Summary
While flowering plants have diversified in virtually every terrestrial clime, climate constrains the distribution of individual lineages. Overcoming climatic constraints may be associated with diverse evolutionary phenomena including whole genome duplication (WGD), gene‐tree conflict, and life‐history changes. Climatic shifts may also have facilitated increases in flowering plant diversification rates. We investigate climatic shifts in the flowering plant order Ericales, which consists of c. 14 000 species with diverse climatic tolerances.
We estimate phylogenetic trees from transcriptomic data, 64 chloroplast loci, and Angiosperms353 nuclear loci that, respectively, incorporate 147, 4508, and 2870 Ericales species. We use these phylogenetic trees to analyse how climatic shifts are associated with WGD, gene‐tree conflict, life‐history, and diversification rates.
Early branches in the phylogenetic trees are extremely short, and have high levels of gene‐tree conflict and at least one WGD. On lineages descended from these early branches, there is a significant association between climatic shifts (primarily out of tropical climates), further WGDs, and life‐history.
Extremely short early branches, and their associated gene‐tree conflict and WGDs, appear to underpin the explosive origin of numerous species rich Ericales clades. The evolution of diverse climatic tolerances in these species rich clades is tightly associated with WGD and life‐history.
Keywords: adaptation, climate, diversification, phylogenetics, whole genome duplication
Introduction
Climate plays a central role in shaping the distribution of biodiversity (e.g. Donoghue, 2008; Crisp et al., 2009). Adaptation to different climates is especially pronounced in angiosperms where different lineages have adapted to almost every terrestrial environment including hot and humid tropical rainforests, arid deserts, and cold polar and alpine conditions. Perhaps because of the sessile nature of plants, and the inability of individuals to move from climatic stress, adaptive traits for different climates in angiosperms are exceptionally diverse, ranging from succulent leaves or stems for arid conditions, cushion growth form in cold conditions, or complex biochemical adaptations for high ultraviolet or cold conditions (Griffith & Yaish, 2004; Eggli & Nyffler, 2009; Folk et al., 2020).
The different climates to which plant lineages have adapted have their own distinct histories. Tropical climates have existed far longer than temperate climates, whilst the varied age of alpine conditions depends on the age of different mountain systems (Hughes & Eastwood, 2006; Favre et al., 2016). The extent of these climatic zones has also changed significantly over time. Interactions between unique climates and the adaptations lineages must undergo to thrive in them underpin key patterns in angiosperm macroevolution. For example, the comparatively recent and rapid diversification of temperate and alpine lineages relative to tropical lineages is likely explained by the younger age and less saturated niche space of many temperate and alpine regions (Hughes & Eastwood, 2006; Zanne et al., 2014; Folk et al., 2019; Tietje et al., 2022).
Evolutionary adaptations, such as those enabling survival in new climates, are associated with diverse molecular evolutionary processes. One of the most important of these processes is whole genome duplication (WGD), which can facilitate the origin of potentially adaptive traits for survival in new climates, or occur as a result of climatic shifts (De Storme et al., 2012; McGrath & Lynch, 2012; Rice et al., 2019; Van de Peer et al., 2021; Feng et al., 2024). Gene‐tree conflict, where individual loci evolve according to different tree topologies, is also associated with the origin of novel traits (Parins‐Fukuchi et al., 2021; Stull et al., 2021). While conflict is not expected to be causally related to the evolution of novel traits, it may be broadly suggestive of shifts in population structure and size (Parins‐Fukuchi et al., 2021).
The angiosperm clade Ericales (comprising 22 families and c. 14 000 species sensu APG IV, 2016) is ideal for investigating adaptation to different climates at a global scale within a diverse and evolutionarily complex clade. Clades within Ericales have adapted to varied climates. For example, Sapotaceae and Lecythidaceae have diversified extensively within tropical climates (Bartish et al., 2010; Rose et al., 2018; Vargas & Dick, 2020), clades within Primulaceae, Polemoniaceae, and Ericaceae have diversified in cold climates (Schwery et al., 2015; Boucher et al., 2016; Larson et al., 2020, 2023), whilst Fouquieriaceae and Euclea (Ebenaceae) have diversified in arid climates (Walnöfer, 2001; Soto‐Trejo et al., 2022). Because Ericales likely originated over 90 million years ago (Ma) (Nixon & Crepet, 1993; Rose et al., 2018), when the global climate was considerably warmer and wetter than today, significant shifts in climatic preference within Ericales are likely to have involved adaptation to colder and drier climates that increased in extent in the Cenozoic. The prevalence of such shifts out of tropical climates has been previously highlighted in Saxifragales and Rosids (Folk et al., 2019; Sun et al., 2020).
Inevitably, a clade of the scale of Ericales incorporates other types of trait variation aside from different climatic preferences. This variation includes diverse life‐history strategies, ranging from often‐large tropical rainforest trees in Sapotaceae and Lecythidaceae, small herbaceous annuals in Polemoniaceae, and shrubs, lianas, and epiphytes distributed in families throughout the clade. Other striking evolutionary adaptations include carnivory in the pitcher plants of Sarraceniaceae, semi‐carnivory in Roridulaceae (where nutrients are sequestered by absorbing the excrement of predatory insects that live on their sticky leaves) mycoheterotrophy in Ericaceae (e.g. Sarcodes), and unique floral morphology in Lecythidaceae where an androecial hood extends over the rest of the flower and restricts pollinator access (Prance, 1976).
The molecular evolution and phylogeny of Ericales are also noteworthy. Early branching events within the order are almost simultaneous, whilst diversification of extant lineages within each family appears far more recent (Rose et al., 2018; Larson et al., 2020). The very short early branches suggest a rapid origination of Ericalean families after the origin of the order. However, incomplete lineage sorting (ILS) or hybridisation may also be prevalent on these early branches and can bias molecular branch length estimates (Mendes & Hahn, 2016). These early branches are also notable for their rapid morphological evolution (Parins‐Fukuchi et al., 2021). By contrast, high species turnover could explain the comparatively recent diversification of extant lineages within each family. In addition, transcriptomic analyses have illustrated the existence of several ancient WGDs within Ericales, including two associated with the early branches, and others occurring within families such as Primulaceae or Actinidiaceae (Larson et al., 2020). Despite knowledge of these varied molecular evolutionary phenomena within Ericales, understanding of the links between them and the striking evolutionary novelties observed throughout Ericales is limited.
We attempt to address this by asking to what extent shifts out of tropical climates in Ericales are associated with: (1) WGDs and gene‐tree conflict? (2) life‐history shifts? (3) diversification rate shifts? We address these questions using a transcriptomic dataset that incorporates 147 Ericales species, and chloroplast, Angiosperms353, and ITS datasets that incorporate over 4500 Ericales species. Incorporation of these datasets into a single study enables complementary insights. The transcriptomic dataset can reveal molecular processes (such as WGDs) that underpin the macroevolution of Ericales, and help to resolve intractable relationships. The species‐level datasets, in combination with climatic and life‐history data, enable insights into when and to what extent clades have adapted to and diversified in different climates, as well as associated life‐history changes.
Materials and Methods
Overview of methods
The methodological structure is as follows. (1) Analyses of transcriptomic data including phylogenetic inference, and estimating potential WGDs by mapping gene duplications and calculating synonymous distances between paralogous gene pairs. Estimates of chromosome numbers (Table S3) and WGDs from previous studies were also reviewed to further evaluate potential WGDs. (2) Phylogenetic inference with the species‐level datasets. WGDs from the previous step were mapped onto the estimated species‐level phylogenetic trees. (3) Estimation of climatic and life‐history shifts on the species‐level phylogenetic trees using evolutionary models, sister‐clade comparison, and analyses of the fossil record. (4) Estimation of lineage‐specific diversification rates in the species‐level phylogenetic trees.
Sampling and sequencing of transcriptomic data
A dataset of 153 transcriptomes was assembled: 97 were newly generated and 56 are publicly available. Outgroups from Asterales, Caryophyllales, Rosales, Santalales, Solanales, and Cornales were included. Overall, 148 taxa were incorporated (Calluna vulgaris (L.) Hull, Monotropa hypopitys L., Barringtonia racemosa (L.) Spreng., Eschweilera coriacea (DC.) S.A.Mori, and Gustavia superba (Kunth) O. Berg were each represented by two samples) representing 18 of the currently recognised 22 Ericales families (Cyrillaceae, Fouquieriaceae, Symplocaceae, and Tetrameristaceae were not sampled).
To generate most new transcriptomes, seeds were obtained from the Missouri Botanical Garden, Millennium Seed Bank (Kew), California Botanic Garden, Florida Museum of Natural History, US Department of Agriculture, and Wake Forest. These seeds were planted at Matthaei Botanical Gardens. When enough tissue was available, young leaves, flowers, or buds were stored in liquid nitrogen at −80°C until RNA was isolated. Eight samples were collected from naturally occurring populations and stored in RNAlater™ until extraction. Some of the newly sequenced samples were prepared with the RNeasy Plant Mini Kit (Qiagen) and the TURBO DNA‐free kit (Thermo Scientific, Waltham, MA, USA) to isolate total RNA and to digest and remove DNA from the sample. Samples were quantified in 1% agarose gel and with a NanoDrop 1000 Spectrophotometer (Thermo Scientific). For newly sequenced samples, library preparation, rRNA depletion, and Illumina HiSeq 2 × 150 bp sequencing was conducted at either GENEWIZ NGS (South Plainfield, NJ, USA) or BGI Genomics (Shenzhen, China).
Homology and orthology inference
Hierarchical clustering based on Yang & Smith (2014) and Walker et al. (2018) was used for detecting homologues. First, an all‐by‐all Blast within user‐defined groups identified initial homologue clusters. User‐defined groups were: (1) outgroups, Balsaminaceae, Marcgraviaceae, Mitrastemonaceae, (2) Ebenaceae, Primulaceae, Sapotaceae (3) Polemoniaceae, (4) Lecythidaceae, (5) Diapensiaceae, Pentaphylacaceae, Sladeniaceae, Styracaceae, Theaceae, (6) Actinidiaceae, Clethraceae, Ericaceae, Roridulaceae, Sarraceniaceae. These groups are consistent with estimated relationships within Ericales from Larson et al. (2020). Sequences were then aligned in Mafft v.7.149 (Katoh et al. 2002; Katoh & Standley, 2013), before estimating initial homologue trees in RAxML v.8 (Stamatakis, 2014). Homologues in the user‐defined groups were then recursively combined, working towards the root of the tree. Homologous sequences were then realigned in Mafft before homologue trees were estimated in RAxML. These steps identified 10 325 homologues.
Orthologues were identified using the rooted tree (RT) method of Yang & Smith (2014). This method accounts for gene duplications and WGDs. Prunus persica (L.) Batsch (Rosaceae) was used as the outgroup. 1752 orthologues were identified.
Species tree estimation with transcriptomic data
Orthologous sequences were realigned in Mafft, and gene trees were re‐estimated in Iq‐Tree2 (Minh et al., 2020), with 100 bootstraps for each gene tree. Sladenia celastrifolia Kurz (Sladeniaceae) was resolved outside Ericales in every gene tree in which it was sampled; it was therefore pruned from all gene trees. Subsequently, any gene tree where Ericales was not monophyletic was removed, leaving a total of 1294 gene trees. Nodes with bootstrap values < 50 were collapsed before a species tree topology was estimated in Astral v5.7.8 (Mirarab et al., 2014).
Molecular branch lengths were then estimated. The 300 loci with gene trees that were most topologically congruent with the species tree, as determined by SortaDate (Smith et al., 2018a), were concatenated. This was the minimum number of loci required to incorporate all species. This concatenated alignment was analysed in RAxML with the topology constrained to the species tree topology and a separate GTR+GAMMA model for each locus.
Mapping gene duplications
Phyparts was used to calculate the number of gene duplications on each branch in the species tree (Smith et al., 2015) using the duplication command. Homologue trees for which the problematic sample for Sladenia celastrifolia was removed were used as the input alongside the species tree topology estimated in Astral.
Calculating synonymous distances (Ks distances) between paralogous gene pairs
Peaks in the distribution of Ks distances for a given species are indicative of WGDs. Ks distances were therefore calculated for each species as per Yang et al. (2015) with the Yang Nielsen method, and using the scripts presented here (https://github.com/tanghaibao/bio‐pipeline/tree/master/synonymous_calculation).
Sampling and sequencing of species‐level datasets
Specimens were sampled from the Missouri Botanical Garden, New York Botanical Garden, University of Michigan, and Florida Museum of Natural History herbaria as set out in Kates et al. (2021). DNA samples were also obtained from Peter Fritsch (Forth Worth Botanical Garden, BRIT). Capture‐Seq sequencing libraries enriched for gene regions targeted by the Angiosperms353 probe set (Johnson et al., 2019) were prepared and sequenced with various Illumina machines to generate either 150‐base pair (bp) or 250‐bp paired‐end reads. Nonenriched Illumina sequencing libraries were also prepared and sequenced as above to generate a whole genome shotgun (WGS) genome skimming sequence data set for each sample. Capture‐Seq and WGS reads were combined into a single pair of read files per sample. Paired reads were trimmed with Trimmomatic (Bolger et al., 2014) to trim adapters, bases with a PHRED score < 20, and discard reads < 50 bases. Reads that became unpaired because of trimming were combined into a single file per sample. Read files were analysed with HybPiper v.1.3.1 (Johnson et al., 2016) under default settings with custom target files designed for the 353 loci in asterids (McLay et al., 2021), collections of Ericales plastome genes, and ITS sequences.
Alignment and phylogenetic inference of Angiosperms353 and ITS data
For Angiosperms353 data, assembled amino acid sequences were analysed, whilst for ITS data nucleotide sequences were analysed. Data were subdivided into families, with family‐level alignments estimated in Mafft. Sequences with a high proportion of gaps in conserved regions (> 20% gaps at sites where fewer than a third of sequences had gaps) were removed before removing any site with > 90% gaps. Family alignments were then merged using ‐‐merge in Mafft. Families comprising a single sequence (often for Sladeniaceae or Mitrastemonaceae) and outgroup sequences (Curtisia dentata (Burm.f.) C.A.Sm. and Grubbia rosmarinifolia P.J.Bergius, Cornales) were then added using ‐‐add and ‐‐keeplength in Mafft. A further filtering step removed sites with > 75% gaps. Gene trees were then estimated in FastTree (Price et al., 2010) with the highest ranked amino acid substitution model for the Angiosperms353 data (with model comparison performed in Iq‐Tree‐2 (Minh et al., 2020)) and a GTR model for the ITS data.
Problematic samples were identified if (1) their corresponding terminal branch or any ancestral branch was abnormally long (defined as six times the mean branch length across each tree), and (2) they were placed outside the clade corresponding to their family in a high proportion of gene trees. Samples identified with criterion 1 were removed from their corresponding gene tree. Samples identified with criterion 2 were removed from every gene tree. For the Angiosperms353 data, sequences corresponding to the remaining samples in each gene tree were then realigned and merged, and a new set of gene trees was estimated as outlined above.
This second set of Angiosperms353 gene trees were pruned of individual samples occurring well outside the main clade for their family, and also pruned of remaining abnormally long branches. A further pruning step then involved retaining one accession for each species with the highest coverage across all genes. Nodes with support values < 0.5 were collapsed, before the 354 gene trees (Angiosperms353+ITS) were used for species tree estimation in ASTRAL.
Molecular branch lengths were then estimated. The 14 Angiosperms353 loci with gene trees that were most topologically congruent with the species tree, as determined by SortaDate, were concatenated with ITS. This concatenated alignment incorporated 99.9% of species in the species tree and was analysed in RAxML with the topology fixed to the species tree topology. The substitution model was partitioned by locus, with a GTR+GAMMA model for the ITS data and a JTT model for each Angiosperms353 locus (the optimal model for each locus).
Alignment and phylogenetic inference of chloroplast data
Alignment and gene‐tree estimation followed similar steps as for the ITS dataset. The resulting 65 gene trees (64 chloroplast gene trees+ITS) were used to estimate a species tree in Astral.
Topological incongruence between gene trees and the species tree was minimal. Therefore, to estimate molecular branch lengths, all loci that incorporated over 25% of tips in the species tree were concatenated. This concatenated alignment was analysed in RAxML, with the topology fixed to the species tree topology. A GTR+GAMMA model was used for each locus.
An additional phylogenetic tree was estimated with only chloroplast data using the same steps as above but excluding the ITS gene tree from the ASTRAL analysis. This species tree incorporated 3638 species, 870 fewer than the chloroplast+ITS tree. Most subsequent analyses use the chloroplast+ITS and Angiosperms353+ITS trees. Some additional analyses use the chloroplast‐only tree to determine the sensitivity of key findings to combining data from different genomes.
Divergence time estimation
Divergence times were estimated using treePL (Smith & O'Meara, 2012) with the estimated species trees. Fossil calibrations were based on a literature search of Ericales fossils in which systematic treatments explicitly described the characters used to assign fossils to clades. Fourteen fossils were used to define minimum age calibrations (Table 1). One maximum constraint of 109 Ma was used at the root node, corresponding to the upper bound for the age of this node in Magallón et al. (2015). A maximum of 125 Ma was trialled (see Zenodo), and had a minimal effect on age estimates within families but led to less stable results. A smoothing value of 1.0 was used. Alternative values had a minimal impact on age estimates.
Table 1.
Information on fossil calibrations used for estimating time‐calibrated phylogenetic trees for Ericales.
| Name | Stratum | Minimum Constraint age (Ma) | Organ | Node calibrated | Reference |
|---|---|---|---|---|---|
| Chrysophyllum tertiarum | Late Paleocene (Thanetian) | 56.0 | Leaf | Crown Sapotaceae | Mehrotra (2000) |
| Cleyera sp. | Middle Eocene | 47.8 | Seed | Stem Cleyera | Collinson et al. (2012) |
| Discoclethra maxima | Late Cretaceous (Maastrichtian) | 66.0 | Seed, fruit | Stem Clethraceae | Knobloch & Mai (1986) |
| Eurya sp. | Late Cretaceous (Santonian) | 83.6 | Seed | Stem Eurya | Knobloch & Mai (1986) |
| Gilisenium hueberi | Middle Eocene | 37.8 | Leaf, root, fruit | Crown Polemoniaceae | Lott et al. (1998) |
| Leucothoe praecox | Late Cretaceous (Maastrichtian) | 66.0 | Fruit | Stem Leucothoe (Ericaceae) | Knobloch & Mai (1986) |
| Pelliciera | Middle Eocene | 37.8 | Pollen | Stem Pelliciera (Tetrameristaceae) | Graham (1977) |
| Pentapetalum trifasciculandricus | Late Cretaceous (Turonian) | 89.8 | Flower | Stem Pentaphylacaceae | Martínez‐Millán (2010) |
| Unnamed Primuloid | Late Cretaceous (Maastrichian) | 66.0 | Flower | Crown Primulaceae | Friis et al. (2010) |
| Rhododendron newburyanum | Paleocene (Woolwich and Reading beds) | 56.0 | Seed | Stem Rhododendron (Ericaceae) | Collinson & Crane (1978) |
| Saurauia antiqua | Late Cretaceous (Santonian) | 83.5 | Seed | Crown Actinidiaceae | Knobloch & Mai (1986) |
| Schima kwangsiensis | Late Oligocene | 23.03 | Fruit | Stem Schima (Theaceae) | Shi et al. (2017) |
| Symplocos nooteboomii | Middle Eocene | 43.0 | Seed | Stem Symplocos (Symplocaceae) | Manchester (1994) |
| Vaccinium minutulum | Late Miocene | 5.33 | Seed | Stem Vaccinium (Ericaceae) | Łańcucka‐Środoniowa (1979) |
Analysis of bioclimatic niche evolution
Occurrence records were downloaded from BIEN and GBIF in March 2023. Records whose geocoordinates were either placed in the ocean or found outside the geographic regions recognised by the WCVP TDWG3 levels were excluded. This resulted in 4020 345 unique localities for Ericales species. Climate data for each record was then obtained from the WorldClim database.
Values that were three SD above or below the mean for each bioclimatic variable were removed. For each taxon, the mean value for each bioclimatic variable was then calculated, before a PCA was performed on the dataset. PC1 accounted for 78% of the variation in the data. Higher values on PC1 correspond to warmer, wetter, and less seasonal climates. The dataset incorporated bioclimatic information for 4271 species in the chloroplast+ITS phylogenetic tree and 2758 species in the Angiosperms353+ITS phylogenetic tree.
Two approaches were used for analysing bioclimatic niche evolution. First, the mean climatic preference of extant species in sister clades was compared. These comparisons were limited to cases where the ‘lower’ clade (the clade with a lower mean value on PC1) contained at least 40 species in the chloroplast+ITS phylogenetic tree, and 25 species in the Angiosperms353+ITS phylogenetic tree (roughly 1% of tips in each case). The analysis therefore focussed on identifying large climatic shifts associated with the origin of diverse clades with low values for PC1.
Alternatively, l1ou (Khabbazian et al., 2016) was used to reconstruct bioclimatic preferences. This method fits an Ornstein–Uhlenbeck (OU) model with multiple optima to phylogenetic trees, enabling estimation of the number, placement, and magnitude of shifts in bioclimatic preference (or optima). l1ou uses a modified version of the BIC (pBIC) for assessing configurations of shifts in OU optima whilst accounting for phylogenetic correlation and avoids overfitting the data (Khabbazian et al., 2016). It also performs a maximum likelihood search for the optimum α (rate of adaptation), exploring values ≥ 0. The search therefore incorporates models equivalent to Brownian motion. When performing analyses, the maximum number of shifts was set to 100. Compared to sister‐clade comparison, l1ou enables a more comprehensive assessment of shifts in bioclimatic preference across the phylogenetic tree, including a direct assessment of directionality. Nonetheless, unlike sister‐clade comparison, inferences depend on accurate divergence time estimates.
Analysis of life‐history evolution
Life history was categorised as tree, shrub, herb, epiphyte, mycoheterotroph, and climber. Data were primarily assembled from POWO (accessed November 2023 – Table S4). Not all categories are mutually exclusive, and therefore were simplified when plotted – species that were both epiphytes, mycoheterotrophs, or climbers, and also trees, shrubs, or herbs, were simply plotted as either epiphytes, mycoheterotrophs, or climbers.
Ancestral life‐history states were reconstructed. A model incorporating all six categories was not used because they are not mutually exclusive, and because of the computational burden of many categories in large phylogenetic trees. Therefore, life‐history reconstruction was limited to woody or herbaceous. Two models were tested as follows: an equal‐rates model assumed rates of transition between each state were equal, whilst an all‐rates‐different model assumed rates were unlinked. In each case, the models were tested with either the root state unknown, or the root state fixed to woody. The log‐likelihood and AIC values for these models were compared using the fitMK function in phytools (Revell, 2024). One‐hundred stochastic character maps were generated with the best fitting model using the make.simmap function from phytools (Revell, 2024).
Diversification rate estimation
Lineage‐specific speciation rates were estimated in Bamm (Rabosky, 2014). Despite previous criticism of Bamm (Moore et al., 2016), the method is reliable in most biological datasets (Rabosky et al., 2017). Further recent criticism of diversification rate estimation (Louca & Pennell, 2020) primarily applies to time‐heterogeneous models rather than lineage‐specific ones (Louca & Pennell, 2020; O'Meara & Beaulieu, 2021). Global and clade‐specific sampling fractions were used. The global sampling fraction corresponded to the proportion of sampled genera in each phylogenetic tree (0.92 for the chloroplast+ITS tree, and 0.82 for the Angiosperms353+ITS tree). The smallest unit over which clade‐specific sampling fractions were specified was individual genera. However, for nonmonophyletic genera, the sampling fraction unit was increased to incorporate all tips for any genus contained within it. For each sampling fraction unit, the sampling fraction was calculated by dividing the number of tips in each unit by the number of accepted species for genera incorporated in each unit (POWO, 2023). The setBAMMpriors function from BAMMtools (Rabosky et al., 2014) was used to define appropriate priors. Shift configurations with the highest posterior probabilities and credible shift sets incorporating 95% of the posterior probability were extracted, enabling uncertainty in the placement of rate shifts to be accounted for. The cor function from the stats R package (R Core Team, 2023) was used to test for a correlation between tip speciation rates and bioclimatic preference of extant species.
Results
Phylogenetic inference
The phylogenetic tree estimated from 1294 orthologues derived from transcriptomic data comprises 151 tips, 143 of which are Ericales species (Fig. 1). Relationships are broadly congruent with Larson et al. (2020): Balsaminaceae and Marcgraviaceae (i.e. the balsaminoid clade) are sister to all other families, and there is a core Ericales clade comprising Ericacaceae, Actinidiaceae, Sarraceniaceae, Theaceae, Styracaceae, Diapensiaceae, and Pentaphylacaceae (Fig. 1). Relationships among families within core Ericales are the same as Larson et al. (2020), although Clethraceae and Mitrastemonaceae are additionally incorporated in this study, and both are resolved in core Ericales. Relationships outside core Ericales differ somewhat from Larson et al. (2020): a clade comprising Primulaceae, Ebenaceae, and Polemoniaceae is recovered here, which contrasts to Larson et al. (2020) who resolve these families as a grade. Nonetheless, relationships outside core Ericales are congruent with a recent Ericales phylogenetic tree estimated from Angiosperms353 data (Larson et al., 2023). Overall, the phylogenetic signal among orthologues for branches defining relationships between families is highly conflicting, so no specific resolutions carry much confidence for classifications (Fig. 1). Conversely, most orthologues support the monophyly of each family.
Fig. 1.
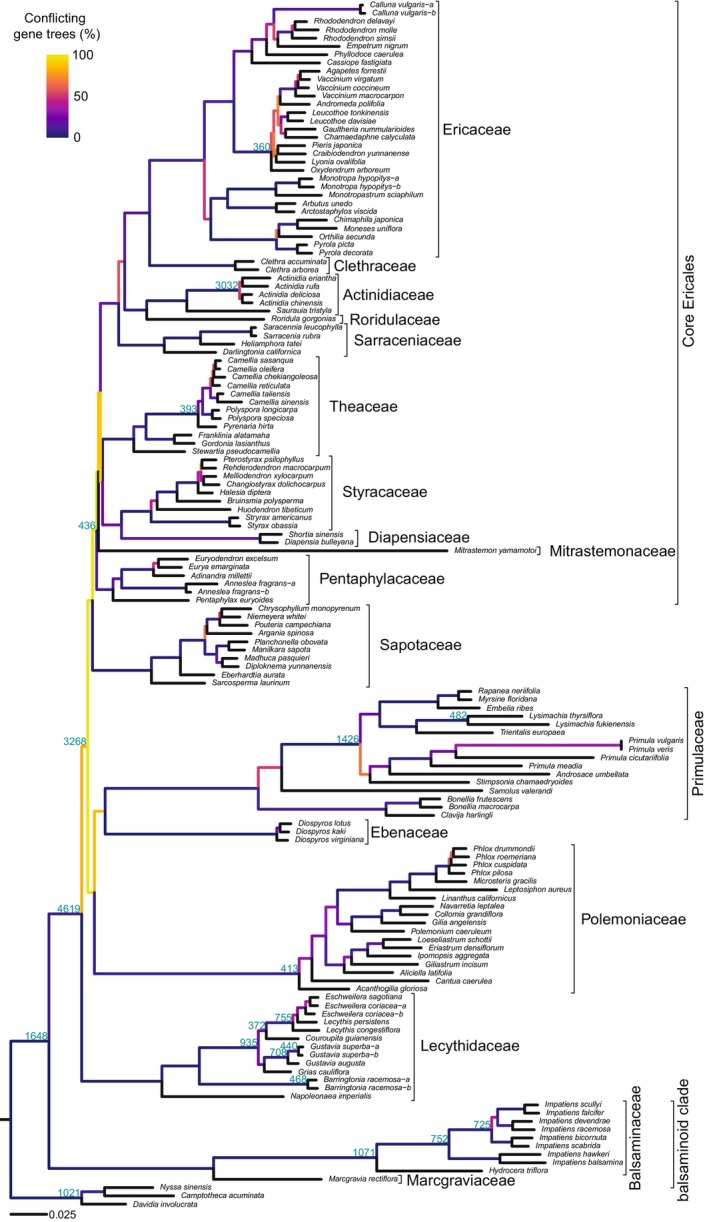
Phylogenetic tree for Ericales estimated from 1294 orthologues derived from transcriptomic data. Blue numbers show the number of gene duplications on each branch. Branch colours show the amount of gene‐tree conflict.
The two species‐level phylogenetic trees estimated from either chloroplast+ITS (Fig. 2) or Angiosperms353+ITS (Fig. S1) incorporate 4508 and 2870 Ericales species, respectively. The Angiosperms353+ITS phylogenetic tree is broadly congruent with the transcriptomic phylogenetic tree, although relationships differed slightly outside core Ericales, with Polemoniaceae no longer occurring in a clade with Primulaceae and Ebenaceae (Fig. S1). The chloroplast+ITS tree is also broadly congruent with these other trees (Fig. 2). However, Pentaphylacaceae is resolved outside core Ericales (Fig. 2), with c. 35% of chloroplast gene trees supporting this alternative placement.
Fig. 2.
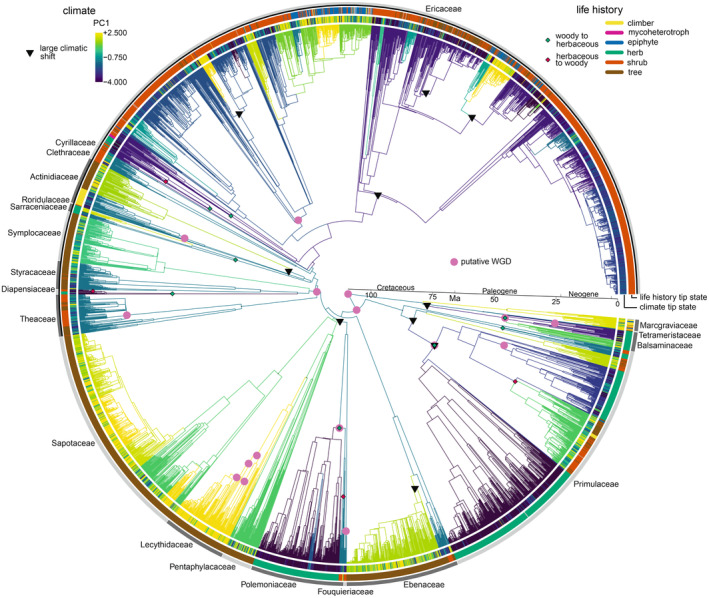
Time‐calibrated phylogenetic tree for Ericales estimated from chloroplast+ITS data and 14 fossil calibrations. Pink circles indicate putative WGDs (there is some uncertainty about the placement of these; see text). Black triangles are large climatic shifts estimated by sister‐clade comparison. Branch colours represent the climatic optima (on PC1) estimated by l1ou, and the observed climatic preference at the tips is shown on the innermost ring around the edge of the phylogenetic tree. These climatic variables correspond to the PC1 scale shown. Diamonds indicate estimated life‐history shifts: green are from woody to herbaceous, and red are from herbaceous to woody. The observed life‐history state at the tips is shown on the second ring around the edge of the phylogenetic tree, with colours corresponding to the key shown. The outermost ring represents the different families. The black line in this ring, which extends from Theaceae to Ericaceae, corresponds to core Ericales.
Divergence time estimation
Divergence times are relatively consistent in the chloroplast+ITS phylogenetic tree and Angiosperms353+ITS phylogenetic tree. Family crown node age estimates range from the late Cretaceous throughout the Tertiary, whilst family stem node age estimates are primarily late Cretaceous (Figs 2, S1, S2). The similar stem node age estimates for each family reflect the very short early branches in the phylogenetic trees (both the time‐calibrated and uncalibrated phylogenetic trees, Figs S3, S4), with major lineages arising over a very short timescale. Nonetheless, stem node ages are consistently older in the Angiosperms353+ITS phylogenetic tree than the chloroplast+ITS phylogenetic tree (Fig. S2). This discrepancy may result from larger effective population sizes for nuclear loci, leading to deeper coalescence times. The duration of family stem lineages varies dramatically, although the extreme length of some of these lineages is a striking feature of both dated phylogenetic trees (Figs 2, S1, S2). Further, some families with comparatively short stem lineages (e.g. Clethraceae, Styracaceae) have very long stem lineages for major intra‐familial clades (Figs 2, S1, S2).
The calibrations at the stem nodes of Cleyera, Eurya, Leucothoe, and Schima; and crown nodes of Actinidiaceae and Sapotaceae (Table 1) are likely to be especially influential. Age estimates for these nodes are < 1 million years (Myr) older than the minimum imposed by the calibration, suggesting a revision of the placement of relevant fossils would likely affect age estimates, at least at these nodes. Nevertheless, re‐running analyses in the absence of these five calibrations did not markedly affect the overall shape of the phylogenetic tree, with short early branches and long family stem branches being retained (see Zenodo). Caution in interpreting these divergence time estimates should also stem from the markedly different substitution rates that likely exist among clades with life histories ranging from trees to herbaceous annuals. Although partially accounted for by treePL, dramatic substitution rate differences may undermine divergence time estimates regardless.
Whole genome duplications
Many gene duplications are mapped onto early branches, including the Ericales stem branch, the branch leading to the clade comprising all families excluding the balsaminoid clade, the branch leading to the clade comprising all families excluding the balsaminoid clade and Lecythidaceae, and the stem branch of core Ericales (Fig. 1). Many gene duplications are also mapped onto more recent branches, including the stem branches of Balsaminaceae, Polemoniaceae, and Actinidia (Actinidiaceae), and clades within Primulaceae, Lecythidaceae, and Theaceae (Fig. 1).
Previous studies, including whole genome analyses, suggest many of these gene duplications result from whole genome duplication (WGD). Several studies have highlighted that a WGD occurred on the short early branches (Shi et al., 2010; Xia et al., 2017; Wei et al., 2018), whilst Leebens‐Mack et al. (2019) potentially detected two WGDs on these branches. The results of Leebens‐Mack et al. (2019) also suggest the many duplicated genes at the origins of Balsaminaceae and Actinidia result from WGD, whilst the results of Shi et al. (2010), Xia et al. (2017), and Wei et al. (2018), suggest WGDs are the cause of the many duplicated genes at the origin of Actinidia, and Camellia+Polyspora (Theaceae).
There are two prominent peaks in the Ks plots at c. 4 and 2 (Fig. S5). These peaks occur for every species, including outgroups, implying WGDs that are ancestral to Ericales. There is also a peak at c. 0.5 for all Ericales species (Fig. S5) that likely corresponds to the putative WGD along the stem branch of the order. A further peak at c. 0.25 is consistently observed in all families aside from the balsaminoid clade and Lecythidaceae and may correspond to an additional WGD on the short early branches.
Chromosome numbers vary considerably within families, and there is little association between chromosome number and the number of duplicated genes or Ks plots (Fig. S5; Table S3). This lack of association is likely due to the phylogenetic trees covering timescales of 10s of Myr, and the potential loss of chromosomes following ancient WGDs over such timescales. Nevertheless, Fouquieriaceae has a relatively young crown age (c. 14 Myr), and a consistently higher chromosome count than its sister family Polemoniaceae (Table S3), suggesting a WGD may have occurred at the origin of Fouquieriaceae. However, no transcriptomic data were analysed for this family. Likewise, Gustavia (Lecythidaceae) generally has high‐chromosome counts (Table S3). Many duplicated genes are also recovered around the origin of Gustavia (Fig. 1), thus supporting the existence of a WGD at or near its origin.
These different lines of evidence suggest at least one WGD has occurred on the short early branches, additional WGDs are associated with the origin of Actinidia, Balsaminaceae, and Camellia+Polyspora (Theaceae), and at least one further WGD has occurred within Lecythidaceae. It is difficult to determine the exact number and placement of WGDs on the short early branches because of the uncertain branching order in this part of the species tree and the limited taxon sampling of previous studies that inferred putative WGDs on these branches. There are also several other branches where the estimated number of duplicated genes is even greater than on the branches discussed here, where WGDs are hypothesised based on multiple lines of evidence (Fig. 1). Additional WGDs are therefore also likely to have occurred on these branches, although additional analyses are required to characterise them.
Based on the aforementioned evidence, putative WGDs are plotted in Figs 2 and S1. Given the difficulties in placing some putative WGDs, neither the placement nor the number of putative WGDs should be overinterpreted, and they are not intended to conflict with earlier studies directly. For example, Zhang et al. (2022) suggest 2 WGDs occurred during the evolutionary history of Theaceae, rather than the four implied here. Instead, the findings here highlight that the short early branches are characterised by unusual molecular evolutionary phenomena that warrant further investigation.
Gene‐tree conflict
Conflict between gene tree and species tree topologies in the transcriptome and Angiosperms353+ITS datasets is highest on the shortest species tree branches (Figs 1, S4). These are early branches in the phylogenetic trees and recent divergences within families. Conflict is relatively lower around the crown nodes of families and longer branches defining relationships within and between families.
Climatic niche evolution, WGDs, and gene‐tree conflict
Five of the ten largest climatic shifts identified by sister‐clade comparison are directly associated with putative WGDs (Figs 2, 3, S1, S6–S8; Tables S5–S7). There would be a 3.6e‐3 probability of sampling a set of nodes as phylogenetically close to putative WGDs by chance alone (i.e. if five nodes satisfying the conditions for inclusion in the sister‐clade comparisons were randomly sampled, there would be a 3.6e‐3 probability that such nodes would be as phylogenetically close to the putative WGDs). These shifts occur near the origin of Balsaminaceae, Primulaceae, Polemoniaceae, and Actinidia (Figs 3, S1, S6–S8; Tables S5–S7). The shifts in Balsaminaceae, Primulaceae, and Polemoniaceae (Fig. 3a–c) represent a switch between the higher and lower peaks for the frequency of climatic preferences accross all Ericales species on PC1 (Fig. 3e; Tables S5–S7). Across these five examples, species in the lower clade constitute over 60% of species with a climatic preference equal to or lower than the lower peak on PC1 (Fig. 3e), and 98% of such species outside Ericaceae.
Fig. 3.
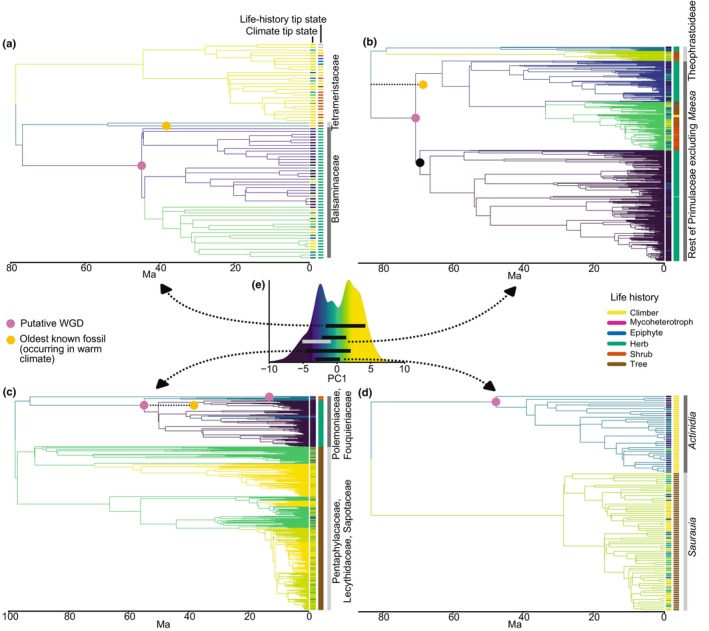
Summary of large climatic shifts down PC1 in Ericales identified by sister‐clade comparison, and their relationship with putative WGDs. In (a–d) branch colours correspond to the colour scale in (e) (and that of Fig. 2) and represent the climatic optima (on PC1), as estimated by l1ou. The observed climatic preference of extant species is shown in the first bar to the right of each clade according to the same colour scale. The observed life‐history state for extant species is shown in the second bar to the right of each clade according to the key shown. The taxa corresponding to each sister‐clade pair are labelled in the third column. Putative WGDs are shown as pink circles. The oldest known fossils in each clade estimated to represent plants that grew in warm climates are indicated by orange circles – these are plotted at their minimum possible age based on stratigraphic assignment. (e) Density plot of climatic preferences for all Ericales species for which climate data was recovered. The black bars indicate the difference in mean climatic preference for extant species in each sister‐clade pair, that is, each end of the bar represents the mean climatic preference of one half of the sister pair. The arrows indicate which sister‐clade pair the bar corresponds to. Note the additional grey bar in (e) that is associated with (b). This bar corresponds to an additional large climatic shift within Primulaceae at the node marked with the black circle in (b).
In these five examples, the WGD occurs near the origin of the clade that is lower on PC1, implying the WGD is associated with the shift down PC1. For Balsaminaceae, Primulaceae, and Polemoniaceae (Fig. 3a–c), this hypothesised directionality from high to low is supported by the l1ou analysis, where shifts down PC1 are estimated near the origin of these clades (Figs S9–S11; Tables S8–S10). The fossil record also supports this hypothesised directionality. The oldest known fossils of Polemoniaceae and a close fossil relative of Balsaminaceae likely represent plants that grew in warm climates in the early to mid‐Eocene given the large fossilised reptiles in associated sediments (Graham, 1977; Lott et al., 1998; Donovan, 2002). Early Primulaceae fossils from Cretaeceous sediments in the Iberian Peninsula are also likely to represent plants that grew in warm climates. However, these climates were arid and fire‐prone (Friis et al., 2010).
Nonetheless, these five examples have unique characteristics (Figs S9, S10). Whilst all are associated with lower temperatures and increased temperature seasonality, the Actinidia shift (Fig. 3d) is characterised by a notably less pronounced decrease in mean annual temperature. It is also the only example where there is no decrease in the mean temperature of the warmest quarter (Fig. S9). This shift (as well as the Balsaminaceae shift) instead shows a pronounced increase in temperature seasonality (Figs S9, S10). Meanwhile, the Polemoniaceae shift is especially characterised by decreased precipitation (Figs S9, S10).
Most of the remaining large climatic shifts identified by sister‐clade comparison (i.e. those not associated with WGDs) occur within Ericaceae (Figs S6–S8; Tables S5–S7). These appear to be shifts from low to high values on PC1 because they involve clades comprising species with high values nested inside large clades with low values, and the l1ou analysis estimated shifts up PC1 near the origin of these clades. Some of these clades with high values for PC1 are geographically restricted, such as two clades within Gaultheria and Rhododendron that primarily consist of Indonesian and Philippine endemics but which are nested within clades occurring widely throughout the Northern Hemisphere (Figs S6–S8; Tables S5–S7). One further large shift identified by sister‐clade comparison, which likely represents a shift down PC1 (Figs S6–S8; Tables S5–S7), occurs near the origin of a clade comprising Euclea and some species of Diospyros (Ebenaceae) that is restricted to arid habitats in southern and south‐eastern Africa. Unlike the probable shifts down PC1 in Fig. 3(a–d), this shift is not a transition between the two peaks on PC1 shown in Fig. 3(e) (Figs S6–S8; Tables S5–S7).
Beyond these large climatic shifts, the l1ou analysis estimated a total of 37 shifts in the chloroplast+ITS phylogenetic tree and 32 shifts in the Angiosperms353+ITS phylogenetic tree (parameter estimates and pBIC scores for competing models are available on Zenodo). In both phylogenetic trees, additional shifts to lower values on PC1 that are not included in the examples above are typically small (Figs S11–S13; Tables S8–S10). Two exceptions are shifts down PC1 near the crown nodes of Diapensiaceae and Ericaceae. Neither is a transition between the two peaks on PC1 shown in Fig. 3(e) (Tables S8–S10; Figs S11–S13) nor directly associated with a putative WGD. There may nonetheless have been a WGD at the crown node of core Ericales (given the duplicated genes mapped to this node (Fig. 1)), which is relatively close to the Ericaceae crown node.
Climatic shifts and gene‐tree conflict have a much less direct link than climatic shifts and WGDs. Gene‐tree conflict concentrated around short early branches (Figs 1, S4) can be interpreted as no more than a precursor to subsequent climatic shifts in descendant lineages. Likewise, gene‐tree conflict around more recent divergences postdates most of the significant climatic shifts estimated here.
Climatic niche evolution and life‐history shifts
The largest climatic shifts down PC1 identified by sister‐clade comparison are each associated with life‐history shifts, which are defined as when the majority sampled life‐history state differs between an ancestral and descendant node (Figs 3, S14, S15, model comparison and parameter estimates available on Zenodo). In Balsaminaceae, Primulaceae, and Polemoniaceae, a woody to herbaceous shift is estimated (Figs S14, S15). In Actinidiaceae, there appears to be a shift from trees to climbers (Fig. 3), although this life‐history shift could not be explicitly estimated. A shift from woody to herbaceous is also estimated at the crown node of Diapensiaceae, where l1ou estimated a large climatic shift down PC1. At least two large climatic shifts up PC1 identified within Ericaceae are associated with shifting from shrubs to epiphytes (Figs 2, S1).
Climatic niche evolution and diversification rates
The credible shift set contained 59–89 shifts in the chloroplast+ITS phylogenetic tree and 31–58 shifts in the Angiosperms353+ITS phylogenetic tree. Rate shifts are distributed throughout Ericales, and show little association with climatic shifts, except around the crown node of Balsaminaceae (Figs S16–19). There was little evidence of a relationship between tip speciation rates and climatic preference, with the chloroplast+ITS and Angiosperms353+ITS trees showing weak positive and negative correlations between these variables, respectively (Figs S20, 21). The existence of rapidly diversifying clades that are both high (e.g. within Sapotaceae) and low (e.g. Primulaceae) on PC1 (Figs S16, 17) and slight differences in sampling between the two trees likely explains this slight difference in correlation.
Discussion
The phylogeny of Ericales is characterised by extremely short early branches that appear to underpin the explosive origin of numerous species rich clades. These clades have undergone repeated large‐scale shifts in climatic preference (Figs 2, 3). The five largest shifts into colder, arid, or more seasonal climates are directly associated with WGDs and often with a woody to herbaceous transition (Figs 2, 3). The short early branches are associated with at least one additional WGD that may have facilitated climatic shifts in descendant lineages. These early branches also have very high gene‐tree conflict, suggesting several interacting processes (WGDs, demographic shifts, hybridisation, and assortment of ancestral polymorphisms among descendant lineages) are associated with the rapid establishment of climatically specialised lineages within Ericales. In contrast to these tight links between WGDs, climatic shifts, and life‐history shifts, and more indirect links between gene‐tree conflict and climatic shifts, more recent shifts in diversification rate appear to have little association with climatic preference. Nevertheless, the extremely short internodes among early branches within Ericales and their association with evolutionary innovation warrants further investigation as to the patterns and processes underpinning major evolutionary events within angiosperms (Parins‐Fukuchi et al., 2021; Stull et al., 2021).
Climatic shifts, whole genome duplication, and life‐history shifts are linked
WGDs are tightly linked to climatic shifts into colder, seasonal, or arid climates, and diversification within these climates over 10s of Myr. Analyses of other groups have also highlighted a close relationship between WGDs and climatic shifts (Rice et al., 2019; Smith et al., 2018b; Wang et al., 2019; Yang et al., 2018), and the importance of WGDs for promoting evolutionary innovation more generally (Soltis & Soltis, 2016; Stull et al., 2021; Van de Peer et al., 2021).
Different mechanisms could explain this link between WGDs and climatic shifts. First, WGDs may facilitate the origin of traits that enable lineages to establish in new climates. Evidence in support of this can be drawn from the several studies that demonstrate how lineages with WGDs exhibit significant differences in environmentally relevant traits such as phenology, transpiration, or water use efficiency (Maherali et al., 2009; Soltis & Soltis, 2016; Van de Peer, 2021). By duplicating genes and altering regulatory networks (e.g. Brockington et al., 2015; Wang et al., 2019; Feng et al., 2024), WGDs can increase the expression of genes that underpin these critical traits, or lead to redundancy within duplicated genomes, facilitating sub/neo‐functionalisation and thus the propensity of a lineage to evolve these traits (McGrath & Lynch, 2012; Van de Peer, 2021).
Alternatively, new environments could increase the frequency of unreduced gametes and thus WGDs (De Storme et al., 2012; Van de Peer et al., 2021), or trigger life‐history shifts that promote the survival of polyploid lineages (Rice et al., 2019). Importantly, these alternatives (i.e. WGDs promoting climatic shifts or climatic shifts increasing the frequency of WGDs and survival of descendant lineages) are not mutually exclusive; whilst stressful environments may trigger WGDs, this does not preclude the WGDs from promoting survival of descendant lineages in novel environments.
In Ericales, the WGD(s) on the short early branches significantly predates reconstructed climatic shifts within the order (Fig. 2) and global cooling and aridification that occurred throughout the Cenozoic (Zachos et al., 2001; Herbert et al., 2016). It is, therefore, reasonable to interpret this WGD as a precursor that may have enabled the origin of diverse traits in descendant lineages, including those that enable survival in diverse climates. In contrast, for climatic shifts associated with Actinidia, Balsaminaceae, Primulaceae, and Polemoniaceae+Fouquieriaceae, the nature of the relationship between WGDs and climatic shifts is less clear. WGDs occur on the same or very similar branches to these large climatic shifts, consistent with either of the scenarios outlined above.
WGDs and climatic shifts are also closely linked to life‐history shifts in Ericales. Therefore, new adaptive traits underpinning shifts in climatic preference within Ericales are likely to be associated with life‐history characteristics (Figs 2, 3). The large climatic shifts in Balsaminaceae, Primulaceae, and Polemoniaceae are associated with shifts to herbaceousness, whilst for Actinidia, there is a shift to woody climbers (Fig. 3). The differences between these life‐history shifts may reflect differences between the climatic shifts occurring in these clades (Figs S9, S10). For example, the climatic shift associated with Actinidia is unique in that it is not linked to any reduction in maximum temperature but is instead linked to increased seasonality.
WGDs may have enabled these life‐history shifts. However, it has also been suggested that life‐history shifts, specifically from trees to herbaceous perennials, may facilitate the survival of lineages with ancestral WGDs (Rice et al., 2019). This may have played a role in causing the association between life‐history shifts and WGDs in Ericales, especially given the preponderance of tropical tree species within the order, for which respective families do not have WGDs near their origins (unlike the examples in Fig. 3). Nonetheless, the relevance of this explanation in Polemoniaceae at least is likely to be limited given the significant proportion of annual species within this family (POWO, 2023).
An indirect link between climatic shifts and gene‐tree conflict
Although gene‐tree conflict has no direct association with any large climatic shift, its concentration around early branches suggests indirect links between conflict and climatic shifts. Conflict is known to be associated with evolutionary innovation (Parins‐Fukuchi et al., 2021; Stull et al., 2021), potentially because it is indicative of dynamic population processes and the reassortment of ancestral polymorphisms among descendant lineages (Parins‐Fukuchi, 2023). Therefore, alongside WGDs (and potentially the horizontal transfer of organellar DNA given the placement of Pentaphylacaceae in the chloroplast phylogenetic tree), gene‐tree conflict (or its underlying causes) may have played a role in enabling descendant lineages to specialise to diverse climates. Additional insights into how these processes have operated in Ericales will require analysis of the function of the genes that have undergone duplication in Ericales and how they have been sorted among descendant lineages.
Climatic shifts and diversification rates are unlinked
The lack of association between climatic shifts (and consequently WGDs) and diversification rate shifts is unsurprising given the lack of direct association between conflict and climatic shifts, but previously identified associations between conflict and diversification rate (Zuntini et al., 2024). Nonetheless, this finding does differ from a recent analysis of gymnosperms (Stull et al., 2021), where increased rates of climatic niche shifts were associated with diversification rate shifts. This difference could reflect general differences between gymnosperms and angiosperms, or more specific characteristics of Ericales. We suggest the latter explanation plays at least some role because (1) the long family stem lineages presumably indicate unusually high species turnover within Ericales, and (2) WGDs have been linked to diversification rate shifts in several other angiosperm lineages (Soltis & Soltis, 2016). If WGDs were associated with diversification rate shifts within Ericales, an association between climatic shifts and diversification rates would also be expected.
The lack of association between tip speciation rates and climatic preferences is more surprising. Numerous recent studies have found that temperate angiosperm lineages diversify faster than their tropical counterparts, especially in recent times (Folk et al., 2019; Sun et al., 2020; Tietje et al., 2022; Dimitrov et al., 2023). This pattern is typically explained by the younger age and climatic dynamism in temperate regions leading to less saturated niche space compared to the relatively stable tropics (Wiens & Donoghue, 2004; Zanne et al., 2014; Folk et al., 2019; Sun et al., 2020). Further research is therefore needed to understand the drivers of recent and rapid diversification of tropical clades in Ericales.
Competing interests
None declared.
Author contributions
Conceptualisation: TC and SAS. Analysis: TC, SAS and DJPG. Investigation: TC, SAS and DJPG. Writing – Original Draft: TC. Writing – Review & Editing: TC, DJPG, PL, ASC, CWD, DAL, DES, PWF, PSS, WNW and SAS. Funding acquisition: CWD, DES, PSS and SAS. Resources: DJPG, PL, ASC, CWD, DAL, DES, PWF, PSS, WNW and SAS.
Supporting information
Fig. S1 The Angiosperms353+ITS time‐calibrated phylogenetic tree summarising climatic shifts, life‐history shifts, and putative WGDs.
Fig. S2 Age estimates for different families in the chloroplast+ITS time‐calibrated phylogenetic tree compared to the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Fig. S3 The chloroplast+ITS species tree. Branch lengths represent substitutions per site.
Fig. S4 The Angiosperms353+ITS species tree. Branch lengths represent substitutions per site, branch colours represent gene‐tree conflict.
Fig. S5 Single species KS plots for species incorporated into the transcriptomic dataset.
Fig. S6 The chloroplast+ITS time‐calibrated phylogenetic tree summarising climatic shifts and putative WGDs.
Fig. S7 The Angiosperms353+ITS time‐calibrated phylogenetic tree summarising climatic shifts and putative WGDs.
Fig. S8 The chloroplast‐only time‐calibrated phylogenetic tree summarising climatic shifts and putative WGDs.
Fig. S9 Further information on the composition of the largest shifts down PC1 as identified by sister‐clade comparison in the chloroplast+ITS time‐calibrated phylogenetic tree and which are associated with putative WGDs.
Fig. S10 Further information on the composition of the largest shifts down PC1 as identified by sister‐clade comparison in the Angiosperms353+ITS time‐calibrated phylogenetic tree and which are associated with putative WGDs.
Fig. S11 Shifts down PC1 estimated by l1ou on the chloroplast+ITS time‐calibrated phylogenetic tree, and the largest 10 differences in climatic preference between sister clades.
Fig. S12 Shifts down PC1 estimated by l1ou on the Angiosperms353+ITS time‐calibrated phylogenetic tree, and the largest 10 differences in climatic preference between sister clades.
Fig. S13 Shifts down PC1 estimated by l1ou on the chloroplast‐only time‐calibrated phylogenetic tree, and the largest 10 differences in climatic preference between sister clades.
Fig. S14 The chloroplast+ITS time‐calibrated phylogenetic tree with life‐history shifts, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S15 The Angiosperms353+ITS time‐calibrated phylogenetic tree with life‐history shifts, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S16 The chloroplast+ITS time‐calibrated phylogenetic tree with estimated speciation rates, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S17 The Angiosperms353+ITS time‐calibrated phylogenetic tree with estimated speciation rates, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S18 Subsample of 9 trees from the credible shift set of speciation rates estimated with BAMM on the chloroplast+ITS time‐calibrated phylogenetic tree.
Fig. S19 Subsample of 9 trees from the credible shift set of speciation rates estimated with BAMM on the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Fig. S20 Climatic preference of extant species on PC1 plotted against tip speciation rates, from the chloroplast+ITS time‐calibrated phylogenetic tree.
Fig. S21 Climatic preference of extant species on PC1 plotted against tip speciation rates, from the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Table S1 Voucher information for transcriptome dataset.
Table S2 Voucher information for species‐level molecular datasets.
Table S3 Chromosome number data.
Table S4 Life‐history information.
Table S5 Largest sister‐clade differences in climatic preference in the chloroplast+ITS time‐calibrated phylogenetic tree.
Table S6 Largest sister‐clade differences in climatic preference in the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Table S7 Largest sister‐clade differences in climatic preference in the chloroplast‐only time‐calibrated phylogenetic tree.
Table S8 Summary of all shifts down PC1 identified by l1ou in the chloroplast+ITS time‐calibrated phylogenetic tree.
Table S9 Summary of all shifts down PC1 identified by l1ou in the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Table S10 Summary of all shifts down PC1 identified by l1ou in the chloroplast‐only time‐calibrated phylogenetic tree.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank the Smith lab for helpful and insightful discussions. Specific thanks also to James Boyko for recommendations about the types of comparative analyses that may be helpful and Keyi Feng for discussing some details in the analysis of the transcriptomic data. TC, SAS, CWD, DJPG, and DL were supported by NSF DEB 1917146. ASC, DES, and PSS were supported by NSF DEB 1916555.
Contributor Information
Tom Carruthers, Email: tomcarr@umich.edu.
Stephen A. Smith, Email: eebsmith@umich.edu.
Data availability
New sequence data is available on the NCBI Sequence Read Archive (PRJNA1076884, PRJNA839108). Supporting Information Tables S1 and S2 provide voucher information. Assembled sequences, target files, analysis outputs, and scripts are available on Zenodo: https://zenodo.org/records/13835677.
References
- APG IV . 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. [Google Scholar]
- Bartish IV, Antonelli A, Richardson JE, Swenson U. 2010. Vicariance of long‐distance dispersal: historical biogeography of the pantropical subfamily Chrysophylloideae (Sapotaceae). Journal of Biogeography 38: 177–190. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher FC, Zimmerman NE, Conti E. 2016. Allopatric speciation with little niche divergence is common among alpine Primulaceae. Journal of Biogeography 43: 591–602. [Google Scholar]
- Brockington SF, Yang Y, Gandia‐Herrero F, Covshoff S, Hibberd JM, Sage RF, Wong GKS, Moore MJ, Smith SA. 2015. Lineage‐specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytologist 207: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson ME, Crane PR. 1978. Rhododendron seeds from the Palaeocene of southern England. Botanical Journal of the Linnean Society 76: 195–205. [Google Scholar]
- Collinson ME, Manchester SR, Wilde V. 2012. Fossil fruits and seeds of the middle Eocene Messel biota, Germany. Abhandlungen der Senckenbergische Naturforschende Gesellschaft 570: 1–249. [Google Scholar]
- Crisp MD, Arroyo MTK, Cook LG, Gandolfo MA, Jordan GJ, McGlone MS, Weston PH, Westoby M, Wilf P, Linder HP. 2009. Phylogenetic biome conservatism on a global scale. Nature 458: 754–756. [DOI] [PubMed] [Google Scholar]
- De Storme N, Copenhaver GP, Geelen D. 2012. Production of diploid male gametes in Arabidopsis by cold‐induced destabilization of postmeiotic radial microtubule arrays. Plant Physiology 160: 1808–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D, Xu X, Su X, Shrestha N, Liu Y, Kennedy JD, Lyu L, Nogués‐Bravo D, Rosindell J, Yang Y et al. 2023. Diversification of flowering plants in space and time. Nature Communications 14: 7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MJ. 2008. A phylogenetic perspective on the distribution of plant diversity. Proceedings of the National Academy of Sciences, USA 105: 11549–11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S. 2002. Field guide to the geology of the Eocene Chapelton Formation (Yellow Limestone Group), western Central Inlier. Caribbean Journal of Earth Sciences 35: 39–47. [Google Scholar]
- Eggli U, Nyffler R. 2009. Living under temporarily arid conditions – succulence as an adaptive strategy. Bradleya 2009: 13–26. [Google Scholar]
- Favre A, Michalak I, Chen CH, Wang JC, Pringle JS, Matuszak S, Sun H, Yuan YM, Struwe L, Muellner‐Riehl AN. 2016. Out‐of‐Tibet: the spatio‐temporal evolution of Gentiana (Gentianaceae). Journal of Biogeography 43: 1967–1978. [Google Scholar]
- Feng K, Walker JF, Marx HE, Yang Y, Brockington SF, Moore MJ, Rabeler RK, Smith SA. 2024. The link between ancient whole‐genome duplications and cold adaptations in the Caryophyllaceae. American Journal of Botany 111: e16350. [DOI] [PubMed] [Google Scholar]
- Folk RA, Siniscalchi CM, Soltis DE. 2020. Angiosperms at the edge: extremity, diversity, and phylogeny. Plant, Cell & Environment 43: 2871–2893. [DOI] [PubMed] [Google Scholar]
- Folk RA, Stubbs RL, Mort ME, Cellinese N, Allen J, Soltis PS, Soltis DE, Guralnick RP. 2019. Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proceedings of the National Academy of Sciences, USA 116: 10874–10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis EM, Pedersen KR, Crane PR. 2010. Cretaceous diversification of angiosperms in the Iberian Peninsula. Review of Palaeobotany and Palynology 162: 341–361. [Google Scholar]
- Graham A. 1977. New records of Pelliceria (Theaceae/Pelliceriaceae) in the tertiary of the Caribbean. Biotropica 9: 48–52. [Google Scholar]
- Griffith M, Yaish MF. 2004. Antifreeze proteins in overwintering plants: a tale of two activities. Trends in Plant Science 9: 399–405. [DOI] [PubMed] [Google Scholar]
- Herbert TD, Lawrence KT, Tzanova A, Cleaveland Peterson L, Caballero‐Gill R, Kelly CS. 2016. Late Miocene global cooling and the rise of modern eco‐systems. Nature Geoscience 9: 843–847. [Google Scholar]
- Hughes CE, Eastwood R. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences, USA 103: 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Pokorny L, Dodsworth S, Botigue LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT et al. 2019. A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k‐medoids clustering. Systematic Biology 68: 594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ. 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high‐throughput sequencing reads using target enrichment. Applications in Plant Sciences 4: apps.1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates HR, Doby JR, Siniscalchi CM, LaFrance R, Soltis DE, Soltis PS, Guralnick RP, Folk RA. 2021. The effects of herbarium specimen characteristics on short‐read NGS sequencing success in nearly 8000 specimens: old, degraded samples have lower DNA yields but consistent sequencing success. Frontiers in Plant Science 12: 669064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. Mafft: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software v.7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabbazian M, Kriebel R, Rohe K, Ané C. 2016. Fast and accurate detection of evolutionary shifts in Ornstein–Uhlenbeck models. Methods in Ecology and Evolution 7: 811–824. [Google Scholar]
- Knobloch E, Mai DH. 1986. Monographie der Früchteund Samen in der Kreide von Mitteleuropa. Rozpravy ústredního ústava geologickénho, Praha 47: 1–219. [Google Scholar]
- Łańcucka‐Środoniowa M. 1979. Macroscopic plant remains from the freshwater Miocene of the Nowy Sacz basin (west Carpathians, Poland). Acta Palaeobotanica 20: 3–117. [Google Scholar]
- Larson D, Chanderbali AS, Maurin O, Gonçalves DJP, Dick CW, Soltis DE, Soltis PS, Fritsch PW, Clarkson JJ, Grall A. 2023. The phylogeny and global biogeography of Primulaceae based on high‐throughput DNA sequence data. Molecular Phylogenetics and Evolution 182: 107702. [DOI] [PubMed] [Google Scholar]
- Larson D, Walker JF, Vargas OM, Smith SA. 2020. A consensus phylogenomic approach highlights palaeopolyploid and rapid radiation in the history of Ericales. American Journal of Botany 107: 773–789. [DOI] [PubMed] [Google Scholar]
- Leebens‐Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S et al. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott TA, Manchester SR, Dilcher DL. 1998. A unique and complete polemoniaceous plant from the middle Eocene of Utah, USA. Review of Palaeobotany and Palynology 104: 39–49. [Google Scholar]
- Louca S, Pennell MW. 2020. Extant timetrees are consistent with a myriad of diversification histories. Nature 580: 502–505. [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez‐Acevedo S, Sánchez‐Reyes LL, Hernández‐Hernández T. 2015. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Manchester SR. 1994. Fruits and seeds of the Middle Eocene nut beds flora, Clarno formation, Oregon. Palaeontographica Americana 58: 1–205. [Google Scholar]
- Martínez‐Millán M. 2010. Fossil record and the age of Asteridae. Botanical Review 76: 83–135. [Google Scholar]
- McGrath CL, Lynch M. 2012. Evolutionary significance of whole‐genome duplication. In: Soltis P, Soltis D, eds. Polyploidy and genome evolution. Berlin, Heidelberg, Germany: Springer. [Google Scholar]
- McLay TGB, Birch JL, Gunn BF, Ning W, Tate JA, Nauheimer L, Joyce EM, Simpson L, Schmidt‐Lebuhn AN, Baker WJ et al. 2021. New targets acquired: improving locus recovery from the Angiosperms353 probe set. Applications in Plant Sciences 9: e11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra RC. 2000. Study of plant megafossils from the Tura Formation of Nangwalibra, Garo Hills, Meghalaya, India. The Palaeobotanist 49: 237–255. [Google Scholar]
- Mendes FK, Hahn MW. 2016. Gene tree discordance causes apparent substitution rate variation. Systematic Biology 65: 711–721. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. Iq‐Tree 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. 2014. Astral: genome‐scale coalescent‐based species tree estimation. Bioinformatics 30: i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. 2016. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proceedings of the National Academy of Sciences, USA 113: 9569–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon KC, Crepet WL. 1993. Late Cretaceous fossil flowers of Ericalean affinity. American Journal of Botany 80: 616–623. [PubMed] [Google Scholar]
- O'Meara B, Beaulieu J. 2021. Potential survival of some, but not all, diversification methods. EcoEvoRxiv. doi: 10.32942/osf.io/w5nvd. [DOI]
- Parins‐Fukuchi C, Stull GW, Smith SA. 2021. Phylogenomic conflict coincides with rapid morphological innovation. Proceedings of the National Academy of Sciences, USA 118: e2023058118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parins‐Fukuchi CT. 2023. Sorting of persistent morphological polymorphisms links palaeobiological pattern to population process. Palaeobiology 50: 1–12. [Google Scholar]
- POWO . 2023. Plants of the world online . Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet. [WWW document] URL https://powo.science.kew.org/ [accessed November 2023].
- Prance GT. 1976. The pollination and androphore structure of some Amazonian Lecythidaceae. Biotropica 8: 235–241. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum‐likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2023. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [WWW document] URL https://www.R‐project.org/. [Google Scholar]
- Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity‐dependence onphylogenetic trees. PLoS ONE 9: e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW, Huang H, Larson JG. 2014. bammtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods in Ecology and Evolution 5: 701–707. [Google Scholar]
- Rabosky DL, Mitchell JS, Chang J. 2017. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi‐rate diversification models. Systematic Biology 66: 477–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell L. 2024. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ 12: e16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A, Šmarda P, Novosolov M, Drori M, Glick L, Sabath N, Meiri S, Belmaker J, Mayrose I. 2019. The global biogeography of polyploid plants. Nature Ecology & Evolution 3: 265–273. [DOI] [PubMed] [Google Scholar]
- Rose JP, Kleist TJ, Löfstrand SD, Drew BT, Schönenberger J, Sytsma KJ. 2018. Phylogeny, historical biogeography, and diversification of angiosperm order Ericales suggest ancient Neotropical and East Asian connections. Molecular Phylogenetics and Evolution 122: 59–79. [DOI] [PubMed] [Google Scholar]
- Schwery O, Onstein RE, Bouchenak‐Khelladi Y, Xing Y, Carter RJ, Linder HP. 2015. As old as the mountains: the radiations of the Ericaceae. New Phytologist 207: 355–367. [DOI] [PubMed] [Google Scholar]
- Shi T, Huang H, Barker MS. 2010. Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Annals of Botany 106: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi XG, Fu QY, Jin JH, Quan C. 2017. Mummified Oligocene fruits of Schima (Theaceae) and their systematic and biogeographic implications. Scientific Reports 7: 4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Brown JW, Walker JF. 2018a. So many genes, so little time: a practical approach to divergence‐time estimation in the genomic era. PLoS ONE 13: e0197433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Brown JW, Yang Y, Bruenn R, Drummond CP, Brockington SF, Walker JF, Last N, Douglas NA, Moore MJ. 2018b. Disparity, diversity, and duplications in Caryophyllales. New Phytologist 217: 836–854. [DOI] [PubMed] [Google Scholar]
- Smith SA, Moore MJ, Brown JW, Yang Y. 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evolutionary Biology 15: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, O'Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Current Opinion in Plant Biology 30: 159–165. [DOI] [PubMed] [Google Scholar]
- Soto‐Trejo F, Magallón S, De‐Nova JA, Dávila P, Sánchez‐González LA, Oyama K. 2022. The evolutionary history of Fouquieriaceae (Ericales): biogeography, growth habit, habitat colonization, and chromosome evolution. Plant Systematics and Evolution 308: 35. [Google Scholar]
- Stamatakis A. 2014. RAxML v.8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull GW, Qu XJ, Parins‐Fukuchi C, Yang YY, Yang JB, Yang ZY, Hu Y, Ma H, Soltis PS, Soltis DE et al. 2021. Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nature Plants 7: 1015–1025. [DOI] [PubMed] [Google Scholar]
- Sun M, Folk RA, Gitzendanner MA, Soltis PS, Chen Z, Soltis DE, Guralnick RP. 2020. Recent accelerated diversification in rosids occurred outside the tropics. Nature Communications 11: 3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietje M, Antonelli A, Baker WJ, Govaerts R, Smith SA, Eiserhardt WL. 2022. Global variation in diversification rate and species richness are unlinked in plants. Proceedings of the National Academy of Sciences, USA 119: e2120662119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Ashman TL, Soltis PS, Soltis DE. 2021. Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell 33: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas OM, Dick CW. 2020. Diversification history of Neotropical Lecythidaceae, an ecologically dominant tree family of Amazon rain forest. In: Rull V, Carnaval AC, eds. Neotropical diversification: patterns and processes. Cham, Switzerland: Springer, 791–809. [Google Scholar]
- Walker JF, Yang Y, Feng T, Timoneda A, Mikenas J, Hutchison V, Edwards C, Wang N, Ahluwalia S, Olivieri J et al. 2018. From cacti to carnivores: improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. American Journal of Botany 105: 446–462. [DOI] [PubMed] [Google Scholar]
- Walnöfer B. 2001. The biology and systematics of Ebenaceae: a review. Annalen des Naturhistorischen Museums Wien 103: 485–512. [Google Scholar]
- Wang N, Yang Y, Moore MJ, Brockington SF, Walker JF, Brown JW, Liang B, Feng T, Edwards C, Mikenas J et al. 2019. Evolution of Portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Molecular Biology and Evolution 36: 112–126. [DOI] [PubMed] [Google Scholar]
- Wei C, Yang H, Wang S, Zhao J, Liu C, Gao L, Xia E, Lu Y, Tai Y, She G et al. 2018. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proceedings of the National Academy of Sciences, USA 115: E4151–E4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ, Donoghue MJ. 2004. Historical biogeography, ecology, and species richness. Trends in Ecology & Evolution 19: 639–644. [DOI] [PubMed] [Google Scholar]
- Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, Zhang Y, Liu Y, Zhu T, Li W et al. 2017. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Molecular Plant 10: 866–877. [DOI] [PubMed] [Google Scholar]
- Yang Y, Moore MJ, Brockington SF, Mikenas J, Olivieri J, Walker JF, Smith SA. 2018. Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytologist 217: 855–870. [DOI] [PubMed] [Google Scholar]
- Yang Y, Moore MJ, Brockington SF, Soltis DE, Wong GKS, Carpenter EJ, Zhang Y, Chen L, Yan Z, Xie Y et al. 2015. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Molecular Biology and Evolution 31: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Smith SA. 2014. Orthology inference in nonmodel organisms using transcriptomes and low‐coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Molecular Biology and Evolution 31: 3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, Fitzjohn RG, McGlinn DJ, O'Meara BC, Moles AT, Reich PB et al. 2014. Three keys to the radiation of angiosperms into freezing environments . Nature 506: 89–92. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhao L, Folk RA, Zhao JL, Zamora NA, Yang SX, Soltis DE, Soltis PS, Gao LM, Peng H et al. 2022. Phylotranscriptomics of Theaceae: generic‐level relationships, reticulation and whole‐genome duplication. Annals of Botany 129: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuntini AR, Carruthers T, Maurin O, Bailey PC, Leempoel K, Brewer GE, Epitawalage N, Françoso E, Gallego-Paramo B, McGinnie C, et al. 2024. Phylogenomics and the rise of the angiosperms. Nature 629: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The Angiosperms353+ITS time‐calibrated phylogenetic tree summarising climatic shifts, life‐history shifts, and putative WGDs.
Fig. S2 Age estimates for different families in the chloroplast+ITS time‐calibrated phylogenetic tree compared to the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Fig. S3 The chloroplast+ITS species tree. Branch lengths represent substitutions per site.
Fig. S4 The Angiosperms353+ITS species tree. Branch lengths represent substitutions per site, branch colours represent gene‐tree conflict.
Fig. S5 Single species KS plots for species incorporated into the transcriptomic dataset.
Fig. S6 The chloroplast+ITS time‐calibrated phylogenetic tree summarising climatic shifts and putative WGDs.
Fig. S7 The Angiosperms353+ITS time‐calibrated phylogenetic tree summarising climatic shifts and putative WGDs.
Fig. S8 The chloroplast‐only time‐calibrated phylogenetic tree summarising climatic shifts and putative WGDs.
Fig. S9 Further information on the composition of the largest shifts down PC1 as identified by sister‐clade comparison in the chloroplast+ITS time‐calibrated phylogenetic tree and which are associated with putative WGDs.
Fig. S10 Further information on the composition of the largest shifts down PC1 as identified by sister‐clade comparison in the Angiosperms353+ITS time‐calibrated phylogenetic tree and which are associated with putative WGDs.
Fig. S11 Shifts down PC1 estimated by l1ou on the chloroplast+ITS time‐calibrated phylogenetic tree, and the largest 10 differences in climatic preference between sister clades.
Fig. S12 Shifts down PC1 estimated by l1ou on the Angiosperms353+ITS time‐calibrated phylogenetic tree, and the largest 10 differences in climatic preference between sister clades.
Fig. S13 Shifts down PC1 estimated by l1ou on the chloroplast‐only time‐calibrated phylogenetic tree, and the largest 10 differences in climatic preference between sister clades.
Fig. S14 The chloroplast+ITS time‐calibrated phylogenetic tree with life‐history shifts, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S15 The Angiosperms353+ITS time‐calibrated phylogenetic tree with life‐history shifts, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S16 The chloroplast+ITS time‐calibrated phylogenetic tree with estimated speciation rates, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S17 The Angiosperms353+ITS time‐calibrated phylogenetic tree with estimated speciation rates, putative WGDs, and the largest 10 differences in climatic preference between sister clades.
Fig. S18 Subsample of 9 trees from the credible shift set of speciation rates estimated with BAMM on the chloroplast+ITS time‐calibrated phylogenetic tree.
Fig. S19 Subsample of 9 trees from the credible shift set of speciation rates estimated with BAMM on the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Fig. S20 Climatic preference of extant species on PC1 plotted against tip speciation rates, from the chloroplast+ITS time‐calibrated phylogenetic tree.
Fig. S21 Climatic preference of extant species on PC1 plotted against tip speciation rates, from the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Table S1 Voucher information for transcriptome dataset.
Table S2 Voucher information for species‐level molecular datasets.
Table S3 Chromosome number data.
Table S4 Life‐history information.
Table S5 Largest sister‐clade differences in climatic preference in the chloroplast+ITS time‐calibrated phylogenetic tree.
Table S6 Largest sister‐clade differences in climatic preference in the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Table S7 Largest sister‐clade differences in climatic preference in the chloroplast‐only time‐calibrated phylogenetic tree.
Table S8 Summary of all shifts down PC1 identified by l1ou in the chloroplast+ITS time‐calibrated phylogenetic tree.
Table S9 Summary of all shifts down PC1 identified by l1ou in the Angiosperms353+ITS time‐calibrated phylogenetic tree.
Table S10 Summary of all shifts down PC1 identified by l1ou in the chloroplast‐only time‐calibrated phylogenetic tree.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
New sequence data is available on the NCBI Sequence Read Archive (PRJNA1076884, PRJNA839108). Supporting Information Tables S1 and S2 provide voucher information. Assembled sequences, target files, analysis outputs, and scripts are available on Zenodo: https://zenodo.org/records/13835677.


