Abstract
The Human Cell Atlas (HCA) is a global partnership “to create comprehensive reference maps of all human cells—the fundamental units of life – as a basis for both understanding human health and diagnosing, monitoring, and treating disease.” (https://www.humancellatlas.org/) The atlas shall characterize cells from diverse individuals across the globe to better understand human biology. HCA proactively considers the priorities of, and benefits accrued to, contributing communities. Here, we lay out principles and action items that have been adopted to affirm HCA’s commitment to equity so that the atlas is beneficial to all of humanity.
Subject terms: Genetics, Molecular biology, Ethics
The Human Cell Atlas (HCA) aims to characterize cells from diverse individuals across the globe to better understand human biology. Here, the authors lay out principles and action items that have been adopted to affirm HCA’s commitment to equity so that the atlas is beneficial to all of humanity.
Introduction
Humans are made of trillions of cells. Although they all share a basic physiological architecture, each is unique because of their own in utero developmental processes, genetic endowment, environmental exposures and life-experiences. These factors can impact cells and are important to consider when people get sick. However, it is not always clear which are the most critical, how they make individuals vulnerable to specific diseases, or how they might be used to select the best treatment. Further, the full list of factors that can influence cells is not well understood.
The Human Cell Atlas (HCA) is a global partnership of individuals spanning different backgrounds—including biologists, clinicians, computer scientists, engineers, ethicists, lawyers, educators, science funders, social workers and others—who are actively collaborating to describe the types and properties of all human cells, using diverse measurement technologies, especially single-cell and spatial genomics. HCA’s ultimate goal is to create reference maps as a foundation to understand how these cells work individually and together to form organs and carry out specific physiological functions essential to life, as well as how to use this information to develop improved preventions, diagnostics and cures for diseases that impact people around the world. By building a comprehensive encyclopaedia of healthy cells and their behaviours in individuals, representing various ages, genders and life-styles, from as many populations and environments as possible, HCA seeks to better understand human biology, as well as the contributions of environment and genetics to the ways in which cells work to keep us well1. However, the task to identify and collect samples from a diversity of populations is daunting. Currently, geographical location of residence of participants of a population is the primary focus, with ongoing efforts to collect from populations with explicitly diverse ancestries. To systematically address the challenge of defining ancestry, the HCA has set up a Task Force to develop recommendations for representing diversity in the HCA project, recording diversity-related metadata and ethically engaging underrepresented populations; the work of this Task Force is in progress. HCA’s Atlas, although only partially complete, is already beginning to provide understanding of health and diagnosis, monitoring, and treatment of disease. For example, in ulcerative colitis, single-cell atlas data enabled the identification of a rare cell type – epithelial M-like cells – which are exceedingly rare in the healthy colon, but expanded significantly in the inflamed, diseased colon2; further examples can be found in Rood et al3.
Until recently, because of technical limitations, it had not been possible to assess comprehensively the fundamental building blocks of the human body – individual cells – in health and disease. With the technologies that were available previously, comprehensive information could only be obtained on the properties of large groups of cells. Recent scientific and technological advances are enabling scientists to obtain information on single cells, and leading to the construction of the atlas. However, at present, there is no adequate understanding of the cell types that make up the different tissues of the human body and how their relative abundance or other properties vary among a diverse set of individuals. It is hoped that the single-cell information that HCA is generating will continue to yield conclusions that provide better management of health to enhance quality of life by pinpointing precisely where things go wrong in disease and how to control them.
In considering how this information may impact individuals around the globe, it is clear that much of the healthcare research performed to date has been limited in its ability to consider the full impacts of variations in age, sex, genetic make-up, environmental exposures or life-experiences4, and that medical breakthroughs have often not been equally beneficial to all5. Unfortunately, some previous scientific studies have even been extractive and exploitative, failing to sufficiently consider the priorities of, or benefits to, contributing communities. Building on knowledge gained from past international research consortia and leveraging the momentum of technological advances in single-cell and spatial genomics, the HCA strives to advance social and scientific benefit by open participation. Furthermore, it is committed to enhancing global engagement by both (i) creating an active outreach and education programme to build local capacity to undertake research projects, and (ii) forging and fostering local collaborative partnerships so that it can benefit from diverse community perspectives and shared approaches to facing complex challenges in human biology. In collaboration with visual and digital artists, inventors, designers, local communities and schoolchildren, the HCA has created an art and science exhibition – One Cell at a Time – for the general public to explore the biological and cellular make-up of the human body6. The HCA considers outreach and education as mission critical–the help of the global research community is needed to get it right.
The path to equitable research
International health research consortia, unfortunately, often build on past history of research practices and in doing so may fail to adequately address questions of equity and diversity, including limited involvement of communities around the world in research design and implementation. These practices have led to scientific bias, as well as inequities and injustices both in capacity-building and benefit-sharing of resources and findings7. Furthermore, inadequate involvement of local communities can lead to underrepresentation of certain populations8 which can, in turn, lead to scientific biases in findings5,9, stigmatization of groups, and, in extreme cases, systemic racism and discrimination10.
In the wake of these realizations, several key players of the research ecosystem, including scientists, funders, publishers and institutions, have called for a reform in the way research is done–to ensure relevant stakeholders are involved throughout the research lifecycle5,11. Trust needs to be built with all involved in research, and local scientists – working in institutions in the regions where a project is being conducted – can act as liaisons to do so by ensuring that science is also relevant to local specificities and needs.
Large international consortia and projects involving collection of large and diverse datasets, such as the HCA, require particular attention. Indeed, equitable research calls for engagement efforts at various levels, including with communities, through the entire period of research; including design, planning, governance and execution10,12. It also requires support and enabling of local scientists – i.e., researchers in a defined geographical region, including resource-poor regions – to undertake their own research projects12,13, which, in turn, contributes to the collection of diverse datasets representative of global populations, and dissemination of discoveries and findings to local populations. Such enabling can also help create jobs and opportunities in science locally, and bring cutting edge approaches to a broader swatch of research than that of the originating consortium per se. We hope that the global information being generated by the HCA will enable a better understanding of the circumstances that are unique to a community, or commonly seen around the world. Information generated from local projects driven by local priorities and conducted by local scientists (i.e., scientists working in institutions in the region where the project is being conducted), and assisted by the information provided by the global collective, will generate conclusions beneficial to local communities.
Setting up the human cell atlas and underlying principles
Objectives of the Atlas: The HCA primarily focuses on cellular variation in healthy tissues as a reference to understand both health and disease. There are many different cell types in the human body. Scientists do not know all of the cell types that compose each organ and their relative proportions. And, even in the same organ, the proportions of cell types can vary between, for example, males and females, children and adults, non-smokers and smokers, Bantu speakers of Africa and Austro-Asiatic speakers of India and southeast Asia, etc. An understanding needs to be developed as to the breadth of cell types, composition and states present when genetic, environmental, and experiential factors vary. This requires collecting cells from various organs from diverse cohorts of people. By explicitly measuring diversity and variation within ‘healthy’ tissues, HCA aims to better understand the range of cells across humans4,10.
The HCA has started to collect cells and study the biological characteristics of each collected cell – for example, which genes are expressed and at what level. This information is enabling scientists to identify the types of cells and their proportions in various organs of the human body, and, to estimate the extent of variation that exists across individuals at a very high resolution – at the level of single cells. Having reference maps with sufficient variation is a first step towards inferring the possible causes of variation across ancestry, demographic variables and environmental exposures, understanding biological mechanisms, and developing disease models and treatments that are more generalizable to the human population as a whole.
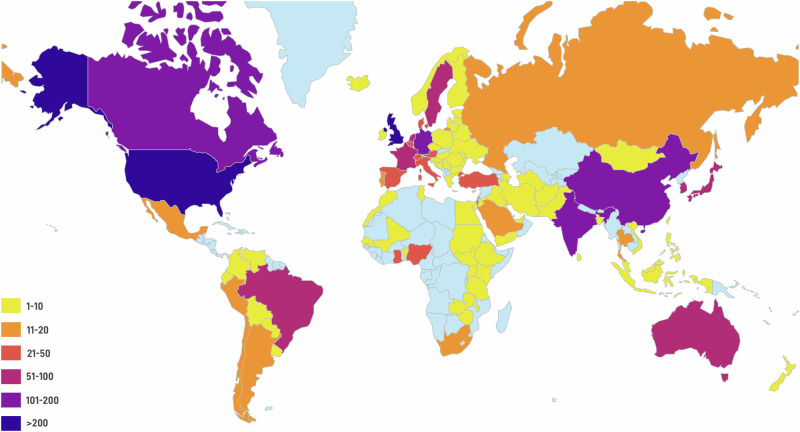
Local involvement and recruitment: HCA investigators are reaching out globally to various communities to undertake projects driven by local scientists. HCA membership is globally open to anyone above 16 years of age who agrees to abide by the ethical standards and principles of the HCA as enshrined in the HCA White Paper (https://arxiv.org/abs/1810.05192) and updates notified on HCA’s website (https://www.humancellatlas.org/). HCA is reaching out far and wide to scientists and other interested persons across communities, ethnicities or ancestries–a group of people who share a common cultural, historical and/or biological background–to become active, contributing members. Currently, there are 3153 members of the HCA from 97 countries located in nearly all regions of the world (Fig. 1; for regularly updated details see https://www.humancellatlas.org/learn-more/hca-metrics/). HCA leadership is committed to empowering, supporting, and working with, local scientists to help facilitate this work; for example, helping to provide adequate information about HCA to the general members of the community to gain their confidence, to train local scientists in methods of single-cell analyses so they can lead research in this area, to engage with local and global funders to help increase the amount of financial resources available for HCA research, to foster research collaborations and to obtain approvals from the local research ethics committees to undertake HCA studies to foster dedicated and vibrant regional networks, including for each of Asia, Latin America, Africa, and the Middle East, and to ensure that local scientists from around the world are foremost members and leaders in HCA’s top governance bodies and key working groups. To facilitate engagement of local communities, the HCA encourages local scientists and other interested persons to become members of the HCA and actively participate in HCA.
Fig. 1. Geographical distribution of locations of members of the Human Cell Atlas.
Geographical distribution of locations of members of the Human Cell Atlas, most of whom are scientists engaged in HCA activities or have expressed an interest to participate in the HCA. (see https://www.humancellatlas.org/learn-more/hca-metrics/ for details).
HCA considers the involvement of local partners as crucial for obtaining tissue samples ethically and studying cells from donors across the world. HCA further considers that building and maintaining trust with communities is critical for recruiting research participants and for the success of HCA. To be clear, the HCA believes that research should be led by local scientists or individuals who wish to be part of the HCA and should include members of the specific communities on whom studies are undertaken. The HCA is committed to the success of these individuals through active partnerships. The HCA is actively engaged in advocacy for international funding in the lesser-endowed global regions so that local scientists can participate in HCA research. Indeed, lack of adequate funding to researchers in many regions of the world is the most serious impediment to their participation in the HCA; this impediment needs to be overcome to enhance inclusivity and diversity. The nature of biological material to be collected should be determined by the local scientific partners and in consultation with the community. The nature of the information to be generated, stored, analyzed, and shared with the community and the modalities of conduct of these activities are also determined in consultation with local scientists, members of the community, ethics committees, and other similar groups of the community. Aside from funding, there are many additional challenges in expanding HCA activities to many global regions. These include restrictions on the import of equipment and reagents, inadequate time and facilities for collection of deep metadata, lack of adequately trained personnel to carry out experimental and computational work for single-cell and spatial genomics, and lack of specialized equipment and appropriate infrastructure. To overcome some of these challenges, HCA has organized several dedicated training workshops and discussion sessions. These include an in-person Computational and Experimental Design Workshop in Bangkok in 2024 attended by researchers from 13 countries of the Asia Pacific region, a hybrid HCA symposium for Latin America with centres in three nodal countries, a Latin America in-person Computational and Experimental Design Workshop in Chile in 2023, a Computational and Experimental Design Workshop in Ghana in 2023; Introduction to Single Cell RNA-Seq Analysis (virtual workshop) in October and November 2022 in Africa; Single Cell Transcriptomics (virtual workshop) in January 2022 in India; and, Single Cell RNA-seq Data Analysis (virtual workshop) in 2021 in Latin America. Overall, since 2019, 13 such events have been organized mostly in LMICs, attended by a total of over 2000 researchers (Table 1). HCA continues to partner with its regional networks on such training efforts.
Table 1.
Meetings, roadshows and workshops organized by the Human Cell Atlas for researchers in various global regions
| Year | Nature of Training | Countries from which Scientists Participated | Theme | No. of Participants |
|---|---|---|---|---|
| 2019 | In-person Roadshow | Brazil | Human Cell Atlas: Scientific foci and equity | 50 |
| 2019 | In-person Meeting (held in Addis Ababa, Ethiopia) | 15 countries from various global regions | Human Cell Atlas: Scientific foci, participation and equity | 58 |
| 2020 | On-line Meeting | 21 countries of Latin America | Human Cell Atlas: Initiation of activities in Latin America | 425 |
| 2021 | On-line Workshop | 24 countries of Latin America | Single-cell RNA_sequencing analysis | 410 |
| 2022 | On-line Workshop | India and Pakistan | Introduction to single-cell transcriptomics methodology | 220 |
| 2022 | On-line Workshop | Various countries of Africa | Introduction to the Human Cell Atlas | 136 |
| 2022 | On-line Symposium | 22 countries of Latin America | Introduction to Human Cell Atlas and single-cell transcriptomics methodology | 170 |
| 2022 | On-line Workshop | 12 countries of Africa | Introduction to single-cell RNA-sequencing analysis | 63 |
| 2022 | On-line Workshop | 18 countries of Africa | Repeat of the previous workshop on Introduction to single-cell RNA-sequencing analysis | 51 |
| 2023 | In-person Workshop | 12 countries of Africa | Single-cell RNA_sequencing: Computational and experimental designs | 60 |
| 2023 | In-person Workshop | 7 countries of South America | Human Cell Atlas Latin America: Computational and Experimental Design Workshop | 42 |
| 2023 | Online Seminar Series | Sri Lanka | Single-Cell Genomics | 163 |
| 2024 | In-person Workshop (held in Bangkok, Thailand) | 13 countries of the Asia Pacific region | Single-Cell Omics: Computational and Experimental Design Workshop | 64 |
| 2024 | Hybrid mode Symposium | 3 nodal countries of Latin America (Chile, Brazil, Mexico) | Human Cell Atlas Latin America Symposium | > 100 |
The leadership of the HCA that initially included a few visionary scientists has been expanded to include prominent scientists from nearly every region of the world. Currently, about a quarter of the HCA leadership (the Organizing Committee) are from Asia, Australia, Latin America and the Middle East. In addition, the Equity Working Group – one of the earliest Working Groups established by the HCA – was also sequentially expanded with members from disparate global regions; currently 11 of the 15 members are from outside of North America or Europe.
The HCA has developed a number of regional networks, including HCA Asia, HCA Latin America and HCA Africa, and research networks—such as, the Genetic Diversity Biological Network—to ensure that the effort remains connected—and relevant—to communities, internationally. These networks efficiently promote engagement of local communities, co-ordination and dissemination.
Consent and sample collection: The process of sharing information about HCA, especially its goals and expected outcomes, to ethically enroll informed study participants involves group discussions with local communities in their own languages. Of course, written informed consent is taken from each volunteer willing to participate in HCA after the volunteer has understood the risks and benefits of participation, prior to collecting samples with adequate safeguards in place, making sure to minimize risks and pain associated with sample collection. The information provided to each individual mainly comprises the intent of the project, the benefit to be accrued in the long run even if not immediately, how the collected biospecimen and the generated data will be used, stored and shared, risks of participation and modalities of later withdrawal from the project. Each collected sample is stored in a vial labelled with a code. The coding process ensures that “direct identifiers” such as name and address are removed, helping to maintain sample donor anonymity.
The nature of samples collected in a HCA project depends on the specific objectives of the project. Indeed, to create a map of the human body, different tissue sampling scenarios must be envisaged depending on the source of the tissue examined and how the sample can be obtained (e.g., post-mortem sampling, use of leftover clinical/surgical tissue, adult participants, pediatric participants, etc.). For example, HCA investigators who are studying the process of infection by SARS-CoV-2 coronavirus have collected nasal and throat swabs from living donors as well as post-mortem lung tissue from deceased individuals who succumbed to COVID-19; those studying the cellular processes associated with childbirth are collecting cord blood and placenta; those studying immune-system development are collecting blood; those studying skin disease are collecting skin tissues, and those creating cell atlases of internal organs such as the lung are collecting tissues using bronchoscopy or organ donations which were ultimately considered inadequate for transplant.
Sample analysis: The collected samples are processed so that information can be collected from single cells and nuclei (for single cell genomics) or tissue sections (for spatial genomics). This process is performed under the strict supervision of well-trained scientists and with great care to minimize wastage of collected samples. At this time, all directly identifying information is removed prior to DNA/RNA analysis of each cell. HCA recommends that, to the extent possible, sample analysis be done in a laboratory close to the community collection site by local scientists. The HCA helps facilitate training to support this. Between 2019 and 2024, the Equity Working Group (EqWG), created to promote equity in the HCA, has conducted roadshows, meetings and training programs in Thailand, Brazil, Ethiopia, India, Ghana, Chile, and two global online training programs on introduction of single-cell RNA sequence analysis. HCA believes that efforts should be made to enable the sample-processing laboratories to be open to visits by study participants and community members. Scientists should provide explanations to such visitors of the various stages of sample analysis.
Data collection: HCA projects are led by scientists who are designated as principal investigators. Data collected from study participants and generated from the experiments on samples collected from them are stored in databases controlled by principal investigators of the study and in publicly-accessible global databases for advancement of science, following strict legal and ethical standards. Only codes–but not “direct identifiers” such as name, address, etc.–are used to mark individual participants in these databases. The data comprise those that are collected from each participant, such as age at the time of collection, gender, information on how the cells were collected, and the primary data generated in the laboratory, such as levels of expression of genes in single cells. Depending on the study, some general information about the study participant’s health (e.g., whether the participant has high blood pressure or has ever been infected with malaria) and environment (e.g., whether the participant lives in a city or near a humid, forested area, or whether the participant typically cooks with gas, charcoal, wood, or another fuel source) might also be recorded. Altogether, such ancillary information helps scientists understand the different factors that might influence the behavior of genes in human cells.
The Human Cell Atlas has created a cloud-based Data Portal that stores data contributed by HCA collaborators. A Data Explorer interface has been created to help researchers find and explore HCA datasets; https://data.humancellatlas.org. As determined by the ethics approval associated with the study, some portions of the data (e.g. the raw sequencing data) may only be accessible by researchers with permission of HCA’s Data Access Committee through HCA’s controlled access repository.
Data dissemination, access and use: HCA data are analysed using statistical, computational and bioinformatic methods. HCA encourages and supports training and engagement of local HCA investigators or other persons from the community to participate equitably in data analysis. HCA researchers have been developing experimental and computational methods that can be effectively deployed in different settings and sharing these by organizing local workshops and scientific consultations. After appropriate analyses of data are completed and conclusions drawn, those general conclusions are conveyed to study participants if they wish to know, or shared with the community in a summary form through local scientists. The HCA strongly advocates that community meetings be organized by local scientists and summary results be explained to the participating communities.
Subsequently, the data – stripped of all direct identifiers but coded to mark individual participants – are placed on a globally-accessible, public-domain database for the advancement of science by not only HCA investigators, but by anyone who may be interested to do so by analyzing the data. Data dissemination in HCA is in accordance with the Fort Lauderdale principles (https://www.genome.gov/Pages/Research/WellcomeReport0303.pdf); details of HCA data release policy are provided on the HCA web site (https://www.humancellatlas.org/wp-content/uploads/2019/07/2019-Jul-09-HCA-Data-Release-Policy-v1.0.pdf). It follows in the footsteps and ethos of other large-scale resources that were built to share and disseminate biomedical datasets (particularly “omics” data) Examples of these databases include the Human Genome Project14, the International HapMap Project15, the 1000 Genomes Project16, the Human Pangenome Project17, the International Cancer Genome Consortium18, as well as local or regional initiatives such as the UK Biobank19, AllofUs20, H3Africa21, to name a few.
In some instances, access controls are used to further heighten the privacy and security guarantees made to research participants. If a researcher contributes unpublished data, on the request of the researcher the data can be “embargoed” until published. Data sharing practices in health sciences generally propose a spectrum ranging from controlled (or managed) access data to open data, where datasets are made publicly available without restrictions. Under a controlled (or managed) access mechanism, datasets are only accessible to researchers having met a certain number of prerequisite conditions (e.g. proposed research is in line with the allowed use of data, research team has the necessary expertise to handle data, ethics approvals are in place, etc.) and in many cases, access requests are governed by a Data Access Committee and subject to a data transfer/sharing agreement. Since its inception, the HCA has favoured an open access data model, whereby datasets with appropriate permissions (e.g. participant consent, institutional approvals, etc.) are released in a public, open access, database. Indeed, this ethos was adopted to enable equitable, rapid, free and unencumbered access to its resources, which are developed collaboratively by researchers around the world22. However, the HCA also recognises that while ultimately, the objective of open access is to reduce the barriers to using and sharing data, it can also unintentionally hinder participation of certain groups, especially where there are concerns of potential stigmatization13,23. Therefore, in an effort to include a wide range of contributions, alongside open access, HCA has also implemented a ‘managed access’ tier whereby datasets that are more sensitive can be deposited, managed and access granted to researchers following approval by a centralized HCA Data Access Committee and signature of a data access agreement between the approved researcher’s institution and the HCA. Since genetic information collected in the RNA and DNA sequences can sometimes be used to predict family relationships between donors, HCA encourages investigators to adopt the best privacy and security practices prevailing (e.g., GA4GH “Data Privacy and Security Policy”24) to maintain the databases and promote responsible use of the HCA data.
Discrimination: Some biological characteristics are observed to be similar within family groups, and among communities with similar ancestries. Regrettably, these similarities and differences have sometimes been used for racial profiling, discrimination and exploitation. Notable historical examples of misuse of research data include, for instance, the Tuskegee Syphilis studies25, the Havasupai Indian Tribe case7, or, more recently, genetic research done without appropriate consent on DNA samples from Xinjiang (Uyghur individuals)26,27 and Tibet28. There are many more examples of data/sample misuse or misinterpretation, often discriminatory in nature, including attempts and approvals to commercialize drugs based on “race”29.
Race is a social construct, not a biological one. HCA strongly opposes the use of biological differences for social discrimination, which is a violation of fundamental human rights and freedoms. Instead, all efforts should be made to explain the scientific meaning of the data and to answer questions from individuals and communities on how information related to biological differences, including genetic, types and numbers of cells, among others, might be used with time to improve the health and well-being of the community. The HCA is aware that misinterpretation of observed biological patterns could lead to genetic discrimination – a source of exclusion and stigmatization. It considers racial profiling or any form of discrimination to be against the spirit of the consortium, the purpose of the data collection, and the ethical conduct that is expected to be followed by the consortium members. Through its governance, HCA places strong emphasis on accountability in the ethical conduct of its scientific research embodying high levels of quality, equity and integrity.
Conclusion: Implications for Future Biomedical Research
The HCA has embedded its commitment to humanity through various means, notably by engaging local scientists and other local persons. It has developed a number of regional networks that facilitate the organization of local meetings and dissemination events.
The Equity Working Group (EqWG) was created by the HCA in order to promote and support progress towards equity in the HCA. The EqWG seeks to engage the global community spanning diverse geographic and ethnic groups to drive inclusive representation and participation, and promote equal benefit from the HCA. Since its inception, the EqWG has adopted an ‘equity in action’ approach – with its activities centred around empowerment to participate in the HCA through outreach, education, and training1.
The Ethics Working Group of the HCA has also supported efforts in implementing the HCA worldwide, through the creation of an ethics toolkit (available online at: https://www.humancellatlas.org/ethics/) This toolkit aims to provide researchers from around the world with templates (e.g., consent form models, information pamphlets) and governance documents (e.g., HCA FAQ, data sharing/material transfer agreements, etc.) in order to implement HCA research in their own communities and institutions. These were developed to be ‘internationally interoperable’ given that legal and ethics standards can vary in different jurisdictions30.
It is hoped that the reference maps created by the HCA will help researchers understand whether every person afflicted with a disease – for example, liver cancer – has common biological or cellular changes in the affected organ. Knowledge gained from this work will result in a better understanding of both health and management and care of individual patients. To achieve this ultimate goal, it is important that the reference maps reflect biological and cellular variation present in various organs among healthy individuals from diverse backgrounds, including variable genetic ancestries, sex, age, geography, environment, and lifestyle. Unless there is an understanding of the extent of variation among healthy persons, it will not be possible to detect changes that may lead to the disease being studied. For example, blood and urine tests could be misleading if they are not calibrated to the natural range of variation observed in a relevant group of individuals. The encyclopaedia of cells that is being created by the HCA will be an important step towards accomplishing the ultimate goal of providing better care to individual patients, immensely beneficial to all of humanity.
The HCA’s action-oriented approach to fostering global partnerships by highlighting the fundamental goal of benefitting humanity (though applying principles of equity, diversity and inclusion) can also serve as a model for other biomedical consortia. Indeed, both its overall philosophy, as well as tools for implementing it – for instance, through working groups, online toolkits (e.g., ethics documentation) and roadshows and training workshops can foster adoption of common strategies and interoperability between similar initiatives.
Acknowledgements
An earlier version of this manuscript was archived on Open Science Forum; DOI 10.17605/OSF.IO/EZVC5; https://osf.io/sk697/. There is considerable overlap in the narratives of this manuscript with the archived version. We have not placed the overlapping text within quotes and have not referenced the archived version in this manuscript. We are grateful to Tracey Andrew, John Randell, Kristine Schwenck, Ellen Todres and Samantha Wynne, for help, suggestions and support. Figure 1 is licensed under Creative Commons CC BY-ND 4.0. This publication is part of the Human Cell Atlas: www.humancellatlas.org/publications/.
Author contributions
P.M., A.K. Shalek, and M.M. conceived of the idea of writing this commentary. The first draft was prepared by them and supplemented by E.K. and B.M.K. All authors contributed to the development and writing of this manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Competing interests
A.R. is an employee of Genentech, a member of the Roche group, and has equity in Roche. A.R. was a co-founder and equity holder of Celsius Therapeutics, is an equity holder in Immunitas, and until July 31, 2020 was an S.A.B. member of Thermo Fisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics and Asimov. A.R. is an inventor on multiple patents related to single cell and spatial genomics. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Majumder, P. P., Mhlanga, M. M. & Shalek, A. K. The human cell atlas and equity: lessons learned. Nat. Med26, 1509–1511 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative Colitis. Cell178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rood, J. E. et al. Impact of the human cell atlas on medicine. Nat. Med28, 2486–2496 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Bustamante, C. D., De La Vega, F. M. & Burchard, E. G. Genomics for the world. Nature475, 163–165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popejoy, A. B. & Fullerton, S. M. Genomics is failing on diversity. Nature538, 161–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.https://www.onecellatatime.org/.
- 7.Mello, M. M. & Wolf, L. E. The Havasupai Indian tribe case—lessons for research involving stored biologic samples. N. Engl. J. Med.363, 204–207 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Lemke, A. A. et al. Addressing underrepresentation in genomics research through community engagement. Am. J. Hum. Genet.109, 1563–1571 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popejoy, A. B. et al. The clinical imperative for inclusivity: race, ethnicity, and ancestry (REA) in genomics. Hum. Mut.39, 1713–1720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Advancing Diverse Participation in Research with Special Consideration for Vulnerable Populations. Am J Hum Genet. 107, 379–380 (2020). [DOI] [PMC free article] [PubMed]
- 11.Seaver, L. H. et al. Points to consider to avoid unfair discrimination and the misuse of genetic information: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med24, 512–520 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Adame, F. Meaningful collaborations can end ‘helicopter research.’ Nature.10.1038/d41586-021-01795-1 (2021). [DOI] [PubMed]
- 13.Improving equity in human genomics research. Commun Biol5, 281 10.1038/s42003-022-03236-9 (2022). [DOI] [PMC free article] [PubMed]
- 14.Lander, E. S. et al. International human genome sequencing consortium. initial sequencing and analysis of the human genome. Nature409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 15.International HapMap Consortium. The international HapMap project. Nature426, 789–796 (2003). [DOI] [PubMed]
- 16.1000 Genomes Project Consortium. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed]
- 17.Wang, T. et al. Human pangenome reference consortium. the human pangenome project: a global resource to map genomic diversity. Nature604, 437–446 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Cancer Genome Consortium. International network of cancer genome projects. Nature464, 993–998 (2010). [DOI] [PMC free article] [PubMed]
- 19.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med381, 668–676 (2019). [DOI] [PMC free article] [PubMed]
- 21.H3Africa Consortium. Research capacity. Enabling the genomic revolution in Africa. Science344, 1346–1348 (2014). [DOI] [PMC free article] [PubMed]
- 22.Amann, R. I. et al. Toward unrestricted use of public genomic data. Science363, 350–352 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Hudson, M. et al. Rights, interests and expectations: Indigenous perspectives on unrestricted access to genomic data. Nat. Rev. Genet21, 377–384 (2020). [DOI] [PubMed] [Google Scholar]
- 24.GA4GH, Global Alliance for Genomics and Health. Data Security and Privacy Policy. https://www.ga4gh.org/product/data-privacy-and-security-policy/#:~:text=Developed%20by%20the%20GA4GH%20Regulatory,privacy%20of%20individuals%2C%20families%2C%20and
- 25.Brandt, A. M. Racism and research: the case of the Tuskegee syphilis study. Hastings Cent. Rep.8, 21–29 (1978). [PubMed] [Google Scholar]
- 26.Molecular Genetics & Genomic Medicine. RETRACTED: Haplotypic diversity and population genetic study of a population in Kashi region by 27 Y-chromosomal short tandem repeat loci. Mol. Genet. Genom.Med.8, e1338 (2020). [DOI] [PMC free article] [PubMed] [Retracted]
- 27.FSI Genetics. Retraction notice to “Analysis of Uyghur and Kazakh populations using the precision ID ancestry panel” FSI Genet66, 102902 (2023). [DOI] [PubMed]
- 28.Molecular Genetics & Genomic Medicine (2021) RETRACTED: Concordance and characterization of massively parallel sequencing at 58 STRs in a Tibetan population. Mol. Genet. Genom. Med.9, e1626 (2021). [DOI] [PMC free article] [PubMed] [Retracted]
- 29.Krimsky, S. The short life of a race drug. Lancet379, 114–115 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Knoppers, B. M., Bernier, A., Bowers, S. & Kirby, E. Open data in the era of the GDPR: lessons from the human cell atlas. Ann. Rev. Genomics Hum. Genet24, 369–391 (2023). [DOI] [PubMed] [Google Scholar]