Abstract
Tuberous sclerosis complex (TSC) is targeted to the lysosomal membrane, where it hydrolyzes RAS homolog–mTORC1 binding (RHEB) from its GTP-bound to GDP-bound state, inhibiting mechanistic target of rapamycin complex 1 (mTORC1). Loss-of-function mutations in TSC cause TSC disease, marked by excessive tumor growth. Here, we overcome a high degree of continuous conformational heterogeneity to determine the 2.8-Å cryo–electron microscopy (cryo-EM) structure of the complete human TSC in complex with the lysosomal recruitment factor WD repeat domain phosphoinositide–interacting protein 3 (WIPI3). We discover a previously undetected amino-terminal TSC1 HEAT repeat dimer that clamps onto a single TSC wing and forms a phosphatidylinositol phosphate (PIP)–binding pocket, which specifically binds monophosphorylated PIPs. These structural advances provide a model by which WIPI3 and PIP-signaling networks coordinate to recruit TSC to the lysosomal membrane to inhibit mTORC1. The high-resolution TSC structure reveals previously unrecognized mutational hotspots and uncovers crucial insights into the mechanisms of TSC dysregulation in disease.
The complete TSC structure reveals how lysosomal targeting proteins and phosphoinositides coordinate mTORC1 inhibition.
INTRODUCTION
Tuberous sclerosis complex (TSC) is a crucial inhibitory checkpoint for the serine and threonine kinase mechanistic target of rapamycin complex 1 (mTORC1). mTORC1 is a central regulator of eukaryotic cell growth pathways, autophagy, and metabolism and is dysregulated in disease and aging (1). TSC functions as a guanosine triphosphatase (GTPase)–activating protein (GAP) and catalyzes the hydrolysis of RAS homolog–mTORC1 binding (RHEB) from its active guanosine 5′-triphosphate (GTP)–bound state to the inactive guanosine diphosphate–bound state (2–5). RHEB allosterically activates mTORC1, and RHEB-GTP hydrolysis by TSC acts as an off switch for cell growth (6). TSC is formed by TSC1 (hamartin), TSC2 (tuberin), and TBC1 domain family member 7 (TBC1D7) (7). Loss-of-function mutation of either TSC1 or TSC2 causes the autosomal dominant genetic disease TSC (affects ~2 million people worldwide) characterized by increased RHEB activation, hyperactive mTORC1 signaling, and tumor growth (8).
TSC functions as a signaling convergence point balancing the cellular response to growth signals and energy stress by regulating RHEB and the activation state of mTORC1 (9). Under conditions of energy stress, adenosine 5′-monophosphate–activated protein kinase (AMPK) and glycogen synthase kinase-3β phosphorylate and activate TSC, triggering RHEB-GTP hydrolysis and inhibiting mTORC1 and cell growth (10, 11). Conversely, under conditions of energy sufficiency, extracellular signal–regulated kinase (12), p90 ribosomal S6 kinase (13), and Akt (14–16) phosphorylate and inhibit TSC, enabling mTORC1 activation.
TSC activity depends on its recruitment to lysosomes, where it interacts with RHEB, which is anchored by C-terminal farnesylation (5). Several proteins appear to modulate TSC targeting to the lysosomal membrane, but it is unclear whether TSC lysosomal recruitment proteins act redundantly, overlap, vary by cell type, or jointly coordinate their actions. Under starved conditions, WD repeat domain phosphoinositide–interacting protein 3 (WIPI3) recruits TSC to lysosomes, a process that is mutually exclusive with active mTORC1, indicating co-regulation of mTORC1 inhibition by TSC and WIPI3 (17). In addition, the RagGTPase complex can act as a switch, modulating the localization of TSC or mTORC1 to the lysosomal membrane (18, 19). LAMP1-bound Ras GTPase-activating protein-binding proteins 1 and 2 (G3BP1/2) also localize TSC to lysosomes with G3BP1 knockout breast cancer cells displaying cell growth defects that phenocopy the loss of TSC component TBC1D7 (20). Despite these insights, the structural and mechanistic basis of TSC lysosomal recruitment remains unclear.
RESULTS
The cryo-EM structure of the complete TSC bound to WIPI3
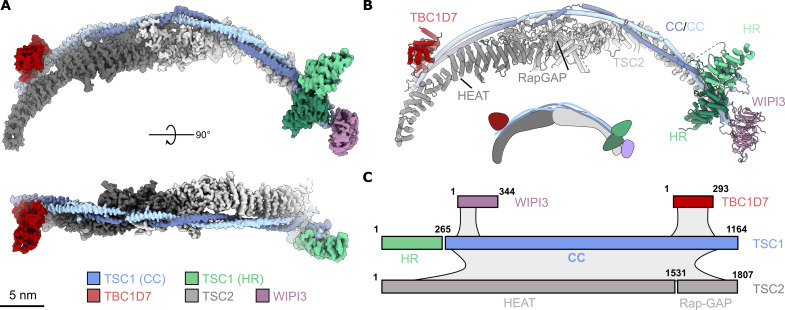
To investigate the mechanism of TSC docking at the lysosomal membrane, we determined the structure of the full-length TSC:WIPI3 complex by single-particle cryo–electron microscopy (cryo-EM) (Fig. 1, figs. S1 to S3, and table S1). The remarkable structural flexibility of TSC, with deviations of ~10 nm between extrema (fig. S2), posed a challenge for high-resolution structure determination. While moderate ~4.5-Å resolution cryo-EM reconstructions have described the stoichiometry and architecture of TSC (21, 22), the overall domain structure and sequence assignment remain incomplete due to substantial conformational flexibility. Here, focused refinement and classification stepped along the 40 nm length of the TSC:WIPI3 complex enabled us to achieve a final composite reconstruction with an average nominal resolution of 2.8 Å (Fig. 1 and fig. S3). The TSC:WIPI3 reconstruction was of sufficiently high resolution to build a complete domain- and residue-assigned model of the human TSC, revealing a previously unresolved TSC1 HEAT repeat (HR) dimer domain that clamps onto one TSC wing (Fig. 1, B and C).
Fig. 1. Overall structure of the TSC:WIPI3 complex.
(A) High-resolution reconstruction (composite) of the TSC:WIPI3 complex. Scale bar, 5 nm. (B) Cartoon model of the TSC:WIPI3 complex [oriented as in (A)]. Below, a two-dimensional (2D) schematic of the TSC:WIPI3 architecture. (C) Schematic showing the domain structure of the TSC components, as well as approximate interfaces and interactions (gray shading) between subunits. CC, coiled coil; HR, HEAT repeat.
Overall, in the TSC:WIPI3 structure, TSC2 forms an extended, arc-shaped dimeric scaffold with C2 symmetry, with a central interface formed by a dimer of the TSC2 Rap-GAP domain (Fig. 1). In contrast, TSC1 forms an asymmetric dimer that extends from one end of the TSC2 arc to the other, breaking the overall apparent symmetry of TSC (Fig. 1). The C-terminal domain of each TSC1 chain forms an extended coiled coil (CC) that runs along the ridge of the TSC2 dimer arc, buttressing the HRs of TSC2. TBC1D7 is bound at the C-terminal region of TSC1, making minor contact with TSC2. A single copy of WIPI3 is bound to TSC at the opposite end to TBC1D7, forming a highly dynamic cap at the TSC1 tip (Fig. 1 and fig. S2).
The TSC1 N terminus forms an HR dimer clamped to one TSC wing
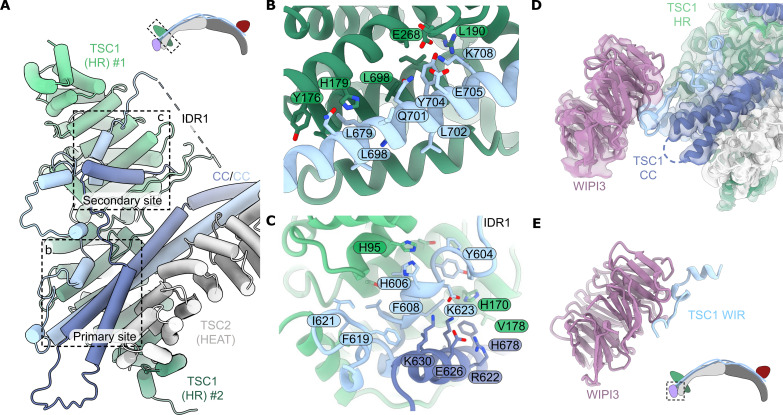
Notably, the previously unresolved TSC1 N terminus forms a large dimer clamped at the opposite tip of TSC from the TBC1D7 subunit (Figs. 1 and 2A). The TSC1 N-terminal dimer consists primarily of HRs (TSC1 HR) with a central pseudosymmetrical interface formed by TSC1 residues 197 to 237 (fig. S4). The interface is extensively hydrophobic, except for two pairs of intermolecular salt bridges (R204:E225) (fig. S4). The TSC1 CC domain binds only a single TSC1 HR subunit interacting at the primary binding site (Fig. 2, A and B). An extensive intrinsically disordered TSC1 loop region (IDR1) connects the N-terminal TSC1 dimer to the TSC1 CC domain to accommodate this conformation (fig. S1). The TSC1 IDR1 also partially occupies the open secondary binding site on the alternate TSC1 dimer subunit (Fig. 2, A and C). Overall, this architecture prevents TSC oligomerization and aligns the TSC1 HR dimer and TBC1D7 at opposite tips of the arc-shaped TSC fold. This structure contrasts with the two-site model proposed from the isolated Chaetomium thermophilum TSC1 N-terminal crystal structure that supported a model of TSC oligomerization (23). Throughout the study, we observed no obvious capacity for the TSC:WIPI3 complex to form extended polymers. In addition, the TSC:WIPI3 structure does not provide an obvious mechanistic explanation for the formation of extended linear polymers by TSC, as seen in endogenous samples (24).
Fig. 2. The TSC1 HR domain and the structural basis of WIPI3 binding.
(A) Focused view of the TSC HR dimer. A single subunit of the TSC1 HR dimer mediates contacts to the TSC1 CC domain and the TSC2 HRs. Two regions of the large extended loop (TSC1 IDR1) shield the exposed second binding site of the TSC1 N-terminal dimer. Top: Schematic of the TSC1 HR dimer in context of TSC. (B) The TSC1 HR dimer clamps onto a hydrophobic helix of the TSC1 CC domain. The second TSC1 helix is hidden for clarity [shown in (A), dark blue]. (C) Key contacts in the TSC1 HR dimer interface mediated by the first TSC1 extended loop (IDR1). (D) Focused view of the WIPI3:TSC1 interaction as resolved by cryo-EM. (E) The 3.17-Å crystal structure of WIPI3 in complex with the TSC1 WIR motif. Bottom: Schematic of the WIPI3 binding site in context of TSC.
The TSC:WIPI3 interaction is mediated by a conserved WIR motif in TSC1
The WIPI3 lysosomal recruitment factor was located bound to a helix-turn-helix at the end of the TSC1 CC domain, providing an anchor point for docking at the TSC tip (Fig. 2D). A single copy of TSC1 coordinates WIPI3 via a moderately conserved WIPI-interacting-region (WIR) motif, positioning WIPI3 such that the WIR motif on the second TSC1 chain is sterically occluded (fig. S4C). The resolution of WIPI3 in the cryo-EM reconstruction was reduced due to its extensive flexibility that resulted in a pivoting motion around the TSC1 anchor point (fig. S2).
To capture the TSC1:WIPI3 interaction at high resolution, we generated several TSC1 truncation constructs for co-crystallization trials with WIPI3 (fig. S5). This approach allowed us to determine the 3.17-Å resolution crystal structure of WIPI3 bound to a truncated TSC1 fragment (residues 467 to 685, Δ500 to 603) (Fig. 2E, fig. S4D, and table S2). Only TSC1 residues 659 to 680 were resolved in the structure with the TSC1 WIR motif mediating the WIPI3 interaction, consistent with the cryo-EM reconstruction. The hydrophobic TSC1 WIR motif meanders across the WIPI3 surface with conserved residues W676 and F679, mediating key hydrophobic interactions that clamp to strand β5 of WIPI3, while the conserved TSC1 H678 forms an electrostatic interaction with WIPI3 D81 (Fig. 2E and fig. S4D). The TSC1:WIPI3-binding mode is broadly analogous to the ATG2A:WIPI3 structure (25), whereby WIPI3 binding is facilitated by a conserved WIR motif in the phospholipid-transfer protein ATG2A (fig. S5). Therefore, to confirm critical TSC1 residues for WIPI3 binding, we conducted alanine scanning across the TSC1 WIR motif, finding that mutation of conserved residues W676, H678, or F679 completely abolished WIPI3 binding (fig. S5, E and F). Previously, the dissociation constant (Kd) for the interaction between the WIPI3 and ATG2A WIR motif was measured at 6.3 ± 0.2 μM (25). Given the strong structural similarity (fig. S5), we propose that the TSC1:WIPI3 interaction likely exhibits a comparable affinity. Overall, this observation would be consistent with a relatively fast rate of TSC dissociation from the endolysosomal membrane. Therefore, we suggest that WIPI3 likely facilitates the stable recruitment of TSC to the endolysosomal membrane through cooperative mechanisms, such as avidity with other binding partners [e.g., phosphatidylinositol phosphates (PIPs)].
Given the flexibility of WIPI3 bound to TSC, we conducted cross-linking mass spectrometry to validate the placement of the WIPI3:TSC1 crystal structure into our cryo-EM reconstruction (fig. S6). We observed excellent agreement between the peptide-derived distance constraints, our cryo-EM TSC:WIPI3 reconstruction, and our TSC:WIP3 crystal structure.
TSC binds monophosphorylated PIP
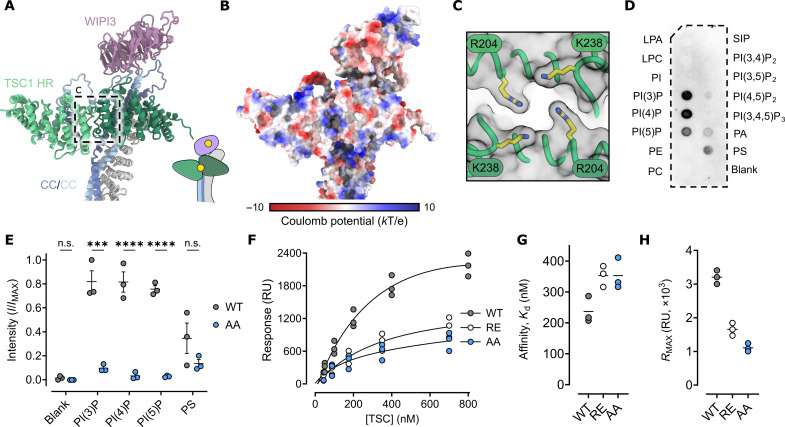
Inspection of the TSC structure revealed a positively charged pocket defined by the dimer interface of the two TSC1 HR domains (Fig. 3, A and B, and fig. S7). Two symmetrically related TSC1 residues, R204 and K238, coordinate to form a positively charged and conserved band that encircles the recessed cleft (Fig. 3C and fig. S7). Given these observations, we measured the binding of recombinant full-length TSC to different lipids using PIP Strip analysis (Fig. 3D). We observed specific binding of TSC to monophosphorylated phosphatidylinositol lipids PI(3)P (phosphatidylinositol 3-phosphate), PI(4)P, and PI(5)P, as well as to phosphatidylserine (PS). In contrast, no substantial binding was detected for di- or tri-phosphorylated PIPs (Fig. 3D). This observation suggests that binding depends on the number of phosphorylation sites. These findings are in contrast to the isolated TSC1 N-terminal dimer that was previously observed to bind PIPs more broadly, with a minor preference for PI(3,4)P2 and PI(4,5)P2 (23). Several surface-exposed charged regions in the isolated N-terminal TSC1 dimer (23) structure are shielded in the context of full-length TSC, suggesting that surface charge differences between the isolated TSC1 N-terminal domain and full-length TSC may explain the observed differences in PIP binding.
Fig. 3. The TSC1 HR dimer is a monophosphorylated PIP selective membrane association domain.
(A) Cartoon rendering of the TSC N-terminal PIP-binding domain and the WIPI3:TSC interaction. Inset: Illustration of the PIP binding sites of WIPI3 and the TSC1 HR dimer. (B) Surface rendering colored by coulomb potential. Several positively charged regions are presented on the surface. (C) The symmetrical arrangement of conserved lysine and arginine residues define an electro-positive recessed pocket. (D) Immunoblot of full length TSC probed against phosphatidylinositol lipid membrane strips illustrating specificity for monophosphorylated phosphatidylinositols. LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PA, phosphatidic acid; PS, phosphatidylserine; SIP, sphingosine-1-phosphate. (E) Densitometry analysis of replicate phosphatidylinositol lipid membrane strips. WT, wild type. AA, TSC K238A, R204A. Symbols show values from independent experimental replicates, the bold line shows the mean, and error bars show SEM; n.s., not significant; P values were determined using ordinary one-way analysis of variance (ANOVA) with Šidák’s multiple comparisons test (***P < 0.0002 and ****P < 0.0001). (F) Steady-state binding of TSC to 5% molar PI(3)P lipid bilayer (L1 chip) with 1,2-dioleoyl-sn-glycero-3-phosphocholine:1,2-dioleoyl-sn-glycero-3-phosphoethanolamine. Symbols show values from independent experimental replicates. RU, response units. (G) Stead-state affinity measurement of wild-type TSC (one-to-one binding model, solid line) estimates an apparent dissociation constant (Kd) of 237 nM, compared to 353 nM for both TSC K238A, R204A (AA) and TSC R204E (RE). (H) Plateau values of maximal response, RMAX, corresponding to model fit (F). Symbols show values from independent biological replicates, and the bold line shows the mean.
To explore the role of the positively charged TSC1 pocket in PIP binding, we mutated TSC1 R204 and K238 to alanine and observed a broad reduction in TSC binding to monophosphorylated PIPs and PS by PIP Strip analysis (Fig. 3E and fig. S7). Next, we used surface plasmon resonance analysis to demonstrate that full-length TSC bound PI(3)P with a relatively high affinity of 237 nM (Fig. 3, F and G, and fig. S7). Substitution of R204 and K238 to alanine showed a loss in PI(3)P binding affinity and a more notable reduction in the maximum observed binding signal (RMAX) (Fig. 3, F to H, and fig. S7). In addition, we tested the binding of TSC with a switched charge TSC1 mutation (R204E) to PI(3)P and observed a similar loss of affinity and reduction in RMAX (Fig. 3, F to H, and fig. S7). The RMAX is dependent on the number of binding sites and the immobilized lipid bilayer. The observed decrease in RMAX is, therefore, consistent with the loss of a lipid binding site in the TSC1 HR domain due to mutation (Fig. 3H and fig. S7). However, given that binding is not completely abolished, these data suggest that TSC may contain additional PI(3)P binding sites beyond the TSC1 HR domain. We note the presence of extensive positively charged surface patches within the full TSC, which may independently mediate interactions with negatively charged lipids.
Together, we conclude that the TSC N-terminal dimer is a membrane association domain that recognizes PI(3)P, PI(4)P, and PI(5)P. Here, the two PIP binding sites, contributed by WIPI3 and the TSC1 N-terminal dimer, together define a single flat surface. This configuration suggests that WIPI3 and TSC1 simultaneously engage PIPs, thereby positioning TSC flush against the lipid bilayer (fig. S7). Given the role of lysosomal PI(3)P and PI(4)P levels in endolysosomal organization and mTORC1 regulation (26, 27), we postulate that the TSC:PIP interaction may provide a mechanism for endolysosomal PIP-signaling pathways to fine-tune TSC lysosomal recruitment and mTORC1 activity.
Disease-associated mutations cluster to the TSC1 N-terminal and TSC2 catalytic domains
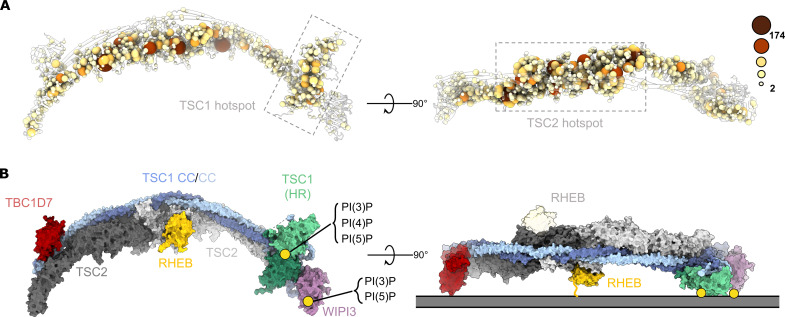
Disease-associated TSC mutations reduce TSC activity, increase the amount of active RHEB-GTP, and drive mTORC1 activation (8). To explore the structural basis for mutation-associated TSC loss of function, we mapped disease-associated TSC mutations across the entire sequence-assigned complex (Fig. 4A and fig. S8). As expected from previous crystal and cryo-EM structures, mutation hotspots cluster to the TSC2 catalytic Rap-GAP domain (Fig. 4A and fig. S8). In addition, we discover a second clear mutation hotspot located at the TSC1 N-terminal HR dimer (Fig. 4A and fig. S8). Several mutations are clustered at the HR dimer PIP binding site and at the interface between the TSC1 HR dimer and the TSC core scaffold (Fig. 4A and fig. S8). Last, mutation hotspots correlate with areas of high surface conservation, further suggesting functional selection pressures on these regions (Fig. 4A and fig. S9). These data, in combination with structural data, suggest that the PIP-binding HR dimer domain plays a critical role in TSC function. Similarly, we find smaller mutational clusters located within the TSC:WIPI3 interface and at key regions within the TBC1D7 and the TBC1D7:TSC1 interface. Together, these data highlight how TSC disease–associated mutations exploit multiple mechanistic pathways to drive TSC loss of function, mTORC1 activation, and tumor growth.
Fig. 4. TSC:WIPI3 disease–associated mutations and model of the TSC:WIPI3:RHEB lysosomal mTORC1 inhibitory complex.
(A) Cα atoms of all disease-associated missense mutations are rendered as spheres of size and color proportional to the frequency of observation (COSMIC, LOVD, and HGMD) (55–57). Disease-associated mutations cluster to the TSC1 HR PIP-binding dimer (left), as well as the TSC2 Rap-GAP central core (right). (B) Surface rendering of the complete human TSC with RHEB modelled according to AlphaFold (58). The binding sites of RHEB, TSC1:PI(3)P, and WIPI3:PI(3)P define a membrane binding plane that is consistent with a singly occupied RHEB-binding model of the TSC:WIPI3:RHEB lysosomal mTORC1 inhibitory complex.
DISCUSSION
The recruitment of TSC to the lysosomal membrane to hydrolyze RHEB-GTP is a pivotal step in regulating eukaryotic cell growth. Despite its importance, the structural basis of this mechanism has remained unclear. Previous studies have determined that WIPI3 binds PI(3)P and PI(3,5)P2 across two distinct binding sites in blades 5 and 6 of the β-propeller fold (28, 29). To gain further insight into TSC lysosomal docking, we performed rigid-body fitting of the WIPI3:PI(3)P structure (29) to our full-length TSC:WIPI3 complex to visualize the overall position of each PIP binding site (fig. S7). Our analysis revealed that the WIPI3:PIP binding site is oriented in the same plane and directly adjacent to the TSC1 HR dimer PIP-binding pocket. These two PIP anchor points align TSC horizontally along the membrane, forming an orientation compatible with RHEB binding (Fig. 4B and fig. S7). We propose that this model represents an active organization of TSC on the lysosomal membrane (Fig. 4B). In this model, the asymmetrical arrangement of the TSC1 dimer acts as a central determinant in providing flexibility to regulate the TSC2 catalytic domains. For example, TSC1 asymmetry enables TBC1D7, the TSC1 HR dimer, and WIPI3 to bind at different positions along the TSC curvature. TSC2 only appears to provide minor contributions to binding these regulatory domains and accessory proteins. In this manner, TSC1 acts as an interaction module regulating the cellular localization and activity of the catalytic TSC2 GAP domains.
Several kinases regulate TSC through phosphorylation, including AMPK, which has been shown to activate TSC, and Akt, which inhibits TSC (9, 10, 14–16). To assess the structural impact of TSC phosphorylation, we mapped known phosphorylation sites onto our model of membrane-bound TSC (fig. S10). Notably, almost all known phosphorylation sites map to unresolved loop regions in TSC1 and TSC2, which are predicted to be intrinsically disordered (figs. S1 and S10). These loops are extensive (40 to 300 residues) and likely of sufficient length to reach the lipid membrane regardless of TSC orientation. Therefore, the phosphorylation state of TSC is unlikely to promote a specific orientation of TSC on the lysosomal membrane. The exact mechanism by which these posttranslational modifications regulate TSC function remains to be determined.
Further layers of TSC regulation at the endolysosomal membrane also remain to be resolved. For example, a recent study determined that mTORC1 cellular localization and activity is regulated by a PI(3)P/PI(4)P switch with increased PI(3)P levels driving mTORC1 lysosomal localization and activity (26). The mTORC1 component Raptor directly binds PI(3)P, providing a potential mechanism for lysosomal localization (26). In agreement, the direct binding of TSC to monophosphorylated PIPs may provide a mechanism to modulate TSC endolysosomal recruitment. Hence, direct PIP binding by TSC may coordinate with lysosomal recruitment factors such as WIPI3 (17) to drive TSC localization, regulation, and potentially compete with mTORC1 for PI(3)P binding. In addition, in our model, TSC orientation positions TBC1D7 to interact with membrane-bound proteins or lipids. Intriguingly, the exposed surface of TBC1D7 is also well conserved, indicating a potential for mediating further functional interactions (fig. S9). TBC1D7 is a nonessential TSC component with TBC1D7 knockout, reducing but not eliminating TSC activity (30). These findings align with a possible role of TBC1D7 in modulating lysosomal TSC positioning. Added layers of TSC regulation are provided by Rag GTPase and G3BP1/2 binding, in addition to TSC phosphorylation (9, 18–20). It will be important for further studies to elucidate whether these multiple layers of TSC regulation act redundantly, have overlapping functions, depend on cell type, or operate in a coordinated manner.
In summary, the structure of the TSC:WIPI3 lysosomal recruitment complex highlights a combined role for protein lysosomal recruitment proteins and phosphoinositide-signaling networks in coordinating TSC lysosomal localization, RHEB hydrolysis, and mTORC1 inhibition. TSC disease–associated mutations cluster at multiple hotspots on the TSC structure, providing several mechanistic pathways to TSC loss of function and tumor growth. A structural understanding of the TSC mutational landscape will guide future endeavors to predict the severity of TSC loss of function from clinical genomic screening.
MATERIALS AND METHODS
Protein expression and purification
TBC1D7 was polymerase chain reaction amplified from pET-28a TBC1D7 (Addgene plasmid no. 32047 was a gift from C. Arrowsmith) and cloned into pcDNA 3.1 (Thermo Fisher Scientific) with a C-terminal FLAG-tag. pRK7 FLAG TSC2 (Addgene plasmid no. 8996) was a gift from J. Blenis, and pcDNA 3.1 myc TSC1 (Addgene plasmid no. 12133) was a gift from C. Walker. The recombinant proteins were co-expressed in Expi293F cells (Thermo Fisher Scientific) grown in Expi293 Expression Medium (Thermo Fisher Scientific) at 37°C in a humidified atmosphere with 5% CO2. Cells were transfected with an equal amount of each TSC component vector (at a final DNA amount of 1.25 μg ml−1) using polyethylenimine (PEI) Max (Polysciences) at a PEI:DNA ratio of 5:1. After 72 hours, cells were harvested by centrifugation and frozen at −80°C before purification. Cells were lysed by sonication in buffer containing 20 mM tris-HCl (pH 8.0), 250 mM NaCl, 2% (v/v) glycerol, 10 mM NaF, and 2 mM dithiothreitol (DTT). Lysate was clarified by centrifugation, filtered at 0.8 μm, and then incubated with anti-FLAG resin (GenScript, L00432-25) for 30 min at 4°C with agitation. The resin was washed with lysis buffer, and TSC was eluted with lysis buffer containing FLAG peptide (0.1 mg ml−1; GenScript). TSC was further purified by anion-exchange chromatography using a Mono Q 5/50 GL (Cytiva) with a linear gradient of 100 to 1000 mM NaCl in 20 mM Hepes (pH 8.0) and 2 mM DDT. Protein was concentrated using a 100-kDa molecular weight cutoff (MWCO) centrifuge concentrator and aliquots snap frozen in liquid nitrogen before storage at −80°C.
To produce WIPI3, an N-terminally His-tagged Δloop construct (residues 12 to 344, Δ75 to 77 and Δ264 to 281) (25) was cloned into the pFastBac Dual (Thermo Fisher Scientific) polyhedron multiple cloning site. Expression of WIPI3 was performed in Sf9 cells for 2.5 days following infection with baculovirus (Bac-to-Bac, Invitrogen). Cell pellets were harvested by centrifugation and resuspended in lysis buffer [20 mM tris-HCl (pH 8), 500 mM NaCl, 10% (w/v) glycerol, and 5 mM β-mercaptoethanol]. Cells were disrupted by sonication and subsequently clarified by centrifugation to remove debris. Cell lysate was supplemented with 10 mM imidazole and batch bound on Ni-NTA for 1 hour at 4°C. Resin was washed with lysis buffer containing 20 mM imidazole and eluted with 400 mM imidazole. Pooled fractions were buffer exchanged, concentrated using a 3-kDa MWCO centrifuge concentrator, and purified by size exclusion chromatography (Superdex75 16/60, Cytiva).
For crystallography and alanine scanning, the N-terminally His-tagged Δloop WIPI3 construct was co-expressed with TSC1. Here, the open reading frame coding for TSC1 (residues 467 to 685, Δ500 to 603) was cloned into the p10 multiple cloning site of the same WIPI3 pFastBac Dual expression construct. His-tagged WIPI3 and TSC1 were co-expressed in Sf9 cells for 2.5 days following infection with baculovirus (Bac-to-Bac, Invitrogen). The TSC1:WIPI3 complex was purified following the same procedure as WIPI3 alone.
Mass photometry
Standard mass photometry landing assays were conducted at 20°C using the TwoMP instrument (Refeyn Ltd., Oxford, UK). Precleaned glass coverslips were purchased (Refeyn Ltd., Oxford, UK). Image series were collected for 60 s with an 8-ms exposure (128 Hz) at 488 nm with a field of view of 12 μm by 17 μm. TSC samples were diluted into 10 mM Hepes (pH 7.5) and 250 mM NaCl for a final concentration of 10 to 20 nM. Standards of bovine serum albumin (Merck) and apoferritin (Merck) were measured on the same day to calibrate extracted particle contrast to mass. Analysis and acquisition were performed using the Refeyn AquireMP and DiscoverMP packages (v2.5), respectively. Frame and pixel binning were applied, with a factor of 3 and 6, respectively, for an effective pixel size of 72 nm.
Cryo-EM sample preparation and data collection
Quantifoil R1.2/1.3 200 mesh Cu grids were glow discharged using a Pelco easiGlow instrument at 30 mA for 30 s. Freshly purified protein (3.5 μl at 0.4 μM) was applied immediately to the discharged grid and vitrified in liquid ethane using a Vitrobot Mk IV (Thermo Fisher Scientific) after blotting by hand using Fisherman Grade 1 filter paper. The temperature was maintained at 4°C with the relative humidity at 100%. Data were collected on a Titan Krios G1 (Thermo Fisher Scientific) operating at 300 kV with a 50-μm C2 aperture. Micrographs were acquired using a Gatan K3 direct electron detector in counting mode at a nominal energy-filtered transmission electron microscopy magnification of ×105,000, corresponding to a calibrated physical pixel size of 0.8234 Å. A Gatan GIF Quantum energy filter was used with a slit width of 10 eV. The electron dose rate was set to 8.505 electrons pixel−1 s−1 with a total exposure time of 3.71 s, yielding a total dose of 46.54 electrons Å−2 distributed across 60 frames. Automated collection was carried out using EPU (v.2.12.1.2782) with beam shift to collect 21 images per stage movement. The average nominal defocus value was −1.4 μm.
Cryo-EM image analysis and model building
A total of 12,807 dose-fractionated movies were corrected for beam-induced motion and compensated for radiation damage within MotionCor2 (v1.1.0) (31). All aligned movie frames were subsequently summed into dose-weighted averages for further processing. The contrast transfer function (CTF) parameters were estimated with CTFFIND (v4.1.8) (32). Multiple rounds of autopicking and blob picking in cryoSPARC (33) were performed, followed by particle duplicate removal and two-dimensional (2D) classification. Clean classes were used to train both TOPAZ (34) and crYOLO (35) models to perform a final round of particle picking. All particles were pooled and included for further processing. These were subjected to multiple rounds of 2D classification in both RELION (v3.1, 4.0b) (36) and cryoSPARC (33), yielding 1,176,292 particles of sufficient quality and homogeneity. These were extracted in a box of 800 × 800 pixels and down-sampled by Fourier cropping to 440 × 440, for a pixel size of 1.5 Å pixel−1. A consensus reconstruction was generated in cryoSPARC (33) by refining this subset using nonuniform refinement with C1 “symmetry” yielding a 3.4-Å map that suffered from a high degree of flexibility and anisotropy due to preferred orientation. Particle polishing was performed in RELION (36), re-extracting particles for a final pixel size of 1.17 Å pixel−1, and the particles were re-refined.
We defined several regions of interest and began signal subtraction followed by 3D classification with angular searches, both in RELION (36). Because of the size of TSC and the degree of flexibility, it was necessary to regenerate consensus reconstructions along the length of the complex after re-centering and re-extracting particles. This process enabled the most accurate signal subtraction and focused classification by reducing the effects of flexibility. Several subsets were obtained for each subregion, each were re-refined using RELION (36) and/or nonuniform refinement in cryoSPARC (33), yielding high-resolution maps. Per-particle CTF parameters were refined in cryoSPARC (33). Conversions between software were performed with EMAN (v2.2) (37), with code written in-house, or by pyem. The fourier shell correlation (FSC) was used to estimate resolution at the 0.143 threshold. Local resolution was estimated by the windowed blocres FSC method (0.5 threshold) as implemented in cryoSPARC (33). Map sharpening in cryoSPARC (33), deepEMhancer (38), and EMReady (39) was performed to assist in residue assignment and model building. Atomic models were built into individual focused reconstructions using rigid-body fitting of AlphaFold (40) predictions, and the TBC1D7 crystal structure (41) and, ultimately, much of the model were built de novo. A combination of Coot (42), ISOLDE (43), and ChimeraX (44) was used for model building, analysis, and figures.
Lastly, a composite representation was generated using Chimera (45). Briefly, each sharpened map was rescaled to have the same mean and SD. Overlapping regions of each focused refinement were used to superimpose and align the fragments. Next, soft masks were applied to remove diffuse signal and regions of poor density (this was performed for reconstructions with overlapping regions, taking those which had higher local resolution). Last, the maps were combined by performing a pairwise voxel comparison (UCSF Chimera “vop maximum”) to discard voxels with lower density. To estimate the resolution of the composite reconstruction, composite half maps were generated by the same procedure applying identical rigid-body transformations to align each half map with the sharpened composite (according to the same procedure as phenix.combine_focused_maps). No scaling, masking, or filtering was performed on the half maps other than pairwise voxel selection by “vop maximum.” A FSC analysis was then performed on the independent half maps with a loose global mask, estimating the global resolution to be 2.8 Å.
Crystallization and structure determination
Final TSC1:WIPI3 crystals were obtained from a complex formed between TSC1 (residues 467 to 685, Δ500 to 603) and a modified WIPI3 construct, in which two loops corresponding to residues 75 to 80 and residues 264 to 281 were removed. This WIPI3 Δloop construct had previously been used to facilitate structural determination of the WIPI3:ATG2A complex (25). In the final TSC1:WIPI3 crystal structure, only TSC1 residues 659 to 680 were observed. These data suggest that the remaining TSC1 residues were likely unstructured and did not participate in complex formation. This conclusion is further supported by site-directed mutagenesis studies that highlight the key role of the TSC1 WIR motif in WIPI3 binding.
TSC1:WIPI3 crystals were grown at 20°C by hanging drop vapor diffusion in 0.2 M ammonium tartrate, 20% (w/v) PEG3350, and 0.25 M sodium acetate. Crystal were flash cooled in liquid nitrogen in mother liquor supplemented with 20% (v/v) glycerol. Crystallographic data were collected at the MX2 microfocus beamline (46) of the Australian Synchrotron at a wavelength of 0.95373 Å and temperature of 100 K. Data were processed and scaled using XDS (47) and programs within the CCP4 suite (48). The high-resolution cutoff was determined by the criteria of CC1/2 > 0.3 (table S2) (49). The structure was solved by molecular replacement using WIPI3 (Protein Data Bank 6IYY) (29) as the search model in Phaser (50). Iterative cycles of refinement and rebuilding were carried out using PHENIX Refine (50) with local rebuilding in Coot (51). The structure had no Ramachandran outliers, with 92.8% of residues in favored regions and 100% in allowed regions and a final MolProbity score of 1.65 (100th percentile) (52).
Cross-linking mass spectrometry
The TSC:WIPI3 complex was assembled at a 1:2 molar ratio using 3 μM TSC, 6 μM WIPI3 in 20 mM Hepes (pH 8.0), 150 mM NaCl, 5 mM MgCl2, and 2 mM DTT. Cross-linking mass spectrometry was then performed by adding 600 μM BS3 (Thermo Fisher Scientific) cross-linker to the complex or respective components and incubated at room temperature for 20 min. Samples were quenched by the addition of 50 mM tris-HCl (pH 8.0) and then snap frozen in liquid nitrogen before further processing. Samples were subsequently denatured for 30 min at 65°C in the presence of 10 mM DTT. Chloroacetamide was added to 40 mM before incubation for 20 min at room temperature. A 1:100 (w/w) ratio of trypsin was added to the samples and further incubated at 37°C overnight. An additional 1:100 (w/w) ratio of trypsin was then added and the samples further incubated at 37°C for 2 hours. Digestion was stopped with the addition of 1% (v/v) formic acid. Samples were subsequently cleaned using OMIX C18 pipette tips (Agilent Technologies) and stored in 0.1% (v/v) formic acid before mass spectrometry.
Samples were analyzed by liquid chromatography–tandem mass spectrometry using a Dionex Ultimate 3000 RSLCnano system coupled onto an Orbitrap Fusion Tribrid instrument (Thermo Fisher Scientific). An Acclaim PepMap RSLC analytical column (75 μm by 50 cm, nanoViper, C18, 2 μm, 100 Å; Thermo Fisher Scientific) and an Acclaim PepMap 100 trap column (100 μm by 2 cm, nanoViper, C18, 5 μm, 100 Å; Thermo Fisher Scientific) were used to separate tryptic peptides by increasing concentrations of 80% (v/v) acetonitrile (can)/0.1% (v/v) formic acid at a flow of 250 nl min−1 for 90 min. The mass spectrometer was operated in data-dependent mode with the following parameters. The cycle time was controlled for 3 s. The MS1 resolution was set at 120,000 and scan range of 375 to 2000 mass/charge ratio. The automatic gain control (AGC) target was set at 1.0 × 106 with an injection time of 118 ms. The MS2 resolution was set at 60,000, and the AGC target was set at 4.0 × 105 with an injection time of 118 ms. pLink and pLink2 were used to identify BS3-cross-linked peptides (53). Each cross-linking dataset is derived from at least duplicate experiments, and cross-linked peptides were analyzed if they were identified at least twice with a P < 0.0001. Visual representations of cross-linked peptides were generated in Circos (54) or UCSF ChimeraX (44).
Surface plasmon resonance
Surface plasmon resonance experiments were performed on a Biacore T200 (GE Healthcare) in running buffer containing 10 mM Hepes (pH 7.5), 250 mM NaCl, 0.1% (w/v) bovine serum albumin (BSA), and 2% (w/v) glycerol at 25°C with a flow rate of 30 μl min−1. To measure binding of TSC to PIPs, we generated lipid bilayers on an L1 sensor chip (GE Healthcare). Briefly, a lipid/chloroform suspension was generated with 1,2-dioleoyl-sn-glycero-3-phosphocholine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, and PI(3)P in a 8:8:1 ratio and dispensed into glass tubes for a total mass of 1 mg. As a negative control, the same composition was used in 1:1 ratio excluding PI(3)P. Chloroform was evaporated under argon gas, and lipid films were resuspended in running buffer (without BSA or glycerol) by vortex and allowed to rehydrate in a sonicating water bath for 1 hour at 25°C. This mixture was passed through a 0.2-μm filter to remove large vesicles and insoluble aggregates.
The L1 chip was prepared by washing with 20 mM CHAPS, 40 mM octyl-β-d-glucopyranoside, and running buffer. Next liposomes were mixed with acetate buffer (pH 4.5) to a final concentration of 100 mM acetate and injected at 5 μl min−1 until the baseline was stabilized. The flow rate was subsequently adjusted to 50 μl min−1 to remove loosely bound material and a final injection of 50 mM NaOH was performed. The flow rate was adjusted to 5 μl min−1, and the chip was left to stabilize for 10 min. Indicated serial dilutions of TSC in running buffer were injected for 180 s, and dissociation was monitored for 1800 s. We carried out three independent analyses for each variant of TSC. Affinity, measured from steady state curves, is expressed as the mean. After each analyses, the sensor surface was regenerated using 20 mM CHAPS and 10 mM NaOH, and liposome deposition was performed again.
Phosphatidylinositol immunoblot
Immobilized phosphatidylinositol lipid strips (Thermo Fisher Scientific, P23750) were blocked with 1% (w/v) skim milk in 20 mM tris-HCl (pH 7.4) and 150 mM NaCl [tris-buffered saline (TBS)] for 1 hour at 25°C. A solution of recombinant TSC (7 μg ml−1) in 1% (w/v) skim milk TBS solution was incubated with the strips for 16 hours at 4°C. After binding, each strip was washed three times in TBS with 0.002% (w/v) Tween 20 for 5 min. The membranes were probed with anti-FLAG M2 antibody (Sigma-Aldrich, F3165) for 1 hour in 1% (w/v) skim milk TBS (1:1000 dilution), washed three times, and subsequently probed with anti-mouse horseradish peroxidase (HRP)–conjugated secondary antibody (Cell Signaling Technology, 7076) for 1 hour in 1% (w/v) skim milk TBS (1:2000 dilution). Triplicate blots were performed for each TSC variant, including wild type, and all blots were developed for the same total duration to enable consistent normalization.
Disease-associated mutation analysis
All mutations and variants identified in TSC1, TSC2, TBC1D7, and WIPI3 were manually curated from the Catalogue of Somatic Mutations in Cancer (COSMIC), Leiden Open Variation Database (LOVD), and Human Gene Mutation Database (HGMD) databases (55–57). Variants corresponding to alterations other than missense mutations were discarded. All variants associated with disease were isolated, and other reported variants were ignored. An in-house python program was used to parse and combine all remaining variants. These were subsequently exported to ChimeraX (44) as residue level attributes to enable visualization in the context of the structural model.
WIPI3:TSC1 mutation analysis
Small-scale protein co-expressions were conducted using the Bac-to-Bac baculovirus system (Invitrogen). WIPI3-His6 and mutants of TSC1-FLAG (residues 467 to 685, Δ500 to 603) were cloned into pFastBac Dual for baculovirus production according to the manufacturer’s protocol (Thermo Fisher Scientific). For WIPI3:TSC1 co-expressions, 50-ml cultures of Sf9 cells at 1.1 × 106 cells ml−1 were infected with 50 μl of baculovirus. After 3 days, cell pellets were harvested by centrifugation, washed in phosphate-buffered saline (PBS; Gibco, no. 10010023), and lysed by sonication. Lysate was clarified by centrifugation at 25,000g and passed through a 0.2-μm centrifugal filter. Lysate protein concentrations were normalized to a total protein content (2 mg ml−1) with PBS. Pull-downs were conducted using paramagnetic Ni–nitrilotriacetic acid (NTA) beads by incubating 100 μl of lysate with 20 μl of Ni-NTA Dynabeads (Thermo Fisher Scientific, no. 10104D) supplemented with 10 mM imidazole to prevent nonspecific binding. Beads were washed with 1 ml of PBS (with 20 mM imidazole) for a total of three washes. The beads were then subjected to 18% (w/v) SDS–polyacrylamide gel electrophoresis. Western blot of the resolved protein species was conducted with anti-FLAG M2 antibody (Sigma-Aldrich, F3165) and anti-mouse HRP-conjugated secondary antibody, with blocking and wash conditions as described above (phosphatidylinositol immunoblots).
Acknowledgments
We acknowledge the use of instruments and assistance at the Monash Ramaciotti Centre for Cryo-Electron Microscopy, a Node of Microscopy Australia. We also acknowledge the office of the Vice-Provost for Research and Research Infrastructure (VPRRI) at Monash University and of Bioplatforms Australia (BPA) as part of the National Collaborative Research Infrastructure Strategy (NCRIS).
Funding: This work was supported by a US Department of Defense Tuberous Sclerosis Complex Research Program Grant (W81XWH-19-1-0182) to A.M.E., an Australian Research Council (ARC) DECRA Fellowship (DE240100992) to C.B.-J., a Viertel Senior Medical Research Fellowship supported by The Cross Family and The Frank Alexander Charitable Trusts to MLH, and a Victorian Department of Health and Human Services Victorian Cancer Agency Mid-Career Fellowship (MCRF21036) to A.M.E. This research was undertaken, in part, using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector.
Author contributions: Conceptualization: C.B.-J., C.J.L., L.D., M.L.H., and A.M.E. Protein purification: L.D., C.B.-J., C.J.L., and Y.-G.C. Cryo-EM data collection: C.J.L. and H.V. Cryo-EM data processing: C.B.-J. Cross-linking experiments: C.B.-J., L.D., Y.-G.C., G.D.J., J.R.S., and R.B.S. Crystallography: L.D. and A.M.E. Surface plasmon resonance and lipid binding: C.B.-J. Writing: C.B.-J., C.J.L., L.D., M.L.H., and A.M.E.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The 3D cryo-EM density maps are deposited into the Electron Microscopy Data Bank (www.ebi.ac.uk/pdbe/emdb/) under the following accession numbers: the final composite reconstruction EMD-45492, the consensus reconstruction EMD-45510, and all focused reconstructions; the core EMD-45511, the TSC1 HR dimer EMD-45512, TBC1D7 alone EMD-45513, the TBC1D7/TSC2 HRs EMD-45514, WIPI3 alone EMD-45515, and the WIPI3 TSC2 HRs EMD-45529. The coordinates are deposited in the Protein Data Bank (www.rcsb.org) with accession numbers 9C9I (TSC1:WIPI3 crystal structure) and 9CE3 (TSC:WIPI3 composite cryoEM reconstruction).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.Laplante M., Sabatini D. M., mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D., Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5, 578–581 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Inoki K., Li Y., Xu T., Guan K. L., Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J., Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Castro A. F., Rebhun J. F., Clark G. J., Quilliam L. A., Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278, 32493–32496 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Duran R. V., Hall M. N., Regulation of TOR by small GTPases. EMBO Rep. 13, 121–128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dibble C. C., Elis W., Menon S., Qin W., Klekota J., Asara J. M., Finan P. M., Kwiatkowski D. J., Murphy L. O., Manning B. D., TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 47, 535–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henske E. P., Jozwiak S., Kingswood J. C., Sampson J. R., Thiele E. A., Tuberous sclerosis complex. Nat. Rev. Dis. Primers. 2, 16035 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Manning B. D., The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 412, 179–190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L., TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Inoki K., Zhu T., Guan K. L., TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P., Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121, 179–193 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J., Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. U.S.A. 101, 13489–13494 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C., Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Inoki K., Li Y., Zhu T., Wu J., Guan K. L., TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Potter C. J., Pedraza L. G., Xu T., Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4, 658–665 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Bakula D., Muller A. J., Zuleger T., Takacs Z., Franz-Wachtel M., Thost A. K., Brigger D., Tschan M. P., Frickey T., Robenek H., Macek B., Proikas-Cezanne T., WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 8, 15637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demetriades C., Doumpas N., Teleman A. A., Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S., Zhang Y., Ting C. Y., Bettedi L., Kim K., Ghaniam E., Lilly M. A., The Rag GTPase regulates the dynamic behavior of TSC downstream of both amino acid and growth factor restriction. Dev. Cell 55, 272–288.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prentzell M. T., Rehbein U., Sandoval M. C., De Meulemeester A. S., Baumeister R., Brohee L., Berdel B., Bockwoldt M., Carroll B., Chowdhury S. R., von Deimling A., Demetriades C., Figlia G.; Genomics England Research Consortium, de Araujo M. E. G., Heberle A. M., Heiland I., Holzwarth B., Huber L. A., Jaworski J., Kedra M., Kern K., Kopach A., Korolchuk V. I., van ‘t Land-Kuper I., Macias M., Nellist M., Palm W., Pusch S., Pittol J. M. R., Reil M., Reintjes A., Reuter F., Sampson J. R., Scheldeman C., Siekierska A., Stefan E., Teleman A. A., Thomas L. E., Torres-Quesada O., Trump S., West H. D., de Witte P., Woltering S., Yordanov T. E., Zmorzynska J., Opitz C. A., Thedieck K., G3BPs tether the TSC complex to lysosomes and suppress mTORC1 signaling. Cell 184, 655–674.e27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H., Yu Z., Chen X., Li J., Li N., Cheng J., Gao N., Yuan H. X., Ye D., Guan K. L., Xu Y., Structural insights into TSC complex assembly and GAP activity on Rheb. Nat. Commun. 12, 339 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramlaul K., Fu W., Li H., de Martin Garrido N., He L., Trivedi M., Cui W., Aylett C. H. S., Wu G., Architecture of the tuberous sclerosis protein complex. J. Mol. Biol. 433, 166743 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzian K., Bruckner A., Brohee L., Zech R., Antoni C., Kiontke S., Gasper R., Linard Matos A. L., Beel S., Wilhelm S., Gerke V., Ungermann C., Nellist M., Raunser S., Demetriades C., Oeckinghaus A., Kummel D., TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation. Mol. Cell 81, 2705–2721.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Dai D. L., Hasan S. M. N., Woollard G., Abbas Y. M., Bueler S. A., Julien J. P., Rubinstein J. L., Mazhab-Jafari M. T., Structural characterization of endogenous tuberous sclerosis protein complex revealed potential polymeric assembly. Biochemistry 60, 1808–1821 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Ren J., Liang R., Wang W., Zhang D., Yu L., Feng W., Multi-site-mediated entwining of the linear WIR-motif around WIPI beta-propellers for autophagy. Nat. Commun. 11, 2702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebner M., Puchkov D., Lopez-Ortega O., Muthukottiappan P., Su Y., Schmied C., Zillmann S., Nikonenko I., Koddebusch J., Dornan G. L., Lucht M. T., Koka V., Jang W., Koch P. A., Wallroth A., Lehmann M., Brugger B., Pende M., Winter D., Haucke V., Nutrient-regulated control of lysosome function by signaling lipid conversion. Cell 186, 5328–5346.e26 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Posor Y., Jang W., Haucke V., Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 23, 797–816 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baskaran S., Ragusa M. J., Boura E., Hurley J. H., Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell 47, 339–348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang R., Ren J., Zhang Y., Feng W., Structural conservation of the two phosphoinositide-binding sites in WIPI proteins. J. Mol. Biol. 431, 1494–1505 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Schrotter S., Yuskaitis C. J., MacArthur M. R., Mitchell S. J., Hosios A. M., Osipovich M., Torrence M. E., Mitchell J. R., Hoxhaj G., Sahin M., Manning B. D., The non-essential TSC complex component TBC1D7 restricts tissue mTORC1 signaling and brain and neuron growth. Cell Rep. 39, 110824 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A. J., Berger B., Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J. M., Sorzano C. O. S., Vargas J., DeepEMhancer: A deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He J., Li T., Huang S. Y., Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun. 14, 3217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senior A. W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Zidek A., Nelson A. W. R., Bridgland A., Penedones H., Petersen S., Simonyan K., Crossan S., Kohli P., Jones D. T., Silver D., Kavukcuoglu K., Hassabis D., Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Qin J., Wang Z., Hoogeveen-Westerveld M., Shen G., Gong W., Nellist M., Xu W., Structural basis of the interaction between tuberous sclerosis complex 1 (TSC1) and Tre2-Bub2-Cdc16 domain family member 7 (TBC1D7). J. Biol. Chem. 291, 8591–8601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Aragao D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N. P., Ericsson D. J., Gee C. L., Macedo S., Mudie N., Panjikar S., Price J. R., Riboldi-Tunnicliffe A., Rostan R., Williamson R., Caradoc-Davies T. T., MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 25, 885–891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Tools for ligand validation in Coot. Acta Crystallogr. D. Struct. Biol. 73, 203–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., Dong M. Q., Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., Marra M. A., Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fokkema I., Kroon M., Lopez Hernandez J. A., Asscheman D., Lugtenburg I., Hoogenboom J., den Dunnen J. T., The LOVD3 platform: Efficient genome-wide sharing of genetic variants. Eur. J. Hum. Genet. 29, 1796–1803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tate J. G., Bamford S., Jubb H. C., Sondka Z., Beare D. M., Bindal N., Boutselakis H., Cole C. G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S. C., Kok C. Y., Noble K., Ponting L., Ramshaw C. C., Rye C. E., Speedy H. E., Stefancsik R., Thompson S. L., Wang S., Ward S., Campbell P. J., Forbes S. A., COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stenson P. D., Mort M., Ball E. V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A. D., Cooper D. N., The human gene mutation database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Punjani A., Fleet D. J., 3D variability analysis: Resolving continuous flexibility and discreteheterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Cook A. S. I., Hurley J. H., Toward a standard model for autophagosome biogenesis. J. Cell Biol. 222, e202304011 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 and S2
References