Abstract
Background
Oncofetal splice variants of extracellular matrix (ECM) proteins present a unique group of target antigens for the immunotherapy of pediatric cancers. However, limited data is available if these splice variants can be targeted with T cells expressing chimeric antigen receptors (CARs).
Methods
To determine the expression of the oncofetal version of tenascin C (TNC) encoding the C domain (C.TNC) in pediatric brain and solid tumors, we used quantitative reverse transcription PCR and immunohistochemistry. Genetically modified T cells were generated from human peripheral blood mononuclear cells and evaluated in vitro and in vivo.
Results
We demonstrate that C.TNC is expressed on a protein level in pediatric tumors, including diffuse intrinsic pontine glioma, osteosarcoma, rhabdomyosarcoma, and Ewing sarcoma. We generate C.TNC-CAR T cells and establish that these recognize and kill C.TNC-positive tumor cells. However, their antitumor activity in vivo is limited. To improve the effector function of C.TNC-CAR T cells, we design a leucine zipper-based chimeric cytokine receptor that activates interleukin-18 signaling pathways (Zip18R). Expression of Zip18R in C.TNC-CAR T cells improves their ability to secrete cytokines and expand in repeat stimulation assays. C.TNC-CAR.Zip18R T cells also have significantly greater antitumor activity in vivo compared with unmodified C.TNC-CAR T cells.
Conclusions
Our study identifies the C domain of the ECM protein TNC as a promising CAR T-cell therapy for pediatric solid tumors and brain tumors. While we focus here on pediatric cancer, our work has relevance to a broad range of adult cancers that express C.TNC.
Keywords: Chimeric antigen receptor - CAR, Immunotherapy, T cell
WHAT IS ALREADY KNOWN ON THIS TOPIC
Oncofetal splice variants of extracellular matrix (ECM) proteins have so far been studied mainly in adult cancers, and the majority of ECM-targeted therapeutic approaches rely on antibody drug, cytokine, or radioisotope conjugates.
WHAT THIS STUDY ADDS
We demonstrate here that the oncofetal version of tenascin C (TNC) encoding the C domain (C.TNC) is expressed on a protein level in both pediatric brain and solid tumors and can be targeted with C.TNC-chimeric antigen receptor T cells.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study highlights that oncofetal splice variants of ECM proteins are viable targets for cellular immunotherapy.
Introduction
Chimeric antigen receptor (CAR) T-cell therapy has shown promise for pediatric hematological malignancies; however, progress in the solid and brain tumor space has been limited.1,3 Although failure of CAR T cells in solid tumors is likely a multifactorial issue, a critical first step to success is target antigen selection.1 2 Most tumor-associated antigens targeted by CAR T cells are surface proteins; however, there have been preclinical successes in targeting secreted proteins such as oncofetal fibronectin and collagen.4,8 We recently developed a computational pipeline to discover cancer-specific exons as targets for the immunotherapy of pediatric solid and brain tumors.8 Top hits included oncofetal splice variants and differentially expressed proteins of the extracellular matrix (ECM), which have been previously described in a broad range of adult cancers.
One of the identified ECM proteins was oncofetal tenascin C (TNC), a hexameric protein that can bind to cells via several cell surface proteins and integrins.9 In normal development, TNC undergoes splicing of nine exons between FN-III domains 5 and 6.10 Changes in isoform expression have been linked to changes in pH, the upregulation of SRSF6, and growth factors such as TGFβ1 and FGF.11,16 The long isoforms of TNC have been previously described in cancer and can be implicated in tumor cell proliferation and migration as well as impairing antitumor immune responses.17,19 Antibody-drug or cytokine conjugates targeting oncofetal TNC for solid and brain tumors have shown promising safety and antitumor activity in preclinical models, and early-phase clinical studies have confirmed the safety of oncofetal TNC as a target for immunotherapy.20
T cells can produce cytokines to self-sustain their effector function; however, in the setting of chronic antigen exposure, cytokine production is limited.21 To engineer this component of T-cell effector function, investigators have genetically modified T cells to co-express CARs with unmodified or membrane-bound cytokines, or chimeric cytokine receptors.2 3 For solid tumor-specific CAR T cells, predominately, JAK/STAT-activating cytokines interleukin (IL)-7, IL-12, IL-15, IL-21, and IL-23 have been investigated.22,31 In addition, we and others have shown that MyD88 signaling, either via IL-18 signaling or including MyD88 as a CAR signaling domain, is beneficial to CAR T cells in the setting of chronic antigen exposure.32,36 Building on these findings, we decided to use our modular leucine zip receptor (ZipR) platform37 to design a constitutively active IL-18 receptor.
In this study, we explored CAR T cells redirected to the C domain, one of the exons expressed in oncofetal TNC (C.TNC-CAR T cells), as an immunotherapeutic for pediatric brain and solid tumors. We developed a second-generation CAR specific for C.TNC based on the G11 monoclonal antibody (mAb), which recognizes the human and murine C domain.38 We found that C.TNC-CAR T cells have antigen-specific cytolysis of target tumor cells but limited efficacy in xenograft models. While altering CAR design did not improve antitumor activity, expressing a constitutively active IL-18 receptor in C.TNC-CAR T cells bolstered their effector function, resulting in improved antitumor activity in vivo.
Methods
Please see online supplemental methods for details on tumor cell lines, immunohistochemistry (IHC), reverse transcription quantitative PCR (RT-qPCR), retroviral vectors, CAR T-cell generation, flow cytometry, co-cultures, MILLIPLEX, and statistical analysis.
Patient-derived xenograft and primary sarcoma tissue samples
Patient-derived xenografts were obtained from the Childhood Solid Tumor Network (CSTN) (https://cstn.stjude.cloud/search/).39 Fresh frozen tissues were hand homogenized in DPBS (Gibco, 14190–144) + 1% FBS (Gibco) and filtered through polystyrene test tubes with cell strainer caps (Falcon, 352235) to generate a single-cell suspension. Formalin-fixed paraffin-embedded tissues were also obtained for IHC purposes. De-identified formalin-fixed paraffin-embedded tissue blocks from clinical patient tumor samples were cut and stained as previously described.8
Single-cell RNA sequencing
Sample collection
C.TNC-CAR.IL-18 receptor-based ZipR (Zip18R) T cells were generated by double transduction as previously described for two biological donors. C.TNC-CAR.Zip18R T cells were collected at baseline or after 12, 24, and 48 hours stimulation with LM7.green fluorescent protein (GFP).firefly luciferase (ffLuc) in the presence of IL-15 (PeproTech; 5 ng/mL). Samples were sorted for viability and hashed for multiplexing with TotalSeq-C anti-human Hashtag Antibodies (BioLegend 394661, 394663, 394665). To add hashing antibodies, up to 5e6 cells were collected and incubated with Human TruStain FcX block (BioLegend), followed by a hashing antibody in a volume of 100 μL.
Library preparation
Equal cell numbers from each hashed sample were pooled before loading onto a Chromium Controller to generate 10,000 single-cell gel beads in emulsion for single-cell RNA sequencing (scRNA-seq). Libraries were prepared using Chromium Next GEM Single Cell 5’ V.2 (Dual index) and Gel Bead Kit (10x Genomics). Complementary DNA (cDNA) was amplified (13 cycles), after which it was used for the preparation of gene expression and cell surface protein libraries. The cDNA content of each sample and library was quality-checked using a high-sensitivity DNA chip with a 2100 Bioanalyzer (Agilent Technologies). The libraries were sequenced on NovaSeq (Illumina) with paired-end reads of 26 cycles for read 1 and 90 cycles for read 2×. Median reads per cell ranged from 117,565 to 132,623 in the baseline cohort and 13,717–31,087 reads for the stimulated cohort.
Data preprocessing and sample integration
C.TNC-CAR and Zip18R sequences were added to the human reference transcriptome (refdata-gex-GRCh38-2020-A) for reads mapping. The Cell Ranger V.7.1.0 Single-Cell software suite (10x Genomics) was used to process scRNA-seq FASTQ files. The “cellranger multi” and “cellranger bamtofastq” commands were performed for sample demultiplexing and extracting sample FASTQ files. The “cellranger count” command was performed to align FASTQ files to reference genome and summarize the data into matrices describing gene read counts (unique molecular identifier (UMI)) per cell. SingleCellExperiment40 object was generated and used for the following analysis. Damaged or dying cells were filtered based on the outlier status of mitochondrial genes expression using scuttle.41 Cells with a detected gene number less than 300 were filtered out. Read counts were normalized using scuttle logNormCounts function.41 Top 2,000 high-variance genes were selected to calculate principal components using scater.41 Harmony42 was used to integrate all the samples for batch correction. To get a pure T-cell population, any cells with no CD3δ expression (zero count) were filtered out.
Differentially expressed gene detection and pathway enrichment analysis
For each sample, the T-cell population was classified into four subpopulations (C.TNC-CAR.Zip18R, C.TNC-CAR, Zip18R, non-transduced) based on whether the CAR or Zip18R was expressed or not (zero count). Then pseudo-bulk data for each subpopulation in each sample was generated using scuttle.41 DESeq2 pipeline43 was used to generate principal component analysis (PCA) plot. Delegate (pseudo-bulk DESeq2 method with Wald test)44 was used to detect differentially expressed genes. ClusterProfiler45 was used to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Particularly, DESeq2 default normalized counts were used to calculate log2 fold change between samples in two conditions (pseudo-count as 10 defined from counts distribution). “gseKEGG” commands were performed with Benjamini-Hochberg (BH) adjustment method.
Statistical analysis
To identify differentially expressed genes from different cell populations on scRNA-seq data, the software Delegate44 was performed with pseudo-bulk DESeq243 method and Wald test. KEGG pathway enrichment analysis for pseudo-bulk T-cell population was performed with “gseKEGG” commands in ClusterProfiler45 and p value was adjusted by BH method.
Data availability
The scRNA-seq data generated in this study has been deposited in the European Genome-Phenome Archive (EGA) under the title “Single-cell RNA sequencing of human IL-18R supported CAR T cells targeting oncofetal Tenascin C”. The raw data can be obtained by a request to EGA. The scRNA-seq expression matrices are available on Gene Expression Omnibus (GEO) under ascension code GSE282046.
Xenograft mouse models
Animal experiments followed a protocol approved by the St. Jude Institutional Animal Care and Use Committee (646–100595). All experiments used NOD-scid IL-2Rgammanull (NSG) mice obtained from St. Jude’s NSG colony. Studies were done in biological duplicate, altering mouse gender between experiments. For imaging, mice were injected in the intraperitoneal (i.p.) cavity with 150 mg/kg of D-luciferin (PerkinElmer) 5–10 min before imaging, anesthetized with isoflurane, and imaged under anesthesia with a Xenogen IVIS-200 imaging system. The photons emitted from the luciferase-expressing cells were quantified using Living Image software (Caliper Life Sciences). Total emitted photon flux (photons per second) was used to evaluate tumor burden. For the i.p. tumor model, 5–6-week-old mice were injected i.p. with 1×106 LM7.GFP.ffLuc cells, and after 7 days, received a single i.p. injection of 1×106 T cells. For the intracranial (i.c.) tumor model, 10–12-week-old mice were injected i.c. with 1×106 DIPG007.YFP.ffLuc cells in 2 µL of 80% Matrigel (Corning) mixed with PBS (Gibco), and after 7 days, received a single i.c. injection of 1 or 2×106 T cells. In both models, mice were imaged to confirm the presence of tumors prior to T-cell injection, and mice were euthanized when they reached (1) two consecutive flux values >1×1010, (2) a single flux value >1×1011, or (3) physical euthanasia criteria (weight loss, signs of distress). For the subcutaneous (s.c). tumor model, 7–9-week-old mice were injected in the right flank s.c. with 2×106 A673 cells, and after 7 days, received a single intravenous injection of T cells. Mice were euthanized when they met physical euthanasia criteria or when the tumor size was greater than 4,000 mm3.
Results
C.TNC is expressed in pediatric solid and brain tumors
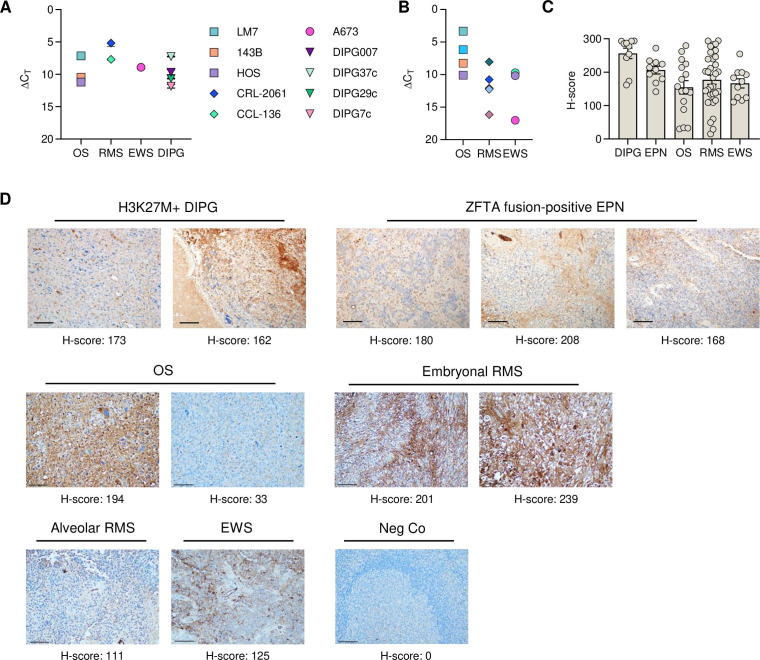
We first set out to establish C.TNC expression in pediatric solid and brain tumor samples. First, we screened pediatric diffuse intrinsic pontine glioma (DIPG) and pediatric sarcoma cell lines using RT-qPCR and confirmed that all samples contained the C.TNC exon at an RNA level (figure 1A). We screened patient-derived xenograft samples and demonstrated high expression of C.TNC in osteosarcoma (OS) samples (figure 1B, online supplemental STable 1). Protein expression was further confirmed by IHC, using the G11 mAb to stain primary samples of H3K27M+DIPG, ZFTA-fusion positive ependymoma, OS, rhabdomyosarcoma, and Ewing sarcoma (figure 1C,D). C.TNC expression, as determined by IHC, was localized to both the cytoplasm and membranes of cells in primary and xenograft tumor samples, and was also observed within the immediate tumor microenvironment, which was interpreted as tumor-associated production and secretion of C.TNC by the evaluated pediatric tumor types. Depending on the xenograft or primary patient samples, neoplasms expressed C.TNC at moderate to strong levels based on their H-score.
Figure 1. C.TNC is expressed in solid and brain tumors. (A,B) Reverse transcription quantitative PCR of (A) pediatric cell lines (n=3 bioreplicates, mean+SEM) and (B) patient-derived xenografts for C.TNC. ΔCT is relative to GAPDH. Acute lymphoblastic leukemia cell line CCRF-CEM had no C.TNC detected. (C) H-scores of primary human H3K27M+DIPG, ZFTA fusion-positive ependymoma (EPN), osteosarcoma (OS), rhabdomyosarcoma (RMS), and Ewing sarcoma (EWS) tumors. (D) Representative immunohistochemistry staining of primary FFPE human tumor samples and tonsils (negative control (neg Co)) with a C.TNC-specific antibody. 20× magnifications, 100 µM scale bar. Images were taken with an Olympus BX46 microscope and a Nikon DS-Fi3 camera at a 20× magnification and edited with Photoshop 25.5.1. C.TNC, tenascin C encoding the C domain; DIPG, diffuse intrinsic pontine glioma.
C.TNC-CAR T cells recognize and kill C.TNC-positive tumor cells
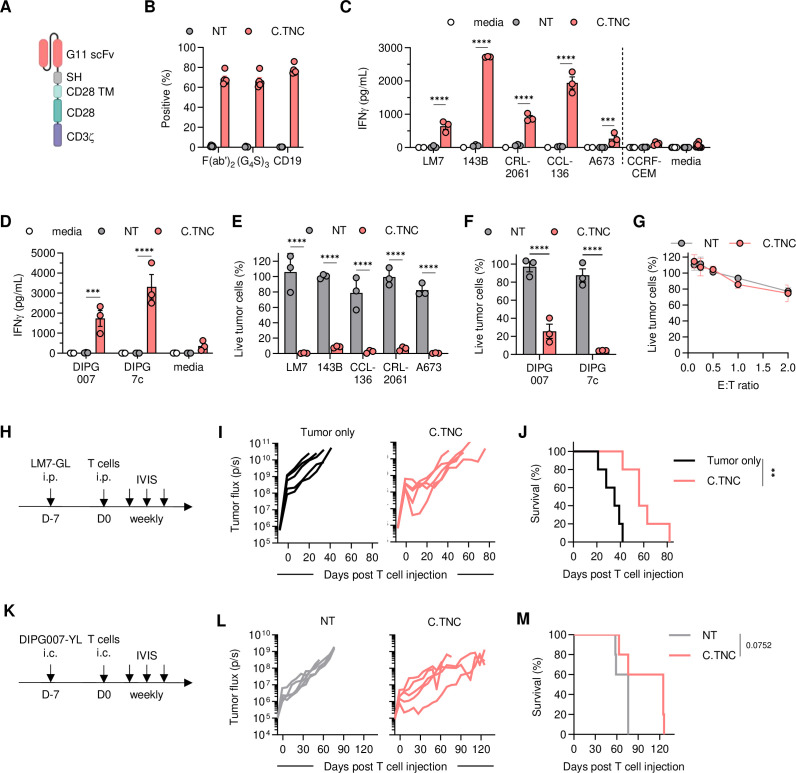
We generated a retroviral vector encoding a C.TNC-CAR consisting of a single chain variable fragment (scFv)-based C.TNC binding domain derived from the G11 mAb, a short hinge, a CD28 transmembrane, and a CD28.zeta endodomain, a self-cleaving T2A sequence, and truncated CD19 (tCD19) (figure 2A, online supplemental SFigure 4A). C.TNC-CAR T cells were generated by standard retroviral transduction and average transduction efficacy was~70% as determined by flow cytometry, detecting either the CAR (scFv or linker) or tCD19 (figure 2B). We evaluated C.TNC-CAR T-cell effector function against C.TNC-positive (sarcoma, DIPG) and C.TNC-negative (CCRF-CEM) cell lines. In co-culture assays, C.TNC-CAR T cells produced significant amounts of interferon (IFN)-γ compared with non-transduced (NT) T cells only in the presence of C.TNC-positive cell lines (figure 2C,D). C.TNC-CAR T cells also had significant cytolytic activity against C.TNC-positive sarcoma cell lines compared with NT T cells (figure 2E,F), and no cytotoxicity against the C.TNC-negative cell line CCRF-CEM (figure 2G). We determined the phenotype of C.TNC-CAR T cells cultured with and without stimulation with C.TNC-positive LM7.GFP.ffLuc cells. C.TNC-CAR expression alone in T cells did not alter the CD4/CD8 ratios or their phenotype (online supplemental SFigure B, C). With antigen stimulation, C.TNC-CAR T cells demonstrated T-cell differentiation as judged by a decline in naïve-like subsets as compared with NT T cells (online supplemental SFigure D, E). To further confirm antigen-specificity, we generated a non-functional C.TNC.mu-CAR that lacked the intracellular CD28 domain and had mutated CD3ζ ITAMS (online supplemental SFigure 4F). C.TNC.mu-CAR T cells were successfully expressed in T cells (online supplemental SFigure 4G) and had no cytolytic activity against LM7.GFP.ffLuc cells compared with NT cells (online supplemental SFigure 4H). Next, we assessed the antitumor activity of C.TNC-CAR T cells in vivo in our OS LM7.GFP.ffLuc and DIPG007.YFP.ffLuc xenograft models (figure 2H,K). In both models, C.TNC-CAR T cells exhibited antitumor activity at early time points as determined by a reduction in tumor flux values as compared with controls (figure 2I,L; online supplemental SFigures 5 and 6). This transient antitumor activity of C.TNC-CAR T cells translated to a significant survival advantage in the OS model, and although this did not reach significance for the mice in DIPG007, the median survival between the groups reached 50 days (figure 2J,M). To evaluate the presence of tumor cells, T cells, and TNC expression post C.TNC-CAR T cell therapy, DIPG007-bearing mice were euthanized 2 weeks after intratumoral injection of NT or C.TNC-CAR T cells. NT T cell-treated tumors, identified by H3K27M-staining, were larger in comparison to tumors injected with C.TNC-CAR T cells (online supplemental SFigure 7). This was mirrored by decreased TNC expression. Human CD3-positive T cells were only detected in mice after C.TNC-CAR T-cell therapy (online supplemental SFigure 7).
Figure 2. C.TNC-CAR T cells have antitumor activity in vitro and in vivo. (A) Schematic of C.TNC-CAR design. (B) Transduction efficiency of healthy donor T cells determined via flow cytometry on day 7 (n=5, mean+SEM). Graph shows the percentage of positive cells stained for each respective antibody for CAR transgene detection. (C,D) IFN-γ production measured by ELISA after 48 hours of co-culture with (C) sarcoma or (D) brain tumor cell lines at 2:1 effector to target (E:T) ratio. Negative values were plotted as zero (mean+SEM, n=3–4 for NT and C.TNC), two-way ANOVA, ***p<0.001, ****p<0.0001. (E,F) Cytotoxicity after 72 hours at a 4:1 E:T ratio determined by a luciferase-based assay (mean+SEM, n=3), two-way ANOVA, ****p<0.0001. (G) Cytotoxicity of C.TNC-negative cell line CCRF-CEM determined by a luciferase-based assay (n=4–5, mean+SEM). (H) Schematic of experiment in 5–6-week-old female NSG mice. 1×106 LM7.green fluorescent protein.firefly luciferase cells were injected intraperitoneally (i.p.), followed by 1×106 T cells injected i.p. 7 days later. (I) Flux values from weekly IVIS images (n=5 per cohort). (J) Overall survival of the mice, Mantel-Cox test, **p<0.01. (K) Schematic of experiment in 10–12-week-old male NSG mice. 1×106 DIPG007.YFP.ffLuc cells were injected intracranially (i.c.), followed by 2×106 T cells injected i.c. 7 days later. (L) Flux values from weekly IVIS images (n=5 per cohort). (M) Overall survival of the mice, Mantel-Cox test. ANOVA, analysis of variance; CAR, chimeric antigen receptor; C.TNC, tenascin C encoding the C domain; DIPG, diffuse intrinsic pontine glioma; IFN, interferon; NT, non-transduced; scFv, single chain variable fragment.
To explore if redesigning the CAR structure could improve C.TNC-CAR T-cell effector function, we designed four additional CARs with changes in the linker, hinge, and transmembrane domains (online supplemental SFigure 8A). All CARs were expressed in T cells (online supplemental SFigure 8B), and all C.TNC-CAR T-cell populations had antitumor activity compared with NT T cells in a 24-hour cytotoxicity assay (online supplemental SFigure 8C). Based on their performance in their cytolytic activity, we selected the C.TNC-SL-CAR and C.TNC-zeta-CAR T cells for in vivo testing (online supplemental SFigure 8D). Neither design improved the antitumor activity of C.TNC-CAR T cells (online supplemental SFigure 9), and we, therefore, continued to use the original CD28.zeta CAR design in future experiments.
An IL-18-based constitutively active cytokine receptor improves the effector function of C.TNC-CAR T cells
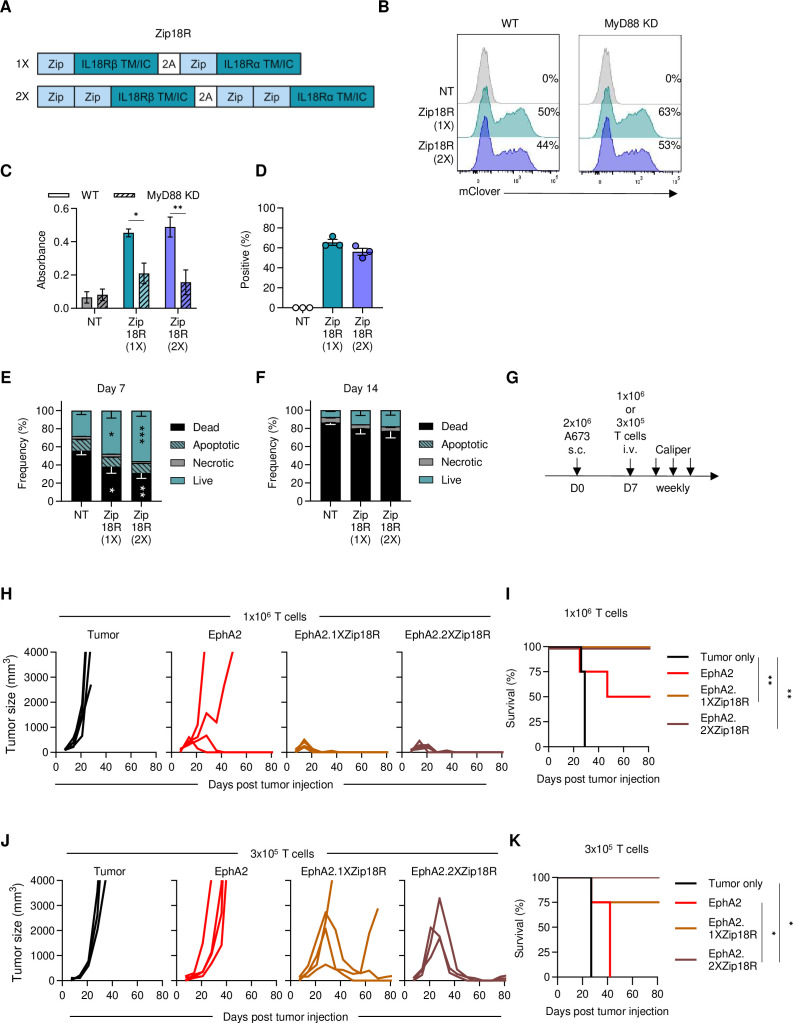
A key requirement for CAR T-cell effector function is the provision of signal 3, or proinflammatory cytokine support.46 47 We and others have demonstrated that MyD88 signaling improves CAR T-cell effector function by inducing type 2 cytokine signaling and enhancing T-cell expansion and proliferation.32,36 We first evaluated if C.TNC-CAR T cells expressing a granulocyte-macrophage colony-stimulating factor (GM-CSF)/IL-18 switch receptor (GM18), which requires GM-CSF binding for IL-18 signaling, could better control tumor growth in our LM7.GFP.ffLuc in vivo model.33 C.TNC-CAR.GM18 T cells had limited antitumor activity compared with NT T cells (online supplemental SFigure 10). We recently published the development of modular leucine zipper-based chimeric cytokine receptors (ZipRs) to provide constitutive JAK/STAT signaling to transduced T cells.37 We decided to explore here if our modular chimeric cytokine receptor platform could be adapted for the IL-18 receptor (Zip18R) for constitutive IL-18 support. We designed two Zip18Rs with one (1X) or two (2X) pairs of leucine zippers (figure 3A) and cloned these into a retroviral vector that also encoded a P2A sequence and mClover.
Figure 3. Zip18R improves CAR T-cell effector function. (A) Schematic of 1X and 2X Zip18Rs, the leucine zippers (Zip) are connected with a (G4S)3 linker. The 1X and 2X Zip18Rs retroviral vectors also encode a P2A skip sequence and mClover. TM/IC: transmembrane and intracellular. (B) Representative transduction efficiency of Ramos-Blue and Ramos-Blue KD-MyD NF-κB/AP-1 reporter cells determined via flow cytometry. (C) Absorbance values of transduced Ramos-Blue NF-κB/AP-1 reporter cell supernatant mixed with QUANTI-Blue reagent (n=3 from two separate transductions). (D) Transduction of T cells with Zip18R (n=3, mean+SEM). (E,F) Populations after (E) 7 days or (F) 14 days of cytokine starvation as determined by flow cytometry (n=3, mean+SEM). Two-way analysis of variance, *p<0.05, **p<0.01, ***p<0.001. (G) Schematic of A673 model. T cells were sorted prior to injection. 7–9-week-old male NSG mice were used. (H) Tumor caliper measurements (n=4 for tumor and CAR only, n=5 for CAR.Zip18Rs). (I) Kaplan-Meier survival curve, Mantel-Cox test, **p<0.01. (J) Tumor caliper measurements. (n=3 for tumor and CAR.2XZip18R, n=4 for CAR and CAR.1XZip18R). (K) Kaplan-Meier survival curve, Mantel-Cox test, *p<0.05. CAR, chimeric antigen receptor; NT, non-transduced; Zip18R, interleukin-18 receptor-based leucine zipper receptor.
Both Zip18Rs were functional and their functionality was dependent on the expression of MyD88 as judged by their ability to activate NFKB/AP-1 only in a reporter cell line in which MyD88 was present (figure 3B,C). T cells could be readily transduced with retroviral vectors encoding Zip18Rs with a transduction efficiency of~50% as judged by mClover expression (figure 3D). Expression of either Zip18R alone enhanced the survival of T cells after 1 week of cytokine starvation but had similar viability when starved for 2 weeks (figure 3E,F). In vivo, the antitumor activity of CAR T cells targeting EphA2, a cell surface antigen,48 was significantly enhanced by expressing either Zip18R (figure 3G–K). Since there was no difference between both Zip18Rs, we selected the 1X Zip18R for further testing.
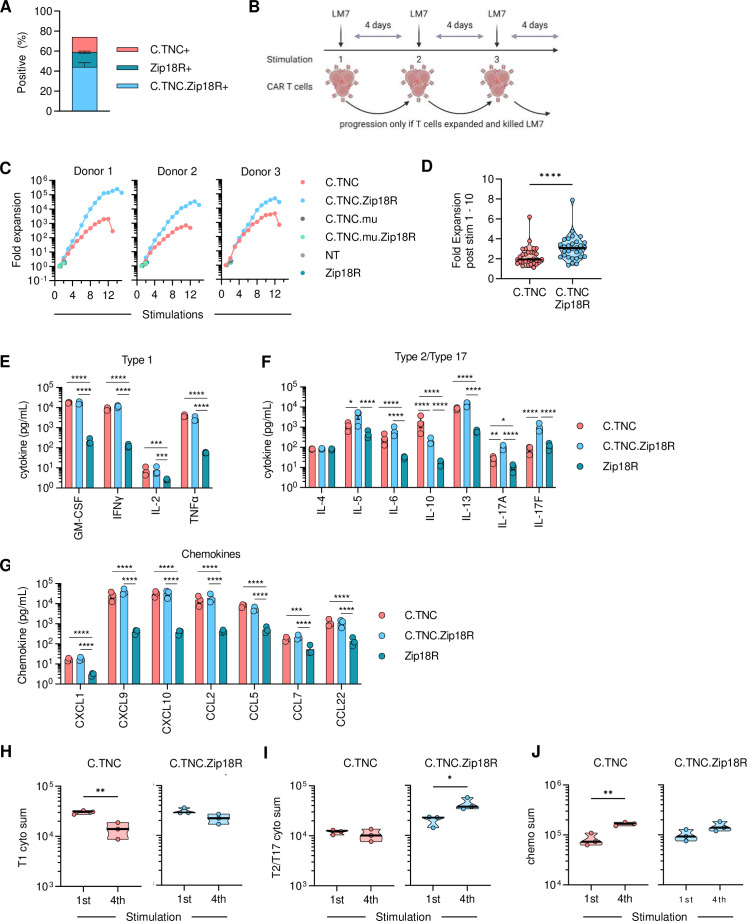
C.TNC-CAR.Zip18R T cells were generated by co-transduction, and the resulting T-cell populations consisted of~44% double-positive cells (figure 4A). We evaluated C.TNC-CAR and C.TNC-CAR.Zip18R T cells against LM7.GFP.ffLuc cells in a repeat stimulation assay, in which T cells are co-cultured with tumor cells at a 2:1 effector to target ratio in the presence of IL-15, and only restimulated if they killed the tumor cells and expanded (figure 4B). Controls included NT, Zip18R, C.TNC.mu-CAR, and C.TNC.mu-CAR.Zip18R T cells. C.TNC-CAR.Zip18R T cell expansion was significantly greater after each stimulation compared with C.TNC-CAR T cells (figure 4C,D). All four control T-cell populations did not kill LM7.GFP.ffLuc cells and did not expand significantly after the first stimulation, confirming antigen-specific C.TNC-CAR and C.TNC-CAR.Zip18R T-cell expansion and demonstrating that expression of Zip18R in T cells does not induce autonomous cell growth (figure 4C). During the repeat stimulation assay, we collected media 48 hours post each stimulation to evaluate cytokine and chemokine production using a 48 Multiplex Assay. After the first stimulation, C.TNC-CAR and C.TNC-CAR.Zip18R T cells produced significant amounts of type 1 (GM-CSF, IFN-γ, IL-2, tumor necrosis factor (TNF)-α), type 2/type 17 (IL-6, IL-10, IL-13, IL-17A), and chemokines (CXCL1, CXCL9, CXCL10, CCL2, CCL5, CCL7, CCL22) compared with Zip18R T cells (figure 4E–G). In addition, C.TNC-CAR.Zip18R T cells produced higher amounts of IL-5, IL-17A, and IL-17F and less IL-10 compared with C.TNC-CAR T cells. C.TNC.mu-CAR.Zip18R, Zip18R, and NT T cells did not produce higher levels of cytokines or chemokines in the presence of tumor cells (online supplemental SFigure 11). There were no significant differences in the cytokine/chemokine expression profile of NT and Zip18R T cells (online supplemental SFigure 11). To evaluate the long-term effects of Zip18R expression on cytokine and chemokine production, we determined cytokine production after the fourth stimulation. After the fourth stimulation, C.TNC-CAR T cells produced significantly lower levels of type 1 cytokines, and higher levels of chemokines (figure 4H–J, online supplemental SFigure 12A). In contrast, there was no significant decrease in type 1 and an increase in type 2/type 17 cytokine production of C.TNC-CAR.Zip18R T cells, and while chemokine production was also increased, this did not reach significance (online supplemental SFigure 12B).
Figure 4. Zip18R bolsters C.TNC-CAR T-cell effector function in vitro. (A) Transduction efficiency of primary T cells was determined via flow cytometry on day 7 post-transduction (n=4, mean+SEM). Determined via F(ab’)2 staining and mClover expression. (B) Repeat stimulation assay schematic. T cells were stimulated with LM7.green fluorescent protein.firefly luciferase at an effector to target ratio of 2:1 in the presence of IL-15 every 4 days. (C) Expansion of T cells in repeat stimulation assay. Each graph represents one donor. (D) Fold change of C.TNC-CAR and C.TNC.CAR.Zip18R T cells after stimulations 1 through 10, paired t-test, ****p<0.0001. (E–G) Quantification of (E) type 1 and (F) type 2/type 17 cytokines, and (G) chemokines 48 hours post first stimulation of the repeat stimulation assay (n=3, mean+SEM), data was log-transformed before statistical analysis, two-way analysis of variance, *p<0.05, **p<0.01, ***:p<0.001, ****p<0.0001. (H–J) Comparison of the sum of (H) type 1 cytokines, (I) type 2/type 17 cytokines, and (J) chemokines produced post first stimulation and fourth stimulation by C.TNC-CAR T cells and C.TNC-CAR.Zip18R T cells from repeat stimulation assay (n=3), unpaired t-test, *p<0.05, **p<0.01. CAR, chimeric antigen receptor; C.TNC, tenascin C encoding the C domain; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; NT, non-transduced; TNF, tumor necrosis factor; Zip18R, interleukin-18 receptor-based leucine zipper receptor.
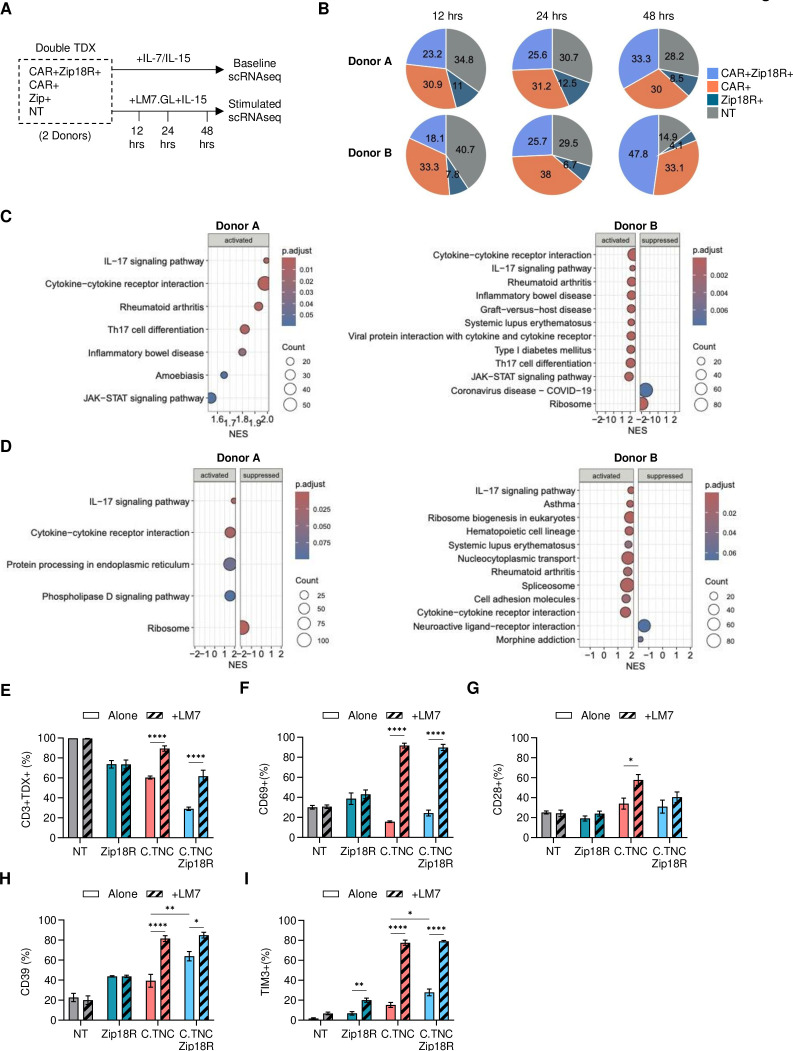
To evaluate the transcriptional changes induced by Zip18R expression, we performed scRNA-seq of C.TNC-CAR.Zip18R T cells. We collected C.TNC-CAR.Zip18R T cells from two donors at baseline and after 12, 24, or 48 hours of co-culture with LM7.GFP.ffLuc cells in the presence of IL-15 (figure 5A). We were able to detect NT, Zip18R+, C.TNC-CAR+, and C.TNC-CAR+Zip18R+cell populations within each CAR T-cell product (online supplemental SFigure 13). Using principal component analysis, we found that the T-cell populations clustered according to the donor (online supplemental SFigure 14A), which in part was due to differences in their gender (online supplemental SFigure 14B). Over the course of tumor cell stimulation, C.TNC+Zip18R+T cells enriched while NT T cells decreased, suggesting antigen-dependent expansion of C.TNC-CAR.Zip18R T cells (figure 5B).
Figure 5. Zip18R signaling alters the transcriptome of C.TNC-CAR T cells. (A) Schematic of experiment. C.TNC-CAR.Zip18R T cells were collected from culture in IL-7/IL-15 and frozen for analysis or freshly collected from a co-culture assay with LM7.GFP.ffLuc (LM7.GL) tumor cells in the presence of IL-15 at 12, 24, and 48 hours post tumor cell stimulation. All four populations are present in the same culture. (B) Percentages of C.TNC-CAR+Zip18R+, C.TNC-CAR+, Zip18R+, and NT T cells at 12, 24, and 48 hours post co-culture. (C,D) Gene Set Enrichment Analysis comparing C.TNC-CAR.Zip18R T cells to C.TNC-CAR T cells at (C) baseline or (D) 48 hours post stimulation with LM7.GFP.ffLuc cells. Top 10 activated or suppressed significant (p.adjust<0.1) KEGG pathways are shown. (E–I) 1×106 NT, Zip18R, C.TNC-CAR, and C.TNC-CAR.Zip18R T cells were cultured in media or against LM7 cells at a 2:1 effector to target ratio for 48 hours and then collected for flow cytometric analysis. (E) Transduction (TDX) percentage of NT, Zip18R, C.TNC-CAR, and C.TNC-CAR.Zip18R T cells alone and after 48 hours of stimulation. CD3+ expression is shown for NT samples. (n=3, mean+SEM), two-way ANOVA, ****p<0.0001. (F–I) Expression within pure isolated populations of T cells gated on CD3+ (NT), mClover+ (Zip18R+), (G4S)3+ (C.TNC-CAR+), and (G4S)3+ mClover+ (C.TNC-CAR+Zip18R+) for (F) CD69, (G) CD28, (H) CD39, and (I) TIM3 (n=3, mean+SEM), two-way ANOVA, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Only showing significance values for each cohort comparing Alone versus +LM7 and C.TNC-CAR versus C.TNC-CAR.Zip18R for both conditions. All p values are reported in online supplemental SFigure 16. ANOVA, analysis of variance; CAR, chimeric antigen receptor; C.TNC, tenascin C encoding the C domain; ffLuc, firefly luciferase; GFP, green fluorescent protein; GM-CSF, granulocyte-macrophage colony-stimulating factor; IL, interleukin; KEGG, Kyoto Encylopedia of Genes and Genomes; NT, non-transduced; TNF, tumor necrosis factor; Zip18R, interleukin-18 receptor-based leucine zipper receptor.
We next used Gene Set Enrichment Analysis to uncover which pathways were activated by Zip18R expression in C.TNC-CAR T cells. At baseline, we found that both donors had IL-17 signaling and cytokine–cytokine receptor interaction pathways upregulated in C.TNC-CAR.Zip18R T cells compared with C.TNC-CAR T cells (figure 5C). Those pathways were also upregulated after 48 hours of antigen stimulation (figure 5D). In addition, for one donor, there were several immune-related disease pathways upregulated in C.TNC-CAR.Zip18R T cells (figure 5C,D (Donor B)). We next evaluated genes involved in MyD88 signaling, T-cell activation, and T-cell exhaustion at baseline and over the time course of antigen stimulation (online supplemental SFigure 15). IL-2RA gene expression was upregulated in C.TNC-CAR.Zip18R T cells at both baseline and over the antigen stimulation course, and IFN-γ gene expression was elevated at baseline for both donors. We did not see significant changes in gene expression for other MyD88-signaling-related genes. At baseline, TIGIT (Donor A) and HAVCR2 (TIM3) (Donor B) gene expression was significantly increased in C.TNC-CAR.Zip18R T cells. Post antigen stimulation, this only persisted for TIM3 at the 48-hour time point (online supplemental SFigure 15).
To confirm our gene expression findings, we used flow cytometry to evaluate three different donors after 48 hours of culture in cytokine-starved media or co-cultured with LM7 cells. We again found that C.TNC-CAR and C.TNC-CAR.Zip18R T cells populations enriched after antigen stimulation (figure 5E). Activation markers CD69 and CD28 were similar across all four T-cell populations at baseline, suggesting that Zip18R constitutive signaling does not alter baseline T-cell activation. After antigen stimulation, we observed a significant increase in CD69-positive C.TNC-CAR and C.TNC-CAR.Zip18R T cells, and only a significant increase in CD28-positive C.TNC-CAR T cells (figure 5F,G). For TIM3 and CD39, markers associated with T-cell exhaustion, we found at baseline that C.TNC-CAR.Zip18R T cells had significantly greater populations of TIM3-positive and CD39-positive T cells compared with C.TNC-CAR T cells (figure 5H,I). On co-culture with LM7 cells, the frequency of TIM3-positive and CD39-positive T cells increased in C.TNC-CAR and C.TNC-CAR.Zip18R T cells with no significant differences between both T-cell populations. We also evaluated lymphocyte activation gene 3 (LAG3), programmed cell death protein 1 (PD-1), and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) expression, which had much lower expression levels (online supplemental SFigure 16). C.TNC-CAR and C.TNC-CAR.Zip18R T cells followed similar trends, with only C.TNC-CAR T cells having an increase in CTLA-4 expression after antigen stimulation, mirroring the CD28 expression data.
Zip18R improves the antitumor activity of C.TNC-CAR T cells in vivo
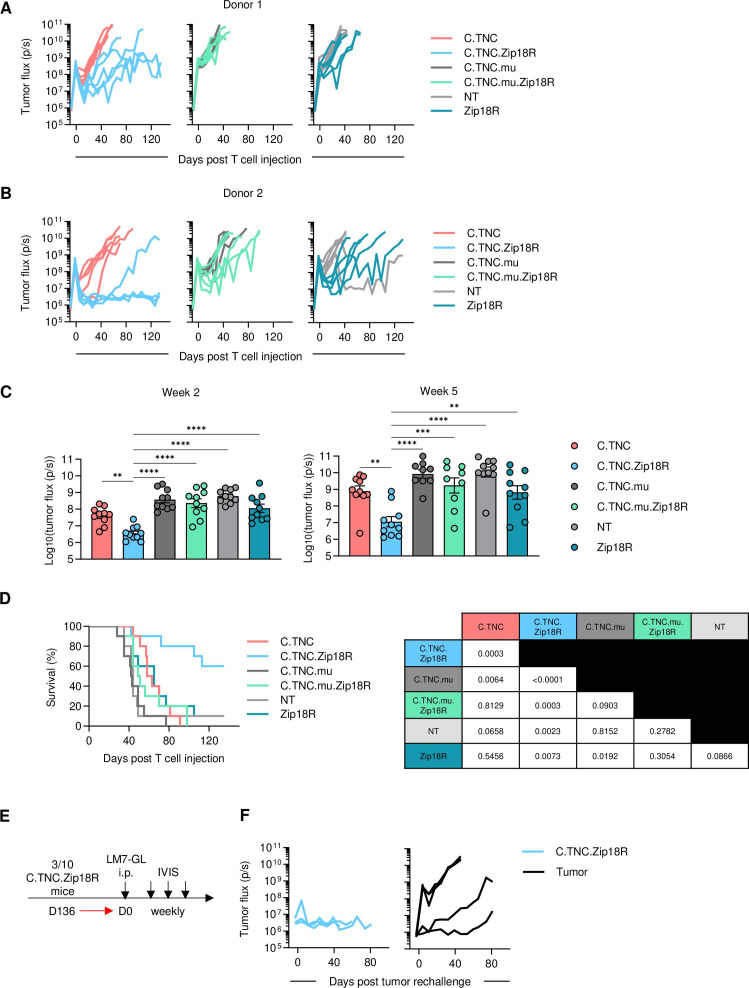
Having established that Zip18R improves the effector function of C.TNC-CAR T cells in vitro, we evaluated if this translates into improved antitumor activity in vivo (figure 6A,B). In the LM7.GFP.ffLuc model, C.TNC-CAR.Zip18R T-cell infusions were well tolerated, and mice continued to gain weight at the same rate as mice that received other CAR or NT T-cell populations (online supplemental SFigure 17). C.TNC-CAR.Zip18R T cells had significantly greater antitumor activity compared with the C.TNC-CAR T cells (figure 6C; online supplemental SFigures 18 and 19), resulting in a significant survival advantage (figure 6D). The gender of mice did not impact the observed benefit in antitumor activity of C.TNC-CAR.Zip18R T cells (online supplemental SFigure 20). The improvement in antitumor activity was dependent on the expression of a functional CAR in T cells since Zip18R expression in C.TNC.mu-CAR or NT T cells did not improve their antitumor activity. Three of the C.TNC-CAR.Zip18R T-cell treated mice (figure 6B) had tumor flux values that returned to baseline. On day 136, we rechallenged these mice with another i.p. injection of LM7.GFP.ffLuc cells to evaluate for functional C.TNC-CAR.Zip18R T-cell persistence (figure 6E). All three mice rejected the tumor, while in control, untreated mice, LM7.GFP.ffLuc tumor cells grew (figure 6F).
Figure 6. C.TNC-CAR.Zip18R T cells have superior antitumor activity in vivo. 1×106 LM7.GFP.ffLuc cells were injected into 5–6-week-old female (Donor 1) or male (Donor 2) NSG mice. Seven days later, mice received an i.p. injection of 1×106 sorted T cells. (A,B) Tumor flux values measured by IVIS imaging. (A) Donor 1, (B) Donor 2 (n=5 per cohort per donor). (C) Combined tumor flux values from both donors at weeks 2 and 5 (n=10, mean+SEM), one-way analysis of variance, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (D) Kaplan-Meier survival curve, Mantel-Cox test. P values listed in the table. (E) Schematic of rechallenge experiment. 5–6-week-old male mice were used for tumor only cohort. 1×106 LM7.GFP.ffLuc cells were injected i.p. into naïve mice or C.TNC-CAR.Zip18R T-cell pretreated mice. (F) Tumor flux values measured by IVIS imaging (n=3–5). CAR, chimeric antigen receptor; C.TNC, tenascin C encoding the C domain; ffLuc, firefly luciferase; GFP, green fluorescent protein; i.p., intraperitoneal; NT, non-transduced; Zip18R, interleukin-18 receptor-based leucine zipper receptor.
We next evaluated if Zip18R could enhance C.TNC-CAR T-cell antitumor activity against DIPG007.YFP.ffLuc i.c. tumors. After 2 weeks post T-cell injections, both C.TNC-CAR and C.TNC-CAR.Zip18R T cells had a significant reduction in tumor burden as compared with NT-treated mice (online supplemental SFigure 21). We found that Zip18R enhanced C.TNC-CAR activity, with a significant difference in tumor flux values between C.TNC-CAR and C.TNC-CAR.Zip18R T-cell treated mice emerging after 3 weeks. However, in this model, 6/15 C.TNC-CAR.Zip18R T-cell treated mice needed to be euthanized in the first week post T-cell injections despite having low tumor flux values.
Discussion
Here, we show that the C domain of TNC is expressed by pediatric solid and brain tumor cells and is also present within the microenvironment of the bulky primary tumor samples. T cells expressing a CAR against C.TNC recognized and killed tumor cell lines in an antigen-dependent fashion and had limited antitumor activity in vivo. While modification of the C.TNC-CAR design did not further improve C.TNC-CAR T-cell activity, expressing a novel constitutively active IL-18 receptor (Zip18R) in C.TNC-CAR T cells significantly improved their effector function, resulting in improved antitumor activity in vivo.
Identifying suitable antigens for solid tumor and brain tumors for CAR T-cell therapy remains a high priority. CAR T cells typically target surface proteins on tumor cells, but these antigens may be heterogeneously expressed or expressed at low levels.49 ECM proteins provide a unique class of target antigens for cell therapy because they are secreted and can bind back to cells within the microenvironment, including bystander cells.50 Additionally, tumor and associated stromal cells can generate cancer-specific ECMs by differentially expressing ECM proteins or secreting alternatively spliced isoforms.8 Indeed, investigators have leveraged tumor-specific ECM proteins as CAR T-cell therapy targets. CAR T cells directed to COL11A1, a collagen protein upregulated in breast cancer and pediatric OS, had robust antitumor activity in human xenograft models.8 51 The extra domains A and B of fibronectin have also been targeted with CAR T cells in preclinical models, demonstrating safety and efficacy.4,752 53
The long isoforms of TNC have been described in adult cancer and present alternatively spliced ECM protein targets.10 38 54 mAbs against the A1 domain (F16), A3/A4/B domains (BC-2), A4/B domains (ST2485), and C/D domains (81C6) of oncofetal TNC conjugated to either IL-2 or radioisotopes have been evaluated in preclinical models, demonstrating tumor localization, safety, and antitumor activity.20 In clinical trials, administration of F16-IL-2, 131I-BC-2, or 131I-81C6 mAb conjugates was safe and associated with promising antitumor activity for metastatic breast cancer and glioblastoma.55,60 We show here that pediatric solid and brain tumors also express the C domain of TNC, and that it is not expressed at the RNA level in non-neoplastic, non-diseased tissues.
We generated T cells expressing a C.TNC-CAR, that consisted of an antigen binding domain derived from the G11 mAb and included a CD28ζ signaling domain.38 C.TNC-CAR T cells recognized and killed C.TNC-expressing tumor cells. However, C.TNC-CAR T cells had limited antitumor activity in xenograft models, and we sought to optimize the CAR design, as the CAR structure is important to its function.61 62 However, none of the alternative CAR designs improved the antitumor activity of C.TNC-CAR T cells. In addition to CARs, several synthetic T-cell receptors (TCRs) have been developed, including synthetic TCRs and antigen receptor and human leukocyte antigen (HLA)-independent T-cell receptors,63 64 which have a higher sensitivity than standard CARs. These receptors could be explored in future studies to evaluate if they improve the effector function of C.TNC-CAR T cells. Here, we decided to focus on improving C.TNC-CAR T-cell effector function via strengthening signal 3.53
IL-18 is part of the IL-1 family of cytokines and activates MyD88 signaling.65 Several groups have demonstrated that transgenic expression of IL-18 or a Fab-based constitutively active IL-18 receptor expression bolstered the antitumor activity of CAR T cells, translating into increased survival in preclinical tumor models,34 36 66 and early phase clinical testing is in progress (NCT04684563).67 However, safety concerns were raised due to weight loss and antigen-independent T-cell expansion in preclinical models.34 35 66 To remedy this potential toxicity concern, we and other investigators have explored activation-dependent IL-18 expression either by using activation-dependent promoters or through a GM18 chimeric cytokine receptor, which signals once GM-CSF is produced by activated T cells.33 35 68
We first investigated if our GM18 receptor could bolster the antitumor activity of C.TNC-CAR T cells. No significant benefit was observed, indicating that C.TNC-CAR.GM18 T cells most likely do not produce enough GM-CSF to induce robust GM18 triggering. To overcome this limitation, we designed a constitutively active cytokine receptor for IL-18 based on our modular ZipR platform to create Zip18R. We created Zip18Rs with one (1X) or two (2X) pairs of leucine zippers, and both activated MyD88 signaling pathways and improved the antitumor activity of EphA2-CAR T cells. This is in contrast to JAK/STAT ZipRs, which require two pairs of leucine zippers,37 indicating the leucine zippers need to be tailored to their cytoplasmic signaling domain. In vitro, Zip18R overall did not improve or decrease the ability of C.TNC-CAR T cells to produce cytokines or chemokines after the first stimulation. However, there were two notable exceptions: C.TNC-CAR.Zip18R T cells produced lower amounts of IL-10 and increased amounts of IL-17A and IL-17F, the latter being consistent with Zip18R-induced MyD88 signaling.3469,71 After the fourth stimulation, type 1 cytokine production was significantly reduced by C.TNC-CAR T cells whereas it was maintained by C.TNC-CAR.Zip18R T cells. Maintenance of the effector function by Zip18R was also evident by a significantly greater expansion of C.TNC-CAR.Zip18R T cells compared with C.TNC-CAR T cells in repeat stimulation assays.
We next evaluated the transcriptional changes that occur in C.TNC-CAR T cells when Zip18R is expressed. Zip18R signaling led to significant upregulation of IL-17 and cytokine–cytokine receptor pathways at baseline and with antigen stimulation, validating our findings from our cytokine multiplex analysis. We noted that for one donor, there were several immune-related disease pathways upregulated. Further evaluation of these disease types shows related pathways such as toll-like receptor signaling, Th17 cell differentiation, cytokine–cytokine receptor signaling, and leukocyte transendothelial migration, which are all hallmarks of T cell and MyD88 signaling. Importantly, we found that Zip18R signaling may contribute to a more activated or more exhausted T-cell phenotype when compared with C.TNC-CAR T cells at baseline; however, differences are not sustained in the setting of antigen stimulation. Thus, our data suggests that Zip18R signaling changes type1/type2/type17 differentiation, resulting in improved effector function.
We found that C.TNC-CAR.Zip18R T-cell safety in vivo was model-dependent. Against LM7.GFP.ffLuc tumors, mice treated with C.TNC-CAR.Zip18R T cells did not show overt toxicity as judged by clinical behavior and weight measurements, and only 1/10 mice died 5 weeks post C.TNC-CAR.Zip18R T-cell therapy without high tumor burden or clinical signs of graft versus host disease. With three mice showing tumor control after rechallenging, this data suggests that C.TNC-CAR.Zip18R T cells were able to persist long-term. Tumors harvested from mice who reached euthanasia requirement were still C.TNC-positive (data not shown). Thus, additional studies are warranted to explore if further genetic modifications of C.TNC-CAR.Zip18R T cells could improve their efficacy, including deleting epigenetic regulators such as DNMT3A or Suv39h1.72 73
In contrast to our LM7.GFP.ffLuc studies, C.TNC-CAR.Zip18R T cells were toxic in a subset of animals after i.c. injection into DIPG007.YFP.ffLuc tumors. Based on the performed IHC analysis, which demonstrated TNC expression only at tumor sites within in the brain, on target/off cancer toxicity is unlikely. Side effects of T cell-induced inflammation within the brain is one likely explanation, which has been observed in mice as well as in humans after the locoregional delivery of CAR T cells.74 75 Clearly, future studies are needed to understand the underlying mechanism and develop genetic engineering approaches to control Zip18R expression in C.TNC-CAR T cells or include safety switches to improve the safety of C.TNC-CAR.Zip18R T cells for the immunotherapy of brain tumors.
In summary, our study identifies the C domain of the ECM protein TNC as a promising CAR T-cell therapy for pediatric solid tumors and brain tumors. C.TNC-CAR T cells expressing a constitutively Zip18R had significant antitumor activity. Although our work focuses on developing CAR T-cell therapies for pediatric cancers, our work has relevance to a broad range of adult cancers, which express C.TNC.
supplementary material
Acknowledgements
The authors would like to thank the members of the department of bone marrow transplantation and cellular therapy for their helpful discussion. We thank the research staff of St. Jude Centers and Cores for their assistance in the conducted experiments. These Centers and Cores include the St. Jude Center for In Vivo Imaging and Therapeutics, Flow Cytometry Core, and Comparative Pathology Core, which are in part supported by National Cancer Institute (NCI) grant P30CA021765. We would like to thank Carmen Coleman for assistance with the in vivo mouse studies, and Liusheng He, Sandy Schwemberger, and Stacie Woolard with flow sorting and/or flow cytometry. In addition, we would like to thank the Center for Translational Immunology and Immunotherapy (CeTI2) and the Center of Excellence for Pediatric Immuno-Oncology (CEPIO) for their support. Figures 2A, 3A, and 4B, and supplemental figure 4F were created with BioRender (Biorender.com), for which we have a license.
Footnotes
Funding: This work was supported by NCI grant (1F31CA257757) to EW, Alex Lemonade Stand Foundation (ALSF) and Cure4Cam Foundation Young Investigator Grant to JW, NCI grant (F31CA250401-01A1) to MB, ALSF grant to SG and GK, NCI grants (R01NS121249, R01NS122859) to GK, the Alliance for Cancer Gene Therapy (ACGT) grant to SG, and the American Lebanese Syrian Associated Charities to GK, JZ, and SG. St. Jude Centers and Cores, including the Center for In Vivo Imaging and Therapeutics, Flow Cytometry Core, and Comparative Pathology Core, are in part supported by National Cancer Institute (NCI) grant P30CA021765. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institutes of Health.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: This study involves human participants and was approved by St. Jude Children’s Research Hospital Institutional Review Board (NCT01050296). Participants gave informed consent to participate in the study before taking part.
Data availability free text: Single-cell RNA sequencing data will be deposited to the European Genome-phenome Archive (EGA) once the manuscript is accepted for publication.
Contributor Information
Elizabeth Wickman, Email: elizabeth.wickman@stjude.org.
Shannon Lange, Email: srapovy@gmail.com.
Jessica Wagner, Email: jessica.wagner@stjude.org.
Jorge Ibanez, Email: JorgeAndres.Ibanez@STJUDE.ORG.
Liqing Tian, Email: liqing.tian@stjude.org.
Meifen Lu, Email: meifen.lu@stjude.org.
Heather Sheppard, Email: heather.tillman@stjude.org.
Jason Chiang, Email: jason.chiang@stjude.org.
Selene C Koo, Email: selene.koo@stjude.org.
Peter Vogel, Email: peter.vogel@stjude.org.
Deanna Langfitt, Email: deanna.langfitt@stjude.org.
S Scott Perry, Email: scott.perry@stjude.org.
Raghuvaran Shanmugam, Email: raghuvaran.shanmugam@stjude.org.
Matthew Bell, Email: matthew.bell@chuv.ch.
Timothy I Shaw, Email: Timothy.Shaw@moffitt.org.
Giedre Krenciute, Email: Giedre.Krenciute@STJUDE.ORG.
Jinghui Zhang, Email: jinghui.zhang@stjude.org.
Stephen Gottschalk, Email: stephen.gottschalk@stjude.org.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1.Globerson Levin A, Rivière I, Eshhar Z, et al. CAR T cells: Building on the CD19 paradigm. Eur J Immunol. 2021;51:2151–63. doi: 10.1002/eji.202049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner J, Wickman E, DeRenzo C, et al. CAR T Cell Therapy for Solid Tumors: Bright Future or Dark Reality? Mol Ther. 2020;28:2320–39. doi: 10.1016/j.ymthe.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labanieh L, Mackall CL. CAR immune cells: design principles, resistance and the next generation. Nat New Biol. 2023;614:635–48. doi: 10.1038/s41586-023-05707-3. [DOI] [PubMed] [Google Scholar]
- 4.Martín-Otal C, Lasarte-Cia A, Serrano D, et al. Targeting the extra domain A of fibronectin for cancer therapy with CAR-T cells. J Immunother Cancer. 2022;10:e004479. doi: 10.1136/jitc-2021-004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie YJ, Dougan M, Jailkhani N, et al. Nanobody-based CAR T cells that target the tumor microenvironment inhibit the growth of solid tumors in immunocompetent mice. Proc Natl Acad Sci U S A. 2019;116:7624–31. doi: 10.1073/pnas.1817147116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Liu C, Yang Z, et al. CAR-T-Cell Therapy for Solid Tumors Positive for Fibronectin Extra Domain B. Cells. 2022;11:2863. doi: 10.3390/cells11182863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner J, Wickman E, Shaw TI, et al. Antitumor Effects of CAR T Cells Redirected to the EDB Splice Variant of Fibronectin. Cancer Immunol Res. 2021;9:279–90. doi: 10.1158/2326-6066.CIR-20-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw TI, Wagner J, Tian L, et al. Discovery of immunotherapy targets for pediatric solid and brain tumors by exon-level expression. Nat Commun. 2024;15:3732. doi: 10.1038/s41467-024-47649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marzeda AM, Midwood KS. Internal Affairs: Tenascin-C as a Clinically Relevant, Endogenous Driver of Innate Immunity. J Histochem Cytochem. 2018;66:289–304. doi: 10.1369/0022155418757443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giblin SP, Midwood KS. Tenascin-C: Form versus function. Cell Adh Migr. 2015;9:48–82. doi: 10.4161/19336918.2014.987587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov. 2011;10:767–77. doi: 10.1038/nrd3554. [DOI] [PubMed] [Google Scholar]
- 12.Borsi L, Allemanni G, Gaggero B, et al. Extracellular pH controls pre-mRNA alternative splicing of tenascin-C in normal, but not in malignantly transformed, cells. Int J Cancer. 1996;66:632–5. doi: 10.1002/(SICI)1097-0215(19960529)66:5<632::AID-IJC9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Jensen MA, Wilkinson JE, Krainer AR. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat Struct Mol Biol. 2014;21:189–97. doi: 10.1038/nsmb.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan L, Yu W, Shen E, et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2019;68:118–29. doi: 10.1136/gutjnl-2017-314983. [DOI] [PubMed] [Google Scholar]
- 15.LaFleur DW, Fagin JA, Forrester JS, et al. Cloning and characterization of alternatively spliced isoforms of rat tenascin. Platelet-derived growth factor-BB markedly stimulates expression of spliced variants of tenascin mRNA in arterial smooth muscle cells. J Biol Chem. 1994;269:20757–63. [PubMed] [Google Scholar]
- 16.Tucker RP, Chiquet-Ehrismann R. The regulation of tenascin expression by tissue microenvironments. Biochim Biophys Acta. 2009;1793:888–92. doi: 10.1016/j.bbamcr.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Hancox RA, Allen MD, Holliday DL, et al. Tumour-associated tenascin-C isoforms promote breast cancer cell invasion and growth by matrix metalloproteinase-dependent and independent mechanisms. Breast Cancer Res . 2009;11:R24. doi: 10.1186/bcr2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parekh K, Ramachandran S, Cooper J, et al. Tenascin-C, over expressed in lung cancer down regulates effector functions of tumor infiltrating lymphocytes. Lung Cancer. 2005;47:17–29. doi: 10.1016/j.lungcan.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Chung CY, Murphy-Ullrich JE, Erickson HP. Mitogenesis, cell migration, and loss of focal adhesions induced by tenascin-C interacting with its cell surface receptor, annexin II. Mol Biol Cell. 1996;7:883–92. doi: 10.1091/mbc.7.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Wu Y, Wang J, et al. Clinical advances in TNC delivery vectors and their conjugate agents. Pharmacol Ther. 2024;253:108577. doi: 10.1016/j.pharmthera.2023.108577. [DOI] [PubMed] [Google Scholar]
- 21.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–9. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 22.Du L, Nai Y, Shen M, et al. IL-21 Optimizes the CAR-T Cell Preparation Through Improving Lentivirus Mediated Transfection Efficiency of T Cells and Enhancing CAR-T Cell Cytotoxic Activities. Front Mol Biosci. 2021;8:675179. doi: 10.3389/fmolb.2021.675179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo H, Su J, Sun R, et al. Coexpression of IL7 and CCL21 Increases Efficacy of CAR-T Cells in Solid Tumors without Requiring Preconditioned Lymphodepletion. Clin Cancer Res. 2020;26:5494–505. doi: 10.1158/1078-0432.CCR-20-0777. [DOI] [PubMed] [Google Scholar]
- 24.Shum T, Omer B, Tashiro H, et al. Constitutive Signaling from an Engineered IL7 Receptor Promotes Durable Tumor Elimination by Tumor-Redirected T Cells. Cancer Discov. 2017;7:1238–47. doi: 10.1158/2159-8290.CD-17-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi K, Kano Y, Nagai T, et al. IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat Biotechnol. 2018;36:346–51. doi: 10.1038/nbt.4086. [DOI] [PubMed] [Google Scholar]
- 26.Krenciute G, Prinzing BL, Yi Z, et al. Transgenic Expression of IL15 Improves Antiglioma Activity of IL13Rα2-CAR T Cells but Results in Antigen Loss Variants. Cancer Immunol Res. 2017;5:571–81. doi: 10.1158/2326-6066.CIR-16-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H, Li A, Dai Z, et al. IL-15 armoring enhances the antitumor efficacy of claudin 18.2-targeting CAR-T cells in syngeneic mouse tumor models. Front Immunol. 2023;14 doi: 10.3389/fimmu.2023.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J, Liu Q, He Y, et al. Combined therapy of CAR-IL-15/IL-15Rα-T cells and GLIPR1 knockdown in cancer cells enhanced anti-tumor effect against gastric cancer. J Transl Med. 2024;22:171. doi: 10.1186/s12967-024-04982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alizadeh D, Wong RA, Yang X, et al. IL15 Enhances CAR-T Cell Antitumor Activity by Reducing mTORC1 Activity and Preserving Their Stem Cell Memory Phenotype. Cancer Immunol Res. 2019;7:759–72. doi: 10.1158/2326-6066.CIR-18-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zannikou M, Duffy JT, Levine RN, et al. IL15 modification enables CAR T cells to act as a dual targeting agent against tumor cells and myeloid-derived suppressor cells in GBM. J Immunother Cancer. 2023;11:e006239. doi: 10.1136/jitc-2022-006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gargett T, Ebert LM, Truong NTH, et al. GD2-targeting CAR-T cells enhanced by transgenic IL-15 expression are an effective and clinically feasible therapy for glioblastoma. J Immunother Cancer. 2022;10:e005187. doi: 10.1136/jitc-2022-005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prinzing B, Schreiner P, Bell M, et al. MyD88/CD40 signaling retains CAR T cells in a less differentiated state. JCI Insight. 2020;5:e136093. doi: 10.1172/jci.insight.136093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange S, Sand LGL, Bell M, et al. A Chimeric GM-CSF/IL18 Receptor to Sustain CAR T-cell Function. Cancer Discov. 2021;11:1661–71. doi: 10.1158/2159-8290.CD-20-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Righi M, Gannon I, Robson M, et al. Enhancing CAR T-cell Therapy Using Fab-Based Constitutively Heterodimeric Cytokine Receptors. Cancer Immunol Res. 2023;11:1203–21. doi: 10.1158/2326-6066.CIR-22-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer-Riepe L, Kailayangiri S, Zimmermann K, et al. Preclinical Development of CAR T Cells with Antigen-Inducible IL18 Enforcement to Treat GD2-Positive Solid Cancers. Clin Cancer Res. 2024;30:3564–77. doi: 10.1158/1078-0432.CCR-23-3157. [DOI] [PubMed] [Google Scholar]
- 36.Jaspers JE, Khan JF, Godfrey WD, et al. IL-18-secreting CAR T cells targeting DLL3 are highly effective in small cell lung cancer models. J Clin Invest. 2023;133:e166028. doi: 10.1172/JCI166028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell M, Lange S, Sejdiu BI, et al. Modular chimeric cytokine receptors with leucine zippers enhance the antitumour activity of CAR T cells via JAK/STAT signalling. Nat Biomed Eng. 2024;8:380–96. doi: 10.1038/s41551-023-01143-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silacci M, Brack SS, Späth N, et al. Human monoclonal antibodies to domain C of tenascin-C selectively target solid tumors in vivo. Protein Eng Des Sel . 2006;19:471–8. doi: 10.1093/protein/gzl033. [DOI] [PubMed] [Google Scholar]
- 39.Stewart E, Federico SM, Chen X, et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature New Biol. 2017;549:96–100. doi: 10.1038/nature23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amezquita RA, Lun ATL, Becht E, et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods. 2020;17:137–45. doi: 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy DJ, Campbell KR, Lun ATL, et al. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–86. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hafemeister C, Halbritter F. Single-cell rna-seq differential expression tests within a sample should use pseudo-bulk data of pseudo-replicates. Bioinformatics. 2023 doi: 10.1101/2023.03.28.534443. Preprint. [DOI] [Google Scholar]
- 45.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A Journal of Integrative Biology . 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curtsinger JM, Schmidt CS, Mondino A, et al. Inflammatory Cytokines Provide a Third Signal for Activation of Naive CD4+ and CD8+ T Cells. J Immunol. 1999;162:3256–62. doi: 10.4049/jimmunol.162.6.3256. [DOI] [PubMed] [Google Scholar]
- 47.Fraietta JA, Lacey SF, Orlando EJ, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24:563–71. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chow KKH, Naik S, Kakarla S, et al. T cells redirected to EphA2 for the immunotherapy of glioblastoma. Mol Ther. 2013;21:629–37. doi: 10.1038/mt.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majzner RG, Mackall CL. Tumor Antigen Escape from CAR T-cell Therapy. Cancer Discov. 2018;8:1219–26. doi: 10.1158/2159-8290.CD-18-0442. [DOI] [PubMed] [Google Scholar]
- 50.Dreyling M, Dickinson M, Martinez Lopez J, et al. Long-Term Clinical Outcomes and Correlative Efficacy Analyses in Patients (Pts) with Relapsed/Refractory Follicular Lymphoma (r/r FL) Treated with Tisagenlecleucel in the Elara Trial. Blood. 2022;140:1459–63. doi: 10.1182/blood-2022-158024. [DOI] [Google Scholar]
- 51.Nallanthighal S, Heiserman JP, Cheon D-J. Collagen Type XI Alpha 1 (COL11A1): A Novel Biomarker and a Key Player in Cancer. Cancers (Basel) 2021;13:935. doi: 10.3390/cancers13050935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang J, Liu N, Zhu Y, et al. CAR-T Therapy Targets Extra Domain B of Fibronectin Positive Solid Tumor Cells. Immunol Invest. 2023;52:985–96. doi: 10.1080/08820139.2023.2264332. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Liu C, Wang M, et al. Treating solid tumors with TCR-based chimeric antigen receptor targeting extra domain B-containing fibronectin. J Immunother Cancer. 2023;11:e007199. doi: 10.1136/jitc-2023-007199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brack SS, Silacci M, Birchler M, et al. Tumor-targeting properties of novel antibodies specific to the large isoform of tenascin-C. Clin Cancer Res. 2006;12:3200–8. doi: 10.1158/1078-0432.CCR-05-2804. [DOI] [PubMed] [Google Scholar]
- 55.Pedretti M, Verpelli C, Mårlind J, et al. Combination of temozolomide with immunocytokine F16-IL2 for the treatment of glioblastoma. Br J Cancer. 2010;103:827–36. doi: 10.1038/sj.bjc.6605832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zalutsky MR, Reardon DA, Akabani G, et al. Clinical experience with alpha-particle emitting 211At: treatment of recurrent brain tumor patients with 211At-labeled chimeric antitenascin monoclonal antibody 81C6. J Nucl Med. 2008;49:30–8. doi: 10.2967/jnumed.107.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Catania C, Maur M, Berardi R, et al. The tumor-targeting immunocytokine F16-IL2 in combination with doxorubicin: dose escalation in patients with advanced solid tumors and expansion into patients with metastatic breast cancer. Cell Adh Migr. 2015;9:14–21. doi: 10.4161/19336918.2014.983785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riva P, Arista A, Sturiale C, et al. Treatment of intracranial human glioblastoma by direct intratumoral administration of 131I-labelled anti-tenascin monoclonal antibody BC-2. Int J Cancer. 1992;51:7–13. doi: 10.1002/ijc.2910510103. [DOI] [PubMed] [Google Scholar]
- 59.Riva P, Arista A, Tison V, et al. Intralesional radioimmunotherapy of malignant gliomas. An effective treatment in recurrent tumors. Cancer. 1994;73:1076–82. doi: 10.1002/1097-0142(19940201)73:3+<1076::aid-cncr2820731347>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 60.Riva P, Arista A, Franceschi G, et al. Local treatment of malignant gliomas by direct infusion of specific monoclonal antibodies labeled with 131I: comparison of the results obtained in recurrent and newly diagnosed tumors. Cancer Res. 1995;55:5952s–6s. [PubMed] [Google Scholar]
- 61.Alabanza L, Pegues M, Geldres C, et al. Function of Novel Anti-CD19 Chimeric Antigen Receptors with Human Variable Regions Is Affected by Hinge and Transmembrane Domains. Mol Ther. 2017;25:2452–65. doi: 10.1016/j.ymthe.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jayaraman J, Mellody MP, Hou AJ, et al. CAR-T design: Elements and their synergistic function. EBioMedicine. 2020;58:102931. doi: 10.1016/j.ebiom.2020.102931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mansilla-Soto J, Eyquem J, Haubner S, et al. HLA-independent T cell receptors for targeting tumors with low antigen density. Nat Med. 2022;28:345–52. doi: 10.1038/s41591-021-01621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Liu G, Wang J, et al. Chimeric STAR receptors using TCR machinery mediate robust responses against solid tumors. Sci Transl Med. 2021;13:eabb5191. doi: 10.1126/scitranslmed.abb5191. [DOI] [PubMed] [Google Scholar]
- 65.Rex DAB, Agarwal N, Prasad TSK, et al. A comprehensive pathway map of IL-18-mediated signalling. J Cell Commun Signal. 2020;14:257–66. doi: 10.1007/s12079-019-00544-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu B, Ren J, Luo Y, et al. Augmentation of Antitumor Immunity by Human and Mouse CAR T Cells Secreting IL-18. Cell Rep. 2017;20:3025–33. doi: 10.1016/j.celrep.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Svoboda J, Gerson JN, Landsburg DJ, et al. Interleukin-18 Secreting Autologous Anti-CD19 CAR T-Cells (huCART19-IL18) in Patients with Non-Hodgkin Lymphomas Relapsed or Refractory to Prior CAR T-Cell Therapy. Blood. 2022;140:4612–4. doi: 10.1182/blood-2022-162393. [DOI] [Google Scholar]
- 68.Chmielewski M, Abken H. CAR T Cells Releasing IL-18 Convert to T-Bethigh FoxO1low Effectors that Exhibit Augmented Activity against Advanced Solid Tumors. Cell Rep. 2017;21:3205–19. doi: 10.1016/j.celrep.2017.11.063. [DOI] [PubMed] [Google Scholar]
- 69.Deason K, Troutman TD, Jain A, et al. BCAP links IL-1R to the PI3K-mTOR pathway and regulates pathogenic Th17 cell differentiation. J Exp Med. 2018;215:2413–28. doi: 10.1084/jem.20171810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Gao L, Lei L, et al. A MyD88-dependent early IL-17 production protects mice against airway infection with the obligate intracellular pathogen Chlamydia muridarum. J Immunol. 2009;183:1291–300. doi: 10.4049/jimmunol.0803075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keestra AM, Godinez I, Xavier MN, et al. Early MyD88-dependent induction of interleukin-17A expression during Salmonella colitis. Infect Immun. 2011;79:3131–40. doi: 10.1128/IAI.00018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prinzing B, Zebley CC, Petersen CT, et al. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci Transl Med. 2021;13:eabh0272. doi: 10.1126/scitranslmed.abh0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.López-Cobo S, Fuentealba JR, Gueguen P, et al. SUV39H1 Ablation Enhances Long-term CAR T Function in Solid Tumors. Cancer Discov. 2024;14:120–41. doi: 10.1158/2159-8290.CD-22-1350. [DOI] [PubMed] [Google Scholar]
- 74.Mount CW, Majzner RG, Sundaresh S, et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat Med. 2018;24:572–9. doi: 10.1038/s41591-018-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahdi J, Dietrich J, Straathof K, et al. Tumor inflammation-associated neurotoxicity. Nat Med. 2023;29:803–10. doi: 10.1038/s41591-023-02276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data generated in this study has been deposited in the European Genome-Phenome Archive (EGA) under the title “Single-cell RNA sequencing of human IL-18R supported CAR T cells targeting oncofetal Tenascin C”. The raw data can be obtained by a request to EGA. The scRNA-seq expression matrices are available on Gene Expression Omnibus (GEO) under ascension code GSE282046.
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.