Abstract
The Butterfly hillstream loach (Beaufortia pingi), an aquatic benthic fish species inhabiting mountain rapids, exhibits exceptional capabilities in movement, adsorption, and desorption processes, enabling it to adhere to smooth and contaminated surfaces in turbulent streams. These attributes make it a significant subject for genetic and evolutionary research. In this study, the genomic sequences of this species were acquired utilizing PacBio sequencing and Hi-C methods. The genome assembly is 459.8 Mb in size with a contig N50 of 5.35 Mb, and the assembled contigs were anchored into 25 chromosomes. BUSCO analysis confirmed a high completeness level with 97.0% gene coverage. A total of 111.47 Mb repetitive sequences (24.25% of the assembled genome), and 22,906 protein-coding genes were identified in the genome. This study represents the first investigation of the species’ genome. The establishment of this genome assembly provides valuable resources for future genetic research and facilitates the study of genetic changes during evolution.
Subject terms: Genome evolution, Comparative genomics
Background & Summary
Beaufortia pingi, endemic to China, is classified within the genus Beaufortia (Cypriniformes: Gastromyzontidae) and is primarily located in the Pearl River basin and various basin on Hainan Island. Commonly referred to as hillstream loaches, species within the Beaufortia genus are native to regions including India, China, and Southeast Asia, encompassing areas such as Sumatra, Java, and Borneo. Predominantly inhabiting river rapids, they adhere to rocky and gravel substrates to feed on fixed algae. Notably, juvenile exhibit the ability to crawl on gravel shortly after hatching. The locomotion, adhesion, and detachment mechanisms of Beaufortia fish are characterized by rapidity, allowing them to maintain their grip on slippery and debris-laden surfaces in fast-flowing aquatic environments1,2. As a result, these fish are frequently employed in the development of adsorption models, and researchers have created biomimetic robots inspired by their distinctive capabilities3,4. Furthermore, Beaufortia species serve as exemplary model for investigating the adaptive evolution of organisms within mountain stream ecosystems5.
Although the mitochondrial genomes of Beaufortia species have been documented in previous studies, comprehensive research on the whole-genome sequencing of these species remains limited. In 2016, a specimen of B. szechuanensis collected from the Yangtze River in China was examined, and the first complete mitochondrial genome report of Beaufortia was obtained6. Subsequently, in 2017, the mitochondrial genome of B. kweichowensis was reported, with phylogenetic analysis indicating that the family Balitoridae could be categorized into two subfamilies: Gastromyzoninae and Homalopterinae7. In 2021, the whole genome of B. kweichowensis was sequenced and assembled, resulting in a genome assembly of 448.52 Mb with an N50 of 5.53 Mb8. In 2023, the complete mitochondrial genome of B. pingi was successively sequenced and analyzed9. B. pingi, in comparison to B. kweichowensis, is characterized by a more circumscribed geographic distribution and exhibits distinct morphological attributes. For instance, the mouth of B. pingi features a small and horseshoe-shaped rather than a large, arc-shaped, and its ventral surface is grayish-white instead of pale yellow. Regrettably, the paucity of high-quality reference genomes has impeded a profound comprehension of the adaptive evolution of specific traits within the Beaufortia species. In this study, we assembled a chromosome-level genome of B. pingi utilizing the PacBio sequencing platform and Hi-C techniques. This study presents the inaugural high-quality genome assembly of B. pingi, which is poised to yield substantial insights into the phylogenetic relationships and the adaptive evolution of Gastromyzontidae fishes. Moreover, this genome assembly will broaden our understanding of the evolutionary relationships of Beaufortia species, thus enabling future exploration of their evolutionary history.
Methods
Sample collection
In May 2023, a one-year-old adult female specimen of B. pingi was collected from Lingyun County (24.51°N, 106.53°E), located in Baise, Guangxi, China (Fig. 1a,b). Muscle tissue from the specimen was collected for DNA extraction, which was conducted using a QIAGEN DNeasy Blood & Tissue Kit (QIAGEN, Shanghai, China), resulting in the acquisition of high-quality genomic DNA. The quality and quantity of the extracted DNA were evaluated using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), while the integrity of the DNA was evaluated through 1% agarose gel electrophoresis10. The electrophoresis was performed with a 1 kb DNA ladder at a voltage of 120 V for 20 minutes. The spectrophotometric analysis yielded an A260/A280 ratio of 1.83 and an A260/A230 ratio of 1.79. Additionally, total RNA was extracted from six tissues of the specimen, i.e., liver, brain, kidney, intestine, skin, and muscle, using the TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, China). The extracted RNA was subsequently evaluated for its quality and quantity.
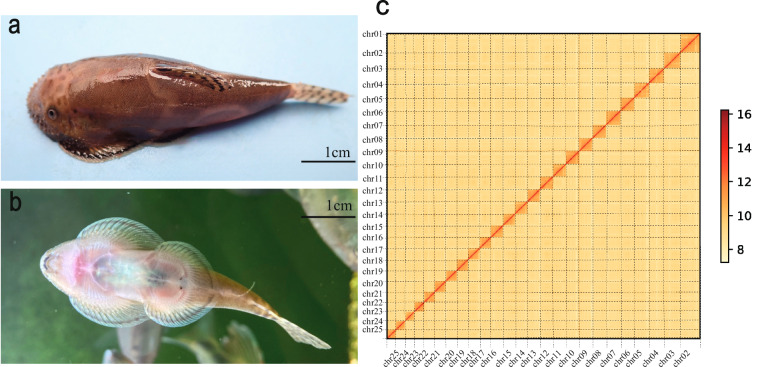
Fig. 1.
Overview of the B. pingi specimen used for sequencing in the study. (a) B. pingi in dorsal view, with broadened craniofacial shape, flattened body; (b) Ventral view: a completely flat exposed abdomen with two suction cups, enlarged paired pectoral and ventral fins; (c) Hi-C interactive heatmap of genome-wide B. pingi. The color blocks represent the interaction strength from yellow (low) to red (high).
Genome and transcriptome sequencing
The DNA libraries were subjected to thorough examination and subsequently sequenced utilizing the PacBio Sequel II platform at Frasergen in Wuhan. The raw sequencing data generated in this study were subjected to a preprocessing regimen employing the CCS program11. A total of 33.22 Gb Circular Consensus Sequencing (CCS) bases were produced, achieving a sequencing depth of 72 ×, which was meticulously calculated in relation to the estimated genome size and the voluminous data output generated. Meanwhile, genomic DNA extracted from the identical specimen was used for the construction of the Hi-C library. The Hi-C library was sequenced using the Illumina Novaseq platform with 150-bp paired-end mode. A total of 59.1 Gb clean reads, corresponding to approximately 128 × coverage, were produced with 150 bp paired-end sequencing reads. An RNA sequencing library was constructed from an equal quantity of high-quality RNA extracted from the liver, brain, kidney, intestine, skin, and muscle, and then sequencing on the PacBio Sequel platform. The polymerase reads were spliced to obtain subread sequences12. Subsequently, subreads derived from the same zero-mode waveguides (ZMW) sequencing well, underwent self-correction to produce highly accurate ccs sequences13. A total of 42.25 Gb raw data was acquired. After self-correction, a total of 378,529 CCS sequences, amounting to 806.39 Mb, were extracted. Subsequent filtering yielded 266,487 full-length non-chimeric (FLNC) sequences, which represented 70.4% of the total. After the elimination of redundant sequences with the CD-HIT v4.8.114 program, a final dataset of 347.8 Mb of clean data was acquired for subsequent genome annotation. This dataset comprised 122,919 high-quality transcripts, with an average length of 2,829.8 bp.
Genome assembly and Hi-C scaffolding
An initial genome assembly was executed utilizing HiFiasm v0.1215 with the CCS reads. We obtained a genome assembly of 459.6 Mb in length with an N50 length of 7.52 Mb, consisting of 341 contigs. To assess the quality of the genome assembly, a quantitative evaluation was performed using the Benchmarking Universal Single-Copy Orthologs (BUSCO) v5.5.016 with the actinopterygii_odb10 geneset, which indicated a completeness level of 96.9%. Following the quality control of the Hi-C reads, Juicer v1.517 was employed to align these reads to the draft genome. Subsequently, 3D-DNA v18092218 was employed to cluster, sort, and orient the contigs or scaffolds, to obtain chromosome-level genomes. Juicebox v1.11.0819 was then utilized for visual inspection and rectification of any errors in the order and orientation of the contigs or assembly errors within the contigs. As a result, the chromosome-level genome of B. pingi was obtained. The genome size was 459.8 Mb with a scaffold N50 of 14.87 Mb and a contigs N50 of 5.35 Mb. The GC content was 38.6%. The 313 contigs (388.5 Mb) were anchored into 25 chromosomes (Fig. 1c, Table 1). The BUSCO analysis indicated that 97.0% (single-copy genes: 95.0%, duplicated genes: 2.0%) of the 3,640 BUSCOs were identified as complete orthologs. These results imply that a high-quality chromosome-level genome of B. pingi has been assembled (Fig. 2a).
Table 1.
Summary of chromosome length of B. pingi genome.
| Pseudo-chromosomes | Length (bp) | Percentage (%) |
|---|---|---|
| Chr01 | 16,516,361 | 3.59% |
| Chr02 | 16,980,721 | 3.69% |
| Chr03 | 11,777,159 | 2.56% |
| Chr04 | 17,418,109 | 3.79% |
| Chr05 | 14,348,534 | 3.12% |
| Chr06 | 12,534,116 | 2.73% |
| Chr07 | 14,872,000 | 3.24% |
| Chr08 | 15,335,500 | 3.34% |
| Chr09 | 16,019,691 | 3.49% |
| Chr10 | 14,255,809 | 3.10% |
| Chr11 | 20,269,816 | 4.41% |
| Chr12 | 12,983,329 | 2.82% |
| Chr13 | 16,036,091 | 3.49% |
| Chr14 | 18,320,117 | 3.99% |
| Chr15 | 14,776,147 | 3.22% |
| Chr16 | 13,465,436 | 2.93% |
| Chr17 | 16,382,927 | 3.56% |
| Chr18 | 18,801,183 | 4.09% |
| Chr19 | 24,319,891 | 5.29% |
| Chr20 | 17,382,782 | 3.78% |
| Chr21 | 15,781,281 | 3.43% |
| Chr22 | 13,557,934 | 2.95% |
| Chr23 | 14,009,566 | 3.05% |
| Chr24 | 11,524,913 | 2.51% |
| Chr25 | 10,832,087 | 2.36% |
| Unmapped | 71,095,472 | 15.47% |
| Total | 459,596,972 | 100.00% |
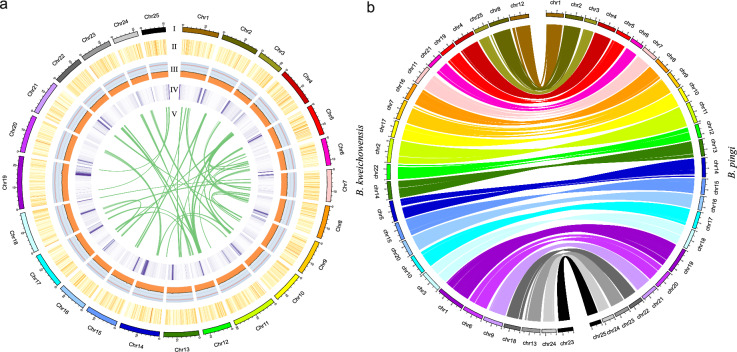
Fig. 2.
Chromosomal organization and synteny in B. pingi. (a) Circos of B. pingi genome characteristics. From outside to inside: chromosome (I), distribution of GC (II), repeat elements (III), gene number (IV), self-collinearity of genes (V). The densities of GC content and genes were calculated in 100-kb windows, while the densities of repeat elements were evaluated in 10-kb windows; (b) Circos of chromosome synteny between B. pingi and B. kweichowensis. Each colored line represents gene model match between two species.
Repeat and protein-coding gene annotation
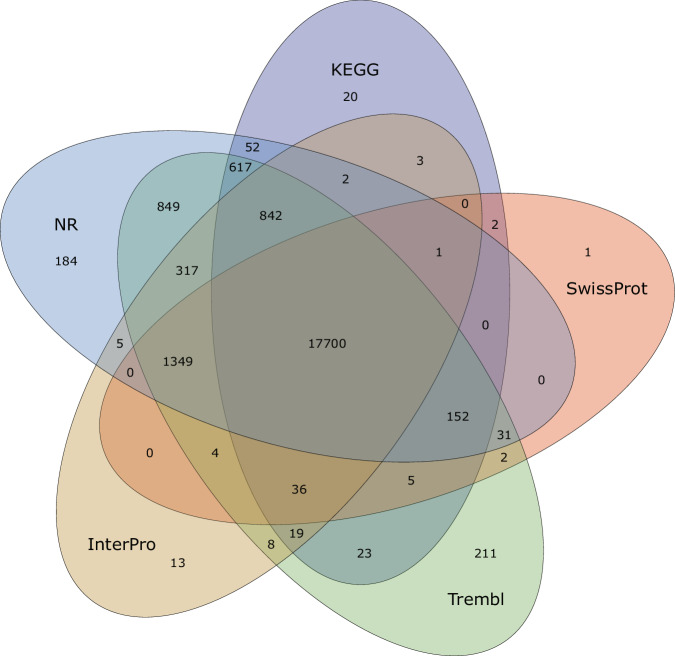
To elucidate the landscape of repeat elements, we employed a homology-based prediction approach utilizing the established repetitive sequence database Repbase with RepeatMasker v4.1.020 and RepeatProteinMask21. A de novo repeat library for the genome was constructed using RepeatModeler v1.0.1122 for ab initio prediction, which was subsequently analyzed with RepeatMasker. Additionally, the Tandem Repeats Finder v4.09 (TRF)23 program was applied to identify tandem repeats in the genome. In total, 111.47 Mb repeat sequences (24.25% of the assembled genome) were identified. Within this total, DNA transposons comprised 7.59%, long interspersed nuclear elements (LINEs) represented 2.07%, and long terminal repeats (LTRs) accounted for 3.10%.The prediction of protein-coding genes was approached with a multifaceted strategy, employing a trio of prediction methodologies: homology-based prediction, transcript-based prediction, and de novo prediction employing Exonerate v2.4.024 and PASA v2.4.125. For the homology-based annotation analysis, protein-coding sequences from related species (B. kweichowensis, Triplophysa tibetana, T. bleekeri, Misgurnus anguillicaudatus, Danio rerio) were utilized alongside transcripts generated via the PacBio platform. The AUGUSTUS v3.2.226 and SNAP27 packages were used for the ab initio prediction. The genesets predicted through these methods were subsequently integrated using MAKER v3.01.0328. Finally, a total of 22,906 genes were predicted as protein-coding genes. The functional annotations were performed with the public databases, including the non-redundant protein database (NR), Kyoto Encyclopedia of Genes and Genomes (KEGG), Swiss-Prot, TrEMBL, and InterPro databases, using Diamond v0.9.30.13129 blastp with the parameters –outfmt 6–max-target-seqs. 1 –evalue 1e-6. A total of 22,448 genes (98% of all predicted genes) were annotated, and of these, 17,700 genes were functionally annotated in all databases (Table 2, Fig. 3).
Table 2.
Number of genes annotated using different databases.
| Database | Number of annotated genes | Percentage |
|---|---|---|
| NR | 22,101 | 96.49% |
| Swiss-Prot | 19,283 | 84.18% |
| KEGG | 19,474 | 85.02% |
| TrEMBL | 22,165 | 96.77% |
| Interpro | 20,299 | 88.62% |
| Total | 22,448 | 98.00% |
Fig. 3.
Venn diagram for functional annotations of B. Pingi protein-coding genes with the public database, including NR, KEGG, Swiss-Prot, TrEMBL, and Interpro.
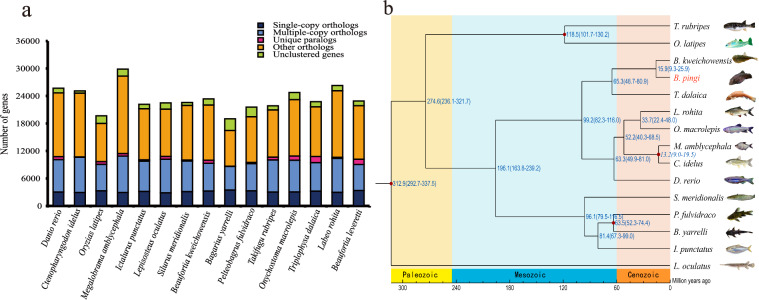
A phylogenetic tree of B. pingi and 14 additional fish species was constructed using 1763 single-copy orthologous genes (Fig. 4a) with RAxML-NG v1.1.030 and the GTR + G + I model. The analysis indicated that the most recent common ancestor of the 14 fish species existed approximately 312.9 million years ago. B. kweichowensis, identified as the closest relative to B. pingi, shared a common ancestor around 15.9 million years ago (Fig. 4b). Additionally, genome-wide collinearity analysis between B. kweichowensis and B. pingi was conducted using JCVI v1.2.731, revealing a conserved collinearity relationship between the chromosomes of B. pingi and those of B. kweichowensis (Fig. 2b).
Fig. 4.
Comparative genomic analysis reveals phylogenetic relationship and genome evolution of B. Pingi. (a) Statistics of orthologous genes in 15 representative fish species; (b) Phylogenetic tree and estimated divergence time of B. pingi and other 14 teleosts, where B. pingi is represented in red font. Tree topology with estimated divergence times (million years ago, MYA, including range) is shown next to each lineage (blue).
Data Records
The Raw data generated by PacBio sequencing platform has been deposited into the NCBI Sequence Read Archive (SRA) with accession number SRR2768495932. The Hi-C reads and RNA-seq reads have been stored in the NCBI Sequence Read Archive (SRA) with the accession numbers SRR2770401733 and SRR2768834534, respectively. The final genome assembly has been deposited in GenBank under accession number JBIQFU00000000035. The genome annotation are available in the FigShare repository36.
Technical Validation
The culminated genome assembly of B. pingi is 459.8 Mb in size, exhibiting a contig N50 of 5.35 Mb and consisting of 25 chromosomes. This assembled genome size is comparable to that of a closely-related species8. Furthermore, the Hi-C chromosome interaction intensity signal corroborates the high quality of genome assembly. Meanwhile, we mapped the Illunima short reads to the assembled sequences, and the mapping ratio was 98.99%, indicating a high completeness of the genome. The BUSCO completeness of the genome annotation was assessed with the predicted protein sequences, which revealed a coverage of 89.06% for complete orthologs, utilizing the actinopterygii_odb10 dataset. This discovery intimates that a significant proportion of the conserved genes are accurately represented in the B. pingi genome. Moreover, synteny analysis between B. kweichowensis and B. pingi demonstrated a substantial degree of synteny conservation, further indicating that the genome assembly and annotation of B. pingi are both comprehensive and of high quality.
Acknowledgements
This study was supported by the Innovation Project of Postgraduate Scientific Research in Huzhou University in 2023 (2023KYCX66).
Author contributions
S. Yi conceived the study. S. Yi and Q. Sheng collected samples. Bioinformatics analysis was performed by X. Zhang, Q. Tang and H. Qi. Q. Shen wrote and revised the original manuscript. All authors have read and approved the final manuscript.
Code availability
All software used in this work is publicly available, and the software versions and parameters used are described in the Methods section. The parameters not mentioned in the analysis were used as default parameters suggested by the developer. No custom script was used in this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qiang Sheng, Email: qsheng@zjhu.edu.cn.
Shaokui Yi, Email: yishaokui@foxmail.com.
References
- 1.Wang, J. et al. An adhesive locomotion model for the rock-climbing fish, Beaufortia kweichowensis. Sci Rep9, 16571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou, J., Wang, J. & Ji, C. The Adhesive System and Anisotropic Shear Force of Guizhou Gastromyzontidae. Sci Rep6, 37221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, J., Xi, Y., Ji, C. & Zou, J. A biomimetic robot crawling bidirectionally with load inspired by rock-climbing fish. J. Zhejiang Univ. Sci. A23, 14–26 (2022). [Google Scholar]
- 4.Wu, J. et al. Light-driven soft climbing robot based on negative pressure adsorption. Chemical Engineering Journal466, 143131 (2023). [Google Scholar]
- 5.Shi, L. et al. Evolutionary relationships of two balitorids (Cypriniformes, Balitoridae) revealed by comparative mitogenomics. Zoologica Scripta47, 300–310 (2018). [Google Scholar]
- 6.Wu, J. et al. The complete mitochondrial genome sequence of Beaufortia szechuanensis (Cypriniformes, Balitoridae). Mitochondrial DNA Part A27, 2535–2536 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Wen, Z.-Y. et al. The complete mitochondrial genome of a threatened loach (Beaufortia kweichowensis) and its phylogeny. Conservation Genet Resour9, 565–568 (2017). [Google Scholar]
- 8.Deng, Y. et al. Genome of the butterfly hillstream loach provides insights into adaptations to torrential mountain stream life. Molecular Ecology Resources21, 1922–1935 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Shen, Z., Sheng, Q., Jin, Z., Zhang, Y. & Lv, H. Mitogenome Characterization of a Vulnerable Gastromyzontid Fish, Beaufortia pingi (Gastromyzontidae): Genome Description and Phylogenetic Considerations. J. Ichthyol.63, 735–746 (2023). [Google Scholar]
- 10.Suganthi, M. et al. A method for DNA extraction and molecular identification of Aphids. MethodsX10, 102100 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao, F., Zhao, Y., Wang, X., Mao, Y. & Jian, X. Comparative transcriptome analysis of dioecious floral development in Trachycarpus fortunei using Illumina and PacBio SMRT sequencing. BMC Plant Biol23, 536 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, R., Duan, Q., Luo, Q. & Deng, L. PacBio Full-Length Transcriptome of a Tetraploid Sinocyclocheilus multipunctatus Provides Insights into the Evolution of Cavefish. Animals13, 3399 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni, P. et al. DNA 5-methylcytosine detection and methylation phasing using PacBio circular consensus sequencing. Nat Commun14, 4054 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondratenko, Y., Korobeynikov, A. & Lapidus, A. Correction to: CDSnake: Snakemake pipeline for retrieval of annotated OTUs from paired-end reads using CD-HIT utilities. BMC bioinformatics21, 362–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uliano-Silva, M. et al. MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinformatics24, 288 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu, J.-J., Han, Y.-W., Lin, C.-F., Cai, J. & Zhao, Y.-P. Benchmarking gene set of gymnosperms for assessing genome and annotation completeness in BUSCO. Horticulture Research10, 165 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand, N. C. et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Systems3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan, S. U. et al. Chromosome-length genome assembly of Teladorsagia circumcincta – a globally important helminth parasite in livestock. BMC Genomics24, 74 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson, J. T. et al. Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Systems6, 256–258.e1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausmann, F. & Kurtz, S. DeepGRP: engineering a software tool for predicting genomic repetitive elements using Recurrent Neural Networks with attention. Algorithms Mol Biol16, 20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Z. et al. Chromosome-level genome assembly of the deep-sea snail Phymorhynchus buccinoides provides insights into the adaptation to the cold seep habitat. BMC Genomics24, 679 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, X., Meyer-Rochow, V. B., Ballantyne, L. & Zhu, X. An Improved Chromosome-Level Genome Assembly of the Firefly Pyrocoelia pectoralis. Insects15, 43 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jo, E. et al. Chromosome-level genome assembly and annotation of the Antarctica whitefin plunderfish Pogonophryne albipinna. Sci Data10, 891 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. J. et al. A chromosome-level reference genome of the Antarctic blackfin icefish Chaenocephalus aceratus. Sci Data10, 657 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia, H. et al. PASA: IDENTIFYING MORE CREDIBLE STRUCTURAL VARIANTS OF HEDOU12. IEEE/ACM Trans. Comput. Biol. and Bioinf.17, 1493–1503 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Brůna, T. et al. Galba: genome annotation with miniprot and AUGUSTUS. BMC Bioinformatics24, 327 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, Y. et al. Chromosome level genome assembly of colored calla lily (Zantedeschia elliottiana). Sci Data10, 605 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuang-Lim, C. et al. Seqping: gene prediction pipeline for plant genomes using self-training gene models and transcriptomic data. BMC bioinformatics18, 1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, H.-S. et al. A chromosome-level genome assembly of Korean mint (Agastache rugosa). Sci Data10, 792 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Togkousidis, A., Kozlov, O. M., Haag, J., Höhler, D. & Stamatakis, A. Adaptive RAxML-NG: Accelerating Phylogenetic Inference under Maximum Likelihood using Dataset Difficulty. Molecular Biology and Evolution40, msad227 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang, H. et al. JCVI: A versatile toolkit for comparative genomics analysis. iMeta3, e211 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR27684959 (2024).
- 33.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR27704017 (2024).
- 34.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR27688345 (2024).
- 35.Shen, Q. et al. NCBI GenBank. The genome assembly of Beaufortia pingihttps://identifiers.org/ncbi/insdc:JBIQFU000000000 (2024).
- 36.Genome annotation of Beaufortia pingi, figshare, 10.6084/m9.figshare.25053224.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR27684959 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR27704017 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR27688345 (2024).
Data Availability Statement
All software used in this work is publicly available, and the software versions and parameters used are described in the Methods section. The parameters not mentioned in the analysis were used as default parameters suggested by the developer. No custom script was used in this study.