ABSTRACT
Interactions between sleep and feeding behaviors are critical for adaptive fitness. Diverse species suppress sleep when food is scarce to increase the time spent foraging. Postprandial sleep, an increase in sleep time following a feeding event, has been documented in vertebrate and invertebrate animals. While interactions between sleep and feeding appear to be highly conserved, the evolution of postprandial sleep in response to changes in food availability remains poorly understood. Multiple populations of the Mexican cavefish, Astyanax mexicanus, have independently evolved sleep loss and increased food consumption compared to surface‐dwelling fish of the same species, providing the opportunity to investigate the evolution of interactions between sleep and feeding. Here, we investigate the effects of feeding on sleep in larval and adult surface fish, and in two parallelly evolved cave populations of A. mexicanus. Larval surface and cave populations of A. mexicanus increase sleep immediately following a meal, providing the first evidence of postprandial sleep in a fish model. The amount of sleep was not correlated to meal size and occurred independently of feeding time. In contrast to larvae, postprandial sleep was not detected in adult surface or cavefish, which can survive for months without food. Together, these findings reveal that postprandial sleep is present in multiple short‐sleeping populations of cavefish, suggesting sleep‐feeding interactions are retained despite the evolution of sleep loss. These findings raise the possibility that postprandial sleep is critical for energy conservation and survival in larvae that are highly sensitive to food deprivation.
Keywords: cavefish, circadian, evolution, feeding, sleep
Summary
Laval Astyanax mexicanus increases sleep following feeding.
Postprandial sleep occurs across the circadian cycle.
Postprandial sleep was not identifiable in adults.
1. Introduction
Sleep and metabolic regulation are highly variable throughout the animal kingdom (Lesku et al. 2006; Joiner 2016; Keene and Duboue 2018; Seebacher 2018). This variability is reflected by the diversity of food availability and foraging strategy, which potently impact the duration and timing of sleep. There is an interaction between sleep and feeding, regardless of life history strategy, that is critical for organismal survival and, therefore, under selection (Capellini et al. 2008; Yurgel et al. 2014; Slocumb et al. 2015; Aulsebrook et al. 2016; Brown et al. 2019). While both of these behavioral processes have been studied in detail, much less is known about interactions between sleep and feeding, particularly in the context of evolution.
In many species, sleep deprivation results in increased food intake, while prolonged periods of food deprivation lead to a reduction in metabolic rate and suppression of sleep (Keene et al. 2010; Arble et al. 2015; Regalado et al. 2017; Stahl et al. 2017; Goldstein et al. 2018). Conversely, animals ranging from the nematode, Caenorhabditis elegans, to humans, increase sleep immediately following a meal, revealing an acute effect of dietary nutrients on sleep regulation (Stahl, Orr, and Bollinger 1983; Murphy et al. 2016; Makino et al. 2021). Defining how evolution has shaped interactions between sleep, metabolic regulation, and feeding is critical to determining the functions of these traits.
The rapidly increasing number of organisms used to study sleep provides new opportunities to study interactions between sleep and metabolism (McNamara, Barton, and Nunn 2009; Anafi, Kayser, and Raizen 2019). Fish have become models to study the biological basis of sleep regulation (Chiu and Prober 2013; Levitas‐Djerbi and Appelbaum 2017; Keene and Appelbaum 2019). Growing evidence suggests the genetic and functional basis of sleep is conserved from fish through humans (Chiu and Prober 2013; Levitas‐Djerbi and Appelbaum 2017; Keene and Appelbaum 2019; Tran and Prober 2022). Further, the small size and amenability to genetic manipulation of these model fish allows for high‐throughput genetic and pharmacological screens to identify novel regulators of sleep (Rihel et al. 2010; Chiu et al. 2016; Kroll et al. 2021). Furthermore, at larval stages, many fish models are transparent, allowing for the mapping of sleep and feeding circuits across the entire brain (Semmelhack et al. 2014; Leung et al. 2019; Wee et al. 2019; Förster et al. 2020). Therefore, zebrafish, Astyanax mexicanus, and other fish models are exceptionally well‐positioned to examine interactions between sleep and feeding.
The adaptation of A. mexicanus to nutrient‐poor cave environments provides the opportunity to examine sleep after fasting and postprandial sleep in an evolutionary context (Jeffery 2009; Gross 2012; McGaugh et al. 2020). Along with sleep loss, multiple cavefish populations have evolved behavioral and physiological differences relative to surface fish, including a reduced metabolic rate and increased feeding behaviors (Duboué, Keene, and Borowsky 2011; Moran, Softley, and Warrant 2014; Aspiras et al. 2015; Yoshizawa 2015; Volkoff 2016). Long‐term starvation has opposing effects on sleep between the surface and cave populations. Starved surface fish suppress sleep, while starved cavefish increase sleep, suggesting that the evolutionary factors shaping the sleep‐feeding interaction differ between populations (Jaggard et al. 2018). However, sleep‐feeding interactions are poorly understood, and postprandial sleep has, to our knowledge, not been identified in any fish model to date. Examining the effects of feeding state on sleep in surface and cave populations of A. mexicanus has the potential to identify whether these behaviors evolved through shared genetic mechanisms and to provide insight into how sleep‐feeding interactions are influenced by adaptation to a nutrient‐poor cave environment. Specifically, we will test whether evolved loss of sleep is accompanied by the loss of sleep‐feeding interactions.
Sleep is a complex phenotype under regulation by circadian and homeostatic systems (Borbély 1982). Fish display robust circadian locomotor behaviors and feeding patterns (Cahill, Hurd, and Batchelor 1998; Moore and Whitmore 2014). In A. mexicanus cavefish, circadian entrainment to light is largely absent, yet animals display a sleep recovery rebound following deprivation (Beale et al. 2013; McGaugh et al. 2020; Mack et al. 2021). These findings suggest A. mexicanus cave populations have lost circadian modulation of behavior yet maintain homeostatic regulation of sleep. While the relationship between feeding and the circadian clock has not been studied in A. mexicanus, food‐entrainable rhythms are present in the Somalian cavefish, Phreatichthys andruzzii (Cavallari et al. 2011), suggesting connections between feeding and the circadian clock may remain intact in the absence of a light‐entrainable oscillator. Therefore, A. mexicanus cave populations provide the opportunity to investigate how the loss of light‐dependent clock entrainment impacts feeding rhythms.
Larval A. mexicanus provide a particularly tractable model for examining the effects of feeding on sleep regulation. The convergent evolution of sleep loss described in these cavefish populations occurs at both the adult and larval stages (Duboué, Keene, and Borowsky 2011; Yoshizawa et al. 2015). However, while adult fish can live for months without food, larval fish live for only a matter of days (Salin et al. 2010; Medley et al. 2022; Pozo‐Morales et al. 2024). Therefore, interactions between feeding and other behaviors may be particularly important for the survival of larvae and young juvenile fish. The amount of nauplii brine shrimp (Artemia fransiscana) consumed by larval fish is readily quantifiable and large numbers of these larval fish can be tested without the need to grow fish to adulthood (Espinasa et al. 2014; Espinasa et al. 2017; Lloyd et al. 2018). The experimental amenability of larval fish allows for efficient characterization of sleep‐feeding interactions across different behavioral and genetic contexts, providing a model to investigate the evolutionary relationship between these processes.
Here, we identify postprandial sleep in surface fish and multiple A. mexicanus cavefish populations. Interestingly, we found postprandial sleep in larval, but not adult fish. In larval fish, we found that feeding promotes sleep, independent of time of day. However, we only found a significant correlation between meal size and the amount of postprandial sleep for larval surface fish in the early morning. Together, these findings reveal interactions between feeding and sleep and provide a model system to examine how these interactions evolved.
2. Results
To investigate the effects of feeding on sleep, we compared sleep in different populations of cavefish immediately following a meal. Briefly, fish were fed a meal, and baseline sleep and activity were measured for 24 hours before sleep and feeding measurements. At Zeitgeber Time (ZT) 0 on the second day, fish were fed ~70 Artemia over 2 hours. At the completion of the assay the number of Artemia consumed within this period was quantified. The feeding assay was followed by a 4‐hour recording of sleep (Figure 1A). In agreement with previous findings, baseline sleep was lower in both Pachón and Tinaja cavefish compared to surface fish (Figure 1B; Duboué, Keene, and Borowsky 2011; Jaggard et al. 2020; O'Gorman et al. 2021). When sleep was measured following a 2‐hour feeding period, surface fish slept significantly more than cavefish from both populations (Figure 1C). Consistent with previous findings, quantification of Artemia consumed during the 2‐hour feeding window revealed significantly greater consumption in Tinaja fish, but not Pachón cavefish, compared to surface fish (Figure 1D; Aspiras et al. 2015; Alié et al. 2018). Taken together, these findings reveal differences in sleep and feeding behavior of larval A. mexicanus populations.
Figure 1.
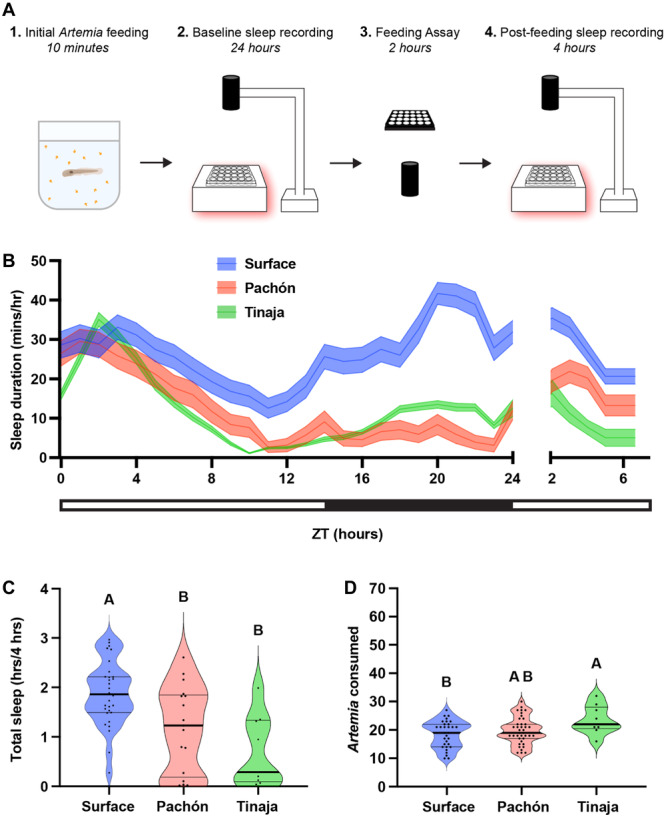
Sleep, feeding, and postprandial sleep behaviors across three populations of wild‐type Astyanax mexicanus. (A) Twenty days post fertilization fish were briefly fed before 24‐hour behavioral sleep recordings. At ZT0 the following day, fish were assayed for feeding behavior until ZT2, immediately after which, we recorded sleep behaviors between ZT2 and ZT6. (B) Sleep profiles of wild‐type surface, Pachón, and Tinaja fish were taken over the course of the experiment. Lines and error bars represent the mean ± SEM. (C) Cross‐population comparison of total sleep duration immediately following the feeding experiment. Letters indicate significant differences between populations. Cavefish slept significantly less than surface fish (ANOVA: F2, 34 = 8.123, p = 0.0013; Tukey's HSD for surface‐Pachón, p = 0.0202; Tukey's HSD for surface‐Tinaja, p = 0.0024). (D) Cross‐population comparison of the number of Artemia eaten during the 2‐h feeding experiment. Letters indicate significant differences between populations. Tinaja ate significantly more than surface fish (ANOVA: F2, 76 = 3.91, p = 0.0242; Tukey's HSD for surface‐Tinaja, p = 0.0178).
It is possible that sleep is elevated across A. mexicanus populations from ZT2 to ZT6 due to postprandial sleep or light‐regulated rest‐activity rhythms. To differentiate between these possibilities, we compared sleep following meals before ZT2, ZT6, and ZT10. Feeding time was limited to half an hour to provide additional resolution for postprandial sleep (Figure 2A–C). Across feeding time courses, surface fish slept more than cavefish populations (Figure 2D–F), supporting the notion that surface fish sleep more than cavefish independent of feeding treatment. To measure postprandial sleep, we compared sleep duration during the 4 hours following feeding to the remaining hours of daytime (excluding the time for the feeding assay) to determine the percent change in sleep postfeeding. Sleep was increased following the meal across all three timepoints, for surface fish and both cavefish populations (Figure 2G–I). Strikingly, for all timepoints tested, there was a significant increase in the amount of postprandial sleep, measured by the increase over the baseline sleep (Figure 2G–I).
Figure 2.
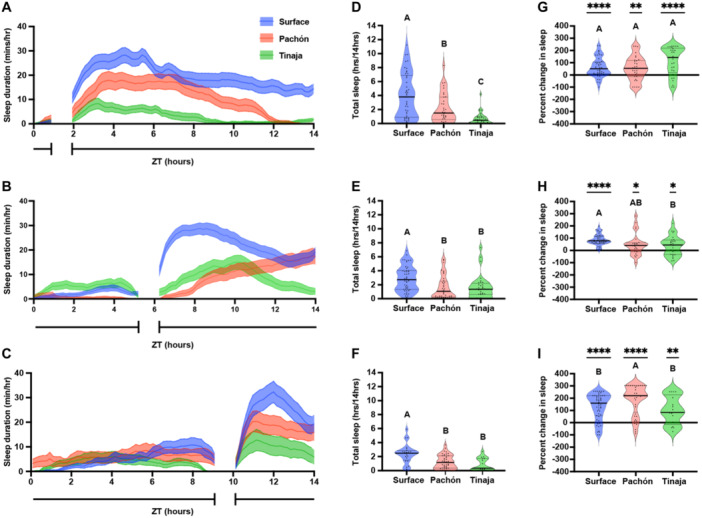
Postfeeding increase in larval Astyanax mexicanus sleep duration is not dependent on daily feeding time. Twenty days post fertilization larvae were fed over a 45‐min window before ZT2 (A, D, G), ZT6 (B, E, H), or ZT10 (C, F, I). (A–C) Sleep profiles of surface, Pachón, and Tinaja larvae, in minutes per hour, averaged across the daylight cycle. Lines and error bars represent the mean ± SEM. (D–F) Cross‐population comparison of total sleep duration in hours over the 14‐hour light cycle. Letters represent significant differences between populations. (D) Total sleep duration around a ZT2 feeding window was significantly different between populations of A. mexicanus (ANOVA: F2, 113 = 20.81, p < 0.0001). (E) Total sleep duration around a ZT6 feeding window was significantly different between surface and cave populations of A. mexicanus (ANOVA: F2, 113 = 8.48, p = 0.0004; Tukey's HSD for Surface‐Pachón, p = 0.001 and Surface‐Tinaja, p = 0.0069). (F) Total sleep duration around a ZT10 feeding window was significantly different between surface and cave populations of A. mexicanus (ANOVA: F2, 81 = 11.64, p < 0.001; Surface‐Pachón, p = 0.0003; Tukey's HSD for surface‐Tinaja, p = 0.0002). (G–I) Percentage change in sleep duration for the 4‐hour period following feeding from total daytime sleep calculated as (proportion of postprandial sleep − proportion of total sleep)/proportion of total sleep. All conditions were significantly different from zero (see Table S1). Letters indicate significant differences between populations. (G) Percent change of postprandial sleep after ZT2 feeding window. There was no significant difference across populations in the percentage of increase in postprandial sleep (ANOVA: F2, 104 = 3.36, p = 0.0417). (H) Percent change of postprandial sleep after ZT6 feeding window. There was no significant different in the percentage of increase in postprandial sleep between surface and Pachón cavefish, but surface fish had a significantly greater increase in sleep than Tinaja cavefish (ANOVA: F2, 96 = 5.758, p = 0.0072; Tukey's HSD for surface‐Tinaja, p = 0.0101). (I) Percent change of postprandial sleep after ZT10 feeding window. Pachón cavefish had a significantly greater percent increase in postprandial sleep than both surface and Tinaja cavefish (ANOVA: F2, 111 = 4.727, p = 0.0107; Tukey's HSD for surface‐Pachón, p = 0.0298; Tukey's HSD for Pachón‐Tinaja, p = 0.0275).
Variation in the degree of postprandial sleep increase across populations was dependent on feeding time. There were no differences in the percent increase in postprandial sleep between populations fed before ZT2 (Figure 2G), but surface fish had a significantly greater increase in postprandial sleep than Tinaja cavefish fed before ZT6 (Figure 2H), and Pachón fish had a significantly greater increase in postprandial sleep than either surface and Tinaja cavefish fed before ZT10 (Figure 2I). Similarly, both surface and Pachón cavefish, but not Tinaja cavefish, experienced a significantly greater increase in postprandial sleep before ZT10 than for the timepoints earlier in the day. Therefore, while postprandial sleep occurs across A. mexicanus populations, the degree to which sleep is increased in each population is dependent on the time of day that feeding occurs. Taken together, these findings reveal the presence of postprandial sleep in surface and cave populations of A. mexicanus.
It is possible that meal size, or its caloric value, contributes to the duration of postprandial sleep. To determine whether the amount of postprandial sleep is related to meal size, we examined the correlation between the number of Artemia consumed and the duration of sleep in the 4 hours following the meal. For surface fish fed before ZT2, there was a significant positive correlation between meal size and post prandial sleep, however there was no significant correlation for surface fish fed before ZT6 and ZT10 (Figure 3A–C). For both Pachón (Figure 3D–F) and Tinaja (Figure 3G–I) cavefish, there was no correlation between Artemia consumed and postprandial sleep. Therefore, postprandial sleep is largely driven by the presence of a meal and does not appear to be directly linked to meal size.
Figure 3.
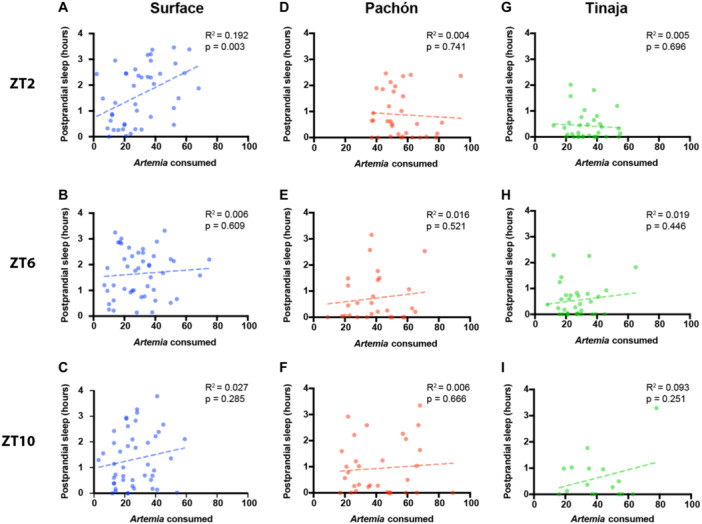
Postprandial sleep in larval Astyanax is only dependent on the amount of food consumed for surface fish fed before ZT2. Correlation of amount of Artemia nauplii consumed with sleep duration in the 4 hours following feeding with a simple linear regression for surface (A–C), Pachón (D–F), and Tinaja (G–I). (A, D, G) Larvae were fed before ZT2. (B, E, H) Larvae were fed before ZT6. (C, F, I) Larvae were fed before ZT10.
Postprandial sleep may provide a mechanism for conserving energy immediately following successful foraging. Larval A. mexicanus survive for only a few days without food (Salin et al. 2010; Medley et al. 2022; Pozo‐Morales et al. 2024), raising the possibility that sleep will be acutely impacted by the feeding state. To directly examine the effects of feeding state on sleep, we compared the sleep in 20 days post fertilization (dpf) fish that were fed from ZT0 to ZT2 to unfed fish that had been starved for the previous 24 hours (Figure 4A–C). Surface fish and both populations of cavefish slept significantly more during the 4 hours following feeding than unfed controls (Figure 4D–F). To further examine the effects of feeding on sleep, we analyzed the activity patterns of fed and unfed fish using a Markov model that predicts sleep and wake propensity, both indicators of sleep drive (Wiggin et al. 2020). Across all three populations, fed fish had a significantly greater sleep propensity P(Doze) and a significantly lower waking propensity P(Wake) than unfed fish, suggesting that sleep drive is increased following feeding (Figure 4G–I). Together, these findings reveal that both surface and cavefish 20 dpf larvae suppress sleep when starved and that starvation‐induced sleep suppression is intact in short‐sleeping cavefish.
Figure 4.
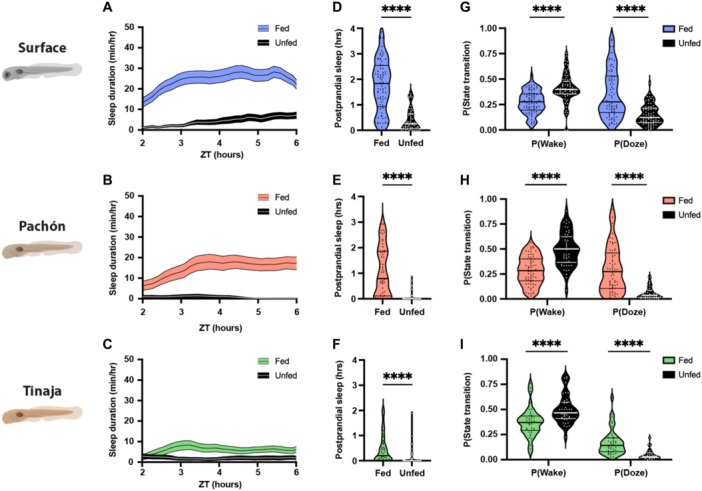
Feeding results in robust increases in sleep duration in larval surface, Pachón, and Tinaja populations of Astyanax mexicanus. (A–C) Four‐hour sleep profiles comparing the sleep of fed (colored) and unfed (black) individuals in each population. Lines and error bars represent the mean ± SEM. (D–F) Fed fish sleep significantly more during the 4 hours following feeding than unfed fish, regardless of the population. (D) Surface: Mann–Whitney U = 524, n fed = 77, n unfed = 55, p < 0.0001. (E) Pachón: Mann–Whitney U = 310.5, n fed = 52, n unfed = 47, p < 0.0001. (F) Tinaja: Mann–Whitney U = 546.5, n fed = 45, n unfed = 49, p < 0.0001. (G–I) Fed fish are less likely to wake while asleep, and more likely to fall asleep while awake, than unfed fish. (G) Surface: P(Wake) Mann–Whitney U = 1317, n fed = 77, n unfed = 76, p < 0.0001; P(Doze) Mann–Whitney U = 1347, n fed = 77, n unfed = 75, p < 0.0001. (H) Pachon: P(Wake) Mann–Whitney U = 663, n fed = 66, n unfed = 52, p < 0.0001; P(Doze) Mann–Whitney U = 802, n fed = 69, n unfed = 52, p < 0.0001. (I) Tinaja: P(Wake) Mann–Whitney U = 369, n fed = 40, n unfed = 38, p < 0.0001; P(Doze) Mann–Whitney U = 229, n fed = 40, n unfed = 34, p < 0.0001. Horizontal lines represent quartiles. Asterisks represent significant differences between fed and unfed groups.
Adult A. mexicanus live months without food and are thought to be highly adapted to survive periods of starvation (Cobham and Rohner 2024). Previously, we have shown that surface fish suppress sleep during periods of prolonged starvation, while cavefish increase sleep (Jaggard et al. 2018). To determine whether differences in sleep response extend to acute behavior following meals, we examined postprandial sleep in adult surface and cavefish. Fish were starved for 5 days before recording to synchronize meal patterns and then fed a blood‐worm meal at ZT6. In agreement with previous findings (Jaggard et al. 2018), control surface fish that were not fed slept significantly more than unfed Pachón and Tinaja cavefish (Figure 5A,C). Similarly, in fish fed at ZT6, surface fish slept significantly more than Tinaja and Pachòn cavefish (Figure 5B,D). To examine whether postprandial sleep is present in adult A. mexicanus, we compared sleep during the 4 hours following feeding to unfed counterparts (Figure 5E–M). Within this 4‐hour duration, there were no significant differences in sleep duration (Figure 5H–J) or sleep propensity (Figure 5K–M) between fed and unfed fish across the three A. mexicanus populations. Therefore, there is no evident postprandial sleep for adults under the conditions tested, supporting the notion that postprandial sleep is less robust at a life stage when fish are more starvation‐resistant.
Figure 5.
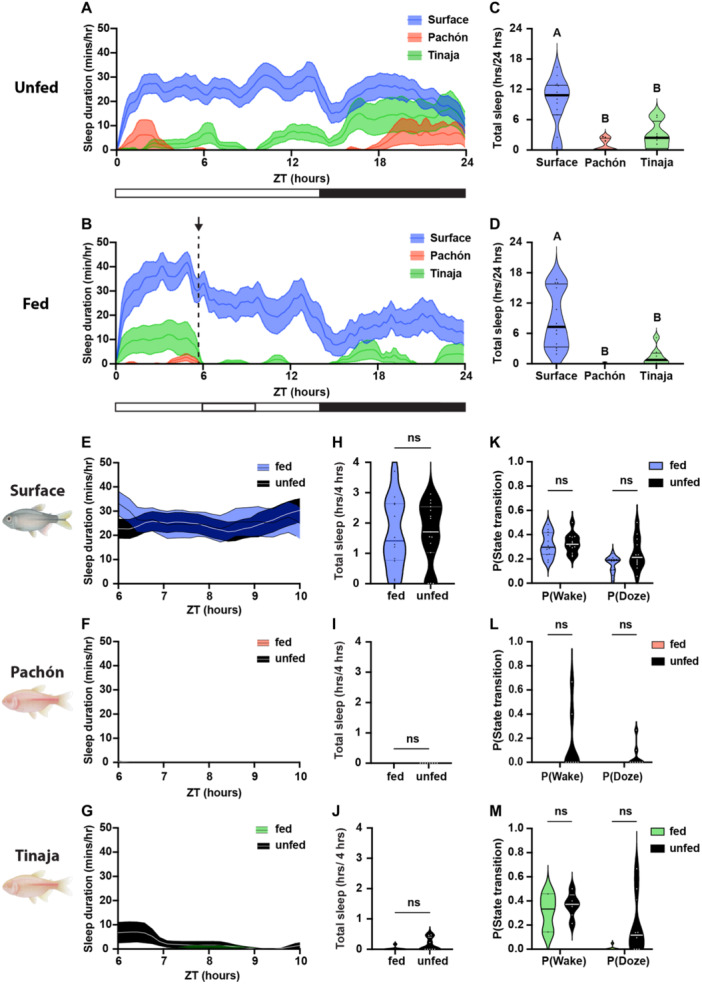
Adult Astyanax do not display postprandial sleep behavior. (A, B) Sleep profiles of adult Surface, Pachón, and Tinaja, in minutes per hour. Lines and error bars represent the mean ± SEM. (A, C) Fish were not fed over the course of the day. (B, D) Fish were provided food from ZT5.5 (indicated by the arrow and dotted black line in B) to ZT6. (C, D) Cross‐population comparison of total sleep duration in hours over the 24‐hour day. Letters represent significant differences between populations. (C) Total sleep duration in 24 hours was significantly different between unfed surface and cave populations of Astyanax mexicanus (ANOVA: F2, 28 = 15.5, p < 0.0001; Tukey's HSD for Surface‐Pachón, p < 0.0001 and Surface‐Tinaja, p = 0.0015). (D) Total sleep duration was significantly different between fed surface and cave populations of A. mexicanus (ANOVA: F2, 25 = 15.04, p < 0.0001; Tukey's HSD for Surface‐Pachón, p < 0.0001 and Surface‐Tinaja, p = 0.0008). (E–G) Four‐hour sleep profiles comparing the sleep of fed (colored) and unfed (black) individuals in each population: surface (E), Pachón (F), and Tinaja (G). Lines and error bars represent the mean ± SEM. (H–J) There were no significant differences in sleep during the 4 hours following feeding, regardless of the population. (H) Surface: Mann–Whitney U = 88, n fed = 12, n unfed = 15, p = 0.9317. (I) Pachon: Mann–Whitney U = 31.5, n fed = 8, n unfed = 8, p > 0.9999. (J) Tinaja: Mann–Whitney U = 22.5, n fed = 8, n unfed = 8, p > 0.2. (K–M) There were no significant differences in activity state transitions between fed and unfed fish. (K) Surface: P(Wake) t = 0.271, df = 22, p = 0.7888; P(Doze) t = 2.041, df = 22, p = 0.054. (L) Pachon: Mann–Whitney U = 24, n fed = 8, n unfed = 8; P(Wake) p = 0.4667; P(Doze) p = 0.4667. (M) Tinaja: Mann–Whitney U = 23, n fed = 8, n unfed = 8; P(Wake) p = 0.5714; P(Doze) p = 0.1319). Horizontal lines represent quartiles.
3. Discussion
To date, five populations of A. mexicanus cavefish have been studied under laboratory conditions, all of which have significantly reduced sleep compared to surface fish populations (Yoshizawa et al. 2015). These findings have led to the speculation that reduced sleep is adaptive in the food‐poor cave environment because it provides more time to forage (Keene, Yoshizawa, and McGaugh 2015; Keene and Duboue 2018). However, nearly all studies to date have examined sleep in fed animals, using daily averages. Therefore, little is known about how sleep differs between populations under natural conditions and in response to feeding. Here, we describe interactions between sleep and feeding behavior in surface fish and two different populations of cavefish. All three populations sleep more following feeding than under food‐deprived conditions, revealing that feeding is required for baseline sleep. Furthermore, all three populations sleep more in the period following a meal as larvae, but not as adults. We cannot conclusively rule out that postprandial sleep is not present in adults, and it is possible that other feeding protocols will reveal sleep‐feeding interactions. Together, these findings suggest that despite robust sleep loss across cavefish populations, sleep‐feeding interactions have remained intact.
Numerous neural mechanisms associated with sleep loss in cavefish have been identified including elevated levels of the wake‐promoting neuropeptide Hypocretin (HCRT) and changes in wake‐promoting catecholamine systems (Duboué, Borowsky, and Keene 2012; Bilandzija et al. 2013; Gallman et al. 2020), providing candidate regulators of postprandial sleep. Similarly, feeding is increased in multiple cave populations of adult A. mexicanus (Aspiras et al. 2015). In agreement with previous findings, we find that feeding is elevated in 20 days postfertilization juvenile cavefish from the Tinaja, but not Pachón population (O'Gorman et al. 2021). In adults, differences in feeding are at least partially attributable to polymorphisms in the GPCR Melanocortin 4 receptor (Mc4r) which is associated with obesity in humans and in animal models (Aspiras et al. 2015). While there is little evidence that MC4R directly regulates sleep, it is thought to contribute to obesity‐induced sleep apnea that, in turn, regulates sleep (Larkin et al. 2010; Pillai et al. 2014). Our findings that postprandial sleep is intact in Tinaja cavefish suggest that Mc4r, and other genes involved in feeding, are likely dispensable for sleep‐feeding interactions. There are also numerous genes that have been identified to regulate sleep or feeding in fish models that are potential regulators of sleep‐metabolism interactions. For example, the orexigenic neuropeptides Neuropetide Y (Npy) and HCRT both induce wakefulness, providing a potential molecular mechanism for feeding‐dependent modulation of sleep (Appelbaum et al. 2009; Penney and Volkoff 2014; Singh, Oikonomou, and Prober 2015; Singh, Rihel, and Prober 2017; Jaggard et al. 2018). Future functional analysis is required to define whether these candidate genes regulate interactions between sleep and feeding.
In A. mexicanus, rhythmic transcription is significantly diminished under dark‐dark conditions, and cavefish have elevated levels of light‐inducible genes (Beale et al. 2013). The circadian clock plays a critical role in the timing of both sleep and feeding, raising the possibility that the circadian clock may be critical for sleep‐feeding interactions. Transcriptome‐wide analysis in larvae reveals a loss of rhythmic gene expression across all cave populations tested (Mack et al. 2021). Therefore, because we identified postprandial sleep in all of the populations tested, across three different timepoints during the day, postprandial sleep may be independent of the time of day and may not require a functioning circadian clock. Differences in the amount of postprandial sleep between populations, across these three timepoints, may reflect differences in circadian regulation of metabolism between surface and cave populations, as well as differences in metabolic regulation and fat storage across Astyanax populations (Moran, Softley, and Warrant 2014; Aspiras et al. 2015; Mack et al. 2021). It is also possible that the link between the amount of food consumed and the duration of postprandial sleep has a circadian component. This correlation was only significant in the surface population, with a fully functioning circadian clock, following feeding at a particular time of day. For the surface fish, early morning may be the most relevant time point for feeding in the wild and coincide with a particular phase of clock gene expression.
Physiological and behavioral responses to differences in diet can provide insights into the regularity and quantity of food available to developing A. mexicanus larvae in the wild. A. mexicanus larvae, like zebrafish, can subsist on a variety of foods including paramecium, rotifers, and fish feed that differ in micronutrients. In this study, A. mexicanus larvae were fed a standard diet of Artemia. Artemia is comprised of macronutrients that include diverse fatty acids, proteins, and carbohydrates. Analysis suggests that Artemia is ~40‐60% protein, raising the possibility that consumption of dietary protein may impact sleep (de Clercq et al. 2005). In Drosophila, dietary protein promotes postprandial sleep, while a loss of dietary protein disrupts sleep depth (Murphy et al. 2016; Brown et al. 2020; Titos et al. 2023). Therefore, it is possible that changes in protein detection, or its downstream targets, regulate the physiology of sleep circuits that are responsible for the different effects of feeding on sleep between Pachón and Tinaja cavefish. It is also possible that differences in protein content between Artemia and blood worms, accompanied by increased energetic demands during development, may underly the presence of postprandial sleep in larval, but not adult fish. Understanding the effects of different diets on sleep, and how individual macronutrients regulate sleep across populations could reveal evolved differences in sleep‐feeding interactions across different A. mexicanus populations.
The identification of postprandial sleep in cavefish provides an avenue for future studies examining the genetic basis of this behavior. In A. mexicanus, gene‐editing technology has been used to functionally interrogate the genetic basis of traits and may provide an avenue for genetic screening (Ma et al. 2015; Stahl et al. 2019). Mapping genetic loci associated with trait variation has been used to identify candidate regulators of many morphological and behavioral traits, including regulators of sleep, activity, feeding posture, and metabolism (Kowalko et al. 2013; Yoshizawa et al. 2015; Carlson et al. 2018; Riddle et al. 2021). Further, population genetic approaches have identified genome‐wide markers of selection across multiple cave populations, and this genetic variation may provide insight into genes impacting sleep‐feeding interactions (Herman et al. 2018; Warren et al. 2021; Moran et al. 2022). Genes with signatures of selection that have previously been implicated in sleep or feeding could provide candidate regulators of postprandial sleep. In A. mexicanus, like zebrafish, CRISPR‐based gene editing has been used to functionally validate genes identified through genomics approaches and could be applied to the investigation of postprandial sleep (Klaassen et al. 2018; Kroll et al. 2021). Genetic studies will require the use of CRISPR for forward genetic screens, or the identification of A. mexicanus with diminished or highly variable postprandial sleep that can be used for genetic mapping studies.
In conclusion, these studies identify postprandial sleep in A. mexicanus and suggest it is under independent genetic regulation from total sleep duration and meal size in surface fish and two populations of cavefish that have evolved in parallel. These studies lay the groundwork for future analysis that apply currently available population genetics, neural anatomical, and genetic screening toolsets in A. mexicanus to examine the integration of feeding and sleep regulation.
4. Materials and Methods
4.1. Husbandry
Throughout this study, we followed previously described standard animal husbandry and breeding for A. mexicanus (Borowsky 2008a; Kozol et al. 2023). All fish were housed under standard temperature (23°C) and lighting conditions (14:10 h light:dark cycle). Feeding was based on established husbandry for A. mexicanus (Kozol et al. 2023). Briefly, larval fish were fed 24‐hour old brine shrimp, A. fransiscana nauplii (Artemia), while adult fish were fed thawed frozen bloodworms. Foods are nutrient‐rich and mimic diets available in the natural environments of the Sierra del Abra region of Mexico where these fish originate (Elipot et al. 2014). With the exception of breeding, fish are fed once daily. Adult fish were bred by increasing water temperature to 27 ± 1°C and feeding thawed frozen bloodworms three times per day (Elipot et al. 2014).
A. mexicanus are classified as larvae, transparent and lacking adult fin rays and spines as well as scales, until at least 90 dpf (Kozol et al. 2023). The fish used in this study were either adults of approximately 1 year of age or 20 dpf larvae. Larvae were fed Artemia ad libitum from 6 to 20 dpf (Borowsky 2008b) and held in small glass bowls until behavioral testing at 20 dpf. All procedures in this study were approved under the Florida Atlantic University and Texas A&M University IACUC.
4.2. Sleep Behavior
These experiments focused on three distinct A. mexicanus morphotypes: the sighted, surface‐dwelling Río Choy, and two blind, cave‐dwelling populations, Pachón and Tinaja. We quantified sleep behavior in these fish using previously described methods (Jaggard et al. 2019) and baseline sleep data (O'Gorman et al. 2021). Briefly, we used Ethovision XT 17.0 software (Noldus Information Technology, Wageningen, the Netherlands) to track locomotor behavior. Raw locomotor behavior was used to calculate sleep behavior parameters using a custom Perl script (Jaggard et al. 2019). We operationally define sleep as 60 s or more of immobility, given that previous studies show both surface and Pachón cavefish exhibit increased arousal thresholds after this period (Jaggard et al. 2019). We defined immobility as a velocity below 6 mm/sec for larval fish and a velocity below 4 cm/sec for adult fish based on previous work that established an increased arousal threshold below these criteria (Yoshizawa et al. 2015). All recordings were performed at 23°C under a 14:10 h light/dark cycle.
4.3. Larval Behavior Recordings
All larval used to quantify sleep behavior were 20 dpf. Fish were fed and then acclimated individually in 24‐well plates for at least 15 hours before behavior recordings. Recordings began at ZT0 and lasted for 24 hours, with interruptions for feeding at specific time points. The 24‐well plates were placed on light boxes made from white acrylic housing infrared (IR) lights (Figure 1A). Basler ace acA1300‐200um Monochrome USB 3.0 Cameras with mounted IR filters were mounted above the well plates and recordings were taken using Pylon Viewer software.
The effects of feeding on sleep were tested throughout the light cycle at time points before ZT0, ZT2, ZT6, and ZT10. No feeding experiments were performed during the dark cycle. Each 24‐well plate was either not fed as a control or fed at a single time point. We conducted two separate feeding experiments. In the first experiment, larvae were fed for 10 min immediately before a 24‐hour recording beginning at ZT0. This 24‐hour recording was followed by a 2‐hour feeding behavior assay (described below) and then another behavior recording for 4 hours from ZT2 to ZT6 (Figure 1). In the second experiment, we recorded behavior for 24 hours around a 45‐min window for feeding before either ZT2, ZT6, or ZT10.
4.4. Larval Feeding Behavior Assay
To quantify the relationship between the amount of food consumption and postprandial sleep duration, we performed feeding assays that allowed us to count the number of Artemia consumed over a given time. The duration of the feeding assay was 2 h for the first experiment, starting at ZT0 following 24 h of recording. The duration of the feeding assay was 30 min for the second experiment, starting before ZT2, ZT6, or ZT10. For the 2‐h feeding assay, fish were given exactly 70 Artemia, for the 30‐min feeding assay, Artemia were provided ad libitum. We filled a new 24‐well plate with Artemia hatched within 24 hours and recorded for at least 1 min before transferring the larval fish from the recording well plate to this new feeding well plate. At the end of the recording duration, fish were removed from the feeding assay, placed back into the original 24‐well recording plate with clean water and returned to the behavior recording. We used FIJI (Schindelin et al. 2012) to count the number of Artemia both before the fish were added to the wells and at the end of the feeding assay. Subtraction of the former from the latter allowed us to determine the amount of Artemia eaten over the duration of the feeding assay.
4.5. Adult Behavior Recordings
Adult fish used for behavior recordings were approximately 1 year old with an equal number of males and females per treatment. Food was withheld for 5 days before recording. Fish were placed in individual glass tanks of approximately 30 × 17 cm in a 2 × 2 grid in front of an IR light board and left to acclimate for at least 24 hours. Recordings began at ZT0 and lasted 24 hours. In the top two tanks, 4 oz of thawed, frozen blood worms were added at ZT5.5 and any uneaten worms were removed after 30 min at ZT6. The fish in the bottom two tanks were not fed as a control.
4.6. Analysis
Statistical analyses were performed in GraphPad Prism (version # 9.5.0) and R (version 4.0.4). When assumptions of normality and equal variances were met, we used parametric t‐tests, ANOVA, and Pearson's r tests, otherwise we used nonparametric Mann–Whitney U, Kruskal–Wallis, and Spearman's ρ tests. Following a significant ANOVA or Kruskal–Wallis test, pairwise comparisons were made using Tukey's HSD or Dunn's test, respectively. Correlation statistics were determined by the goodness of fit from a linear regression.
To quantify the percent change in sleep duration during the 4 hours following feeding, we calculated proportions of total daylight sleep and postprandial sleep. The proportion of total daylight sleep was determined by dividing the total daylight sleep duration by the total daylight recording time. The proportion of postprandial sleep was determined by dividing the duration of sleep in the 4 hours after eating by 4 hours. We then calculated percent change as the proportion of postprandial sleep minus the proportion of total daylight sleep divided by the proportion of total daylight sleep.
Sleep propensity was quantified using a hidden Markov model described previously (Wiggin et al. 2020). Briefly, sleep/wake behavior was divided into 1‐min bins across the 4‐h postprandial sleep interval. Waking propensity or P(Wake) was calculated as the number of times the animal transitioned from asleep to awake divided by the number of bins where the animal was asleep, except the last bin. If there were no bins where the animal was asleep, the transition probability was considered undefined. Sleep propensity or P(Doze) was calculated as the number of times the animal transitioned from awake to asleep divided by the number of bins the animal was awake, except the last bin.
Supporting information
Supporting information.
Acknowledgments
This work was supported by NIH Grants NIH 1R01GM127872 to Nicolas Rohner, and Alex C. Keene; R24 OD030214 to Wes Warren, Nicolas Rohner, and Alex C. Keene; R21 NS122166 to Alex C. Keene and Johanna E. Kowalko; 1DP2AG071466‐01 to Nicolas Rohner; and NSF Grant IOS 2202359 to Johanna E. Kowalko and S.E.M. The authors are grateful for technical assistance from Kaya Harper and Lawal Agboola.
Morgan O'Gorman and Pierce Hutton contributed equally to this study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Alié, A. , Devos L., Torres‐Paz J., et al. 2018. “Developmental Evolution of the Forebrain in Cavefish, From Natural Variations in Neuropeptides to Behavior.” eLife 7: e32808. 10.7554/eLife.32808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anafi, R. C. , Kayser M. S., and Raizen D. M.. 2019. “Exploring Phylogeny to Find the Function of Sleep.” Nature Reviews Neuroscience 20, no. 2: 109–116. 10.1038/s41583-018-0098-9. [DOI] [PubMed] [Google Scholar]
- Appelbaum, L. , Wang G. X., Maro G. S., et al. 2009. “Sleep‐Wake Regulation and Hypocretin‐Melatonin Interaction in Zebrafish.” Proceedings of the National Academy of Sciences 106: 21942–21947. 10.1073/pnas.906637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble, D. M. , Bass J., Behn C. D., et al. 2015. “Impact of Sleep and Circadian Disruption on Energy Balance and Diabetes: A Summary of Workshop Discussions.” Sleep 38, no. 12: 1849–1860. 10.5665/sleep.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiras, A. C. , Rohner N., Martineau B., Borowsky R. L., and Tabin C. J.. 2015. “Melanocortin 4 Receptor Mutations Contribute to the Adaptation of Cavefish to Nutrient‐Poor Conditions.” Proceedings of the National Academy of Sciences 112, no. 31: 9668–9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulsebrook, A. E. , Jones T. M., Rattenborg N. C., Roth T. C., and Lesku J. A.. 2016. “Sleep Ecophysiology: Integrating Neuroscience and Ecology.” Trends in Ecology & Evolution 31, no. 8: 590–599. 10.1016/j.tree.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Beale, A. , Guibal C., Tamai T. K., et al. 2013. “Circadian Rhythms in Mexican Blind Cavefish Astyanax mexicanus in the Lab and in the Field.” Nature Communications 4: 2769. 10.1038/ncomms3769. [DOI] [PubMed] [Google Scholar]; http://www.ncbi.nlm.nih.gov/pubmed/24225650.
- Bilandzija, H. , Ma L., Parkhurst A., and Jeffery W.. 2013. “A Potential Benefit of Albinism in Astyanax Cavefish: Downregulation of the oca2 Gene Increases Tyrosine and Catecholamine Levels As an Alternative to Melanin Synthesis.” PLoS One 8, no. 11: e80823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbély, A. A. , Daan S., Wirz‐Justice A., and Deboer T.. 2016. “The Two‐Process Model of Sleep Regulation: A Reappraisal.” Journal of Sleep Research 25, no. 3: 131–143. 10.1111/jsr.12371. [DOI] [PubMed] [Google Scholar]
- Borowsky, R. 2008a. “Breeding Astyanax mexicanus Through Natural Spawning.” Cold Spring Harbor Protocols 3, no. 11: 10–12. 10.1101/pdb.prot5091. [DOI] [PubMed] [Google Scholar]
- Borowsky, R. 2008b. “Handling Astyanax mexicanus Eggs and Fry.” Cold Spring Harbor Protocols 3, no. 11. 10.1101/pdb.prot5093. [DOI] [PubMed] [Google Scholar]
- Brown, E. B. , Shah K. D., Faville R., Kottler B., and Keene A. C.. 2020. “Drosophila Insulin‐Like Peptide 2 Mediates Dietary Regulation of Sleep Intensity.” PLoS Genetics 16, no. 3: e1008270. 10.1371/journal.pgen.1008270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E. B. , Slocumb M. E., Szuperak M., et al. 2019. “Starvation Resistance Is Associated With Developmentally Specified Changes in Sleep, Feeding and Metabolic Rate.” Journal of Experimental Biology 222, no. 3: jeb191049. 10.1242/jeb.191049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill, G. M. , Hurd M. W., and Batchelor M. M.. 1998. “Circadian Rhythmicity in the Locomotor Activity of Larval Zebrafish.” Neuroreport. 9: 3445–3449. [DOI] [PubMed] [Google Scholar]
- Capellini, I. , Barton R. A., McNamara P., Preston B. T., and Nunn C. L.. 2008. “Phylogenetic Analysis of the Ecology and Evolution of Mammalian Sleep.” Evolution 62: 1764–1776. 10.1111/j.1558-5646.2008.00392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, B. M. , Klingler I. B., Meyer B. J., and Gross J. B.. 2018. “Genetic Analysis Reveals Candidate Genes for Activity QTL in the Blind Mexican Tetra, Astyanax mexicanus .” PeerJ 6: e5189. 10.7717/peerj.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallari, N. , Frigato E., Vallone D., et al. 2011. “A Blind Circadian Clock in Cavefish Reveals That Opsins Mediate Peripheral Clock Photoreception.” PLoS Biology 9, no. 9: e1001142. 10.1371/journal.pbio.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, C. N. , and Prober D. A.. 2013. “Regulation of Zebrafish Sleep and Arousal States: Current and Prospective Approaches.” Frontiers in Neural Circuits 7 (April): 58. 10.3389/fncir.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, C. N. , Rihel J., Lee D. A., et al. 2016. “A Zebrafish Genetic Screen Identifies Neuromedin U As a Regulator of Sleep/Wake States.” Neuron 89, no. 4: 842–856. 10.1016/j.neuron.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Clercq, P. , Arijs Y., van Meir T., et al. 2005. “Nutritional Value of Brine Shrimp Cysts As a Factitious Food for Orius laevigatus (Heteroptera: Anthocoridae).” Biocontrol Science and Technology 15, no. 5: 467–479. 10.1080/09583150500086706. [DOI] [Google Scholar]
- Cobham, A. E. , and Rohner N.. 2024. “Unraveling Stress Resilience: Insights From Adaptations to Extreme Environments by Astyanax mexicanus Cavefish.” Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 342: 178–188. 10.1002/jez.b.23238. [DOI] [PubMed] [Google Scholar]
- Duboué, E. R. , Borowsky R. L., and Keene A. C.. 2012. “β‐Aadrenergic Signaling Regulates Evolutionarily Derived Sleep Loss in the Mexican Cavefish.” Brain, Behavior and Evolution 80, no. 4: 233–243. 10.1159/000341403. [DOI] [PubMed] [Google Scholar]
- Duboué, E. R. , Keene A. C., and Borowsky R. L.. 2011. “Evolutionary Convergence on Sleep Loss in Cavefish Populations.” Current Biology 21, no. 8: 671–676. 10.1016/j.cub.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Elipot, Y. , Legendre L., Père S., Sohm F., and Rétaux S.. 2014. “Astyanax Transgenesis and Husbandry: How Cavefish Enters the Laboratory.” Zebrafish 11, no. 4: 291–299. 10.1089/zeb.2014.1005. [DOI] [PubMed] [Google Scholar]
- Espinasa, L. , Bibliowicz J., Jeffery W. R., and Rétaux S.. 2014. “Enhanced Prey Capture Skills in Astyanax Cavefish Larvae Are Independent From Eye Loss.” Evodevo 5, no. 1: 35. 10.1186/2041-9139-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinasa, L. , Bonaroti N., Wong J., Pottin K., Queinnec E., and Rétaux S.. 2017. “Contrasting Feeding Habits of Post‐Larval and Adult Astyanax Cavefish.” Subterranean Biology 21, no. 1: 1–17. 10.3897/subtbiol.21.11046. [DOI] [Google Scholar]
- Förster, D. , Helmbrecht T. O., Mearns D. S., Jordan L., Mokayes N., and Baier H.. 2020. “Retinotectal Circuitry of Larval Zebrafish Is Adapted to Detection and Pursuit of Prey.” eLife 9: e58596. 10.7554/eLife.58596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallman, K. , Fortune Rivera D., and Soares D.. 2020. “Differences in Behavior Between Surface and Cave Astyanax Mexicanus May be Mediated by Changes in Catecholamine Signaling.” Journal of Comparative Neurology 528, no. 16: 2639–2653. 10.1101/72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, N. , Levine B. J., Loy K. A., et al. 2018. “Hypothalamic Neurons That Regulate Feeding Can Influence Sleep/Wake States Based on Homeostatic Need.” Current Biology 28, no. 23: 3736–3747.e3. 10.1016/j.cub.2018.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, J. B. 2012. “The Complex Origin of Astyanax Cavefish.” BMC Evolutionary Biology 12: 105. 10.1186/1471-2148-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, A. , Brandvain Y., Weagley J., et al. 2018. “The Role of Gene Flow in Rapid and Repeated Evolution of Cave‐Related Traits in Mexican Tetra, Astyanax mexicanus .” Molecular Ecology 27: 4397–4416. 10.1111/mec.14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggard, J. B. , Lloyd E., Lopatto A., Duboue E. R., and Keene A. C.. 2019. “Automated Measurements of Sleep and Locomotor Activity in Mexican Cavefish.” Journal of Visualized Experiments 145: 1–8. 10.3791/59198. [DOI] [PubMed] [Google Scholar]
- Jaggard, J. B. , Lloyd E., Lopatto A., Duboue E. R., and Keene A. C.. 2019. “Automated Measurements of Sleep and Locomotor Activity in Mexican Cavefish.” Journal of Visualized Experiments, no. 145: e59198. 10.3791/59198. [DOI] [PubMed] [Google Scholar]
- Jaggard, J. B. , Lloyd E., Yuiska A., et al. 2020. “Cavefish Brain Atlases Reveal Functional and Anatomical Convergence Across Independently Evolved Populations.” Science Advances 6, no. 38: eaba3126. 10.1126/sciadv.aba3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggard, J. B. , Stahl B. A., Lloyd E., Prober D. A., Duboue E. R., and Keene A. C.. 2018. “Hypocretin Underlies the Evolution of Sleep Loss in the Mexican Cavefish.” eLife 7: e32637. 10.7554/eLife.32637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery, W. R. 2009. “Regressive Evolution in Astyanax Cavefish.” Annual Review of Genetics 43: 25–47. 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner, W. J. 2016. “Unraveling the Evolutionary Determinants of Sleep.” Current Biology 26, no. 20: R1073–R1087. 10.1016/j.cub.2016.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, A. C. , and Appelbaum L.. 2019. Sleep in Fish Models. Elsevier. 10.1016/B978-0-12-813743-7.00024-4. [DOI]
- Keene, A. C. , and Duboue E. R.. 2018. “The Origins and Evolution of Sleep.” Journal of Experimental Biology 221: jeb159533. 10.1242/jeb.159533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, A. C. , Duboué E. R., McDonald D. M., et al. 2010. “Clock and Cycle Limit Starvation‐Induced Sleep Loss in Drosophila.” Current Biology 20, no. 13: 1209–1215. 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, A. C. , Yoshizawa M., and McGaugh S. E.. 2015. Biology and Evolution of the Mexican Cavefish.
- Klaassen, H. , Wang Y., Adamski K., Rohner N., and Kowalko J. E.. 2018. “CRISPR Mutagenesis Confirms the Role of oca2 in Melanin Pigmentation in Astyanax mexicanus .” Developmental Biology 441, no. 2: 313–318. 10.1016/j.ydbio.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Kowalko, J. E. , Rohner N., Linden T. A., et al. 2013. “Convergence in Feeding Posture Occurs Through Different Genetic Loci in Independently Evolved Cave Populations of Astyanax mexicanus .” Proceedings of the National Academy of Sciences 110, no. 42: 16933–16938. 10.1073/pnas.1317192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozol, R. A. , Yuiska A., Han J. H., et al. 2023. “Novel Husbandry Practices Result in Rapid Rates of Growth and Sexual Maturation Without Impacting Adult Behavior in the Blind Mexican Cavefish.” Zebrafish 20, no. 2: 86–94. 10.1089/zeb.2023.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, F. , Powell G. T., Ghosh M., et al. 2021. “A Simple and Effective F0 Knockout Method for Rapid Screening of Behaviour and Other Complex Phenotypes.” eLife 10: e59683. 10.7554/eLife.59683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, E. K. , Patel S. R., Goodloe R. J., et al. 2010. “A Candidate Gene Study of Obstructive Sleep Apnea in European Americans and African Americans.” American Journal of Respiratory and Critical Care Medicine 182, no. 7: 947–953. 10.1164/rccm.201002-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesku, J. A. , Roth T. C. II, Amlaner C. J., and Lima S. L.. 2006. “A Phylogenetic Analysis of Sleep Architecture in Mammals: The Integration of Anatomy, Physiology, and Ecology.” American Naturalist 168, no. 4: 441–453. 10.1086/506973. [DOI] [PubMed] [Google Scholar]
- Leung, L. C. , Wang G. X., Madelaine R., et al. 2019. “Neural Signatures of Sleep in Zebrafish.” Nature 571, no. 7764: 198–204. 10.1038/s41586-019-1336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitas‐Djerbi, T. , and Appelbaum L.. 2017. “Modeling Sleep and Neuropsychiatric Disorders in Zebrafish.” Current Opinion in Neurobiology 44: 89–93. 10.1016/j.conb.2017.02.017. [DOI] [PubMed] [Google Scholar]
- Lloyd, E. , Olive C., Stahl B. A., et al. 2018. “Evolutionary Shift Towards Lateral Line Dependent Prey Capture Behavior in the Blind Mexican Cavefish.” Developmental Biology 441: 328–337. 10.1016/j.ydbio.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Jeffery W. R., Essner J. J., and Kowalko J. E.. 2015. “Genome Editing Using TALENs in Blind Mexican Cavefish, Astyanax mexicanus .” PLoS One 10, no. 3: e0119370. 10.1371/journal.pone.0119370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, K. L. , Jaggard J. B., Persons J. L., et al. 2021. “Repeated Evolution of Circadian Clock Dysregulation in Cavefish Populations.” PLoS Genetics 17, no. 7: e1009642. 10.1371/journal.pgen.1009642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, M. , Ulzii E., Shirasaki R., Kim J., and You Y.‐J.. 2021. “Regulation of Satiety Quiescence by Neuropeptide Signaling in Caenorhabditis elegans .” Frontiers in Neuroscience 15: 678590. 10.3389/fnins.2021.678590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh, S. E. , Kowalko J. E., Duboué E., et al. 2020. “Dark World Rises: The Emergence of Cavefish as a Model for the Study of Evolution, Development, Behavior, and Disease.” Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 334, no. 7–8: 397–404. 10.1002/jez.b.22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh, S. E. , Passow C. N., Jaggard J. B., Stahl B. A., and Keene A. C.. 2020. “Unique Transcriptional Signatures of Sleep Loss Across Independently Evolved Cavefish Populations.” Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 334: 22949. 10.1002/jez.b.22949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, P. , Barton R., and Nunn C.. 2009. Evolution of Sleep: Phylogenetic and Functional Perspectives. http://www.lavoisier.fr/notice/frTWOL6K6ASRW3AO.html. [Google Scholar]
- Medley, J. K. , Persons J., Biswas T., et al. 2022. “The Metabolome of Mexican Cavefish Shows a Convergent Signature Highlighting Sugar, Antioxidant, and Ageing‐Related Metabolites.” eLife 11: e74539. 10.7554/eLife.74539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, H. A. , and Whitmore D.. 2014. “Circadian Rhythmicity and Light Sensitivity of the Zebrafish Brain.” PLoS One 9, no. 1: e86176. 10.1371/journal.pone.0086176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, D. , Softley R., and Warrant E. J.. 2014. “Eyeless Mexican Cavefish Save Energy by Eliminating the Circadian Rhythm in Metabolism.” PLoS One 9, no. 9: e107877. 10.1371/journal.pone.0107877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, R. , Richards E., Ornelas‐Garcia C., et al. 2023. “Selection‐Driven Trait Loss in Independently Evolved Cavefish Populations.” Nature Communications 14: 2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K. R. , Deshpande S. A., Yurgel M. E., et al. 2016. “Postprandial Sleep Mechanics in Drosophila.” eLife 5: e19334. 10.7554/eLife.19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman, M. , Thakur S., Imrie G., et al. 2021. “Pleiotropic Function of the oca2 Gene Underlies the Evolution of Sleep Loss and Albinism in Cavefish.” Current Biology 31, no. 16: 3694–3701.e4. 10.1016/J.CUB.2021.06.077. [DOI] [PubMed] [Google Scholar]
- Penney, C. C. , and Volkoff H.. 2014. “Peripheral Injections of Cholecystokinin, Apelin, Ghrelin and Orexin in Cavefish (Astyanax fasciatus mexicanus): Effects on Feeding and on the Brain Expression Levels of Tyrosine Hydroxylase, Mechanistic Target of Rapamycin and Appetite‐Related Hormones.” General and Comparative Endocrinology 196: 34–40. [DOI] [PubMed] [Google Scholar]
- Pillai, S. , Nandalike K., Kogelman Y., Muzumdar R., Balk S. J., and Arens R.. 2014. “Severe Obstructive Sleep Apnea in a Child With Melanocortin‐4 Receptor Deficiency.” Journal of Clinical Sleep Medicine 10, no. 1: 99–101. 10.5664/jcsm.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo‐Morales, M. , Cobham A. E., Centola C., et al. 2024. “Starvation‐Resistant Cavefish Reveal Conserved Mechanisms of Starvation‐Induced Hepatic Lipotoxicity.” Life Science Alliance 7, no. 5: e202302458. 10.26508/lsa.202302458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado, J. M. , Cortez M. B., Grubbs J., Link J. A., van der Linden A., and Zhang Y.. 2017. “Increased Food Intake After Starvation Enhances Sleep in Drosophila melanogaster .” Journal of Genetics and Genomics 44, no. 6: 319–326. 10.1016/j.jgg.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, M. R. , Aspiras A., Damen F., McGaugh S., Tabin J. A., and Tabin C. J.. 2021. “Genetic Mapping of Metabolic Traits in the Blind Mexican Cavefish Reveals Sex‐Dependent Quantitative Trait Loci Associated With Cave Adaptation.” BMC Ecology and Evolution 21, no. 1: 94. 10.1186/s12862-021-01823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel, J. , Prober D. A., Arvanites A., et al. 2010. “Zebrafish Behavioral Profiling Links Drugs to Biological Targets and Rest/Wake Regulation.” Science 327, no. 5963: 348–351. 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salin, K. , Voituron Y., Mourin J., and Hervant F.. 2010. “Cave Colonization Without Fasting Capacities: An Example With the Fish Astyanax fasciatus mexicanus .” Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 156, no. 4: 451–457. 10.1016/j.cbpa.2010.03.030. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras I., Frise E., et al. 2012. “Fiji: An Open‐Source Platform for Biological‐Image Analysis.” Nature Methods 9, no. 7: 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebacher, F. 2018. “The Evolution of Metabolic Regulation in Animals.” Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 224: 195–203. 10.1016/j.cbpb.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Semmelhack, J. L. , Donovan J. C., Thiele T. R., Kuehn E., Laurell E., and Baier H.. 2014. “A Dedicated Visual Pathway for Prey Detection in Larval Zebrafish.” eLife 3: e04878. 10.7554/eLife.04878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, C. , Oikonomou G., and Prober D. A.. 2015. “Norepinephrine Is Required to Promote Wakefulness and for Hypocretin‐Induced Arousal in Zebrafish.” eLife 4 (September): e070000. 10.7554/eLife.07000.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, C. , Rihel J., and Prober D. A.. 2017. “Neuropeptide Y Regulates Sleep by Modulating Noradrenergic Signaling.” Current Biology 27, no. 24: 3796–3811.e5. 10.1016/j.cub.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocumb, M. E. , Regalado J. M., Yoshizawa M., et al. 2015. “Enhanced Sleep Is an Evolutionarily Adaptive Response to Starvation Stress in Drosophila.” PLoS One 10, no. 7: e0131275. 10.1371/journal.pone.0131275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, B. A. , Jaggard J. B., Chin J. S. R., Kowalko J. E., Keene A. C., and Duboué E. R.. 2019. “Manipulation of Gene Function in Mexican Cavefish.” Journal of Visualized Experiments 00: e59093. 10.3791/59093. [DOI] [PubMed]
- Stahl, B. A. , Slocumb M. E., Chaitin H., DiAngelo J. R., and Keene A. C.. 2017. “Sleep‐Dependent Modulation of Metabolic Rate in Drosophila.” Sleep 40, no. 8: zsx084. 10.1093/sleep/zsx084. [DOI] [PMC free article] [PubMed]
- Stahl, M. L. , Orr W. C., and Bollinger C.. 1983. “Postprandial.” Sleep 6, no. 1: 29–35. http://www.ncbi.nlm.nih.gov/pubmed/6844795. [DOI] [PubMed] [Google Scholar]
- Tran, S. , and Prober D. A.. 2022. “Validation of Candidate Sleep Disorder Risk Genes Using Zebrafish.” Frontiers in Molecular Neuroscience 15: 873520. 10.3389/fnmol.2022.873520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkoff, H. 2016. “Feeding Behavior, Starvation Response, and Endocrine Regulation of Feeding in Mexican Blind Cavefish (Astyanax fasciatus mexicanus).” In Biology and Evolution of the Mexican Cavefish, 1st Edition , edited by Keene A., Yoshizawa M., and McGaugh S. E., 269–290. Amsterdam, Boston, Heidelberg, London, New York, Oxford, Paris, San Diego, San Franscisco, Sydney, Tokyo: Elsevier. [Google Scholar]
- Warren, W. C. , Boggs T. E., Borowsky R., et al. 2021. “A Chromosome‐Level Genome of Astyanax mexicanus Surface Fish for Comparing Population‐Specific Genetic Differences Contributing to Trait Evolution.” Nature Communications 12, no. 1: 1447. 10.1038/s41467-021-21733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee, C. L. , Song E. Y., Johnson R. E., et al. 2019. “A Bidirectional Network for Appetite Control in Larval Zebrafish.” eLife 8: e43775. 10.7554/eLife.43775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggin, T. D. , Goodwin P. R., Donelson N. C., et al. 2020. “Covert Sleep‐Related Biological Processes Are Revealed by Probabilistic Analysis in Drosophila.” Proceedings of the National Academy of Sciences 117, no. 18: 10024–10034. 10.1073/pnas.1917573117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa, M. 2015. “Behaviors of Cavefish Offer Insight Into Developmental Evolution.” Molecular Reproduction and Development 82, no. 4: 268–280. 10.1002/mrd.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa, M. , Robinson B. G., Duboué E. R., et al. 2015. “Distinct Genetic Architecture Underlies the Emergence of Sleep Loss and Prey‐Seeking Behavior in the Mexican Cavefish.” BMC Biology 13, no. 13: 15. 10.1186/s12915-015-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurgel, M. , Masek P., DiAngelo J. R., and Keene A.. 2014. “Genetic Dissection of Sleep‐Metabolism Interactions in the Fruit Fly.” Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology 201: 869–877. 10.1007/s00359-014-0936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


