |
| MR Egger | −0.100935812 | 0.356448699 | 0.803665238 | ||
| C3 | 9 | Inverse variance weighted | -0.078582584 | 0.088645338 | 0.37535732 |
| MR Egger | -0.293898703 | 0.195053304 | 0.17559864 | ||
| IL-1β | 2 | Inverse variance weighted | −0.006112501 | 0.100996165 | 0.951739808 |
ARHL GWAS Data
Related to ARHL genetic information acquired from public Finngen GWAS summary data, these Numbers According to the can be accessed by GWAS directory (https://gwas.mrcieu.ac.uk/). The study’s recently published GWAS included 198,327 participants (1735 and 196,592 controls).
GWAS Data on Markers of Inflammation
CRP data were derived from the most extensive meta-analysis of the European Individual genome-wide Association Study (GWAS). The cohort study drew on 88 previous pooled statistics, including 204,402 people with a SNP number of 2,414,379. See Published GWAS. In addition, the SNP package for TNF-α, IL-1β was derived from GWAS collected from European subjects in the SuhreK study, with a number of SNPS of 501,428. The SNP package for C3 was derived from the European Individual Genome-Wide Association Study (GWAS), which resulted in a dataset containing 6240,610 SNPS from 3672 individuals with European genetic ancestry.
Statistical Analysis
We performed a two-sample random Mendelian effect analysis to assess causal effects between CRP, TNF-α, IL-1β, C3, and ARHL. First, we selected the genetic variance of exposure (CRP, TNF-α, IL-1β, C3) to the outcome (genetic variation of ARHL susceptibility), each snp represents a data point, and after reconciling the GWAS effect alleles of both before and after, we used two MR Methods to determine the MR Estimate. That is, IVW and MR-Egger because they have different basic assumptions about horizontal pleiotropy. P<0.05 was taken as a significant value in the primary analysis. The estimate produced by this method is expected to exceed the Wald ratio estimate of the variance, so we chose it as the primary method for this MR.15
In addition, MR-Egger can also be used as a supplement to IVW for sensitivity analysis to identify any deviations in the assessment of MR Assumptions. The MR-Egger analysis allows for pleiotropy of all genetic variants, but the magnitude of pleiotropy (from genetic variation to outcome, bypassing exposure) should be separated from the magnitude of the primary effect (from genetic variation to exposure). In addition, we also apply the MR-Egger intercept test to detect unbalanced horizontal pleiotropy. The intercept obtained from MR-Egger regression is an indicator of directional pleiotropy (p<0.05 is considered to be the existence of directional pleiotropy).16 For meaningful predictions, we use Cochrane Q values to assess heterogeneity by plotting the inverse distribution of the standard error for each SNP around the MR Estimates.17 In addition, we performed a leave-one analysis to assess whether MR Estimates were driven by a single SNP or biased. In the remain-one method, each SNP was sequentially removed, and the remaining SNPS were used to calculate the causal effect of gene predictive exposure (CRP, TNF-α, IL-1β, C3) on the outcome (ARHL).18
All MR Analyses in this study were performed using R software (version 4.2.1) and the “TwoSampleMR” software package (version 0.5.6).
Results
The results of ABR Indicate That Aging Causes Severe Hearing Loss in C57 Mice
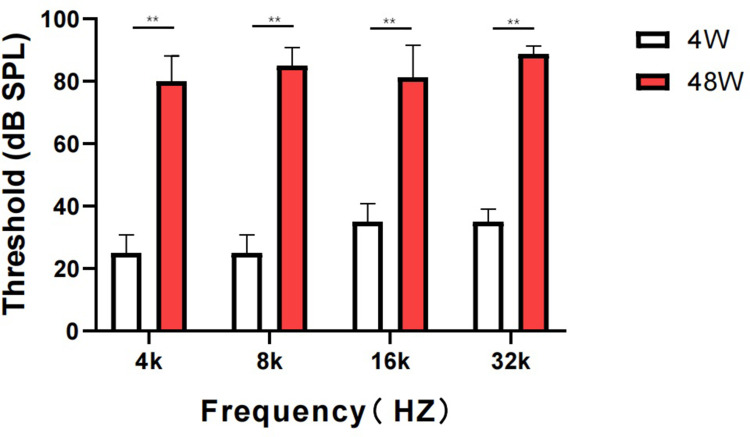
Aging causes around a 60 dB hearing loss between the youth group and old-aged group. The ABR threshold results for the youth group (4W) and old-aged group (48W) are shown in Figure 2. The average auditory thresholds of young mice were 20±5 dB at 4 kHz, 20±5 dB at 8 kHz, 25±5 dB at 16 kHz, and 30±5 dB at 32 kHz. For old-aged mice, the average thresholds were 70±5 dB at 4 kHz, 80±5 dB at 8 kHz, 90±5 dB at 16 kHz, and 90±5 dB at 32 kHz.
Figure 2.
ABR audiometry. Hearing thresholds were measured in mice aged 4 weeks and 48 weeks with different interference conditions using an auditory brain-stem response test. **P<0.005 compared with normal control 4 weeks mice at SPL, sound pressure level.
We observed that ARHL affected the threshold of the full-frequency hearing threshold, causing a severe hearing loss condition, showing a significant difference between aged and young mice.
qPCR Expression Results of Inflammatory Factors in Cochlear Nucleus and Microglia
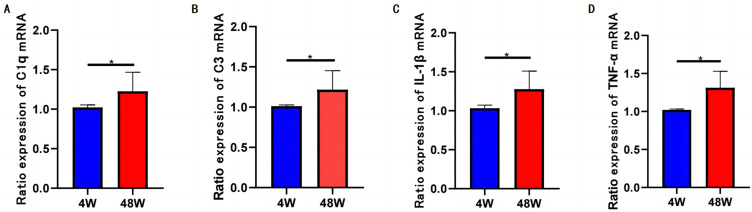
The expression levels of complement C1q factor, TNF-α, IL-1β and complement C3 factor (mainly secreted by astrocytes) in cochlear nucleus were significantly higher in the aged group than in the young group, indicating inflammatory deposition. As shown in Figure 3.
Figure 3.
The relative expression levels of cochlear inflammatory factors in 4W and 48W groups. C1q (A), complement C3 factor (B), IL-1β (C) and TNF-α (D).
This suggests the presence of inflammatory factor deposition and complement system activation in the auditory center of the brain, which appears to contribute to the mechanism of ARHL development.
Accumulation of Neuroinflammation in Cochlear Nucleus
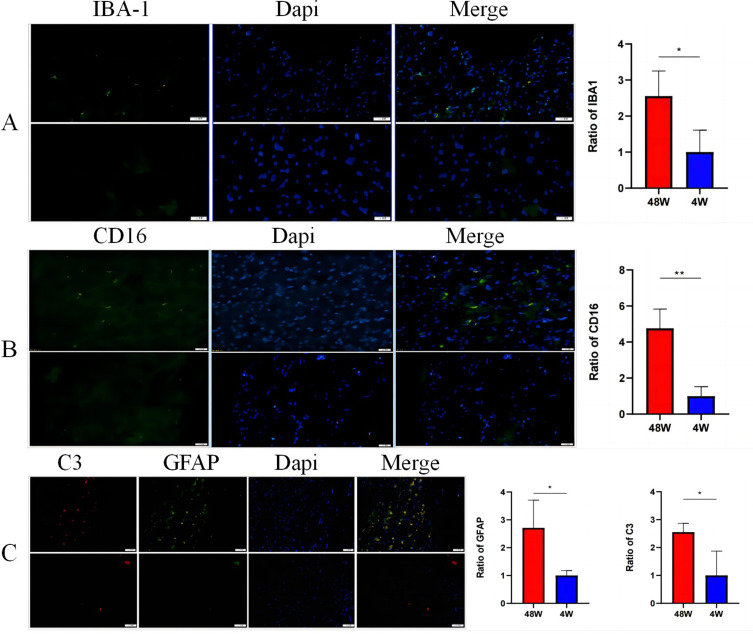
A number of studies have shown that neuroinflammation of microglia and astrocytes can cause the occurrence and development of neurodegenerative diseases. In this study, immunofluorescence detection showed that the immunofluorescence expression levels of iBA1 (microglial surface marker), CD16 (M1 microglial surface marker), GFAP (astrocyte surface marker) and C3 (mainly secreted by astrocytes) in the cochlear nucleus of the elderly group were much higher than those of the youth group. This suggests that neuroinflammation can affect the development of ARHL. As shown in Figure 4.
Figure 4.
Expression of markers in microglia and astrocytes. Comparison of IBA1 (A), CD16 (B), GFAP and C3 (C). (*P<0.05 and **P<0.005).
Clinical Data Verified the Serum Expression of CRP
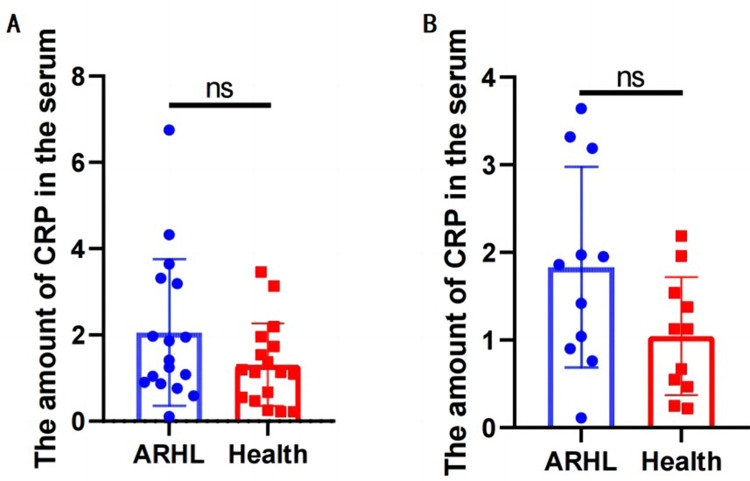
We conducted a t-test analysis on data from 20 ARHL patients and 20 control subjects. After excluding three outliers, the results are shown in Figure 5A. Considering the impact of gender on hearing,8 we excluded female patients from both groups and performed the t-test again, with results shown in Figure 5B. Both statistical analyses indicate that the average serum CRP levels in ARHL patients are higher than those in the healthy control group. However, due to the limited sample size, the statistical results are not significant, necessitating further large-scale analysis.
Figure 5.
Clinical data verified the serum expression of CRP. (A) Comparison of CRP content between ARHL group and normal elderly group. (B) After excluding the data of female patients, comparison of CRP content between ARHL group and normal elderly group.
So far, the relationship between inflammatory factors and ARHL remains unclear. Most previous epidemiological studies have been case-control designs, which fail to clarify causality due to ambiguous timelines. Even in prospective observational studies, some diagnosed cases of ARHL may have reverse effects, where undiagnosed ARHL could trigger inflammation-related factors. Thus, these factors may not predict the development and survival of ARHL but could be a result of it. In summary, there is no doubt that Mendelian randomization is needed to determine causality.
Causal Effects from CRP to ARHL
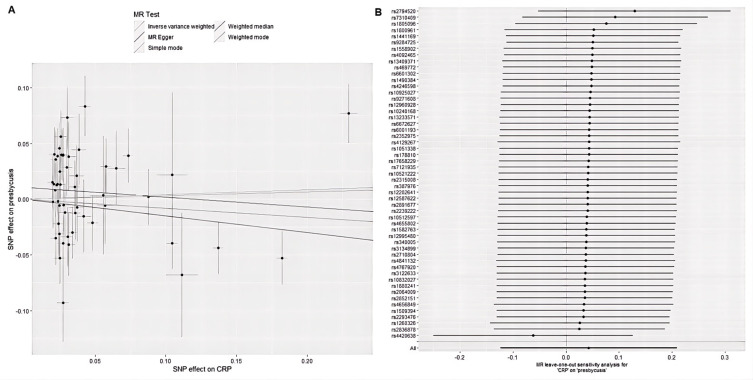
Using 56 SNPS associated with CRP, we found weak evidence of a potential causal effect of CRP on the risk of ARHL with no significant statistical significance (OR = 1.04, 95% CI = 0.88–1.23, P = 0.62). Meanwhile, a similar risk estimate was obtained using MR-Egger regression (OR = 1.24, 95% CI = 0.49–3.09, p = 0.653). Although the association was not statistically significant. However, we observed heterogeneity, with a p-value of 1.69×10−5 derived from the Cochran Q test and a P-value of 3.65×10−5 derived from the MR-Egger. But there was no evidence of significant interception (interception = 0.011, SE = 0.007, p = 0.148), indicating that no directed pleiotropy was observed, and pleiotropy tested p, indicating that genetically predicted CRP increases were not significantly associated with ARHL risk (Figure 6). The relationship between CRP and ARHL is shown in the figure below.
Figure 6.
Causal effects from CRP to ARHL. (A) Forest plot and (B) Scatter plot of the potential effects of CRP to ARHL.
In fact, no significant causal relationship was found between CRP and ARHL. In the sensitivity analysis, no single SNP strongly violated the overall effect of CRP on ARHL. Additionally, in the pleiotropy test, the p-values were symmetric, indicating the absence of pleiotropy.
Causal Effects from IL-1β, TNF-α, and C3 to ARHL
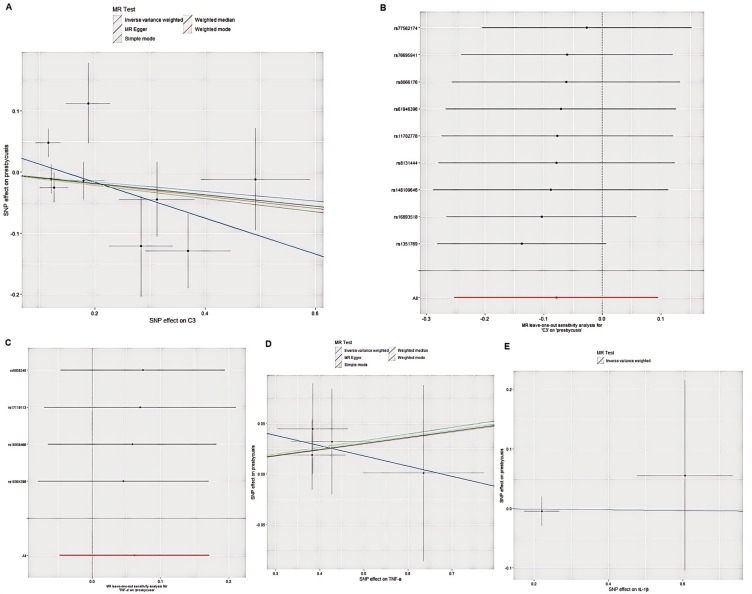
Since only two SNPS were found to be significantly and independently associated with IL-1β, only the IVW method was applied to them. We identified two SNPS (rs6102001 and rs7298175) that were significantly and independently associated with ARHL. The results of IVW showed that there was no causal relationship between TNF-α, IL-1β and C3 on ARHL (OR = 0.99, 95% CI = 0.82–1.21, P = 0.95). The P-value of Cochran Q test was 0.363, indicating that no significant heterogeneity was observed. Four SNPS were significantly independently associated with TNF-α. Nine SNPS were independently correlated with C3, and IVW and MR-Egger were used for statistical analysis. The results of IVW showed no causal relationship between TNF-α and C3 on ARHL (OR = 1.06, 95% CI = 0.95–1.19, P = 0.27; OR = 0.92, 95% CI = 0.78–1.10, P = 0.38). As shown in Figure 7.
Figure 7.
Causal effects from IL-1β, TNF-α, and C3 to ARHL. (A) Forest plot and (B) scatter plot of the potential effects of C3 to ARHL. (C) Scatter plot and (D) Forest plot of the potential effects of TNF-α to ARHL. (E) Forest plot of the potential effects of IL-1β to ARHL.
Causal Effects of CRP, IL-1β, TNF-α and C3 on Potential ARHL Risk Factors
A recent cohort study suggests that chronic inflammation may play a different role in ARHL incidence. To solve this problem, we used two sets of genetic instruments to perform MR Estimation, combined with basic experimental and clinical data, and selected representative inflammatory indicators, inflammatory factors and complement factors for analysis. The results showed that CRP, IL-1β, TNF-α and C3 had no causal relationship with the incidence of ARHL.
To determine whether the MR Association between genetically determinative inflammatory related factors and ARHL is violated by a pleiotropic pathway associated with ARHL. No causal effect of C3, IL-1β and TNF-α on potential risk factors for ARHL was observed. We used a two-sample MR Approach to comprehensively assess whether inflammatory factors have a causal effect on the incidence of ARHL, and found no clear evidence to support genetic prediction of the causal effect of inflammatory factors on the risk of ARHL. At the same time, we noted that Zhao Jingbo et al retrospectively analyzed 165 Chinese patients through an epidemiological study and were progressively screened by multivariate Logistic regression: age (OR=3.473,95% CI:=2.553–4.726), education level (OR=0.524, 95% CI=0.360–0.762), high total cholesterol (OR=4.271, 95% CI=1.853–9.844) and family history of deafness (OR=7.208,95% CI=3.846–13.509) were associated with the occurrence of ARHL. The results of Li Huan’s retrospective analysis of 1018 Chinese patients showed: old age (OR=2.356;95% CI=1.942–2.858), hyperlipidemia (OR=1.386;95% CI=1.012–1.896); Smoking (OR=1.636;95% CI=1.140–2.348), alcohol consumption (OR=1.985;95% CI =1.426–2.764), hypertension (OR=1.379;95% CI=1. 046–1.818) was a risk factor for presbycusis.
In summary, blood lipids, blood pressure, smoking, and age are closely related to age-related hearing loss (ARHL). Through the screening of relevant SNPs, we found that the SNPs associated with C3, IL-1β, and TNF-α showed no significant correlation with these exposure factors, indicating that our study results were not confounded by pleiotropy.
Discussion
Our study has several advantages. First, using the MR Design, our study can simulate a randomized controlled trial in an observational setting. Randomized controlled traits are widely accepted when studying causation, but are quite expensive and often impractical. But MR Studies can effectively avoid confounding bias in SNPS randomly assigned at the time of conception. In contrast to other observational studies, MR Can also avoid reverse causal effects. Second, our findings may have implications for healthcare policy on inflammation-related factors and ARHL. Given the high prevalence of inflammatory factors and ARHL in the general population, uncovering a causal relationship between inflammatory factors and ARHL could influence public health policies regarding early prevention and timely intervention. Our findings imply that enhanced screening for ARHL in patients with genetic predictors of inflammation-related factors may be useless. More attention should be paid to the association between environmentally determined inflammatory factors and the prognosis of ARHL.
Chronic inflammation not only contributes to the development of the disease, but also affects its prognosis. As the disease progresses, it increases the likelihood of microvascular damage and ischemia, promoting atherosclerosis.19 In the periphery, the cochlear is supplied by a single blood vessel (the labyrinth artery), and the hair cells of the cochlear are susceptible to hypoxia, which may lead to cochlear dysfunction.20 In the center, chronic neuroinflammation stimulates morphological changes in cochlear nucleus microglia, leading to neuronal degeneration and promoting disease development. CRP, a widely studied acute phase protein, may participate in the pathogenesis of atherosclerosis by activating complement, stimulating monocyte chemotaxis and inhibiting neutrophil chemotaxis through classical pathways.21 TNF-α has been identified as a representative of one of the most important regulators of the immune system, which can affect the activation of many intracellular signaling pathways through various pathways, ultimately leading to cell survival, cell migration, apoptosis, and necrosis.22 The cytokine IL-1β is a key mediator of inflammatory response. It is essential for host response and resistance to pathogens, but can also exacerbate damage during chronic disease and acute tissue injury.23 C3 is an important part of the innate immune system, which combines with other complement proteins to form the main mechanism of host detection and elimination of potential pathogens.24 These four factors that respond to inflammation and complement system function can all be detected in the blood, and the results of this work are consistent with previous studies, and the levels of CRP and TNF-α in ARHL patients are higher than those in the general population.8 However, in a recent study on hearing loss, only CRP was associated with the outcome of idiopathic hearing loss,25 which also suggests that inflammatory factors still have high research value in the field of hearing loss and need to be further studied. However, because most routine studies to date have been observational or cohort studies, susceptible to potential confounders and cause-and-effect reversals, as well as bias due to insufficient sample sizes, different diagnostic criteria for disease, and diverse populations that limit interpretation of results, It is therefore still unknown whether these three inflammatory markers play a role in triggering the risk of ARHL.
In summary, this is the first study to combine basic experimental, clinical and genetic data to study the complex relationship between CRP, TNF-α, IL-1β, C3 and ARHL, which has high research value and corroborates each other. The strength of this study is that it analyzed the correlation between these three factors and ARHL risk using large-scale GWAS data from European populations, reducing bias due to population stratification and making our statistical power very strong and convincing. The two-sample MR Design was also used to provide causal evidence that CRP was a risk factor for ARHL, while IL-1β, C3, and TNF-α were not significantly associated with ARHL, largely eliminating the influence of conventional study limitations.
Based on the results of this two-sample Mendelian randomization study, we can conclude that there is no direct causal relationship between major markers of chronic inflammation and ARHL. Increasing the screening of nucleotide sequences for chronic inflammation markers does not significantly aid in the prevention of ARHL. Extending this line of thought, we can consider whether these results might serve as a reference for predicting similar mechanisms in other conditions.
Although idiopathic sudden sensorineural hearing loss (SSNHL) is relatively rare, it poses significant challenges in diagnosis and treatment, potentially leading to severe consequences. Both ARHL and SSNHL share common inflammatory mechanisms, including biomarkers such as CRP, IL-1β, and TNF-α.26 Given that no causal relationship was found between certain inflammatory biomarkers and ARHL, it may be valuable to explore other inflammatory pathways or markers that could play a role in the progression of both ARHL and SSNHL. For example, the NF-kB pathway and the complement pathway may be worth exploring. The NF-kB pathway plays a crucial role in regulating inflammatory responses and could influence the progression of hearing loss. Similarly, the complement pathway is associated with immune responses and may contribute to inner ear damage. Investigating how these pathways affect ARHL and SSNHL could uncover new therapeutic targets and mechanisms, thereby providing novel insights for improving diagnosis and treatment.
However, some limitations in this study need to be considered. First, the GWAS participants in the study were limited to Europe, where big data is available, and it remains highly doubtful whether the same results can be extrapolated to other ethnic groups. Second, the lack of stratified data such as gender and age in existing aggregated statistical sets prevents us from conducting a comprehensive and more refined analysis. If more detailed publicly available stratified data becomes available in the future, this will allow new MR Analyses to be further normalized. Finally, the GWAS sample sizes of TNF-α, IL-1β and C3 were relatively small, which may lead to some bias in the results. Therefore, we should be cautious when interpreting negative results on the effects of TNF-α and IL-1β and C3 on ARHL risk. It is undeniable that conducting Mendelian randomization analysis on these inflammatory markers (TNF-α, IL-1β and C3) with relatively small GWAS sample sizes may affect the robustness of the conclusions drawn. We should note the diversity of patients with ARHL. Inflammation-related factors may have a causal relationship with certain ARHL. A more extensive study including the ARHL subgroup could be considered in the future.
Conclusion
In summary, this study uses Mendelian randomization to provide causal evidence that genetic chronic inflammation factors do not increase the risk of ARHL. However, clinical results indicate that patients with ARHL exhibit changes associated with chronic inflammation, and foundational experiments suggest that chronic inflammation may also affect the auditory center. These findings offer new insights into the relationship between inflammatory markers and ARHL, highlighting the need for further research to validate these observations.
Funding Statement
This work was supported in part by the National Natural Science Foundation of China (#82371153), the China Postdoctoral Science Foundation (#2023M731845), the Natural Science Foundation of Shandong Province (#ZR2021MH378 and #ZR2022QH073), and the Yantai Science and Technology Innovation Development Project (#2022YD009, #2023YD050).
Ethics Approval and Consent to Participate
This study met the ethical standards and was approved by the Ethics Committee of Yantai Yuhuangding Hospital. Approval NO is 2024-140. Ethical approval paper to upload as attachment content.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Stucky SR, Wolf KE, Kuo T. The economic effect of age-related hearing loss: national, state, and local estimates, 2002 and 2030. J Am Geriatr Soc. 2010;58:618–619. doi: 10.1111/j.1532-5415.2010.02746.x [DOI] [PubMed] [Google Scholar]
- 2.Bowl MR, Dawson SJ. Age-related hearing loss. Cold Spring Harb Perspect Med. 2019;9(8):a033217. doi: 10.1101/cshperspect.a033217 PMID: 30291149; PMCID: PMC6671929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keithley EM. Pathology and mechanisms of cochlear aging[J]. J Neurosci Res. 2020;98(9):1674–1684. doi: 10.1002/jnr.24439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu T, Zhou J, Qiu J, et al. Tumor necrosis factor-α mediated inflammation versus apoptosis in age-related hearing loss[J]. Front Aging Neurosci. 2022;14:956503. doi: 10.3389/fnagi.2022.956503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson N, Ding B, Zhu X, Frisina RD. Chronic inflammation - inflammaging - in the ageing cochlea: a novel target for future presbycusis therapy. Ageing Res Rev. 2017;40:142–148. [Epub 2017 Oct 7. PMID: 29017893; PMCID: PMC5675822]. doi: 10.1016/j.arr.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seicol BJ, Lin S, Xie R. Age-related hearing loss is accompanied by chronic inflammation in the cochlea and the cochlear nucleus. Front Aging Neurosci. 2022;14:846804. doi: 10.3389/fnagi.2022.846804 PMID: 35418849; PMCID: PMC8995794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verschuur C, Agyemang-Prempeh A, Newman TA. Inflammation is associated with a worsening of presbycusis: evidence from the MRC national study of hearing. Int J Audiol. 2014;53(7):469–475. doi: 10.3109/14992027.2014.891057 [DOI] [PubMed] [Google Scholar]
- 8.Verschuur CA, Dowell A, Syddall HE, et al. Markers of inflammatory status are associated with hearing threshold in older people: findings from the Hertfordshire ageing study. Age Ageing. 2012;41(1):92–97. doi: 10.1093/ageing/afr140 [DOI] [PubMed] [Google Scholar]
- 9.Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27(11):3253–3265. doi: 10.1681/ASN.2016010098 Epub 2016 Aug 2. PMID: 27486138; PMCID: PMC5084898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li HS, Borg E. Age-related loss of auditory sensitivity in two mouse genotypes. Acta Otolaryngol. 1991;111(5):827–834. doi: 10.3109/00016489109138418 PMID: 1759567. [DOI] [PubMed] [Google Scholar]
- 11.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. EPIC-InterAct consortium. using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Euro J Epidemiol. 2015;30:543–552. doi: 10.1007/s10654-015-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skrivankova VW, Richmond RC, Woolf BAR, et al.Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. 2021;326:1614–1621. doi: 10.1001/jama.2021.18236 [DOI] [PubMed] [Google Scholar]
- 13.Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahola-Olli AV, Würtz P, Havulinna AS, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100:40–50. doi: 10.1016/j.ajhg.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zha L-F, Dong J-T, Wang J-L, et al. Effects of insomnia on peptic ulcer disease using Mendelian randomization. Oxid Med Cell Longev. 2021;2021:2216314. doi: 10.1155/2021/2216314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosa M, Chignon A, Li Z, et al. A Mendelian randomization study of IL6 signaling in cardiovascular diseases, immune-related disorders and longevity. NPJ Genom Med. 2019;4:23. doi: 10.1038/s41525-019-0097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden J, Smith GD, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dziedzic T. Systemic inflammation as a therapeutic target in acute ischemic stroke. Exp Rev Neurother. 2015;15:523–531. doi: 10.1586/14737175.2015.1035712 [DOI] [PubMed] [Google Scholar]
- 20.Masuda M, Kanzaki S, Minami S, et al.Correlations of inflammatory biomarkers with the onset and prognosis of idiopathic sudden sensorineural hearing loss. Otol Neurotol. 2012;33(9):1142–1150. doi: 10.1097/MAO.0b013e3182755fbb [DOI] [PubMed] [Google Scholar]
- 21.Fransén K, Pettersson C, Hurtig-Wennlöf A. CRP levels are significantly associated with CRP genotype and estrogen use in the lifestyle, biomarker and atherosclerosis (LBA) study. BMC Cardiovasc Disord. 2022;22:170. doi: 10.1186/s12872-022-02610-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y, Zeng X, Tu W-J, Zhao J. Tumor necrosis factor-α: the next marker of stroke. Dis Mark. 2022;2022:2395269. doi: 10.1155/2022/2395269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011;22(4):189–195. doi: 10.1016/j.cytogfr.2011.10.001. Epub 2011 Oct 22. PMID: 22019906; PMCID: PMC3714593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delanghe JR, Speeckaert R, Speeckaert MM. Complement C3 and its polymorphism: biological and clinical consequences. Pathology. 2014;46(1):1–10. doi: 10.1097/PAT.0000000000000042 PMID: 24300728. [DOI] [PubMed] [Google Scholar]
- 25.Zhou T, Chen M, Yuan Z, et al. Inflammatory markers and the risk of idiopathic sudden sensorineural hearing loss: a Mendelian randomization study. Front Neurol. 2023;14:1111255. doi: 10.3389/fneur.2023.1111255 PMID: 36908593; PMCID: PMC9992207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frosolini A, Franz L, Daloiso A, Lovato A, de Filippis C, Marioni G. Digging into the role of inflammatory biomarkers in sudden sensorineural hearing loss diagnosis and prognosis: a systematic review and meta-analysis. Medicina. 2022;58(7):963. doi: 10.3390/medicina58070963 PMID: 35888682; PMCID: PMC9324865. [DOI] [PMC free article] [PubMed] [Google Scholar]