Abstract
NLR receptor is suggested as a component of plant nonhost resistance (NHR). However, the evolutionary process of how plants develop receptors for recognizing broad-spectrum pathogens is still elusive. Here, we observe that multiple RxLR effector families including 12 reported avirulence effectors of Phytophthora infestans are broadly conserved across the Phytophthora species. We select 69 effectors distributed into 8 families from 6 Phytophthora species, and confirm that 60.87% of the tested effectors are recognized by Solanum NLRs according to their defined families. Furthermore, we confirm that expression of R1, R8, and Rpi-amr1 confer broad-spectrum resistance against multiple Phytophthora species. Combined results suggest that conserved effector families of Phytophthora species allow solanaceous plants to recognize broad-spectrum pathogens via NLRs that originally reported to recognize P. infestans. Thus, NLR-mediated recognition would contribute to NHR against pathogens that possess similar repertoires of effectors. Moreover, this homology-based approach would be applicable to other plant-pathogen systems and provide an alternative strategy of genetic mapping to identify functional NLRs against various crop-threatening pathogens.
Subject terms: Effectors in plant pathology, Molecular engineering in plants, Virulence
Genus Phytophthora threat various plant species including fruit, vegetable, and flower crops. The authors report that some NLRs recognize conserved effector families of multiple Phytophthora species and confer broad-spectrum resistance in plant.
Introduction
In nature, while plants are surrounded by numerous pathogenic microorganisms, only a portion of those pathogens which are adapted to a given plant are able to colonize the plant. In this context, nonhost resistance (NHR) could be simply defined as the resistance of a plant species against non-adapted pathogens1. To date, a number of studies have investigated molecular mechanisms of NHR, and currently, NHR is thought to rely on the cooperation of multiple layers of the plant immune system2, including pre-formed physico-chemical barriers and receptor-mediated induced immune responses3,4. The molecular mechanism of NHR is quite depend on context, and evolutionary distance between original host and nonhost plant of a given pathogen have been suggested as a key factor of determining which component of plant immune system majorly contribute to the each circumstance4,5. In particular, cytoplasmic immune receptor proteins (NLRs) are also suggested as a component of NHR1. Various loss-of-function approaches6–8 and multiple cases of transferring NLRs derived from nonhost plant into host plant9–13 support that NLR-mediated immunity is associated with NHR. However, several questions still remain regarding the NLR-mediated NHR, such as ‘How and why does plant retain receptors for recognizing non-adapted pathogens?’. Considering that pattern recognition receptors (PRRs) recognize broadly conserved pathogen-associated molecular patterns (PAMPs), it seems quite possible that PRRs recognize PAMPs from both evolutionarily distant adapted and non-adapted pathogens of a given plant14–16. However, NLRs recognize effectors that are relatively less conserved and generally known to have lineage-specific sequence diversity17,18. Thus, it is relatively hard to imagine how NLRs contribute to recognition of a wide range of pathogen species including non-adapted pathogens of a given plant.
Several recent studies have provided clues on the mechanism by which NLRs contribute to the recognition of effectors from a wide range of pathogens. In a bacterial system, a screen of 529 effectors distributed into 70 families of multiple Pseudomonas syringae pathovars revealed that multiple Arabidopsis NLRs are able to recognize orthologous effectors of closely related Pseudomonas syringae pathovars19. Similarly, in oomycetes, Rpi-amr3 (resistance gene against P. infestans) is reported to recognize Avramr3 orthologs of multiple Phytophthora species20. These studies suggest that certain effectors that are conserved across both adapted and non-adapted pathogens of a given plant could be similarly recognized by NLRs and may contribute to broad-spectrum resistance.
The Phytophthora genus is composed of multiple subclades and more than one hundred species21. Each Phytophthora species is adapted to infect its host plant species. Thus, Phytophthora genus threaten a wide range of plants, including vegetables (Solanaceae, Cucurbitaceae, Fabaceae), fruits, orchids, and even trees22–24. Since various Phytophthora isolates that tolerant to fungicide have been continuously reported24,25, there has always been a demand for breeding resistant crops against Phytophthora pathogens. Indeed, approximately 2–30 NLR type resistance genes against P. infestans have been identified in the wild Solanum species26,27. However, only a few resistance genes against most other Phytophthora species have been reported.
In this context, we assumed that not only Avramr320, but also other effectors might be conserved across the evolutionarily related Phytophthora species, and some of these conserved effectors could also be similarly recognized by the previously reported Solanum NLRs that originally known to recognize P. infestans effectors. To test this hypothesis, we focused on the RxLR (conserved N-terminal Arg-Xaa-Leu-Arg motif) effectors of multiple Phytophthora species21,28. We discovered that multiple effector families, including 12 well-characterized P. infestans avirulence effectors are conserved across Phytophthora species. In addition, 42 out of the 69 tested homologous effectors were recognized by Solanum NLRs which were originally reported to recognize avirulence effectors of P. infestans according to their defined families. Finally, we confirmed that three Solanum NLRs, R1, R8, and Rpi-amr1 are capable of conferring broad-spectrum resistance against multiple Phytophthora species in addition to P. infestans. These results reveal the landscape of how effectors are conserved across the Phytophthora genus and provide evidence supporting the role of NLRs in resistance against a certain range of pathogens by recognizing those effectors.
Breeding disease resistant crops through classical approaches requires a number of genetic resources and enormous costs. Since resistance genes against most Phytophthora species that threaten various crop species, such as soybean, chili pepper, pumpkin, cacao, apple, strawberry, orchid, and tobacco, have never been identified, the NLRs identified in this study support that homology-based reverse genetic screening could be an alternative strategy of genetic mapping to identify functional NLRs against a wide range of pathogens. In addition, these resources could also be exploited for improving crop resistance against multiple Phytophthora species.
Results
Multiple RxLR effector families are conserved across Phytophthora species
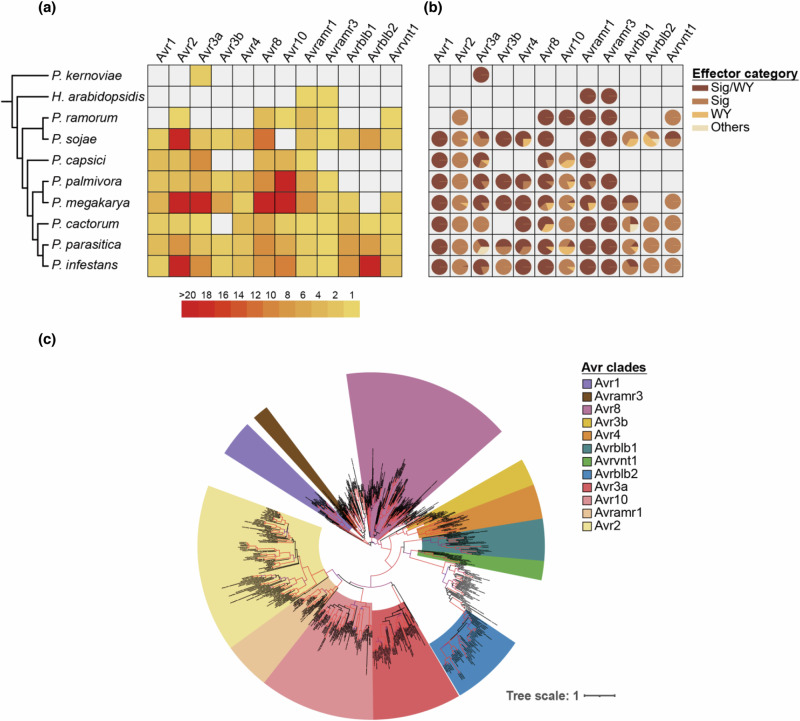
To investigate the conservation of RxLR effectors across plant pathogenic oomycetes, we searched for homologs of 12 reported P. infestans avirulence effectors, including Avr129,30, Avr231, Avr3a32, Avr3b, Avr433, Avr834, Avr10, Avramr135, Avramr320, Avrblb136, Avrblb237, and Avrvnt138 in the protein database of 12 oomycete species (Supplementary Table 1, 2). We clustered 243,558 proteins from 12 oomycetes into 19,039 orthogroups, and obtained 647 homologous effector candidates from 34 orthogroups (Fig. 1a, Supplementary Fig. 1, 2, and Supplementary Table 2). Candidate effectors were categorized through motif and domain analyses to determine the structural features of RxLR effector (Fig. 1b), and these effectors were further classified into each family through phylogenetic analyses (Fig. 1c). Subsequently, 514 homologous effector candidates of Phytophthora species remained after excluding non-homologous candidates from the phylogenetic analysis (Supplementary Fig. 1, Supplementary Data 1). The majority of investigated P. infestans effectors were conserved within the closely related Phytophthora species, and some families such as Avr3a and Avramr1 were conserved beyond Hyaloperonospora arabidopsidis (Fig. 1b). Moreover, by comparing the conservation of effector families based on phylogenetic (patristic) distances between the reference P. infestans avirulence effectors and their closest homologs in each Phytophthora species, Avr1, Avr3a, Avr8, Avramr1, and Avramr3 were found to be more conserved than other families in multiple Phytophthora species (Supplementary Fig. 3).
Fig. 1. Multiple avirulence effector families are conserved across Phytophthora species.
a The number of homologs of each reference avirulence effector of P. infestans in multiple Oomycetes species. Light gray boxes indicate absence of homologous effector. Full species names are described in Supplementary Table 2. b The proportion of categories based on the signal peptide (Sig) and WY domain (WY) of homologous effectors. Others represent effector candidates without Sig and WY. The detailed domain and motif information of homologous effectors is provided in Supplementary Data 1. c The phylogenetic relationships of homologous effectors against reference avirulence effectors. The range of colors indicates the clade of each reference avirulence effector. The red branches represent ultra-fast bootstrap value above 95.
We also performed protein structure prediction and structure-based clustering of 514 homologous effector candidates to strengthen our phylogeny-based classification of effector families. The accuracy of the predicted protein structure was evaluated using the predicted local distance difference test (pLDDT) score, and most effectors with WY domain structures had a high pLDDT scores (Supplementary Fig. 4). Structure-based clustering revealed that most effector family members, except for Avrblb2, were structurally conserved within multiple Phytophthora species (Supplementary Fig. 5). In most cases, a positive correlation was observed between the protein sequence similarity and the structural homology of each homologous effector candidate relative to the corresponding reference P. infestans effector (Supplementary Fig. 6). The combined results suggest that multiple effector families are conserved among Phytophthora species.
Homologous effectors of multiple Phytophthora species are recognized by Solanum R genes
Given that Avramr3 and its orthologs in multiple Phytophthora species were recognized by Rpi-amr320, we expected that the other conserved effector families of multiple Phytophthora species could also be similarly recognized by their putative corresponding Solanum NLRs according to their defined families (Fig. 1a, Supplementary Fig. 2, 3, 5). To test whether the homologous effectors of multiple Phytophthora species could be similarly recognized by their cognate Solanum NLRs, we minimized the candidate pool to the proper size for the experimental validation. We selected effectors that exhibited higher sequence and structural similarity against each reference avirulence effector of P. infestans (Avr1, Avr2, Avr3a, Avr8, Avr-amr1, Avr-blb1, Avr-blb2, and Avr-vnt1) by comparing bit-scores and pTM scores (Supplementary Fig. 6). Candidate effectors were obtained from P. parasitica, P. palmivora, P. cactorum, P. capsici, and P. sojae which are able to colonize the Solanaceae model plant N. benthamiana, except for P. sojae (Supplementary Table 3).
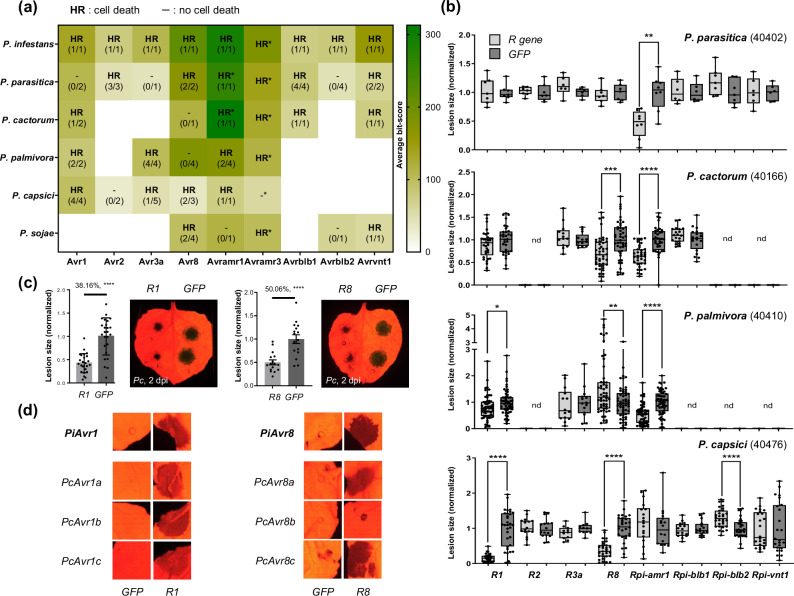
Among the 514 candidates, we successfully cloned 69 effectors distributed into 8 families (Supplementary Data 2) into an expression vector (Supplementary Data 3). Cloned effectors were co-expressed with their putative corresponding Solanum NLRs (R139, R240, R3a41, R842, Rpi-amr143, Rpi-blb136,44, Rpi-blb245, and Rpi-vnt138,46) in N. benthamiana. As a result, 60.87% (42/69) of the tested effectors induced cell death upon being co-expressed with their putative corresponding Solanum NLR (Fig. 2a, Supplementary Table 4, Supplementary Data 2). To our surprise, all the tested Solanum NLRs induced cell death against multiple homologs derived from multiple Phytophthora species, with the exception of Rpi-blb2 (Fig. 2a, Supplementary Data 2). Considering that Rpi-amr1/3 are reported to recognize Avramr1/3 orthologs of multiple Phytophthora species20,43, eight of the nine investigated effector families (89%) of Phytophthora species are conserved enough to be recognized by the same Solanum NLRs (Fig. 2a, Supplementary Fig. 7b, 9b, 10b, 12b, Supplementary Data 2). Based on these results, NLR-mediated recognition of conserved effectors of Phytophthora species may also be applicable to other effector families that were not functionally validated in our investigation due to the lack of identified cognate NLRs.
Fig. 2. Solanum NLRs are able to recognize conserved effectors of multiple Phytophthora species and confer resistance in Nicotiana benthamiana.
a Eight Solanum NLRs, except for Rpi-amr3 (results presented with asterisk mark as HR*/-* were previously reported in Witek et al.43; Lin et al.20), were co-expressed with their putative corresponding effectors of multiple Phytophthora species in N. benthamiana leaves. The number of HR positive/tested effectors are presented as fraction for each case (blank space means not tested). Average bit-scores obtained by comparing 69 cloned effectors against each reference avirulence effector of P. infestans are presented as heatmap. b Average lesion size of four Phytophthora species on N. benthamiana leaves expressing one of the eight Solanum NLRs and GFP (used as negative control, normalized to 1.0) are presented as box plot, the dots show individual values, while the boxes display the first quartile, median, and third quartile, with whiskers extending to the smallest and largest values. Statistical significances were calculated from unpaired t-test (two-tailed *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; nd: non-determined). The number of replicates is presented in the corresponding Supplementary Figs. 7–12. c Representative image of reduced lesion of P. capsici (40476) on R1-expressed half compared to the GFP-expressed area in N. benthamiana leaf and bar graph presenting average lesion size (left), image and graph for the case of R8 (right). Statistical significances were calculated from unpaired t-test (two-tailed ****P < 0.0001) and presented with the error bands (percentiles indicate average lesion size of R1 or R8/GFP-expressed leaves). d Representative images of R1-mediated cell death when co-expressed with Avr1 homologs of P. capsici (left), and R8-mediated cell death against Avr8 homologs of P. capsici (right).
Solanum NLRs confer resistance against multiple Phytophthora species by recognizing conserved effectors
To determine whether the Solanum NLR-mediated cell death phenotypes against conserved effector families of multiple Phytophthora species are also associated with resistance against multiple Phytophthora species, we transiently expressed each Solanum NLR in N. benthamiana and inoculated four Phytophthora species (P. parasitica or Pp, P. cactorum or Pcac, P. palmivora or Ppal, and P. capsici or Pc). As a result, although not all the Solanum NLRs that induce HR cell death against effectors of certain Phytophthora species could reduce lesion size of each pathogen, we observed that the expression of R1, R8, or Rpi-amr1 reduced lesion size of multiple Phytophthora species in addition to P. infestans (Fig. 2b, c, Supplementary Figs. 7–12, Supplementary Data 2).
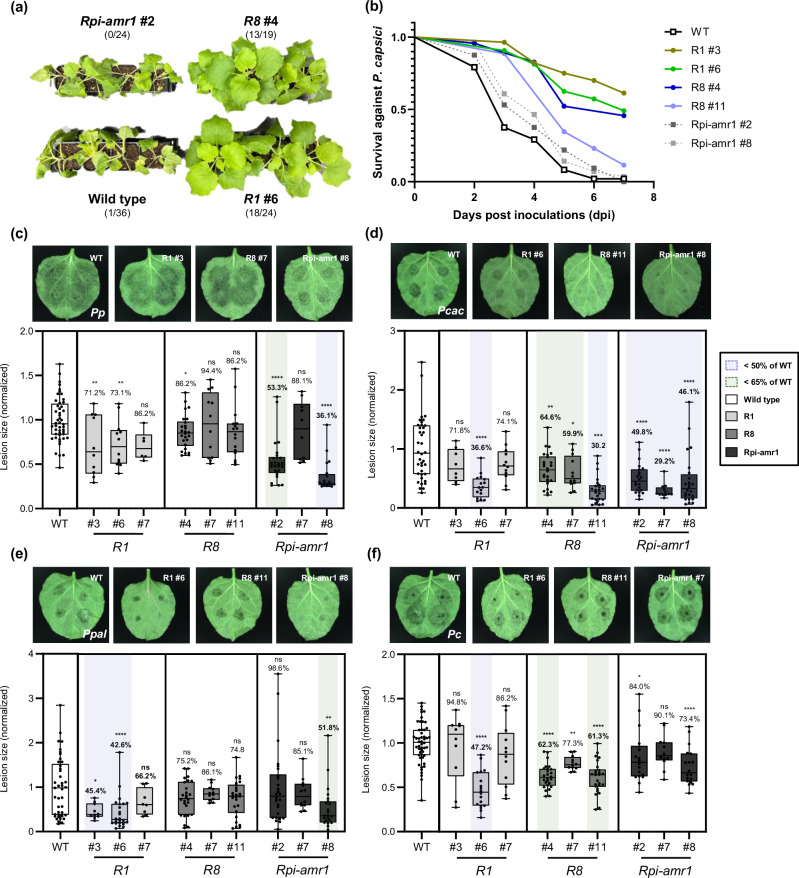
To validate Solanum NLR-mediated resistance against multiple Phytophthora species using transgenic plants, we generated transgenic N. benthamiana expressing R1, R8, or Rpi-amr1, through Agrobacterium-mediated transformation. We obtained 30, 15, and 21 T0 plants that survived on kanamycin media, which were thought to express R1, R8, or Rpi-amr1. To select T0 plants that properly expressed R1, R8, or Rpi-amr1 by observing effector-triggered cell death phenotypes, Avr1, Avr8, or Avramr1 was transiently expressed in the leaves of each T0 plant (Supplementary Fig. 13a, 13b). T1 seeds were harvested from the confirmed lines that induced cell death against transient expression of Avr1, Avr8, or Avramr1, respectively (Supplementary Fig. 13c).
The roots of T1 transgenic plants expressing R1, R8, or Rpi-amr1 were inoculated with Pc zoospores for the whole plant disease resistance assay (Fig. 3a, Supplementary Figs. 14, 15, 16) and detached leaves of each plant were inoculated with zoospores of Pp, Pcac, Ppal, or Pc (Fig. 3c–f). From the detached leaves assay (DLA), similar results were observed compared to the transient expression-based DLA result (Figs. 2b, 3). Using transgenic N. benthamiana, we additionally found that expression of R1 slightly reduced lesion size of Pcac (Fig. 3d). Indeed, the results obtained using transgenic plants were more reliable with HR screening test, because R1 induced cell death in response to the transient expression of PcacAvr1 (Fig. 2a, Supplementary Fig. 9a). Similar to the DLA results for Pc (Figs. 2b, c, and 3f), R1 and R8 transgenic plants exhibited significant resistance against root infection caused by soil drenching of Pc zoospores, while wild type and Rpi-amr1 transgenic plants exhibited complete wilt and shrivel phenotypes (Fig. 3a, b, Supplementary Figs. 14, 15, 16).
Fig. 3. Transgenic N. benthamiana expressing R1, R8, or Rpi-amr1 conferred resistance against multiple Phytophthora species.
a Representative images for the P. capsici root infection assay. T1 plants expressing R1 or R8 exhibited significant resistance against P. capsici compared to the wild type or T1 Rpi-amr1 plants. The number of healthy/tested plants are described as fraction. b Survival rate (healthy/tested plants) of each transgenic line in P. capsici root infection assay (combined results obtained from three independent trials) is presented as line graph. Phenotypes were observed until all wild type plants were completely wilt. c–f Representative images of (c) P. patasitica 40402 (Pp); (d) P. cactorum 40166 (Pcac); (e) P. palmivora 40410 (Ppal); and (f) P. capsici 40476 (Pc) lesions inoculated on wild type (WT), R1, R8, or Rpi-amr1 transgenic plants (upper) and the average lesion size of each case is presented with box plot, the dots show individual values, while the boxes display the first quartile, median, and third quartile, with whiskers extending to the smallest and largest values. Average lesion size of each pathogen on the WT plant were normalized to 1.0, and the ratio of average lesion size on transgenic/WT plant are marked as percentage above each box (for the cases of average lesion size of transgenic/WT plant < 50% and < 65%, are marked with blue and green shades, respectively). Statistical significances were calculated from unpaired t-test (two-tailed *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
In conclusion, four of the nine investigated Solanum NLRs, including Rpi-amr3, which has previously been reported to confer resistance against P. infestans, P. parasitica, and P. palmivora20, conferred broad-spectrum resistance against multiple Phytophthora species (Figs. 2, 3). These results suggest that effectors which are conserved across multiple Phytophthora species would enable plants to recognize and fend off a wide range of pathogens through NLRs.
Sequence- and structure-based identification of homologous effectors enable to search broadly conserved effector families across the Phytophthora species
The combined results of Solanum NLR-mediated HR cell death against homologous Phytophthora effectors and resistance against multiple Phytophthora species stimulated us to determine the correlation between the sequence or structural similarities of the homologous effectors and the possibility of being recognized by corresponding Solanum NLRs. Specifically, we tried to identify the critical sequence or structural features of effectors that are recognized by the same Solanum NLRs.
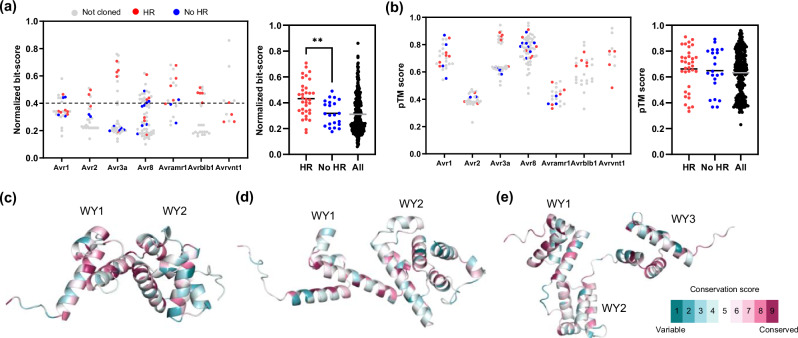
To determine common threshold of sequence and structural similarities in multiple effector families recognized by Solanum NLRs, we compared the normalized bit-scores and pTM scores of HR-positive and negative effectors to their corresponding reference effectors of P. infestans (Fig. 4a, b, Supplementary Fig. 17). We found that the overall structural conservation of candidate effectors (average pTM score: HR-positive = 0.66, HR-negative = 0.64) was higher than the sequence conservation (average normalized bit-scores: HR-positive = 0.43, HR-negative = 0.32), however, a statistically significant difference was observed only in sequence similarity between the HR-positive and HR-negative effectors (Fig. 4a, b). The average normalized bit-score of HR-positive effectors was significantly higher than the average of HR-negative effectors. Notably, 20 of 26 (76.92%) tested effectors with a 0.4 normalized bit-score were HR positive (Fig. 4a). However, the average pTM score of HR-positive and negative effectors were not significantly different (Fig. 4b).
Fig. 4. Sequence and structure-based identification of homologous effectors enable to search broadly conserved effector families across the Phytophthora species.
a Normalized bit-score of 316 homologous effectors (of P. infestans, P. parasitica, P. cactorum, P. palmivora, and P. sojae) against corresponding reference P. infestans effectors are plotted (left). Red dots, blue dots, and gray dots represent HR positive, negative, and not cloned effectors, respectively. Average normalized bit-score of HR positive, negative, and 316 homologous effectors presented as black and gray bar (right). Average normalized bit-score of HR positive effectors (n = 34) was significantly higher than HR negative effectors (n = 22). Statistically significance is analyzed with unpaired t-test (two-tailed **P < 0.01). b pTM score of 316 homologous effectors against corresponding reference P. infestans effectors are plotted (left). Average pTM score of HR positive, negative, and 316 homologous effectors presented as black and gray bar (right). Average pTM score of HR positive effectors was not significantly different from HR negative effectors. Statistically significance is analyzed with unpaired t-test. c–e Predicted effector domain structures of PiAvr1 (c), PiAvr8 (d), and PiAvramr1 (e). The WY domain is represented by four α-helix units. The amino acid conservation scores of structures were calculated in ConSurf server using sequence alignment of HR-positive effectors.
After observing the overall trends, we focused on several specific effectors which were not recognized by their putative corresponding Solanum NLR despite exhibiting high normalized bit-scores or pTM scores compared to reference P. infestans effectors. In particular, we compared the sequences and structures of the HR-positive and negative homologs of Avr1, Avr8, and Avramr1 families (Fig. 4c–e, and Supplementary Figs. 18, 19). Since the WY domain is known to be functionally critical and structurally conserved through effectors of Peronosporales species47, we predicted WY domain structure of HR-positive/negative effectors and observed that all the defined members of Avr1, Avr8, and Avramr1 families possess 2, 2, and 3 conserved WY domains, respectively (Fig. 4c–e, and Supplementary Fig. 18). However, structural comparison showed no significant differences between the WY domain of HR positive (Supplementary Fig. 18a, c, e) and negative (Supplementary Fig. 18b, d, f) effectors compared to the reference P. infestans effectors. But contrary, protein sequence alignment revealed that several residues were differently conserved between the HR-positive and negative homologs (Supplementary Fig. 19). In particular, PpalAvramr1a_1 and PpalAvramr1a_2 exhibited only several amino acid differences (94.25% identical) within the WY domains, despite of their opposite responses to Rpi-amr1 (Supplementary Data 2). These results suggest that several residual changes in the WY domain of homologous effectors of Phytophthora species may enable effectors to avoid from Solanum NLR-mediated recognition without significant changes in the overall structure of the effectors.
In conclusion, the combined results indicate that both sequence- and structure-based methods for defining homologous effector could be used to identify candidate Phytophthora effectors that could be similarly recognized by the same Solanum NLRs. However, in our dataset, a sequence similarity-based method would be better for identifying candidates with a higher probability of being recognized by NLRs because significant structural differences are hardly observed between the homologous effectors of Phytophthora species which were already defined through phylogenetic analysis and sequence-based clustering. Thus, additional functional validations and sequence/structure-based comparisons using less homologous (compared to the tested effectors used in this study) effector candidates could provide more accurate thresholds to be similarly recognized by Solanum NLRs. We expect the criteria used in this study to identify homologous effectors would contribute to search for functionally homologous effectors and identification of additional NLR-effector combinations in other plant-pathogen systems as well.
Discussion
This study suggests that multiple homologous effectors recognized by similar NLRs, could be identified through sequence-based comparisons and homology-based screening in evolutionarily related pathogens such as Phytophthora species. These evolutionarily conserved effectors of Phytophthora species enable plants to recognize a wide range of pathogens and would contribute to NHR. Furthermore, this study suggest that homology-based reverse genetic approach could be an alternative strategy for identifying functional plant resistance genes against pathogens that pose a threat to various crops, instead of the forward genetics breeding program that requires diverse genetic resources and long-term planning27. In addition, it is worth noting that resistance genes against most Phytophthora species, including P. cactorum and P. capsici, have not yet been identified.
NLRs recognizing effectors of broad-spectrum pathogens have a potential to be exploited to confer durable resistance in crops
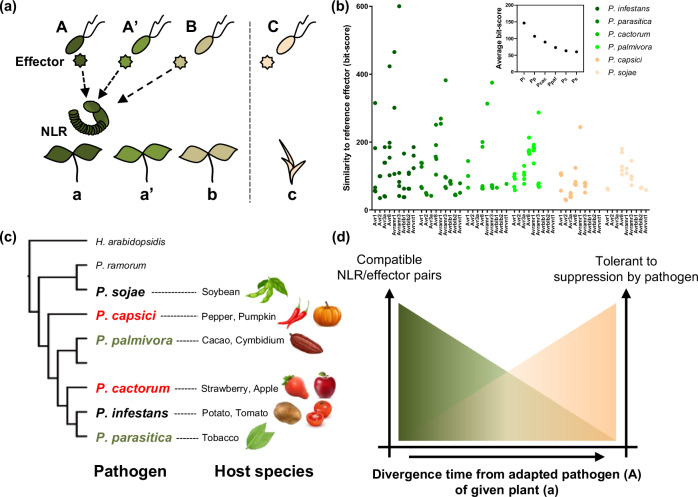
The conserved NLRs and effectors are thought to have originated from common ancestral plant and pathogen species that have struggled with each other in the past. As plant species diverge, each pathogen species also diverges and adapts to each host plant species. This evolutionary process is accompanied by changes in repertoires of NLRs and effectors (Fig. 5a). A previous study suggested that the relative contribution of NLRs to ‘how NHRs are mediated’ depends on the evolutionary distance between host and nonhost plants of the given pathogen species5. Similarly, this study showed that plants are possibly recognize effectors of pathogens which are recently diverged from the adapted pathogen of a given plant (pathogen A in Fig. 5a), because a similar repertoire of effectors are conserved in recently diverged pathogen species (Fig. 5a–c). Moreover, the number of functional NLRs that recognize effectors of evolutionarily distant pathogens (pathogen B or C in Fig. 5a) would decrease proportionally with the divergence time between pathogen species.
Fig. 5. NLR-mediated recognitions of conserved effectors would contribute to resistance against a wide range of pathogens.
a Pathogens diverged from common ancestral species adapt to its host plant species (with the same color) and this adaptation is accompanied with the changes in repertoire and sequence of effectors. However, effectors are conserved between the recently diverged pathogen species. Thus, a portion of conserved effectors could be recognized by same NLRs. b Bit-scores obtained by comparing homologous effectors (69 cloned effectors, Avramr3 and its homologs are included) against each reference avirulence effector of P. infestans are presented as dot graph, and average bit-score of whole tested effectors of each species is presented in a small boxed graph (above). Conservation of P. infestans reference avirulence effectors across the Phytophthora species gradually decreases with increasing divergence time from P. infestans. c Resistance genes against most Phytophthora species have never been identified (red: no NLRs were reported; green: few NLRs were reported) though they cause serious disease in various crops. d Considering that conservation of effectors decreased with increasing of divergence time between pathogen species, the number of compatible NLR/effector pairs would also similarly decrease. Simultaneously, pathogens are less-likely to suppress NLRs derived from evolutionarily distant plants from its host species.
Simultaneously, the NLRs that function against pathogen species that are more divergent from the adapted pathogen of a given plant species might be less possibly to be overcome by the pathogen (Fig. 5a and d). Indeed, among the tested Solanum NLRs, only Rpi-amr143 and Rpi-amr320 conferred significant resistance against P. parasitica, although six of the nine tested Solanum NLRs induced cell death against the cognate effector homologs of P. parasitica (Figs. 2, 3). Considering that S. americanum is evolutionarily more distant from potato compared to the other wild Solanum species such as S. demissum or S. bulbocastanum, we could assume that P. parasitica is relatively well adapted to potato and its closely related species but not to S. americanum. Thus, it may enable P. parasitica to colonize N. benthamiana expressing NLRs derived from closely related wild Solanum species, despite possessing multiple effectors that could be recognized by corresponding Solanum NLRs. Moreover, we observed a similar phenomenon that P. infestans was able to suppress Rpi-blb2 of S. bulbocastanum but could not suppress Rpi-blb2 homolog (CaRpi-blb2a) derived from chili pepper48, which plant is more diverged from potato compared to S. bulbocastanum. In this context, introducing NLRs from the evolutionarily more distant plant species could be a promising strategy for conferring durable resistance against pathogens adapted to a target crop.
Disparity between HR cell death against effectors and resistance against pathogens
All the tested Solanum NLRs, except for Rpi-blb2, were able to recognize effectors of multiple Phytophthora species (Fig. 2a). However, only R1, R8, and Rpi-amr1 conferred resistance to multiple Phytophthora pathogens (Figs. 2b, 3). These disparities between HR cell death and resistance phenotype may be due to the complexity of the virulence mechanisms of pathogen. When pathogens infect plants, they secrete hundreds of effectors into plant cells. Hence, while certain Solanum NLRs could induce HR cell death against Phytophthora species by recognizing the conserved effectors of pathogens, they may fail to confer resistance against the particular pathogen if the pathogen is able to suppress immune response of N. benthamiana by using numerous other effectors that target the essential components for NLR-mediated immune signaling49, such as NRC, or by directly suppressing the transferred sensor NLR. Furthermore, we cannot exclude the possibility that pathogens would transcriptionally regulate recognizable effectors or suppress them from being secreted, because the experiments performed in this study were mostly based on Agrobacterium-based overexpression. In addition, we observed increased lesions of Phytophthora pathogen in NLR-expressed N. benthamiana leaves in some cases (Fig. 2b). We assume that both NLRs (R8 and Rpi-blb2) cannot function as resistance gene (because R8 were not able to induce cell death against PpalAvr8 and no Avrblb2 in Pc as shown in Fig. 2a) against each pathogen but the unknown physiological changes occurred by over-expressing those NLRs provide more infectious environment in N. benthamiana leaves to each pathogen.
Therefore, obtaining a more comprehensive understanding of the detailed molecular mechanisms and host proteins that are associated with each NLR-mediated resistance and pathogen effectors would provide a strategy to harness the potential of these HR-inducing Solanum NLRs that cannot confer resistance in this study.
Transferring NLRs to evolutionarily distant plant species
In order to transfer functional NLRs to distantly related plant species, genetic compatibility between NLRs and their signaling components, such as NLRs and guardee/decoy proteins or helper and sensor NLRs, should be considered. Indeed, several studies have reported that each component of EDS1-PAD4-ADR1/EDS1-SAG101-NRG1 signaling complex is not fully compatible between tomato and Arabidopsis50. Similarly, CaRpi-blb2a is compatible with NRC8 of pepper but not with NRC4 of N. benthamiana48.
Considering that R1, R8, and Rpi-amr1 are NRC-dependent sensor NLRs43,51, transferring these NLRs beyond the solanaceous plants would be a complex task. However, transferring these NLRs into other solanaceous plants could be regarded as a feasible strategy. Thus, we expect that generating R1/R8-expressing pepper/tomato or Rpi-amr1-expressing tobacco/eggplant would be promising strategies to fend off P. capsici and P. parasitica, respectively.
Beyond Phytophthora species and solanaceous plants
Similar to this study, several cases of conserved effectors recognized by similar NLRs have been reported in different kingdoms of pathogens. For instance, various CCG effectors conserved in Albugo candida races are recognized by Arabidopsis WRR4A and WRR4B52, and Avramr1/3 homologs of multiple Phytophthora species are recognized by Rpi-amr1/320,43. Additionally, multiple effector families of Pseudomonas syringae, a bacterial pathogen, are also recognized by corresponding Arabidopsis NLRs19. In this context, we anticipate that the NLR-mediated recognition of conserved effectors between plant and pathogen species would also be applied beyond the Solanaceae-Phytophthora interaction. Therefore, identification of more NLRs recognizing broadly conserved effectors would contribute to reducing damage caused by a wide range of continuously evolving plant pathogenic microorganisms beyond Phytophthora species.
Methods
Plant materials and growth conditions
N. benthamiana were grown in a controlled chamber at 24–26 °C and 40–60% relative humidity with a 16-h light/8-h dark cycle. Commercial topsoil for general/horticultural (product of Seoulbio®, http://www.seoulbio.co.kr/product/product_04_01.asp) was used and additional nutrients were not added.
Maintenance conditions for pathogenic oomycete materials and sporulation methods
P. infestans (T30-4) were grown on rye sucrose agar plate media in a dark chamber at 17–19 °C for 7–9 days. P. parasitica (40164, 40402, 40906 strains were provided from RDA, Korea), P. cactorum (40166, 40183), P. palmivora (40402, 40410), P. capsici (40476) and P. sojae (48989, 40468, 40412) were grown on V8 juice plate media in dark chamber at 23–25 °C for 7–9 days. Except for P. infestans, 7 days old plate were flooded with 4 ml of TDW (triple distilled water) and placed under continuous fluorescent light during 2 days (1 days for P. capsici) for the sporulation (in case of P. infestans, 7 ~ 9 days old pate were flooded with chilled 8 ml of TDW and incubated at 4 °C for an hour and additional 30 min without sporulation step). Zoospores were harvested from flooded (pouring 6–8 ml of chilled TDW and rubbing mycelia) plates after being incubated at 4 °C for an hour and additional 30 min at 25 °C before used for inoculation.
Extraction of effector homologs against functional avirulence effectors
Protein sequences of the 12 oomycete genomes were downloaded from the NCBI GenBank database (P. cactorum [GCA_016864655.1], P. megakarya [GCA_002215365.1], and P. palmivora var. palmivora [GCA_002911725.1]) and the Oomycete Gene Order Browser (OGOB, https://ogob.ie/v1/gob/data.html)53. The full list of species information used in this study was described in Supplementary Table 2. The longest primary protein sequences were clustered into orthogroups by OrthoFinder v2.5.4 with -S diamond_ultra_sens parameter54. The known functional RXLR effectors in P. infestans were collected from a literature survey and UniProtKB/Swiss-Prot database55 using an “Avr” keyword search. To identify known effector orthogroups, a similarity search was performed between the 12 known effectors as query and the 12 oomycetes proteins as subject using BLASTp with a cutoff value of query coverage above 80%. Orthogroups with proteins matched in the similarity search were used for downstream analysis. The effector signature was identified based on a previously reported study56. Briefly, the signal peptide was predicted using SignalP v4.157 with sensitive mode (-u 0.34 -U 0.34). The RXLR, highly degenerate RXLR (hdRXLR), and EER motifs were searched in the first 100 amino acids of protein sequence as the following regular expressions: [RQGH]XLR for RXLR, [RKHGQ][X] {0,1} [LMYFIV][RNK] for hdRXLR, and [DE][DE][KR] for EER56. The WY domain was predicted using HMMer v3.1b58 with the best 1 domain score above 0 based on an HMM file constructed by 721 protein sequences obtained from previous study59.
Phylogenetic analyses and classification of Phytophthora RxLR effectors
To identify effector homologs, protein sequences of orthogroups with similarity to known effectors were aligned using MAFFT v7.407 with --maxiterate 1000 --globalpair60, and the positions containing gaps in >30% were trimmed using trimlAl61 v1.4.rev22 with -gt 0.3 parameter. A maximum-likelihood phylogenetic tree was reconstructed using IQ-TREE62 v1.6.12 with -bb 1000 -alrt 1000 -safe parameters. The best-fit substitution model JTT + F + R7 was selected by ModelFinder63 implemented in IQ-TREE. The avirulence effector clades were classified according to ultra-fast bootstrap value64 over 95% and taxonomic distribution. The phylognetic distances from each reference avirulence effector in P. infestans to its nearest-neighbor homologs in other Phytophthora species were calculated and visualized by a previously reported Python script (https://github.com/slt666666/Phylogenetic_distance_plot2)65.
Structure-based clustering of homologs of P. infestans avirulence effectors
Effector domains were extracted from a defined alignment for each phylogenetic-based avirulence effector clade. The effector domain region was defined as the sequence following the RxLR or RxLR dEER motifs based on the reference avirulence protein of each clade. Proteins with a miss rate exceeding 80% in the alignment of the effector domain region were filtered out for downstream analysis. Effector domain structures were predicted using Colabfold_batch v1.5.266 with --num-recycle 12 --num-seeds 3 parameters. For each protein, the best model defined by the pLDDT score was selected, and structures with a pLDDT score of < 50 were considered inaccurate and filtered out. Out of a total of 514 effector homologs, 473 successfully predicted the structures of effector domain. Using 473 predicted protein structures, structure-based clustering was performed in three main steps: (1) all-against-all alignment, (2) network construction, and (3) community detection. First, all predicted structures were aligned using US-align v2023060967 with default parameter, and the TM-score and alignment length were calculated for each pair. Next, a protein structure network was constructed based on the pairwise structural similarities. We connected two protein structures in the network if the average of their reciprocal TM-score and alignment coverage were both > 0.5. Finally, the structure clustering was performed using Louvain community detection algorithm68 implemented in networkx v3.169. Cytoscape v3.9.170 was used for network visualization.
Comparison of sequence and structure similarity between avirulence effectors and their homologs
We calculated the sequence and structural similarity of the effector domain between avirulence effectors of P. infestans and their homologs in each avirulence effector clade. For sequence similarity analysis, bit-scores were calculated using blastp 2.15.0+ and normalized them by dividing by the bit-score of each reference avirulence effector. For structure similarity analysis, we used the average score of the reciprocal TM-scores calculated from structure clustering.
Selection of sequence- and structure-based homologous effectors for cloning
Eight avirulence effectors of P. infestans including, Avr1, Avr2, Avr3a, Avr8, Avramr1, Avrblb1, Avrblb2, and Avrvnt1 were queried to protein database of P. parasitica, P. cactorum, P. palmivora, P. capsici, and P. sojae reference strains. We selected effectors with higher similarity against each reference avirulence effector by sorting bit-score and pTM score (cut off was normalized bit-score > 0.2; pTM > 0.35). At least one and max five of each family were targeted.
Cloning of homologous avirulence effectors into expression vector
Effector domains (after RxLR dEER motif, in case of no dEER, just after RxLR) were defined based on protein sequence database of each Phytophthora species (P. infestans, P. parasitica, P. cactorum, P. palmivora, P. capsici, and P. sojae). Then, predicted coding sequence (from start to end codons) of each effector domain were amplified using Primestar GXL (TAKARA®) enzyme from genomic DNA of each Phytophthora species. Among the initial candidates, 69 were successfully amplified from the genomic DNA of each Phytophthora species. Each amplicon was cloned into the pICH31160 vector (pKW, PVX virus coat protein promoter) using the ligation-independent cloning (LIC) method and the cloned sequences were validated by bi-directional sequencing71,72. Total 69 effectors were successfully cloned into expression vector.
Screening of R / Avr interactions through agroinfiltration
Agrobacterium strain GV3101 (pMP90) containing each construct was cultured in YEP medium for 1 day. Pellets were collected by centrifugation (10 min, 3000 rpm), resuspended into agroinfiltration buffer (10 mM MES, 10 mM MgCl2, 150 μM acetosyringone, pH 5.6). Agrobacterium expressing Solanum R genes and candidate effectors were mixed into 1:1 ratio after being adjusted to OD600 at 0.7 and 0.3 respectively. 4 ~ 5 weeks old N. benthamiana leaves were used for agroinfiltration. We observed infiltrated leaves until the 5 dpi, and took images using Fluorescence in vivo imaging system (FOBI, CELLGENTEK). Cell death phenotypes were simply recorded as positive or negative with picture.
Detached leaf assay using Solanum NLRs against Phytophthora species
For the detached leaf assay, the abaxial side of the detached leaves of 4 ~ 5 weeks old N. benthamiana were inoculated with 10 ~ 11 μl of zoospore solution (1.0 ~ 2.0 × 105 zoospores/ml) and placed in SPL® rectangular plates with wet tissue paper to maintain 100% relative humidity. Inoculated leaves were incubated at 23–25 °C in dark chamber until further examination. We observed until 2 ~ 4 dpi, and took images using FOBI machine.
Generation of transgenic plants
Thirty days old N. benthamiana leaves were cut into 3 cm x 3 cm pieces and sterilized with NaClO (0.1%) solution for 5 min. Agrobacterium strain GV3101 (pMP90) containing R1, R8, and Rpi-amr1 were cultured in YM media (D-mannitol: 10 g/L, yeast extract: 0.4 g/L, K2HPO4: 0.328 g/L, MgSO4 7H2O: 0.2 g/L, NaCl: 0.1 g/L) for 24 h, respectively, and co-incubated with cut leaves for 20 min in shaking incubator. Incubated leaves were wiped and moved to MS media (with acetosyringon) and co-cultured for 2 days in controlled chamber (23–25 °C, dark condition). Incubated leaves were washed out for 3 times with washing buffer (liquid MS media with benzyladenine and cefotaxime, pH 5.7) and moved to MS media (3% sucrose, kanamycin: 100 mg/L, BAP cytokinin 1.5 mg/L) for 4 weeks under the continuous light. Shoots were cut from the callus and moved to MS media (without cytokinin) and cultured until the root generation.
Root infection assay using Solanum NLRs against Phytophthora species
For the root infection assay, 3 ~ 4 weeks old N. benthamiana plants expressing Solanum R1, R8, or Rpi-amr1 were inoculated with 500 = ul of zoospore solution (1.0 ~ 2.0 × 105 zoospores/ml) of Phytophthora species and placed in controlled chamber (23–25 °C,16 h/8 h of light/dark cycle) until the further examination. Phenotypes were categorized into three groups (healthy; wilt: wilting leaves; dead: shrink, and main stem bends) until the wild type plants were completely dead (takes 4 ~ 9 days). Unsterilized commercial topsoil for general/horticultural (product of Seoulbio®, http://www.seoulbio.co.kr/product/product_04_01.asp) were used and additional nutrition was not added.
Sequence and structural comparisons of cloned homologous avirulence effectors
Sequences and structures of cloned effectors from the Avr1, Avr8, and Avramr1 clades were aligned based on their respective reference avirulence proteins. Sequence alignment was performed using MAFFT v7.407 with --maxiterate 1000 --globalpair60. Aligned sequences were visualized by Jalview v2.11.3.273. To confirm the evolutionary conservation of the sequence, a conservation score was calculated based on the aligned sequences of HR-positive effectors and each reference Avirulence protein using the ConSurf server74. Comparison of protein structures was performed using the jFATCAT75-flexible mode of pairwise structure alignment in the RCSB PDB database and visualized using PyMol v2.5.576.
Statistical information
Statistics in this paper were performed on the all replicates for Figs. 2b, c, 3c–f, 4a, b, and Supplementary Figs. 7–12. For the all cases, we performed unpaired t-test, using a P-value threshold for a significance of two-tailed *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Individual P-values are presented in Supplementary Data 4.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description Of Additional Supplementary File
Acknowledgements
We thank Y. H. Lee, C. Segonzac, and H. A. Lee for helpful suggestions, KACC of RDA Korea for providing materials, and S. J. Yoon and Y. J. Yeom for technical support. This project was supported by the National Research Foundation of Korea (2022R1C1C2011450; RS-2024-00333777; RS-2024-00333225), Plant Immunity Research Center, SRC (2018R1A5A1023599).
Author contributions
S.O., M.K., and D.C. designed the research. S.O., H.K., and T.K. performed the experiments. S. O., M.K., J.K. analyzed the data. S.O., and M.K. draw the figures. S.O., M.K., and D.C. wrote the paper.
Peer review
Peer review information
Nature Communications thanks Kirankumar Mysore, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Data supporting the findings of this work are available within the paper and its Supplementary Information files. All the resources described are available from the corresponding authors upon request. Source data are deposited in Figshare (10.6084/m9.figshare.26421439).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Soohyun Oh, Myung-Shin Kim.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54452-2.
References
- 1.Oh, S. & Choi, D. Receptor-mediated nonhost resistance in plants. Essays Biochem.66, 435–445 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Lee, H. A. et al. Current understandings of plant nonhost resistance. Mol. Plant Microbe. Interact.30, 5–15 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Panstruga, R. & Moscou, M. J. What is the molecular basis of nonhost resistance? Mol. Plant-Microbe Interact.33, 1253–1264 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Schulze-Lefert, P. & Panstruga, R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci.16, 117–125 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Lipka, V. et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science310, 1180–1183 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Moreau, M. et al. EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora. Mol. Plant Microbe Interact.25, 421–430 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Peart, J. R. et al. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl Acad. Sci. USA.99, 10865–10869 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, B. Y. et al. The Rxo1/Rba1 locus of maize controls resistance reactions to pathogenic and non-host bacteria. Theor. Appl. Genet.109, 71–79 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Borhan, M. H. et al. WRR4 Encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Mol. Plant Microbe Interact.21, 757–768 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Kawashima, C. G. et al. A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol.34, 661–665 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Bettgenhaeuser, J. et al. The barley immune receptor Mla recognizes multiple pathogens and contributes to host range. Nat. Commun.12, 6915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witek, K. et al. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol.34, 656–660 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Felix, G., Duran, J. D., Volko, S. & Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J.18, 265–276 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Lacombe, S. et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol.28, 365–369 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Schoonbeek, H. J. et al. Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. N. Phytol.206, 606–613 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Dong, S. et al. Effector specialization in a lineage of the Irish potato famine pathogen. Science343, 552–555 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Dong, S., Raffaele, S. & Kamoun, S. The two-speed genomes of filamentous pathogens: Waltz with plants. Curr. Opin. Genet. Dev.35, 57–65 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Laflamme, B. et al. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science367, 763–768 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Lin, X. et al. A potato late blight resistance gene protects against multiple Phytophthora species by recognizing a broadly conserved RXLR-WY effector. Mol. Plant15, 1457–1469 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Kroon, L. P. N. M., Brouwer, H., De Cock, A. W. A. M. & Govers, F. The genus Phytophthora anno 2012. Phytopathology102, 348–364 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G. & Brasier, C. M. A molecular phylogeny of phytophthora and related oomycetes. Fungal Genet. Biol.30, 17–32 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Kamoun, S. et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol.16, 413–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausbeck, M. K. & Lamour, K. H. Phytophthora capsici on vegetable crops: Research progress and management challenges. Plant Dis.88, 1292–1303 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Fry, W. Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol.9, 385–402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vleeshouwers, V. G. A. A. et al. Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol.49, 507–531 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Haverkort, A. J. et al. Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Res59, 35–66 (2016). [Google Scholar]
- 28.Hansen, E. M., Reeser, P. W. & Sutton, W. Phytophthora beyond agriculture. Annu. Rev. Phytopathol.50, 359–378 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Van Der Lee, T., Robold, A., Testa, A., Van’T Klooster, J. W. & Govers, F. Mapping of avirulence genes in Phytophthora infestans with amplified fragment length polymorphism markers selected by bulked segregant analysis. Genetics157, 949–956 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du, Y., Mpina, M. H., Birch, P. R. J., Bouwmeester, K. & Govers, F. Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol.169, 1975–1990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders, D. G. O. et al. Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell24, 3420–3434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong, M. R. et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl Acad. Sci. USA.102, 7766–7771 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, J., Jiang, R. H. Y., Kamphuis, L. G. & Govers, F. A cDNA-AFLP based strategy to identify transcripts associated with avirulence in Phytophthora infestans. Fungal Genet. Biol.43, 111–123 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Jo, K. R. Unveiling and Deploying Durability of Late Blight Resistance in Potato; From Natural Stacking to Cisgenic Stacking.https://research.wur.nl/en/publications/unveiling-and-deploying-durability-of-late-blight-resistance-in-p (2013).
- 35.Lin, X. et al. Identification of Avramr1 from Phytophthora infestans using long read and cDNA pathogen-enrichment sequencing (PenSeq). Mol. Plant Pathol.21, 1502–1512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vleeshouwers, V. G. A. A. et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE3, e2875 (2008). [DOI] [PMC free article] [PubMed]
- 37.Oh, S. K. et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell21, 2928–2947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pel, M. A. et al. Mapping and cloning of late bright resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol. Plant Microbe Interact.22, 601–615 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Ballvora, A. et al. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J.30, 361–371 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Lokossou, A. A. et al. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol. Plant. Microbe Interact.22, 630–641 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Huang, S. et al. The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol. Plant-Microbe Interact.17, 428–435 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Vossen, J. H. et al. The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor. Appl. Genet.129, 1785–1796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witek, K. et al. A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. Nat. Plants7, 198–208 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Der Vossen, E. et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J.36, 867–882 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Van Der Vossen, E. A. G. et al. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J.44, 208–222 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Foster, S. J. et al. Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol. Plant Microbe Interact.22, 589–600 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Win, J. et al. Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog.8, 8–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh, S. et al. Nucleotide-binding leucine-rich repeat network underlies nonhost resistance of pepper against the Irish potato famine pathogen Phytophthora infestans. Plant Biotechnol. J.21, 1361–1372 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson, R. G., Deb, D., Fedkenheuer, K. & Mcdowell, J. M. Recent progress in RXLR effector research. 28, 1063–1072 (2015). [DOI] [PubMed]
- 50.Lapin, D. et al. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell31, 2430–2455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, C. H. et al. NLR network mediates immunity to diverse plant pathogens. Proc. Natl Acad. Sci. USA.114, 8113–8118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redkar, A. et al. The Arabidopsis WRR4A and WRR4B paralogous NLR proteins both confer recognition of multiple Albugo candida effectors. N. Phytol.237, 532–547 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGowan, J., Byrne, K. P. & Fitzpatrick, D. A. Comparative analysis of oomycete genome evolution using the oomycete gene order browser (OGOB). Genome Biol. Evol.11, 189–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emms, D. M. & Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol.20, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Consortium, T. U. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res.51, 523–531 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood, K. J. et al. Effector prediction and characterization in the oomycete pathogen Bremia lactucae reveal host-recognized WY domain proteins that lack the canonical RXLR motif. PLoS Pathog.16, 1–29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen, T. N., Brunak, S., Von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed]
- 59.Boutemy, L. S. et al. Structures of phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J. Biol. Chem.286, 35834–35842 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol.30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol.35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harant, A., Pai, H., Sakai, T., Kamoun, S. & Adachi, H. A vector system for fast-forward studies of the HOPZ-ACTIVATED RESISTANCE1 (ZAR1) resistosome in the model plant Nicotiana benthamiana. Plant Physiol.188, 70–80 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, C., Shine, M., Pyle, A. M. & Zhang, Y. US-align: universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nat. Methods19, 1109-1115 (2022). [DOI] [PubMed]
- 68.Blondel, V. D., Guillaume, J. L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 10.1088/1742-5468/2008/10/P10008 (2008).
- 69.Schult, D. La-Ur- Exploring Network Structu Exploring Network Structure, Dynamics, and Function Using Networkx.https://www.osti.gov/biblio/960616 (1943).
- 70.Shannon, P. et al. Cytoscape: A software environment for integrated models. Genome Res.13, 426 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aslanidis, C. & Jong, P. J. De. Ligation-independent cloning of PCR products (LIC-PCR). 18, 6069–6074 (1990). [DOI] [PMC free article] [PubMed]
- 72.Oh, S. K., Kim, S. B., Yeom, S. I., Lee, H. A. & Choi, D. Positive-selection and ligation-independent cloning vectors for large scale in planta expression for plant functional genomics. Mol. Cells30, 557–562 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res.44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li, Z., Jaroszewski, L., Iyer, M., Sedova, M. & Godzik, A. FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res.48, W60–W64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. PyMOL by Schrödinger. The PyMOL Molecular Graphics System, Version 2.5.5, Schrödinger, LLC.https://www.pymol.org/ (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description Of Additional Supplementary File
Data Availability Statement
Data supporting the findings of this work are available within the paper and its Supplementary Information files. All the resources described are available from the corresponding authors upon request. Source data are deposited in Figshare (10.6084/m9.figshare.26421439).