Abstract
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are a family of small conserved eukaryotic proteins that mediate membrane fusion between organelles and with the plasma membrane. SNAREs are directly or indirectly anchored to membranes. Prior to fusion, complementary SNAREs assemble between membranes with the aid of accessory proteins that provide a scaffold to initiate SNARE zippering, pulling the membranes together and mediating fusion. Recent advances have enabled the construction of detailed models describing bilayer transitions and energy barriers along the fusion pathway and have elucidated the structures of SNAREs complexed in various states with regulatory proteins. In this Review, we discuss how these advances are yielding an increasingly detailed picture of the SNARE-mediated fusion pathway, leading from first contact between the membranes via metastable non-bilayer intermediates towards the opening and expansion of a fusion pore. We describe how SNARE proteins assemble into complexes, how this assembly is regulated by accessory proteins and how SNARE complexes overcome the free energy barriers that prevent spontaneous membrane fusion.
Introduction
Biological membranes are bilayers formed by membrane lipids, with a hydrophobic core and hydrophilic surfaces. Membranes are generally stable in an aqueous environment and form diffusion barriers that segregate aqueous reaction spaces from the surroundings. However, it is essential that membrane-enclosed spaces such as cells and intracellular organelles can both merge (termed ‘fusion’) and split (known as ‘fission’) without becoming leaky in the process. Such transitions do not occur spontaneously because they are prevented by free energy barriers. To overcome these barriers, bilayers need to be perturbed, which frequently involves dedicated protein complexes undergoing exergonic conformational changes.
Membrane fusion and fission describe the forward and backward direction of a reversible reaction, but the two biological processes underlying these changes are fundamentally different. Although some steps of each process are reversible, both pathways employ different proteins that ensure that the reaction is unidirectional. Fusion proteins operate at the proximal contact site between the membranes, bringing them together and establishing hydrophobic connections. In contrast, fission proteins form either constricting helices and rings around a tubular neck of a budding vesicle (dynamins, discussed in ref. 1) or spirals on a flat membrane surface (ESCRT-III polymers, reviewed in ref. 2), extruding vesicles away from the assembly site. Consequently, the membrane geometry and, thus, the membrane curvatures of the intermediate steps are different between fusion and fission. Note, however, that members of the dynamin superfamily also mediate fusion, most notably in mitochondria3, suggesting that in certain cases fusion and fission may share structurally related steps in the reaction pathway.
In the last decades we have witnessed enormous progress in the understanding of protein-mediated membrane fusion. The first fusion proteins to be characterized were surface glycoproteins of enveloped viruses such as haemagglutinin of influenza virus or Env of the human immunodeficiency virus (for review see refs. 4,5). In contrast, the protein machineries responsible for both extracellular and intracellular membrane fusions in eukaryotic cells remained enigmatic for a long time. This changed with the discovery of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins 30 years ago6,7. SNAREs are a protein superfamily whose members are responsible for almost all fusion events of both the exocytotic and endocytic branches of the secretory pathway. These include reactions that are highly diverse in structure and timing, such as exocytosis of synaptic vesicles that occurs at a millisecond timescale8, or exocytosis of giant vesicles containing viscous material such as endothelial secretory granules termed Weibel–Palade bodies that require minutes to be released9. Moreover, SNAREs are conserved across the eukaryotic kingdom and are found in plants and animals inhabiting the entire biosphere10. Since the first description of SNAREs, additional and structurally unrelated eukaryotic fusion proteins have been discovered that function in fusion events not covered by SNARE proteins. These include, for instance, the fusion between mitochondria or the fusion between entire cells as it occurs during fertilization or during formation of syncytia (reviewed elsewhere11–13). In many cases, it is not yet understood by which mechanisms these proteins operate, and thus, in addition to viral membrane fusion, our understanding of SNARE-mediated fusion is most advanced.
Advances were also made in our understanding of the physical principles governing membrane fusion. Experimentally, it is still difficult to access the transition states of fusion reactions, particularly at molecular resolution, and thus simplified physical models are required in which all structural parameters are defined and which can be treated in accordance with the laws of physics. The structures of all intermediates are determined by the hydrophobic effect that creates forces aimed at minimizing the exposure of non-polar surfaces to the aqueous surroundings. Physical models describing the fusion of protein-free bilayers have advanced significantly and are increasingly supported by experiments as described in more detail in the next section. Thus, we presently have a much better understanding of the reaction pathway facilitated by fusion proteins such as SNAREs.
In this Review, we discuss the function of SNAREs in the context of modern physical concepts of lipid bilayer fusion. We will first describe the transitions that purely lipidic membranes undergo along the fusion pathway. Fusion proceeds through an ordered sequence of molecular rearrangements, termed the reaction coordinate, in which the positions of the participating molecules in the three-dimensional space are changed in a defined manner, beginning with the membranes approaching and contacting each other and ending with the opening of an aqueous fusion pore. Only if the reaction coordinate of the fusion pathway with its intermediate steps and the associated free energy barriers and valleys are known can the precise function of the SNARE machinery be determined, which we discuss in the following sections. For more comprehensive recent reviews on other aspects of SNARE structure and function, in particular the structural details of SNARE assembly and disassembly and the role of accessory proteins, see refs. 14–17.
Energy landscape of lipid bilayer fusion
Protein-free lipid membranes do not fuse spontaneously in aqueous media under physiological conditions. However, fusion can be induced if the energy barriers are overcome. This is investigated using both various types of models and experiments. We will first discuss fusion pathways of lipid bilayers (that is, a protein-free environment) as these models allowed researchers to define the tasks performed by fusion proteins.
Despite the structural and mechanistic diversity of fusion events, the reaction pathway of membrane fusion involves a series of common and sequential steps (Fig. 1). As mentioned above, the mechanisms by which fusion proteins such as SNAREs operate cannot be understood without detailed knowledge of the free energy profile of protein-free membranes moving along the fusion pathway, which is governed by the hydrophobic effect, in combination with electrostatic, solvation and steric forces. Once the reaction pathway(s) can be precisely described in all geometric and molecular detail, the free energy profile can be calculated using only the laws of physics, that is, independent of model-dependent assumptions. However, due to the complexity and fluid disorder of the transition states it is far from trivial to define a reaction coordinate, as the precise position of each atom is not experimentally accessible and is still difficult to predict by simulation models. Even variations that appear ‘intuitively’ minor with respect to geometry or molecular composition, such as local rearrangements of a few lipid molecules, may have drastic effects on the energy profile (see refs. 18,19 for a more detailed discussion). Thus, although frequently referred to, a ‘general’ fixed value (kT) for the overall free energy barrier for membrane fusion, which fusion proteins must overcome, cannot be given. This requires a fully defined reaction coordinate, which may vary widely between different fusion reactions.
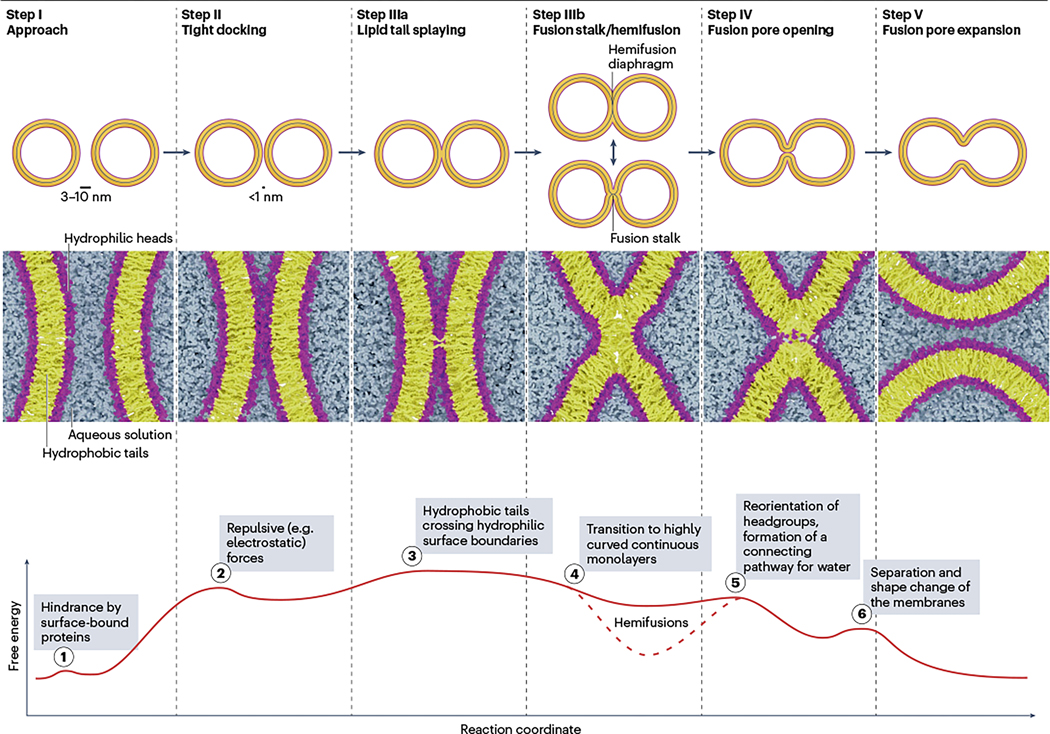
Fig. 1 |. Reaction pathway of fusion between two lipid vesicles.
Steps involved in the fusion of two lipid vesicles, with the two phospholipid monolayers of the membrane indicated (black line in the middle) (top panel). Molecular details of the intermediate steps, modelled by integrating information derived from various coarse-grained simulation studies (see text). Coarse-grained models of the phospholipids of the membrane, and the surrounding water molecules, are shown (middle panel). Hypothetical free energy profile of the fusion pathway in the absence of proteins, with the energy barriers indicated (encircled Arabic numbers) (bottom panel). Vesicle fusion is divided into steps, beginning with the approach of two membranes, whereby bilayers destined to fuse become close to each other (3–10 nm, Step I). Energy barrier 1 indicates that surface-bound proteins may hinder a close approach. Once the membranes are in close proximity, a tight contact is formed between the two hydrophilic bilayer surfaces, with headgroups being less than 1 nm apart (Step II). Major repulsive (mainly electrostatic) forces need to be overcome, constituting the major energy barrier 2, but once this is achieved, tight contact is stabilized by adhesive forces. The subsequent step involves lipid tail splaying, where aliphatic tails of individual membrane lipids cross the hydrophilic gap between the connected proximal monolayers, forming a hydrophobic link between the bilayer cores (Step IIIa). This state is thought to be unstable (energy barrier 3) and rapidly proceeds towards the fusion stalk or hemifusion stage (Step IIIb), but it is also possible that an additional small barrier (energy barrier 4) needs to be overcome. In hemifusion, the proximal lipid monolayers are merged whereas the distal monolayers are still separated. Stalks may develop into extended hemifusion diaphragms with increased stability (dotted line in the energy profile) but may also overcome the next energy barrier 5 towards fusion pore opening (Step IV), which allows formation of an aqueous connection, which can only commence in areas where the two distal bilayers are in direct contact. Fusion pores are still not well understood. For instance, it is unclear which structural changes are involved in rapid opening–closing events referred to as fusion pore flickering. Finally, pores expand and re-establish stable bilayers (step V). Energy barrier 6 indicates that full opening may be hindered by transitions from a highly curved to a flatter state, which may involve major lateral rearrangements of membrane lipids.
Originally, the free energy profile of the fusion pathway was calculated using elastic continuum models in which the properties of membranes were approximated by modelling them as elastic and bendable sheets characterized by parameters such as membrane curvature, membrane stiffness or electrostatic repulsion (see for example refs. 20–22). Although these models were instrumental for describing fusion pathways in accordance with physical principles, it became clear early on that the free energy barriers along the reaction coordinate cannot be accurately described without incorporating molecular details21,23–26. To account for these parameters, simplified molecular models in which groups of atoms are consolidated into particles for simulation (termed ‘coarse-grained’) were developed that are becoming increasingly sophisticated and that are beginning to bridge between atomistic simulation and elastic continuum models19,27–30. Such coarse-grained models proved instrumental for our understanding of the molecular structure and the free energy profiles of the intermediate steps.
Figure 1 shows a hypothetical free energy profile along the fusion reaction coordinate, with molecular models of possible fusion intermediates (see for example refs. 31,32), that is discussed below in more detail32. As mentioned above, both free energies and numbers of the peaks and valleys depend on the specific reaction coordinate of a given fusion reaction. A barrier along the pathway means kinetic retardation of the reaction, which may result in the accumulation of metastable intermediates that can be trapped, thus becoming accessible to experimental characterization.
Step I: approach
At a distance of 3–10 nm the repulsive forces between bilayers are low and are easily overcome by thermal fluctuations. Thus, liposomes in a test tube spontaneously collide with each other. The transient contact distance can be very short and is governed by factors such as surface charge (electrostatic repulsion), hydrophilicity, charge shielding by counter ions, temperature and size of the contact area33,34. In biological membranes, close bilayer contact by diffusive collisions is sterically hindered by surface proteins or protein domains (Fig. 1, energy barrier 1), which can be overcome by proteins connecting the membranes, such as tethering factors.
Step II: tight docking
In order to proceed to the next stage, which involves intermediates where the bilayer structure is disrupted (Step III, see below), the membranes need to approach each other very closely, below a distance of 1 nm. To achieve this, water must be removed from between the surfaces of the membranes and electrostatic repulsive forces need to be overcome35. For most fusion reactions, the free energy difference between the preceding minimum and this second barrier is likely to be the highest along the entire pathway.
How can this energy barrier be reduced? Aside from electrostatic shielding of fixed negative surface charges by monovalent and divalent cations36, one option is to minimize the contact area, which implies highly curved regions at the contact site. Moreover, positive curvature at the contact site increases hydrophobicity as the hydrophilic headgroups become spaced farther apart, resulting in hydrophobic ‘defects’ in the hydrated membrane surface. This facilitates merging of the monolayers, which results in the formation of an hourglass-shaped intermediate termed the fusion stalk (Step III; see Fig. 1 and below). Indeed, the fusion-promoting effect of membrane lipids with small headgroups (cone-shaped lipids such as phosphatidylethanolamine) is usually explained by stabilizing negative curvature in the distal monolayer (that is, opposite to the positively curved contact site) during transition to the stalk (reviewed elsewhere23). However, the fusion-promoting effect of positive curvature may also be explained by increased interfacial hydrophobicity, which due to increased spacing of the charges reduces repulsion by electrostatic and dehydration forces and increases the probability of lipid tail fluctuations and, thus, stalk formation (Step III) at the surface of the contacting proximal monolayers19. Moreover, polypeptide chains that have a propensity to form amphipathic structures partition into membrane interfaces where they may perturb the bilayer structure and thereby lower the energy barrier, even up to the point that fusion occurs spontaneously. This is the case with certain amphiphilic viral fusion peptides37,38 or peptides corresponding to transmembrane domains (TMDs) capped with hydrophilic residues39.
Despite early experimental evidence to the contrary (see for example ref. 34), it was thought for many years that membranes cannot form tight connections due to the strong repulsive electrostatic forces, unless the contact sites are limited to very small areas involving local protrusions23. Thus, tight contacts were considered to represent an energy maximum along the pathway. Recently, however, tightly connected membranes were captured by cryo-electron microscopy (cryo-EM) as metastable intermediates after reconstituting several protein-mediated fusion reactions in vitro, including membrane fusion facilitated by SNAREs40–42. Such contacts persist after SNARE disassembly and may expand due to adhesive forces, contain negatively charged headgroups, are free of proteins and do not depend on (albeit being stabilized by) divalent cations. Simulations showed that at the contact site, the phospholipids are packed more tightly because of headgroup tilting, resulting in measurable thickening of the proximal monolayers43.
Step III: lipid tail splaying and fusion stalk formation
The emerging consensus in the field is that the first hydrophobic connection between the two apposed membranes is mediated by the ‘splaying’ of phospholipid side chains (Step IIIa) (see for example refs. 44–46). As discussed above, the experimental capture of tightly docked intermediates suggests that, at least under certain circumstances, transition to this stage requires overcoming a free energy barrier (Fig. 1, energy barrier 3). However, it is also possible that there are fusion pathways in which a tightly docked metastable intermediate is bypassed, that is, that the transition from two separate membranes to the stalk involves only a single free energy barrier. In any case, perturbation of the hydrophilic–hydrophobic boundary in either of the monolayers that reduces the energetic penalty of hydrophobic lipid side chain exposure will lower this barrier. Once the first hydrophobic connection is established, other hydrophobic side chains reorient along the template provided by the splayed side chains, moving downhill on the free energy scale towards a fusion stalk in which the proximal leaflets are forming a highly curved continuous monolayer (Fig. 1, Step IIIb). Although stalks were originally predicted by elastic continuum models to be transient20,23, they can be stabilized by mild dehydration of stacked membranes, representing a separate (termed rhombohedral) phase that is accessible to experimental characterization47–50.
The free energies of the transition towards the stalk (energy barrier 4) and of the stalk itself can be reduced by lowering negative curvature stress. This can be achieved, for example, by introducing lipids with small headgroups (cone-shaped lipids, see above) with increased surface hydrophobicity23, by decreasing membrane rigidity51 and/or by perturbations of the hydrophobic–hydrophilic boundary of either membrane interface (see above). This transition is the critical step for the final ‘triggering’ of fusion reactions as from here onwards the reaction generally proceeds downhill on the free energy scale, except for fusion pore opening that may require additional input of energy (see below).
Although stalks are metastable, and thus can be experimentally trapped, they can relax into extended hemifusion diaphragms in which the two distal monolayers connect by forming a bilayer (Fig. 1, Step IIIb). Such diaphragms may progress to fusion, but they may also extend, getting trapped in an energy minimum (Fig. 1, Step IIIb, dotted line in the energy profile) and likely represent dead-end off-pathways52. Intriguingly, although hemifusion diaphragms can be induced by experimental manipulation of fusion proteins resulting in the abortion of fusion52, it is debated whether they occur during biological fusions in intact cells (see ref. 53 for a more comprehensive discussion). It needs to be taken into consideration that the formation of extended hemifusion diaphragms creates significant imbalances between the surface areas of the two leaflets that need to be compensated, for example by unidirectional flipping or selective removal of membrane lipids from the proximal monolayers of the contacting membranes.
Step IV: fusion pore opening
Fusion pores can only open at sites where the two distal leaflets are in contact, for example, by forming a stalk or a (possibly very small) patch of a bilayer diaphragm as described above. The development of a fusion pore requires the formation of a pathway for water across the bilayer, which depends on reorientation of the lipid headgroups until the headgroups of the proximal and distal membranes meet to form a hydrophilic channel, thus overcoming energy barrier 5.
Fusion pores are probably initiated by local defects in side chain packing at the centre of the stalk or at the rim of a hemifusion diaphragm. Even though single water molecules may penetrate through hydrophobic side chains, reorientation of lipid headgroups must precede any flux of water molecules and, thus, the initial formation of a fusion pore18,54 (Fig. 1). It remains to be established whether the so-called ‘flickering’ of fusion pores, describing the repetitive opening and closing of an aqueous permeation pathway for ions carrying electric charge (which, although experimentally observed only in fusion of protein-free bilayers55, appears to be a hallmark of protein-mediated fusion as well, see below), represents a reversal equilibrium of the pore opening or is governed by more complex mechanisms. To our knowledge, physical models that accurately describe the features and the parameters governing reversible fusion pores between bilayers are still lacking.
Step V: fusion pore expansion
Once the reorientation of the lipid headgroups is completed and an aqueous channel lined by headgroups is formed, the bilayer becomes stable again, and fusion pore opening is governed by minimizing curvature stress and lateral membrane tension. It is unclear whether, for expansion, an additional energy barrier needs to be overcome after initial fusion pore opening (energy barrier 6) or whether such barriers are dependent on proteins (see below).
The assembly–disassembly cycle of SNARE proteins
SNAREs represent a unique class of fusion proteins. Single SNARE molecules are not active and cannot fuse membranes even if present at high concentrations. For fusion, SNARE proteins residing in the two membranes destined to fuse need to interact in a highly regulated assembly reaction that connects the membranes and guides them along the fusion pathway. After fusion, these SNARE complexes are disassembled by a dedicated ATPase termed NEM-sensitive factor (NSF; also known as NEM-sensitive fusion protein), thus regenerating the SNARE proteins for another round of membrane fusion.
Domain structure of SNAREs
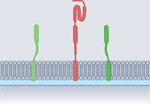
SNARE proteins represent a superfamily of small and mostly membrane-anchored proteins with a common domain structure (Fig. 2). The most characteristic feature is the SNARE motif, a stretch of 60–70 amino acids arranged in heptad repeats present in all SNAREs56. There are four different variants of SNARE motifs termed Qa, Qb, Qc and R, respectively, that were first distinguished by their conserved position in the structure of the assembled SNARE complex and later shown to be conserved in all eukaryotes using in-depth bioinformatic sequence analysis56–59. The other domains of SNAREs exhibit more diversity. For instance, although many SNAREs possess a carboxy-terminal hydrophobic TMD, connected to the SNARE motif by a short linker, there are some in which a classical TMD is lacking. These SNAREs may contain post-translational hydrophobic modifications or phospholipid binding domains, or lack membrane anchors altogether (reviewed elsewhere10). Examples include the members of the synaptosomal-associated protein 25 (SNAP25) subfamily, which contain a Qb motif and a Qc motif (see below)60, synaptobrevin homologue Ykt6 (containing an R-type motif)61 or the yeast SNARE vacuolar morphogenesis protein 7 (Vam7; with a Qc motif)62–64.
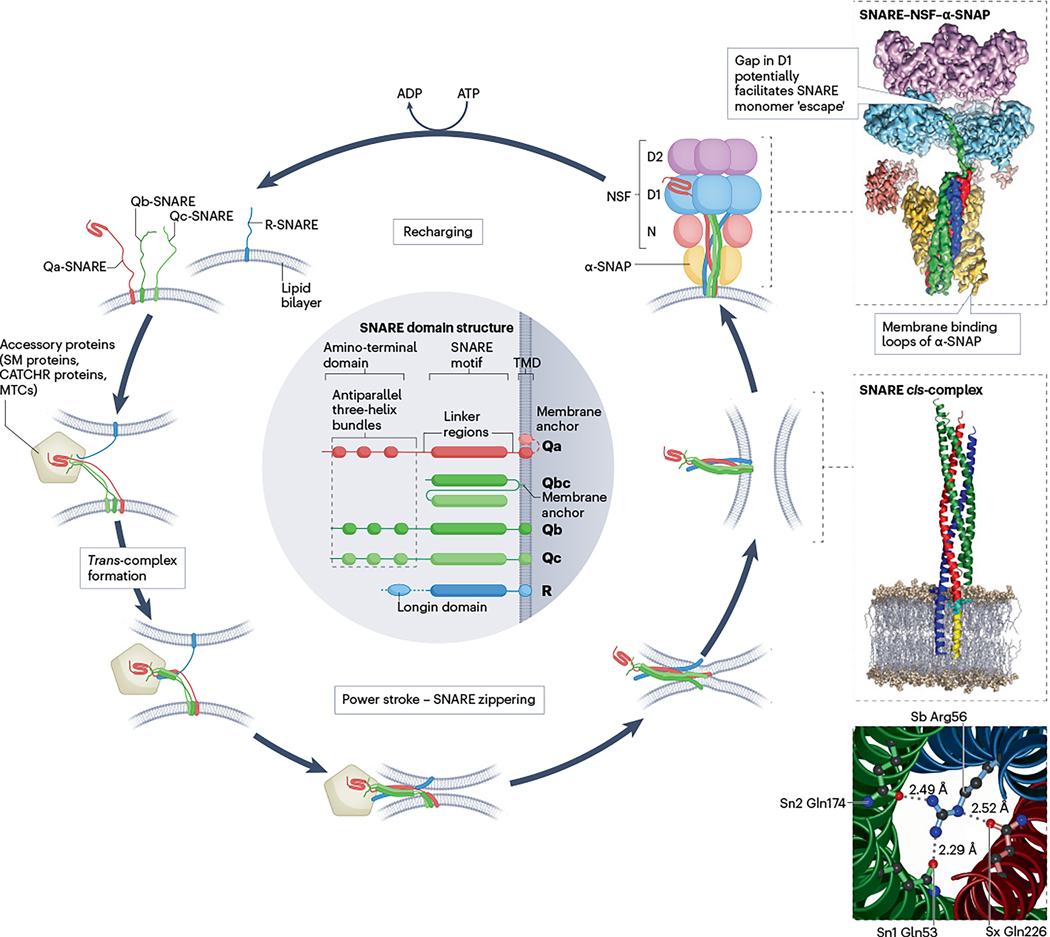
Fig. 2 |. SNARE assembly–disassembly cycle.
The cycle is depicted using an example in which three Q-SNAREs (Qa, Qb and Qc), each with its own transmembrane domain (TMD), form a trans-complex with an R-SNARE in the opposing membrane. This process is supported by accessory proteins such as Sec1/Munc18-like (SM) proteins and complexes associated with tethering containing helical rods (CATCHR) proteins, which may be part of multisubunit tethering complexes (MTCs) containing additional proteins. Once trans-assembly is initiated at the amino-terminal ends of the SNARE motifs, zippering of SNARE proteins progresses spontaneously towards the carboxy-terminal membrane anchors (‘power stroke’, with the gradual formation of the α-helical bundle indicated by progressively thicker lines), pulling the two membranes together and allowing fusion. After fusion, the SNAREs are all aligned in parallel (SNARE cis-complex). Disassembly of SNARE cis-complexes requires the vesicle-fusing ATPase NEM-sensitive factor (NSF), which is composed of D2, D1 and N domains, and several copies of α-soluble NSF attachment protein (α-SNAP) that link NSF to SNARE complexes. Structures on the right show, from top to bottom, an assembled SNARE complex with bound α-SNAP and NSF [PDB:6MDM] (ref. 149), a model of a membrane-anchored neuronal SNARE cis-complex including linkers and TMDs of syntaxin 1 and synaptobrevin 2 (based on [PDB:3HD7] (ref. 76)), and an ionic layer of interacting side chains in the middle of the neuronal SNARE complex (based on [PDB:1SFC] (ref. 233)). This layer is highly conserved across all SNAREs, contains three glutamines (Q) and one arginine (R) and defines the four SNARE subfamilies Qa-SNAREs, Qb-SNAREs, Qc-SNAREs and R-SNAREs, respectively57,233. SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor.
In addition, many — but not all — SNAREs possess independently folded amino-terminal domains. Most commonly, the N terminus consists of antiparallel three-helix bundles connected to the SNARE motif by a short linker. Three-helix bundles are typical for Qa-SNAREs but are also observed in some Qb-SNAREs and Qc-SNAREs. Other structures include, for instance, a profilin-like domain called the long in domain that is present in some R-SNAREs (reviewed elsewhere10) (Fig. 2). Some SNAREs lacking a TMD contain two SNARE motifs (usually Qb or Qc motifs) that are separated by a linker of variable length and that may contain palmitoyl residues acting as a membrane anchor (Qbc-SNAREs, reviewed elsewhere60). Another exception includes the Qa-SNARE syntaxin 17 that contains two adjacent TMDs forming a hairpin, targeting the protein to autophagosomes65. Note that complete or partial SNARE motifs are also found in a few soluble proteins which may regulate SNARE function by interacting with complementary SNAREs. Examples include the neuronal proteins tomosyn (also known as syntaxin-binding protein 5)66,67, amisyn (also known as syntaxin-binding protein 6)68 and, although with lower similarity, lethal(2) giant larvae protein homologue 1 (LgL) and its homologues Sro7 and Sro77 (ref. 69).
SNARE assembly
Assembly of SNAREs between membranes is the key step in SNARE-mediated membrane fusion. In the following sections, we first give an overview over the general rules governing SNARE assembly in different fusion reactions. Next, we discuss structural details of the various interactions between free SNAREs, largely based on in vitro studies with purified proteins. We then outline how conserved accessory proteins that are recruited to the fusion sites are thought to control this conformational landscape for initiating assembly. The section is concluded by a discussion of the specialist proteins controlling the final steps of SNARE zippering in regulated exocytosis of neurons.
Common properties of SNARE complexes.
SNARE assembly requires the participation of four different SNARE motifs, one from each subfamily (Fig. 2). Starting from dissociated, largely unstructured monomers they assemble into a coiled-coil bundle of extraordinary stability, which consists of four α-helices aligned in parallel, one of each SNARE subfamily (the QabcR complex, reviewed elsewhere10,16,17). The number of SNARE complexes required for fusion is thought to range between one and three (at least in neuronal exocytosis), but whether this is generally the case is still debated70–72. Assembly is initiated at the N-terminal ends and progresses towards the C termini of the α-helices (termed zippering, see for example refs. 73–75). Although assembly does occur spontaneously when purified SNAREs are mixed, its initiation is controlled by accessory proteins that are required for SNARE function in vivo (see below). For fusion, SNAREs that are anchored to opposing membranes assemble into a complex (‘trans-complex’), with the progression of zippering pulling the membranes together until the membranes merge. In the fully assembled complex (‘cis-complex’) the linker and the TMDs are aligned in parallel and also form α-helices, thus extending the helices of the SNARE motifs into the membrane76 (Fig. 2).
The evolutionary success of the SNARE fusion engine is probably due not only to its structural simplicity but also to an enormous flexibility and robustness, which allows for adjustments to the specific needs of individual fusion reactions. For instance, only two SNAREs need to possess TMDs for fusion, and even replacement of one of these TMDs with lipid anchors maintains fusion, at least to some extent77. Accordingly, the SNAREs providing the other SNARE motifs can be structurally more variable. Thus, SNARE complexes contain not only ‘canonical’ SNAREs with TMDs but also SNAREs lacking them, with their function being limited to contributing additional helices required for the formation of the stable QabcR four-helix bundle (reviewed elsewhere78). Similarly, there is flexibility in the distribution of the SNARE subfamilies between the membranes before fusion: whereas in neuronal exocytosis the vesicle contains the R-SNARE and the plasma membrane contributes the Qa-SNARE, Qb-SNARE and Qc-SNARE motifs to the complex6, a different topology is observed during the fusion of a transport vesicle generated at the endoplasmic reticulum (characterized by a COPII coat) with the cis-Golgi membrane — here, the vesicle contains Qb-SNAREs, Qc-SNAREs and R-SNAREs, whereas the target membrane contains the Qa-SNARE (reviewed elsewhere79). Although in these cases one membrane contributes three SNAREs and the other one SNARE, it is unclear whether this is always the case: at least in vitro, fusion is also observed if each membrane contributes two SNAREs80. Moreover, one of the essential yeast R-SNAREs (Ykt6) lacks a TMD61, requiring trans-assembly between membrane-anchored Q-SNAREs for fusion81. Thus, multiple permutations are possible. For instance, liposomes containing only Qc-SNAREs (carrying a TMD) fuse with their endogenous counterparts when injected into cells82. Moreover, SNAREs belonging to the same subfamily can substitute for each other, albeit only to a certain extent (for review see ref. 10), which provides additional flexibility due to partial redundancy and explains why deletion of some SNAREs in yeast results in surprisingly mild phenotypes79,81.
Assembly is highly exergonic, with the resulting QabcR-SNARE complex being of extraordinary stability83, and is considered to represent the main energy source (‘power stroke’) driving fusion. However, it has been far from trivial to determine the free energy of the assembly reaction. This is due to strong hysteresis that is observed when SNARE complexes are first dissociated by heat and then cooled for reassembly, suggesting that assembly and disassembly are not in equilibrium but, rather, follow different pathways83. Therefore, indirect approaches are needed such as single-molecule force experiments84; however, these are limited by spatial constraints of the force trajectories (when pulling SNAREs apart and allowing for their reassembly), which may not represent those of membrane-anchored SNAREs. Binding energies determined by such methods for the neuronal SNARE complex range from 30 to 85 kT (for discussion see refs. 15,85).
Assembly of free SNAREs.
Assembly of SNAREs is a complex molecular reaction that requires four different SNARE motifs on three or four separate proteins to interact and align in the correct sequence and spatial orientation. In vitro studies using purified SNAREs either in solution or after reconstitution in artificial membranes yielded structural details of intermediate complexes and associated conformational transitions, thus providing a framework for understanding how the accessory proteins (discussed in the next section) channel the reaction. Most in vitro studies of SNARE assembly were carried out using the SNAREs mediating neuronal exocytosis. These include syntaxin 1 (Qa-SNARE) and SNAP25 (Qbc-SNARE), both localized at the plasma membrane, and vesicle-associated membrane protein 2 (VAMP2; also known as synaptobrevin 2) (R-SNARE), localized at the synaptic vesicle. Although these SNAREs spontaneously assemble into QabcR-SNARE complexes86, assembly is very slow, requiring hours for completion, regardless of whether assembly proceeds in solution87 or whether it proceeds between complementary SNAREs reconstituted in liposomes74. This may be caused by the tendency of the SNARE motifs to reversibly form homo-oligomeric and hetero-oligomeric complexes of varying stoichiometry and composition, resulting in a panoply of metastable helical assemblies that are facilitated by the propensity of many SNARE motifs for helix formation88–92. For instance, neuronal syntaxin 1 forms homo-oligomers of varying stoichiometry, both in solution and in membranes93–95. In addition, syntaxin 1 and SNAP25 form stable binary complexes with a 2:1 stoichiometry86,96,97. Moreover, individual SNARE motifs of SNAP25 (Qb, Qc) can associate with syntaxin alone or in combination with synaptobrevin98. Several of these complexes represent four-helix bundles that are reminiscent of the QabcR complex although less stable. Binary complexes between synaptobrevin and SNAP25, and also between synaptobrevin and syntaxin 1, have been reported, but they are of very low affinity (see ref. 99 and references therein). Finally, some Qa-SNAREs can flip between a ‘closed’ and an ‘open’ conformation100. In the closed conformation, the N-terminal domain is folded back onto the N-terminal portion of the SNARE motif, preventing binding to other SNARE motifs101.
Although the formation of SNARE complexes is likely to proceed in sequential steps, the diversity of such oligomeric assemblies and conformational transitions renders it difficult to distinguish which of these complexes are intermediates of the assembly pathway and which are kinetically trapped side reactions102. Intriguingly, it appears that trapping in such side reactions may not be a general feature of all SNAREs as it is not, or at least not to this extent, observed in other (non-neuronal) SNAREs. For instance, endosomal SNARE complexes spontaneously assemble and fuse much faster in vitro than the neuronal SNAREs, with the reaction completed within minutes80,103 and with no evidence for helical off-pathway interactions104.
Why are endosomal SNAREs more reactive than neuronal SNAREs, particularly when considering that neuronal SNAREs catalyse exocytosis of synaptic vesicles, the arguably fastest fusion reaction in our body? Although one can only speculate about the reasons, it needs to be considered that any accidental contact between SNARE-containing membranes may lead to spontaneous SNARE ‘firing’ and fusion unless prevented by kinetic barriers. The membrane concentration of neuronal SNAREs is extremely high, more than two orders of magnitude higher than that of endosomal SNAREs105,106. The low reactivity of the neuronal SNAREs may thus be a safeguard mechanism preventing uncontrolled exocytosis at contact sites crowded with SNAREs. In contrast, endosomes contain only very few copies of SNAREs106, rendering it highly unlikely that four different SNAREs are accidentally present at such contact sites.
In conclusion, although free SNARE proteins are able to assemble spontaneously and mediate fusion in the absence of other proteins, their reactivity is low. Rather, regulatory proteins are required for activation, making sure that SNAREs only mediate fusion at predefined sites.
Proteins regulating initiation of assembly.
In the secretory pathway where the SNAREs operate, membranes destined to fuse first need to recognize and bind to each other. Initial connection between the membranes is mediated by diverse sets of so-called tethering factors that recognize molecular markers in both membranes. Such markers include specific Rab GTPases and phosphorylated variants of the membrane lipid phosphatidylinositol (reviewed elsewhere107). Efficient tethering only takes place if both membranes contain the appropriate combination of molecular markers107,108. Many (but not all) tethering factors form multiprotein particles, referred to as multisubunit tethering complexes (MTCs), and they are usually specific for a single intracellular trafficking step (for review see ref. 108). However, tethering is not sufficient for initiating SNARE assembly and fusion — it requires, in addition, specific regulatory proteins.
Two structurally conserved protein families are primarily responsible for controlling SNARE assembly. These are the Sec1/Munc18-like (SM) proteins109,110 and proteins characterized by bundles of helical rods, referred to as complexes associated with tethering containing helical rods (CATCHR) proteins111,112. These proteins act either as monomers that are separately recruited to SNAREs, such as the well-characterized neuronal proteins Munc18 (SM) or Munc13 (CATCHR), or they are integral components (subunits) of MTCs. Examples for the latter include the SM protein vacuolar protein sorting-associated protein 33 (Vps33) that is part of the homotypic fusion and protein sorting (HOPS) as well as the class C core vacuole/endosome tethering (CORVET) complex. Another example is provided by the CATCHR protein Tip20 that is part of the Dsl1 complex (also known as the syntaxin 18 complex).
Although SM and CATCHR proteins interact primarily with Qa-SNAREs, their main role probably consists of providing a scaffold for the ordered and sequential alignment of the other SNARE motifs. Accordingly, assembly proceeds through structurally defined intermediate states that not only prevent the SNAREs from entering off-pathway complexes but also result in a metastable acceptor site for the final SNARE motif completing the four-helix bundle, thus triggering efficient SNARE zippering and fusion. Indeed, it is possible to artificially create such highly reactive acceptor complexes using R-SNARE fragments without the need for SM or CATCHR proteins. For instance, if a Qabc-SNARE acceptor complex is stabilized by binding of a C-terminal R-SNARE fragment, yielding a free N-terminal acceptor site, binding of an intact R-SNARE is greatly accelerated, with rapid fusion occurring while the stabilizing C-terminal fragment is displaced113. Similarly, rapid zippering and fusion is observed if the R-SNARE is split into two fragments that each possess their own membrane anchor114.
It is not known whether SNAREs need to be monomeric for the initial binding of SM and/or CATCHR proteins. In that case, kinetically trapped oligomers (see above) or inactive cis-complexes that are known to spontaneously form in membranes would first need to be dissociated by the ATPase NSF (see ref. 106) (Fig. 3) that disassembles all homo-oligomeric and hetero-oligomeric SNARE complexes (described in more detail below). In addition, it is still unclear exactly how assembly is initiated. This is at least partially due to the vexing properties of SM proteins that exhibit two distinct SNARE binding modes. In the first mode, the SNARE is kept in the closed conformation (with the N-terminal domain folded back on the SNARE motif, see above), being stabilized by binding to a cleft of the SM protein, essentially rendering the SNARE inactive. In the second mode, a short stretch at the N-terminal end of the Qa-SNARE binds to the outer surface of the SM protein, with no major changes in the structure of the SM protein. The relative distributions between the two binding modes vary between different Qa-SNAREs, and not all Qa-SNAREs may be able to adopt a closed conformation (for review see refs. 15,109,110).
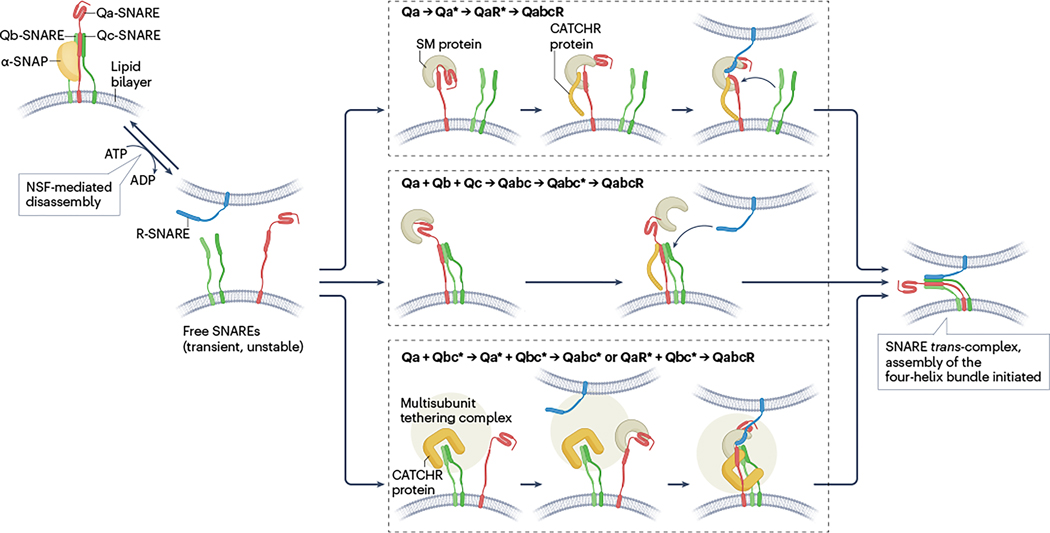
Fig. 3 |. Pathways for trans-SNARE assembly prior to fusion.
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) zippering requires the coordinated assembly of four conformationally flexible SNARE motifs between membranes facing each other (‘trans’ configuration). Before assembly, NEM-sensitive factor (NSF)-driven disassembly may be required to free SNAREs that are trapped in various homomeric and heteromeric complexes. trans-assembly is guided by Sec1/Munc18-like (SM) proteins and complexes associated with tethering containing helical rods (CATCHR) proteins. Their sequential or simultaneous recruitment is thought to allow for an ordered SNARE trans-assembly by stabilizing reactive intermediate complexes (indicated by asterisks). Three possible assembly pathways are shown that are not necessarily exclusive. According to one model (top panel), the SM protein first binds to the Qa-SNARE, yielding a closed conformation that is then opened by the subsequent binding of the CATCHR protein. The resulting SM–CATCHR–Qa-SNARE protein complex serves as a binding site for the R-SNARE, followed by binding of the Qb-SNARE and Qc-SNARE motifs that initiates zippering15,116,117. According to a second model (middle panel), SM proteins form a metastable complex with Qa-SNARE, Qb-SNARE and Qc-SNARE motifs, with the CATCHR protein possibly providing further stabilization, which allows for binding of the R-SNARE as the final step in initiating zippering102,118,119. The third model (bottom panel) accounts for variations that may occur if either the SM or the CATCHR protein are integrated into multisubunit tethering complexes (MTCs). Shown is the example of the Dsl1 complex that contains a CATCHR protein and operates in the fusion of retrograde Golgi vesicles with the endoplasmic reticulum. The complex first binds to the Qb-SNAREs and Qc-SNAREs, and then associates with the Qa-SNARE that binds the SM protein separately. The final step again involves recruitment of the R-SNARE to initiate zippering120,121. It is still unclear at which point during (or after) SNARE zippering and in which sequence the accessory proteins dissociate or whether they remain bound until fusion is completed. α-SNAP, α-soluble NSF attachment protein.
How can these features be integrated into a working model of SNARE assembly? One scenario implies that the two binding modes occur consecutively, with the switch from closed to open possibly being mediated by simultaneous binding of a CATCHR protein (see Fig. 3, top panel), creating a metastable SM–CATCHR–Qa-SNARE acceptor complex15,16,115. The next steps are still unclear and may not be the same for all known SNARE complexes. Three possible scenarios, each supported by experimental evidence, are discussed (Fig. 3). According to the first model, the R-SNARE binds to a metastable SM–CATCHR–Qa-SNARE acceptor complex (Fig. 3, top panel), thereby connecting the membranes and initiating trans-SNARE assembly. This view is supported by reports showing structures of SM proteins bound to either Qa-SNAREs or R-SNAREs (reviewed elsewhere15), or to both simultaneously116,117. Such a complex would then form a template for subsequent binding of Qb-SNAREs and Qc-SNAREs15,117. In a second model, SM proteins stabilize reactive Qabc-SNARE instead of QaR-SNARE complexes (Fig. 3, middle panel), providing an N-terminal and transient three-helix template for the rapid binding of the R-SNARE, possibly without directly contacting the SM protein102,118,119. According to the third model, CATCHR proteins, as part of MTCs, first bind to Qbc-SNAREs in one of the membranes before connecting with the target membrane, which is then followed by trans-SNARE assembly (Fig. 3, bottom panel). For instance, in the fusion of Golgi-derived vesicles with the endoplasmic reticulum, the CATCHR protein Tip20, as part of the Dsl1 tethering complex, binds to Qb-SNAREs and Qc-SNAREs120,121, thus holding a SNARE acceptor complex in place. This assembly then interacts with the Qa-SNARE and its SM protein and recruits the incoming vesicle containing the R-SNARE. This process may involve some rearrangement of the SNAREs, possibly involving a SM protein-mediated QaR intermediate as in the first model, before SNARE zippering is initiated (for a more thorough discussion see refs. 15,16).
Why is it so difficult to identify the intermediates of the SNARE assembly pathway? Most probably, the intermediates are of low stability in order to maximize the energy gain from SNARE zippering for fusion (see for example ref. 122), rendering it highly challenging to isolate such states and to capture their structure123. For instance, stabilized neuronal SNARE complexes were created for the binding of the accessory proteins in which the SNAREs were either fully zippered or partly zippered by the use of truncated SNAREs, resulting in shortened bundles124–126 (see Box 1). Although insightful, it remains to be established to what extent such structures resemble conformational intermediates in the fusion pathway (discussed elsewhere123,127). Moreover, several consecutive steps may be involved in which bound SNARE motifs may need to be rearranged before final nucleation, for example by dissociation–association cycles of individual SNARE motifs (reviewed elsewhere15,16). Finally, the activity of SNAREs towards assembly appears to be regulated at additional levels including, for instance, phosphorylation by an array of kinases of side chains within or adjacent to the SNARE motifs (for review see refs. 128,129).
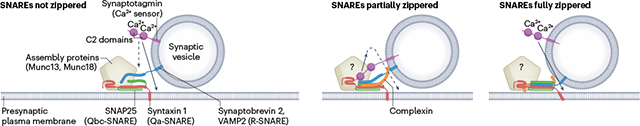
Box 1. Pre-fusion arrest in neuronal exocytosis: how far are the SNAREs zippered?
Although Ca2+-triggered fusion of neuronal soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) has been reconstituted in artificial membranes using neuronal SNAREs, synaptotagmin and various combinations of the Sec1/Munc18-like (SM) protein Munc18, the complexes associated with tethering containing helical rods (CATCHR) protein Munc13 and complexin (see for example refs. 168,217,221,225,234), it has been difficult to determine the status of SNARE assembly before fusion and the mechanism by which synaptotagmin triggers progression to fusion. Three scenarios are discussed for the release-ready state that are not mutually exclusive:
According to the first model, SNAREs are not zippered. The Q-SNAREs syntaxin 1 and synaptosomal-associated protein 25 (SNAP25) are pre-assembled in an acceptor complex exposing a binding site for the R-SNARE vesicle-associated membrane protein 2 (VAMP2; also known as synaptobrevin 2) to ‘snap’ in, for which the assembly proteins provide a scaffold. Alternatively, syntaxin and synaptobrevin may be bound to Munc18, with SNAP25 zippering not yet initiated (see Fig. 3). Synaptotagmin triggers zippering indirectly by pulling the vesicle closer to the target membrane235. Here, the full SNARE power stroke is available for the final fusion step. Complexin only binds after SNARE zippering is triggered in this model, which then cannot explain its clamping function (below). Also, docking of release-ready vesicles at a distance of 1–2nm requires SNAREs, arguing for SNARE trans-contact in this state131.
According to a second model, SNARE zippering is halted halfway. Arrest may be caused by complexin that can bind to partially assembled Qabc complexes236 or may interfere with the carboxy-terminal zippering of synaptobrevin237, and/or by binding of synaptotagmin238. In this state, Munc18 and Munc13 are still thought to be bound. According to one model, the block is released by Ca2+-dependent dissociation of the C2B domain from the surface of the SNARE complex, probably switching instead to the plasma membrane (‘release-of-inhibition model’238). This model is supported by high-resolution structures showing SNARE complexes with bound C2 domains, Munc18, Munc13 and complexin in various combinations, sometimes in symmetrical arrangements117,124,126,239. Alternatively, Ca2+-triggered binding of synaptotagmin to ionic lipids may change the lipid order and thus, indirectly, the conformation of the Q-SNARE acceptor complex by lifting the Qa-SNARE, thus pulling it towards the vesicle membrane168. Other models are also discussed, such as oligomerization of synaptotagmin between the vesicle and the plasma membrane which is then dissociated upon calcium binding240. Arrest in the partially zippered state is an attractive model as it preserves part of the SNARE power stroke for the last step but does not require major rearrangements of the SNAREs after Ca2+-induced triggering. Moreover, distinct steps in the force profile are observed when SNARE complexes are pulled apart with optical tweezers and allowed to reassemble, supporting an energy barrier upon zippering halfway237,241. However, it has been difficult to show Ca2+-dependent binding or dissociation of C2 domains to or from SNAREs, or any direct effect of C2 domains on zippering, and synaptotagmin–SNARE interactions are abolished at physiological salt and metabolite concentrations242. Moreover, the arrested complex with bound synaptotagmin, complexin and assembly proteins is very large and difficult to fit close to the fusion site.
A third model postulates that SNAREs are fully zippered throughout the SNARE motifs and, possibly, into the linker regions, resulting in a tightly docked intermediate to which complexin is bound, requiring only minor energy input for fusion completion (reviewed elsewhere134) (Fig. 1, Step II). In this scenario, triggering by the C2 domains is mediated by perturbing the hydrophilic–hydrophobic boundary at the membrane contact site, for example by creating locally highly curved protrusions, perhaps assisted by the C-terminal final zippering steps into the membrane and/or rotational twisting of the SNARE complex, which may be carried out by synaptotagmin bound to the surface of the SNARE complex (alternative model, not shown). It is not clear whether, at this stage, accessory proteins such as Munc18 and Munc13 are still bound.
SNARE zippering and its regulation by late-acting proteins.
Once SNARE assembly is initiated at the N-terminal end of the SNARE motifs, zippering proceeds downhill on an energy gradient but may be controlled by late-acting regulatory proteins. This is particularly conspicuous in neurons where Ca2+-mediated triggering of synaptic vesicle exocytosis proceeds with a delay time of less than a millisecond130. Vesicles are docked in a ‘release-ready’ state only 1–2 nm apart from the plasma membrane (corresponding to Step II; see Fig. 1) before fusion131. The receptor for the Ca2+ ions is synaptotagmin, an abundant transmembrane protein of synaptic vesicles. Synaptotagmin contains two adjacent Ca2+-binding C2 domains, termed C2A and C2B, that each possess several Ca2+-binding sites but are somewhat different in their relative affinities132,133. Ca2+ binding does not appear to induce major conformational changes of the stably folded C2 domains but, rather, triggers electrostatic binding to clusters of negative charges such as acidic phospholipids in the plasma membrane or the acidic surface of (Q-)SNAREs, or both (reviewed elsewhere16,17,134). Effective triggering also requires complexin, a small protein that binds to the surface of the partially or fully assembled SNARE complex (reviewed elsewhere135,136). Note that synaptotagmins and complexins represent special adaptations of neuronal exocytosis, but are not part of the universal SNARE fusion machine. Importantly, phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)P2), a membrane lipid specifically associated with the plasma membrane and found to cluster in the proximity of syntaxin137, strongly modulates the binding of synaptotagmin, and probably plays a critical role in the final assembly of the SNAREs and progression to fusion138,139.
How does Ca2+ binding to synaptotagmin trigger the final step of SNARE zippering and fusion? Whereas binding of synaptotagmin 1 and complexin to both SNAREs and lipids seems to be required, the underlying mechanisms and the assembly status of the SNAREs before the final triggering are still controversially discussed (see Box 1 and ref. 134 for more details). In particular, the interaction of one of the two calcium-binding C2 domains (C2B) with the surface of the SNARE complex appears to be essential for this step. Although C2 domains were shown to bind at several sites of the assembled SNARE complex, one of them (referred to as the primary binding site) appears to be critical as any interference with this interaction, for example by mutagenesis or small-molecule inhibitors, impairs calcium-dependent triggering of SNARE assembly (reviewed elsewhere127). However, it remains to be established how this interaction, or the lack thereof, is connected to the fusion reaction (Box 1).
SNARE disassembly
As discussed above, assembly of individual SNAREs into helical bundles is a highly exergonic reaction that allows for overcoming energy barriers along the fusion pathway. After fusion, the SNAREs within the complex are all aligned in the same membrane (cis-complexes) and need to be reactivated by dissociation. This reaction requires metabolic energy in the form of ATP. It is mediated by NSF140, a member of the AAA+ superfamily of proteins, whereby AAA refers to their designation as ATPases associated with diverse cellular activities. These proteins are nanomechanical devices that are capable of driving major conformational changes in highly diverse client proteins and nucleic acids. Unlike some other AAA+ proteins, NSF does not directly interact with its substrate but requires soluble NSF attachment proteins (SNAPs) as adaptors (see below).
AAA+ proteins share a structurally conserved ATPase domain (also referred to as the AAA+ core domain) that forms ring-like or spiral-like oligomers141,142. NSF belongs to the type II ATPases that are hexameric. Each of the monomers contains three domains, two of which are represented by homologous ATPase core domains, referred to as D1 and D2, that are fused together and assembled into two stacked hexameric rings (Fig. 2). The third domain, referred to as the N domain, is independently folded and mediates contact with the client proteins, that is, SNARE complexes bound to SNAP proteins. The D2 ring binds ATP with very high affinity, which is not cleaved during the regular catalytic cycle and is responsible for stabilizing the hexameric assembly of the monomers. The catalytically active D1 domain undergoes conformational changes during ATP cleavage, which are transferred to the N domain. As a result, the N domain undergoes large movements that are propagated through the SNAP intermediates to the SNARE complex, driving its disassembly (reviewed elsewhere142).
Both NSF and SNAPs are evolutionarily highly conserved and operate on all SNARE complexes studied so far (reviewed elsewhere143). Unlike NSF, of which only a single variant is expressed in most species, SNAPs are represented by three isoforms in mammals, termed α-SNAP, β-SNAP and γ-SNAP144. SNAPs are composed of stiff, twisted sheets of stacked α-helices connected to a C-terminal helix bundle that is negatively charged and, as shown in an elegant cryo-EM study, connects to a positively charged groove in the N domain of NSF145. Binding of SNAP to the grooves of the SNARE complex is dependent on a pattern of positive and negative surface charges that appears to be conserved in all SNARE complexes146. Moreover, SNAPs possess an extended loop containing exposed phenylalanine residues that mediate membrane binding, thus directing the disassembly machinery to membrane-anchored SNARE complexes147. As in many other AAA+ proteins, ATPase activity of NSF is stimulated by substrate binding148.
It is still unclear how exactly the concerted action of NSF and SNAP leads to unwinding and dissociation of the SNARE helical bundle. Whereas in several other AAA+ proteins the substrates are threaded through the central hole of the hexameric ATPase ring, this does not appear to be the case in the NSF–SNAP–SNARE interaction, even though conserved pore-lining residues are likely involved in the catalytic cycle145. Rather, there is a conspicuous gap between the first and last protomers of the D1 ring, which may allow for sideways escape of dissociated SNARE monomers during disassembly145,149. Moreover, the number of SNAP molecules required for disassembly appears to be variable, with estimates ranging between two and four molecules (see ref. 142 for review). Finally, single-molecule experiments have revealed that disassembly occurs within 10 ms in a single and concerted round of ATP hydrolysis rather than by sequential disassembly of individual subunits150. The reaction involves only a single unfolding intermediate in which the membrane-adjacent C-terminal part of the SNARE complex is dissociated whereas the N-terminal parts of the SNAREs are still joined.
The NSF–SNAP disassembly machinery dissociates not only fully assembled SNARE complexes but also various off-pathway hetero-oligomeric and even homo-oligomeric SNARE complexes (see above) in which SNAREs otherwise may remain trapped. Moreover, in vitro experiments suggest that NSF can attack SNARE trans-complexes that are not yet fully zippered, that is, before fusion is completed151. Such complexes may be shielded by the scaffold and accessory proteins that compete with SNAP proteins for binding to the SNARE complex, preventing premature abortion of fusion152,153. Intriguingly, an opposite role of α-SNAP was also suggested: the protein was reported to bind after nucleation of SNARE assembly to recruit SM proteins154 and promote full zippering in the absence of NSF155. Presently, it is unclear how these two views can be reconciled and to what extent assembling SNARE complexes are protected from attack by NSF before fusion is completed.
The function of SNAREs along the fusion pathway
As detailed above (Fig. 1), the pathway of membrane fusion involves sequential steps separated by energy barriers. Fusion proteins such as SNAREs transfer conformational energy onto the membranes and change the reaction coordinate in such a way that the fusion reaction follows a downhill energy gradient, giving directionality to the pathway. Moreover, they lower the energy barriers of transition states, allowing fusion to be completed instead of becoming trapped in one of the intermediate states. Experimental manipulations of SNAREs, by studying fusion both in vitro and in intact cells and organisms, suggest that SNARE assembly is mechanistically involved in all of the steps depicted in Fig. 1. However, despite a multitude of approaches for observing SNARE-mediated membrane fusion (see Table 1), it is experimentally far from trivial to differentiate the steps between the first transition towards close apposition (Fig. 1, Step II) and the opening of an aqueous fusion pore (Step IV). Thus, it is still controversially discussed how exactly SNARE zippering and its control by the regulatory proteins is associated with the progression towards fusion, that is, a unified model for the function of SNAREs along the fusion pathway is not yet available. SNAREs are conserved across the entire eukaryotic kingdom and function in a wide range of highly diverse fusion reactions. It is conceivable that the built-in flexibility of the SNARE machine and the fluid dynamic nature of lipid bilayers allow for multiple different reaction pathways in a complex free energy landscape. This landscape may resemble energy funnels with many valleys and obstructions along their slopes to eventually merge into a common energy minimum of the fully fused state, akin to a concept that is now frequently used to describe the energetics of protein folding156. Moreover, the free energy landscape is dependent on the overall lipid composition and local variations in lipid content. SNAREs and some of the accessory proteins may alter the local lipid composition, for example by de-mixing or binding lipids such as PtdIns species or phosphatidylserine137, thus influencing free energy barriers along the reaction coordinate.
Table 1 |.
Overview of techniques commonly used to study SNARE-mediated membrane fusion by in vitro reconstitution
| Membrane type | Fusion partner | Fusion assays | Refs. (selection) |
|---|---|---|---|
 ‘Native’ vesicles (purified from cells or tissues) For example, synaptic vesicles, neuroendocrine granules, endosomes |
Native vesicles, SUVs, LUVs, GUVs, supported bilayers | Lipid mixing – measured using fluorescence dequenching, microscopic spreading of labelled lipids at the fusion site Formation of SNARE complexes |
212–215 |

Nanodiscs |
Cells Liposomes | Fusion pore opening and closing – measured using electrophysiological methods, fluorescence microscopy | 71,205,207, 209,216 |
 Liposomes In solution or immobilized on solid support SUVs (30–60 nm) LUVs (70–300 nm) GUVs (>1 μm) |
Native vesicles SUVs, LUVs, GUVs | Lipid mixing – shown using FRET, FCCS, fluorescence dequenching, microscopic spreading of labelled lipids or proteins Content mixing – measured using various fluorescence assays Formation of SNARE complexes– shown using FRET, biochemical assays |
74,166, 217–226 |
 Supported bilayers Immobilized on solid support |
Native vesicles SUVs, LUVs | Lipid mixing – measured using FRET, FCCS, fluorescence dequenching, microscopic spreading of labelled lipids or proteins Content release – measured using fluorescence spreading/dequenching Formation of SNARE complexes |
227–231 |
|
Suspended bilayers Black lipid membranes Pore-spanning membranes |
Native vesicles SUVs, LUVs | Lipid mixing – measured using FRET, FCCS, fluorescence dequenching, microscopic spreading of labelled lipids or proteins, or by monitoring current flow with electrophysiological methods using vesicles containing pores Content release – measured by fluorescence spreading or dequenching |
207,232 |
FCCS, fluorescence cross-correlation spectroscopy; FRET, fluorescence resonance energy transfer; GUV, giant unilamellar vesicle; LUV, large unilamellar vesicle; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; SUV, small unilamellar vesicle.
Despite this diversity, some common features are beginning to emerge, and increasingly sophisticated molecular models are being developed for SNARE action in individual steps of the pathway (see for example ref. 157).
Step I: SNARE action during membrane approach
As already discussed above, initial contact between membranes destined to fuse is mediated by tethering factors that interact with both polyphosphoinositides and Rab GTPases108,158,159. Although this step precedes activation and trans-interaction of SNAREs, it is becoming clear that SNAREs are instrumental in orchestrating tethering sites. For instance, at least in the plasma membrane, SNARE proteins are concentrated in nanodomains that may serve as beacons marking prospective fusion sites for incoming vesicles160–164. Moreover, most multisubunit tethering factors bind to SNAREs not only by means of their SM or CATCHR protein subunits but also by means of other subunits, thus recruiting them to the prospective fusion sites (reviewed elsewhere165). Linking the recruitment of SNAREs to tethering ensures that the SNAREs are in place when membrane contact is established, thus facilitating subsequent SNARE assembly.
Step II: SNARE zippering promotes tight membrane docking
Bringing the membranes sufficiently close to allow for lipid tail splaying is arguably the essential function of SNARE assembly. Indeed, SNAREs are perfectly designed as force-generating ‘zippering’ machines capable of overcoming large repulsive electrostatic and hydration forces.
Except in the case of regulated exocytosis in neurons and related cells in which SNARE assembly, and thus progress towards fusion, is arrested in a tightly docked state (Box 1), such intermediates appear to be transient and rapidly progress towards fusion. Although these intermediates can be stabilized in vitro, cryo-EM is required to distinguish this state from more loosely docked precursor states. So far, only few studies are available131,166,167 and it is unclear how far SNARE zippering needs to proceed to reach this state, that is, how much of the energy of the SNARE power stroke is required. If the SNARE motifs fully assemble close to the TMDs, these may become tilted (see Box 1). Straightening out the TMDs in alignment with the SNARE complex bundle will buckle up the membranes and force them towards each other, as proposed in a recent model in which SNARE conformations respond to the action of Ca2+-activated synaptotagmin and, thus, facilitate transition to Step III168. Polyunsaturated membrane lipids may facilitate the formation of such high curvature protrusions as they appear to reduce membrane rigidity and increase surface hydrophobicity51.
Tight contact between the membranes at the prospective fusion site imposes steric constraints on the fusion proteins, which are excluded from the contact site43. Indeed, the small size of the SNAREs allows for their positioning directly adjacent to the protein-free contact area but this may be limited by bulky accessory proteins (such as SM and CATCHR proteins) which are thought to remain bound during zippering (see ref. 169 for review) (Box 1). Small contact areas are advantageous as they reduce the energy barrier, and thus may explain why, as mentioned above, neuronal exocytosis requires only one to three SNARE complexes for fusion70–72,170. It is likely that biological fusion reactions generally proceed through small contact sites even if the participating membranes are mostly flat (such as yeast vacuoles)171. Apparently, this is more difficult to reproduce in vitro where the contact area, and thus repulsion, scales inversely with curvature, explaining why larger vesicles need more SNARE complexes for fusion40. Once formed, however, tight contact zones expand in vitro, probably due to adhesive forces involving both electrostatic and hydrophobic components43,166. It is unclear whether such expanded tight contact areas exist in intact cells. Here, accessory proteins may prevent adhesive expansion and limit the contact area, which has also been suggested by a recent reconstitution study167. Note that in neurons, tight docking of synaptic vesicles is reversible172. This means that in that state SNARE zippering can still be undone, either by maintaining the SNAREs in an only loosely assembled state that is reversible or by dissociating zippered SNARE trans-complexes with the aid of NSF (see above).
Step III: SNARE-mediated perturbations promote fusion stalk formation
The closer the membrane surfaces are, the lower the energy barrier for the next step — the formation of a lipid stalk that establishes a hydrophobic connection between the core of the two membranes. Even minor surface perturbations may suffice, such as transient dislocations of membrane lipids caused by tilting or outward pulling of the SNARE TMDs. Both the linkers connecting the SNARE motifs with the TMDs and the TMDs themselves appear to be critical for this transition: molecular dynamics simulations and various in vitro and in vivo experiments document that manipulations of SNARE TMDs, for example by mutations in the hydrophobic part leading to increased helicity or by adding charges to the C terminus, inhibit progression to fusion19,173–175 (see also below). Similar impairments of fusion, albeit to different degrees, are observed when SNARE TMDs are truncated or replaced with lipid anchors (see for example refs. 77,173,176,177). Thus, it is possible that the motion associated with C-terminal zippering of the SNARE motif enhances TMD-mediated perturbations at the contact sites, causing lipid tail splaying and progression to hemifusion in a single continuous step.
Once tight contact is established by SNARE zippering, however, progress towards hemifusion may also be induced by other factors. For instance, it is well established that docked synaptic vesicles that are ready to be released can be driven towards exocytosis by increasing membrane tension (and thus surface hydrophobicity), for example by mechanical stretching178 or osmotic pressure179. Mechanical stretching and osmotic pressure increase hydrophobicity at the surface of the bilayer, which, as discussed above, reduces the energy barrier for lipid tail splaying. This happens also when pure lipid bilayers are induced to fuse in the presence of polyethyleneglycol, which induces osmotic pressure by reducing the water activity at the membrane surface180. Moreover, the fusion-promoting effect of Ca2+-activated synaptotagmin may be exerted by insertion of C2 domain(s) into the proximal monolayer of the plasma membrane, perhaps attracted to the contact site by PtdIns(4,5)P2 clusters around the TMD of the SNARE syntaxin (see Box 1 and above). Also, the presence of bulky accessory proteins such as MTCs close to the contact area may cause membrane deformations that contribute to overcoming the final energy barrier169,181. Similarly, recruitment of the NSF adaptor SNAP to such states promotes fusion even if the SNAREs are not fully zippered, which appears to depend on membrane binding caused by the abovementioned hydrophobic loop region of SNAP182 (Fig. 2).
Thus, the conserved and universal function of SNAREs in fusion is embedded in the zippering reaction. Its initiation is guided by protein machineries containing conserved components, followed by zippering that brings the membrane surfaces very close. Progression towards hemifusion requires less energy and little structural specificity. Although completion of SNARE zippering alone can be sufficient for driving the reaction towards full fusion, there may be fusion reactions where this is not the case, requiring other proteins to control the final step(s).
Steps IV and V: SNAREs shape fusion pore opening and expansion
Opening and subsequent expansion of an aqueous fusion pore are the final steps in the fusion trajectory. Originally, based on electrophysiological signatures reminiscent of ion channels, fusion pores were considered to be mainly lined by proteins183. However, fusion pore-like signatures such as rapid sequences of pore opening and closing (referred to as pore flickering) were also observed when fusion was forced to occur between protein-free membranes55 (see also above). Presently, the emerging consensus is that fusion pores are primarily lipidic but may contain TMDs of proteins184.
Whereas in many systems fusion is rapidly completed after the initial opening of the first aqueous connection, a more complex pattern emerged from measuring exocytosis of secretory vesicles at high resolution using various electrophysiological and imaging approaches185–187. Accordingly, initial pores are small and may linger for a while before they either expand or close again. Of these, re-closure is the least understood, and it is still unclear whether it involves mechanistic reversal of fusion, that is, a reversal of SNARE zippering, particularly in cases where closure occurs only milliseconds after opening (fusion pore flickering)188. For a long time, it was debated whether synaptic vesicles recycle by direct recapture after such brief opening of a fusion pore (termed ‘kiss-and-run’189) or by endocytosis at sites away from the fusion site (for review see ref. 190). Using an elegant combination of optogenetics and time-resolved cryo-EM, the latter hypothesis was proved to be correct191. Recapture of only partially fused vesicles by the endocytic machinery at the site of exocytosis was reported192,193, but in these cases the pores remained open for much longer (seconds to minutes) and lipid mixing occurred before recapture194.
Numerous studies have shown that SNAREs are instrumental in shaping the dynamic properties of fusion pores such as the size of the initial pore or the delay and kinetics of its expansion. For example, changes in SNARE expression levels or mutations affecting SNARE zippering, post-translational modifications or minor changes in the TMDs such as conservative substitutions of single amino acids may have profound effects on these parameters (for review see refs. 169,184,195–199). In addition, fusion pores are governed by membrane tension or stresses exerted by curvature, or by other means such as osmotic pressure caused by osmotic swelling of vesicle contents174,200. Evidently, the local composition of membrane lipids, particularly the cholesterol content201,202, has a profound influence on these factors. For instance, addition of cholesterol promotes fusion pore opening while preventing stalling of fusion in a hemifusion state203. The lifetime and expansion of fusion pores can be readily determined from a shape analysis of single-vesicle fusion profiles measured by total internal reflection fluorescence microscopy, which also showed that addition of negative curvature-inducing phosphatidylethanolamine lipids in the cis leaflet, but not in the trans leaflet, increased the lifetime of fusion pores204.
In intact cells the fusion apparatus is embedded in the physiological regulatory context, with a multitude of interacting proteins in the vicinity of the fusion site, complicating the interpretation of molecular perturbations. Molecular mechanisms governing fusion pores cannot be deduced from even the most sophisticated kinetic measurements — their interpretation is limited to supporting plausible models. Thus, in such experiments it is difficult to discern whether SNAREs or other proteins govern fusion pore dynamics by directly interacting with membrane lipids within the pore structure, for instance with the SNARE TMDs lining the pore rim, or whether their effects are exerted indirectly by affecting membrane tension from a distance, with the pore itself being lipidic. In recent years, in vitro assays for monitoring SNARE-mediated fusion involving ab initio reconstitution have become highly sophisticated (see Table 1), allowing for monitoring fusion pore opening using imaging or electrophysiology with high temporal resolution. For instance, SNAREs reconstituted in nanodiscs can be fused with liposomes, planar membranes or even intact cells (using artificially ‘flipped’ SNAREs)71,205–208 (for review see refs. 196,209). Although complete fusion cannot be achieved due to the spatial constraints of lateral lipid movement in nanodiscs, initial opening states can be captured with this approach, thus allowing for dissecting parameters affecting pore features such as lipid composition, membrane tension, structure of TMDs or protein stoichiometries.
Concluding remarks
Since the discovery of the SNARE fusion machinery about 30 years ago, major progress was made not only in the detailed understanding of the SNARE machinery with its complex conformational association–dissociation cycle and the multitude of proteins controlling this cycle but also in understanding the physical principles of fusion itself. Indeed, due to the structural simplicity of the SNARE ‘core engine’, SNAREs were instrumental in the development of refined models of fusion trajectories that are compatible with physical principles. There are still major unresolved questions concerning the structure and dynamics of the transition states governing the non-bilayer intermediates of the SNARE-mediated fusion pathway. For example, the structure of the neuronal pre-fusion complex that is activated by Ca2+ ions is still unclear, despite recent advances described above. Similarly, it is unclear to what extent the SNARE TMDs and the short linkers connecting them to the SNARE motifs are part of transition states such as hemifusion stalks and initial fusion pores. Moreover, in vitro reconstitutions of SNARE-mediated fusion have considerably improved in recent years and exhibit features approximating those of biological fusion reactions including Ca2+-regulated exocytosis. Although these reconstitutions were crucial in achieving the insights discussed in this Review, there are still major uncertainties regarding to what extent findings obtained in such systems can be transferred to in vivo reactions. However, we expect that due to rapid technical advances, particularly in obtaining time-resolved high-resolution structures by cryo-EM and cryo-electron tomography and in the parallel development of increasingly sophisticated physical models (see for example refs. 117,127,168,210,211), a complete molecular description of the fascinating SNARE nanomachines appears to be within reach in the foreseeable future.
Acknowledgements
The authors thank H. Grubmüller, M. Müller, S. Pribićević and A. Stein (all Max-Planck Institute for Multidisciplinary Sciences, Göttingen) for critical comments, and A. Chizhik (Third Institute of Physics, University of Göttingen) for help in preparing Fig. 1. Work in the authors’ laboratory was supported by Program Project Grant 2 P01 GM072694 from the National Institutes of Health (NIH) awarded to L.K.T., R.J. and D.S.C.
Glossary
- Black lipid membranes
An experimental model in which single bilayers span a hole 10–100 μm wide created in a Teflon sheet immersed in aqueous media. They appear black because light reflected off the back of the membrane interferes with light reflected from the front
- Content mixing
Mixing of the aqueous interior of two vesicles undergoing fusion. Only occurs after opening of a fusion pore
- Content release
Release of the enclosed content of a vesicle into the environment upon fusion of a vesicle with a planar membrane
- Hysteresis
The state of a system depends on the history (or pathway) by which the state has been reached. Hysteretic systems therefore cannot be modelled using equilibrium thermodynamics where a system can only assume a single state under a defined set of conditions, independent of how it got there
- kT
The product of the Boltzmann constant k with the absolute temperature T, a measure of the thermal energy per molecule. Can be converted into other energy units such as calories, joules or electronvolts
- Lipid mixing
Mixing of membrane lipids during fusion, often describing the stage when the contacting lipid leaflets have mixed and the two distal leaflets remain separated
- Secretory pathway
A system that connects most intracellular membranes, including the plasma membrane of eukaryotic cells, by vesicles that dissociate from the ‘donor’ membrane and then fuse with the target membrane, including the plasma membrane, resulting in secretion of the vesicle content
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Kalia R & Frost A Open and cut: allosteric motion and membrane fission by dynamin superfamily proteins. Mol. Biol. Cell 30, 2097–2104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfitzner AK, von Filseck JM & Roux A Principles of membrane remodeling by dynamic ESCRT-III polymers. Trends Cell Biol. 31, 856–868 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Westermann B Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol 11, 872–884 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Harrison SC Viral membrane fusion. Virology 479, 498–507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White JM, Ward AE, Odongo L & Tamm LK Viral membrane fusion: a dance between proteins and lipids. Annu. Rev. Virol 10, 10.1146/annurev-virology-111821-093413 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Söllner T et al. SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Südhof TC & Rothman JE Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Südhof TC The synaptic vesicle cycle. Annu. Rev. Neurosci 27, 509–547 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Schillemans M, Karampini E, Kat M & Bierings R Exocytosis of Weibel–Palade bodies: how to unpack a vascular emergency kit. J. Thromb. Haemost 17, 6–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jahn R & Scheller RH SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol 7, 631–643 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Brukman NG, Uygur B, Podbilewicz B & Chernomordik LV How cells fuse. J. Cell Biol 218, 1436–1451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrany MJ & Millay DP Cell fusion: merging membranes and making muscle. Trends Cell Biol. 29, 964–973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S & Hu J Mitochondrial fusion: the machineries in and out. Trends Cell Biol. 31, 62–74 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Wickner W & Rizo J A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell 28, 707–711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y & Hughson FM Chaperoning SNARE folding and assembly. Annu. Rev. Biochem 90, 581–603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizo J Molecular mechanisms underlying neurotransmitter release. Annu. Rev. Biophys 51, 377–408 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunger AT, Choi UB, Lai Y, Leitz J & Zhou Q Molecular mechanisms of fast neurotransmitter release. Annu. Rev. Biophys 47, 469–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bubnis G & Grubmüller H Sequential water and headgroup merger: membrane poration paths and energetics from MD simulations. Biophys. J 119, 2418–2430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smirnova YG, Risselada HJ & Müller M Thermodynamically reversible paths of the first fusion intermediate reveal an important role for membrane anchors of fusion proteins. Proc. Natl Acad. Sci. USA 116, 2571–2576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlov MM & Markin VS Possible mechanism of membrane fusion [Russian]. Biofizika 28, 242–247 (1983). [PubMed] [Google Scholar]
- 21.Kuzmin PI, Zimmerberg J, Chizmadzhev YA & Cohen FS A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl Acad. Sci. USA 98, 7235–7240 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozlov MM & Chernomordik LV Membrane tension and membrane fusion. Curr. Opin. Struct. Biol 33, 61–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernomordik LV & Kozlov MM Protein–lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem 72, 175–207 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Cohen FS & Melikyan GB The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol 199, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Fan ZA, Tsang KY, Chen SH & Chen YF Revisit the correlation between the elastic mechanics and fusion of lipid membranes. Sci. Rep 6, 31470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chernomordik LV & Kozlov MM Mechanics of membrane fusion. Nat. Struct. Mol. Biol 15, 675–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marrink SJ & Mark AE The mechanism of vesicle fusion as revealed by molecular dynamics simulations. J. Am. Chem. Soc 125, 11144–11145 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Beaven AH, Sapp K & Sodt AJ Simulated dynamic cholesterol redistribution favors membrane fusion pore constriction. Biophys. J 122, 2162–2175 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johner N, Harries D & Khelashvili G Implementation of a methodology for determining elastic properties of lipid assemblies from molecular dynamics simulations. BMC Bioinform. 17, 161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice A, Zimmerberg J & Pastor RW Initiation and evolution of pores formed by influenza fusion peptides probed by lysolipid inclusion. Biophys. J 122, 1018–1032 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grafmuller A, Shillcock J & Lipowsky R The fusion of membranes and vesicles: pathway and energy barriers from dissipative particle dynamics. Biophys. J 96, 2658–2675 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamoto S, Klein ML & Shinoda W Coarse-grained molecular dynamics study of membrane fusion: curvature effects on free energy barriers along the stalk mechanism. J. Chem. Phys 143, 243112 (2015). [DOI] [PubMed] [Google Scholar]
- 33.McLaughlin S The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem 18, 113–136 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Lipowsky R The conformation of membranes. Nature 349, 475–481 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Rand RP & Parsegian VA Physical force considerations in model and biological membranes. Can. J. Biochem. Cell Biol 62, 752–759 (1984). [DOI] [PubMed] [Google Scholar]
- 36.Israelachvili JN Surface forces. In The Handbook of Surface Imaging and Visualization (ed. Hubbard AT) Ch. 24, 793–817 (Taylor & Francis, 1995). [Google Scholar]
- 37.Tamm LK & Han X Viral fusion peptides: a tool set to disrupt and connect biological membranes. Biosci. Rep 20, 501–518 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Risselada HJ et al. Line-tension controlled mechanism for influenza fusion. PLoS ONE 7, e38302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langosch D et al. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J. Mol. Biol 311, 709–721 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Hernandez JM, Kreutzberger AJB, Kiessling V, Tamm LK & Jahn R Variable cooperativity in SNARE-mediated membrane fusion. Proc. Natl Acad. Sci. USA 111, 12037–12042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brandt T, Cavellini L, Kühlbrandt W & Cohen MM A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. eLife 5, e14618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gui L, Ebner JL, Mileant A, Williams JA & Lee KK Visualization and sequencing of membrane remodeling leading to influenza virus fusion. J. Virol 90, 6948–6962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witkowska A, Heinz LP, Grubmüller H & Jahn R Tight docking of membranes before fusion represents a metastable state with unique properties. Nat. Commun 12, 3606 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasson PM, Lindahl E & Pande VS Atomic-resolution simulations predict a transition state for vesicle fusion defined by contact of a few lipid tails. PLoS Comput. Biol 6, e1000829 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsson P & Kasson PM Lipid tail protrusion in simulations predicts fusogenic activity of influenza fusion peptide mutants and conformational models. PLoS Comput. Biol 9, e1002950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheidt HA et al. Light-induced lipid mixing implies a causal role of lipid splay in membrane fusion. Biochem. Biophys. Acta Biomembr 1862, 183483 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Yang L & Huang HW Observation of a membrane fusion intermediate structure. Science 297, 1877–1879 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Aeffner S, Reusch T, Weinhausen B & Salditt T Energetics of stalk intermediates in membrane fusion are controlled by lipid composition. Proc. Natl Acad. Sci. USA 109, E1609–E1618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salditt T & Aeffner S X-ray structural investigations of fusion intermediates: lipid model systems and beyond. Semin. Cell Dev. Biol 60, 65–77 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Qian S & Rai DK Grazing-angle neutron diffraction study of the water distribution in membrane hemifusion: from the lamellar to rhombohedral phase. J. Phys. Chem. Lett 9, 5778–5784 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Tiberti ML, Antonny B & Gautier R The transbilayer distribution of polyunsaturated phospholipids determines their facilitating effect on membrane deformation. Soft Matter 16, 1722–1730 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Chernomordik LV & Kozlov MM Membrane hemifusion: crossing a chasm in two leaps. Cell 123, 375–382 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Sharma S & Lindau M The fusion pore, 60 years after the first cartoon. FEBS Lett. 592, 3542–3562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risselada HJ, Bubnis G & Grubmüller H Expansion of the fusion stalk and its implication for biological membrane fusion. Proc. Natl Acad. Sci. USA 111, 11043–11048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chanturiya A, Chernomordik LV & Zimmerberg J Flickering fusion pores comparable with initial exocytotic pores occur in protein-free phospholipid bilayers. Proc. Natl Acad. Sci. USA 94, 14423–14428 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kienle N, Kloepper TH & Fasshauer D Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol. Biol 9, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fasshauer D, Sutton RB, Brunger AT & Jahn R Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA 95, 15781–15786 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kloepper TH, Kienle CN & Fasshauer D An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18, 3463–3471 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma D, Chen ZH, He ZP & Huang XQ A SNARE protein identification method based on iLearnPlus to efficiently solve the data imbalance problem. Front. Genet 12, 818841 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kadkova A, Radecke J & Sorensen JB The SNAP-25 protein family. Neuroscience 420, 50–71 (2019). [DOI] [PubMed] [Google Scholar]
- 61.McNew JA et al. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum–Golgi transport. J. Biol. Chem 272, 17776–17783 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Weimbs T et al. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl Acad. Sci. USA 94, 3046–3051 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ungermann C & Wickner W Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 17, 3269–3276 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato TK, Darsow T & Emr SD Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell Biol 18, 5308–5319 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Itakura E & Mizushima N Syntaxin 17: the autophagosomal SNARE. Autophagy 9, 917–919 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masuda ES, Huang BC, Fisher JM, Luo Y & Scheller RH Tomosyn binds t-SNARE proteins via a VAMP-like coiled coil. Neuron 21, 479–480 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Pobbati AV, Razeto A, Boddener M, Becker S & Fasshauer D Structural basis for the inhibitory role of tomosyn in exocytosis. J. Biol. Chem 279, 47192–47200 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Scales SJ, Hesser BA, Masuda ES & Scheller RH Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. J. Biol. Chem 277, 28271–28279 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Hattendorf DA, Andreeva A, Gangar A, Brennwald PJ & Weis WI Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature 446, 567–571 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Sinha R, Ahmed S, Jahn R & Klingauf J Two synaptobrevin molecules are sufficient for vesicle fusion in central nervous system synapses. Proc. Natl Acad. Sci. USA 108, 14318–14323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi L et al. SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science 335, 1355–1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van den Bogaart G et al. One SNARE complex is sufficient for membrane fusion. Nat. Struct. Mol. Biol 17, 358–364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanson PI, Roth R, Morisaki H, Jahn R & Heuser JE Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/ deep-etch electron microscopy. Cell 90, 523–535 (1997). [DOI] [PubMed] [Google Scholar]
- 74.Weber T et al. SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Walter AM, Wiederhold K, Bruns D, Fasshauer D & Sorensen JB Synaptobrevin N-terminally bound to syntaxin–SNAP-25 defines the primed vesicle state in regulated exocytosis. J. Cell Biol 188, 401–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stein A, Weber G, Wahl MC & Jahn R Helical extension of the neuronal SNARE complex into the membrane. Nature 460, 525–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou P, Bacaj T, Yang X, Pang ZP & Sudhof TC Lipid-anchored SNAREs lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron 80, 470–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hong W SNAREs and traffic. Biochim. Biophys. Acta 1744, 120–144 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Pelham HR SNAREs and the secretory pathway — lessons from yeast. Exp. Cell Res 247, 1–8 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Zwilling D et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 26, 9–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burri L & Lithgow T A complete set of SNAREs in yeast. Traffic 5, 45–52 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Koike S & Jahn R SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat. Commun 10, 1608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fasshauer D, Antonin W, Subramaniam V & Jahn R SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol 9, 144–151 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y Energetics, kinetics, and pathway of SNARE folding and assembly revealed by optical tweezers. Protein Sci. 26, 1252–1265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiederhold K & Fasshauer D Is assembly of the SNARE complex enough to fuel membrane fusion? J. Biol. Chem 284, 13142–13152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fasshauer D, Otto H, Eliason WK, Jahn R & Brunger AT Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem 272, 28036–28041 (1997). [DOI] [PubMed] [Google Scholar]
- 87.Nicholson KL et al. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat. Struct. Biol 5, 793–802 (1998). [DOI] [PubMed] [Google Scholar]
- 88.Ellena JF et al. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc. Natl Acad. Sci. USA 106, 20306–20311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang B, Kiessling V & Tamm LK Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc. Natl Acad. Sci. USA 110, 19384–19389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liang B, Dawidowski D, Ellena JF, Tamm LK & Cafiso DS The SNARE motif of synaptobrevin exhibits an aqueous-interfacial partitioning that is modulated by membrane curvature. Biochemistry 53, 1485–1494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lakomek NA, Yavuz H, Jahn R & Perez-Lara A Structural dynamics and transient lipid binding of synaptobrevin-2 tune SNARE assembly and membrane fusion. Proc. Natl Acad. Sci. USA 116, 8699–8708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stief T et al. Intrinsic disorder of the neuronal SNARE protein SNAP25a in its pre-fusion conformation. J. Mol. Biol 435, 168069 (2023). [DOI] [PubMed] [Google Scholar]
- 93.Lerman JC, Robblee J, Fairman R & Hughson FM Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry 39, 8470–8479 (2000). [DOI] [PubMed] [Google Scholar]
- 94.Misura KM, Scheller RH & Weis WI Self-association of the H3 region of syntaxin 1A. Implications for intermediates in SNARE complex assembly. J. Biol. Chem 276, 13273–13282 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Sieber JJ et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317, 1072–1076 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Xiao W, Poirier MA, Bennett MK & Shin YK The neuronal t-SNARE complex is a parallel four-helix bundle. Nat. Struct. Biol 8, 308–311 (2001). [DOI] [PubMed] [Google Scholar]
- 97.Margittai M, Fasshauer D, Pabst S, Jahn R & Langen R Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J. Biol. Chem 276, 13169–13177 (2001). [DOI] [PubMed] [Google Scholar]
- 98.Misura KM, Gonzalez LC Jr., May AP, Scheller RH & Weis WI Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a. J. Biol. Chem 276, 41301–41309 (2001). [DOI] [PubMed] [Google Scholar]
- 99.Hesselbarth J & Schmidt C Mass spectrometry uncovers intermediates and off-pathway complexes for SNARE complex assembly. Commun. Biol 6, 198 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dulubova I et al. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 18, 4372–4382 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Margittai M et al. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc. Natl Acad. Sci. USA 100, 15516–15521 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fasshauer D & Margittai M A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem 279, 7613–7621 (2004). [DOI] [PubMed] [Google Scholar]
- 103.Furukawa N & Mima J Multiple and distinct strategies of yeast SNAREs to confer the specificity of membrane fusion. Sci. Rep 4, 4277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Antonin W et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19, 6453–6464 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilhelm BG et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Bethani I et al. The specificity of SNARE pairing in biological membranes is mediated by both proof-reading and spatial segregation. EMBO J. 26, 3981–3992 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Behnia R & Munro S Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Ungermann C & Kummel D Structure of membrane tethers and their role in fusion. Traffic 20, 479–490 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Archbold JK, Whitten AE, Hu SH, Collins BM & Martin JL SNARE-ing the structures of Sec1/Munc18 proteins. Curr. Opin. Struct. Biol 29, 44–51 (2014). [DOI] [PubMed] [Google Scholar]
- 110.Toonen RF & Verhage M Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 13, 177–186 (2003). [DOI] [PubMed] [Google Scholar]
- 111.Yu IM & Hughson FM Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol 26, 137–156 (2010). [DOI] [PubMed] [Google Scholar]
- 112.Santana-Molina C, Gutierrez F & Devos DP Homology and modular evolution of CATCHR at the origin of the eukaryotic endomembrane system. Genome Biol. Evol 13, evab125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pobbati AV, Stein A & Fasshauer D N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313, 673–676 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Liu Y et al. SNARE zippering is suppressed by a conformational constraint that Is removed by v-SNARE splitting. Cell Rep. 34, 108611 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gerber SH et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science 321, 1507–1510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andre T et al. The interaction of Munc18–1 helix 11 and 12 with the central region of the VAMP2 SNARE motif is essential for SNARE templating and synaptic transmission. eNeuro 7, 10.1523/ENEURO.0278-20.2020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stepien KP, Xu J, Zhang X, Bai XC & Rizo J SNARE assembly enlightened by cryo-EM structures of a synaptobrevin–Munc18–1–syntaxin-1 complex. Sci. Adv 8, eabo5272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dawidowski D & Cafiso DS Munc18–1 and the syntaxin-1 N terminus regulate open-closed states in a t-SNARE complex. Structure 24, 392–400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jakhanwal S, Lee CT, Urlaub H & Jahn R An activated Q-SNARE/SM protein complex as a possible intermediate in SNARE assembly. EMBO J. 36, 1788–1802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kraynack BA et al. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell 16, 3963–3977 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tripathi A, Ren Y, Jeffrey PD & Hughson FM Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat. Struct. Mol. Biol 16, 114–123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Magdziarek M et al. Re-examining how Munc13–1 facilitates opening of syntaxin-1. Protein Sci. 29, 1440–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rizo J, David G, Fealey ME & Jaczynska K On the difficulties of characterizing weak protein interactions that are critical for neurotransmitter release. FEBS Open Bio 12, 1912–1938 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kummel D et al. Complexin cross-links prefusion SNAREs into a zigzag array. Nat. Struct. Mol. Biol 18, 927–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou QJ et al. Architecture of the synaptotagmin–SNARE machinery for neuronal exocytosis. Nature 525, 62–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou QJ et al. The primed SNARE–complexin–synaptotagmin complex for neuronal exocytosis. Nature 548, 420–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brunger AT & Leitz J The core complex of the Ca2+-triggered presynaptic fusion machinery. J. Mol. Biol 435, 167853 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Snyder DA, Kelly ML & Woodbury DJ SNARE complex regulation by phosphorylation. Cell Biochem. Biophys 45, 111–123 (2006). [DOI] [PubMed] [Google Scholar]
- 129.Warner H, Mahajan S & van den Bogaart G Rerouting trafficking circuits through posttranslational SNARE modifications. J. Cell Sci 135, jcs.260112 (2022). [DOI] [PubMed] [Google Scholar]
- 130.Sabatini BL & Regehr WG Timing of synaptic transmission. Annu. Rev. Physiol 61, 521–542 (1999). [DOI] [PubMed] [Google Scholar]
- 131.Imig C et al. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron 84, 416–431 (2014). [DOI] [PubMed] [Google Scholar]
- 132.Pang ZP & Sudhof TC Cell biology of Ca2+-triggered exocytosis. Curr. Opin. Cell Biol 22, 496–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wolfes AC & Dean C The diversity of synaptotagmin isoforms. Curr. Opin. Neurobiol 63, 198–209 (2020). [DOI] [PubMed] [Google Scholar]
- 134.Park Y & Ryu JK Models of synaptotagmin-1 to trigger Ca2+-dependent vesicle fusion. FEBS Lett. 592, 3480–3492 (2018). [DOI] [PubMed] [Google Scholar]
- 135.Mohrmann R, Dhara M & Bruns D Complexins: small but capable. Cell Mol. Life Sci 72, 4221–4235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lottermoser JA & Dittman JS Complexin membrane interactions: implications for synapse evolution and function. J. Mol. Biol 435, 167774 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.van den Bogaart G et al. Membrane protein sequestering by ionic protein–lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bai J, Tucker WC & Chapman ER PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol 11, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- 139.Li LY et al. Phosphatidylinositol phosphates as co-activators of Ca2+ binding to C-2 domains of synaptotagmin 1. J. Biol. Chem 281, 15845–15852 (2006). [DOI] [PubMed] [Google Scholar]
- 140.Söllner T, Bennett MK, Whiteheart SW, Scheller RH & Rothman JE A protein assembly–disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 141.Puchades C, Sandate CR & Lander GC The molecular principles governing the activity and functional diversity of AAA plus proteins. Nat. Rev. Mol. Cell Biol 21, 43–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khan YA, White KI & Brunger AT The AAA plus superfamily: a review of the structural and mechanistic principles of these molecular machines. Crit. Rev. Biochem. Mol 57, 156–187 (2021). [DOI] [PubMed] [Google Scholar]
- 143.Wickner W & Schekman R Membrane fusion. Nat. Struct. Mol. Biol 15, 658–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Clary DO, Griff IC & Rothman JE SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell 61, 709–721 (1990). [DOI] [PubMed] [Google Scholar]
- 145.Zhao ML et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature 518, 61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vivona S et al. Disassembly of all SNARE complexes by N-ethylmaleimide-sensitive factor (NSF) is initiated by a conserved 1:1 interaction between α-soluble NSF attachment protein (SNAP) and SNARE complex. J. Biol. Chem 288, 24984–24991 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Winter U, Chen X & Fasshauer D A conserved membrane attachment site in α-SNAP facilitates N-ethylmaleimide-sensitive factor (NSF)-driven SNARE complex disassembly. J. Biol. Chem 284, 31817–31826 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cipriano DJ et al. Processive ATP-driven substrate disassembly by the N-ethylmaleimide-sensitive factor (NSF) molecular machine. J. Biol. Chem 288, 23436–23445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.White KI, Zhao M, Choi UB, Pfuetzner RA & Brunger AT Structural principles of SNARE complex recognition by the AAA plus protein NSF. eLife 7, e38888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ryu JK et al. Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science 347, 1485–1489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yavuz H et al. Arrest of trans-SNARE zippering uncovers loosely and tightly docked intermediates in membrane fusion. J. Biol. Chem 293, 8645–8655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu H, Jun Y, Thompson J, Yates J & Wickner W HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 29, 1948–1960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Prinslow EA, Stepien KP, Pan YZ, Xu JJ & Rizo J Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and αSNAP. eLife 8, e38880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lobingier BT, Nickerson DP, Lo SY & Merz AJ SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife 3, e02272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zick M, Orr A, Schwartz ML, Merz AJ & Wickner WT Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc. Natl Acad. Sci. USA 112, E2290–E2297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Finkelstein AV, Bogatyreva NS, Ivankov DN & Garbuzynskiy SO Protein folding problem: enigma, paradox, solution. Biophys. Rev 14, 1255–1272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Manca F et al. SNARE machinery is optimized for ultrafast fusion. Proc. Natl Acad. Sci. USA 116, 2435–2442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dubuke ML & Munson M The secret life of tethers: the role of tethering factors in SNARE complex regulation. Front. Cell Dev. Biol 4, 42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Baker RW & Hughson FM Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol 17, 465–479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chamberlain LH, Burgoyne RD & Gould GW SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl Acad. Sci. USA 98, 5619–5624 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lang T et al. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20, 2202–2213 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Barg S, Knowles MK, Chen X, Midorikawa M & Almers W Syntaxin clusters assemble reversibly at sites of secretory granules in live cells. Proc. Natl Acad. Sci. USA 107, 20804–20809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li ML, Oh TJ, Fan HX, Diao JJ & Zhang K Syntaxin clustering and optogenetic control for synaptic membrane fusion. J. Mol. Biol 432, 4773–4782 (2020). [DOI] [PubMed] [Google Scholar]
- 164.Gandasi NR & Barg S Contact-induced clustering of syntaxin and munc18 docks secretory granules at the exocytosis site. Nat. Commun 5, 3914 (2014). [DOI] [PubMed] [Google Scholar]
- 165.Koike S & Jahn R SNARE proteins: zip codes in vesicle targeting. Biochem. J 479, 273–288 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hernandez JM et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science 336, 1581–1584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ginger L et al. Arrangements of proteins at reconstituted synaptic vesicle fusion sites depend on membrane separation. FEBS Lett. 594, 3450–3463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kiessling V et al. A molecular mechanism for calcium-mediated synaptotagmin-triggered exocytosis. Nat. Struct. Mol. Biol 25, 911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Risselada HJ & Mayer A SNAREs, tethers and SM proteins: how to overcome the final barriers to membrane fusion? Biochem. J 477, 243–258 (2020). [DOI] [PubMed] [Google Scholar]
- 170.Domanska MK, Kiessling V & Tamm LK Docking and fast fusion of synaptobrevin vesicles depends on the lipid compositions of the vesicle and the acceptor SNARE complex-containing target membrane. Biophys. J 99, 2936–2946 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang L, Seeley ES, Wickner W & Merz AJ Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108, 357–369 (2002). [DOI] [PubMed] [Google Scholar]
- 172.Rizzoli SO & Betz WJ The structural organization of the readily releasable pool of synaptic vesicles. Science 303, 2037–2039 (2004). [DOI] [PubMed] [Google Scholar]
- 173.Pieren M, Desfougeres Y, Michaillat L, Schmidt A & Mayer A Vacuolar SNARE protein transmembrane domains serve as nonspecific membrane anchors with unequal roles in lipid mixing. J. Biol. Chem 290, 12821–12832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dhara M et al. Synergistic actions of v-SNARE transmembrane domains and membrane-curvature modifying lipids in neurotransmitter release. eLife 9, e55152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hu YR, Zhu L & Ma C Structural roles for the juxtamembrane linker region and transmembrane region of synaptobrevin 2 in membrane fusion. Front. Cell Dev. Biol 8, 609708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Grote E, Baba M, Ohsumi Y & Novick PJ Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J. Cell Biol 151, 453–466 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chang CW, Chiang CW, Gaffaney JD, Chapman ER & Jackson MB Lipid-anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J. Biol. Chem 291, 2848–2857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ucar H et al. Mechanical actions of dendritic-spine enlargement on presynaptic exocytosis. Nature 600, 686–689 (2021). [DOI] [PubMed] [Google Scholar]
- 179.Rosenmund C & Stevens CF Definition of the readily releasable pool of vesicles at hippocampal synapses. Neuron 16, 1197–1207 (1996). [DOI] [PubMed] [Google Scholar]
- 180.Chakraborty H, Tarafdar PK, Bruno MJ, Sengupta T & Lentz BR Activation thermodynamics of poly(ethylene glycol)-mediated model membrane fusion support mechanistic models of stalk and pore formation. Biophys. J 102, 2751–2760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.D’Agostino M, Risselada HJ, Lurick A, Ungermann C & Mayer A A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature 551, 634–638 (2017). [DOI] [PubMed] [Google Scholar]
- 182.Orr A, Song H & Wickner W Fusion with wild-type SNARE domains is controlled by juxtamembrane domains, transmembrane anchors, and Sec17. Mol. Biol. Cell 33, ar38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lindau M & Almers W Structure and function of fusion pores in exocytosis and ectoplasmic membrane fusion. Curr. Opin. Cell Biol 7, 509–517 (1995). [DOI] [PubMed] [Google Scholar]
- 184.Chang CW, Chiang CW & Jackson MB Fusion pores and their control of neurotransmitter and hormone release. J. Gen. Physiol 149, 301–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Spruce AE, Breckenridge LJ, Lee AK & Almers W Properties of the fusion pore that forms during exocytosis of a mast cell secretory vesicle. Neuron 4, 643–654 (1990). [DOI] [PubMed] [Google Scholar]
- 186.Chow RH, von Ruden L & Neher E Delay in vesicle fusion revealed by electrochemical monitoring of single secretory events in adrenal chromaffin cells. Nature 356, 60–63 (1992). [DOI] [PubMed] [Google Scholar]
- 187.Takahashi N, Kishimoto T, Nemoto T, Kadowaki T & Kasai H Fusion pore dynamics and insulin granule exocytosis in the pancreatic islet. Science 297, 1349–1352 (2002). [DOI] [PubMed] [Google Scholar]
- 188.Lindau M & Alvarez DT The fusion pore. Biochim. Biophys. Acta 1641, 167–173 (2003). [DOI] [PubMed] [Google Scholar]
- 189.Fesce R, Grohovaz F, Valtorta F & Meldolesi J Neurotransmitter release: fusion or ‘kiss-and-run’? Trends Cell Biol. 4, 1–4 (1994). [DOI] [PubMed] [Google Scholar]
- 190.Alabi AA & Tsien RW Perspectives on kiss-and-run: role in exocytosis, endocytosis, and neurotransmission. Annu. Rev. Physiol 75, 393–422 (2013). [DOI] [PubMed] [Google Scholar]
- 191.Watanabe S et al. Ultrafast endocytosis at mouse hippocampal synapses. Nature 504, 242–247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Holroyd P, Lang T, Wenzel D, De Camilli P & Jahn R Imaging direct, dynamin-dependent recapture of fusing secretory granules on plasma membrane lawns from PC12 cells. Proc. Natl Acad. Sci. USA 99, 16806–16811 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Taraska JW, Perrais D, Ohara-Imaizumi M, Nagamatsu S & Almers W Secretory granules are recaptured largely intact after stimulated exocytosis in cultured endocrine cells. Proc. Natl Acad. Sci. USA 100, 2070–2075 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Taraska JW & Almers W Bilayers merge even when exocytosis is transient. Proc. Natl Acad. Sci. USA 101, 8780–8785 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jackson MB & Chapman ER Fusion pores and fusion machines in Ca2+-triggered exocytosis. Annu. Rev. Biophys. Biomol. Struct 35, 135–160 (2006). [DOI] [PubMed] [Google Scholar]
- 196.Wu Z, Thiyagarajan S, O’Shaughnessy B & Karatekin E Regulation of exocytotic fusion pores by SNARE protein transmembrane domains. Front. Mol. Neurosci 10, 315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hastoy B et al. A central small amino acid in the VAMP2 transmembrane domain regulates the fusion pore in exocytosis. Sci. Rep 7, 2835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dhara M, Mohrmann R & Bruns D v-SNARE function in chromaffin cells. Pflug. Arch 470, 169–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhang Y, Ma L & Bao H Energetics, kinetics, and pathways of SNARE assembly in membrane fusion. Crit. Rev. Biochem. Mol. Biol 57, 443–460 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bretou M, Anne C & Darchen F A fast mode of membrane fusion dependent on tight SNARE zippering. J. Neurosci 28, 8470–8476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Stratton BS et al. Cholesterol increases the openness of SNARE-mediated flickering fusion pores. Biophys. J 110, 1538–1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wu L, Courtney KC & Chapman ER Cholesterol stabilizes recombinant exocytic fusion pores by altering membrane bending rigidity. Biophys. J 120, 1367–1377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kreutzberger AJ, Kiessling V & Tamm LK High cholesterol obviates a prolonged hemifusion intermediate in fast SNARE-mediated membrane fusion. Biophys. J 109, 319–329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kreutzberger AJB et al. Asymmetric phosphatidylethanolamine distribution controls fusion pore lifetime and probability. Biophys. J 113, 1912–1915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wu ZY et al. Nanodisc-cell fusion: control of fusion pore nucleation and lifetimes by SNARE protein transmembrane domains. Sci. Rep 6, 27287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bao H et al. Exocytotic fusion pores are composed of both lipids and proteins. Nat. Struct. Mol. Biol 23, 67–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bao H et al. Dynamics and number of trans-SNARE complexes determine nascent fusion pore properties. Nature 554, 260–263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Das D, Bao H, Courtney KC, Wu L & Chapman ER Resolving kinetic intermediates during the regulated assembly and disassembly of fusion pores. Nat. Commun 11, 231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Karatekin E Toward a unified picture of the exocytotic fusion pore. FEBS Lett. 592, 3563–3585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sharma S & Lindau M Molecular mechanism of fusion pore formation driven by the neuronal SNARE complex. Proc. Natl Acad. Sci. USA 115, 12751–12756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Grushin K, Kalyana Sundaram RV, Sindelar CV & Rothman JE Munc13 structural transitions and oligomers that may choreograph successive stages in vesicle priming for neurotransmitter release. Proc. Natl Acad. Sci. USA 119, e2121259119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Holt M, Riedel D, Stein A, Schuette C & Jahn R Synaptic vesicles are constitutively active fusion machines that function independently of Ca2+. Curr. Biol 18, 715–722 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Park Y et al. Controlling synaptotagmin activity by electrostatic screening. Nat. Struct. Mol. Biol 19, 991–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kreutzberger AJB et al. Reconstitution of calcium-mediated exocytosis of dense-core vesicles. Sci. Adv 3, e1603208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kiessling V et al. Rapid fusion of synaptic vesicles with reconstituted target SNARE membranes. Biophys. J 104, 1950–1958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bello OD, Auclair SM, Rothman JE & Krishnakumar SS Using ApoE nanolipoprotein particles to analyze SNARE-induced fusion pores. Langmuir 32, 3015–3023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Malsam J et al. Complexin arrests a pool of docked vesicles for fast Ca2+-dependent release. EMBO J. 31, 3270–3281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Schuette CG et al. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl Acad. Sci. USA 101, 2858–2863 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nickel W et al. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc. Natl Acad. Sci. USA 96, 12571–12576 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Diao J et al. Single-vesicle fusion assay reveals Munc18–1 binding to the SNARE core is sufficient for stimulating membrane fusion. ACS Chem. Neurosci 1, 168–174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kyoung M et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc. Natl Acad. Sci. USA 108, E304–E313 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yoon TY, Okumus B, Zhang F, Shin YK & Ha T Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl Acad. Sci. USA 103, 19731–19736 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cypionka A et al. Discrimination between docking and fusion of liposomes reconstituted with neuronal SNARE-proteins using FCS. Proc. Natl Acad. Sci. USA 106, 18575–18580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bhalla A, Chicka MC, Tucker WC & Chapman ER Ca2+-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat. Struct. Mol. Biol 13, 323–330 (2006). [DOI] [PubMed] [Google Scholar]
- 225.Ma C, Su L, Seven AB, Xu Y & Rizo J Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tareste D, Shen J, Melia TJ & Rothman JE SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc. Natl Acad. Sci. USA 105, 2380–2385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fix M et al. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl Acad. Sci. USA 101, 7311–7316 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bowen ME, Weninger K, Brunger AT & Chu S Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J 87, 3569–3584 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Karatekin E et al. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc. Natl Acad. Sci. USA 107, 3517–3521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Domanska MK, Kiessling V, Stein A, Fasshauer D & Tamm LK Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J. Biol. Chem 284, 32158–32166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kiessling V, Domanska MK & Tamm LK Single SNARE-mediated vesicle fusion observed in vitro by polarized TIRFM. Biophys. J 99, 4047–4055 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schwenen LL et al. Resolving single membrane fusion events on planar pore-spanning membranes. Sci. Rep 5, 12006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sutton RB, Fasshauer D, Jahn R & Brunger AT Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 234.Tucker WC, Weber T & Chapman ER Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science 304, 435–438 (2004). [DOI] [PubMed] [Google Scholar]
- 235.van den Bogaart G et al. Synaptotagmin-1 may be a distance regulator acting upstream of SNARE nucleation. Nat. Struct. Mol. Biol 18, 805–812 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zdanowicz R et al. Complexin binding to membranes and acceptor t-SNAREs explains its clamping effect on fusion. Biophys. J 113, 1235–1250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li F et al. A half-zippered SNARE complex represents a functional intermediate in membrane fusion. J. Am. Chem. Soc 136, 3456–3464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Brunger AT et al. The pre-synaptic fusion machinery. Curr. Opin. Struct. Biol 54, 179–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li X et al. Symmetrical organization of proteins under docked synaptic vesicles. FEBS Lett. 593, 144–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhu J et al. Synaptotagmin rings as high-sensitivity regulators of synaptic vesicle docking and fusion. Proc. Natl Acad. Sci. USA 119, e2208337119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gao Y et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337, 1340–1343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Park Y et al. Synaptotagmin-1 binds to PIP2-containing membrane but not to SNAREs at physiological ionic strength. Nat. Struct. Mol. Biol 22, 815–823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]