Abstract
Human neural organoids, generated from pluripotent stem cells in vitro, are useful tools to study human brain development, evolution and disease. However, it is unclear which parts of the human brain are covered by existing protocols, and it has been difficult to quantitatively assess organoid variation and fidelity. Here we integrate 36 single-cell transcriptomic datasets spanning 26 protocols into one integrated human neural organoid cell atlas totalling more than 1.7 million cells1–26. Mapping to developing human brain references27–30 shows primary cell types and states that have been generated in vitro, and estimates transcriptomic similarity between primary and organoid counterparts across protocols. We provide a programmatic interface to browse the atlas and query new datasets, and showcase the power of the atlas to annotate organoid cell types and evaluate new organoid protocols. Finally, we show that the atlas can be used as a diverse control cohort to annotate and compare organoid models of neural disease, identifying genes and pathways that may underlie pathological mechanisms with the neural models. The human neural organoid cell atlas will be useful to assess organoid fidelity, characterize perturbed and diseased states and facilitate protocol development.
Subject terms: Neural stem cells, Transcriptomics, Developmental neurogenesis, Data integration
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, showing potential to assess organoid fidelity and facilitate protocol development.
Main
Human neural organoids, self-organizing three-dimensional human neural tissues grown in vitro, are becoming powerful tools for studying the mechanisms of human brain development, evolution and disease31–33. They can be generated using external patterning factors (for example, morphogens) to guide their development towards certain brain regions or to drive the emergence of specific cell types (guided protocols)7,11,18,34,35. Conversely, unguided protocols rely on the self-patterning capacity of organoids to generate diverse cell types and states36,37.
Single-cell RNA sequencing (scRNA-seq) is a powerful technology to characterize cell type heterogeneity in complex tissues, and has illuminated a remarkable heterogeneity of diverse progenitor, neuronal and glial cell types that can develop within neural organoids2–4,37,38, as well as differentiation trajectories of certain neural lineages. The data also enable the comparison of human neural organoid cells to those in the primary human brain, and most analyses have revealed strong similarity in molecular signatures6,18,25,39. Substantial differences have also been reported, including differential gene expression linked to media components39 and perturbed metabolic signatures associated with glycolysis3,10,23,24,38. Nevertheless, analysis of organoid tissues supports a useful recapitulation of early brain development, and scRNA-seq methods have been applied to study the molecular basis of neural cell type fate determination20, evolutionary differences in primates3,38,40,41 and pathological changes in neural disorders16,26,42,43. However, it is unclear which portions of the developing central nervous system can be generated with existing protocols and which ones are still lacking. It has also remained challenging to systematically quantify the transcriptomic fidelity of neural organoid cells compared to their primary counterparts.
In this study, we address these challenges by combining 36 scRNA-seq datasets covering numerous human neural organoid protocols into an integrated transcriptomic cell atlas. We establish an analytical pipeline that allows for the comprehensive and quantitative comparison of the organoid atlas to reference atlases of the developing human brain27. We harmonize annotations of cell populations in the primary and organoid systems, estimate the capacity and precision of different neural organoid protocols to generate different brain regions, and identify primary cell populations that are under-represented in neural organoids. We estimate transcriptomic fidelity of neurons in neural organoids, and identify previously described cell stress3,10,23,24 as a universal factor distinguishing metabolic states of in vitro neurons from primary neurons without strongly affecting core identities of neuronal cell types. We map the data of a neural organoid morphogen screen44 to the integrated atlas to assess regional specificity and generation of new states. We also collect 11 scRNA-seq datasets modelling 10 different neural diseases, and map the integrated data to the neural organoid atlas for cell type annotation and differential expression (DE) analysis. Finally, we show that the atlas can be expanded by projecting new data to the current atlas. Together, our work provides a rich resource and a new framework to assess the fidelity of neural organoids, characterize perturbed and diseased states and streamline protocol development.
Data curation, harmonization and integration
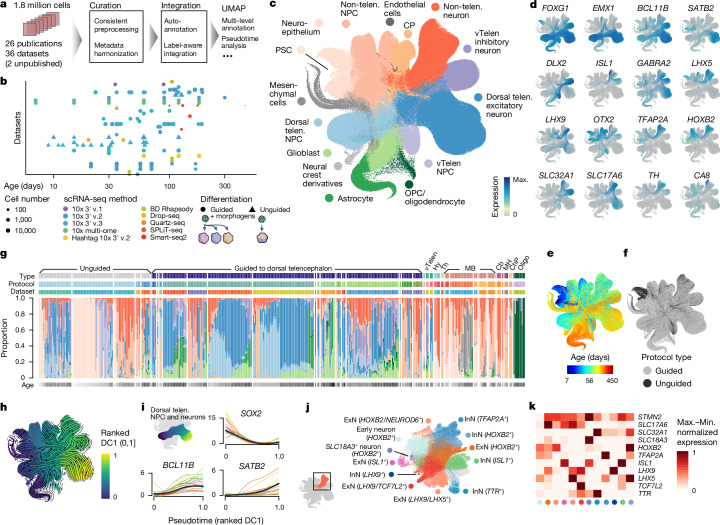
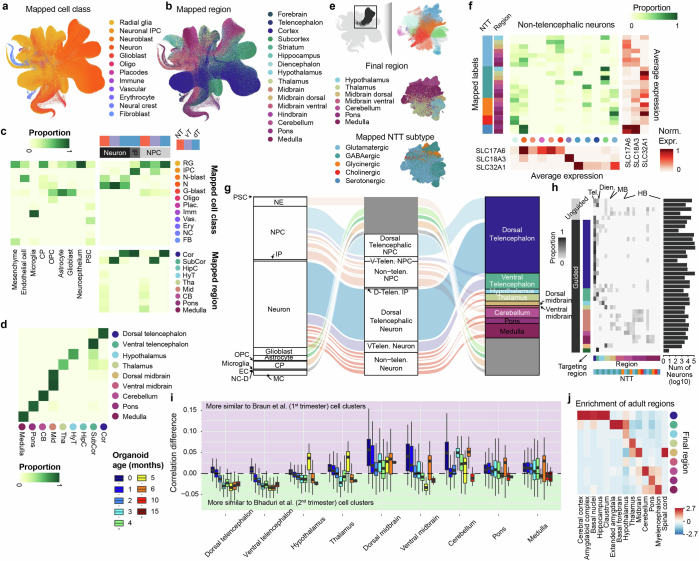
To build a transcriptomic human neural organoid cell atlas (HNOCA), we collected scRNA-seq data and detailed, harmonized technical and biological metadata from 36 datasets, including 34 published1–26 and two as yet unpublished ones (Supplementary Table 1), accounting for 1.77 million cells after consistent preprocessing and quality control (Fig. 1a). The HNOCA represents cell types and states generated with 26 distinct neural organoid differentiation protocols, including three unguided and 23 guided ones, at time points ranging from 7 to 450 days (Fig. 1b). To remove batch effects, we implemented a three-step integration pipeline. First, we projected the HNOCA to a single-cell atlas of the developing human brain27 using reference similarity spectrum (RSS)3. Then, we developed snapseed (Methods) to perform preliminary marker-based hierarchical cell type annotation. Last, we used scPoli45 for label-aware data integration based on the snapseed annotations. Evaluation of different integration approaches using a previously established benchmarking pipeline46 showed that scPoli had the best performance for these datasets (Extended Data Fig. 1). We performed clustering on the basis of the scPoli representation and annotated clusters on the basis of canonical marker gene expression, organoid sample age and the auto-generated cell type labels. A uniform manifold approximation and projection (UMAP) embedding highlighted three neuronal differentiation trajectories corresponding to dorsal telencephalic, ventral telencephalic and non-telencephalic populations as well as trajectories leading from progenitors to glial cell types such as astrocytes and oligodendrocytes precursors (Fig. 1c–e and Extended Data Fig. 2). Cells from both unguided and guided protocols were distributed across all trajectories (Fig. 1f).
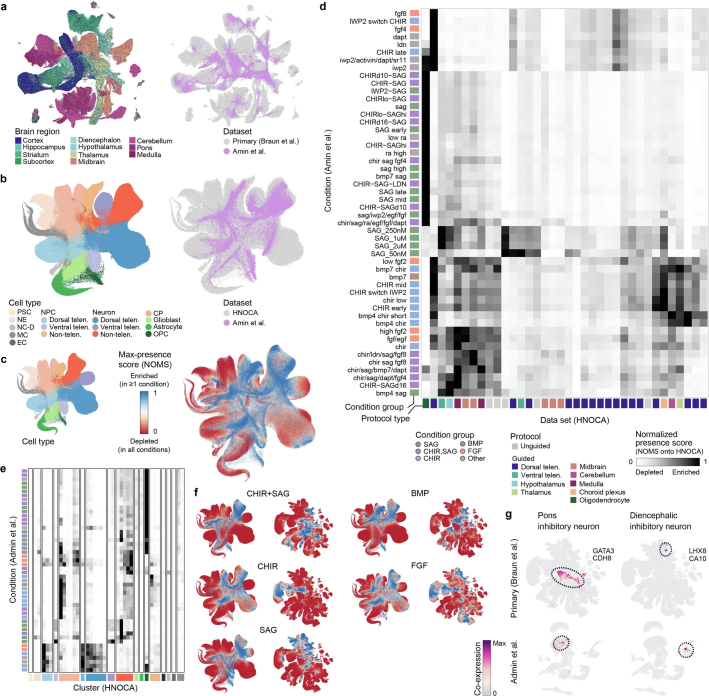
Fig. 1. Integrated HNOCA.
a, Overview of HNOCA construction pipeline. b, Metadata of biological samples included in HNOCA. c–f, UMAP of the integrated HNOCA, coloured by level 2 cell type annotations (c), gene expression profiles of selected markers (d), sample ages (e) and differentiation protocol types (f). g, Proportions of cells assigned to different cell types in the HNOCA. Every stacked bar represents one biological sample, grouped by datasets and ordered by increasing sample ages. Top bars show 36 datasets, organoid differentiation protocols, protocol types. Bottom bars show the sample age. h, UMAP of the integrated HNOCA coloured by top-ranked diffusion component (DC1) on the real-time-informed transition matrix between cells. The stream arrows visualize the inferred flow of cell states toward more mature cells. i, Marker gene expression profiles along cortical pseudotime. j, UMAP of non-telencephalic neurons, coloured and labelled by clusters. k, Heatmap showing relative expression of selected genes across different non-telencephalic neuron clusters. Coloured dots show cluster identities as shown in j. Cb, cerebellum; ChP, choroid plexus; CP, choroid plexus; Hy, hypothalamus; max., maximum; MB, midbrain; MH, medulla; min., minimum; Oligo, oligodendrocyte; OPC, oligodendrocyte progenitor cell; PSC, pluripotent stem cell; telen., telencephalon; Th, thalamus; vTelen, ventral telencephalon.
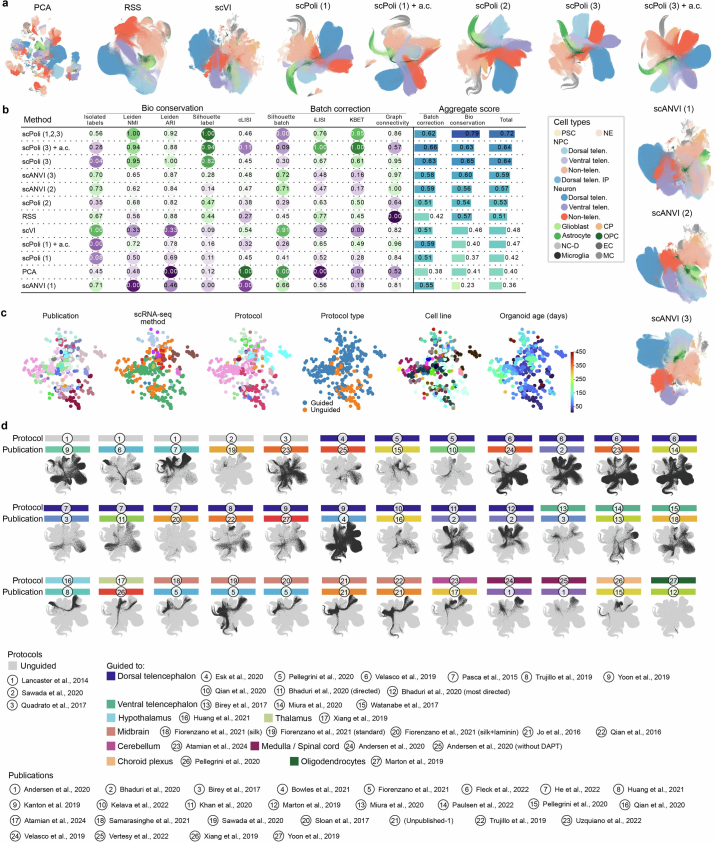
Extended Data Fig. 1. Benchmark of data integration.
(a) UMAPs of HNOCA, either without any data integration (PCA) or with different data integration methods applied. Number in parenthesis indicates which level of RSS-based snapseed annotation labels were provided as input to the model for methods which support semi-supervised data integration. Dots in all UMAP embeddings, each of which represents a cell, are colored by the cell type annotation introduced in Fig. 1. a.c. = aggrecell algorithm (b) scIB benchmarking metrics on all tested integration methods. (c) PCA of the scPoli sample embeddings from the final scPoli integration of HNOCA presented throughout the manuscript, colored by publications, scRNA-seq methods, organoid protocols, protocol types, cell lines, and sample ages. (d) UMAPs of HNOCA based on the final scPoli integration, each with one data set highlighted. Here, one data set is defined as data representing one protocol in one publication. The protocol and publication of each data set are shown by the color bar and indices on top of the UMAP.
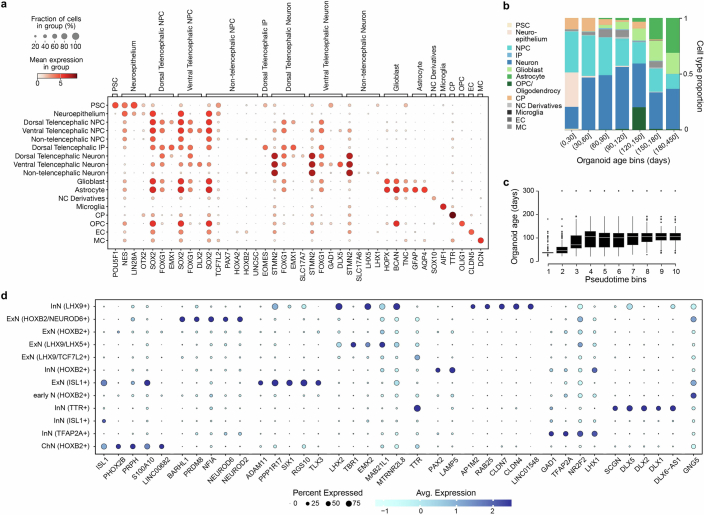
Extended Data Fig. 2. Characterization of HNOCA.
(a) Expression of selected marker genes used in the semi-automatic annotation of cell types for Fig. 1. (b) Mean cell type proportion over all data sets per organoid age bin. (c) Distribution of sample real-time age in days over deciles of computed pseudotime. (d) Expression of top markers in different non-telencephalic neural cell types. Markers are defined as genes with AUC > 0.7, in-out detection rate difference>20%, in-out detection rate ratio>2 and fold change>1.2. When more than 5 markers are found, only the top-5 (with the highest in-out detection rate ratio) are shown.
To elucidate the dynamics and transitions of cell states and types, we reconstructed a real-age-informed pseudotime of HNOCA cells on the basis of neural optimal transport47 using moscot48 (Fig. 1h). Focusing on the dorsal telencephalic neural trajectory, we observed consistent pseudotemporal expression profiles of marker genes such as SOX2 (neural progenitor cells (NPCs)), BCL11B (deeper layer cortical neurons) and SATB2 (upper layer cortical neurons) (Fig. 1i). To further resolve heterogeneity among non-telencephalic neurons, we performed subclustering of this population, which revealed numerous neuronal populations characterized by distinct marker gene expression (Fig. 1j,k).
HNOCA projection to a human developing brain atlas
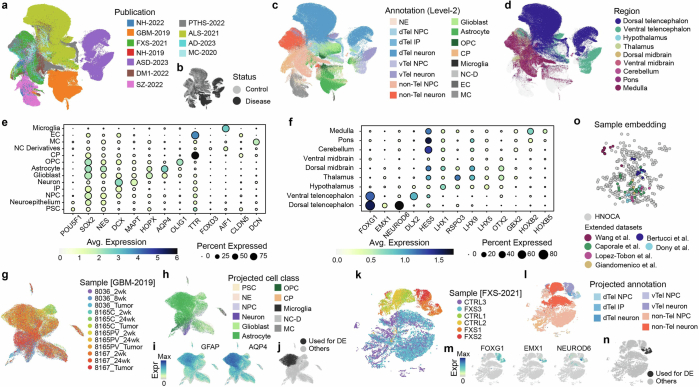
To assess our cell type annotation, and more precisely annotate the heterogeneous non-telencephalic neuronal populations, we compared the HNOCA to a recently published single-cell transcriptomic atlas of the developing human brain27 (Fig. 2a). We applied scVI49 and scANVI50 to the primary reference atlas, and used scArches51 to project the HNOCA to the same latent space. The shared latent space allowed us to reconstruct a bipartite weighted k-nearest-neighbour (wkNN) graph between cells in the HNOCA and the primary reference atlas, which was used to transfer the ‘CellClass’ and ‘Subregion’ labels, as well as the neurotransmitter transporter (NTT) information of neuroblasts and neurons to the HNOCA. The transferred labels are strongly consistent with our assigned labels (Extended Data Fig. 3) and allowed us to refine the regional annotation of HNOCA non-telencephalic NPCs and neurons, as well as the NTT annotation of the non-telencephalic neurons (Fig. 2b), resulting in the final hierarchical HNOCA cell type annotation (Extended Data Fig. 3).
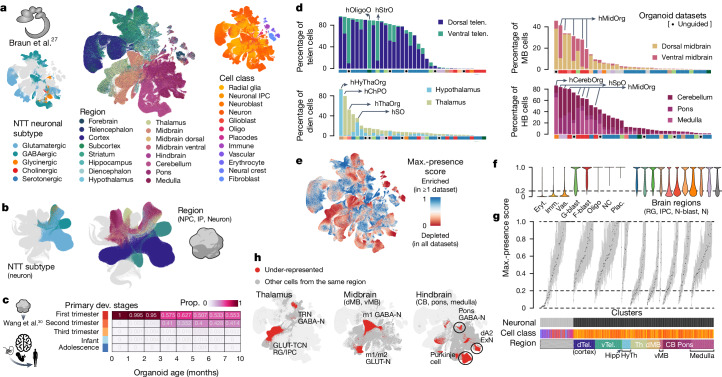
Fig. 2. Projection of HNOCA to primary developing human brain cell atlases assists organoid neural cell type annotation and estimation of primary cell type representation.
a, UMAP of a human developing brain cell atlas27, coloured by NTT subtypes (left), region (middle) and annotated cell classes (right). b, UMAP of HNOCA, coloured by the mapped neuron NTT subtypes (left) and regional labels of NPCs, intermediate progenitor cells (IP) and neurons. c, Heatmap showing proportions of cells from organoids of different ages matched to cells from different primary developmental (dev.) stages. d, Percentages of neural cells representing different regions (telencephalon, diencephalon, midbrain and hindbrain) in different datasets. The x axes show datasets, descendingly ordered by the total proportions (bar height). Datasets based on unguided differentiation protocols are marked by dots underneath. The bars at the bottom of each panel show organoid protocol types. e, UMAP of the human developing brain cell atlas27 coloured by cell population presence within HNOCA datasets (max presence score). A low score denotes under-representation of cell state in HNOCA datasets. f, Distribution of max presence scores of different cell classes in the human reference atlas27. Eryt., erythrocyte; Imm., immune; Vas., vascular; G-blast, glioblast; F-blast, fibroblast; NC, neural crest; Plac., placodes; RG, radial glia; IPC, intermediate progenitor cell; N-blast, neuroblast; N, neuron. g, Box plots showing distribution of max presence scores in different primary reference cell clusters. Bottom side bars show neuronal versus non-neuronal, cell class, region information of primary reference. h, UMAP of human developing brain atlas showing primary neural cell types or states under-represented in HNOCA (in red). Hippo, hippocampus; HyTh, hypothalamus; d, dorsal; v, ventral; CB, cerebellum.
Extended Data Fig. 3. Mapping-assisted annotation refinement of HNOCA.
(a-b) UMAP of HNOCA colored by the mapped (a) cell classes and (b) brain regions, both from the human developing brain cell atlas as the primary reference. (c) Comparison of the HNOCA cell type annotation with the primary reference mapping-based transferred cell class and brain region labels. Darkness of cells indicates proportions of each HNOCA cell type being assigned to different cell class and brain region categories. Brain region labels are only shown for the HNOCA neural cell types. (d) Comparison of the simple majority-voting-based regional label transfer and the hierarchical regional label transfer with random-walk-with-restart-based smoothening. Only cells annotated as NPCs, IPs and neurons are included. (e) UMAP of non-telencephalic neurons, colored by clusters (upper), mapped brain regions (middle) and mapped neurotransmitter transporter (NTT) subtypes (bottom). (f) Comparison of non-telencephalic neural cell types, defined as the concatenation of the mapped brain region and NTT subtype, with the clusters. The middle heatmap shows contributions of different clusters to different neural cell types. The sidebar on the left shows the neural cell types; dots under the heatmap show clusters. The heatmaps on the bottom and on the right show the average expression of three neurotransmitter transporters SLC17A6, SLC18A3 and SLC32A1 in clusters (bottom) and neural cell types (right). (g) Overview of the HNOCA cell type composition for the first two levels of the cell annotation (left - level-1, middle - level-2), and the refined regional annotation assisted by mapping of non-telencephalic NPC and neurons to the primary reference (right). (h) neural cell type compositions of different data sets (rows). Darkness of the heatmap shows the proportions of different neural cell types per HNOCA data set. Sidebars on the left show organoid protocol types of different data sets. Sidebars on the bottom show neural cell types. Bars on the right show total neuron numbers across data sets. (i) Distribution of transcriptomic similarity differences of NPCs and neurons in HNOCA with the primary neuronal populations in the first trimester (represented by Braun et al.27) and the second trimester (represented by Bhaduri et al.29). Cells are firstly grouped by regional identities, followed by organoid ages (in months). Colors of boxes indicate organoid ages. (j) Heatmap shows the enrichment of adult regional identities (columns) for HNOCA NPCs and neurons with different estimated regional identities (rows).
We also sought to compare organoid cells to stages of human brain development beyond the first trimester. Focusing on dorsal telencephalon, we compared the transcriptomic profile of HNOCA NPCs and neurons with cells in a primary atlas of human cortex development spanning the first trimester to adolescence30. We observed a transition from cell states observed in the first trimester to more mature states observed in the second-trimester cortex (Fig. 2c), and did not detect substantial matching to later stages. We extended the comparison to other brain regions using two primary atlases27,29 representing the first and second trimester, respectively. We confirmed increased similarity to second-trimester cell states in older organoids for other brain regions (Extended Data Fig. 3).
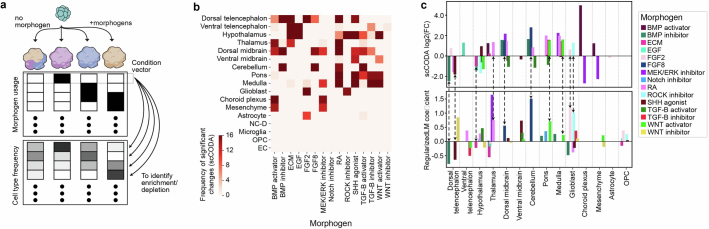
We evaluated the capacity of each neural organoid protocol to generate neural cells of different brain regions (Fig. 2d, Extended Data Figs. 3 and 4 and Supplementary Table 2). Datasets of unguided neural organoids contain cells across all brain regions with proportions varying across datasets, indicating the capacity of unguided protocols to generate many brain regions but with high variability. By contrast, datasets derived from guided organoid protocols are strongly enriched for cells of the targeted brain region, but often show an increased proportion of cells of the brain regions neighbouring the targeted regions. For example, several datasets derived from midbrain organoid protocols also show high proportions of hindbrain neurons, indicating an imprecision of morphogen guidance.
Extended Data Fig. 4. Relationship between morphogen usage and cell type as well as regional composition.
(a) Schematic of estimating cell type enrichment with different morphogen usages. (b) This heat map indicates in how many of the 17 iterations scCODA was executed (using each of the 17 regional cell identity as a reference once) the respective morphogen was found to lead to compositional changes with respect to the reference regional cell identity. A morphogen effect was called significant in this consensus approach if it had a significant effect on cell type composition with respect to more than half of the reference cell types. (c) Effect of different morphogens on regional organoid composition in HNOCA. Positive values correspond to a higher abundance of cells from the indicated regional cell identity in cases where the respective morphogen was used in the differentiation protocol. Top: log2-fold-effect sizes of morphogens per regional cell identity as computed by the scCODA model. Bottom: L1-regularized linear model coefficients. The dashed arrows show consistent enrichment/depletion identified by the two methods.
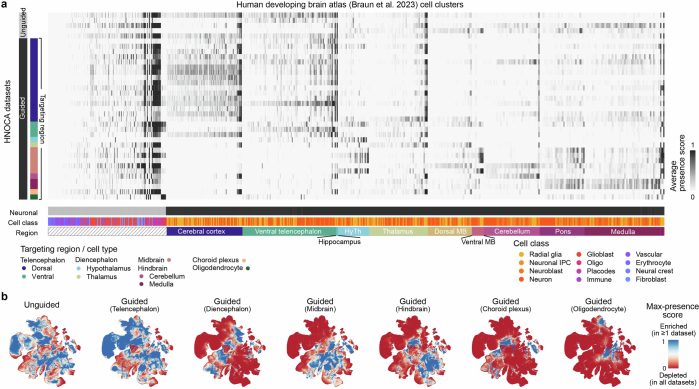
To comprehensively evaluate how well organoid protocols represented by the HNOCA generate primary brain cell types, we estimated presence scores for every primary cell type in each HNOCA dataset (Methods). A large presence score indicates high frequency and likelihood that cells of a similar type are observed in the HNOCA dataset. By normalizing the scores per organoid dataset (Extended Data Fig. 5 and Supplementary Table 3), we obtained a metric to describe how well each primary cell type is represented in at least one HNOCA dataset (Fig. 2d). This analysis confirmed the absence of erythrocytes, immune cells and vascular endothelial cells in the HNOCA, all of which are derived from non-neuroectodermal germ layers (Fig. 2e). As expected, telencephalic cell types are most strongly represented in HNOCA. By contrast, cell types of the thalamus, midbrain and cerebellum are least represented, including thalamic reticular nucleus GABAergic neurons, dorsal midbrain m1-derived GABAergic neurons and m1/m2-derived glutamatergic neurons, and cerebellar Purkinje cells (Fig. 2f,g). It is worth noting that, even though these cell types are less abundant in HNOCA datasets than in the primary atlas, certain organoid protocols can generate some of these under-represented cell types (for example, Purkinje cells in posterior brain organoid protocols).
Extended Data Fig. 5. Presence scores per HNOCA data set.
(a) Average normalized presence scores of different HNOCA data sets (rows) in different cell clusters in the primary reference of the human developing brain atlas27 (columns). Sidebars on the left show organoid differentiation protocol types of HNOCA data sets. Sidebars underneath show cell class and the commonest region information of the cell clusters in the primary reference (HyTh - hypothalamus, MB - midbrain). (b) UMAP of the primary reference, colored by the max presence scores across different HNOCA data subsets, split by organoid protocol types. A high max presence score suggests enrichment of the corresponding primary cell state in at least one HNOCA data set among the data sets based on the specific type of organoid protocols, with a low score meaning under-representation of the cell state in all data sets in the subset.
Transcriptomic fidelity organoid cell types
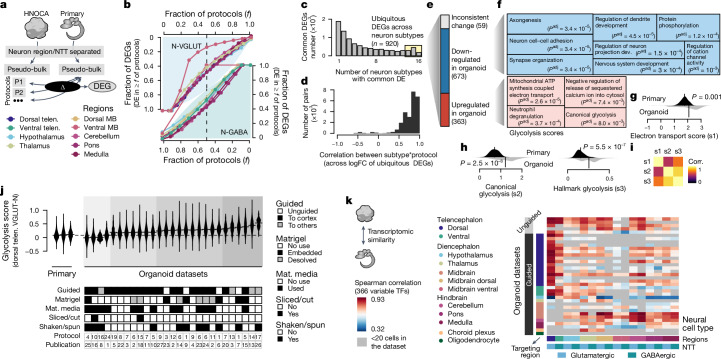
We next aimed to understand the transcriptomic similarities and differences between organoids generated by distinct differentiation protocols as well as between organoids and primary brain tissue. We identified differentially expressed genes (DEGs), comparing neural cell types in the HNOCA with their primary counterparts27 (Fig. 3a and Supplementary Table 4). We found that for most neural cell types, more than one-third (mean 34.4%, standard deviation 12.1%) of DEGs were shared across at least half of the protocols (protocol-common DEGs), suggesting that many transcriptomic differences between organoid and primary cells were independent of organoid protocol (Fig. 3b). We verified our results using an extra primary human cortex scRNA-seq dataset28 (Extended Data Fig. 6 and Supplementary Table 5). We next assessed differential transcriptomic programmes that were shared across regional neural cell types, and identified a total of 920 ubiquitous, protocol-common DEGs (uDEGs) that were differentially expressed in at least 14 out of the 16 neural cell types (Fig. 3c). These uDEGs showed consistent fold changes (r > 0.8) across neuron types and protocols (Fig. 3d), and represent consistent molecular differences between neurons in organoids and those in primary tissues regardless of protocol or neuronal cell type. Out of all 920 uDEGs, 363 genes were consistently upregulated and 673 genes were consistently downregulated, with only 59 genes (6%) inconsistently differentially expressed across subtypes or protocols (Fig. 3e).
Fig. 3. Transcriptomic comparison between organoid neurons and their primary counterpart reveals universal cell stress in organoids.
a, Schematic of DE analysis comparing neural cell types in different protocols in HNOCA to their primary counterparts27. b, Proportions of expressed genes in different neural cell types that show DE in certain fractions of protocols that generate the corresponding subtypes. Top left, glutamatergic neurons; bottom right, GABAergic neurons. Colour shows the brain region. c, Numbers of protocol-common DEGs (DE in at least half of protocols), grouped by the number of neural cell types in which a gene is DE. d, Distribution of expression log-fold-change (logFC) correlation of ubiquitous DEGs among different neuron subtype*protocol (that is, each of the neural cell types generated by each of the different protocols). e, Numbers of DEGs per category. f, Gene ontology enrichment analysis of downregulated (upper, blue) and upregulated (lower, red) ubiquitous DEGs. Sizes of the squares correlate with −log-transformed adjusted P values. g,h, Distribution of the mitochondrial ATP synthesis-coupled electron transport module scores (g), canonical glycolysis module scores (h, left) and the Molecular Signatures Database hallmark glycolysis module scores (h, right), in primary neural cell types (upper, dark) and organoid counterparts (lower, light). P values, significance of a two-sided Wilcoxon test. i, Heatmap shows pairwise correlation (corr.) of the three module scores. j, Hallmark glycolysis score of dorsal telencephalic excitatory neurons (dTelen VGLUT-N), split by the three primary developing human brains and 27 organoid datasets with at least 20 dTelen VGLUT-N. The lower panel shows selected features of differentiation protocols that may be relevant to cell stress. The protocol and publication indices are shown in Extended Data Fig. 1. Mat. media, maturation media. k, Spearman correlations between gene expression profiles of neural cell types in HNOCA and those in the human developing brain atlas27, across the variable transcription factors (TFs). Datasets are in the same order as in Supplementary Table 1.
Extended Data Fig. 6. Robustness of organoid-primary DEGs against primary reference, and across organoid data set.
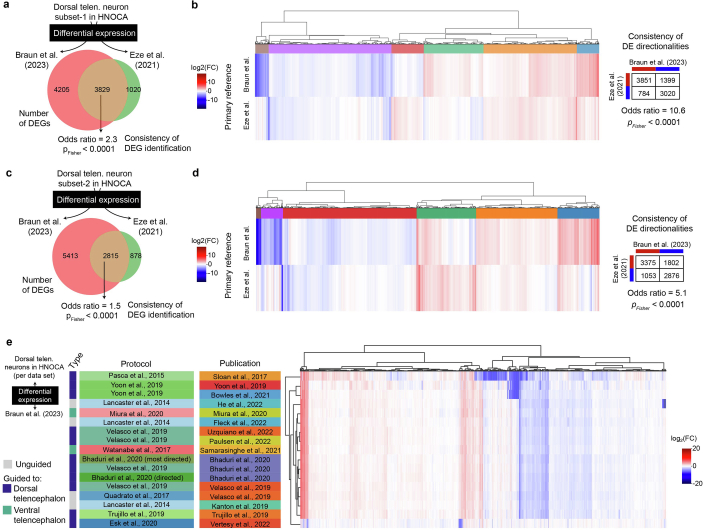
(a) Number of DEGs between organoid Dorsal Telencephalic Neurons NT-VGLUT generated using the Velasco et al.6 protocol (10×3’ v2 chemistry only) and primary fetal cortical neurons from Braun et al.27 (10×3’ v2 chemistry only) or Eze et al.28 respectively. Of the 3829 shared DEGs, 3423 genes had an aligned direction of fold-change while 406 genes had an opposite direction of fold-change. (b) Heatmap of log2-transformed fold changes (log2FC) across all 9054 DEGs between Dorsal Telencephalic Neurons NT-VGLUT from Velasco et al. and either primary fetal cortical neurons from Braun et al. (10×3’ v2 chemistry only) or Eze et al. The dendrogram shows the hierarchical clustering of DEGs based on their log2FC against the two primary data. (c) Number of DEGs between organoid Dorsal Telencephalic Neurons NT-VGLUT generated using the Lancaster et al.36 protocol (10×3’ v2 chemistry only) and primary fetal cortical neurons from Braun et al.27 (10×3’ v2 chemistry only) or Eze et al.28 respectively. Of the 2815 shared DEGs, 2375 genes had an aligned direction of fold-change while 440 genes had an opposite direction of fold-change. (d) Heatmap of log2-transformed fold changes (log2FC) across all 9106 DEGs between dorsal telencephalic neurons from Lancaster et al. and either primary fetal cortical neurons from Braun et al. (10×3’ v2 chemistry only) or Eze et al. The dendrogram shows the hierarchical clustering of DEGs based on their log2FC against the two primary data. (e) Heatmap showing the mean log-fold change per gene across organoid publications for Dorsal Telencephalic Neurons NT-VGLUT compared to the expression in the matching cell type from the Braun et al.27 primary atlas. Shown are all genes that are significantly differentially expressed compared to primary cells in the data from at least one publication.
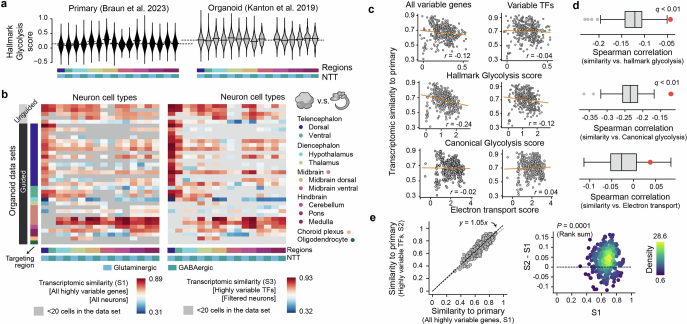
Using gene ontology enrichment analysis52,53 on the uDEGs, we found downregulated uDEGs enriched in neurodevelopmental processes including neuron cell–cell adhesion and synapse organization (Fig. 3f). Upregulated uDEGs were enriched in many metabolism-associated terms including mitochondrial ATP synthesis-coupled electron transport (electron transport in short) and canonical glycolysis (Fig. 3f). An enrichment of energy-associated pathways has previously been associated with metabolic changes caused by the limitations of current culture conditions10,24. Also, the Molecular Signatures Database gene set hallmark glycolysis54,55 has previously been used to define metabolic states in neural organoids23. Scoring mitochondrial electron transport, canonical glycolysis and hallmark glycolysis gene sets across the HNOCA and the primary reference atlas27, we found that all three terms showed significant separation of organoid and primary cells (Fig. 3g,h). Using the datasets from refs. 3 and 27 as representative examples, we identified a similar distribution of glycolysis scores across all neural cell types with an overall increased score in organoid cells (Extended Data Fig. 7). Focusing on dorsal telencephalic neurons, we compared the distribution of glycolysis scores across organoid differentiation protocols and identified several protocol features that correlated with metabolic cell stress. For instance, the usage of maturation media, slicing or cutting of organoids and, to a lesser extent, shaking or spinning of organoids led to overall lower glycolysis scores (Fig. 3h). Mean glycolysis score and transcriptomic similarity of organoid and primary reference cell types27 across differentiation protocols were negatively correlated10,24. The correlation was significantly reduced when considering only variable transcription factors, indicating that the metabolic changes in organoids have limited impact on the core molecular identity of neuronal cell types (Extended Data Fig. 7). This observation is consistent with previous studies23,24 of distinct metabolic states of cells in neural organoids relative to the primary tissue, which were shown to not affect neuron fate specification and maturation.
Extended Data Fig. 7. Transcriptomic fidelity of neurons and cell stress.
(a) Hallmark glycolysis scores of different neural cell types in primary (left, Braun et al.27) and a selected organoid data set (right, Kanton et al.3). (b) Spearman correlation between average gene expression profiles of neural cell types in HNOCA and those in the primary reference of human developing brain atlas27, across either all the variable genes (left, S1) or variable transcriptional factors (TFs) (right, S3). The average gene expression profile per neural cell type was calculated with all cells (S1) or cells with low glycolysis scores (glycolysis score <0.6, S3). (c) Correlation between different average metabolic scores (up - hallmark glycolysis score, middle - canonical glycolysis score, low - electron transport score) and transcriptomic similarities (Spearman correlation) to primary counterparts. Each dot represents one neural cell type generated by one protocol. The correlation is calculated based on either all variable genes (left, S1) or variable TFs (right, S2). (d) The correlation between hallmark and canonical glycolysis scores and transcriptomic similarities to primary is significantly weaker when only TFs are taken into consideration, while electron transport scores show no correlation with transcriptomic similarities. The boxes show the distributions of correlation when a random subset of variable genes, with the same number as the variable TFs, are used. The red dots show the correlation using variable TFs. (e) Core transcriptomic fidelity of organoid neurons (S2, shown in Fig. 3) which only considers TFs, is higher than the global transcriptomic fidelity (S1) which considers all the highly variable genes. Core transcriptomic fidelity and global transcriptomic fidelity are highly correlated (left, x-axis - S1, y-axis - S2, each dot represents one neural cell type in one HNOCA data set), while core transcriptomic fidelity is significantly higher (right, x-axis: S1, y-axis: S2-S1, dots are colored by density estimated with Gaussian kernel). P-value shows the Wilcoxon test significance.
Next, we focused on the expression of 366 variable transcription factors to calculate the correlation between corresponding neuronal cell types in the HNOCA datasets and the primary reference atlas27. We found that both guided and unguided organoid differentiation protocols generated neuronal cell types with comparable similarity to the corresponding primary reference cell types. However, we observed brain region-dependent differences in transcriptomic similarity. For example, organoid neurons from the dorsal parts of most brain regions showed higher similarity to their primary counterparts across organoid datasets than cell types derived from the ventral parts of most brain regions (Fig. 3i).
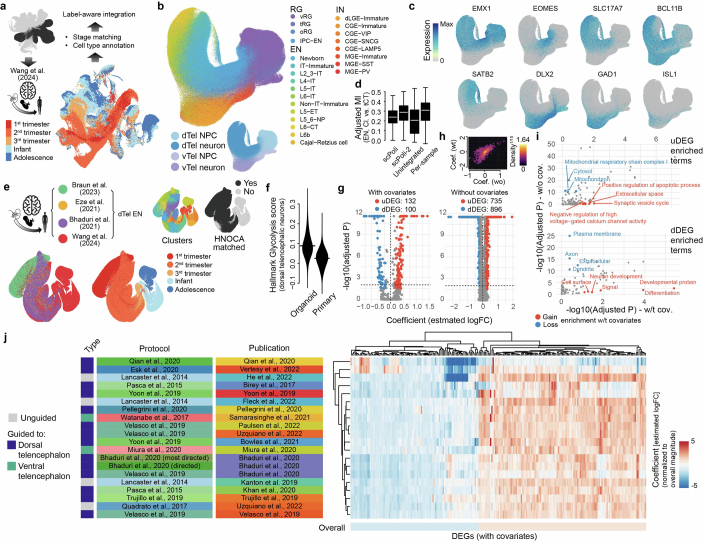
To identify molecular features other than metabolic state that decreased organoid fidelity, we incorporated dorsal telencephalic glutamatergic neurons from four different primary developing human brain atlases27–30 as an integrated primary reference, and identified neuron subtype and maturation state heterogeneity (Extended Data Fig. 8). Projection of dorsal telencephalic neurons in the HNOCA to the primary atlases revealed the corresponding heterogeneity in neural organoids. Considering metabolic state, maturation state and cell subtype as covariates during DE analysis3 significantly reduced the number of DEGs, supporting the idea that these are the major factors differentiating organoid and primary brain cells (Extended Data Fig. 8 and Supplementary Table 6). We observed enriched biological processes that included synaptic vesicle cycle and negative regulation of high voltage-gated calcium channel activity (Extended Data Fig. 8), suggesting that organoids are deficient in these processes. Of note, these differences are observed across organoid protocols, and highlight areas of consistent transcriptomic divergence between in vitro and primary counterparts.
Extended Data Fig. 8. Heterogeneity of telencephalic NPCs and neurons and its incorporation to differential expression analysis between dorsal telencephalic neurons in HNOCA and primary developing human brains.
(a) Overview of mapping the telencephalic NPCs and neurons in HNOCA to the human neocortical developmental atlas30 for cell type annotations. (b) UMAP of cells from the HNOCA telencephalic trajectories, colored by the transferred cell types from the human neocortical developmental atlas (upper) and the HNOCA annotation. (c) UMAP of HNOCA telencephalic cells colored by expression levels of selected cell type markers. (d) Distributions of adjusted mutual information across dorsal telencephalic neurons in different HNOCA samples, between the transferred cell type labels and cluster labels generated with four different representations: 1) the original scPoli (scPoli-1), 2) the re-computed telencephalon-only scPoli based on given the transferred labels; 3) unintegrated PCA of the merged data; 4) PCA and clustering sample-wise. (e) The joint atlas of human neocortical development, colored by data sets, developmental stages, clusters, and whether there is any counterpart in HNOCA dorsal telencephalic neurons. (f) Distribution of the hallmark glycolysis scores in HNOCA and the primary atlas. (g) Volcano plots show the F-test-based DE analysis results, with (left) and without (right) the glycolysis scores and matched cluster labels as covariates. The identified DEGs are colored by red (increased expression in HNOCA) or blue (decreased expression in HNOCA). (i) Changes of functional term enrichment by DAVID for DEGs based on the analysis with or without covariates. The top panel shows enrichments for the up-regulated DEGs (uDEG) in organoids, and the lower panel shows enrichments for the down-regulated DEGs (dDEG). Each dot indicates one functional term with raw P-value < 0.05 for both DEG sets. Red dots indicate functional terms gaining enrichment with DEGs with covariates (with-covariate adjusted Pwt < 0.1, and without-covariate adjusted Pwo>Pwt). Blue dots indicate functional terms losing enrichment with DEGs without covariates (Pwt>0.1 and Pwo < 1 × 10−10). (j) Heatmap shows normalized coefficient (estimated logFC normalized by the overall logFC magnitude) of each DEG per data set. Dendrograms show hierarchical clustering of DEGs and data sets. Rows represent data sets. Side bars on the left are colored based on the types of protocols, individual protocols, and publications corresponding to the data sets. Columns represent DEGs.
HNOCA facilitates organoid protocol evaluation
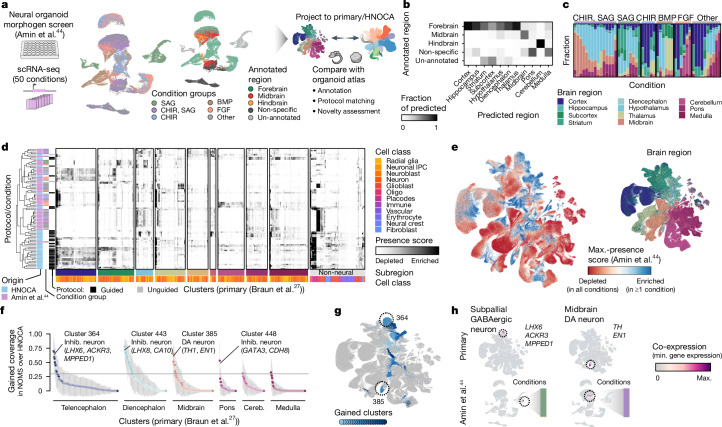
The HNOCA, as well as the analytical pipeline we established, provides a framework to query new neural organoid scRNA-seq datasets not included in the HNOCA. To showcase this application, we retrieved scRNA-seq data from a recently published multiplexed neural organoid morphogen screen44 and projected them to the HNOCA and primary reference27 latent spaces (Fig. 4a, Extended Data Fig. 9 and Supplementary Table 7). We transferred regional labels and found high consistency with the provided regional annotation, but with higher resolution within each of the broad brain sections of forebrain, midbrain and hindbrain (Fig. 4b). Our transferred annotation therefore allowed a more comprehensive assessment of the effects of different morphogen conditions on generating neurons of different brain regions (Fig. 4c). We further calculated presence scores for reference cells in each screen condition and compared the data of the different screen conditions with the 36 HNOCA datasets. Using hierarchical clustering on average presence scores revealed distinct presence score profiles for many screen conditions (Fig. 4d), suggesting regional cell type composition distinct from the HNOCA datasets. Next, we summarized the max presence scores for the whole morphogen screen data (Fig. 4e), and compared them to those for the HNOCA data to identify primary reference cell types with increased presence in the screen (Fig. 4f). This analysis highlighted several reference cell clusters with significant abundance increase under certain screen conditions (Fig. 4g) such as LHX6/ACKR3/MPPED1 triple-positive GABAergic neurons in the ventral telencephalon and dopaminergic neurons in ventral midbrain. In summary, the projection of the morphogen screen query data to HNOCA and primary reference allowed a refined annotation of the morphogen screen data, as well as a comprehensive and quantitative evaluation of the value of new differentiation protocols to generate neuronal cell types previously under-represented or lacking in neural organoids.
Fig. 4. Projection of neural organoid morphogen screen scRNA-seq data to HNOCA and human developing brain atlas allows cell type annotation and organoid protocol evaluation.
a, Schematic of projecting neural organoid morphogen screen44 scRNA-seq data to the HNOCA, and a human developing brain reference atlas27. UMAPs show screen condition groups (left, using morphogens SAG (sonic hedgehog signaling agonist), CHIR, BMP and FGF) and regional annotation of screen data (right). b, Comparison of regional annotation of screen data (rows) and scArches-transferred regional labels from the primary reference. c, Proportions of cells assigned to different regions on the basis of reference projection. Every stacked bar represents one screen condition. d, Clustering of HNOCA datasets with conditions in the screen data on the basis of average presence scores of clusters in the primary reference. The heatmap shows average presence scores per cluster in the primary reference (columns). e, UMAP of primary reference coloured by the dissected regions (right) and the maximum presence scores across the screen conditions (left). f, Gain of cell cluster coverage of screen conditions relative to HNOCA datasets, with negative values trimmed to zero. The grey horizontal line shows the threshold (0.3) to define gained clusters in screen data. g, UMAP of the primary reference, with gained clusters highlighted in shades of blue. Dashed circles highlight two clusters with highest gain of coverage in telencephalon and midbrain, respectively. h, Coexpression scores of cluster marker genes of the two clusters highlighted in g, in the primary reference (upper) and screen dataset (lower). DA, dopaminergic.
Extended Data Fig. 9. Reference mapping of the neural organoid morphogen screen data to HNOCA and the human developing brain atlas.
(a) UMAP embedding of the human developing brain atlas and neural organoid morphogen screen44 data sets based on the joint scANVI latent space colored by brain region (left) and data set (right). (b) UMAP embedding of HNOCA and the screen data sets based on the joint scPoli latent space colored by annotated cell type (left) and data set (right). (c) scPoli UMAP embedding of the HNOCA colored by cell type (left) and max presence score across all data sets (right). (d) Heatmap showing min-max scaled average presence scores of each condition in the screen data set in HNOCA data sets. (e) Heatmap showing min-max scaled average presence scores of each condition in the screen data set in each leiden cluster in HNOCA, ordered by annotated cell type. (f) UMAP embeddings of HNOCA (left) and the human developing brain atlas (right) colored by presence scores for each condition group in the screen data set. (g) UMAP embeddings of the human developing brain atlas (upper) and screen data set (lower) colored by coexpression scores of clusters with gained coverage in the screen data set.
HNOCA facilitates disease model interpretation
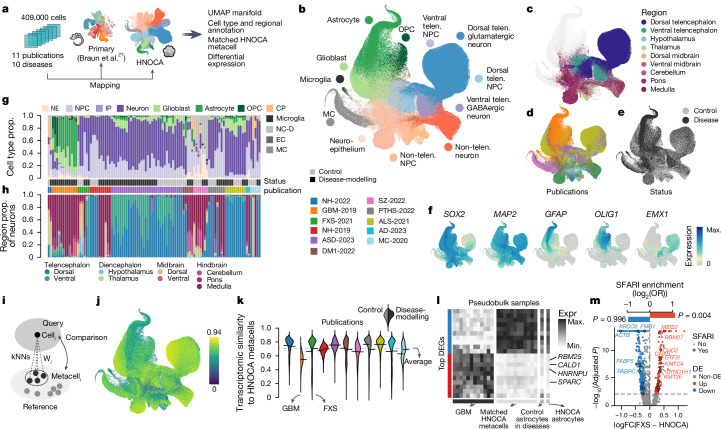
We next tested whether the integrated HNOCA can serve as a control cohort for assessing organoid models of neural disease. We collected 11 scRNA-seq datasets from 10 neural organoid disease models and their respective controls (microcephaly56, amyotrophic lateral sclerosis43, Alzheimer’s disease57, autism42, fragile-X syndrome (FXS)58, schizophrenia59, neuronal heterotopia60,61, Pitt–Hopkins syndrome62, myotonic dystrophy63 and glioblastoma64) (Fig. 5a, Extended Data Fig. 10 and Supplementary Table 8). We projected the data to the HNOCA and the primary reference atlas to transfer annotations (Fig. 5b–f). We found differences in cell type and brain regional composition between disease model organoids and their respective, study-specific control organoids for most studies (Fig. 5g,h). These differences might represent disease phenotypes, but could also be the consequence of cell line variability. It is therefore important to properly annotate the cell type and regional composition of disease and control organoids to identify disease phenotypes, particularly when analysing disease-associated transcriptomic alterations in a given cell type.
Fig. 5. The HNOCA as a control cohort to facilitate cell type annotation and transcriptomic comparison for neural organoid disease-modelling data.
a, Overview of disease-modelling neural organoid atlas construction, and projection to primary atlas27 and HNOCA for downstream analysis. b–f, UMAP of integrated disease-modelling neural organoid atlas coloured by predicted cell type annotation (b), predicted regional identities of NPCs, intermediate progenitor cells and neurons (c), publications (d), disease status (e) and marker gene expression (f). g,h, Proportions (prop.) of cells assigned to different cell classes (g) and regions (h). Every stacked bar represents one biological sample. Side bars show disease status and publication. i, Schematic of reconstructing matched HNOCA metacell for each cell in the disease-modelling neural organoid atlas. j, UMAP of disease-modelling neural organoid atlas, coloured by transcriptomic similarity with the matched HNOCA metacells. k, Violin plot indicates distribution of estimated transcriptomic similarities, split by publication. Left, distribution in control cells and right, distribution in disease cells. l, Heatmap showing expression of top DEGs between the AQP4+ population in the GBM-2019 dataset and their matched HNOCA metacells. Rows show DEGs with the ten strongest decreased and increased expressions. Columns show average expression in the AQP4+ population of disease-modelling samples (first panel), the matched HNOCA metacells per sample (second panel), all predicted control astrocytes and all astrocytes in HNOCA. m, Volcano plot shows DE analysis between dorsal telencephalic cells in the FXS-2021 dataset and their matched HNOCA metacells. DEGs coloured in red (increased in FXS) and blue (decreased in FXS). Encircled dots show DEGs annotated in SFARI database. Top bars show the log-transformed odds ratio of SFARI gene enrichment in the increased (red) and decreased (blue) DEGs. GBM, glioblastoma.
Extended Data Fig. 10. Disease-modeling neural organoid scRNA-seq atlas and data projection based extension of HNOCA.
(a-c) UMAP of the unintegrated disease-modeling neural organoid atlas, colored by (a) publications, (b) disease status, (c) transferred level-2 annotation from HNOCA, and (d) transferred regional identities from HNOCA. (e) Dot plot shows expression of selected cell type markers in cells with different transferred cell class labels (level-1) from HNOCA. (f) Dot plot shows expression of selected regional markers in the predicted NPCs and neurons in the disease-modeling atlas with different transferred regional identities from HNOCA. In both (e) and (f), sizes of dots represent percentages of cells expressing the gene, and colors of dots represent the average expression levels. (g-j) UMAP of the glioblastoma GBM-2019 data set, colored by (g) samples, (h) predicted cell class labels (level-1) from the HNOCA projection, (i) expression of astrocyte markers GFAP and AQP4, and (j) the AQP4+ population selected for DE analysis with HNOCA. (k-n) UMAP of the fragile X syndrome FXS-2021 data set, colored by (k) samples, (l) predicted cell type annotation (level-2) from the HNOCA projection, (m) expression of dorsal telencephalic cell markers FOXG1, EMX1 and NEUROD6, (n) the dorsal telencephalic NPC and neuron subset for DE analysis with HNOCA. (o) PCA of the scPoli sample embeddings of samples in HNOCA and five additional data sets projected to HNOCA.
We developed a wkNN-based strategy to generate matched HNOCA metacells for every cell in each disease model organoid scRNA-seq dataset (Fig. 5i), and quantified their transcriptomic similarity (Fig. 5j). The dataset of glioblastoma organoids64 showed substantially lower similarity to their primary counterpart than the other disease models (Fig. 5k). To assess these transcriptomic differences, we performed DE analysis between glioblastoma and matched control metacells. Focusing on the AQP4+ population (Extended Data Fig. 10), we identified 1,951 DEGs in glioblastoma cells compared to matched HNOCA metacells (Supplementary Table 9) and found increased expression of genes such as RBM25 (ref. 65) CALD1 (ref. 66), HNRNPU67 and SPARC68 (Fig. 5l), all of which have been reported to be relevant to glioblastoma.
Next, we focused on the organoid model of FXS58, in which NPCs and neurons in the control organoids were of non-telencephalic identities whereas the disease model organoids mainly contained telencephalic cells (Fig. 5h and Extended Data Fig. 10). The integrated HNOCA provides the opportunity to perform DE analysis for FXS neocortical neurons with matched HNOCA metacells, which identified 444 DEGs. DEGs higher expressed in FXS cells (122 genes) were enriched for autism-associated genes annotated in the Simons Foundation Autism Research Initiative (SFARI) database. One such gene, CHD2, was reported in the original publication58 as a key regulator of FXS with increased protein level, but its expression change on messenger RNA (mRNA) level change could not be detected in a bulk RNA-seq experiment. We also detected decreased expression of FMR1, whose loss-of-function mutation causes FXS69.
Extending the HNOCA through data projection
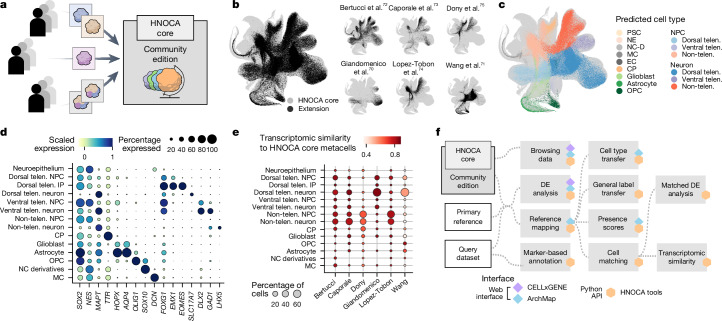
New scRNA-seq datasets of human neural organoids continue to be generated, and it will be important to continuously extend and update the HNOCA with this extra data. We therefore established a computational toolkit to project new scRNA-seq data to the HNOCA (Fig. 6a). We demonstrate the use of the toolkit by incorporating scRNA-seq data from six more studies70–75 into the HNOCA (HNOCA-extended; Fig. 6b and Supplementary Table 10), using query-to-reference mapping. We harmonized cell type annotations using wkNN-based label transfer, and placed the cells in the context of the existing organoid single-cell transcriptomic landscape as represented by the HNOCA (Fig. 6c–e). Mapping further datasets to the HNOCA using our approach enhances the atlas by increasing its coverage over existing neural organoid protocols and neural cell types generated in organoids.
Fig. 6. Extending the HNOCA by means of projection of extra datasets.
a, Schematic of projecting further scRNA-seq data by the community to extend the HNOCA. b, UMAP shows the dataset composition of the current extended HNOCA. c, UMAP shows the projected cell type annotation of cells in the five extended datasets. NE, neuroepithelium; NC-D, neural crest derivatives; MC, mesenchymal cell; EC, endothelial cell. d, Dot plot shows the expression of selected cell type and regional markers across projected cell types in the extended HNOCA datasets. e, Dot plot shows cell type composition and average similarity to the matched HNOCA metacells of the extended datasets. f, Schematic shows the analytical pipelines and varied interfaces to facilitate analysing scRNA-seq data of neural organoids for the community.
To enable researchers to use the HNOCA in their own analysis, we provide various options for exploration and interaction with the atlas (Fig. 6f). The HNOCA can be browsed through an online portal76, enabling visualization of gene expression and discovery of marker genes. We also provide the HNOCA through an online interface (http://www.archmap.bio/) for the interactive mapping of new datasets, enabling label transfer, presence score computation and metabolic scoring of cell states. Finally, we have developed HNOCA-tools, a Python package implementing all central analysis approaches presented in this paper, such as annotation, reference mapping, label transfer and DE testing methods.
Discussion
In this study, we built a large-scale integrated cell atlas of human neural organoids, the HNOCA, by integrating 1.8 million cells spanning 36 scRNA-seq datasets generated by 15 different laboratories worldwide using 26 different differentiation protocols as well as diverse scRNA-seq technologies. The resulting atlas revealed the high complexity of neuronal, glial and non-neural cell types that can develop in neural organoids grown under existing protocol conditions. Mapping the HNOCA data to various human developing brain cell reference atlases27–30 allowed comprehensive evaluation of neural organoid protocols to generate cell types of different brain regions. We found that organoids in the first 3 months of culture best match to first-trimester primary data, whereas organoids around 3 months of culture and older best match second-trimester primary cell states. We did not observe significant neuronal maturation and diversification signatures matching older developmental stages, suggesting a limitation of neuronal maturation in current neural organoid protocols.
We performed DE analysis between organoid neuron types and their primary counterparts to evaluate transcriptomic fidelity, and identified metabolic changes related to the glycolysis pathway as a main factor that distinguishes organoid and primary cell states, consistent with previous reports. Despite the negative effects of metabolic stress on overall transcriptomic fidelity, the molecular identity of regional cell types is maintained as evidenced by transcription factor coexpression patterns that are highly consistent with primary counterparts.
We showcased the mapping of query data, a recently published single-cell transcriptomic neural organoid morphogen screen, to the HNOCA and the primary reference, which enabled a refined cell type annotation, as well as a compositional comparison with existing neural organoid datasets. Our powerful framework will facilitate quantitative and comparative analysis of scRNA-seq data of human neural organoids, and for the benchmarking of new neural organoid protocols.
Consistent with earlier reports3,77, we find that unguided protocols generate neural cells with high brain regional variability, which is useful when studying broader fate determination during neurodevelopment. Guided protocols resulted in a strong enrichment of the targeted brain regions. We also note that some guided protocols, particularly those targeting midbrain, show relatively low specificity and generate neural cells from the nearby brain regions. This issue may be due to a differential response of neural stem cells in the organoid to the same morphogen cue, or to the lack of a full understanding of the timing, concentration and combinations of morphogens required to precisely define cells of the deeper regions in the central nervous system.
The integrated HNOCA is also an excellent resource for analysis of disease-modelling neural organoid data. It facilitates cell type annotation and provides a large control cohort of single-cell transcriptomes for comparison. For example, we observed discrepancy of cell type and regional composition between control and disease model samples in many studies. At the same time, the HNOCA provides the opportunity to identify disease-specific molecular features against a multi-line multi-protocol large-scale control cohort.
We demonstrate how the HNOCA can be extended and updated by projecting extra single-cell transcriptomic data of neural organoids to the atlas. Further, we have developed a computational toolkit, HNOCA-tools, which will enable other researchers to recapitulate the analytic framework applied in our study. Together, we imagine that the HNOCA will be kept up to date and continue to reflect the landscape of human neural cell states generated in organoids in vitro, serving as a living resource for the neural organoid community that enables the assessment of organoid fidelity, the characterization of perturbed and diseased states and the development of new protocols.
Methods
Metadata curation and harmonization of human neural organoid scRNA-seq datasets
We included 33 human neural organoid data from a total of 25 publications1–24,26 plus three unpublished datasets in our atlas (Supplementary Table 1). We curated all neural organoid datasets used in this study through the sfaira78 framework (GitHub dev branch, 18 April 2023). For this, we obtained scRNA-seq count matrices and associated metadata from the location provided in the data availability section for every included publication or directly from the authors in case of unpublished data. We harmonized metadata according to the sfaira standards (https://sfaira.readthedocs.io/en/latest/adding_datasets.html) and manually curated an extra metadata column organoid_age_days, which described the number of days the organoid had been in culture before collection.
We next removed any non-applicable subsets of the published datasets: diseased samples or samples expressing disease-associated mutations (refs. 14–16,18,19,26), fused organoids (ref. 1), primary fetal data (refs. 10,23), hormone-treated samples (ref. 22), data collected before neural induction (refs. 3,20) and share-seq data (ref. 23). We harmonized all remaining datasets to a common feature space using any genes of the biotype ‘protein_coding’ or ‘lncRNA’ from ensembl79 release 104 while filling any genes missing in a dataset with zero counts. On average, 50% of the full gene space (36,842 genes) was reported in each of the constituent datasets. We then concatenated all remaining datasets to create a single AnnData80 object.
Preprocessing of the HNOCA scRNA-seq data
All processing and analyses were carried out using scanpy81 (v.1.9.3) unless indicated otherwise. For quality control and filtering of HNOCA, we removed any cells with fewer than 200 genes expressed. We next removed outlier cells in terms of two quality control metrics: the number of expressed genes and percentage mitochondrial counts. To define outlier cells on the basis of each quality control metric, z-transformation is first applied to values across all cells. Cells with any z-transformed metric less than −1.96 or greater than 1.96 are defined as outliers. For any dataset collected using the v.3 chemistry by 10X Genomics, which contains more than 500 cells after the filtering, we fitted a Gaussian distribution to the histogram denoting the number of expressed genes per cell. If a bimodal distribution was detected, we removed any cell with fewer genes expressed than defined by the valley between the two maxima of the distribution. We then normalized the raw read counts for all Smart-seq2 data by dividing it by the maximum gene length for each gene obtained from BioMart. We next multiplied these normalized read counts by the median gene length across all genes in the datasets and treated those length-normalized counts equivalently to raw counts from the datasets obtained with the help of unique molecular identifiers in our downstream analyses.
As a next step we generated a log-normalized expression matrix by first dividing the counts for each cell by the total counts in that cell and multiplying by a factor of 1,000,000 before taking the natural logarithm of each count + 1. We computed 3,000 highly variable features in a batch-aware manner using the scanpy highly_variable_genes function (flavor = ‘seurat_v3’, batch_key = ‘bio_sample’). Here, bio_sample represents biological samples as provided in the original metadata of the datasets. On average, 72% of the 3,000 highly variable genes were reported in each of the constituent HNOCA datasets. We used these 3,000 features to compute a 50-dimensional representation of the data using principal component analysis (PCA), which in turn we used to compute a k-nearest-neighbour (kNN) graph (n_neighbors = 30, metric = ‘cosine’). Using the neighbour graph we computed a two-dimensional representation of the data using UMAP82 and a coarse (resolution 1) and fine (resolution 80) clustering of the unintegrated data using Leiden83 clustering.
Hierarchical auto-annotation with snapseed
Snapseed is a scalable auto-annotation strategy, which annotates cells on the basis of a provided hierarchy of cell types and the corresponding cell type markers. It is based on enrichment of marker gene expression in cell clusters (high-resolution clustering is preferred), and data integration is not necessarily required.
In this study, we used snapseed to obtain initial annotations for label-aware integration. First, we constructed a hierarchy of cell types including progenitor, neuron and non-neural types, each defined by a set of marker genes (Supplementary Data 1). Next, we represented the data by the RSS3 to average expression profiles of cell clusters in the recently published human developing brain cell atlas27. We then constructed a kNN graph (k = 30) in the RSS space and clustered the dataset using the Leiden algorithm83 (resolution 80). For both steps, we used the graphical processing unit (GPU)-accelerated RAPIDS implementation that is provided through scanpy81,84.
For all cell type marker genes on a given level in the hierarchy, we computed the area under the receiver operating characteristic curve (AUROC) as well as the detection rate across clusters. For each cell type, a score was computed by multiplying the maximum AUROC with the maximum detection rate among its marker genes. Each cluster was then assigned to the cell type with the highest score. This procedure was performed recursively for all levels of the hierarchy. The same procedure was carried out using the fine (resolution 80) clustering of the unintegrated data to obtain cell type labels for the unintegrated dataset that were used downstream as a ground-truth input for benchmarking integration methods.
This auto-annotation strategy was implemented in the snapseed Python package and is available on GitHub (https://github.com/devsystemslab/snapseed). Snapseed is a light-weight package to enable scalable marker-based annotation for atlas-level datasets in which manual annotation is not readily feasible. The package implements three main functions: annotate() for non-hierarchical annotation of a list of cell types with defined marker genes, annotate_hierarchy() for annotating more complex, manually defined cell type hierarchies and find_markers() for fast discovery of cluster-specific features. All functions are based on a GPU-accelerated implementation of AUROC scores using JAX (https://github.com/google/jax).
Label-aware data integration with scPoli
We performed integration of the organoid datasets for HNOCA using the scPoli45 model from the scArches51 package. We defined the batch covariate for integration as a concatenation of the dataset identifier (annotation column ‘id’), the annotation of biological replicates (annotation column ‘bio_sample’) as well as technical replicates (annotation column ‘tech_sample’). This resulted in 396 individual batches. The batch covariate is represented in the model as a learned vector of size five. We used the top three levels of the RSS-based snapseed cell type annotation as the cell type label input for the scPoli prototype loss. We chose the hidden layer size of the one-layer scPoli encoder and decoder as 1,024, and the latent embedding dimension as ten. We used a value of 100 for the ‘alpha_epoch_anneal’ parameter. We did not use the unlabelled prototype pretraining. We trained the model for a total of seven epochs, five of which were pretraining epochs.
Benchmark of data integration methods
To quantitatively compare the organoid atlas integration results from several tools, we used the GPU-accelerated scib-metrics46,85 Python package (v.0.3.3) and used the embedding with the highest overall performance for all downstream analyses. We compared the data integration performance across the following latent representations of the data: unintegrated PCA, RSS3 integration, scVI49 (default parameters except for using two layers, latent space of size 30 and negative binomial likelihood) integration, scANVI50 (default parameters) integrations using snapseed level 1, 2 or 3 annotation as cell type label input, scPoli45 (parameters shown above) integrations using either snapseed level 1, 2 or 3 annotation or all three annotation levels at once as the cell type label input, scPoli45 integrations of metacells aggregated with the aggrecell algorithm (first used as ‘pseudocell’3) using either snapseed level 1 or 3 annotation as the cell type label input to scPoli. We used the following scores for determining integration quality (each described in ref. 46): Leiden normalized mutual information score, Leiden adjusted rand index, average silhouette width per cell type label, isolated label score (average silhouette width-scored) and cell type local inverse Simpson’s index to quantify conservation of biological variability. To quantify batch-effect removal, we used average silhouette width per batch label, integration local inverse Simpson’s index, kNN batch-effect test score and graph connectivity. Integration approaches were then ranked by an aggregate total score of individually normalized (into the range of [0,1]) metrics. Before we carried out the benchmarking, we iteratively removed any cells from the dataset that had an identical latent representation to another cell in the dataset until no latent representation contained any more duplicate rows. This procedure removed a total of 3,293 duplicate cells (0.002% of the whole dataset) and was required for the benchmarking algorithm to complete without errors. We used the snapseed level 3 annotation computed on the unintegrated PCA embedding as ground-truth cell type labels in the integration.
Pseudotime inference
To infer a global ordering of differentiation state, we sought to infer a real-time-informed pseudotime on the basis of neural optimal transport47 in the scPoli latent space. We first grouped organoid age in days into seven bins ((0, 15], (15, 30], (30,60], (60, 90], (90, 120], (120, 150], (150, 450]). Next, we used moscot48 to solve a temporal neural problem. To score the marginal distributions on the basis of expected proliferation rates, we obtained proliferation and apoptosis scores for each cell with the method score_genes_for_marginals(). Marginal weights were then computed with
The optimal transport problem was solved using the following parameters: iterations = 25,000, compute_wasserstein_baseline = False, batch_size = 1,024, patience = 100, pretrain = True, train_size = 1. To compute displacement vectors for each cell in age bin i, we used the subproblem corresponding to the [i, i + 1] transport map, except for the last age bin, where we used the subproblem [i − 1,i]. Displacement vectors were obtained by subtracting the original cell distribution from the transported distribution. Using the velocity kernel from CellRank86 we computed a transition matrix from displacement vectors and used it as an input for computing diffusion maps87. Ranks on negative diffusion component 1 were used as a pseudotemporal ordering.
Preprocessing of the human developing brain cell atlas scRNA-seq data
The cell ranger-processed scRNA-seq data for the primary atlas27 were obtained from the link provided on its GitHub page (https://storage.googleapis.com/linnarsson-lab-human/human_dev_GRCh38-3.0.0.h5ad). For further quality control, cells with fewer than 300 detected genes were filtered out. Transcript counts were normalized by the total number of counts for that cell, multiplied by a scaling factor of 10,000 and subsequently natural-log transformed. The feature set was intersected with all genes detected in the organoid atlas and the 2,000 most highly variable genes were selected with the scanpy function highly_variable_genes using ‘Donor’ as the batch key. An extra column of ‘neuron_ntt_label’ was created to represent the automatic classified neural transmitter transporter subtype labels derived from the ‘AutoAnnotation’ column of the cell cluster metadata (https://github.com/linnarsson-lab/developing-human-brain/files/9755350/table_S2.xlsx).
Reference mapping of the organoid atlas to the primary atlas
To compare our organoid atlas with data from the primary developing human brain, we used scArches51 to project it to the above mentioned primary human brain scRNA-seq atlas27. We first pretrained a scVI model49 on the primary atlas with ‘Donor’ as the batch key. The model was constructed with following parameters: n_latent = 20, n_layers = 2, n_hidden = 256, use_layer_norm = ‘both’, use_batch_norm = ‘none’, encode_covariates = True, dropout_rate = 0.2 and trained with a batch size of 1,024 for a maximum or 500 epochs with early stopping criterion. Next, the model was fine-tuned with scANVI50 using ‘Subregion’ and ‘CellClass’ as cell type labels with a batch size of 1,024 for a maximum of 100 epochs with early stopping criterion and n_samples_per_label = 100. To project the organoids atlas to the primary atlas, we used the scArches51 implementation provided by scvi-tools88,89. The query model was fine-tuned with a batch size of 1,024 for a maximum of 100 epochs with early stopping criterion and weight_decay = 0.0.
Bipartite weighted kNN graph reconstruction
With the primary reference27 and query (HNOCA) data projected to the same latent space, an unweighted bipartite kNN graph was constructed by identifying 100 nearest neighbours of each query cell in the reference data with either PyNNDescent or RAPIDS-cuML (https://github.com/rapidsai/cuml) in Python, depending on availability of GPU acceleration. Similarly, a reference kNN graph was also built by identifying 100 nearest neighbours of each reference cell in the reference data. For each edge in the reference-query bipartite graph, the similarity between the reference neighbours of the two linked cells, defined as A and B, respectively, is represented by the Jaccard index:
The square of Jaccard index was then assigned as the weight of the edge, to get the bipartite weighted kNN graph between the reference and query datasets.
wkNN-based primary developing brain atlas label transfer to HNOCA cells
Given the wkNN estimated between primary reference27 and query (HNOCA), any categorical metadata label of reference can be transferred to query cells by means of majority voting. In brief, for each category, its support was calculated for each query cell as the sum of weights of edges that link to reference cells in this category. The category with the largest support was assigned to the query cell.
To get the final regional labels for the non-telencephalic NPCs and neurons, as well as the NTT labels for non-telencephalic neurons, constraints were added to the transfer procedure. For regional labels, only the non-telencephalic regions, namely diencephalon, hypothalamus, thalamus, midbrain, midbrain dorsal, midbrain ventral, hindbrain, cerebellum, pons and medulla, were considered valid categories to be transferred. The label-transfer procedure was only applied to the non-telencephalic NPCs and neurons in HNOCA. Before any majority voting was done, the support scores of each valid category across all non-telencephalic NPCs and neurons in HNOCA were smoothed with a random-walk-with-restart procedure (restart probability alpha, 85%). Next, a hierarchical label transfer, which takes into account the structure hierarchy, was applied. First, the considered regions were grouped into diencephalon, midbrain and hindbrain, with a support score of each structure as its score summed up with scores of its substructures. Majority voting was applied to assign each cell to one of the three structures. Next, a second majority voting was applied to only consider the substructures under the assigned structure (for example, hypothalamus and thalamus for diencephalon).
For NTT labels, we first identified valid region-NTT label pairs in the reference on the basis of the provided NTT labels in the reference neuroblast and neuron clusters and their most common regions. Here, the most common regions were re-estimated in a hierarchical manner to the finest resolution mentioned above. Next, when transferring NTT labels, for each non-telencephalic neuron with the regional label transferred, only NTT labels that were considered valid for the region were considered during majority voting.
Stage-matching analysis
To match telencephalic NPCs and neurons in HNOCA to developmental stages, we used the recently published human neocortical development atlas30 as the reference. The processed single nucleus RNA-seq data were obtained from its data portal (https://cell.ucsf.edu/snMultiome/). Given the ‘class’, ‘subclass’ and ‘type’ labels in the provided metadata as annotations, and ‘individual’ as the batch label, scPoli was applied for label-aware data integration. Next, data representing different developmental stages were split. For each stage, Louvain clustering based on the scPoli latent representation (resolution, 5) was applied. Clusters of all stages were pooled, and highly variable genes were identified on the basis of coefficient of variations as described in this page: https://pklab.med.harvard.edu/scw2014/subpop_tutorial.html. Finally, every one of HNOCA telencephalic NPCs and neurons were correlated to each cluster across the identified highly variable genes. The stage label of the best-correlated cluster was assigned to the query HNOCA cell.
To extend the analysis to other neuronal cell types, the second-trimester multiple-region human brain atlas29 was also introduced. The processed count matrices and metadata were obtained from the NeMO data portal (https://data.nemoarchive.org/biccn/grant/u01_devhu/kriegstein/transcriptome/scell/10x_v2/human/processed/counts/). Given the ‘cell_type’ label of the provided metadata as the annotation and ‘individual’ as the batch label, scPoli was run for label-aware data integration. Louvain clustering was applied to the scPoli latent representation to identify clusters (resolution, 20). Similarly, Louvain clustering with a resolution of 20 was also applied to the first-trimester multiple-region human brain atlas27 on the basis of the scANVI latent representation we generated earlier. Average expression profiles were calculated for all the clusters, and highly variable genes were identified using the same procedure as above for clusters of the two primary atlases combined. Next, every NPC and neuron in HNOCA was correlated to the average expression profiles of those clusters. The best-correlated first- and second-trimester clusters, as well as the correlations, were identified. The differences between the two correlations were used as the metrics to indicate the stage-matching preferences of NPCs and neurons in HNOCA.
Presence scores and max presence scores of cells in the primary developing brain atlas
Given a reference dataset and a query dataset, the presence score is a score assigned to each cell in the reference, which describes the frequency or likelihood of the cell type or state of that reference cell appearing in the query data. In this study, we calculated the presence scores of primary atlas cells in each HNOCA dataset to quantify how frequently we saw a cell type or state represented by each primary cell in each of the HNOCA datasets.
Specifically, for each HNOCA dataset, we first subset the wkNN graph to only HNOCA cells in that dataset. Next, the raw weighted degree was calculated for each cell in the primary atlas, as the sum of weights of the remaining edges linked to the cell. A random-walk-with-restart procedure was then applied to smooth the raw scores across the kNN graph of the primary atlas. In brief, we first represented the primary atlas kNN graph as its adjacency matrix (A), followed by row normalization to convert it into a transition probability matrix (P). With the raw scores represented as a vector s0, in each iteration t, we generated st as
This procedure was performed 100 times to get the smooth presence scores that were subsequently log transformed. Scores lower than the 5th percentile or higher than the 95th percentile were trimmed. The trimmed scores were normalized into the range of [0,1] as the final presence scores in the HNOCA dataset.
Given the final presence scores in each of the HNOCA datasets, the max presence scores in the whole HNOCA data were then easily calculated as the maximum of all the presence scores for each cell in the primary atlas. A large (close to one) max presence score indicates a high frequency of appearance for the cell type or state in at least one HNOCA dataset whereas a small (close to zero) max presence score suggests under-representation in all the HNOCA datasets.
Cell type composition comparison among morphogen usage using scCODA
To test the cell type compositional changes on admission of certain morphogens from different organoid differentiation protocols, we used the pertpy90 implementation of the scCODA algorithm91. scCODA is a Bayesian model for detecting compositional changes in scRNA-seq data. For this, we have extracted the information about the added morphogens from each differentiation protocol and grouped them into 15 broad molecule groups on the basis of their role in neural differentiation (Supplementary Table 1). These molecule groups were used as a covariate in the model. The region labels transferred from the primary atlas were used as labels in the analysis (cell_type_identifier). For cell types without regional identity, the cell type labels presented in Fig. 1c were used. Pluripotent stem cells and neuroepithelium cells were removed from the analysis because they are mainly present in the early organoid stages. We used bio_sample as the sample_identifier. We ran scCODA sequentially with default parameters, using No-U-turn sampling (run_nuts function) and selecting each cell type once as a reference. We used a majority vote-based system to find the cell types that were credibly changing in more than half of the iterations.
Cell type composition comparison among morphogen usage using regularized linear regression
To complement the composition analysis conducted with scCODA, we devised an alternative approach to test for differential composition using regularized linear regression. We fit a generalized linear model with the region composition matrix as the response Y and molecule usage as independent variables X:
The model was fit with lasso regularization (alpha = 1) using Gaussian noise and an identity link function. The regularization parameter lambda was automatically determined through cross-validation as implemented in the function cv.glmnet() from the glmnet92 R package. All non-zero coefficients β were considered as indications of enrichment and depletion.
DE analysis between HNOCA neural cell types and their primary counterparts and functional enrichment analysis
To study the transcriptomic differences between organoid and primary cells, we subset HNOCA using the final level 1 annotation to cells labelled ‘Neuron’. We furthermore subset the human developing brain atlas to cells that had been assigned a valid label in the neuron_ntt_label annotation column. We added an extra two datasets of fetal cortical cells from ref. 39 and ref. 28. For the data from ref. 39, we subset the data to cells labelled ‘fetal’ and estimated transcripts per million reads for each gene in each cell using RSEM93 given the STAR94 mapping results. We then computed a PCA, a kNN graph, UMAP and Leiden clustering (resolution 0.2) using scanpy. We then selected the cluster with the highest STMN2 and NEUROD6 expression as the cortical neuron cluster and used only those cells. For the data from ref. 28 we subset the datasets to cells annotated as ‘Neuronal’ in Supplementary Table 5 (‘Cortex annotations’) of their publication and computed a PCA, neighbourhood graph and UMAP to visualize the dataset. We found that only samples from the individuals CS14_3, CS20, CS22 and CS20 contained detectable expression of STMN2 and NEUROD6 so we subset the dataset further to only cells from those individuals.
To compute DE between HNOCA cells and their primary counterparts, we first aggregated cells of the same regional neural cell type into pseudobulk samples by summing the counts for every sample (annotation columns, ‘batch’ for HNOCA; ‘SampleID’ for the human developing brain atlas; ‘sample’ for ref. 39 and ‘individual’ for ref. 28) using the Python implementation of decoupler95 (v.1.4.0) while discarding any samples with fewer than ten cells or 1,000 total counts. We then subsetted the feature space to the intersection of features of all datasets and removed any cells with fewer than 200 genes expressed. We further removed any genes expressed in less than 1% of neurons in HNOCA and any genes located on the X and Y chromosomes. Out of the remaining 11,636 genes, on average, 99% were reported in each of the constituent HNOCA datasets. For each regional neural cell type, we removed any sample from the pseudobulk data that was associated with an organoid differentiation assay with fewer than two total samples or fewer than 100 total cells. We next used edgeR96 to iteratively compute DE genes between each organoid differentiation protocol and primary cells of the matching regional neural cell types for every regional neural cell type while correcting for organoid age in days, number of cells per pseudobulk sample, median and standard deviation of the number of detected genes per pseudobulk sample. We used the data from ref. 27 (the human developing brain atlas mentioned above), ref. 28 and ref. 39 as primary data for the DE comparison in the cell type ‘Dorsal Telencephalic Neuron NT-VGLUT’, whereas for all other cell types we used the human developing brain atlas as the fetal dataset. We used the edgeR genewise negative binomial generalized linear model with quasi-likelihood F-tests. We deemed a gene significantly DE if its false-discovery rate (Benjamini–Hochberg) corrected P value was smaller than 0.05 and it had an absolute log2-fold change above 0.5. We used the GSEApy97 Python package to carry out functional enrichment analysis in our DE results using the ‘GO_Biological_Process_2021’ gene set.
To evaluate the effect of different primary datasets on the DE results, we computed the DE between Dorsal Telencephalic Neuron NT-VGLUT from the HNOCA subset generated with the protocol from ref. 6 and the matching cell type from the Braun et al.27 primary dataset as well as the data from ref. 28. To prevent technology effects to affect this analysis, we only used cells generated with the 10X Genomics 3′ v.2 protocol in this comparison. We generate pseudobulk samples as described above and corrected organoid age in days and number of cells per pseudobulk sample in the DE comparison. We used the same edgeR-based procedure and cut-offs as described above. We used the scipy fcluster method to cluster genes on the basis of their log-fold changes in the two primary datasets. We grouped clusters to represent consistently upregulated, consistently downregulated and three different inconsistently regulated groups of genes. We computed functional enrichment of each gene group as described above.
To evaluate the effect of different organoid datasets on the protocol-based DE analysis, we computed DE between Dorsal Telencephalic Neuron NT-VGLUT of every organoid publication (further split by protocol, where more than one protocol was used in a publication) and the matching cell type in the dataset from ref. 27. We computed pseudobulk samples and carried out the DE analysis using the same procedure and cut-offs as in the protocol-based DE analysis.
Transcriptomic similarity between HNOCA neural cell types and their primary counterparts in the human developing brain atlas
To estimate the transcriptomic similarity between neurons in HNOCA and the human developing brain atlas27, we first summarized the average expression of each neural cell type in the primary reference, as well as in each dataset of HNOCA. For each HNOCA dataset, only neural cell types with at least 20 cells were considered. Highly variable genes were identified across the neural cell types in the primary reference using a Chi-squared test-based variance ratio test on the generalized linear model with Gamma distribution (identity link), given coefficient of variance of transcript counts across neural cell types as the response and the reciprocal of average transcript count across neural cell types as the independent variable. Genes with Benjamini–Hochberg adjusted P values less than 0.01 were considered as highly variable genes. Similarity between one neural cell type in the primary atlas and its counterpart in each HNOCA dataset was then calculated as the Spearman correlation coefficient across the identified highly variable genes.
To estimate the similarity of the core transcriptomic identity, which is defined by the coexpression of transcription factors, the highly variable genes were subset to only transcription factorsfor calculating Spearman correlations. The list of transcription factors was retrieved from the AnimalTFDB v.4.0 database98.
To identify metabolically stressed cells in the datasets, we used the scanpy score_genes function with default parameters to score the ‘canonical glycolysis’ gene set obtained from the enrichR GO_Biological_Process_2021 database across all neuronal cells from HNOCA and refs. 27,28,39.
To estimate the significance of the difference between the correlation of glycolysis scores and whole transcriptomic similarities, and the correlation of glycolysis scores and core transcriptomic identity similarities, we generated 100 subsets of highly variable genes, each with the same size as the highly variable transcription factor. Transcriptomic similarities were calculated on the basis of those subsets, and then correlated with the glycolysis scores.
Heterogeneity of the telencephalic trajectories
To characterize heterogeneity of telencephalic NPCs and neurons in HNOCA, we first transferred the cell type labels (as indicated as the ‘type’ label in the given metadata) from the human neocortical development atlas to the HNOCA telencephalic NPCs, intermediate progenitor cells and neurons, on the basis of transcriptomic correlation. In brief, each primary atlas cluster we obtained as mentioned above was assigned to a cell type as the most abundant cell type among cells in the cluster. The label of the best-correlated primary cluster was then transferred to every query cell. Given the transferred label, together with the level 2 cell type annotation shown in Fig. 1c, as the annotation label, scPoli was applied to the telencephalic subset of HNOCA for data integration.
To benchmark how well different integration strategies recover the neuron subcell type heterogeneity, we generated four different clustering labels: (1) Louvain clustering (resolution, 2) with the original scPoli latent representation; (2) Louvain clustering (resolution, 2) with the updated scPoli representation; (3) Louvain clustering (resolution, 2) with PCA of HNOCA telencephalic subset (based on scaled expression of 3,000 highly variable genes of the telencephalic subset with flavor = ‘seurat’) and (4) Louvain clustering (resolution, 1) for each sample separately (each with 3,000 highly variable genes identified with flavor = ‘seurat’, followed by data scale and PCA). Next, for each sample with at least 500 dorsal telencephalic neurons, the adjusted mutual information scores were calculated between each of those four clustering labels with the transferred cell type label mentioned above as the gold standard, across the dorsal telencephalic neurons as annotated as the level 2 annotation.
To create a comprehensive primary atlas of dorsal telencephalic neurons for DE analysis between neural organoids and primary tissues, we subset dorsal telencephalic neurons or neocortical neurons from four different primary atlases27–30. For ref. 28, cells in five author-defined clusters (60, 57, 79, 45, 65) with high expression of MAP2, DCX and NEUROD6 were selected. For ref. 29, cells with the following ‘clusterv2 - final’ labels were selected: ‘Neuron_28’, ‘Neuron_34’, ‘GW19_2_29NeuronNeuron’, ‘Neuron_30’, ‘Neuron_66Neuron’, ‘GW18_2_42NeuronNeuron’, ‘Neuron_33’, ‘Neuron_39Neuron’, ‘Neuron_35’, ‘Neuron_63Neuron’, ‘Neuron_9’, ‘Neuron_11’, ‘Neuron_20’, ‘Neuron_22’, ‘Neuron_5Neuron’, ‘Neuron_21’, ‘Neuron_18’, ‘Neuron_101Neuron’, ‘Neuron_17’, ‘Neuron_19’, ‘Neuron_16’, ‘Neuron_50Neuron’, ‘Neuron_12’, ‘Neuron_13’, ‘Neuron_68Neuron’, ‘Neuron_100Neuron’, ‘Neuron_25’, ‘Neuron_27’, ‘Neuron_53Neuron’, ‘Neuron_23’, ‘Neuron_26’, ‘Neuron_24’, ‘Neuron_102Neuron’, ‘Neuron_72Neuron’, ‘Neuron_15’, ‘Neuron_29’ and ‘Neuron_35Neuron’ on the basis of their high expression of NEUROD6 and FOXG1. For ref. 27, cells dissected from dorsal telencephalon that were annotated as neurons with and only with the VGLUT NTT label were selected. For ref. 30, cells annotated as excitatory neurons were selected. The curated clusters of the Wang et al. primary atlas, as described earlier, were also subset to those with excitatory neuron labels. The selected dorsal telencephalic neuron subsets of the atlases were merged into the joint neocortical neuron atlas.
Next, cells in the joint neocortical neuron atlas were correlated with the average expression profile of each excitatory neuron cluster of the Wang et al. atlas30. The cluster label of the best-correlated cluster was assigned to each cell in the joined neocortical neuron atlas, so that cell cluster labels were harmonized for all cells in the atlas. Label-aware data integration was then performed using scPoli45. On the basis of the scPoli latent representation, Louvain clustering was performed on the joint neocortical neuron atlas (resolution, 1). This cluster label was transferred to the dorsal telencephalic neurons in HNOCA with max-correlation manner across highly variable genes defined on average transcriptomic profiles of clusters in the joint neocortical neuron atlas.
Reference mapping of the neural organoid morphogen screen scRNA-seq data to the human developing brain atlas and HNOCA
We used scArches to map scRNA-seq data from the neural organoid morphogen screen to both the scANVI model of the human developing brain atlas27 and the scPoli model of the HNOCA. In both cases, the ‘dataset’ field of the screen data was used as the batch covariate, which indicates belonging to one of the three categories: ‘organoid screen’, ‘secondary organoid screen’ or ‘fetal striatum 21pcw’. For mapping to the primary reference, we used the scvi-tools implementation of scArches without the use of cell type annotations and trained the model for 500 epochs with weight_decay of 0 and otherwise default parameters. For mapping to HNOCA we used scArches through scPoli and trained the model for 500 epochs without unlabelled prototype training.
Retrieval and harmonization of disease-modelling human neural organoid scRNA-seq datasets
We included 11 scRNA-seq datasets of neural organoids, which were designed to model 10 different neural diseases including microcephaly56, amyotrophic lateral sclerosis43, Alzheimer’s disease57, autism42, FXS58, schizophrenia59, neuronal heterotopia60,61, Pitt–Hopkins syndrome62, myotonic dystrophy63 and glioblastoma64. Count matrices and metadata were directly downloaded for the ten datasets with processed data provided in the Gene Expression Omnibus or ArrayExpress. For the dataset with only FASTQ files available56, we downloaded the FASTQ files and used Cell Ranger (v.4.0) to map reads to the human reference genome and transcriptome retrieved from Cell Ranger website (GRCh38 v.3.0.0) for gene expression quantification. All datasets were concatenated together with anndata in Python (join = ‘inner’). For each dataset, samples were grouped into either ‘disease’ or ‘control’ as their disease status, with ‘disease’ representing data from patient cell lines, mutant cell lines with disease-related alleles, cells carrying targeting guide RNAs (gRNAs) in CRISPR-based screen and tumour-derived organoids. and ‘control’ representing data from healthy cell lines, mutation-corrected cell lines and cells carrying only non-targeting gRNAs in a CRISPR-based screen.
Projection and label transfer-based annotation of the disease-modelling dataset
To compare the disease-modelling atlas with the integrated HNOCA, we used scArches51 to project it to the HNOCA as well as the first-trimester primary human brain scRNA-seq atlas27. For projecting to the primary atlas, the same implementation as mentioned above to map HNOCA to the atlas was used. For projecting to HNOCA, the query model was based on the scPoli model pretrained with the HNOCA data, and fineturned with a batch size of 16,384 for a maximum of 30 epochs with 20 pretraining epochs. A nearest neighbour graph was created for the disease-modelling atlas on the basis of the projected latent representation to HNOCA with scanpy (default parameters), with which a UMAP embedding was created with scanpy (default parameters).
Next, for both HNOCA and the disease-modelling atlas, cells were represented by the concatenated representation of HNOCA-scPoli and primary-scANVI models. A bipartite wkNN graph was then reconstructed as mentioned above, by identifying 50 nearest neighbours in HNOCA for each disease-modelling atlas cell. On the basis of the bipartite wkNN, the majority voting-based label transfer was applied to transfer the four levels of hierarchical cell type annotation and regional identity to the disease-modelling atlas.
Reconstruction of matched HNOCA metacells
For each cell in the disease-modelling atlas, a matched HNOCA metacell was reconstructed on the basis of the above mentioned bipartite wkNN. In brief, for a query cell i and a gene j measured in HNOCA, its matched metacell expression of j, denoted as , is calculated as:
Here, Ni represents all HNOCA nearest neighbours of the query cell ci, wik represents the edge weight between query cell i and reference cell k, and ekj represents expression level of gene j in reference cell k.
Given the matched HNOCA metacell transcriptomic profile, the similarity between a query cell and its matched cell state in HNOCA is then calculated as the Spearman correlation between the query cell transcriptomic profile and its matched HNOCA metacell transcriptomic profile.
Re-analysis of GBM-2019 and FXS-2021 datasets
To analyse the glioblastoma organoid dataset (GBM-2019), cells from the publication were subset from the integrated disease-modelling atlas. Using scanpy, highly variable genes were identified with default parameters. The log-normalized expression values of the highly variable genes were then scaled across cells, the truncated PCA was performed with the top 20 principal components used for the following analysis. Next, harmonypy, the Python implementation of harmony99, was applied to integrate cells from different samples. On the basis of the harmony-integrated embeddings, the neighbour graph was reconstructed. UMAP embeddings and Louvain clusters (resolution, 0.5) were created on the basis of the nearest neighbour graph. Among the 12 identified clusters, cluster-7 and cluster-0, the two clusters with the highest AQP4 expression, were selected for the following DE analysis.
To analyse the FXS dataset (FXS-2021), cells from the publication were subset from the integrated disease-modelling atlas. The same procedure of highly variable gene identification, data scaling and PCA as the GBM-2019 dataset was applied. Next, the nearest neighbour graph was created directly on the basis of the top 20 principal components. UMAP embeddings and Louvain clusters (resolution, 1) were then created on the basis of the reconstructed nearest neighbour graph. Among the 30 clusters, cluster-17 and cluster-23, which express EMX1 and FOXG1 and were largely predicted to be dorsal telencephalic NPCs and neurons according to the transferred labels from HNOCA, were selected for the following DE analysis.
F-test-based DE analysis for paired transcriptome
To compare expression levels of two groups of paired cells, the expression difference per gene of each cell pair is first calculated on the basis of the log-normalized expression values. Next, for each gene to test for DE, its variance over the calculated expression difference per cell pair (σ2) is compared with the sum of squared of expression differences (di for gene i) normalized by the number of cell pairs:
Here, an F-test is applied for the comparison, with f = σ2/s20, d.f.1 = n − 1 and d.f.2 = n.
Construction of the HNOCA Community Edition by query-to-reference mapping
To construct the HNOCA-CE, we first collected raw count matrices and associated metadata of five more neural organoid studies. For two publications71,75, we obtained them from the sources listed in the ‘Data availability’ section of the paper. For the remaining three publications72–74, count matrices and associated metadata were provided directly by the authors. We subset each dataset to the healthy control cells and removed any cells with fewer than 200 genes expressed. We subset the gene space of every dataset to the 3,000 HVGs of HNOCA while filling the expression of missing genes in the community datasets with zeros. On average, 23% of genes with zero expression were added per dataset. We instantiated a mapping object from the HNOCA-tools package (at commit fe38c52) using the saved scPoli45 model weights from the HNOCA integration. Using the map_query method of the mapper instance, we projected the community datasets to HNOCA. We used the following training hyperparameters: retrain = ‘partial’, batch_size = 256, unlabeled_prototype_training = False, n_epochs = 10, pretraining_epochs = 9, early_stopping_kwargs = early_stopping_kwargs, eta = 10, alpha_epoch_anneal = 10. We computed the wkNN graph using the compute_wknn method of the mapper instance with k = 100. We transferred the final level_2 cell type labels from HNOCA to the community datasets using this neighbour graph. To obtain the combined representation of HNOCA-CE, we projected HNOCA together with the added community datasets through the trained model and computed a neighbour graph and UMAP from the resulting latent representation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08172-8.
Supplementary information
YAML file of cell type markers and hierarchy used for snapseed.
Supplementary Tables 1–10.
Acknowledgements
We thank C. De Donno for his support in improving our data integration efforts using scPoli. We thank D. Klein, P. Weiler and M. Lange for insightful discussions on the moscot framework, (neural) optimal transport and real-time-informed pseudotime analyses. We thank C. Bright for customizing the ArchMap tool to meet the requirements of this project. We thank F. Sanchis-Calleja, S. Jansen and F. Zenk for insightful comments on summarizing neural organoid protocols. We thank P. Lönnerberg and S. Linnarsson for insightful discussions on the application of the human developing brain atlas in this study. We thank the Human Cell Atlas Organoid Biological Network, in particular F. Birey, J. Andersen, S. A. Sloan, A. R. Muotri, S. Velasco, P. Arlotta, Y. Xiang, I.-H. Park, A. Bhaduri, A. R. Kriegstein, L. Pellegrini, M. A. Lancaster, G.-L. Ming, T. Sawada, T. Kato, O. Revah, K. R. Bowles, A. M. Goate, S. Temple, A. Fiorenzano, M. Parmar, R. Samarasinghe, B. G. Novitch, I. Kelava, J. A. Knoblich, G. Testa, T. Bertucci, R. Shyti, E. B. Binder, F. H. Gage and C. Bock for their support on data and metadata retrieval. This work was supported by Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation (grant nos. CZF2019-002440 and CZF2021-237566, to J.G.C. and B.T.). This work was cofunded by the Swiss National Science Foundation (project grant no. 310030_192604, to B.T.), the European Union (European Research Council (ERC), DeepCell grant no. 101054957, to A.S. and F.J.T.; ERC, Organomics grant no. 758877, to B.T.; H2020, Braintime grant no. 874606, to B.T.; ERC, Anthropoid grant no. 803441, to J.G.C.) and the Roche Institute for Human Biology (Z.H., H.C.L., B.T.). Views and opinions expressed are however those of the authors only and do not necessarily reflect those of the European Union or the ERC. Neither the European Union nor the granting authority can be held responsible for them. This work was supported by the Bavarian Ministry of Science and the Arts in the framework of the Bavarian Research Association ForInter (Interaction of Human Brain Cells) (to F.J.T.). This work was supported by the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (grant nos. 031A532B, 031A533A, 031A533B, 031A534A, 031A535A, 031A537A, 031A537B, 031A537C, 031A537D, 031A538A) (to L.D., A.S., K.X.L. and I.S.). This work was supported through a Fulbright grant of the German-American Fulbright Commission (to K.X.L.). L.D. acknowledges support by the Joachim Herz Foundation. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications/).
Extended data figures and tables
Author contributions
A.S., K.X.L. and I.S. contributed equally. Z.H., L.D. and J.S. collected and retrieved the scRNA-seq data involved in HNOCA, with suggestions from S.P.P., J.G.C. and B.T. H.-C.L. and M.S. generated the unpublished midbrain organoid data. A.A. and G.Q. generated and shared the cerebellar organoid data before its publication. J.S.F. developed snapseed. Z.H. and J.S.F. curated cell type hierarchy with the support from L.D. L.D., K.X.L., I.S. and A.S. performed HNOCA data curation and metadata harmonization. L.D., with the support from K.X.L. and I.S., performed HNOCA data preprocessing and integration using the pipeline developed by Z.H., L.D., J.S.F. and A.S. L.D. and K.X.L. performed the benchmark of integration methods. Z.H. did HNOCA cell type annotation. K.X.L. and J.S.F. performed the real-time-informed pseudotime analysis. J.S.F. performed reference mapping of HNOCA to the human developing brain atlas with support from A.S. Z.H. developed and performed label transfer and presence score estimation. Z.H. performed stage-matching analysis of HNOCA cells. I.S., J.S.F. and L.D. performed morphogen analysis, with the organoid protocols summarized by Z.H. L.D. and Z.H. with support from I.S. K.X.L. performed DE and transcriptomic comparison analysis. Z.H. performed the heterogeneity analysis of telencephalic cells and cell-level DE analysis with covariates. J.S.F. and A.S. performed reference mapping of organoid morphogen screen dataset to HNOCA and the human developing brain atlas and the follow-up analysis. Z.H. collected, retrieved and analysed the scRNA-seq data of disease-modelling neural organoids, and developed the procedure to compare with HNOCA. L.D. curated extra datasets and performed reference mapping to expand HNOCA. J.S.F. developed the HNOCA-tools Python package implementing analysis approaches developed in the study. Z.H., J.G.C., F.J.T. and B.T. designed the project. Z.H., L.D., J.S.F., A.S., I.S., S.P.P., J.G.C., F.J.T. and B.T. wrote the paper with input from all the coauthors. All authors read and approved the final manuscript.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
All curated individual HNOCA datasets are available for easy access through the sfaira Python tool78. The integrated HNOCA data is available at Zenodo (10.5281/zenodo.11203684)100 and the CellxGene Discover Census (https://cellxgene.cziscience.com/collections/de379e5f-52d0-498c-9801-0f850823c847). The extended HNOCA Community Edition Atlas is also available through the CellxGene Discover Census (same URL as above). Both versions of HNOCA are available for reference mapping through the ArchMap web interface (https://www.archmap.bio/). The HNOCA-tools package provides a Python interface for annotation, reference mapping and central downstream analysis steps and is available at https://github.com/devsystemslab/HNOCA-tools. More information on the available tools and a documentation of HNOCA-tools is available at https://devsystemslab.github.io/HNOCA-tools. Jupyter notebooks and scripts to reproduce the analysis are available at https://github.com/theislab/neural_organoid_atlas.
Competing interests
F.J.T. consults for Immunai Inc., Singularity Bio B.V., CytoReason Ltd, Cellarity, and has ownership interest in Dermagnostix GmbH and Cellarity. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhisong He, Leander Dony, Jonas Simon Fleck
A list of authors and their affiliations appears at the end of the paper
Change history
12/11/2024
A Correction to this paper has been published: 10.1038/s41586-024-08482-x
Contributor Information
Zhisong He, Email: zhisong.he@bsse.ethz.ch.
J. Gray Camp, Email: jarrettgrayson.camp@unibas.ch.
Fabian J. Theis, Email: fabian.theis@helmholtz-munich.de
Barbara Treutlein, Email: barbara.treutlein@bsse.ethz.ch.
Human Cell Atlas Organoid Biological Network:
Neal D. Amin, Kevin W. Kelley, Taylor Bertucci, Sally Temple, Kathryn R. Bowles, Nicolò Caporale, Emanuele Villa, Giuseppe Testa, Cristiana Cruceanu, and Elisabeth B. Binder
Extended data
is available for this paper at 10.1038/s41586-024-08172-8.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08172-8.
References
- 1.Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sloan, S. A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron95, 779–790.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanton, S. et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Marton, R. M. et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci.22, 484–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trujillo, C. A. et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell25, 558–569.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature570, 523–527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang, Y. et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell24, 487–497.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon, S.-J. et al. Reliability of human cortical organoid generation. Nat. Methods16, 75–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen, J. et al. Generation of functional human 3D cortico-motor assembloids. Cell183, 1913–1929.e26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhaduri, A. et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature578, 142–148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura, Y. et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol.38, 1421–1430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellegrini, L. et al. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science369, eaaz5626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian, X. et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell26, 766–781.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawada, T. et al. Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol. Psychiatry25, 2695–2711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan, T. A. et al. Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat. Med.26, 1888–1898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowles, K. R. et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell184, 4547–4563.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorenzano, A. et al. Single-cell transcriptomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nat. Commun.12, 7302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, W.-K. et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell28, 1657–1670.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samarasinghe, R. A. et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat. Neurosci.24, 1488–1500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleck, J. S. et al. Inferring and perturbing cell fate regulomes in human brain organoids. Nature10.1038/s41586-022-05279-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, Z. et al. Lineage recording in human cerebral organoids. Nat. Methods19, 90–99 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelava, I., Chiaradia, I., Pellegrini, L., Kalinka, A. T. & Lancaster, M. A. Androgens increase excitatory neurogenic potential in human brain organoids. Nature602, 112–116 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzquiano, A. et al. Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell185, 3770–3788.e27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vértesy, Á. et al. Gruffi: an algorithm for computational removal of stressed cells from brain organoid transcriptomic datasets. EMBO J.41, e111118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atamian, A. et al. Human cerebellar organoids with functional Purkinje cells. Cell Stem Cell31, 39–51.e6 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulsen, B. et al. Autism genes converge on asynchronous development of shared neuron classes. Nature602, 268–273 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun, E. et al. Comprehensive cell atlas of the first-trimester developing human brain. Science382, eadf1226 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Eze, U. C., Bhaduri, A., Haeussler, M., Nowakowski, T. J. & Kriegstein, A. R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci.24, 584–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhaduri, A. et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nature598, 200–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, L. et al. Molecular and cellular dynamics of the developing human neocortex at single-cell resolution. Preprint at bioRxiv10.1101/2024.01.16.575956 (2024).
- 31.Velasco, S., Paulsen, B. & Arlotta, P. 3D Brain organoids: studying brain development and disease outside the embryo. Annu. Rev. Neurosci.43, 375–389 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Sidhaye, J. & Knoblich, J. A. Brain organoids: an ensemble of bioassays to investigate human neurodevelopment and disease. Cell Death Differ.28, 52–67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pașca, S. P. et al. A nomenclature consensus for nervous system organoids and assembloids. Nature609, 907–910 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods12, 671–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jo, J. et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell19, 248–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature545, 48–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollen, A. A. et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell176, 743–756.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA112, 15672–15677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mora-Bermúdez, F. et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife5, e18683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benito-Kwiecinski, S. et al. An early cell shape transition drives evolutionary expansion of the human forebrain. Cell184, 2084–2102.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, C. et al. Single-cell brain organoid screening identifies developmental defects in autism. Nature621, 373–380 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szebényi, K. et al. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci.24, 1542–1554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amin, N. D. et al. Generating human neural diversity with a multiplexed morphogen screen in organoids. Preprint at bioRxiv10.1101/2023.05.31.541819 (2023). [DOI] [PubMed]
- 45.De Donno, C. et al. Population-level integration of single-cell datasets enables multi-scale analysis across samples. Nat. Methods20, 1683–1692 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods19, 41–50 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eyring, L. et al. Unbalancedness in neural monge maps improves unpaired domain translation. In Proc. Twelfth International Conference on Learning Representationshttps://iclr.cc/virtual/2024/poster/19548 (2024).
- 48.Klein, D. et al. Mapping cells through time and space with moscot. Preprint at bioRxiv10.1101/2023.05.11.540374 (2023).
- 49.Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, C. et al. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst. Biol.17, e9620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lotfollahi, M. et al. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol.40, 121–130 (2022). [DOI] [PMC free article] [PubMed]
- 52.Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet.25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aleksander, S. A. et al. The Gene Ontology knowledgebase in 2023. Genetics224, iyad031 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L. et al. Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat. Commun.11, 4038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanova, T. et al. Cerebral organoids derived from patients with Alzheimer’s disease with PSEN1/2 mutations have defective tissue patterning and altered development. Cell Rep.42, 113310 (2023). [DOI] [PubMed] [Google Scholar]
- 58.Kang, Y. et al. A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat. Neurosci.24, 1377–1391 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Notaras, M. et al. Schizophrenia is defined by cell-specific neuropathology and multiple neurodevelopmental mechanisms in patient-derived cerebral organoids. Mol. Psychiatry27, 1416–1434 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jabali, A. et al. Human cerebral organoids reveal progenitor pathology in EML1-linked cortical malformation. EMBO Rep.23, e54027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klaus, J. et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat. Med.25, 561–568 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Papes, F. et al. Transcription Factor 4 loss-of-function is associated with deficits in progenitor proliferation and cortical neuron content. Nat. Commun.13, 2387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morelli, K. H. et al. MECP2-related pathways are dysregulated in a cortical organoid model of myotonic dystrophy. Sci. Transl. Med.14, eabn2375 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacob, F. et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell180, 188–204.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng, Q. et al. CALD1 modulates gliomas progression via facilitating tumor angiogenesis. Cancers13, 2705 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavlyukov, M. S. et al. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell34, 119–135.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rich, J. N. et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res.65, 4051–4058 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Mila, M., Alvarez-Mora, M. I., Madrigal, I. & Rodriguez-Revenga, L. Fragile X syndrome: an overview and update of the FMR1 gene. Clin. Genet.93, 197–205 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Giandomenico, S. L. et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci.22, 669–679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, M. et al. Morphological diversification and functional maturation of human astrocytes in glia-enriched cortical organoid transplanted in mouse brain. Nat. Biotechnol.10.1038/s41587-024-02157-8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertucci, T. et al. Improved protocol for reproducible human cortical organoids reveals early alterations in metabolism with mutations. Preprint at bioRxiv10.1101/2023.07.11.548571 (2023).
- 73.Caporale, N. et al. Multiplexing cortical brain organoids for the longitudinal dissection of developmental traits at single cell resolution. Preprint at bioRxiv10.1101/2023.08.21.553507 (2023). [DOI] [PubMed]
- 74.López-Tobón, A. et al. dosage regulates neuronal differentiation and social behavior in 7q11.23 neurodevelopmental disorders. Sci. Adv.9, eadh2726 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dony, L. et al. Chronic exposure to glucocorticoids amplifies inhibitory neuron cell fate during human neurodevelopment in organoids. Preprint at bioRxiv10.1101/2024.01.21.576532 (2024).
- 76.CZI Single-Cell Biology Program et al. CZ CELL×GENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. Preprint at bioRxiv10.1101/2023.10.30.563174 (2023). [DOI] [PMC free article] [PubMed]
- 77.Qian, X., Song, H. & Ming, G.-L. Brain organoids: advances, applications and challenges. Development146, dev166074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fischer, D. S. et al. Sfaira accelerates data and model reuse in single cell genomics. Genome Biol.22, 248 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res.50, D988–D995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Virshup, I., Rybakov, S., Theis, F. J., Angerer, P. & Wolf, F. A. anndata: annotated data. Preprint at bioRxiv10.1101/2021.12.16.473007 (2021).
- 81.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw.3, 861 (2018). [Google Scholar]
- 83.Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep.9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nolet, C. et al. Accelerating single-cell genomic analysis with GPUs. Preprint at bioRxiv10.1101/2022.05.26.493607 (2022).
- 85.YosefLab/scib-metrics: accelerated, Python-only, single-cell integration benchmarking metrics. GitHubhttps://github.com/YosefLab/scib-metrics (2024).
- 86.Lange, M. et al. CellRank for directed single-cell fate mapping. Nat. Methods19, 159–170 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haghverdi, L., Buettner, F. & Theis, F. J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics31, 2989–2998 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Gayoso, A. et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol.40, 163–166 (2022). [DOI] [PubMed] [Google Scholar]
- 89.Virshup, I. et al. The scverse project provides a computational ecosystem for single-cell omics data analysis. Nat. Biotechnol.41, 604–606 (2023). [DOI] [PubMed] [Google Scholar]
- 90.Heumos, L. et al. Pertpy: an end-to-end framework for perturbation analysis. Preprint at bioRxiv10.1101/2024.08.04.606516 (2024).
- 91.Büttner, M., Ostner, J., Müller, C. L., Theis, F. J. & Schubert, B. scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun.12, 6876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tay, J. K., Narasimhan, B. & Hastie, T. Elastic net regularization paths for all generalized linear models. J. Stat. Softw.106, 1–31 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf.12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Badia-I-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv.2, vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics39, btac757 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shen, W.-K. et al. AnimalTFDB 4.0: a comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res.51, D39–D45 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He, Z. Dony, L. & Fleck, J. S. An integrated transcriptomic cell atlas of human neural organoids: cleaned datasets. Zenodo10.5281/zenodo.11203684 (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
YAML file of cell type markers and hierarchy used for snapseed.
Supplementary Tables 1–10.
Data Availability Statement
All curated individual HNOCA datasets are available for easy access through the sfaira Python tool78. The integrated HNOCA data is available at Zenodo (10.5281/zenodo.11203684)100 and the CellxGene Discover Census (https://cellxgene.cziscience.com/collections/de379e5f-52d0-498c-9801-0f850823c847). The extended HNOCA Community Edition Atlas is also available through the CellxGene Discover Census (same URL as above). Both versions of HNOCA are available for reference mapping through the ArchMap web interface (https://www.archmap.bio/). The HNOCA-tools package provides a Python interface for annotation, reference mapping and central downstream analysis steps and is available at https://github.com/devsystemslab/HNOCA-tools. More information on the available tools and a documentation of HNOCA-tools is available at https://devsystemslab.github.io/HNOCA-tools. Jupyter notebooks and scripts to reproduce the analysis are available at https://github.com/theislab/neural_organoid_atlas.