Abstract
Bacteria are typically isolated on rich media to maximise isolation success, removing them from their native evolutionary context. This eliminates selection pressures, enabling otherwise deleterious genomic events to accumulate. Here, we present a cautionary tale of these ‘quiet mutations’ which can persist unnoticed in bacterial culture lines. We used a combination of microbiological culture (standard and minimal media conditions), whole genome sequencing and metabolic modelling to investigate putative Klebsiella pneumoniae L-histidine auxotrophs. Additionally, we used genome-scale metabolic modelling to predict auxotrophies among completed public genomes (n = 2637). Two sub-populations were identified within a K. pneumoniae frozen stock, differing in their ability to grow in the absence of L-histidine. These sub-populations were the same ‘strain’, separated by eight single nucleotide variants and an insertion sequence-mediated deletion of the L-histidine biosynthetic operon. The His− sub-population remained undetected for > 10 years despite its inclusion in independent laboratory experiments. Genome-scale metabolic models predicted 0.8% public genomes contained ≥ 1 auxotrophy, with purine/pyrimidine biosynthesis and amino acid metabolism most frequently implicated. We provide a definitive example of the role of standard rich media culture conditions in obscuring biologically relevant mutations (i.e. nutrient auxotrophies) and estimate the prevalence of such auxotrophies using public genome collections. While the prevalence is low, it is not insignificant given the thousands of K. pneumoniae that are isolated for global surveillance and research studies each year. Our data serve as a pertinent reminder that rich-media culturing can cause unnoticed wild-type domestication.
Keywords: Metabolism, Klebsiella pneumoniae, Attenuation, Whole genome sequencing, Nutrient auxotrophy
Introduction
Microbial species often display high levels of genetic diversity [1, 2], which can drive ecological and pathogenic differentiation. Whole genome sequencing of environmental and clinical microbes is crucial for understanding microbial diversity and distribution [3, 4], predicting antimicrobial resistance and virulence [5]. Prior to sequencing, isolates are enriched using nutrient-dense growth media like Luria–Bertani (LB) to maximise isolation success in large-scale studies [3, 4]. However, what often goes unacknowledged is that isolation and sequencing represent a snapshot of an organism. Genomes and populations are in constant flux, guided by evolutionary selection pressures such as competition for space [6] and nutrients [7], bacteriophage predation [8] and antibiotics [9]. Growth in rich media removes these pressures, permitting deleterious genomic events to accumulate. These ‘quiet mutations’ may go unnoticed or incorrectly accepted as wild-type characteristics. In this study, we present a cautionary tale of quiet mutations, how they can be present in primary frozen stocks and persist unnoticed. We also estimate the frequency of these events across public Klebsiella pneumoniae Species Complex (KpSC) genomes.
Methods
Bacterial Strains and Culture
Two KpSC isolates collected from hospitalised patients in Melbourne, Australia were used in this study: K. pneumoniae INF018 and K. variicola subsp. variicola INF232, both causing urinary tract infections [5]. INF018 was part of a reported transmission cluster [10] and was used in two subsequent studies focusing on metabolism [5, 11] and one exploring passage through an in vitro bladder model [12], from which additional genome sequences were generated (Fig. 1A). Isolates were grown as previously reported [13]. Briefly, 5 mL cultures were grown at 37 °C, shaking at 200 RPM overnight in broth: M9 minimal media (Sigma) containing either 20 mM D-glucose, 20 mM L-histidine or both. Isolates were plated onto LB or M9 plates containing the same substrates.
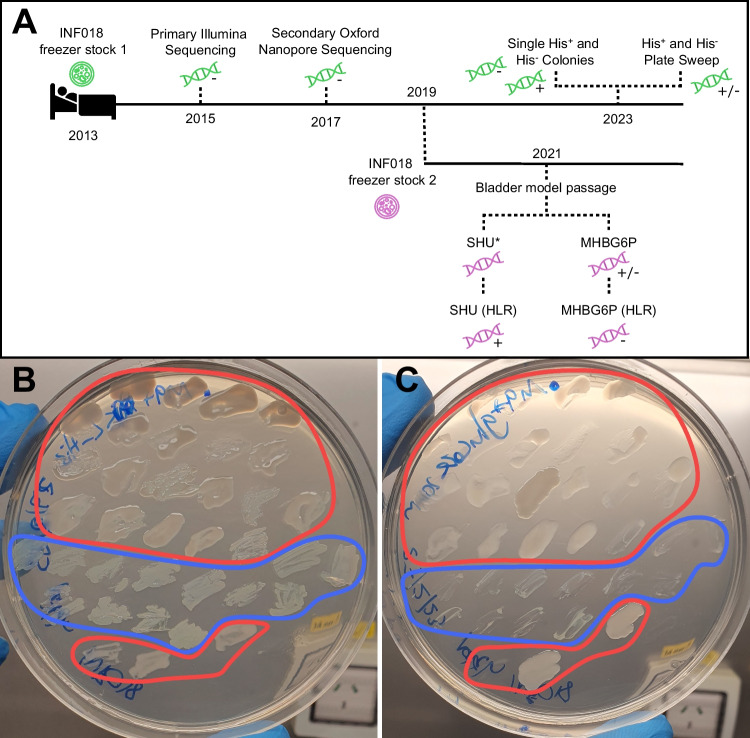
Fig. 1.
a Timeline of K. pneumoniae INF018, with dark lines representing different frozen stocks (green and purple) and dotted lines showing independent subculturing events. Sequencing is represented with a DNA icon coloured by the frozen stock they originated from, with a “ + ” indicating a His+ colony, “ − ” indicating a His− colony and “ + / − ” indicating mixed colonies. Sequencing for the in vitro bladder model was done using INF018 passaged in either cation-adjusted Mueller–Hinton broth (MHBG6P) or synthetic human urine (SHU), including high-level Fosfomycin-resistant (HLR) subpopulations from each media [12]. Sequencing data for INF018 SHU (labelled with an asterisk) is unavailable as the genome failed quality control [12]. B and C Patch plates showing heterogeneous population of His+ and His− sub-populations within the 2013 frozen stocks. Colonies within the red boundaries show K. pneumoniae INF018, whereas the blue boundary shows K. variicola subsp. variicola INF232. Colonies match positions on each plate. B M9 agar + 20 mM L-histidine. C M9 agar + 20 mM D-glucose
DNA Isolation and Sequencing
Overnight cultures were pelleted at 8000 × g and DNA extracted using GenFind V3 (Beckman Coulter). DNA libraries were constructed using Illumina DNA Library Prep Kit, (M) Tagmentaton (Illumina) and sequenced on the Illumina NovaSeq 6000 at 2 × 250 bp. Reads and assemblies were deposited at Genbank (BioProject PRJNA1115910, further details on Figshare 10.6084/m9.figshare.25864864).
Genomic Analysis
Genomes were assembled using Unicycler v0.4.7 with the –keep 0 option [14] and annotated with Bakta v1.8.1 [15] using the ‘–gram – ‘ option and v1.8.1 database. Kaptive 3 [16, 17] was used for determining K/O locus intactness. Read and contig mapping to a completed genome representing a His+ isolate from the same transmission cluster (accession: GCA_904863215.1) was performed with minimap2 v2.26 [18], using SAMtools v1.19 [19] for depth estimation. Genome-wide single nucleotide variant (SNV) distances were calculated via read mapping and variant calling against the completed INF018 genome (accession: SAMEA3356961, His−) using RedDog v1beta.11 (https://github.com/katholt/RedDog). ISEScan v1.7.2.3 [20] was used to identify Insertion Sequences.
Completed public Klebsiella genomes (n = 2637) were obtained from Genbank, accessed: 29/01/2024 (10.6084/m9.figshare.25864864). Bactabolize [21] v1.0.2 was used to construct metabolic models (using the KpSC pan v2.0.2 reference) and predict growth phenotypes [13].
Results
Heterogeneous Sub-Population in Clinical Isolate Sample
We previously reported metabolic models [13] which unexpectedly predicted L-histidine auxotrophies for two isolates (K. pneumoniae INF018 and K. variicola INF232). L-histidine biosynthesis is essential for cellular growth and protein production and is conserved across all bacteria [22]. Auxotrophic strains are unable to produce L-histidine and therefore cannot grow in environments lacking this substrate. Both genomes were complete [5], ruling out misassembly as an explanation, and both isolates were subsequently tested in vitro to confirm L-histidine auxotrophy [13]. In parallel, another team member, unaware of these data, performed an independent experiment using the same 2013 INF018 frozen stock, which demonstrated growth in M9 + D-glucose without L-histidine.
When these inconsistencies were realised, the 2013 frozen stocks were plated onto LB and colonies patched onto M9 + D-glucose and M9 + L-histidine (Fig. 1B, C). All colonies grew on L-histidine, 19/23 INF018 colonies grew on D-glucose without L-histidine, while 0/13 INF232 colonies were able to grow on D-glucose without L-histidine. This confirmed the 2013 INF018 frozen stock was a heterogeneous population, consisting of His+ and His− sub-populations, while INF232 was entirely His−. Compared to His+, His− mutants displayed a flatter, paler colony morphology. It was unclear if these morphological differences were driven by the direct impact of the auxotrophy, or by independent genetic changes.
Genomics Confirmed Deletion of L-Histidine Biosynthetic Operon
Single INF018 His+ and His− colonies from the 2013 frozen stock were whole-genome sequenced to confirm the heterogeneous population (Fig. 1). Compared to the complete INF018 genome [5], the INF018 His+ genome had eight SNVs, while INF018 His− had zero, indicating they were the same strain [23]. Gorrie et al. had previously reported the INF018 frozen stock contained an ESBL plasmid (blaCTX-M-15), which was lost during culture and original sequencing [5], but we recovered the plasmid in both His+ and His− colony genomes.
Read mapping indicated the INF018 His− sub-population had a 19,388 bp deletion spanning the L-histidine biosynthetic operon (his) and part of the adjacent O antigen biosynthetic locus (Fig. 2). The chromosomal deletion event was likely mediated by insertion of IS5_222 introduced into the genome on the ESBL plasmid, and found exclusively on plasmids in closely related His+ K. pneumoniae strains [5]. The loss of O-antigen would prevent K-antigen anchoring, which may have contributed to the colony morphology variation (Fig. 1B, C). Notably, the K and O antigens are also major Klebsiella pathogenicity determinants and targets for novel anti-Klebsiella vaccines [24]. Antigen variant prioritisation is informed by genomic analyses which would have predicted a misleading O-antigen deficiency in this case.
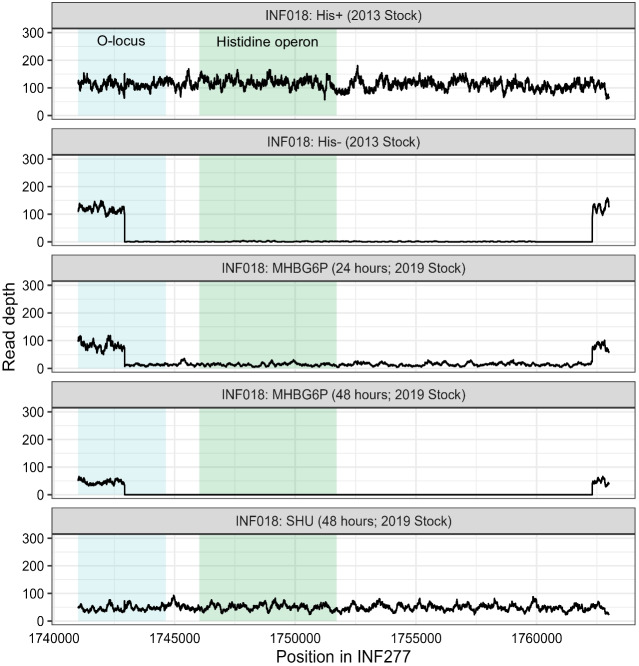
Fig. 2.
Read mapping of experimentally verified INF018 His+ and His− strains (2013 frozen stock) as well as INF018 reads from the in vitro bladder model experiment (accessions: SAMN17846711, SAMN17846713 and SAMN21465650) [12] wherein INF018 (2019 frozen stock) was passaged in cation-adjusted Mueller–Hinton broth (MHBG6P) or synthetic human urine (SHU), prior to sequencing at 24 and 48-h time-points. High-level Fosfomycin-resistant (HLR) subpopulations from each passage were also sequenced [12]. Reads were aligned to K. pneumoniae INF277 (accession: SAMN06112222, His +), which is part of the same transmission cluster as INF018 [5]. The green box highlights the location of the L-histidine biosynthesis operon, and the blue box highlights the 3′ region of the O-antigen locus. Mean sequencing depth for 2013 INF018 genomes: His+ (108.9x), His.− (113.88x); 2019 INF018 genomes: MHBG6P 24 h (82.11x), MHBG6P 48 h (49.48x), SHU 48 h (52.27x)
Prior to this work, INF018 was used in an in vitro bladder model experiment [12] where it was passaged in cation-adjusted Mueller–Hinton broth (MHBG6P) or synthetic human urine (SHU) with/without fosfomycin and subjected to de novo whole-genome sequencing (Fig. 1A). Re-analysis suggests that a His+ isolate was used for SHU experiments while a His− isolate was used for MHBG6P, suggesting that the second INF018 freezer stock also contained a mixed population (Fig. 1A and 2). However, his operon reads in the MHBG6P 24-h passaged genome were absent in the 48-h passaged genome, indicating a gradual loss in the operon over time (Fig. 2).
Estimating Auxotrophies from Public Genome Data
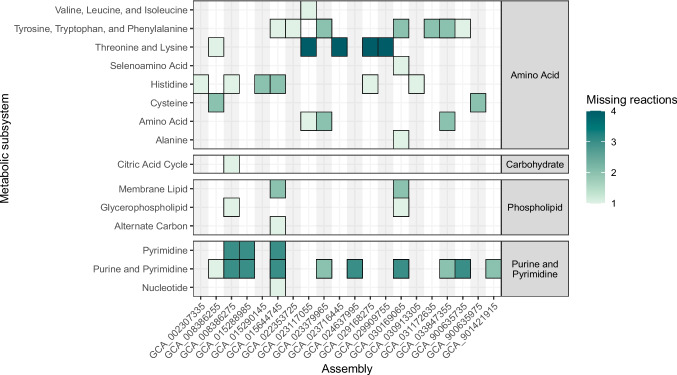
Given that we had identified two L-histidine auxotrophs among a collection of just 507 isolates (0.4%) [13], we sought to estimate the general prevalence of such auxotrophs among public genome collections. We constructed metabolic models for all complete [21] KpSC genomes in Genbank (n = 2637). Twenty (0.8%) were predicted as auxotrophs on M9 + D-glucose media, with the most frequent auxotrophies being purine and pyrimidine biosynthesis and amino acid metabolism (Fig. 3).
Fig. 3.
Metabolism auxotrophies identified in the 20 putatively auxotrophic isolates. Counts of individual missing reactions are shown by colour. Auxotrophs are split by the subsystem and general metabolic class. Median missing reactions resulting in substrate auxotrophism was 3 (range 1 to 25)
Discussion
Our data highlights how the use of rich culture media can facilitate and conceal genetic mutations that impact bacterial growth capabilities and clinically relevant traits. This has implications for the broad suite of research and diagnostics processes that rely on the use of rich medias, and which may be misled by analysis of mutant strains that do not reflect wild-type characteristics. Example undesirable outcomes may include misrepresentations of ecological roles, molecular epidemiology or antimicrobial susceptibilities, which are known to be influenced by metabolic state [25, 26].
Loss of plasmids during culturing [5, 27, 28] and spontaneous mutants with varying colony morphology [29, 30] have been described previously, as have passage-induced mutations in historical and globally distributed laboratory strains [31, 32]. For example, passage of Burkholderia pseudomallei strain K96243 in LB broth for 28 days impacted virulence, protein expression and regulation of metabolism [32]. However, to our knowledge there are few discussions of spontaneous nutrient auxotrophs occurring over the timescale of standard clinical microbiological diagnostic protocols and/or overnight cultures for DNA extractions.
Current recommendations for Klebsiella isolation include growth in LB + ampicillin then Simmons Citrate + Inositol [33] or MacConkey/CHROMagar/horse blood agar [34]. The two isolates used in this study (K. pneumoniae INF018 and K. variicola INF232) were originally collected from patient samples plated onto MacConkey then grown in LB broth prior to sequencing in 2013 [5]. These rich medias would support the within-isolate heterogeneity we observed, eventually resulting in the complete loss of His+ sub-populations. However, it is also possible that both sub-populations were present in the patient (urine is histidine rich), but in either case the use of rich media enabled the His− population to persist and outcompete His+ populations undetected [12]. We therefore propose that these spontaneous auxotrophies act like ‘quiet mutations’—a phenotypic parallel to silent mutations. These occur when DNA changes cause a phenotype alteration that remains unnoticed in standard culture conditions.
Our analysis predicted at least 0.8% of completed, high-quality KpSC genomes represent isolates with ≥ 1 auxotrophy (Fig. 3), consistent with data published from a smaller analysis of ~ 230 genomes [35] and indicating that the phenomenon is rare but not insignificant considering the scale of modern genomic sequencing. i.e. 100,000 K. pneumoniae genomes are now available in NCBI Pathogens. Similar analyses of four other Gram-negative genera (Escherichia, Pseudomonas, Salmonella and Yersinia) predicted between 1.2 and 4.5% were auxotrophic for one or more nutrients [35]. However, it is not possible to determine whether these reflect ‘quiet’ mutations acquired during laboratory culture or otherwise, or genuine biological adaptations. We acknowledge that adaptive loss of biosynthetic pathways has been reported in E. coli and other bacterial species (36, 37), especially in specific host-species combinations.
Like the previous study, our investigation of public genomes leveraged reference-based metabolic reconstructions, which may not capture the true extent of metabolic capabilities and associated genetic diversity in the population. As a result, we may have overestimated the prevalence of nutrient auxotrophies. However, we argue that we have likely underestimated the total burden of quiet mutations because our approach focused solely on metabolic gene content, but we expect that non-metabolic gene content would also be impacted. The likelihood of propagation and detection of these mutations is highly dependent on media and growth conditions. Our cautionary tale has served as a good self-reminder when studying microbial ecology to ensure we are using the most appropriate conditions to observe the wild-type organism.
Author Contribution
Conceptualization: BV, HBC, KLW Methodology: BV, HBC, KLW Validation: BV, HBC Formal analysis: BV, HBC Investigation: JAW, MHP Resources: JAW, MHP, AWJJ, KEH Writing—Original Draft: BV, HBC, KLW Writing—Review & Editing: BV, HBC, JAW, MHP, AWJJ, KEH, KLW Visualization: BV, HBC, KLW Supervision: KLW Project administration: KLW Funding acquisition: KEH, KLW.
Funding
This research was supported by an Australian Government Research Training Program (RTP) Scholarship to HBC and an Australian Research Council Discovery Project award (DP200103364, to KLW and KEH). KLW is supported by a National Health and Medical Research Council of Australia Investigator Grant (APP1176192).
Data Availability
Reads and assemblies were deposited at Genbank (BioProject PRJNA1115910). Additional supporting code and details found at Figshare (10.6084/m9.figshare.25864864).
Declarations
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ben Vezina and Helena B. Cooper contributed equally to this work.
Contributor Information
Ben Vezina, Email: benjamin.vezina@monash.edu.
Helena B. Cooper, Email: helena.cooper@monash.edu
Kelly L. Wyres, Email: Kelly.wyres@monash.edu
References
- 1.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, et al (2008) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 190(20):6881–93 [DOI] [PMC free article] [PubMed]
- 2.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D et al (2015) Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci 112(27):E3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorpe HA, Booton R, Kallonen T, Gibbon MJ, Couto N, Passet V, et al (2022) A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings. Nature Microbiology. 7(12):2054–67 [DOI] [PMC free article] [PubMed]
- 4.Calland JK, Haukka K, Kpordze SW, Brusah A, Corbella M, Merla C et al (2023) Population structure and antimicrobial resistance among Klebsiella isolates sampled from human, animal, and environmental sources in Ghana: a cross-sectional genomic One Health study. The Lancet Microbe 4(11):e943–e952 [DOI] [PubMed] [Google Scholar]
- 5.Gorrie CL, Mirčeta M, Wick RR, Judd LM, Lam MMC, Gomi R et al (2022) Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen. Nat Commun 13(1):3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyte KZ, Tabuteau H, Gaffney EA, Foster KR, Durham WM (2017) Microbial competition in porous environments can select against rapid biofilm growth. Proc Natl Acad Sci U S A 114(2):E161–E170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culp EJ, Goodman AL (2023) Cross-feeding in the gut microbiome: ecology and mechanisms. Cell Host Microbe 31(4):485–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erken M, Lutz C, McDougald D (2013) The rise of pathogens: predation as a factor driving the evolution of human pathogens in the environment. Microb Ecol 65(4):860–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christaki E, Marcou M, Tofarides A (2020) Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol 88(1):26–40 [DOI] [PubMed] [Google Scholar]
- 10.Gorrie CL, Mirceta M, Wick RR, Judd LM, Wyres KL, Thomson NR et al (2018) Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis 67(2):161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vezina B, Cooper HB, Rethoret-Pasty M, Brisse S, Monk JM, Holt KE, et al (2024) A metabolic atlas of the Klebsiella pneumoniae species complex reveals lineage-specific metabolism that supports persistent co-existence of diverse lineages. bioRxiv. 2024.07.24.605038.
- 12.Abbott IJ, van Gorp E, Wyres KL, Wallis SC, Roberts JA, Meletiadis J et al (2022) Oral fosfomycin activity against Klebsiella pneumoniae in a dynamic bladder infection in vitro model. J Antimicrob Chemother 77(5):1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper HB, Vezina B, Hawkey J, Passet V, López-Fernández S, Monk JM, Brisse S, Holt KE, Wyres KL (2024) A validated pangenome-scale metabolic model for the Klebsiella pneumoniae species complex. Microb Genom 10(2) [DOI] [PMC free article] [PubMed]
- 14.Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13(6):e1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A (2021) Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7(11) [DOI] [PMC free article] [PubMed]
- 16.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE (2016) Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2(12) [DOI] [PMC free article] [PubMed]
- 17.Lam MMC, Wick RR, Judd LM, Holt KE, Wyres KL (2022) Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb Genom 8(3) [DOI] [PMC free article] [PubMed]
- 18.Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Z, Tang H (2017) ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33(21):3340–3347 [DOI] [PubMed] [Google Scholar]
- 21.Vezina B, Watts SC, Hawkey J, Cooper HB, Judd LM, Jenney AWJ, et al (2023) Bactabolize is a tool for high-throughput generation of bacterial strain-specific metabolic models. eLife. 12:RP87406. [DOI] [PMC free article] [PubMed]
- 22.Fondi M, Emiliani G, Liò P, Gribaldo S, Fani R (2009) The evolution of histidine biosynthesis in archaea: insights into the his genes structure and organization in LUCA. J Mol Evol 69(5):512–526 [DOI] [PubMed] [Google Scholar]
- 23.Gorrie CL, Mirčeta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA et al (2017) Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 65(2):208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dangor Z, Benson N, Berkley JA, Bielicki J, Bijsma MW, Broad J, et al. (2024) Vaccine value profile for Klebsiella pneumoniae. Vaccine. 42(19s1):S125-s41. [DOI] [PubMed]
- 25.Stokes JM, Lopatkin AJ, Lobritz MA, Collins JJ (2019) Bacterial metabolism and antibiotic efficacy. Cell Metab 30(2):251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loose M, Naber KG, Coates A, Wagenlehner FME, Hu Y (2020) Effect of different media on the bactericidal activity of colistin and on the synergistic combination with azidothymidine against mcr-1-positive colistin-resistant Escherichia coli. Front Microbiol 11:54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daum RS, Syriopoulou VP, Smith AL, Scheifele DW, Willard JE (1981) Loss of plasmid DNA coding for beta-lactamase during experimental infection with Haemophilus influenzae type b. J Infect Dis 143(4):548–553 [DOI] [PubMed] [Google Scholar]
- 28.Joris M-A, Verstraete K, De Reu K, De Zutter L (2011) Loss of vtx genes after the first subcultivation step of verocytotoxigenic Escherichia coli O157 and non-O157 during isolation from naturally contaminated fecal samples. Toxins [Internet]. 3(6):[672–7 pp.]. [DOI] [PMC free article] [PubMed]
- 29.Gao S, Zhang Z, Xu X, Zhou H, Zhu H, Zhang Y et al (2022) Characteristics of a capnophilic small colony variant of Escherichia coli co-isolated with two other strains from a patient with bacteremia in China. Arch Microbiol 204(6):333 [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Zheng X, Huang M, Liu L (2020) A comparative genomic analysis of small-colony variant and wild-type Burkholderia pseudomallei in a patient with bacterial liver abscess. J Glob Antimicrob Resist 21:16–21 [DOI] [PubMed] [Google Scholar]
- 31.Achtman M, Van den Broeck F, Cooper KK, Lemey P, Parker CT, Zhou Z (2021) Genomic population structure associated with repeated escape of Salmonella enterica ATCC14028s from the laboratory into nature. PLoS Genet 17(9):e1009820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duangurai T, Reamtong O, Rungruengkitkun A, Srinon V, Boonyuen U, Limmathurotsakul D et al (2020) In vitro passage alters virulence, immune activation and proteomic profiles of Burkholderia pseudomallei. Sci Rep 10(1):8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sylvain Brisse, Virginie Passet, Rodrigues C (2019) Isolation of Klebsiella strains from human or animal fecal material. . protocolsio.
- 34.Leber AL (ed) (2016) Clinical microbiology procedures handbook, 4th edn. ASM Press
- 35.Seif Y, Choudhary KS, Hefner Y, Anand A, Yang L, Palsson BO (2020) Metabolic and genetic basis for auxotrophies in Gram-negative species. Proc Natl Acad Sci 117(11):6264–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X-J, Walker DH, Liu Y, Zhang L (2009) Amino acid biosynthesis deficiency in bacteria associated with human and animal hosts. Infect Genet Evol 9(4):514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Souza G, Waschina S, Pande S, Bohl K, Kaleta C, Kost C (2014) Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68(9):2559–2570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reads and assemblies were deposited at Genbank (BioProject PRJNA1115910). Additional supporting code and details found at Figshare (10.6084/m9.figshare.25864864).