Abstract
Adaptation to high-altitude hypoxia is characterized by systemic and organ-specific metabolic changes. This study investigates whether intestinal metabolic rewiring is a contributing factor to hypoxia adaptation. We conducted a longitudinal analysis over 108 days, with seven time points, examining fecal metabolomic data from a cohort of 46 healthy male adults traveling from Chongqing (a.s.l. 243 m) to Lhasa (a.s.l. 3,658 m) and back. Our findings reveal that short-term hypoxia exposure significantly alters intestinal metabolic pathways, particularly those involving purines, pyrimidines, and amino acids. A notable observation was the significantly reduced level of intestinal uric acid, the end product of purine metabolism, during acclimatization (also called acclimation) and additional two long-term exposed cohorts (Han Chinese and Tibetans) residing in Shigatse, Xizang (a.s.l. 4,700 m), suggesting that low intestinal uric acid levels facilitate adaptation to high-altitude hypoxia. Integrative analyses with gut metagenomic data showed consistent trends in intestinal uric acid levels and the abundance of key uric acid-degrading bacteria, predominantly from the Lachnospiraceae family. The sustained high abundance of these bacteria in the long-term resident cohorts underscores their essential role in maintaining low intestinal uric acid levels. Collectively, these findings suggest that the rewiring of intestinal uric acid metabolism, potentially orchestrated by gut bacteria, is crucial for enhancing human resilience and adaptability in extreme environments.
Keywords: high-altitude hypoxia, intestinal UA, metabolic rewiring, UA-degrading bacteria, human acclimatization
Introduction
The challenge of high-altitude hypoxia presents a unique opportunity to observe the remarkable adaptability of human metabolism. The metabolic rewiring at both systemic and organ levels has been documented to confer significant benefits during hypoxia adaptation (Matheson et al. 1991; Baibas et al. 2005; Castillo et al. 2007). Native Tibetans, residing at elevations exceeding 3,000 m, demonstrate distinctive physiological adaptations (Zhou et al. 2023), such as stable glucose homeostasis (Braun 2008; Woolcott et al. 2015), reduced oxidative stress (Quindry et al. 2016; Horscroft et al. 2017), low cardiovascular disease mortality (Faeh et al. 2009), and fortified intestinal barrier function (McKenna et al. 2022), collectively mitigating the risks of metabolic syndrome in high-altitude hypoxic conditions. Adaptation extends beyond the respiratory, cardiovascular, and hematological systems, which adjust to ensure a continuous oxygen supply to metabolic tissues (Ramirez et al. 2007; Storz et al. 2010). In skeletal muscle, adaptation manifests as a rewiring of the tricarboxylic acid cycle (TCA) (Capitanio et al. 2017), while in the liver, it involves a shift in tryptophan metabolism, decreasing kynurenine synthesis in favor of tryptamine production (Mohapatra et al. 2021). These examples underscore the critical role of metabolic rewiring in systemic and organ-level adaptation to hypoxia (Midha et al. 2023). Yet, the intestinal metabolic rewiring and its role in promoting hypoxia acclimatization/adaptation remain largely unexplored.
The intestine, as a core metabolic site, plays an indispensable role in human adaptation to high-altitude environments by ensuring the integrity of the barrier and metabolic homeostasis. High-altitude–induced systemic hypoxia can precipitate ischemia of the intestinal mucosa, compromise the intestinal barrier, and disrupt the gut microbiota balance, leading to metabolic disorders (Suzuki et al. 2019). Consequently, individuals ascending to high altitudes frequently encounter gastrointestinal issues such as anorexia, abdominal discomfort, indigestion, acid reflux, and infectious diarrhea (Adak et al. 2014), which can exacerbate the risk of altitude-related diseases (Karl et al. 2018; McKenna et al. 2022).
In light of these challenges, our study analyzed fecal metabolomic data from 46 participants over seven time points to reveal the metabolic rewiring in the intestine triggered by systemic hypoxia. Our findings indicate that (i) hypoxia exposure significantly alters fecal metabolites and various metabolic pathways, including those of nucleotides (including purines and pyrimidines) and amino acids, with purine metabolism showing a particularly strong signal. (ii) The end product of purine metabolism, uric acid (UA), increased initially but declined during later exposure stages. Observations in additional cohorts, including six long-term Han Chinese populations (∼5 to 60 months) and native Tibetans living at high altitudes (Shigatse, Xizang, a.s.l. 4,700 m), consistently showed intestinal UA levels below detection thresholds. This consistent pattern indicates that lower intestinal UA concentrations may be advantageous for physiological adaptation to high-altitude hypoxia. (iii) Through gut metagenomic data analysis targeting UA gene clusters, we identified a notable presence of UA-degrading bacteria within both short-term and long-term Han Chinese cohorts, as well as native Tibetans. These bacteria, largely from the Lachnospiraceae family, increased in abundance under hypoxia exposure, a trend that paralleled the changes in intestinal UA levels. Our findings reveal that intestinal metabolism, particularly UA metabolism, undergoes adaptive rewiring likely driven by gut bacteria, thereby facilitating human acclimatization/adaptation to high-altitude hypoxia.
Results
Significant Alterations in Fecal Metabolites Occurred with High-Altitude Hypoxia Exposure
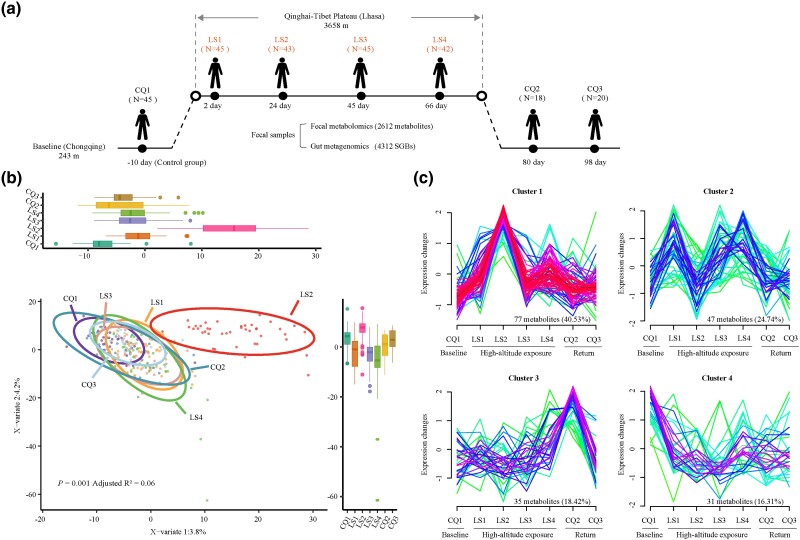
We collected 260 fecal samples at seven distinct time points from 46 participants who transitioned from the lowland plains (a.s.l. 243 m) to the Qinghai–Tibet Plateau (a.s.l. 3,658 m), where they spent 73 days. Subsequently, 20 participants returned to the lowland areas, as illustrated in Fig. 1a.
Fig. 1.
Study design and fecal metabolite dynamics during high-altitude hypoxia. a) Diagrammatic representation of the study design, illustrating exposure to high-altitude hypoxia and the ensuing comprehensive longitudinal analysis of fecal metabolomics and metagenomics. b) PLS-DA score plot of 2311 identified fecal metabolites from 260 samples. Each point denotes an individual sample, with color coding reflecting different time points, and coordinates values for each sample at each time point depicted in boxplots. c) Cluster analysis of 190 significant fecal metabolites (fold change > 1, VIP > 1, FDR < 0.2), illustrating their temporal patterns associated with hypoxic exposure.
To minimize the confounding influences of diet and lifestyle on our findings, we employed rigorous controls: Participants refrained from antibiotic use for at least 1 month and ceased probiotic product intake 2 weeks prior to the initial sampling (CQ1, control group). Meals were standardized across participants, provided in a unified canteen, and consisted of traditional Chinese cuisine, including rice, vegetables, and meat, with alcohol consumption prohibited. Additionally, participants shared uniform accommodation and maintained comparable daily routines. Subsequently, our metabolomic data yielded an abundance profile of 2,612 metabolites. After applying quality control protocols outlined in the Materials and Methods, 2,311 metabolites were retained for in-depth study.
Partial least-squares discriminant analysis (PLS-DA) indicated that high-altitude hypoxia significantly influenced the fecal metabolite profile (PERMANOVA P = 0.001; Fig. 1b). The distinct separation of the LS2 time point from others suggests a substantial correlation between fecal metabolites and hypoxia exposure, independent of potential dietary changes postarrival at high altitude. To further explore the dynamics of fecal metabolites under hypoxia, 190 significantly differential fecal metabolites (SDFMs) [fold change > 1, variable importance in projection (VIP) > 1, false discovery rate (FDR) < 0.2] were categorized into four clusters based on their longitudinal trajectories (Fig. 1c; supplementary fig. S1 and table S1, Supplementary Material online). Some metabolites exhibited initially upregulated (LS1 and LS2) followed by a recovery trend (from LS3-CQ3) (Cluster 1), while others increased during initial exposure (LS1) and continued to fluctuate (since LS2) (Cluster 2). The remaining metabolites remained stable during exposure and surged upon return to lowland (Cluster 3) or consistently decreased from the onset of exposure (Cluster 4). The majority of fecal metabolites (65.2%) fell into Cluster 1 (n = 77, 40.5%) and Cluster 2 (n = 47, 24.7%), indicating a clear influence of hypoxia exposure on fecal metabolite dynamics.
The SDFMs Were Enriched Mainly in Purine, Pyrimidine, and Amino Acid Metabolism Pathways
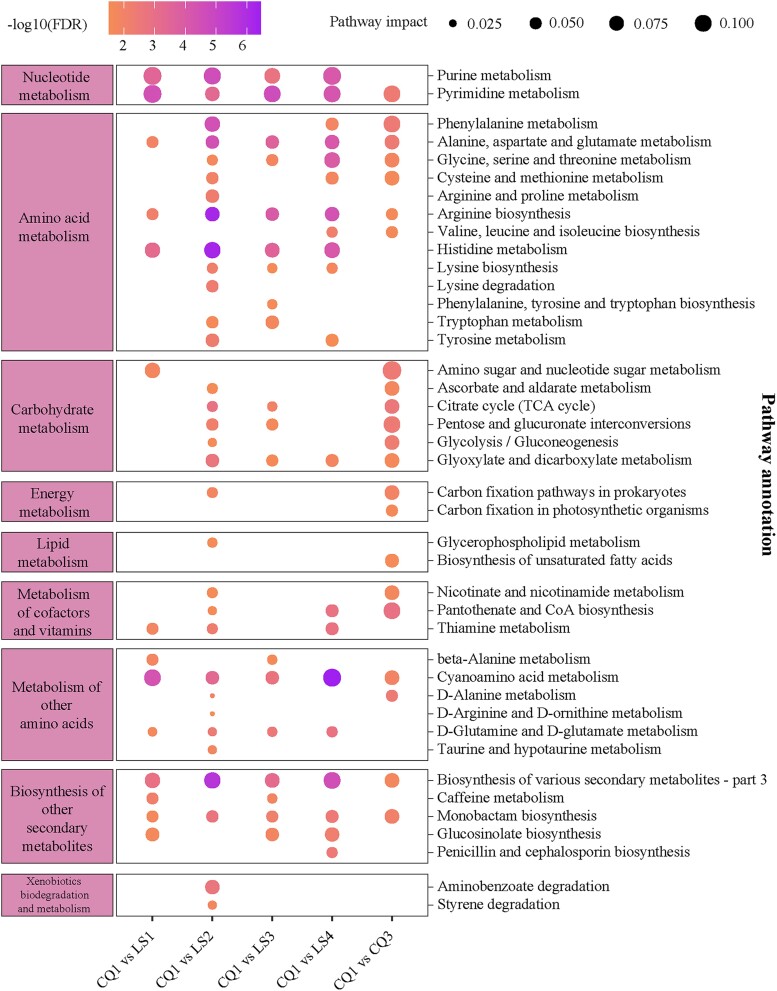
To elucidate the metabolic pathways that are altered under hypoxic conditions, enrichment analysis was performed utilizing the SDFMs (Fig. 2; supplementary table S2, Supplementary Material online). Our analysis revealed significant enrichment of SDFMs within nucleotide metabolism pathways (including purine and pyrimidine metabolism), amino acid metabolism (such as histidine metabolism, arginine biosynthesis, and the metabolism of alanine, aspartate, and glutamate), and carbohydrate metabolism (FDR < 0.05). Notably, upon descent to lowland conditions (CQ3), these pathways continued to show significant enrichment (FDR < 0.05) compared to the baseline, particularly in pyrimidine metabolism and amino acid metabolism. These persistent modifications suggest a potential biological significance in the acclimatization to hypoxic environments.
Fig. 2.
KEGG pathway-based functional profiling of fecal metabolites. A bubble plot illustrates the significantly enriched KEGG pathways at various time points. The CQ2 time point is omitted due to the absence of significant pathway enrichment compared to the baseline. The left and right panels categorize the pathways according to KEGG metabolic categories: Levels B and C, respectively. In the central panel, bubble colors indicate the significance of enrichment relative to the baseline, while bubble sizes reflect the proportion of metabolites associated with each pathway. Enrichment significance (FDR) is calculated using Fisher's exact test (see “Materials and Methods”).
Pathways demonstrating significant enrichment (FDR < 0.05) across at least two exposure time points were indicative of robust alterations under hypoxia (Butterfield et al. 2006; Torres et al. 2020). Our study identified 14 such metabolic pathways and 64 metabolites from the SDFMs (supplementary table S3, Supplementary Material online). Remarkably, three organic acids (fumarate, oxoglutaric acid, and pyruvate), and several amino acids, such as L-glutamine and glycine, were involved in a broad spectrum of metabolic pathways, encompassing both amino acid and nucleotide metabolism.
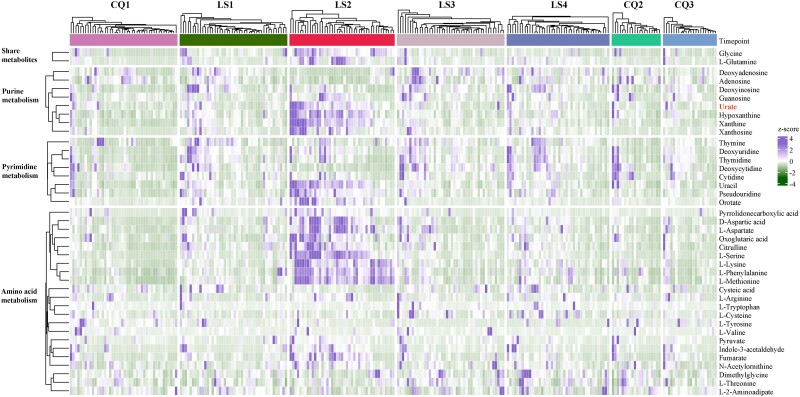
Significant shifts in fecal metabolites related to purine, pyrimidine, and amino acid metabolism primarily occurred during the exposure period (supplementary figs. S2 and S3 and table S2, Supplementary Material online). A total of 40 metabolites within these pathways demonstrated a high degree of consistency across individuals, particularly LS2 (Fig. 3). Within purine metabolism, metabolites such as urate (also known as UA), hypoxanthine, xanthosine, and xanthine were categorized under Cluster 1, while deoxyinosine and deoxyadenosine were in Cluster 2 and adenosine in Cluster 3. In pyrimidine metabolism, uracil, orotate, and pseudouridine were in Cluster 1, while deoxyuridine, thymidine, and thymine in Cluster 2 and deoxycytidine in Cluster 3. In amino acid metabolism, the majority of metabolites, including nine amino acids (L-glutamine, D-aspartic acid, L-aspartate, L-methionine, L-phenylalanine, L-serine, L-lysine, glycine, and citrulline) were in Cluster 1, with only L-tyrosine in Cluster 4. The trajectories of three key organic acids (fumarate, oxoglutaric acid, and pyruvate) were also in Cluster 1 (supplementary table S1, Supplementary Material online).
Fig. 3.
Core metabolites dynamics under high-altitude hypoxia. This heatmap illustrates the longitudinal trajectory of key metabolites implicated in purine, pyrimidine, and amino acid metabolism, which are pivotal for high-altitude adaptation in humans. The left panel categorizes the pathways (Level B) enriched by these 40 notable metabolites. In the central panel, each grid corresponds to the relative abundance of a specific metabolite within a given sample, with the grid color denoting the z-score of the metabolite's relative abundance. On the right-hand panel, there corresponds a name for each metabolite. For additional insights, refer to supplementary figs. S2 and S3 and table S7, Supplementary Material online.
Overall, during the exposure period, the majority of metabolites were enriched in the three pathways: those involved in purine and pyrimidine metabolism were mainly in Cluster 1 or 2 (12/14), while amino acids and organic acids were predominantly in Cluster 1 (11/12). These findings suggest that the alterations of 40 core metabolites in these pathways occur predominantly during the early stages of exposure, as evidenced by the rapid increase from LS1 to LS2.
Lower Levels of Intestinal UA Were Observed in Cohorts with Short-Term or Long-Term Exposure to High-Altitude Hypoxia
Our pathway enrichment analysis indicated significant alterations in metabolic pathways when compared to baseline levels under hypoxic conditions. Specifically, purine metabolism, pyrimidine metabolism, and cyanoamino acid (other amino acids) metabolism pathways showed the most substantial enrichment (pathway impact score > 0.1 and FDR < 0.001). Purine metabolism, in particular, was consistently upregulated across all four exposure time points (FDR < 0.05; supplementary table S2, Supplementary Material online), with the highest pathway impact values observed at LS1 and LS4. These findings imply a critical role for intestinal purine metabolism in the physiological alterations under hypoxia and in facilitating adaptability to high-altitude conditions.
In line with these findings, we noted an initial increase in the abundance of UA, at LS2, followed by a reduction to baseline levels at LS3 and LS4 within our short-term exposure cohort. This pattern suggests the importance of maintaining low intestinal UA levels during later stages of exposure as the host acclimates to high-altitude hypoxia. Further validation was sought through nontargeted fecal metabolomic data from long-term exposure cohorts, including 163 Han residents (residing at high altitudes for above 5 to 60 months) and 28 native Tibetans from Shigatse, Xizang (a.s.l. 4,700 m). Remarkably, both cohorts displayed intestinal UA levels below the detection threshold, indicating a near absence of UA among long-term residents and natives adapted to high-altitude living. These results underscore the potential adaptive advantage of low intestinal UA levels under chronic high-altitude hypoxic conditions.
Lower Intestinal UA Levels Significantly Correlated with the Degradation of Gut Bacteria
Recent research has pinpointed a key gene cluster in gut bacteria crucial for degrading UA, thereby regulating UA levels in the host's intestine and blood (Kasahara et al. 2023; Liu, Xu, et al. 2023; Liu, Jarman, et al. 2023). Our study sought to detect this gene cluster in our cohorts, examining its link to intestinal UA levels under hypoxic conditions.
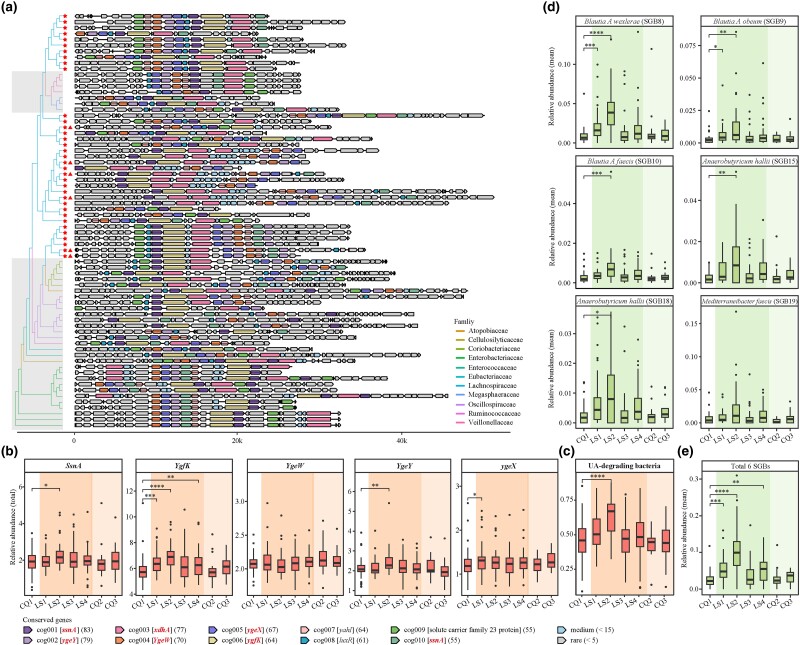
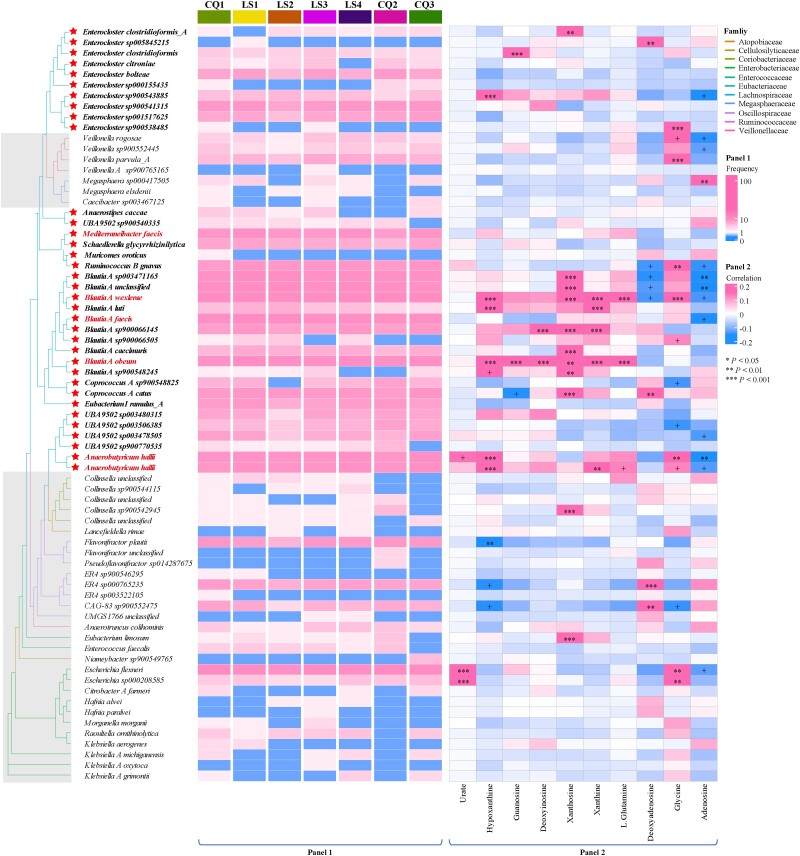
Analysis revealed 71 gene clusters spanning 5 phyla, 11 families, and 33 genera (Fig. 4a; supplementary table S4, Supplementary Material online). The Lachnospiraceae family within Firmicutes A was notably prevalent, representing 49.2% across seven time points (Fig. 5, Panel 1), with the Blautia A and Enterocloster genera each contributing 14%. Extending this analysis to 163 fecal metagenomic samples from six long-term exposure cohorts in Shigatse, Xizang (a.s.l. 4,700 m), we predicted 153 UA gene clusters (supplementary table S5, Supplementary Material online). Lachnospiraceae was again the most prominent family, particularly the Collinsella (17.6%), Blautia A (10.4%), and Enterocloster (7.1%) genera (supplementary fig. S4, Supplementary Material online). These patterns suggest a potential role for Lachnospiraceae in modulating intestinal UA levels under hypoxic stress.
Fig. 4.
Characteristics of UA gene cluster. a) The phylogenetic landscape of the UA gene cluster is depicted, with asterisks denoting SGBs from the Lachnospiraceae family and triangles highlighting the six indicator species in d). The shaded gray region delineates families distinct from Lachnospiraceae. See also supplementary table S4, Supplementary Material online. The term “cog” symbolizes the cluster orthogroup, with gene symbol IDs encased in square brackets. IDs highlighted in bold italic font signify UA-inducible genes validated by prior studies (Kasahara et al. 2023; Liu, Xu, et al. 2023; Liu, Jarman, et al. 2023), while the remaining IDs pertain to genes identified in our present study. Numbers in parentheses indicate the frequency of each gene within its respective cluster orthogroup. b) The total relative abundance of five UA-inducible genes in each sample, based on 1,079 SGBs. UA-inducible genes are styled in italicized text. Each point corresponds to an individual sample, with color gradients reflecting distinct exposure stages. c) The total abundance changes of UA-degrading bacteria in each sample. Here, “UA-degrading bacteria” refers to bacteria that harbor all five UA-inducible genes. d, e) The dynamic shifts in the relative abundance of the six indicator SGBs at the LS2 time point and their cumulative relative abundances, respectively. Statistical significance is denoted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001 by Wilcoxon rank-sum test.
Fig. 5.
Gene clusters from Lachnospiraceae exhibit the most stable and abundant capacity for UA consumption. Panel 1: Gene clusters from the Lachnospiraceae family demonstrate a significant and consistent capacity for UA consumption. The asterisk on the left highlights SGBs belonging to the Lachnospiraceae family. The red font emphasizes the six indicator species outlined in Fig. 4. The squares represent the prevalence of UA gene clusters across individuals at sequential time points, with frequencies above ten highlighted in the same color. Panel 2: Spearman's correlation coefficients detail the relationships between the relative abundance of SGBs, where the gene clusters are sourced, and ten fecal metabolites linked to purine metabolism. Correlations with absolute values exceeding 0.2 are indicated in the same color. Statistical significance is denoted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Then, we quantified the total abundance of five UA-inducible genes, observing a significant increase in three at LS2 (Fig. 4b). Notably, the UA-degrading bacteria count was significantly higher only at LS2 (Fig. 4c). Further analysis revealed a significant positive correlation between purine-enriched metabolites and 71 UA-degrading bacteria, predominantly Lachnospiraceae, at LS2 (Fig. 5, Panel 2; supplementary fig. S5, Supplementary Material online). Concurrently, fecal UA levels reached the peaks at this time point, coinciding with a marked increase in indicator SGBs with UA gene clusters (Fisher's exact test, P < 0.05), comprising over 10% of the bacterial abundance (Fig. 4d and e; supplementary table S6, Supplementary Material online). This trend suggests that UA may encourage the growth of UA-degrading bacteria, supporting their role in UA regulation, particularly in the initial 2 weeks of hypoxic exposure.
Discussion
This study deciphers the rewiring of intestinal metabolism in lowlanders during high-altitude hypoxia, focusing on its association with the host's adaptability to hypoxia. Short-term hypoxia exposure induced significant metabolic shifts in purine, pyrimidine, and amino acid pathways (hallmarks of hypoxia). These changes peak early (LS1-LS2) and then return to baseline, indicating a rapid initial impact on fecal metabolites followed by a recovery stage. This suggests that gut metabolism is regulated in response to high-altitude hypoxia, highlighting the intestine's essential role in maintaining cellular function and ensuring host survival in such conditions (Rocchetti et al. 2022; Zhao et al. 2023). Notably, the increase of multiple amino acids may protect cells from the challenge of hypoxia (Weinberg et al. 2016), like L-glutamine protects against hypoxia stress (Weinberg et al. 2016) and supports the intestinal barrier (Dos et al. 2020; Schumacher et al. 2021), vital for physiological and immunological functions, and microbial diversity (Xu et al. 2014). Similarly, glycine can protect cell function during hypoxia (Weinberg et al. 2016). Phenylalanine helps establish effective energy metabolism (Lin et al. 2016) and promote lung function under hypoxic conditions (Førli et al. 2000). Moreover, the conversion of amino acid and pyrimidine end-products into TCA cycle intermediates like fumarate, oxoglutaric acid, and pyruvate boosts energy metabolism, crucial for host endurance in high altitudes (Rocchetti et al. 2022; Zhao et al. 2023).
Purine metabolism, in particular, displayed a pronounced enrichment during hypoxia exposure. Intestinal UA, its end product, showed a significant initial increase followed by a return to baseline levels in individuals exposed to short-term hypoxia. Intriguingly, populations long resident at high altitudes, such as the Han and native Tibetans, presented with undetectable UA levels. Generally, hypoxia disrupts energy metabolism, accelerating ATP degradation and subsequently enhancing purine metabolite production (Tran-Thi et al. 1994). Hypoxia also stimulates erythropoiesis, increasing the enucleation of mature red blood cells, which further elevates purine metabolism (Anderson et al. 2018; Semenza 2022). These processes ultimately lead to increased synthesis and accumulation of blood UA, peaking around 30 days of hypoxic exposure (Sinha et al. 2009; Bartziokas et al. 2014; Du et al. 2022). Interestingly, we found that 40.53% of fecal metabolites under exposure belong to Cluster 1, the most abundant cluster type that changes significantly at the LS2 time point. This suggests that the critical time point for gut metabolic rewiring under high-altitude exposure likely occurred around 1 month. Nevertheless, UA is not directly utilized by the body and is excreted via the kidneys (2/3 load) and intestines (1/3 load) (Yanai et al. 2021). Under hypoxic conditions, renal clearance is impaired, and elevated blood lactate levels competitively inhibit UA excretion, reducing renal clearance by 70% to 80%, leading to UA accumulation (Vij et al. 2005; Mao et al. 2024). This accumulation is associated with oxidative stress and cardiovascular diseases (Hayden and Tyagi 2004; Hurtado-Arestegui et al. 2018), as well as endothelial dysfunction and mitochondrial damage (Yin et al. 2019; Liu, Xu, et al. 2023; Liu, Jarman, et al. 2023). Additionally, it significantly impacts intestinal health and potentially leads to hyperuricemia and gout (Sahebjami and Scalettar 1971; Vij et al. 2005; Lv et al. 2020, 2021). These findings underscore the significance of intestinal UA clearance in hypoxic environments and suggest an adaptive rewiring of UA metabolism in the intestine, facilitating effective adaptation to high altitudes.
Further investigation disclosed a strong link between UA metabolic rewiring and gut microbiota composition. The gut microbiota regulates intestinal UA levels through various mechanisms, including influencing host metabolic pathways and immune responses, promoting gut health, and facilitating UA excretion. Certain gut bacteria have been reported to enhance UA breakdown, thereby reducing its concentration in the body. For instance, some bacteria can convert UA into nucleotides that support bacterial DNA synthesis (Goncheva et al. 2022) and improve hyperuricemia by inhibiting xanthine oxidase, a key enzyme in UA synthesis (Wang et al. 2019). Additionally, compared to healthy individuals, gout patients exhibit significantly lower levels of microbial metabolism related to butyrate biosynthesis, which can enhance the intestinal immune barrier, reduce inflammation, promote the growth of beneficial microbes, and facilitate UA excretion and clearance (Cleophas et al. 2016; Guo et al. 2016).
Here, we identified a proliferation of UA-degrading bacteria, predominantly from the Lachnospiraceae family, known for their anti-inflammatory properties (Zhang et al. 2019). These bacteria were significantly more abundant during short-term hypoxia exposure and maintained high levels among long-term high-altitude residents. This observation, in combination with previous studies, suggests their potential to degrade UA by (i) consuming UA, (ii) reducing intestinal inflammation and influencing host metabolism through bacterial metabolites like butyrate, ultimately enhancing UA transport and excretion (Wang and Ye 2024), and (iii) compensating for the loss of uricase to maintain UA homeostasis (Wang and Ye 2024). Furthermore, in line with the most significant changes in the gut microbiome at LS2 time point (Su et al. 2024), there is a simultaneous peak in both UA-degrading bacteria and UA levels at LS2, thereby reinforcing the gut microbiota's integral part in the intestinal UA metabolic rewiring.
In conclusion, our study has unveiled a strategic metabolic rewiring of UA in the intestine, representing an essential adaptation strategy that extends beyond a mere response to hypoxia. This rewiring appears to enable the host to regulate UA balance through gut bacteria, offering a foundation for microbiota-focused interventions in managing and preventing altitude-related ailments. Further investigation is needed to verify these relationships and mechanisms.
Materials and Methods
Study Population
From 2019 July 11 to October 26, we recruited 46 healthy male students, aged 21 to 27, with an average body mass index of 22.23 ± 1.97 kg/m2 (mean ± SD), for health examination at Lhasa Hospital. These participants were selected based on their lack of cardiopulmonary, metabolic, dyslipidemic, or gastrointestinal conditions. They provided the required information, written informed consent, and biospecimens, including fresh morning fecal samples at seven time points. The study received ethical approval from and the Ethics Committee at Kunming Institute of Zoology, Chinese Academy of Sciences (approval number: KIZRKX-2021-010).
Fecal Sample Collection
Fecal samples were systematically collected from 46 male participants aged 21 to 27 years across seven time points: 10 days prior to departure (CQ1, serving as the control) and at 2 (LS1), 24 (LS2), 45 (LS3), and 66 (LS4) days following arrival in Lhasa. Additional collections were conducted 1 week (80 days; CQ2) and 3 weeks (98 days; CQ3) after their return to lowland (Fig. 1a). None of the participants had been to a high altitude within 6 months before the study began. Participants were required to fast overnight for at least 8 h before sample collection. In total, 260 stool samples were collected from participants in a fasting state in the morning, including 44 at CQ1, 44 at LS1, 43 at LS2, 44 at LS3, 43 at LS4, 20 at CQ2, and 22 at CQ3, corresponding to the seven time points. All samples were immediately preserved at −80 °C. The genomic DNA of all fecal samples (from the seven time points) was extracted at the same time in the lab.
Metabolome Profiling of Fecal Samples
Fecal metabolite extraction was conducted using a 1:1 acetonitrile:methanol solution containing an isotopically labeled internal standard mixture. Analysis was performed on a UHPLC-QTOF-MS/MS system (Vanquish, Thermo Fisher Scientific, Bremen, Germany). The separation of metabolite extracts was achieved using a Waters Amide column (2.1 mm × 100 mm, 1.7 μm). We injected 2 µL of the metabolite solution and performed the elution using a mobile phase composed of phase A (25 mmol/L ammonium acetate and 25 mmol/L ammonium hydroxide in water, pH = 9.75) and phase B (acetonitrile), with a flow rate of 0.5 mL/min. The gradient was as follows: 95% B for 0 to 0.5 min, a linear gradient from 95% to 65% B over 0.5 to 7 min, 65% to 40% B over 7 to 8 min, held at 40% B for 8 to 9 min, a sharp increase from 40% to 95% B over 9 to 9.1 min, and then held at 95% B for 9.1 to 12 min. Metabolic fragments were acquired using a Q Exactive HFX mass spectrometer (Orbitrap MS, Thermo) coupled with electrospray ionization (ESI). ESI source settings were MS/MS resolution at 7,500, capillary temperature at 320 °C, and full MS resolution at 60,000. Ion spray voltage floating was set at 3.5 kV in positive mode and −3.2 kV in negative mode. A total of 7,059 peaks in positive mode and 4,599 peaks in negative mode were detected. Raw data were converted to mzXML format using ProteoWizard (v3.0) and processed with the R package XCMS for peak detection, extraction, alignment, and integration (Want et al. 2006). Metabolite annotations were performed using public databases (METLIN, HMDB, KEGG, CHEBI, mzCloud, mzVault, and LIPID) with a cutoff of 0.3, and duplicates were filtered based on LC-MS/MS scores.
Fecal volatile organic compounds were analyzed using an Agilent 7890 gas chromatograph with a DB-5MS capillary column (30 m × 250 μm × 0.25 μm, J&W Scientific, Folsom, CA, USA). MS-DIAL software (v4.0) was utilized for raw peak extraction, baseline filtering and calibration, peak alignment, deconvolution, identification, and integration of peak areas (Tsugawa et al. 2015). The Fiehn BinBase database was employed for metabolite identification, which included mass spectral and retention time index matching (Kind et al. 2009). After excluding QC samples with peak detection below 0.5 (Dunn et al. 2018), 1,175 fecal metabolite peaks were identified, and 2,612 features were detected in experimental samples. Of these, 2,311 metabolites with a high MS/MS similarity score (>60) were retained for further analysis.
Differential metabolites were identified using the Wilcoxon rank-sum test and adjusted by the FDR method. Criteria for screening included a VIP score of >1.0 and an FDR of <0.2 (Li et al. 2019; Fu et al. 2020). In total, 190 distinct fecal metabolites were identified when comparing baseline to the other six time points (supplementary table S1, Supplementary Material online). Metabolite annotation was conducted using the KEGG database (https://www.genome.jp/kegg/pathway.html), and VIP scores were calculated with MetaboAnalyst (v5.0) (Pang et al. 2021). Pathway enrichment (KEGG level C pathways) was analyzed using MBROLE3 (http://csbg.cnb.csic.es/mbrole3/index.php) (Huang et al. 2012), and metabolism-related functions were further examined. Graphs were generated in R (v4.0.2).
Dynamic Change Analysis (Mfuzz)
Fuzzy c-means clustering, a soft clustering approach, was executed using the R package Mfuzz (v2.54.0) (Kumar and Futschik 2007). We clustered differential fecal metabolites with the “Mufzz” function to delineate dynamic patterns of change. The optimal number of clusters was determined using the “WSS” and “Gap statistic” methods in Mfuzz (supplementary fig. S1, Supplementary Material online).
UA-Inducible Gene Prediction Based on Fecal Metagenomics
The five UA-inducible genes (ygeX, ygeY, ygeW, ygfK, and ssnA) were predicted from 1,079 high-quality species-level genomes (SGBs) derived from fecal metagenomic data of the same cohort (Su et al. 2024). These genes were chosen for their role in encoding enzymes that utilize UA and contribute to the maintenance of host UA homeostasis (Kasahara et al. 2023; Liu, Xu, et al. 2023; Liu, Jarman, et al. 2023). The conserved protein sequences of these five genes across different species were retrieved from the InterPro database (https://www.ebi.ac.uk/interpro/) (Paysan-Lafosse et al. 2023). Using BITACORA (v1.4.1) (Vizueta et al. 2020), we predicted the distribution of these genes with parameters set to an E-value of 1e−3 and a filter length of 50. Out of 586 SGBs, a total of 94 contigs displayed complete UA metabolic potential (encompassing all five UA-inducible genes). These 94 contigs are considered potential UA gene clusters. For further analysis, 71 contigs (supplementary table S4, Supplementary Material online) met stringent quality criteria: (i) genes classified as outliers, exceeding 1.5 times the interquartile range [q75 to q25], were excluded; and (ii) sequences 10-kb upstream and downstream of the five genes were retained, ensuring the total length did not surpass 50 kb.
The Validation Populations Resided at High Altitude for a Long Time
To further investigate the association between UA consumption by gut microbiota and adaptation to high-altitude hypoxia, we expanded our analysis to include fecal metagenomic and metabolomic data from 163 independent Han Chinese residents of Shigatse, Xizang (a.s.l. 4,700 m), with residency durations ranging from 5 to 60 months. We also incorporated fecal metabolomic data from 28 native Tibetans residing in the same region. The analytical approach for fecal metabolomic and UA gene cluster identification was aligned with the previously described methods. A total of 1,166 high-quality SGBs were selected for UA gene cluster prediction and analysis. However, the abundance of UA and urate in the feces were below the detection threshold of the Q Exactive HFX mass spectrometer; therefore, the fecal metabolomic results are not reported here.
Statistical Analysis
PLS-DA for the metabolome and microbiome data was conducted using the “plsda” function from the R package mixOmics (v6.18.1). Indicator species analysis was used to evaluate the dominant gut microbiota at various stages of high-altitude exposure, considering P < 0.05 as statistically significant (Leff et al. 2018). To assess significant differences in analytes between time points, we employed the Wilcoxon rank-sum test (paired). The KEGG pathway enrichment was analyzed using Fisher's exact test, focusing on the ratio of annotated metabolite counts to the total counts within specific pathways.
Supplementary Material
Acknowledgment
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research (2019QZKK0607 and 2019QZKK0503), the Yunnan Fundamental Research Projects (202401AS070073 and 202201AW070012), the Major Science and Technology Project in Yunnan Province of China (No. 202001BB050001), the CAS “Light of West China” Program of the Chinese Academy of Sciences (Y.-C.L.), the Digitalization, Development, and Application of Biotic Resource Program (202002AA100007), the High-level Talent Promotion and Training Project of Kunming (Spring City Plan; 2020SCP001), the Yunling Scholar of Yunnan Province (Q.-P.K. and Z.Z.), the Yunnan Ten Thousand Talents Plan Young & Elite Talents Project (Y.-C.L.), the Chinese National Natural Science Foundation (No. U2002206 and 31970571), the central government guides local science and technology development funds (202407AA110009), and the Hainan Provincial Natural Science Foundation of China (ZDYF2021SHFZ227). We also thank the Advanced Computing Center of Yunnan University for providing computational resources, as well as to Shanghai Biotree Biotech Co., Ltd. for the technical support.
Contributor Information
Qian Su, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yu-Chun Li, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Dao-Hua Zhuang, State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, School of Life Sciences, Yunnan University, Kunming, Yunnan 650091, China.
Xin-Yuan Liu, Department of Military Medical Geography, Army Health Service Training Base, Army Medical University, Chongqing 400038, China.
Han Gao, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Dong Li, College of Life Science, Sichuan Agricultural University, Ya’an, Sichuan 625014, China.
Yu Chen, Department of Military Medical Geography, Army Health Service Training Base, Army Medical University, Chongqing 400038, China.
Ming-Xia Ge, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Yi-Ming Han, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Zong-Liang Gao, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Fan-Qian Yin, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Long Zhao, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Yun-Xia Zhang, The Second Affiliated Hospital, the School of Basic Medicine and Life Sciences, Hainan Medical University, Hainan 570102, China.
Li-Qin Yang, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Qin Zhao, Hospital of Chengdu Office of People's Government of Tibetan Autonomous Region, Chengdu, Sichuan 610041, China.
Yong-Jun Luo, Department of Military Medical Geography, Army Health Service Training Base, Army Medical University, Chongqing 400038, China.
Zhigang Zhang, State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, School of Life Sciences, Yunnan University, Kunming, Yunnan 650091, China.
Qing-Peng Kong, Key Laboratory of Genetic Evolution & Animal Models (Chinese Academy of Sciences), Key Laboratory of Healthy Aging Research of Yunnan Province, Kunming Key Laboratory of Healthy Aging Study, KIZ/CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650201, China; CAS Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
Q.-P.K. designed and supervised the study, while Y.-C.L., Y.-J.L., Y.C., M.-X.G., X.-Y.L., F.-Q.Y., Y.-M.H., Z.-L.G., Y.-X.Z., Q.Z., and L.Z. were responsible for the sample collection. Z.-L.G., L.Z., and L.-Q.Y. performed total DNA extraction from the samples. Data analysis was carried out by Q.S., D.-H.Z., T.-Y.X., Q.S., and D.-H.Z. contributed to writing the manuscript. Y.-C.L., Q.-P.K., and Z.Z. provided valuable suggestions and made modifications to the manuscript. All authors reviewed and approved the final version of the manuscript.
Ethics Approval and Consent to Participate
This study was approved by the Institute Research Medical Ethics Committee of the Army Medical University, the Ethics Committee at Kunming Institute of Zoology, Chinese Academy of Sciences. A written informed consent was obtained from each participant.
Data Availability
All metagenomic sequencing data generated and analyzed in the present study have been deposited in the Genome Sequence Archive in the National Genomics Data Center (https://bigd.big.ac.cn/gsa/; accession number: subHRA005186).
References
- Adak A, Maity C, Ghosh K, Mondal KC. Alteration of predominant gastrointestinal flora and oxidative damage of large intestine under simulated hypobaric hypoxia. Z Gastroenterol. 2014:52(2):180–186. 10.1055/s-0033-1336007. [DOI] [PubMed] [Google Scholar]
- Anderson HL, Brodsky IE, Mangalmurti NS. The evolving erythrocyte: red blood cells as modulators of innate immunity. J Immunol. 2018:201(5):1343–1351. 10.4049/jimmunol.1800565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baibas N, Trichopoulou A, Voridis E, Trichopoulos D. Residence in mountainous compared with lowland areas in relation to total and coronary mortality. A study in rural Greece. J Epidemiol Commun H. 2005:59(4):274–278. 10.1136/jech.2004.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartziokas K, Papaioannou AI, Loukides S, Papadopoulos A, Haniotou A, Papiris S, Kostikas K. Serum uric acid as a predictor of mortality and future exacerbations of COPD. Eur Respir J. 2014:43(1):43–53. 10.1183/09031936.00209212. [DOI] [PubMed] [Google Scholar]
- Braun B. Effects of high altitude on substrate use and metabolic economy: cause and effect? Med Sci Sport Exer. 2008:40(8):1495–1500. 10.1249/MSS.0b013e3181729dd3. [DOI] [PubMed] [Google Scholar]
- Butterfield LH, Ribas A, Dissette VB, Lee Y, Yang JQ, De la Rocha P, Duran SD, Hernandez J, Seja E, Potter DM, et al. A phase I/II trial testing immunization of hepatocellular carcinoma patients with dendritic cells pulsed with four alpha-fetoprotein peptides. Clin Cancer Res. 2006:12(9):2817–2825. 10.1158/1078-0432.CCR-05-2856. [DOI] [PubMed] [Google Scholar]
- Capitanio D, Fania C, Torretta E, Viganò A, Moriggi M, Bravatà V, Caretti A, Levett DZH, Grocott MPW, Samaja M, et al. TCA cycle rewiring fosters metabolic adaptation to oxygen restriction in skeletal muscle from rodents and humans. Sci Rep. 2017:7(1):9723. 10.1038/s41598-017-10097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo O, Woolcott OO, Gonzales E, Tello V, Tello L, Villarreal C, Méndez N, Damas L, Florentini E. Residents at high altitude show a lower glucose profile than sea-level residents throughout 12-hour blood continuous monitoring. High Alt Med Biol. 2007:8(4):307–311. 10.1089/ham.2007.8407. [DOI] [PubMed] [Google Scholar]
- Cleophas MCP, Crişan TO, Lemmers H, Toenhake-Dijkstra H, Fossati G, Jansen TL, Dinarello CA, Netea MG, Joosten LAB. Suppression of monosodium urate crystal-induced cytokine production by butyrate is mediated by the inhibition of class I histone deacetylases. Ann Rheum Dis. 2016:75(3):593–600. 10.1136/annrheumdis-2014-206258. [DOI] [PubMed] [Google Scholar]
- Dos SQM, Souza W, Lemos VA, Caris AV, Thomatieli-Santos RV. The possible importance of glutamine supplementation to mood and cognition in hypoxia from high altitude. Nutrients. 2020:12(12):3627. 10.3390/nu12123627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Qi M, Wang W, Chen B. Effect of high-altitude hypoxia environment on uric acid excretion, desmin protein level in podocytes, and Na+-K+- ATPase activity. Cell Mol Biol. 2022:68(6):84–91. 10.14715/cmb/2022.68.6.14. [DOI] [PubMed] [Google Scholar]
- Dunn WB, David B, Paul B, Eva Z, Sue FM, Nadine A, Marie B, Knowles JD, Antony H, Haselden JN. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2018:6(7):1060–1083. 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Faeh D, Gutzwiller F, Bopp M. Lower mortality from coronary heart disease and stroke at higher altitudes in Switzerland. Circulation. 2009:120(6):495–501. 10.1161/circulationaha.108.819250. [DOI] [PubMed] [Google Scholar]
- Førli L, Pedersen JI, Bjørtuft, Vatn M, Kofstad J, Boe J. Serum amino acids in relation to nutritional status, lung function and energy intake in patients with advanced pulmonary disease. Resp Med. 2000:94(9):868–874. 10.1053/rmed.2000.0830. [DOI] [PubMed] [Google Scholar]
- Fu M, Zhang X, Liang Y, Lin S, Qian W, Fan S. Alterations in vaginal microbiota and associated metabolome in women with recurrent implantation failure. mBio. 2020:11(3):e03242–e03219. 10.1128/mBio.03242-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncheva MI, Chin D, Heinrichs DE. Nucleotide biosynthesis: the base of bacterial pathogenesis. Trends Microbiol. 2022:30(8):793–804. 10.1016/j.tim.2021.12.007. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, Su X, Qiao J, Zheng Y, Wang L, et al. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep. 2016:6(1):20602. 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MR, Tyagi SC. Uric acid: a new look at an old risk marker for cardiovascular disease, metabolic syndrome, and type 2 diabetes mellitus: the urate redox shuttle. Nutr Metab. 2004:1(1):10. 10.1186/1743-7075-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horscroft JA, Kotwica AO, Laner V, West JA, Hennis PJ, Levett D, Howard DJ, Fernandez BO, Burgess SL, Ament Z, et al. Metabolic basis to Sherpa altitude adaptation. Proc Natl Acad Sci U S A. 2017:114(24):6382–6387. 10.1073/pnas.1700527114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XD, Tan HY, Long R, Liang JB, Wright AG. Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai–Tibetan plateau, China. BMC Microbiol. 2012:12(1):237. 10.1186/1471-2180-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Arestegui A, Plata-Cornejo R, Cornejo A, Mas G, Carbajal L, Sharma S, Swenson ER, Johnson RJ, Pando J. Higher prevalence of unrecognized kidney disease at high altitude. J Nephrol. 2018:31(2):263–269. 10.1007/s40620-017-0456-0. [DOI] [PubMed] [Google Scholar]
- Karl JP, Berryman CE, Young AJ, Radcliffe PN, Branck TA, Pantoja-Feliciano IG, Rood JC, Pasiakos SM. Associations between the gut microbiota and host responses to high altitude. Am J Physiol-Gastr L. 2018:315(6):G1003–G1015. 10.1152/ajpgi.00253.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Kerby RL, Zhang Q, Pradhan M, Mehrabian M, Lusis AJ, Bergström G, Bäckhed F, Rey FE. Gut bacterial metabolism contributes to host global purine homeostasis. Cell Host Microbe. 2023:31(6):1038–1053. 10.1016/j.chom.2023.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009:81(24):10038–10048. 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007:2(1):5–7. 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff J, Bardgett R, Wilkinson A, Jackson B, Pritchard W, De Long J, Oakley S, Mason K, Ostle N, Johnson D, et al. Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 2018:12(7):1794–1805. 10.1038/s41396-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li J, Wang H, Qi LW, Zhu Y, Lai M. Tyrosine and glutamine-leucine are metabolic markers of early-stage colorectal cancers. Gastroenterology. 2019:157(1):257–259. 10.1053/j.gastro.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Lin X, Zhao L, Tang S, Zhou Q, Lin Q, Li X, Zheng H, Gao H. Metabolic effects of basic fibroblast growth factor in streptozotocin-induced diabetic rats: a 1H NMR-based metabolomics investigation. Sci Rep. 2016:6(1):36474. 10.1038/srep36474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Xu G, Sun B, Wu G, Chen J, Gao Y. Clinical and biochemical indices of people with high-altitude experience linked to acute mountain sickness. Travel Med Infect Di. 2023:51:102506. 10.1016/j.tmaid.2022.102506. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jarman JB, Low YS, Augustijn HE, Huang S, Chen H, DeFeo ME, Sekiba K, Hou BH, Meng X, et al. A widely distributed gene cluster compensates for uricase loss in hominids. Cell. 2023:186(16):3400–3413. 10.1016/j.cell.2023.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Xu D, Ma J, Wang Y, Yang X, Zhao P, Ma L, Li Z, Yang W, Liu X, et al. Uric acid drives intestinal barrier dysfunction through TSPO-mediated NLRP3 inflammasome activation. Inflamm Res. 2021:70(1):127–137. 10.1007/s00011-020-01409-y. [DOI] [PubMed] [Google Scholar]
- Lv Q, Xu D, Zhang X, Yang X, Zhao P, Cui X, Liu X, Yang W, Yang G, Xing S. Association of hyperuricemia with immune disorders and intestinal barrier dysfunction. Front Physiol. 2020:11:524236. 10.3389/fphys.2020.524236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang J, Zhou Q, He X, Zheng Z, Wei Y, Zhou K, Lin Y, Yu H, Zhang H, et al. Hypoxia induces mitochondrial protein lactylation to limit oxidative phosphorylation. Cell Res. 2024:34(1):13–30. 10.1038/s41422-023-00864-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson GO, Allen PS, Ellinger DC, Hanstock CC, Gheorghiu D, McKenzie DC, Stanley C, Parkhouse WS, Hochachka PW. Skeletal muscle metabolism and work capacity: a 31P-NMR study of Andean natives and lowlanders. J Appl Physiol. 1991:70(5):1963–1976. 10.1152/jappl.1991.70.5.1963. [DOI] [PubMed] [Google Scholar]
- McKenna ZJ, Gorini PF, Gillum TL, Amorim FT, Deyhle MR, Mermier CM. High-altitude exposures and intestinal barrier dysfunction. Am J Physiol Regul Integr Comp Physiol. 2022:322(3):R192–R203. 10.1152/ajpregu.00270.2021. [DOI] [PubMed] [Google Scholar]
- Midha AD, Zhou Y, Queliconi BB, Barrios AM, Haribowo AG, Chew B, Fong C, Blecha JE, VanBrocklin H, Seo Y, et al. Organ-specific fuel rewiring in acute and chronic hypoxia redistributes glucose and fatty acid metabolism. Cell Metab. 2023:35(3):504–516. 10.1016/j.cmet.2023.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra SR, Sadik A, Sharma S, Poschet G, Gegner HM, Lanz TV, Lucarelli P, Klingmüller U, Platten M, Heiland I, et al. Hypoxia routes tryptophan homeostasis towards increased tryptamine production. Front Immunol. 2021:12:590532. 10.3389/fimmu.2021.590532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Chong J, Zhou G, de Lima MD, Chang L, Barrette M, Gauthier C, Jacques PÉ, Li S, Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021:49(W1):W388–W396. 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, et al. Interpro in 2022. Nucleic Acids Res. 2023:51(D1):D418–D427. 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quindry J, Dumke C, Slivka D, Ruby B. Impact of extreme exercise at high altitude on oxidative stress in humans. J Physiol. 2016:594(18):5093–5104. 10.1113/JP270651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JM, Folkow LP, Blix AS. Hypoxia tolerance in mammals and birds: from the wilderness to the clinic. Annu Rev Physiol. 2007:69(1):113–143. 10.1146/annurev.physiol.69.031905.163111. [DOI] [PubMed] [Google Scholar]
- Rocchetti G, Ghilardelli F, Carboni E, Atzori AS, Masoero F, Gallo A. Milk metabolome reveals pyrimidine and its degradation products as the discriminant markers of different corn silage-based nutritional strategies. J Dairy Sci. 2022:105(11):8650–8663. 10.3168/jds.2022-21903. [DOI] [PubMed] [Google Scholar]
- Sahebjami H, Scalettar R. Effects of fructose infusion on lactate and uric acid metabolism. Lancet. 1971:1(7695):366–369. 10.1016/s0140-6736(71)92208-2. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Pothof J, Vijg J, Hoeijmakers JHJ. The central role of DNA damage in the ageing process. Nature. 2021:592(7856):695–703. 10.1038/s41586-021-03307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Breakthrough science: hypoxia-inducible factors, oxygen sensing, and disorders of hematopoiesis. Blood. 2022:139(16):2441–2449. 10.1182/blood.2021011043. [DOI] [PubMed] [Google Scholar]
- Sinha S, Singh SN, Ray US. Total antioxidant status at high altitude in lowlanders and native highlanders: role of uric acid. High Alt Med Biol. 2009:10(3):269–274. 10.1089/ham.2008.1082. [DOI] [PubMed] [Google Scholar]
- Storz JF, Scott GR, Cheviron ZA. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 2010:213(24):4125–4136. 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Zhuang D, Li Y, Chen Y, Wang X, Ge M, Xue T, Zhang Q, Liu X, Yin F, et al. Gut microbiota contributes to high-altitude hypoxia acclimatization of human populations. Genome Biol. 2024:25(1):232. 10.1186/s13059-024-03373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, Martins FM, Nachman MW. Altitudinal variation of the gut microbiota in wild house mice. Mol Ecol. 2019:28(9):2378–2390. 10.1111/mec.14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, Strauss R, Li XJ, Laird RM, Gutierrez RL, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology. 2020:159(1):96–104. 10.1053/j.gastro.2020.03.007. [DOI] [PubMed] [Google Scholar]
- Tran-Thi T, Holstege A, Decker K. Effects of hypoxia on the oxygen-dependent metabolism of prostaglandins and adenosine in liver cells. J Hepatol. 1994:20(5):570–579. 10.1016/S0168-8278(05)80342-3. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, Vandergheynst J, Fiehn O, Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015:12(6):523–526. 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij AG, Dutta R, Satija NK. Acclimatization to oxidative stress at high altitude. High Alt Med Biol. 2005:6(4):301–310. 10.1089/ham.2005.6.301. [DOI] [PubMed] [Google Scholar]
- Vizueta J, Sánchez-Gracia A, Rozas J. Bitacora: a comprehensive tool for the identification and annotation of gene families in genome assemblies. Mol Ecol Resour. 2020:20(5):1445–1452. 10.1111/1755-0998.13202. [DOI] [PubMed] [Google Scholar]
- Wang H, Mei L, Deng Y, Liu Y, Wei X, Liu M, Zhou J, Ma H, Zheng P, Yuan J, et al. Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutrition. 2019:62:63–73. 10.1016/j.nut.2018.11.018. [DOI] [PubMed] [Google Scholar]
- Wang L, Ye J. Commentary: gut microbiota reduce the risk of hyperuricemia and gout in the human body. Acta Pharm Sin B. 2024:14(1):433–435. 10.1016/j.apsb.2023.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want EJ, O’Maille G, Abagyan R, Siuzdak G, Smith CA. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006:78(3):779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Weinberg JM, Bienholz A, Venkatachalam MA. The role of glycine in regulated cell death. Cell Mol Life Sci. 2016:73(11-12):2285–2308. 10.1007/s00018-016-2201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcott OO, Ader M, Bergman RN. Glucose homeostasis during short-term and prolonged exposure to high altitudes. Endocr Rev. 2015:36(2):149–173. 10.1210/er.2014-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CL, Sun R, Qiao XJ, Xu CC, Shang XY, Niu WN. Protective effect of glutamine on intestinal injury and bacterial community in rats exposed to hypobaric hypoxia environment. World J Gastroentero. 2014:20(16):4662–4674. 10.3748/wjg.v20.i16.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Adachi H, Hakoshima M, Katsuyama H. Molecular biological and clinical understanding of the pathophysiology and treatments of hyperuricemia and its association with metabolic syndrome, cardiovascular diseases and chronic kidney disease. Int J Mol Sci. 2021:22(17):9221. 10.3390/ijms22179221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Zhou Q, OuYang S, Chen Y, Gong Y, Liang Y. Uric acid regulates NLRP3/IL-1β signaling pathway and further induces vascular endothelial cells injury in early CKD through ROS activation and K+ efflux. BMC Nephrol. 2019:20(1):319. 10.1186/s12882-019-1506-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Song L, Wang Y, Liu C, Zhang L, Zhu S, Liu S, Duan L. Beneficial effect of butyrate-producing lachnospiraceae on stress-induced visceral hypersensitivity in rats. J Gastroen Hepatol. 2019:34(8):1368–1376. 10.1111/jgh.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Yang L, Zhang T, Zhuang D, Wu Q, Yu J, Tian C, Zhang Z. Gut microbiome signatures of extreme environment adaption in Tibetan pig. NPJ Biofilms Microbi. 2023:9(1):27. 10.1038/s41522-023-00395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, He R, Shen Z, Zhang Y, Gao X, De JQZ, Xiao X, Zhang T, Yang D, Wang Y, et al. Altitude and metabolic syndrome in China: beneficial effects of healthy diet and physical activity. J Glob Health. 2023:13:4061. 10.7189/jogh.13.04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All metagenomic sequencing data generated and analyzed in the present study have been deposited in the Genome Sequence Archive in the National Genomics Data Center (https://bigd.big.ac.cn/gsa/; accession number: subHRA005186).