Abstract
Background
Previous studies have demonstrated that asthma is closely associated with bronchiectasis, however, the causal relationship between asthma and bronchiectasis has not been investigated in depth. Therefore, this study aims to explore the causal relationship and to identify potential factors that mediate between these two diseases.
Method
All the necessary summarized information were obtained from publicly available genome-wide association study (GWAS). Two-sample Mendelian randomization (two-sample MR) was employed to explore the causal relationship between asthma and bronchiectasis, with an additional dataset used for validation. Heterogeneity and pleiotropy analyses were utilized to verify the robustness of the results. Subsequently, mediation MR analyses were performed to identify potential mediating factors. Lastly, a retrospective observational study was conducted to validate the findings.
Result
Preliminary inverse-variance weighted (IVW) results indicated there was a causal effect of asthma on bronchiectasis (odds ratio [OR] = 1.228, 95% confidence interval [CI]: 1.077–1.400, P = 0.002). Repetition validation yielded a consistent result. Mediation MR analysis demonstrated that the presence of nasal polyps (OR = 1.063, 95% CI: 1.015–1.113, mediation ratio = 30.492%, P = 0.009), acute sinusitis (OR = 1.062, 95% CI: 1.009–1.118, mediation ratio = 30.157%, P = 0.018), chronic sinusitis (OR = 1.085, 95% CI: 1.024–1.150, mediation ratio = 40.677%, P = 0.005), and peripheral eosinophil counts (OR = 1.013, 95% CI: 1.000–1.026, mediation ratio = 6.514%, P = 0.042) served as significant mediators in the occurrence and development of bronchiectasis induced by asthma. Furthermore, a retrospective observational study observed that bronchiectasis patients with asthma had a higher prevalence of sinusitis (5.043% vs 2.971%, P < 0.001), nasal polyps (0.536% vs 0.152%, P < 0.001), and rhinitis (13.197% vs 1.860%, P < 0.001). The ratio (1.950 (0.500, 5.600) vs 1.500 (0.500, 2.600), P = 0.006) and counts (0.125 (0.040, 0.363) vs 0.090 (0.030, 0.160), P < 0.001) of peripheral blood eosinophils were also elevated in bronchiectasis patients with asthma.
Conclusion
The MR analysis uncovered a notable genetic association between asthma and bronchiectasis, which was partially mediated by sinusitis, nasal polyps, and eosinophils. A subsequent retrospective study provided further evidence by demonstrating that bronchiectasis patients with asthma had a higher prevalence of sinusitis, nasal polyps, an elevated proportion of eosinophils, and higher eosinophil counts.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12931-024-03034-3.
Keywords: Asthma, Bronchiectasis, Mendelian randomization, Sinusitis, Nasal polyp, Eosinophil
Introduction
Bronchiectasis is a chronic respiratory disease characterized by abnormal and permanent dilation of the bronchi, which can be caused by various factors [1, 2]. The etiology of bronchiectasis is complex, often regarded as the common consequence for the destruction of airway structures resulting from multiple diseases [3]. In recent years, there has been a significant increase in the prevalence of bronchiectasis. As of 2013, the prevalence rate of bronchiectasis had escalated to 485.5 per 100,000 cases in men and 566.1 per 100,000 cases in women [4]. The incidence rate of bronchiectasis in Spain was approximately 48.1 per 100,000 cases in 2012 [5]. There was a 2.31-fold increase in the prevalence of adult bronchiectasis in China from 2013 to 2017 [6]. Nevertheless, the etiology and pathogenesis behind bronchiectasis are still not fully understood, which greatly restricts its prevention and treatment.
Asthma is a chronic inflammatory disease of the airways, with the principal characteristics of airway hyperresponsiveness, type 2 inflammation and airway remodeling [7]. Bronchiectasis and asthma are frequently co-existing diseases [8]. A previous study revealed that up to 50% patients with severe asthma were found to have evidence of bronchiectasis using high resolution computed tomography (HRCT) scans [1]. European bronchiectasis registry (EMBARC) reported that the complication rate of asthma in patients with bronchiectasis was as high as 31% [9]. Our research team also pioneered the concept of bronchiectasis-asthma overlap syndrome and demonstrated that asthma is an independent risk factor for worsening bronchiectasis [10]. Despite numerous studies indicating a strong association between asthma and bronchiectasis, the causal relationship between the two conditions remains to be elucidated [11].
The upper and lower respiratory tracts are considered to be a unified morphological and functional unit. The concept of “upper and lower airway comorbidity” has gained significant recognition in recent years [12]. Previous studies had shown that over 80% of individuals diagnosed with asthma had rhinitis, while approximately 10–40% of those rhinitis patients experienced asthma [13]. Specific allergens and infections were found to be common risk factors for both allergic rhinitis and asthma, with allergic rhinitis being an independent risk factor for asthma [14]. It was demonstrated that the treatment of upper airway inflammation could alleviate the clinical manifestations of lower airway inflammation and reduce the frequency of acute episodes [15]. Another study revealed that the expression characteristics of upper airway ciliary markers (DNAH5, DNAI1, and RSPH9) in bronchiectasis patients were largely consistent with those in the lower airway [16]. These findings strongly support the existence of an interaction between the upper and lower respiratory tracts. In addition, eosinophilic inflammation plays an important role in the occurrence and development of asthma, as well as related upper respiratory tract diseases, such as sinusitis and rhinitis [17–19]. In recent years, eosinophilic bronchiectasis had also been widely reported [20–22]. Therefore, it is worth exploring whether upper respiratory diseases and eosinophils mediate the impact of asthma on bronchiectasis.
Mendelian randomization (MR) is a method used in genetic epidemiology that explores the causal relationship between risk factors and outcomes [23]. This study aims to investigate the causal relationship and potential mediating factors between asthma and bronchiectasis through MR analysis, as well as to validate this phenomenon through a retrospective observational study.
Methods
Study design
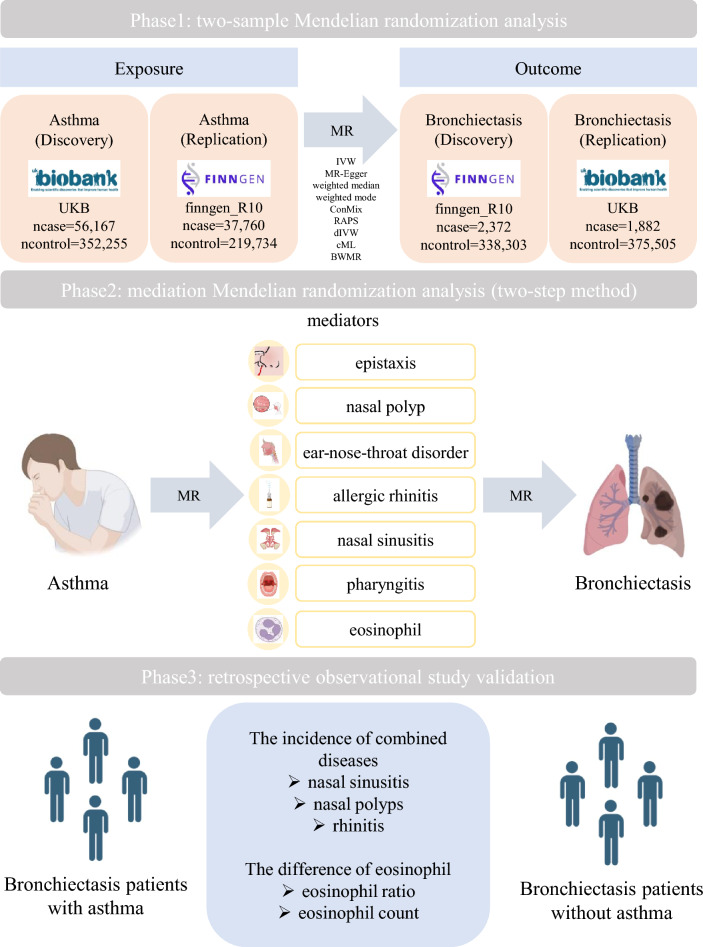
This study comprises three broad components (Fig. 1). Two-sample MR was used first to analyze the genome-wide association results between asthma and bronchiectasis. Subsequently, reverse MR was also proceeded. Heterogeneity and pleiotropy analyses were performed to explore the stability of the findings. Simultaneously, another pair of dataset was used for cross validation. In addition, mediation MR analysis was conducted to investigate the potential mediating role of upper respiratory diseases and eosinophils between asthma and bronchiectasis. Lastly, a retrospective study was conducted to validate this phenomenon.
Fig. 1.
Diagram of Mendelian randomization hypothesis and study design
Data sources
The dataset used in this study was retrieved from publicly accessible GWAS datasets. Specifically, the GWAS summary datasets of the European population were utilized, including information regarding asthma, bronchiectasis, upper respiratory diseases (such as nasal polyps, acute sinusitis and so on), and eosinophils. These necessary data are sourced from the OPEN GWAS website (https://gwas.mrcieu.ac.uk/) [24], the UK Biobank (UKB), FinnGen database [25] or other referenced studies [26–29]. Detailed data sources can be found in Table 1, including GWAS ID, source and sample size.
Table 1.
GWAS summary datasets used in this study
| Explore or Outcome | GWAS ID | Source | Population | Sample size | Case | Control |
|---|---|---|---|---|---|---|
| Asthma | ebi-a-GCST90014325 | Valette K et al. | European | 408,442 | 56,167 | 352,255 |
| Asthma (only as main-diagnosis) | finngen_R10_J10_ASTHMA_MAIN_EXMORE | FinnGen | European | 257,494 | 37,760 | 219,734 |
| Bronchiectasis | finngen_R10_J10_BRONCHIECTASIS | FinnGen | European | 340,675 | 2,372 | 338,303 |
| Bronchiectasis | ukb-saige-496.3 | Taliun D et al. | European | 377,387 | 1,882 | 375,505 |
| Epistaxis | ukb-b-11412 | MRC-IEU | European | 463,010 | 1,512 | 461,498 |
| Nasal polyp | finngen_R10_J10_NASALPOLYP | FinnGen | European | 315,298 | 6,841 | 308,457 |
| Ear-nose-throat disorder | ebi-a-GCST90038669 | Dönertaş HM et al. | European | 484,598 | 17,317 | 467,281 |
| Allergic rhinitis | finngen_R10_ALLERG_RHINITIS | FinnGen | European | 404,309 | 12,240 | 392,069 |
| Acute sinusitis | finngen_R10_J10_SINUSITIS | FinnGen | European | 357,879 | 22,847 | 335,032 |
| Chronic sinusitis | finngen_R10_J10_CHRONSINUSITIS | FinnGen | European | 326,444 | 17,987 | 308,457 |
| Acute pharyngitis | finngen_R10_J10_PHARYNGITIS | FinnGen | European | 340,327 | 5,295 | 335,032 |
| Chronic rhinitis, nasopharyngitis and pharyngitis | finngen_R10_J10_CHRONRHINITIS | FinnGen | European | 320,425 | 11,968 | 308,457 |
| Eosinophill count | ebi-a-GCST90013985 | Mbatchou J et al. | European | 395,949 | N/A | N/A |
| Neutrophill count | ebi-a-GCST90013984 | Mbatchou J et al. | European | 395,949 | N/A | N/A |
Selection of genetic instrumental variables
The genetic variation as an instrumental variable (IV) for MR analysis was used in our study. These IVs satisfied three core assumptions: (1) the relevance hypothesis, representing a reliable association between genetic variation and exposure; (2) the independence hypothesis, indicating that genetic variation is not related to any known or unknown confounding factors; (3) the exclusion restriction hypothesis, implying that genetic variation influences the outcome only through the exposure. A rigorous quality control procedure was applied to identify IVs that met the MR assumptions [30]. Specifically, single nucleotide polymorphisms (SNPs) were identified at a genome-wide significance threshold (P < 5 × 10− 8) [31]. In detail, the threshold was P < 5 × 10− 8 when analyzed the effect of asthma on bronchiectasis. Due to insufficient SNPs for bronchiectasis, the threshold was adjusted to P < 5 × 10− 6 when investigated the effect of bronchiectasis on asthma. Eligible SNPs were included by clumping for linkage disequilibrium (within 10,000 kb and r2 > 0.01). The F-statistic, F = r2(n − 2)/ (1 − r2), which was used to evaluate the strength of IVs, and those with F < 10 were excluded to avoid the risk of weak instrument bias in MR analysis [32]. Furthermore, LDtrait was used to remove confounding factors [33]. MR-Steiger filtration method was used to exclude variables that were more related to outcome than exposure, which could improve the reliability of conclusions. All final IVs included were detailed in Table S1-4.
MR analysis
This research mainly involved nine different MR methods for the analysis, including inverse-variance weighted (IVW), MR-Egger, weighted median, weighted mode, contamination mixture (ConMix), robust adjusted profile score (RAPS), debiased inverse-variance weighted (dIVW), constrained maximum likelihood (cML) based Mendelian randomization and Bayesian weighted Mendelian randomization (BWMR). The characteristic of IVW is that the existence of an intercept term is not considered in the regression, and the reciprocal of outcome variance is used for fitting [34]. MR-Egger adds an intercept term to assess horizontal pleiotropy [35]. Weighted median uses the majority of SNPs to determine whether causal relationships exist [36]. The weighted mode method weights the causal effects of different genetic variations on traits, and then takes the weighted mode as the final causal effect estimate [37]. The ConMix method excels at analyzing hundreds of instrumental variables and can provide causal estimates even when some instruments are invalid [38]. RAPS allows the inclusion of weak instrumental variables, through which robust statistical estimation of MR can be made [39]. The dIVW method eliminates the weak instrument bias of the IVW method and has stronger robustness under many weak instruments. BWMR can help determine the causal effects of risk factors on complex traits or diseases [40]. Among these methods, IVW was the main analysis method, while other approaches were used to further support our findings [41]. The reverse MR analysis was performed using the same process.
Heterogeneity and pleiotropy analyses
There may be heterogeneity in MR analysis results due to different analysis platforms, selected populations, and instrumental variables, leading to bias in the estimation of causal effects. The Cochran’s Q test was used to evaluate the heterogeneity of instrumental variables [42]. Horizontal pleiotropy occurs when genetic variants affect outcomes other than the exposure. Therefore, MR-Egger regression was applied to verify the existence of horizontal pleiotropy [35]. The level of validity and the elimination of outliers were identified by utilizing MR-PRESSO [37]. In addition, leave-one-out analysis was employed to evaluate the impact of a single SNP on the regression coefficients [43]. The IVW method was used to evaluate the impact of SNPs on overall estimates by eliminating these individual SNPs. R software (version 4.3.2) was used to perform MR analyses with the TwoSampleMR (version 0.6.0), MendelianRandomization (version 0.8.0), RMediation (version 1.2.2), and MRPRESSO (version 1.0) package.
Retrospective study validation
A total of 13,745 hospitalized patients diagnosed with bronchiectasis from January 1, 2017 to May 15, 2024 were retrospectively collected from the hospitalization system in Shanghai Pulmonary Hospital, Shanghai, China. Ethical approval for this study was obtained from the Ethics Committees of Shanghai Pulmonary Hospital, Tongji University (No. K18-167). Bronchiectasis and asthma were diagnosed by senior physicians from Shanghai Pulmonary Hospital according to the international consensus recommendations [44] and Global Initiative for Asthma (GINA) [45], respectively. Upper respiratory diseases such as nasal polyps, sinusitis, and rhinitis were diagnosed by otolaryngologists based on relevant clinical examinations. When patients came to the department of respiratory and critical care medicine for treatment, these diseases were self-reported by patients when the doctors asked about their medical history. The earliest records were uniformly chosen for patients with multiple hospitalizations, and then the patients were divided into two groups: bronchiectasis patients with asthma (BE + A) and bronchiectasis patients without asthma (BE). The incidence of upper respiratory diseases such as sinusitis, nasal polyps, and rhinitis in both groups was analyzed using χ2 test. In terms of eosinophils, a stratified sampling approach was conducted according to the year of admission, and a total of 1000 patients were selected proportionally. The differences in the eosinophil ratio and eosinophil counts were compared by using Mann–Whitney U test.
Results
Causal effects of asthma on bronchiectasis
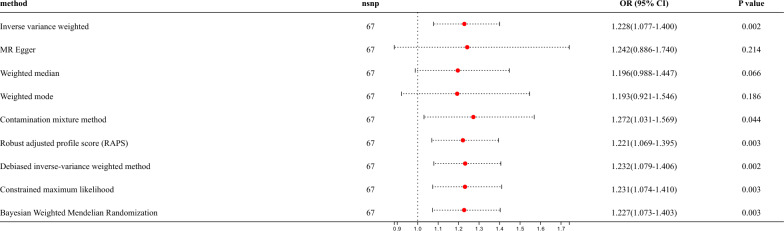
In this study, asthma (GWAS ID: ebi-a-GCST90014325) was designated as the exposure factor, while bronchiectasis (GWAS ID: finngen_R10_J10_BRONCHIECTASIS) was identified as the outcome. A total of 70 SNPs were included in this research. After removing the confounding factors (rs7936312, rs479844 and rs5743618), 67 SNPs significantly associated with asthma were identified as IVs finally. The F-values of these IVs exceeded 10, indicating the absence of weak IVs. All these IVs were used for further two-sample MR analysis. The IVW results showed the causal effect of asthma on bronchiectasis (OR = 1.228, 95% CI: 1.077–1.400, P = 0.002) (Fig. 2, Fig.S1A-B). In addition, multiple methods, such as ConMix, RAPS, dIVW, cML and BWMR, yielded consistent results, supporting the causal effect of asthma on bronchiectasis. Leave-one-out analyses indicated that the causal relationship between asthma and bronchiectasis was not driven by a single SNP (Fig.S1C). Through Cochran’s Q heterogeneity test, there was no significant heterogeneity (P = 0.206) (Table S5). The funnel plot also showed the absence of heterogeneity (Fig.S1D). Horizontal pleiotropy was detected by the MR Egger intercept, which showed that there was no horizontal pleiotropy in MR analysis (P = 0.946) either (Table S5). Afterwards, another dataset was selected for cross validation, specifically choosing asthma data from the FinnGen database (GWAS ID: finngen.R10_J10_ASTHMA_MAIN_EXMORE) as exposure and bronchiectasis data from the ukb cohort (GWAS ID: ukb saige-496.3) as outcome. The IVW results suggested a consistent result (OR = 1.619, 95% CI: 1.310–2.000, P < 0.001) (Fig.S2). The results of Cochran’s Q heterogeneity test and MR Egger intercept both indicated there was no evidence of heterogeneity and horizontal pleiotropy in asthma and bronchiectasis (P > 0.05) (Table S5).
Fig. 2.
Putative bidirectional causality of asthma on bronchiectasis
Reverse MR analysis
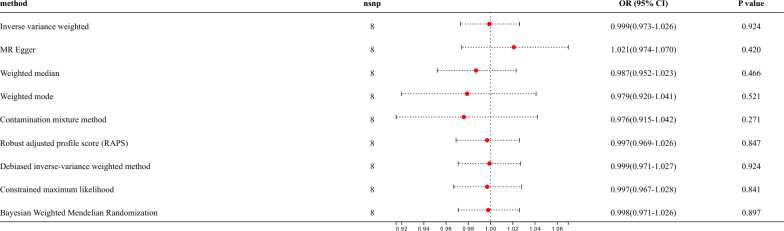
Reverse MR analysis was used to investigate the effect of bronchiectasis on the occurrence of asthma. The threshold was adjusted to P < 5 × 10−6, due to insufficient SNPs for bronchiectasis, and finally, 8 SNPs tightly related to bronchiectasis were included. However, the IVW analysis showed there was no causal effect of bronchiectasis on asthma (OR = 0.999, 95% CI: 0.973–1.026, P = 0.924) (Fig. 3, Fig.S3A-B). Leave-one-out analyses were conducted, and the findings showed the absence of any non-specific nucleotide polymorphisms that could potentially impact the results of the causal estimation (Fig. S3C). Heterogeneity testing was conducted using Cochran’s Q, and there was no significant heterogeneity (P = 0.606) between bronchiectasis and asthma (Table S5). The funnel plot also revealed no heterogeneity existed (Fig.S3D). The results of the MR Egger method showed that no horizontal pleiotropy (P = 0.309) was present (Table S5). A total of 8 SNPs were included in the replication datasets, nevertheless, rs17260332 were excluded as outlier value through MR-PRESSO. 7 SNPs were ultimately identified as IVs significantly associated with bronchiectasis. MR analysis indicated the same conclusion (OR = 1.007, 95% CI: 0.961–1.054, P = 0.772) (Fig.S4).
Fig. 3.
Putative bidirectional causality of bronchiectasis on asthma
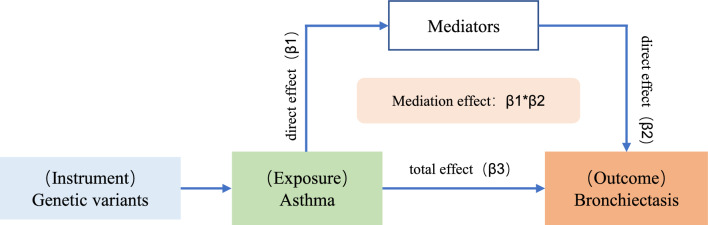
Mediational role of upper respiratory diseases and eosinophils
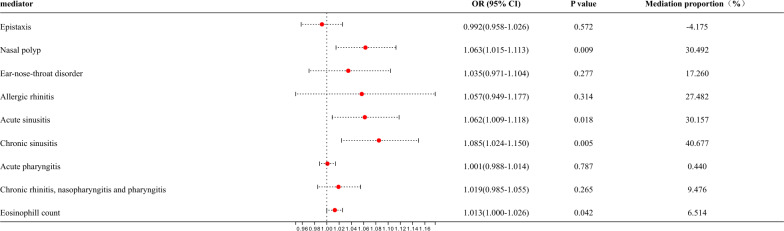
This study focused primarily on whether upper respiratory diseases and eosinophils could act as mediators. Mediation MR analysis on various upper respiratory diseases (epistaxis, nasal polyp, ear-nose-throat disorder, allergic rhinitis, acute sinusitis, chronic sinusitis, acute pharyngitis, chronic rhinitis, nasopharyngitis, and pharyngitis), peripheral eosinophil counts (Fig. 4). Results suggested that nasal polyps (OR = 1.063, 95% CI:1.015–1.113, mediation ratio = 30.492%, P = 0.009), acute sinusitis (OR = 1.062, 95% CI: 1.009–1.118, mediation ratio = 30.157%, P = 0.018), chronic sinusitis (OR = 1.085, 95% CI: 1.024–1.150, mediation ratio = 40.677%, P = 0.005), and peripheral eosinophil counts (OR = 1.013, 95% CI: 1.000–1.026, mediation ratio = 6.514%, P = 0.042) were identified as the mediators increasing the risk of bronchiectasis in asthma status (Fig. 5). Other factors have no potential as mediators. Considering the important role of neutrophils in asthma and bronchiectasis, the mediation MR analysis was conducted on neutrophils. It was found that neutrophils were not a mediating factor in the effect of asthma on bronchiectasis (OR = 1.000, 95% CI: 0.996–1.004, mediation ratio = −1.888%, P = 0.934) (Fig.S5A).
Fig. 4.
Diagram of intermediate Mendelian randomization analysis
Fig. 5.
The mediating role of upper respiratory diseases and eosinophils in the causal effect of asthma on bronchiectasis
Retrospective observational study validation
A total of 13,745 hospitalized patients diagnosed with bronchiectasis were enrolled through the hospitalization system of Shanghai Pulmonary Hospital from January 1, 2017 to May 15, 2024. Among them, 1864 patients were diagnosed with asthma (BE + A group, 13.561%), while 11,881 patients did not have asthma (BE group, 86.439%) (Table 2). In these two groups of patients, 5.043% of the BE + A patients had concurrent sinusitis, which was higher compared to the BE group (2.971%, P < 0.001). Similarly, the proportion of nasal polyps and rhinitis in the BE + A group (0.536% and 13.197%) was significantly higher than BE group (0.152% and 1.860%, P < 0.001). A total of 1000 bronchiectasis patients were selected proportionally, including 136 (13.600%) patients with asthma and 864 (86.400%) patients without asthma. Peripheral blood eosinophil ratio and eosinophil counts were analyzed respectively. The results indicated that the BE + A group had a higher ratio of eosinophil (1.950 (0.500, 5.600) vs 1.500 (0.500, 2.600), P = 0.006), as well as a higher eosinophil count (0.125 (0.040, 0.363) vs 0.090 (0.030, 0.160), P < 0.001) compared with the BE group. However, there was no significant statistical difference in the ratio and counts of neutrophil between the two groups (Fig.S5B-C). The findings of this retrospective study confirmed the results of MR analysis, suggesting that the prevalence rate of upper respiratory tract diseases such as sinusitis, nasal polyps and rhinitis, as well as eosinophil ratio and eosinophil counts may play potential mediating effects in the development of asthma into bronchiectasis.
Table 2.
Combined rates of upper respiratory disease (sinusitis, nasal polyps and rhinitis) and eosinophils in BE + A group and BE group
| Variables | Bronchiectasis with asthma (n = 1864) | Bronchiectasis without asthma (n = 11,881) | P value |
|---|---|---|---|
| Demographics | |||
| Age (y), mean ± sd | 57.241 ± 13.003 | 59.519 ± 13.195 | < 0.001 |
| Sex, female, no.(%) | 1071 (57.457) | 5916 (49.794) | < 0.001 |
| Comorbidity, no.(%) | |||
| Nasal sinusitis | 94 (5.043) | 353 (2.971) | < 0.001 |
| Nasal polyps | 10 (0.536) | 18 (0.152) | < 0.001 |
| Rhinitis | 246 (13.197) | 221 (1.860) | < 0.001 |
| *Eosinophils, median (IQR) | |||
| Blood eosinophil ratio (%) | 1.950 (0.500, 5.600) | 1.500 (0.500, 2.600) | = 0.006 |
| Blood eosinophil count (cells·109/L) | 0.125 (0.040, 0.363) | 0.090 (0.030, 0.160) | < 0.001 |
*Regarding eosinophils, clinical data were missing for two patients in the BE group
Discussion
This study is the first to comprehensively explore the causal relationship between asthma and bronchiectasis. The association was examined using two-sample MR, which revealed a causal effect of asthma on bronchiectasis. To further investigate the mechanism behind this association, mediation MR analysis was conducted, which was found that upper respiratory diseases (including nasal polyps, acute sinusitis, chronic sinusitis) and eosinophil counts played mediating roles in the causal relationship between asthma and bronchiectasis. Additionally, a retrospective observational study was conducted to assess the upper respiratory complications and eosinophils of bronchiectasis patients with or without asthma. The conclusions drawn from the MR analysis were validated, and it was observed that compared to the BE group, the BE + A group had a higher proportion of complications including sinusitis, nasal polyps, and rhinitis, as well as higher proportion and count of peripheral blood eosinophils.
Many patients have been observed to have the coexistence of bronchiectasis and asthma [46]. Previous studies conducted by our research team discovered that the presence of asthma was independently associated with an increased risk of exacerbation of bronchiectasis [10]. This study further explored the causal relationship between asthma and bronchiectasis based on MR analysis. According to the study by Säynäjäkangas et al., asthma was common in hospitalized patients with bronchiectasis and appeared to be the consequence of bronchiectasis [47]. However, our findings differed from this hypothesis, as it was found that bronchiectasis was not the cause of asthma. Our results were also supported by previous research. Data from the EMBARC, which included 16,963 patients with bronchiectasis, revealed that patients with asthma had a higher incidence of sinusitis, nasal polyps, and elevated peripheral eosinophil counts compared to those without asthma [9]. Schwartz et al. demonstrated that chronic rhinosinusitis was strongly associated with an increased risk of bronchiectasis, with chronic rhinosinusitis (CRS) being detected on average more than 6 years before the onset of bronchiectasis [48]. Interestingly, this study also demonstrated that allergic rhinitis was not related to bronchiectasis, which was consistent with our findings indicating that allergic rhinitis did not mediate the relationship between asthma and bronchiectasis. Another prospective cohort study found that CRS status was associated with increased odds of new onset of non-cystic fibrosis bronchiectasis [49]. These two studies strongly suggested that CRS was a significant contributor to the development of bronchiectasis. The research by Shteinberg’s team found that compared to bronchiectasis patients without sinusitis, bronchiectasis with sinusitis had higher level of peripheral blood eosinophils [50]. Eosinophilic inflammation is commonly observed when bronchiectasis is accompanied by type 2 airway inflammation such as asthma, allergic bronchopulmonary aspergillosis, CRS with nasal polyps [51]. The typical features of bronchiectasis are recurrent airway infections and neutrophil airway inflammation [52]. Neutrophil airway inflammation plays a very important role in refractory asthma, most of these patients were severe asthma [53]. In this study, we did not find the important role of neutrophils in the relationship between asthma and bronchiectasis, probably because most of the patients included did not have severe asthma. These studies suggested the mutual interaction among these factors. Considering the higher co-incidence of upper respiratory tract diseases and higher eosinophil counts in asthma patients, these factors seem to make reasonable sense as intermediaries between asthma and bronchiectasis.
According to the “united airways” theory, the upper and lower respiratory tracts are anatomically connected and have similar pathophysiology, thus our conclusion is biologically reasonable [54]. There may be multiple possible hypotheses regarding the specific mechanism of bronchiectasis caused by upper respiratory tract diseases such as sinusitis and nasal polyps [48]. Firstly, the nose and sinuses play an important role in filtering and humidifying air. Long-term chronic inflammation, along with repeated stimulation of pus, can result in pathological changes in the nasal mucosa [55]. Meanwhile, it has also been confirmed that these factors induce defects in epithelial barrier function and decrease expression of epithelial tight junctions, which may generate increased inflammatory responses caused by various stimuli or infection [56, 57]. Secondly, nasal congestion and inflammation may lead to oral breathing, which can alter the oral-nasopharyngeal microbiota and promote the inhalation of microorganisms, pollutants, and allergens into the lower respiratory tract [58]. Finally, long-term postnasal drip, which is the purulent secretion produced by the inflamed area flowing back through the nasal cavity into the lower respiratory tract, leads to infection or exacerbation of inflammation in the lower respiratory tract. Previous studies have reported that even in individuals without lung disease, chronic sinusitis and nasal polyps are still associated with subclinical lower respiratory flow restriction [59]. The similar inflammation or pathogens in the upper and lower respiratory tracts have been confirmed by numerous studies. It has been discovered that nasal allergen stimulation can cause bronchoconstriction and airway hyperresponsiveness, leading to asthma-like allergic inflammatory reactions in the lungs [60]. Experimental rhinovirus 16 infection caused lower respiratory symptoms in addition to upper respiratory symptoms, with rhinovirus RNA being detected in both the nose and lung [61, 62]. In patients with bronchiectasis, there was a strong correlation between bacterial culture in the upper and lower airways, particularly involving Pseudomonas aeruginosa [63]. These studies alluded to the important influence that upper respiratory tract disorders exert in lower respiratory tract diseases.
In recent years, the treatable characteristics of bronchiectasis have gradually received attention. Previous work by our research team found that psychological states such as depression were closely related to acute exacerbation and hospitalization risk in patients with bronchiectasis, suggesting that mental health was an important treatable aspect in bronchiectasis [64]. This study suggests that active intervention in asthma, upper respiratory diseases (such as sinusitis and nasal polyps), and elevated peripheral blood eosinophils may also be important treatable features in some patients with bronchiectasis. It has been shown that intranasal corticosteroids could significantly improve clinical symptoms in patients with allergic rhinitis and asthma [15]. Therefore, follow-up interventions for these symptoms may be considered. It is also worth noting that although findings from EMBARC’s data are consistent with this study, the incidence of upper respiratory diseases (such as sinusitis and nasal polyps) is higher. This may be attributed to the absence of routine specialized examinations for these diseases in the department of respiratory and critical care medicine. Therefore, it is necessary to explore ways to improve relevant examinations as much as possible in the future follow-up clinical practice.
This study possesses several advantages. Firstly, a comprehensive analysis of the causal relationship between asthma and bronchiectasis using GWAS data was conducted. This association from the perspective of the “upper and lower airway comorbidity” hypothesis was further elucidated. Secondly, a rigorous approach with an F statistic exceeding 10 was employed to mitigate bias arising from weak instruments. Simultaneously, sensitivity and heterogeneity analyses were performed to enhance the reliability of the findings. a diverse array of MR analysis methods was utilized to enhance the robustness of the findings. Thirdly, different datasets for replication were used to validate our conclusion. Finally, a retrospective observational study involving the Chinese population afflicted with bronchiectasis was conducted. Consequently, GWAS data from European samples were validated within the Chinese population, illustrating the universality of the findings. However, our study also has some limitations. Firstly, variations in sequencing and analysis methods of GWAS data encompassing asthma, upper respiratory diseases, and bronchiectasis may yield disparate results. Secondly, due to the quality of GWAS data and the absence of demographic information in the study, additional subgroup analyses concerning confounding factors such as age and gender for asthma and bronchiectasis remain unexplored. Thirdly, the retrospective observational study solely engaged a single clinical center, subsequent studies require more comprehensive validation from multiple clinical centers. Fourthly, unconventional specialized examinations for upper respiratory diseases like sinusitis and nasal polyps are not routinely performed in contemporary clinical practice, thereby resulting in a relatively low detection rate of associated diseases, potentially inducing corresponding deviations. Additionally, MR design itself has certain limitations, such as the potential for unmeasured confounding factors, the existence of pleiotropy, weak instrumental variable bias, and reverse causality. This study circumvented these issues through the aforementioned methods to ensure the accuracy of the results, however, the above issues still need further improvement. Lastly, present endeavors have been limited to retrospective observational studies, necessitating future prospective studies within the asthma population cohort to ascertain whether upper respiratory diseases (sinusitis and nasal polyps) and eosinophils (eosinophil ratio and eosinophil count) exert a predisposing influence on bronchiectasis occurrence.
Conclusion
MR analysis revealed significant causal correlations between asthma and bronchiectasis. Mediation analysis unveiled those upper respiratory diseases such as sinusitis and nasal polyps, as well as peripheral blood eosinophil counts were identified as the primary mediators. From a retrospective study, it was demonstrated that bronchiectasis patients with asthma had a higher prevalence of sinusitis, nasal polyps, and elevated proportion of eosinophils and eosinophil counts compared to those without asthma. These findings provide genetic evidence warranting further mechanistic and clinical investigations, which elucidate the association between asthma and bronchiectasis.
Supplementary Information
Additional file 1: Figure S1. Scatter plot (A), forest plot (B), leave-one-out analyses (C) and funnel plot (D) illustrating the causal effect of asthma (ebi-a-GCST90014325) on bronchiectasis (finngen_R10_J10_BRONCHIECTASIS).
Additional file 2: Figure S2. Putative bidirectional causality (A), Scatter plot (B), forest plot (C), leave-one-out analyses (D) and funnel plot (E) illustrating the causal effect of asthma (finngen_R10_J10_ASTHMA_MAIN_EXMORE) on bronchiectasis (ukb-saige-496.3).
Additional file 3: Figure S3. Scatter plot (A), forest plot (B), leave-one-out analyses (C) and funnel plot (D) illustrating the causal effect of bronchiectasis (finngen_R10_J10_BRONCHIECTASIS) on asthma (ebi-a-GCST90014325).
Additional file 4: Figure S4. Putative bidirectional causality (A), Scatter plot (B), forest plot (C), leave-one-out analyses (D) and funnel plot (E) illustrating the causal effect of bronchiectasis (ukb-saige-496.3) on asthma (finngen_R10_J10_ASTHMA_MAIN_EXMORE).
Additional file 5: Figure S5. (A) The mediating role of neutrophils in the causal effect of asthma on bronchiectasis; Statistical chart of neutrophil ratio (B) and counts (C) in BE + A group and BE group.
Additional file 6: Table S1. IVs in asthma (GWAS ID: ebi-a-GCST90014325). Table S2. IVs in bronchiectasis (GWAS ID: finngen_R10_J10_BRONCHIECTASIS). Table S3. IVs in asthma (GWAS ID: finngen.R10_J10_ASTHMA_MAIN_EXMORE). Table S4. IVs in bronchiectasis (GWAS ID: ukb saige-496.3). Table S5. Measures of heterogeneity and pleiotropy test.
Acknowledgements
We are exceptionally grateful to the FinnGen study, the UKB study, and all other researchers and participants for selflessly and generously making the summary GWAS data publicly available. We would like to express our deepest gratitude to all the patients who participated in this study.
Author contributions
JFX and WJC conceived and designed the study. RF, HQ and YS performed the study and analyzed the data. RF, HQ, JYX, JYW and JWY collected clinical data. RF and HQ wrote the manuscript, JFX, WJC and FJ supervised the study and critically reviewed the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81925001; 82330070; 82100055); the Innovation Program of Shanghai Municipal Education Commission (202101070007-E00097); Program of Shanghai Municipal Science and Technology Commission (21DZ2201800) and Program of Shanghai Shenkang Development Center (SHDC12023110); Medical Research Project of Jiangsu Provincial Health Commission (M2022101).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Ethical approval for this study was obtained from the Ethics Committees of Shanghai Pulmonary Hospital, Tongji University (No. K18-167). The anonymous nature of the data allowed the requirement for informed consent from the patients to be waived.
Consent for publication
Not applicable.
Competing interests
All authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rui Fan, Hao Qian, Jia-Yan Xu and Jia-Yi Wang contributed equally to this work.
Contributor Information
Wei-Jun Cao, Email: weijuncao@126.com.
Jin-Fu Xu, Email: jfxucn@163.com.
References
- 1.O’Donnell AE. Bronchiectasis—a clinical review. N Engl J Med. 2022;387(6):533–45. [DOI] [PubMed] [Google Scholar]
- 2.Elborn JS, et al. Bronchiectasis and inhaled tobramycin: a literature review. Respir Med. 2022;192: 106728. [DOI] [PubMed] [Google Scholar]
- 3.Flume PA, Chalmers JD, Olivier KN. Advances in bronchiectasis: endotyping, genetics, microbiome, and disease heterogeneity. Lancet (London, England). 2018;392(10150):880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quint JK, et al. Changes in the incidence, prevalence and mortality of bronchiectasis in the UK from 2004 to 2013: a population-based cohort study. Eur Respir J. 2016;47(1):186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monteagudo M, et al. Prevalence and incidence of bronchiectasis in Catalonia, Spain: a population-based study. Respir Med. 2016;121:26–31. [DOI] [PubMed] [Google Scholar]
- 6.Feng J, et al. Increasing prevalence and burden of bronchiectasis in urban Chinese adults, 2013–2017: a nationwide population-based cohort study. Respir Res. 2022;23(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu T, Prescott WG, Zhou X. Advances in Non-Type 2 Asthma in the Severe Cases: from molecular insights to novel treatment strategies. Eur Respir J. 2024;64:2300826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendien SA, et al. Real-world effectiveness of IL-5/5Ra targeted biologics in severe eosinophilic asthma with comorbid bronchiectasis. J Allergy Clin Immunol Pract. 2023;11(9):2724-2731.e2. [DOI] [PubMed] [Google Scholar]
- 9.Polverino E, et al. Bronchiectasis and asthma: data from the European Bronchiectasis Registry (EMBARC). J Allergy Clin Immunol. 2024;153(6):1553–62. [DOI] [PubMed] [Google Scholar]
- 10.Mao B, et al. Asthma and bronchiectasis exacerbation. Eur Respir J. 2016;47(6):1680–6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XX, et al. Bacteria and viruses and clinical outcomes of asthma-bronchiectasis overlap syndrome: a cohort study. Clin Transl Allergy. 2024;14(1): e12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giavina-Bianchi P, et al. United airway disease: current perspectives. J Asthma Allergy. 2016;9:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bousquet J, et al. Allergic rhinitis and its impact on asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen). Allergy. 2008;63(Suppl 86):8–160. [DOI] [PubMed] [Google Scholar]
- 14.Shaaban R, et al. Rhinitis and onset of asthma: a longitudinal population-based study. Lancet. 2008;372(9643):1049–57. [DOI] [PubMed] [Google Scholar]
- 15.Lohia S, Schlosser RJ, Soler ZM. Impact of intranasal corticosteroids on asthma outcomes in allergic rhinitis: a meta-analysis. Allergy. 2013;68(5):569–79. [DOI] [PubMed] [Google Scholar]
- 16.Zhang RL, et al. Motile ciliary disorders of the nasal epithelium in adults with bronchiectasis. Chest. 2023;163(5):1038–50. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura H, et al. Galectin-10 in serum extracellular vesicles reflects asthma pathophysiology. J Allergy Clin Immunol. 2024;153(5):1268–81. [DOI] [PubMed] [Google Scholar]
- 18.Lubner RJ, et al. Particulate matter exposure is associated with increased inflammatory cytokines and eosinophils in chronic rhinosinusitis. Allergy. 2024;79(5):1219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Lans R, et al. Eosinophils are the dominant type2 marker for the current indication of biological treatment in severe uncontrolled chronic rhinosinusitis with nasal polyps. Rhinology. 2024;62(3):383–4. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-García MA, et al. Reliability of blood eosinophil count in steady-state bronchiectasis. Pulmonology. 2024. [DOI] [PubMed]
- 21.Pollock J, Goeminne PC. Eosinophils in bronchiectasis: a U-turn for bronchiectasis management. Chest. 2023;164(3):561–3. [DOI] [PubMed] [Google Scholar]
- 22.Shoemark A, et al. Characterization of eosinophilic bronchiectasis: a European multicohort study. Am J Respir Crit Care Med. 2022;205(8):894–902. [DOI] [PubMed] [Google Scholar]
- 23.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck T, et al. GWAS Central: an expanding resource for finding and visualising genotype and phenotype data from genome-wide association studies. Nucleic Acids Res. 2023;51(D1):D986-d993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurki MI, et al. Author Correction: FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;615(7952):E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dönertaş HM, et al. Common genetic associations between age-related diseases. Nat Aging. 2021;1(4):400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mbatchou J, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53(7):1097–103. [DOI] [PubMed] [Google Scholar]
- 28.Valette K, et al. Prioritization of candidate causal genes for asthma in susceptibility loci derived from UK Biobank. Commun Biol. 2021;4(1):700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590(7845):290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skrivankova VW, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375: n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadimitriou N, et al. Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat Commun. 2020;11(1):597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018; 7. [DOI] [PMC free article] [PubMed]
- 35.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbanck M, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burgess S, et al. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11(1):376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K, et al. Assessment of bidirectional relationships between brain imaging-derived phenotypes and stroke: a Mendelian randomization study. BMC Med. 2023;21(1):271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, et al. Bayesian weighted Mendelian randomization for causal inference based on summary statistics. Bioinformatics. 2020;36(5):1501–8. [DOI] [PubMed] [Google Scholar]
- 41.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64. [DOI] [PubMed] [Google Scholar]
- 42.Kulinskaya E, Dollinger MB, Bjørkestøl K. On the moments of Cochran’s Q statistic under the null hypothesis, with application to the meta-analysis of risk difference. Res Synth Methods. 2020;11(6):920. [DOI] [PubMed] [Google Scholar]
- 43.Jardim LL, et al. Prediction of inhibitor development in previously untreated and minimally treated children with severe and moderately-severe hemophilia A using a machine-learning network. J Thromb Haemost. 2024. [DOI] [PubMed]
- 44.Aliberti S, et al. Criteria and definitions for the radiological and clinical diagnosis of bronchiectasis in adults for use in clinical trials: international consensus recommendations. Lancet Respir Med. 2022;10(3):298–306. [DOI] [PubMed] [Google Scholar]
- 45.Global Initiative for Asthma (GINA). Global strategy for asthma management and prevention. 2023.
- 46.Tiotiu A, et al. Does asthma-bronchiectasis overlap syndrome (ABOS) really exist? J Asthma. 2023;60(11):1935–41. [DOI] [PubMed] [Google Scholar]
- 47.Säynäjäkangas O, et al. Links between hospital diagnoses of bronchiectasis and asthma. Allergy. 1997;52(11):1120–2. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz BS, et al. Strong and consistent associations of precedent chronic rhinosinusitis with risk of non-cystic fibrosis bronchiectasis. J Allergy Clin Immunol. 2022;150(3):701-708.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirsch AG, et al. Risk of new-onset and prevalent disease in chronic rhinosinusitis: a prospective cohort study. Int Forum Allergy Rhinol. 2023;13(9):1715–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shteinberg M, et al. Upper airway involvement in bronchiectasis is marked by early onset and allergic features. ERJ Open Res. 2018; 4(1). [DOI] [PMC free article] [PubMed]
- 51.Guan WJ, et al. Significance and potential role of eosinophils in non-cystic fibrosis bronchiectasis. J Allergy Clin Immunol Pract. 2023;11(4):1089–99. [DOI] [PubMed] [Google Scholar]
- 52.Nomura N, et al. Nationwide survey of refractory asthma with bronchiectasis by inflammatory subtypes. Respir Res. 2022;23(1):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray A, Kolls JK. Neutrophilic inflammation in asthma and association with disease severity. Trends Immunol. 2017;38(12):942–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guilemany JM, et al. United airways again: high prevalence of rhinosinusitis and nasal polyps in bronchiectasis. Allergy. 2009;64(5):790–7. [DOI] [PubMed] [Google Scholar]
- 55.Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol. 2017;12:331–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soyka MB, et al. Defective epithelial barrier in chronic rhinosinusitis: the regulation of tight junctions by IFN-γ and IL-4. J Allergy Clin Immunol. 2012;130(5):1087-1096.e10. [DOI] [PubMed] [Google Scholar]
- 57.Castellani S, et al. NHERF1 and CFTR restore tight junction organisation and function in cystic fibrosis airway epithelial cells: role of ezrin and the RhoA/ROCK pathway. Lab Invest. 2012;92(11):1527–40. [DOI] [PubMed] [Google Scholar]
- 58.Fan C, et al. Alterations in oral-nasal-pharyngeal microbiota and salivary proteins in mouth-breathing children. Front Microbiol. 2020;11: 575550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SY, et al. Influence of chronic sinusitis and nasal polyp on the lower airway of subjects without lower airway diseases. Allergy Asthma Immunol Res. 2014;6(4):310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braunstahl GJ, et al. Nasal allergen provocation induces adhesion molecule expression and tissue eosinophilia in upper and lower airways. J Allergy Clin Immunol. 2001;107(3):469–76. [DOI] [PubMed] [Google Scholar]
- 61.Grünberg K, et al. Effect of experimental rhinovirus 16 colds on airway hyperresponsiveness to histamine and interleukin-8 in nasal lavage in asthmatic subjects in vivo. Clin Exp Allergy. 1997;27(1):36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gern JE, et al. Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am J Respir Crit Care Med. 1997;155(3):1159–61. [DOI] [PubMed] [Google Scholar]
- 63.Ramakrishnan VR, et al. Upper and lower airways associations in patients with chronic rhinosinusitis and bronchiectasis. Int Forum Allergy Rhinol. 2013;3(11):921–7. [DOI] [PubMed] [Google Scholar]
- 64.Gao YH, et al. The impact of depression and anxiety on the risk of exacerbation in adults with bronchiectasis: a prospective cohort study. Eur Respir J. 2023;61:2201695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Scatter plot (A), forest plot (B), leave-one-out analyses (C) and funnel plot (D) illustrating the causal effect of asthma (ebi-a-GCST90014325) on bronchiectasis (finngen_R10_J10_BRONCHIECTASIS).
Additional file 2: Figure S2. Putative bidirectional causality (A), Scatter plot (B), forest plot (C), leave-one-out analyses (D) and funnel plot (E) illustrating the causal effect of asthma (finngen_R10_J10_ASTHMA_MAIN_EXMORE) on bronchiectasis (ukb-saige-496.3).
Additional file 3: Figure S3. Scatter plot (A), forest plot (B), leave-one-out analyses (C) and funnel plot (D) illustrating the causal effect of bronchiectasis (finngen_R10_J10_BRONCHIECTASIS) on asthma (ebi-a-GCST90014325).
Additional file 4: Figure S4. Putative bidirectional causality (A), Scatter plot (B), forest plot (C), leave-one-out analyses (D) and funnel plot (E) illustrating the causal effect of bronchiectasis (ukb-saige-496.3) on asthma (finngen_R10_J10_ASTHMA_MAIN_EXMORE).
Additional file 5: Figure S5. (A) The mediating role of neutrophils in the causal effect of asthma on bronchiectasis; Statistical chart of neutrophil ratio (B) and counts (C) in BE + A group and BE group.
Additional file 6: Table S1. IVs in asthma (GWAS ID: ebi-a-GCST90014325). Table S2. IVs in bronchiectasis (GWAS ID: finngen_R10_J10_BRONCHIECTASIS). Table S3. IVs in asthma (GWAS ID: finngen.R10_J10_ASTHMA_MAIN_EXMORE). Table S4. IVs in bronchiectasis (GWAS ID: ukb saige-496.3). Table S5. Measures of heterogeneity and pleiotropy test.
Data Availability Statement
No datasets were generated or analysed during the current study.