Abstract
Introduction
The delineation of organs-at-risk and lymph node areas is a crucial step in radiotherapy, but it is time-consuming and associated with substantial user-dependent variability in contouring. Artificial intelligence (AI) appears to be the solution to facilitate and standardize this work. The objective of this study is to compare eight available AI software programs in terms of technical aspects and accuracy for contouring organs-at-risk and lymph node areas with current international contouring recommendations.
Material and methods
From January–July 2023, we performed a blinded study of the contour scoring of the organs-at-risk and lymph node areas by eight self-contouring AI programs by 20 radiation oncologists. It was a single-center study conducted in radiation department at the Lorraine Cancer Institute. A qualitative analysis of technical characteristics of the different AI programs was also performed. Three adults (two women and one man) and three children (one girl and two boys) provided six whole-body anonymized CT scans, along with two other adult brain MRI scans. Using a scoring scale from 1 to 3 (best score), radiation oncologists blindly assessed the quality of contouring of organs-at-risk and lymph node areas of all scans and MRI data by the eight AI programs. We have chosen to define the threshold of an average score equal to or greater than 2 to characterize a high-performing AI software, meaning an AI with minimal to moderate corrections but usable in clinical routine.
Results
For adults CT scans: There were two AI programs for which the overall average quality score (that is, all areas tested for OARs and lymph nodes) was higher than 2.0: Limbus (overall average score = 2.03 (0.16)) and MVision (overall average score = 2.13 (0.19)). If we only consider OARs for adults, only Limbus, Therapanacea, MVision and Radformation have an average score above 2. For children CT scan, MVision was the only program to have a average score higher than 2 with overall average score = 2.07 (0.19). If we only consider OARs for children, only Limbus and MVision have an average score above 2. For brain MRIs: TheraPanacea was the only program with an average score over 2, for both brain delineation (2.75 (0.35)) and OARs (2.09 (0.19)). The comparative analysis of the technical aspects highlights the similarities and differences between the software. There is no difference in between senior radiation oncologist and residents for OARs contouring.
Conclusion
For adult CT-scan, two AI programs on the market, MVision and Limbus, delineate most OARs and lymph nodes areas that are useful in clinical routine. For children CT-scan, only one IA, MVision, program is efficient. For adult brain MRI, Therapancea,only one AI program is efficient.
Trial registration: CNIL-MR0004 Number HDH434.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13014-024-02554-y.
Introduction
The delineation of organs-at-risk (OARs) and lymph node areas is an important aspect of radiotherapy treatment. This step is crucial to define the healthy tissue that will be spared. Although there are many contouring guidelines for OARs and lymph node areas that we use in our department to standardize practices [1–6], there is still great intra- and inter-user variability [7–9]. Some radiotherapy teams have partially or totally delegated the preparation to the contouring of the OARs to radiotherapy therapists (RTTs), dosimetrists or residents with a final medical validation made by the senior radiation oncologist, which is time-consuming but mandatory in clinical practice. Single-atlas, multiple-atlas and model-based solutions were developed to simplify, speed up and improve manual contouring [10]. However, there are still limitations, and these technological advances were also limited by the availability of segmented data and computer power [11]. Artificial intelligence (AI) encompasses a set of programs capable of simulating human intelligence, and machine learning, deep learning and convolutional neural networks (CNNs) are used for automated contouring [12, 13]. Artificial intelligence (AI) refers to computer models designed to solve complex problems that lack a clear mathematical solution or a defined set of rules, much like how the human brain tackles real-world challenges. Machine learning (ML), a subset of AI, focuses on developing models that can recognize patterns in high-dimensional data and make predictions based on new information. The goal of ML is to enable computers to learn how to achieve specific objectives without being explicitly programmed with the steps to reach those objectives (Meyer et al.). Within ML, deep learning (DL) models are based on neural networks—multi-layered, interconnected networks capable of adapting their pathways to integrate new data and identify patterns. A specialized type of DL model is the convolutional neural network (CNN), which is specifically designed for image recognition and computer vision tasks.
Many studies have recently been published on the emergence of AI in the field of auto-contouring in radiotherapy (the first published by Ibragimov in 2017, using CNNs for delineation of head and neck OARs [14]). These studies have shown the efficacy and efficiency of auto-contouring compared to a gold standard (manual delineation by an expert radiation oncologist) or compared with techniques based on atlases. However, they are often limited to a single locations, such as head and neck cancer [15–17], lung cancer [18–20], prostate cancer [21–23], rectal cancer [24, 25] or breast cancer [26] and there are few studies comparing multiple anatomical regions among themselves [27, 28]. Moreover, there are currently only a few studies that compare so many self-contouring AI systems with each other (for example, Doolan et al. compared 5 solutions [27] and Heilemann et al. compared 3 solutions [28]).
Today there are several auto-contouring programs available on the European market with strong competition and regular developmental updates. Therefore, the goal of this study is to compare eight AI programs available in terms of technical aspects and accuracy for contouring OARs and lymph node areas compared.
Methods
Study design
First, we compared in a table the technical characteristics for the eight different AI auto-contouring programs for OARs and lymph nodes. Second, 20 radiation oncologists (12 seniors and 8 juniors) performed a single-center blinded analysis of the contour scoring of the OARs and lymph node areas carried out by AI. All evaluators assessed the contours independently and had not previously outlined them. This analysis was carried out by the radiation oncology team at the Lorraine Cancer Institute from January–July 2023. CT scans of selected patients were sent to radiotherapy centers in France that had the different AI programs or to the software manufacturer. They performed the auto-contouring with their software and the completed scans with delineated OARs and lymph node areas then we imported all the contours into our contouring software RayStation.
Patient data
We chose three patients scheduled for total body irradiation with a whole-body anonymized scan (two women aged 28 and 36 with acute lymphoid leukemia (ALL) and aplastic anemia and a 67-year-old man with ALL) and two other patients scheduled for brain irradiation with anonymized centering MRI. To test the software on scans of children, we also chose three anonymized scans of whole-body irradiation of children (a 15-year-old girl with ALL and two boys aged 7 and 15 with medulloblastoma and ALL). Simulation contrast CT data were acquired on a Brilliance CT Big Bore (Philips Healthcare, Best, the Netherlands) system set on helical scan mode without contrast enhancement. CT images were reconstructed using a matrix size of 512 × 512 and thickness of 1 mm for stereotactic irradiation or 2 mm for other irradiations. All patients were supine with usual immobilization system. MRI scans of cerebral localizations were acquired to compare MRI self-contouring for software with this functionality (Mvison, Limbus and TheraPanacea).
Intervention: automatic contouring/contour content
We analyzed all the OARs and all lymph node areas used in clinical practice, divided into four subgroups: 1. Head and neck; 2. thorax and breast; 3. abdomen and pelvis; 4. central nervous system on MRI. The evaluated software were: Raystation version 12A by Reasearch laboratories, MVision version 1.2.3, Limbus version 1.7.0, TheraPanacea version 1.11.2, MIRADA version 1.8.6.44363, Radformation version 2.0.19, Mim version 7.2.7 and Varian/Siemens version A50 (pre-release version).
Measures
The quality of OAR and lymph node area contouring of all scans and MRI data of the eight AI programs were blinded, checked and scored by 20 radiation oncologists in various locations in accordance with international contouring recommendations [1–6]. 5 seniors and 3 juniors evaluated the head and neck region, 6 seniors and 4 juniors evaluated the thoraco-abdominopelvic region, and 1 senior and 1 junior conducted the comprehensive analysis. No contour corrections were made. The scoring criteria were as follows: 3 points (no correction, major time saving); 2 points (moderate corrections, moderate correction on 1 or a few cuts taking a few seconds, moderate time saving); 1 point (major corrections; no time saving, it’s easier for the radiation oncologist to completely manually redo the OAR) or NR (not perform by IA software). A figure has been added in supplementary data to illustrate the scoring criteria (Appendix 1).
Statistics
The average score per observer was described as mean for each AI. Then overall average score was described by calculating mean of all observers score and standard deviation. We have chosen to define the threshold of an average score equal to or greater than 2 to characterize a high-performing AI software, meaning an AI with minimal to moderate corrections but usable in clinical routine. Scores equal to or greater than 2 were compared using a one-tailed paired-sample Student's t-test. All analysis were performed using Microsoft Excel 2016 (Microsoft corporation, Redmond, Washington, USA).
Human ethics approval and consent to participation
This study was approved by ethics and conducted in accordance with the ethical standards of the Declaration of Helsinki (as revised in 2013). This study was approved by Ethics committee named the French National Commission of Informatics and Liberty (CNIL) (CNIL-MR0004 Number HDH434). The present study has been approved by the French Health Data Institute (Health DataHub) as the number HDH301. All methods were carried out in accordance with relevant guidelines and regulations. All participants have signed informed consent to the use of their data for research purposes.
Results
Technical considerations
Table 1 summarizes the technical aspects of the different AI programs currently available.
Table 1.
Comparative table of the different technical aspects between the 8 artificial intelligence systems
| Limbus | Thera-panacea | Mvision | Mirada | Rad-formation | Raysearch | Varian siemens | MIM | |
|---|---|---|---|---|---|---|---|---|
| Functionality | ||||||||
| Algorithm type | Deep learning | Deep learning | Deep learning | Deep learning | Deep learning | Deep learning | Deep learning | Deep learning |
| Version | 1.7.0 | 1.11.2 | 1.2.3 | 1.8.6.44363 | 2.0.19 | 12A | A50a | 7.2.7 |
| Automatic contouring (via Dicom tag) | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes |
| Based on international contouring guidelines | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Contouring time(min) | 1–3 | 1–2 (up to 10) | 1–10 | 15 | 0.30 | 1–2 | 1–2 | < 1–10 |
| Settings | ||||||||
| Changes the name, color and order of structures | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Contouring 1 cut/2 | Yes | No | Yes | Yes | Next version | No | No | Yes |
| Segmentation of prostheses | Next version | No | Yes | No | No | No | No | No |
| Generation of templates with empty structures | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes |
| Generation of structures with margins/Boolean operation | Next version | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Software language | EN/FR/DE/Eb | EN/FRb | EN/FRb | ENb | ENb | ENb | 7 languages including EN/FR/DEb | EN/FRb |
| Hardware | ||||||||
| Cloud version | No | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Data anonymization | No | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Locally installed | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Hardware requirements |
Works on a local PCc Windows 10 No GPUd card needed Can be installed on a computer virtual machine |
Server |
Intel/AMD × 86 processor 2 GBe RAMf Hard disk 32 GBe HDD/SSDg |
Server (with remote access, with citrix access, with thin or thick clients) |
CPU⁸-based Google Cloud server |
Server |
Multi-software physical server or virtualized server Windows environment |
Server |
| Technical needs | ||||||||
| Can be used on several sites | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Update | 4 per year | 2–3 per year | 2 per year | No | 2–3 per year | 1 per year | 1–2 per year | 2–3 per year |
|
Installation Formation Technical support |
Online or onsite training 24h technical support |
User training Response in 6 working hrs Online technical support |
User training Hotline in France |
Onsite or video training Technical support via email and hotline |
Video training Hotline European technical support by email |
-On-site training - On-site installation with local ITa⁰ team |
Support by IT11 technical team Large hotline Online training |
On-site training in French Technical support available and responsive |
aVersion A50 teamplay Organs RT (pre-release version); bEN/FR/DE/E: English/French/German/Español; cPersonal Computer; dGraphics Processing Unit; eGigabytes; fRandom Access Memory; gHard Disk Drive/Solid State Srive; hCentral Processing Unit; iInformation Technology
All the AI programs used deep learning, including six with automatic contouring via Tag Dicom. All the AI programs are based on theoretical guidelines to train their model, and the automatic contouring time is less than 15 min for all the programs, depending on the number of OARs and lymph node areas contoured. All the AI programs can create templates with empty structures except TheraPanacea. Most AI programs include a “double version”; that is, a version with Cloud and data anonymization and a version with local installation on a server or a local PC etc. Six AI programs are accessible by Cloud, seven have local accessibility, and five have both. Limbus and Raysearch, for example, do not have Cloud versions, whereas Radformation is not installed locally. According to the contracts, all software can be used on several sites with, in general, several updates per year, from a minimum of one update per year and up to 4 per year for Limbus; the exception Mirada, which does not receive updates because this software is no longer commercially available. A list of all the OARs and lymph node areas produced by each software can be found in Appendices 2–6.
OARs and lymph node contouring by AI
OARs and lymph node contouring by AI in adults
There were two AI programs for which the overall average quality score (that is, all areas tested for OARs and lymph nodes) was higher than 2.0: Limbus (overall average score = 2.03 (0.16)) and MVision (overall average score = 2.13 (0.19)). If we only consider OARs for adults, only Limbus, Therapanacea, MVision and Radformation have an average score above 2. MVision had the best overall average quality score for OARs and lymph node areas, but Limbus had a better score for the delineation of OARs alone (average score = 2.42 (0.20)) (p = 0.01). For head and neck localization, MVision had a better overall average score of 2.21(0.20), than Limbus (2.03(0.14) (p < 0.001); particularly in the delineation of lymph node areas with an average score of 2.15 (0.33), versus 1.80 (0.24) for Limbus (p < 0.001). For thorax and breast localization, Limbus scored higher than MVision for the delineation of OARs with average score of 2.42 (0.30) (p < 0.001), but MVision has a higher score for lymph node areas with average score of 1.65 (0.40) (p < 0.001) For the abdomen and pelvis localization, Limbus scored higher than MVision again for the delineation of OARs (average score = 1.99 (0.26)) (p < 0.001) and not statistically different from MVision for lymph node areas (average score = 1.66 (0.45)) versus 1.70 (0.36) (p = 0.4) (Table 2).
Table 2.
Table of average scoring for Organs-at-Risk (OARs) and lymph node areas assessments across 8 AI Systems by each location (head and neck, thorax and breast, abdomen and pelvis) in adult CT Scans
| Adults | Limbusa | Therapanacea | Mvision | Mirada | Radformation | Raysearch | Siemens | MIM | T test |
|---|---|---|---|---|---|---|---|---|---|
| OARs | |||||||||
| Head and necka (n = 750) | 2.25 (0.13) | 1.91 (0.10) | 2.28 (0.10) | 1.62 (0.1) | 1.94 (0.14) | 1.86 (0.11) | 1.35 (0.18) | 1.72 (0.15) | p < 0.001 |
| Thorax and breasta (n = 456) | 2.42 (0.30) | 2.01 (0.23) | 2.37 (0.24) | 1.57 (0.14) | 2.06 (0.20) | 1.59 (0.08) | 1.51 (0.36) | 1.41 (0.09) | p < 0.001 |
| Abdomen and pelvisa (n = 492) | 2.33 (0.20) | 2.04 (0.17) | 2.07 (0.14) | 1.72 (0.16) | 1.92 (0.18) | 1.64 (0.12) | 1.87 (0.37) | 1.26 (0.04) | p < 0.001 |
| TOTALSa (n = 1698) | 2.42 (0.20) | 2.00 (0.15) | 2.28 (0.12) | 1.71 (0.17) | 2.02 (0.17) | 1.80 (0.12) | 1.54 (0.34) | 1.56 (0.16) | p < 0.01 |
| LNs | |||||||||
| Head and neck (n = 480) | 1.80 (0.24) | 1.69 (0.19) | 2.15 (0.33) | 1 (0) NRb | 1.69 (0.15) | 1 (0) NRb | 1.45 (0.29) | 1 (0) NRb | p < 0.01 |
| Thorax and breast (n = 216) | 1.25 (0.24) | 1.40 (0.32) | 1.65 (0.40) | 1.03 (0.05) | 1.36 (0.33) | 1.54 (0.33) | 1.31 (0.39) | 1 (0) NRb | p < 0.001 |
| Abdomen and pelvis (n = 180) | 1.66 (0.45) | 1.63 (0.32) | 1.70 (0.36) | 1 (0) NRb | 1.57 (0.31) | 1 (0) NRb | 1.47 (0.54) | 1 (0) NRb | p < 0.045 |
| Totals (n = 876) | 1.59 (0.29) | 1.58 (0.24) | 1.88 (0.38) | 1.01 (0.02) | 1.57 (0.27) | 1.14 (0.17) | 1.43 (0.37) | 1 (0) NRb | p < 0.001 |
| Totals = OARs + LNs by anatomical locations | |||||||||
| Head and neck (n = 1230) | 2.03 (0.14) | 1.80 (0.13) | 2.21 (0.20) | 1.31 (0.05) | 1.81 (0.10) | 1.43 (0.05) | 1.40 (0.22) | 1.36 (0.07) | p < 0.001 |
| Thorax and breast (n = 672) | 1.90 (0.20) | 1.70 (0.26) | 2.01 (0.27) | 1.30 (0.09) | 1.71 (0.25) | 1.56 (0.20) | 1.41 (0.36) | 1.20 (0.02) | p < 0.001 |
| Abdomen and pelvis (n = 672) | 1.99 (0.26) | 1.84 (0.23) | 1.88 (0.22) | 1.36 (0.08) | 1.74 (0.22) | 1.32 (0.06) | 1.67 (0.44) | 1.13 (0.02) | p < 0.013 |
| Totals = OARs + LNs | |||||||||
| Overall (n = 2574) | 2.03 (0.16) | 1.84 (0.16) | 2.13 (0.19) | 1.39 (0.12) | 1.82 (0.14) | 1.50 (0.10) | 1.48 (0.32) | 1.29 (0.08) | p < 0.02 |
Data are presented as: average (± SD); a Organs at risk; b Not Realized
OARs and lymph node contouring by AI in children
For children, MVision was the only program to have a average score higher than 2 with overall average score = 2.07 (0.19). If we only consider OARs for children, only Limbus and MVision have an average score above 2. Limbus overall average score was slightly below 2 (overall average score = 1.93 (0.13), and statistically lower than MVision score (p = 0.001), but Limbus scored better for OARs delineation alone, with an average score of 2.28(0.17) (p = 0.03) (Table 3).
Table 3.
Table of average scoring for Organs-at-Risk (OARs) and lymph node areas assessments across 8 AI Systems by each location (head and neck, thorax and breast, abdomen and pelvis) in children CT Scans
| Children | Limbus | Therapanacea | Mvision | Mirada | Radformation | Raysearch | Siemens | MIM | T test |
|---|---|---|---|---|---|---|---|---|---|
| OARs | |||||||||
| Head and necka (n = 750) | 2.24 (0.19) | 1.93 (0.17) | 2.32 (0.12) | 1.62 (0.18) | 1.89 (0.24) | 1.87 (0.12) | 1.25 (0.16) | 1.72 (0.14) | p = 0.02 |
| Thorax and breasta (n = 444) | 2.35 (0.18) | 2.0 (0.20) | 2.16 (0.12) | 1.43 (0.06) | 1.74 (0.19) | 1.48 (0.08) | 1.26 (0.20) | 1.36 (0.09) | P = 0.02 |
| Abdomen and pelvisa (n = 516) | 1.92 (0.17) | 1.88 (0.16) | 1.62 (0.15) | 1.35 (0.14) | 1.61 (0.13) | 1.44 (0.10) | 1.34 (0.20) | 1.28 (0.05) | NA |
| TOTALSa (n = 1710) | 2.28 (0.17) | 1.98 (0.15) | 2.15 (0.20) | 1.57 (0.17) | 1.83 (0.20) | 1.72 (0.18) | 1.29 (0.19) | 1.54 (0.17) | P = 0.03 |
| LNs | |||||||||
| Head and neck (n = 480) | 1.71 (0.24) | 1.62 (0.17) | 2.10 (0.18) | 1 (0) NRb | 1.40 (0.06) | 1 (0) NRb | 1.26 (0.20) | 1 (0) NRb | NA |
| Thorax and breast (n = 216) | 1.07 (0.10) | 1.20 (0.19) | 1.95 (0.32) | 1.07 (0.13) | 1.12 (0.15) | 1.61 (0.28) | 1.18 (0.31) | 1 (0) NRb | NA |
| Abdomen and pelvis (n = 180) | 1.69 (0.36) | 1.61 (0.42) | 1.61 (0.32) | 1 (0) NRb | 1.18 (0.14) | 1 (0) NRb | 1.18 (0.22) | 1 (0) NRb | NA |
| Totals (n = 876) | 1.51 (0.26) | 1.50 (0.25) | 1.92 (0.27) | 1.02 (0.05) | 1.27 (0.17) | 1.16 (0.18) | 1.22 (0.24) | 1 (0) NRb | P < 0.001 |
| Totals = OARs + LNs by anatomical locations | |||||||||
| Head and neck (n = 1230) | 1.97 (0.15) | 1.78 (0.15) | 2.21 (0.12) | 1.31 (0.09) | 1.64 (0.13) | 1.44 (0.06) | 1.26 (0.18) | 1.36 (0.07) | NA |
| Thorax and breast (n = 660) | 1.76 (0.11) | 1.60 (0.18) | 2.05 (0.21) | 1.25 (0.09) | 1.43 (0.16) | 1.55 (0.18) | 1.22 (0.23) | 1.18 (0.04) | NA |
| Abdomen and pelvis (n = 696) | 1.80 (0.25) | 1.75 (0.25) | 1.61 (0.22) | 1.17 (0.07) | 1.39 (0.12) | 1.22 (0.5) | 1.26 (0.20) | 1.14 (0.02) | NA |
| Totals = OARs + LNs | |||||||||
| Overall (n = 2586) | 1.93 (0.13) | 1.78 (0.15) | 2.07 (0.19) | 1.32 (0.10) | 1.58 (0.14) | 1.47 (0.10) | 1.25 (0.19) | 1.28 (0.08) | NA |
Data are presented as: average (± SD); a Organs at risk; b Not realized. NA Not applicable
OARs contouring of brain MRIs
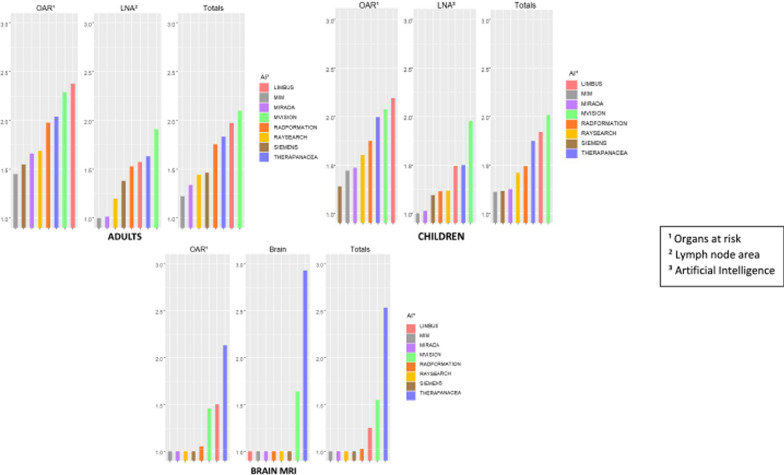
TheraPanacea was the only program with an average score over 2, for both brain delineation (2.75 (0.35)) and OARs (2.09 (0.19)). (Table 4). Figure 1 summarizes the results of this section. Note that we did not compare the contouring quality differences between CT and MRI when an OAR could be generated interchangeably from either a CT scan or an MRI.
Table 4.
Table of average scoring for Organs-at-Risk (OARs) delineations in cerebral MRI scans across the 8 AI systems
| MRI | Limbus | Therapanacea | Mvision | Mirada | Radformation | Raysearch | Siemens | MIM |
|---|---|---|---|---|---|---|---|---|
| Brain | 1.00 (0) | 2.75 (0.35) | 1.60 (0.61) | NRb | 1.00 (0) | NRb | NRb | NRb |
| OARsa (n = 300) | 1.54 (0.12) | 2.09 (0.19) | 1.45 (0.13) | NRb | 1.00 (0) | NRb | NRb | NRb |
Data are presented as: average (± SD); a Organs at risk; b Not Realized
Fig. 1.
Summary of quality scoring of AI contouring programs for adult CT scan, children CT scan and adult brain MRI. Footnotes: 1 Organs at risk; 2 Lymph node area; 3 Artificial Intelligence
Comparison between senior radiation oncologist and residents
Overall (OARs and LNs) for senior physicians, the mean is 1.63 and 1.66 for residents (p = 0.03). If we consider only OARs, the mean for senior physicians is 1.93 versus 1.90 for residents (p = 0.12). If we consider only LNs, the mean is 1.37 for senior physicians and 1.43 for residents (p = 0.004).
Discussion
Four AI programs, MVision, Limbus, TheraPanacea and Radformation, successfully delineated most OARs and lymph nodes areas useful in clinical routine for the head and neck, thorax and breast, and abdomen and pelvis in adults and children, with a higher than average score. All AI programs required manual corrections from the radiation oncologist.
Wong et al. [29] evaluated Limbus using 29 experts and found minor corrections were required for most head and neck OARs, with mean satisfaction score of 4.8 for lymph node areas (5 was the highest overall satisfaction score). This agrees with the results of our study whereby Limbus has a higher mean score for both OARs and lymph node areas than the overall averages. Grégoire et al. [30] studied TheraPanacea and scored 15 head and neck OARs by of 5 experts, finding 98% of the auto-contouring classified as relevant; we found TheraPanacea to be an accurate AI program for contouring OARs in the head and neck. In the thorax and breast, Almberg et al. [31] evaluated deep learning and found either no correction or minor corrections were required for 14% and 71% of clinical target volumes, respectively, and 72% and 26% of OARs, respectively. Major corrections accounted for only 15% of clinical target volumes and 2% of OARs. In our study, for the OARs, Limbus, MVision, TheraPanacea and Radformation had higher mean scores than the overall average, suggesting these may be a good clinical choice. For the lymph node areas, in our study MVision, TheraPanacea, Radformation and Raysearch scored better than average. In the abdomen and pelvis localization, Azria et al. [32] showed that 79% of OARs and target volumes (prostate and seminal vesicles) contoured by TheraPanacea were acceptable versus 69% of contours produced by experts. In our study, TheraPanacea also scored highly in terms of contouring quality in OARs.
None of the AI programs studied here has a specific option for auto-contouring OARs and lymph node areas in children, but this is a greater challenge, in particular because of child growth. Bondiau et al. [33] found that 90.5% of the contours produced on brain MRI scans by TheraPanacea of 39 children aged 0–15 years are clinically acceptable, although they also note that in younger children (0–5 years) there are fewer acceptable contours than in older children. In our study, we did not compare brain MRIs of children, but we also found a tendency for delineation in body scans of children to be worse than in adults, with more contouring errors, as demonstrated by the lower scores in children versus adults for all delineations, especially in the abdominal and pelvic regions.
As far as cerebral MRI is concerned, in both delineation of the brain and for all the OARs, TheraPanacea scored highly. However, at the time of the study, the only programs with a specific brain MRI option were Limbus, MVision, Radformation and TheraPanacea; TheraPanacea has the largest number of OARs delineation options, as shown in Appendix 6. We have carried literature reviews of the localizations of interest for head and neck (Appendix 7), abdomen and pelvis (Appendix 8) and thorax, breast and children (Appendix 9).
This robustness of the study was enhanced by the direct comparison of most AI software available on the market, by a substantial number of blinded evaluators, making it possible to remain fair and for the data to be reproducible in clinical practice. Thus, we were able to compare a large number of OARs and lymph node areas (80 OARs and lymph node areas) from full-body CT and MRI although the sample size may appear limited, it corresponds to a large amount of data, which ensures statistical significance. We not only carried out a quantitative analysis, which is interesting in terms of clinical applicability, but also an analysis of the technical characteristics of each software in terms of implementation in the department [11, 34].
Within the limits of this study, we did not measure time-saving with AI contouring compared with manual contouring as we wanted to emphasize the applicability and practical nature of this study. Nevertheless, all the AI programs are fast, and the generation time of OARs no longer seems to be an issue for its use in clinical routine [34]. AI contouring programs are time-saving for not only radiation oncologists [35] but also for RTTs whose time can be redeployed on other activities. The workflow can be improved with automatic generation of OARs as soon as the CT scan is produced. In addition, there are other clinically important measurements that we have not determined here, such as a dosimetric study with contour deviations [36–38]. Some, like Gooding et al. [39] or Sherer et al. [40], emphasize the significance of quantitative, geometrical, dosimetric parameters, or time-related data to evaluate an AI system. However, in our study, we opted to conduct only a qualitative assessment due to the vast amount of data we have and for practical implementation purposes within the department. We used a 3-point scale because it seemed sufficiently discriminating, while remaining simple, pragmatic and easy to appropriate for all evaluators. It is based on the medical time savings for the contouring of OARs. It is necessarily a little subjective. To date, there is no validated scale to evaluate the contouring of OARs by AI software. We are not the first to publish this type of study with a 3-point scale, even if other authors have indeed since proposed 4- or 5-point scales. Unfortunately, we were unable to test self-contouring on pelvic MRI for technical reasons (data sending incompatibility). Comparing senior radiation oncologists and residents, There is a statistically significant difference for LN contouring but it remains very small in absolute value. We do not consider these differences to be clinically relevant. On the other hand, there was no statistically significant difference for OAR contouring. The residents who participated in this study were advanced residents in their training curriculum for whom we already considered in practice that their level of skills was sufficient in clinical routine for the contouring of OARs and LNs. It is for this reason that they were able to participate as evaluators in this study.
Owing to the fast evolution of AI, further studies will soon be required for the information to remain up to date; we also note that MIRADA software has stopped the development and marketing of its AI in auto-contouring. There is also the question of the validation of these tools in practice (especially when new versions are released) and the role of scientific societies to propose validation sets (Question of the Gold Standard for contouring).
We note that the AI can be affected by certain clinical situations: for example, in our study almost all the programs (except Raysearch) were disturbed by a bronchial syndrome for the delineation of the lungs. We also saw this for anatomical atypia such as kidney cysts or catheters in vessels. We notice that in children, some OARs such as sexual organs or femoral heads are not well performed by the AI, which is probably owing to product development being based on adults, and improvements are still to be made. This could also be explained by an insufficient number of stress tests on the algorithm, although we did not address it in this study. It has already been demonstrated that a large number of training models leads to better contours [41]. Kanwar et al. [42], Kumar and al. [43] or Bibault et al. [44] also highlight the importance of diversity in datasets, particularly the inclusion of pediatric data in training segmentation models to achieve robust systems. Delineation of some OARs are not available in these programs, even though they are essential in clinical practice such as the pulmonary artery, the constrictor muscle of the pharynx or the duodenum. And conversely, some available OARs are more a matter of research at this stage, for example spleen, pancreas and arytenoids. However, the automatic generation of OARs not performed in clinical routine will make it possible to optimize dosimetry on more OARs with a clinical benefit for the patient. It will also be possible to document the doses delivered to these OARs in a prospective manner for the purpose of research.
With the continuous improvement of AI software through increased use creating better data, there is potential for various future applications. One such possibility is the development of automated contouring of volumes in brachytherapy with applicators, optimizing volumes in dosimetry, volumes on MRI or PET scanners, and potentially even targeting volumes with tumors in place. This opens doors for the progress of adaptive radiotherapy and online replanning. By utilizing its pattern-recognition capabilities, AI can identify details that may be imperceptible or unclear to the human eye, thereby surpassing human capabilities. Technical progress has often created mistrust in society, including the fear of the replacement of man by machines [45], but today it is still necessary to make a manual correction to automatic contouring. The emergence of AI therefore raises the question of its use in training. Indeed, doctors could use AI to the extent they lose technical skills. For the training of residents in contouring of OARs and lymph node areas, over-reliance on AI could lead to lack development of manual contouring skills, and an inability to correct automated contouring.. In addition, some AI solutions, such as MVision, are developing applications for contouring training for residents to precisely compensate for lack of technical skill in manual contouring.
Radiation oncologists will have to refine their technique for verifying AI automated contouring, which involves a different intellectual approach from the delineation of OARs by the radiation oncologists themselves.
Conclusion
Our qualitative analysis of the AI softwares is not enough for implementing any automatic segmentation method in routine practice. Geometric parameters (DSC…) were not measured. AI contouring programs are advanced enough today to be implemented in clinical routine. We found four AI programs to be particularly efficient, although it is still necessary to carry out manual corrections in all cases and the radiation oncologist's skill remains as necessary and relevant as ever. For a future study, it might be worthwhile to evaluate the dosimetric impact of the OARs defined by artificial intelligence versus those corrected manually, to quantify the clinical significance of the correction.
Supplementary Information
Additional file 1. Summary of quality scoring of AI contouring programs for adult CT scan, children CT scan and adult brain MRIFootnotes: ¹ Organs at risk; ² Lymph node area; ³ Artificial Intelligence.
Additional file 2. Comparative table of Organs-at-Risk (OARs) delineations performed in head and neck as well as cerebral regions using CT scans by the 8 AI systemsFootnotes: Y = available and N = unavailable.
Additional file 3. Comparative table of Organs-at-Risk (OARs) delineations performed in thoracic and breast regions using CT scans by the 8 AI systemsFootnotes: Y = available and N = unavailable.
Additional file 4. Comparative table of Organs-at-Risk (OARs) delineations performed in abdominal and pelvic regions using CT scans by the 8 AI systemsFootnotes: Y = available and N = unavailable; 1High Dose Rate.
Additional file 5. Comparative table of lymph node areas delineated in head and neck, breast, abdominal and pelvic, and gynecological region using CT scans by the 8 AI systems29Footnotes: Y = available and N = unavailable; 1Radiation Therapy Oncology Group; 2 Clinical Target Volume Tumor.
Additional file 6. Comparative table of Organs-at-Risk (OARs) delineations achieved using MRI for cerebral and pelvic regions by the 8 AI systemsFootnotes: Y = available and N = unavailable.
Additional file 7. Literature Review on AI-Based Auto-contouring of Organs at Risk (OARs) in Head and Neck RegionFootnotes: 1Organs at risk; 2Dice score coefficient; 3Average symmetric surface distance; 4Pharyngeal constrictor muscle; 5Hausdorff distance; 6Head and neck; 7Central nervous system; 8Clinical Target Volume.
Additional file 8. Literature Review on AI-Based Auto-contouring of Organs at Risk (OARs) and Clinical Target Volume (CTV) in Abdominal and Pelvic RegionsFootnotes: 1Organs at risk; 2Clinical target volume; 3Volumetric Dice score coefficient; 4Surface-Dice score coefficient; 5Added path length; ⁶Dice score coefficient; ⁷Hausdorff distance (95%); 6Mean surface distance; 7Seminal vesicles.
Additional file 9. Literature review on AI-based auto-contouring of organs at risk (OARs) and Clinical Target Volume in the thoracic and breast regions and in pediatrics Footnotes: 1Organs at risk; 2Non-small cell lung cancer; 3Dice score coefficient; 4Hausdorff distance; 5Clinical target volume; 6Head and Neck.
Acknowledgements
None.
Author contributions
Concept and design: Céline Meyer, Sandrine Huger, Jean-Christophe Faivre, Jean-François Py. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: Céline Meyer, Jean-Christophe Faivre Critical revision of the manuscript for important intellectual content: all authors Statistical analysis: Céline Meyer, Marie Bruand, Jean-Christophe Faivre Administrative, technical, or material support: Sandrine Huger, Jérémy Palisson, Paul Rétif, Thomas Sarrade, Anais Barateau Supervision: Céline Meyer, Jean-Christophe Faivre, Sandrine Huger, Jean-François Py.
Funding
No funding support was provided.
Availability of data and materials
Céline Meyer and Jean-Christophe Faivre had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. This study database is open to the scientific and medical community upon request to the steering committee. All data will be made available (anonymizedzed participant data, participant data with identifiers, data dictionary, or other specified data set) depending on the collaboration in place. Study protocol, statistical analysis plan, informed consent forms, and consortium statuses are available upon request. Proposals should be addressed to Dr Jean-Christophe Faivre at: jc.faivre@nancy.unicancer.fr. The steering committee will evaluate the pertinence of the request before sending the database to any academic partners. After agreement of the steering committee, data requestors will have to sign a data access agreement to gain access to the database. The steering committee as the sponsor will be vigilant regarding the General Data Protection Regulation (GDPR) compliance of the requestors. No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was approved by ethics and conducted in accordance with the ethical standards of the Declaration of Helsinki (as revised in 2013). This study was approved by Ethics committee named the French National Commission of Informatics and Liberty (CNIL) (CNIL-MR0004 Number HDH434). The present study has been approved by the French Health Data Institute (Health DataHub) as the number HDH301. All methods were carried out in accordance with relevant guidelines and regulations. All participants have signed informed consent to the use of their data for research purposes.
Consent for publication
Not applicable.
Competing interests
The authors have declared no conflicts of interest. The Lorraine Cancer Institute used MVision AI software until January 1, 2023.
Footnotes
The original version of this article was revised: an error in the author names of Olivier Cravéreau and Alizée Renan (transposed given names and family names) was corrected.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/22/2025
A Correction to this paper has been published: 10.1186/s13014-025-02586-y
References
- 1.Grégoire V, Ang K, Budach W, Grau C, Hamoir M, Langendijk JA, et al. Delineation of the neck node levels for head and neck tumors: A 2013 update DAHANCA, EORTC, HKNPCSG, NCIC CTG, NCRI, RTOG, TROG consensus guidelines. Radiother Oncol. 2014;110(1):172–81. [DOI] [PubMed] [Google Scholar]
- 2.Jabbour SK, Hashem SA, Bosch W, Kim TK, Finkelstein SE, Anderson BM, et al. Upper abdominal normal organ contouring guidelines and atlas: a radiation therapy oncology Group consensus. Pract Radiat Oncol mars. 2014;4(2):82–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gay HA, Barthold HJ, O’Meara E, Bosch WR, El Naqa I, Al-Lozi R, et al. Pelvic normal tissue contouring guidelines for radiation therapy: a radiation therapy oncology group consensus panel atlas. Int J Radiat Oncol. 2012;83(3):e353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Offersen BV, Boersma LJ, Kirkove C, Hol S, Aznar MC, Biete Sola A, et al. ESTRO consensus guideline on target volume delineation for elective radiation therapy of early stage breast cancer. Radiother Oncol. 2015;114(1):3–10. [DOI] [PubMed] [Google Scholar]
- 5.Scoccianti S, Detti B, Gadda D, Greto D, Furfaro I, Meacci F, et al. Organs at risk in the brain and their dose-constraints in adults and in children: a radiation oncologist’s guide for delineation in everyday practice. Radiother Oncol. 2015;114(2):230–8. [DOI] [PubMed] [Google Scholar]
- 6.Brouwer CL, Steenbakkers RJHM, Bourhis J, Budach W, Grau C, Grégoire V, et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG oncology and TROG consensus guidelines. Radiother Oncol. 2015;117(1):83–90. [DOI] [PubMed] [Google Scholar]
- 7.Brouwer CL, Steenbakkers RJ, van den Heuvel E, Duppen JC, Navran A, Bijl HP, et al. 3D variation in delineation of head and neck organs at risk. Radiat Oncol. 2012;7(32):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H, et al. Vision 20/20: Perspectives on automated image segmentation for radiotherapy. Med Phys. 2014;41(5):050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Veen J, Gulyban A, Nuyts S. Interobserver variability in delineation of target volumes in head and neck cancer. Radiother Oncol. 2019;137:9–15. [DOI] [PubMed] [Google Scholar]
- 10.Chen A, Niermann KJ, Deeley MA, Dawant BM. Evaluation of multiple-atlas-based strategies for segmentation of the thyroid gland in head and neck CT images for IMRT. Phys Med Biol. 2012;57(1):93–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in auto-segmentation. Semin Radiat Oncol. 2019;29(3):185–97. [DOI] [PubMed] [Google Scholar]
- 12.Meyer P, Noblet V, Mazzara C, Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med. 2018;98:126–46. [DOI] [PubMed] [Google Scholar]
- 13.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 14.Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44(2):547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Veen J, Willems S, Deschuymer S, Robben D, Crijns W, Maes F, et al. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother Oncol. 2019;138:68–74. [DOI] [PubMed] [Google Scholar]
- 16.Men K, Chen X, Zhang Y, Zhang T, Dai J, Yi J, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017;7:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys. 2018;45(10):4558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126(2):312–7. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Wang C, Zhan W, Jia Y, Ruan F, Qiu L, et al. A comparative study of auto-contouring softwares in delineation of organs at risk in lung cancer and rectal cancer. Sci Rep. 2021;11(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mak RH, Endres MG, Paik JH, Sergeev RA, Aerts H, Williams CL, et al. Use of crowd innovation to develop an artificial intelligence-based solution for radiation therapy targeting. JAMA Oncol. 2019;5(5):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cha E, Elguindi S, Onochie I, Gorovets D, Deasy JO, Zelefsky M, et al. Clinical implementation of deep learning contour autosegmentation for prostate radiotherapy. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2021;159:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savenije MH, Maspero M, Sikkes GG, et al. Clinical implementation of MRI-based organs-at-risk auto-segmentation with convolutional networks for prostate radiotherapy. Radiat Oncol. 2020;15(104):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin S, Rodrigues G, Patil N, Bauman G, D’Souza D, Sexton T, et al. A multiphase validation of atlas-based automatic and semiautomatic segmentation strategies for prostate MRI. Int J Radiat Oncol. 2013;85(1):95–100. [DOI] [PubMed] [Google Scholar]
- 24.Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44(12):6377–89. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Kang K, Han C, Wang S, Chen Q, Chen Y, et al. A blind randomized validated convolutional neural network for auto-segmentation of clinical target volume in rectal cancer patients receiving neoadjuvant radiotherapy. Cancer Med. 2022;11(1):166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schreier J, Attanasi F, Laaksonen H. A full-image deep segmenter for CT images in breast cancer radiotherapy treatment. Front Oncol. 2019;9(677):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doolan PJ, Charalambous S, Roussakis Y, Leczynski A, Peratikou M, Benjamin M, et al. A clinical evaluation of the performance of five commercial artificial intelligence contouring systems for radiotherapy. Front Oncol. 2023;13:1213068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilemann G, Buschmann M, Lechner W, Dick V, Eckert F, Heilmann M, et al. Clinical Implementation and evaluation of auto-segmentation tools for multi-site contouring in radiotherapy. Phys Imaging Radiat Oncol. 2023;28: 100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong J, Huang V, Wells D, Giambattista J, Giambattista J, Kolbeck C, et al. Implementation of deep learning-based auto-segmentation for radiotherapy planning structures: a workflow study at two cancer centers. Radiat Oncol. 2021;16(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grégoire V, Blanchard P, Allajbej A, Petit C, Milhade N, Nguyen F, et al. OC-0681: deep learning auto contouring of OAR for HN radiotherapy: a blinded evaluation by clinical experts. Radiother Oncol. 2020;152:379–80. [Google Scholar]
- 31.Almberg SS, Lervåg C, Frengen J, Eidem M, Abramova TM, Nordstrand CS, et al. Training, validation, and clinical implementation of a deep-learning segmentation model for radiotherapy of loco-regional breast cancer. Radiother Oncol. 2022;173:62–8. [DOI] [PubMed] [Google Scholar]
- 32.Azria D, Boldrini L, De Ridder M, Fenoglietto P, Gambacorta MA, Gevaert T, et al. OC-0463 AI surpassing human expert: a multi-centric evaluation for organ at risk delineation. Radiother Oncol. 2022;170:408–10. [Google Scholar]
- 33.Bondiau P, Bolle S, Escande A, Duverge L, Demoor C, Rouyar-Nicolas A, et al. PD-0330 AI-based OAR annotation for pediatric brain radiotherapy planning. Radiother Oncol. 2022;170:S293–5. [Google Scholar]
- 34.Vandewinckele L, Claessens M, Dinkla A, Brouwer C, Crijns W, Verellen D, et al. Overview of artificial intelligence-based applications in radiotherapy: recommendations for implementation and quality assurance. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2020;153:55–66. [DOI] [PubMed] [Google Scholar]
- 35.Vaassen F, Hazelaar C, Vaniqui A, Gooding M, van der Heyden B, Canters R, et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Phys Imaging Radiat Oncol. 2020;13:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dijk LV, Van den Bosch L, Aljabar P, Peressutti D, Both S, Steenbakkers Roel JHM, et al. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiother Oncol. 2020;142:115–23. [DOI] [PubMed] [Google Scholar]
- 37.van Rooij W, Dahele M, Ribeiro Brandao H, Delaney AR, Slotman BJ, Verbakel WF. Deep learning-based delineation of head and neck organs at risk: geometric and dosimetric evaluation. Int J Radiat Oncol. 2019;104(3):677–84. [DOI] [PubMed] [Google Scholar]
- 38.Yan C, Guo B, Keller LM, Suh JH, Xia P. Dosimetric quality of artificial intelligence based organ at risk segmentation. Int J Radiat Oncol. 2023;117(2): e493. [Google Scholar]
- 39.Gooding MJ, Smith AJ, Tariq M, Aljabar P, Peressutti D, Van Der Stoep J, et al. Comparative evaluation of autocontouring in clinical practice: a practical method using the Turing test. Med Phys. 2018;45(11):5105–15. [DOI] [PubMed] [Google Scholar]
- 40.Sherer MV, Lin D, Elguindi S, Duke S, Tan LT, Cacicedo J, et al. Metrics to evaluate the performance of auto-segmentation for radiation treatment planning: a critical review. Radiother Oncol juill. 2021;160:185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung SY, Chang JS, Kim YB. Comprehensive clinical evaluation of deep learning-based auto-segmentation for radiotherapy in patients with cervical cancer. Front Oncol. 2023;13:1119008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanwar A, Merz B, Claunch C, Rana S, Hung A, Thompson RF. Stress-testing pelvic autosegmentation algorithms using anatomical edge cases. Phys Imaging Radiat Oncol. 2023;25: 100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar K, Yeo AU, McIntosh L, Kron T, Wheeler G, Franich RD. Deep learning auto-segmentation network for pediatric computed tomography data sets: can we extrapolate from adults? Int J Radiat Oncol. 2024;119:1297–306. [DOI] [PubMed] [Google Scholar]
- 44.Bibault JE, Giraud P. Deep learning for automated segmentation in radiotherapy: a narrative review. Br J Radiol. 2024;97(1153):13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lahmi L, Mamzer MF, Burgun A, Durdux C, Bibault JE. Ethical aspects of artificial intelligence in radiation oncology. Semin Radiat Oncol. 2022;32(4):442–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Summary of quality scoring of AI contouring programs for adult CT scan, children CT scan and adult brain MRIFootnotes: ¹ Organs at risk; ² Lymph node area; ³ Artificial Intelligence.
Additional file 2. Comparative table of Organs-at-Risk (OARs) delineations performed in head and neck as well as cerebral regions using CT scans by the 8 AI systemsFootnotes: Y = available and N = unavailable.
Additional file 3. Comparative table of Organs-at-Risk (OARs) delineations performed in thoracic and breast regions using CT scans by the 8 AI systemsFootnotes: Y = available and N = unavailable.
Additional file 4. Comparative table of Organs-at-Risk (OARs) delineations performed in abdominal and pelvic regions using CT scans by the 8 AI systemsFootnotes: Y = available and N = unavailable; 1High Dose Rate.
Additional file 5. Comparative table of lymph node areas delineated in head and neck, breast, abdominal and pelvic, and gynecological region using CT scans by the 8 AI systems29Footnotes: Y = available and N = unavailable; 1Radiation Therapy Oncology Group; 2 Clinical Target Volume Tumor.
Additional file 6. Comparative table of Organs-at-Risk (OARs) delineations achieved using MRI for cerebral and pelvic regions by the 8 AI systemsFootnotes: Y = available and N = unavailable.
Additional file 7. Literature Review on AI-Based Auto-contouring of Organs at Risk (OARs) in Head and Neck RegionFootnotes: 1Organs at risk; 2Dice score coefficient; 3Average symmetric surface distance; 4Pharyngeal constrictor muscle; 5Hausdorff distance; 6Head and neck; 7Central nervous system; 8Clinical Target Volume.
Additional file 8. Literature Review on AI-Based Auto-contouring of Organs at Risk (OARs) and Clinical Target Volume (CTV) in Abdominal and Pelvic RegionsFootnotes: 1Organs at risk; 2Clinical target volume; 3Volumetric Dice score coefficient; 4Surface-Dice score coefficient; 5Added path length; ⁶Dice score coefficient; ⁷Hausdorff distance (95%); 6Mean surface distance; 7Seminal vesicles.
Additional file 9. Literature review on AI-based auto-contouring of organs at risk (OARs) and Clinical Target Volume in the thoracic and breast regions and in pediatrics Footnotes: 1Organs at risk; 2Non-small cell lung cancer; 3Dice score coefficient; 4Hausdorff distance; 5Clinical target volume; 6Head and Neck.
Data Availability Statement
Céline Meyer and Jean-Christophe Faivre had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. This study database is open to the scientific and medical community upon request to the steering committee. All data will be made available (anonymizedzed participant data, participant data with identifiers, data dictionary, or other specified data set) depending on the collaboration in place. Study protocol, statistical analysis plan, informed consent forms, and consortium statuses are available upon request. Proposals should be addressed to Dr Jean-Christophe Faivre at: jc.faivre@nancy.unicancer.fr. The steering committee will evaluate the pertinence of the request before sending the database to any academic partners. After agreement of the steering committee, data requestors will have to sign a data access agreement to gain access to the database. The steering committee as the sponsor will be vigilant regarding the General Data Protection Regulation (GDPR) compliance of the requestors. No datasets were generated or analysed during the current study.