Abstract
Background
Gastric carcinomas (GC) are aggressive malignancies, and only ~15% of patients respond to anti-programmed cell death (ligand) 1 (PD-(L)1) monotherapy. However, Epstein-Barr virus (EBV)-associated GCs (~5–10% of GCs) often harbor PD-L1 and PD-L2 chromosomal amplifications and robust CD8+ T cell infiltrates, and respond at a high rate to anti-PD-1. The current study compares the tumor immune microenvironments (TiMEs) of EBV+ versus EBV(−) GCs.
Methods
Over 1000 cases of primary invasive GCs were screened to identify 25 treatment-naïve specimens for study (11 EBV+, 14 EBV(−)). Quantitative immunohistochemistry (IHC) was conducted for markers of immune cell subsets and co-regulatory molecules. Gene expression profiling (GEP) was performed on RNAs isolated from macrodissected areas of CD3+ T cell infiltrates abutting PD-L1+ stromal/tumor cells, using multiplex quantitative reverse transcriptase PCR for a panel of 122 candidate immune-related genes.
Results
IHC revealed that 17/25 GCs contained PD-L1+ stromal cells, with no significant difference between EBV+/- specimens; however, only 3/25 specimens (all EBV+) contained PD-L1+ tumor cells. CD8+ T cell densities were higher in EBV+ versus EBV(−) tumors (p=0.044). With GEP normalized to the pan-leukocyte marker PTPRC/CD45, EBV+ GCs overexpressed ITGAE (CD103, marking intraepithelial T cells and a dendritic cell subset) and the interferon-inducible genes CXCL9 and IDO1. In contrast, EBV(−) tumors overexpressed several functionally-related gene groups associated with myeloid cells (CD163, IL1A, NOS2, RIGI), immunosuppressive cytokines/chemokines (CXCL2, CXCR4, IL10, IL32), coinhibitory molecules (HAVCR2/TIM-3 and VSIR/VISTA), and adenosine pathway components (ENTPD1/ CD39 and NT5E/CD73). Notably, compared with EBV+ GCs, EBV(−) GCs also overexpressed components of the cyclooxygenase 2 (COX-2)/prostaglandin E2 (PGE2) pathway associated with cancer-promoting inflammation, including PTGS2/COX-2 (most highly upregulated gene, 32-fold, p=0.005); prostaglandin receptors PTGER1 (EP1; up 21-fold, p=0.015) and PTGER4 (EP4; up twofold, p=0.022); and the major COX-2-inducing cytokine IL1B (up 11-fold, p=0.019). Consistent with these findings, COX-2 protein expression trended higher in EBV(−) versus EBV+ GCs (p=0.068).
Conclusions
While certain markers of immunosuppression are found in the GC TiME regardless of EBV status, EBV(−) GCs, which are much more common than EBV+ GCs, overexpress components of the COX-2/PGE2 pathway. These findings provide novel insights into the immune microenvironments of EBV+ and EBV(−) GC, and offer potential targets to overcome resistance to anti-PD-(L)1 therapies.
Keywords: Immunotherapy, Gastric Cancer, Tumor microenvironment - TME, Gene expression profiling - GEP
WHAT IS ALREADY KNOWN ON THIS TOPIC
Epstein-Barr virus (EBV)-associated gastric cancers (GCs), comprising ~5–10% of all GCs, are reported to have a much higher response rate to anti-programmed cell death 1 (PD-1) therapy than the more common EBV(−) GC subset.
WHAT THIS STUDY ADDS
This study comparing the tumor immune microenvironments (TiMEs) of EBV+ versus EBV(−) GCs revealed a more highly immunosuppressive TiME in EBV(−) GCs, in which upregulation of the cyclooxygenase 2 (COX-2)/prostaglandin E2 tumor-promoting pathway was a prominent feature.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These results nominate the COX-2 pathway as a co-target for enhancing the effects of anti-PD-1 immunotherapy in EBV(−) GC.
Introduction
Gastric cancer (GC) is an aggressive malignancy and is one of the most common causes of cancer-related mortality worldwide.1 Although GC can be cured in its early stages with locoregional procedures, the median survival of patients with metastatic disease receiving chemotherapy is less than 1 year.2 Fluoropyrimidine-containing and platinum-containing chemotherapies are used in the initial management of advanced metastatic GC. Recently, nivolumab, a monoclonal antibody blocking the immune inhibitory receptor programmed cell death 1 (PD-1), received Food and Drug Administration approval for first-line management of advanced or metastatic GC and gastroesophageal junction cancer (GEJC) in combination with chemotherapy, based on improvements in overall survival (OS) and progression-free survival compared with chemotherapy alone. Subsequently, the PD-1 inhibitor pembrolizumab was authorized for use with chemotherapy in advanced GC and GEJC if human epidermal growth factor 2-negative. Although these approvals established chemoimmunotherapy as a new standard of care for advanced GC, with response rates ~50%, only 10–20% of patients respond to anti-PD-1 monotherapies.3 4 Patients with GCs expressing programmed cell death ligand 1 (PD-L1) may be somewhat more likely to benefit, but the response rate to anti-PD-(L)1 monotherapy in patients with PD-L1 positive tumors remains less than 20%.4 Notably, in GCs that are associated with Epstein-Barr virus (EBV) infection (~5–10% of GCs),5 6 response rates to anti-PD-(L)1 monotherapy exceed 50%, nominating EBV positivity as a predictive biomarker for anti-PD-(L)1 response.7 8
EBV-associated GC (EBVaGC) is a distinct subtype of GC with characteristic clinicopathological and molecular features. As compared with sporadic GC, EBVaGC occurs more often in younger male individuals, frequently arises in the gastric cardia or corpus, and typically has diffuse histology and a lower frequency of lymph node metastasis.9 The precise mechanism of oncogenesis by latent EBV infection remains unclear, but EBV latent genes have previously been shown to disrupt various cellular processes and signaling pathways.10 11 Whole-genome sequencing and comprehensive molecular profiling of EBVaGC have revealed a strong association with CpG island methylator phenotype, CDKN2A/p16 gene silencing, PIK3CA mutations, and frequent amplification of JAK2, PD-L1 and PD-L2.12,15 The frequent amplification of PD-L1 and PD-L2 in EBVaGC suggests that immune evasion may be significant in EBVaGC oncogenesis.
Virus-associated cancers may have a distinct immune phenotype from their virus-negative counterparts, providing a rationale to investigate the tumor immune microenvironment (TiME) in EBVaGC and non-EBVaGC. In virus-associated cancers, tumor-specific proteins encoded by viral open reading frames may serve as strong immune stimulants, generating antigen-specific T cells but also inducing adaptive immune resistance through immune checkpoint pathways.16 For example, human papillomavirus (HPV)-positive head and neck squamous cell carcinomas (HNSCC) have a more extensive T-cell infiltrate, higher levels of immune activation, and higher expression of the immune checkpoint cytotoxic T-lymphocyte associated protein 4 than HPV-negative HNSCC.17 18 Similarly, the presence of Merkel cell polyomavirus in Merkel cell carcinoma is associated with a robust immune infiltrate and increased tumor cell PD-L1 expression.19 20 Several studies previously reported that EBVaGC has a more extensive lymphocyte infiltrate than EBV-negative (EBV(−)) GC,21,26 but trends in forkhead box P3 (FOXP3)+ cell infiltration and PD-1/PD-L1 expression are less consistent.21 Such studies have been hampered by the relative rarity of EBVaGC. Identifying unique features in the TiME of EBV(−) GC, which is much less responsive to anti-PD-(L)1 than EBVaGC, may support the development of next-generation clinical trials of treatment combinations targeting specific immune inhibitory pathways. We, therefore, undertook the current study comparing the TiMEs of EBV(−) GC versus EBVaGC, finding that EBV(−) GCs are distinguished by significant overexpression of genes associated with the cyclooxygenase 2 (COX-2) pathway, which has the potential to dampen antitumor immunity and thereby offers a potential co-target with anti-PD-(L)1-based immunotherapy.
Methods
Patients and tumor specimens
After screening over 1,000 pathology records of GCs excised at Johns Hopkins Hospital in 2000–2014, 11 EBV+ and 14 EBV(−) primary GC specimens were identified from treatment-naïve patients (online supplemental table S1). EBV status was determined by EBV-encoded RNA in situ hybridization (EBER-ISH) conducted on whole tissue sections or tissue microarrays. EBV(−) cases were selected in reverse chronological order, beginning with the most recent pathology specimens. Specimens were included in this study if there was sufficient material for immunohistochemical (IHC) and gene expression analyses. An approximately equal number of EBV+ cases were then selected within the same time period.
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissue sections were stained with H&E and with antibodies specific for markers of immune cell subsets and immunomodulatory molecules, including CD3, CD4, CD8, CD20, CD68, FOXP3, PD-1, PD-L1, colony stimulating factor 1 receptor (CSF-1R), glucocorticoid-induced tumor necrosis factor receptor family-related protein (GITR), lymphocyte activating gene 3 (LAG-3), indoleamine 2,3-dioxygenase 1 (IDO-1), and COX-2. IHC for CD3, CD4, CD8, CD20, FOXP3, CSF-1R and COX-2 was performed according to standard automated methods. IHC for CD68, PD-1, PD-L1, LAG-3, IDO-127 and GITR28 was performed manually, as previously described (online supplemental table S2). For digital analysis, expression of CD8, CD20, CD68, FOXP3, PD-1, CSF-1R, and GITR was quantified as cell density (number of positive cells per mm2) using HALO software (Indica Labs, Albuquerque, New Mexico, USA) as described.18 Manual scoring for CD3, CD4, PD-L1, LAG-3, IDO-1, and COX-2 expression was conducted by a pathologist (RAA) using a compound light microscope. PD-L1 expression was determined on tumor cells (membranous cell surface staining pattern, among all tumor cells) or stromal cells (cell surface and cytoplasmic staining in non-tumor, non-lymphocyte cells, among the total number of nucleated stromal cells) and reported at 5% intervals. For COX-2, the percentage of positive tumor cells was recorded. IDO-1 staining was reported as positive tumor and normal gastric epithelial cells, with attention to prominent expression at the tumor:stromal interface. CD3 was scored semi-quantitatively as 0 (none), 1 (focal), 2 (moderate) or 3 (marked) infiltration of positively stained cells. The presence of CD4+ cells was scored as the ratio of CD4+ to CD8+ cells, reported as 0.5 (1:2), 1 (1:1) or 2 (2:1). Expression of LAG-3 was estimated as the percentage of positive cells among CD3+ tumor-infiltrating lymphocytes.
Multiplex real-time quantitative reverse transcriptase PCR
Macrodissection (manual scraping of annotated slide areas with a scalpel) was conducted on 5 µm-thick FFPE tissue sections, capturing areas of CD3+ infiltrates in the TiME and, if present, neighboring PD-L1+ stromal and/or tumor cells. Following total RNA isolation, the expression of 122 unique immune-related genes and three reference genes (18S, GUSB, PTPRC) was evaluated with quantitative reverse transcriptase PCR (qRT-PCR) using Taqman low-density array cards (Life Technologies, Carlsbad, California, USA) as previously reported.28 29 Gene expression by EBV+ versus EBV(−) tumors was normalized to either PTPRC (CD45, pan-leukocyte marker) or 18S ribosomal RNA using the 2−ΔΔCt method.30 Undetermined cycle thresholds were assigned a value of 40 for computational purposes. Volcano plots were created with GraphPad software (La Jolla, California, USA). Heat maps displaying unsupervised clustering of genes and specimens were performed by Cluster 3.031 32 using city-block distance with average linkage. Heat maps were generated using Java TreeView V.1.1.6r4.32,34 The Cluster parameters EORDER (for samples) and GORDER (for genes) were used to govern the (otherwise arbitrary) respective top/bottom, left/right order at each pair of subcluster joins in the dendrograms of the heat maps.
TCGA analysis
Data from The Cancer Genome Atlas (TCGA) were downloaded from https://www.cancer.gov/tcga. Trimmed mean of M-values-normalized log2TPM (transcripts per million) tumor RNA sequencing data were used in the analysis. Clinical data pertaining to TCGA GC specimens were downloaded from cBioPortal (http://www.cbioportal.org). Among 415 GC samples in TCGA, 30 were EBV+ and 385 were EBV(−).15 Expression of PTGS2 (COX-2) as a single gene, as well as a 13-gene inflammation signature, were compared between EBV+ and EBV(−) GCs. The inflammation signature comprised CCL2, CCL3, CCL4, CD8A, CXCL9, CXCL10, GZMK, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, ICOS, and IRF1.35 The signature was calculated as the average log2TPM of these 13 genes. All analyses were performed using R statistics software and validated. For overall survival analysis, 407 patients with GC with available survival data were divided into four groups by expression level of PTGS2. Kaplan-Meier analysis of the lowest and highest quartiles of patients was conducted using the R survminer package (https://cran.r-project.org/web/packages/survminer/index.html), and p values were generated from the log rank test with default settings in the package. PTGS2 expression was also analyzed across 25 cancer types in TCGA having at least 100 samples each, including 9,712 primary, 394 metastatic and 42 recurrent tumors.
Statistical analysis
For analysis of qRT-PCR and IHC data, p values were obtained using the Wilcoxon rank-sum test, via the wilcox_test function from the R “coin” package (V.1.2–2)36 with distribution=“exact”. All p values are two-sided except where noted. When running the Wilcoxon rank-sum test on qRT-PCR data, all delta Ct values coming from Ct values of 40 were set to the same (tie) value, chosen larger than any other delta Ct (so that all delta Ct values coming from undetermined Ct values have the same highest rank).
Results
Characteristics of patients and tumor specimens
25 primary treatment-naïve GC specimens with sufficient tissue for protein and gene expression studies were identified by screening >1,000 pathology records from resections conducted at the Johns Hopkins Hospital from 2000 to 2014 (online supplemental table S1). Among them, 11 GCs were determined to be EBV+ and 14 EBV(−) by EBER-ISH. Patients had American Joint Committee on Cancer (AJCC) stage 1A – 4 disease at the time of resection. Patients with EBV+ GC were significantly younger than those with EBV(−) GC, as anticipated (median age 68 vs 77 years, respectively, p=0.0284). However, there were no significant differences in gender (data not shown), ethnicity, or AJCC stage.
Expression of candidate immune-related markers by IHC
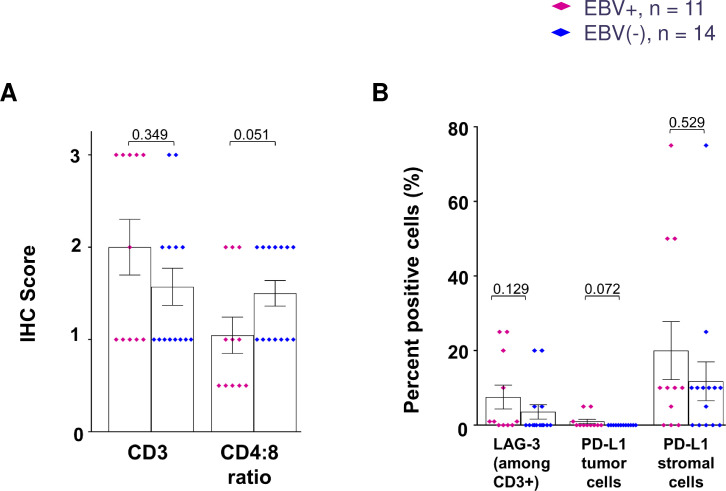
To assess and compare the characteristics of the TiME in EBV+ versus EBV(−) GCs, we first conducted IHC for select markers of immune cell subsets and immunoregulatory receptors and ligands (figure 1). We found a significantly higher density of CD8+ cells infiltrating EBV+ versus EBV(−) GCs (845 cells/mm2 vs 375 cells/mm2 respectively, p=0.044). Consistent with this, we found lower CD4:CD8 ratios in EBV+ tumors using semi-quantitative IHC scoring (p=0.051; figure 2A). Although densities of tumor-infiltrating CD20+ B cells, FOXP3+ regulatory T (Treg) cells, and CD68+ macrophages were numerically higher in EBV+ versus EBV(−) GCs, these differences did not achieve statistical significance (figure 1B). Likewise, densities of cells expressing the immunomodulatory molecules PD-1, GITR, CSF-1R, and IDO-1 were not significantly different between the two groups (figure 1B), nor was the percentage of CD3+ cells expressing LAG-3 (figure 2B). The expression pattern of the immunosuppressive enzyme IDO-1 was notable in that it was observed intracellularly in macrophages, tumor cells, intratumoral endothelial cells, and/or normal gastric epithelium (when present) overlying tumor areas, regardless of EBV status (online supplemental figure S1). Heightened IDO-1 expression was observed at the tumor:stromal interface in some specimens. Consistent with previous findings in other gastrointestinal cancers, the immune checkpoint ligand PD-L1 was expressed mainly by infiltrating immune cells and not by tumor cells themselves in GC specimens37 38 (figure 2B). There was no significant difference in the proportion of stromal cells expressing PD-L1 between EBV+ and EBV(−) GCs (mean 20% vs 12%, p=0.529). Thus, both EBV+ and EBV(−) GCs were characterized by immunosuppressive features upon IHC analysis of candidate markers, although EBV+ tumors contained a significantly higher density of CD8+ T cell infiltrates, implying the potential for antitumor immune effector functions.
Figure 1. Quantitative IHC analysis of candidate immune-related markers in EBV+ and EBV(−) GC. (A) Representative photomicrographs are displayed for each marker with IHC in different EBV+ and EBV(−) specimens. Scale bar, 100 µm. (B) Densities of immune cell subsets and co-regulatory molecules in EBV+ versus EBV(−) GCs, determined by HALO image analysis. Some specimens with limited amounts of tissue were not tested for every marker. Bars indicate mean values and SEM. P values, Wilcoxon rank-sum test. CSF-1R, colony stimulating factor 1 receptor; EBV, Epstein-Barr virus; FOXP3, forkhead box P3; GC, gastric cancer; GITR, glucocorticoid-induced tumor necrosis factor receptor family-related protein; IDO-1, indoleamine 2,3-dioxygenase 1; IHC, immunohistochemistry; PD-1, programmed cell death 1.
Figure 2. Semi-quantitative IHC analysis of immune cell subsets and checkpoint molecules in EBV+ versus EBV(−) GC. (A) While CD3 scores were similar in the two GC subtypes, the CD4:CD8 ratio was lower in EBV+ GC, indicating a higher density of CD8+ T cells. (B) There were no significant differences in the expression of LAG-3 among CD3+ T cells, or PD-L1 by tumor or stromal cells, in EBV+ versus EBV(−) GCs. PD-L1 expression was more prevalent on stromal cells than on tumor cells. Percent positive cells were estimated visually in 5% increments. Bars indicate mean values and SEM. P values, Wilcoxon rank-sum test. EBV, Epstein-Barr virus; GC, gastric cancer; IHC, immunohistochemistry; LAG-3, lymphocyte activating gene 3; PD-L1, programmed cell death ligand 1.
Expression of immune-related genes in the TiME of EBV+ versus EBV(−) GCs
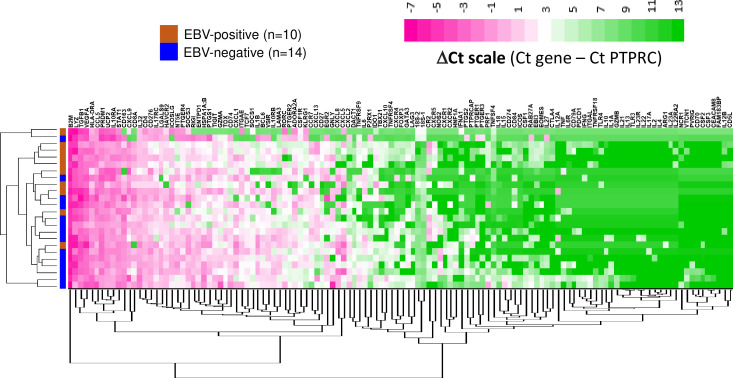
We further characterized and compared the TiMEs of EBV+ versus EBV(−) GCs by performing gene expression profiling (GEP), using multiplex qRT-PCR to detect 122 unique genes of interest and two reference genes (18S, and the pan-immune cell marker PTPRC/CD45). With unsupervised hierarchical heat map clustering, specimens from each GC subtype tended to cluster together, suggesting distinct features between these transcriptomes (figure 3 and online supplemental figure S2). For some genes, robust expression was detected across all specimens regardless of EBV status; this included genes related to immune activation and antigen presentation (B2M, HLA-DRA, STAT1, UCP2) as well as others involved in immunosuppression (CD276 (B7-H3), TGFB1, VEGFA). Conversely, certain genes were poorly expressed across all samples, including those involved in immune activation (GZMB, IFNG, IL2, IL12B) or associated with a Th17 profile (IL17A, IL22, IL22RA2, IL23A, IL23R). Of particular interest were the genes that were significantly differentially expressed (figure 4 and online supplemental figure S3). When normalized to PTPRC, three genes were overexpressed in EBV+ compared with EBV(−) GCs: ITGAE (CD103, marking intraepithelial T cells and a dendritic cell subset; up threefold, p=0.036), and two interferon (IFN)-gamma inducible genes—CXCL9 (chemokine involved in cytotoxic T lymphocyte recruitment; up fourfold, p=0.005) and IDO1 (immunosuppressive enzyme; up sixfold, p=0.030). In contrast, EBV(−) tumors overexpressed several functionally-related gene groups, including those characterizing myeloid cells (CD163, IL1A, NOS2, RIGI); immunosuppressive cytokines/chemokines such as CXCL2, CXCR4, IL10 and IL32; and coinhibitory molecules such as HAVCR2 (TIM-3), VSIR (VISTA), and components of the adenosine pathway (ENTPD1 (CD39) and NT5E (CD73)). Notably, compared with EBV+ GCs, EBV(−) GCs highly overexpressed PTGS2 (the most highly upregulated single gene, 32-fold, p=0.005) encoding the COX-2 enzyme which mediates prostaglandin E2 (PGE2) synthesis and tumor-promoting inflammation; the prostaglandin receptors PTGER1 (EP1; up 21-fold, p=0.015) and PTGER4 (EP4; up twofold, p=0.022), and the major COX-2-inducing cytokine IL1B (up 11-fold, p=0.019). PTGS2, PTGER1, and IL1B were also significantly overexpressed in EBV(−) GCs when gene expression was normalized to the 18S ribosomal subunit (online supplemental figure S3).
Figure 3. Expression of candidate immune-related genes in primary gastric cancer specimens using quantitative reverse transcriptase PCR. Heat map displays unsupervised clustering of ΔCt values normalized to immune cell content (Ct gene – Ct PTPRC) in EBV+ (n=10) and EBV(−) (n=14) GCs. Clustering on genes and samples was performed by Cluster 3.0 using city-block distance with average linkage. 122 immune-related genes and 18S are depicted. EBV, Epstein-Barr virus.
Figure 4. Differential expression of candidate immune-related genes in EBV+ (n=10) versus EBV(−) (n=14) primary GCs. Data were normalized to PTPRC expression using the ΔΔCt method. The vertical hatched lines represent a twofold expression difference. The horizontal hatched line represents p value=0.1, as determined by the Wilcoxon rank-sum test. EBV(−) GCs overexpressed genes associated with the cyclooxygenase 2/prostaglandin E2 pathway, including PTGS2, PTGER1, PTGER4, and IL1B. Two genes that failed to amplify in any specimen (FAM183B/CFAP144P1 and IL4) were excluded from this analysis. EBV, Epstein-Barr virus; GC, gastric cancer.
Expression of COX-2 protein in GC
We next wanted to determine if heightened PTGS2 (COX-2) gene expression in EBV(−) versus EBV+ GCs translated into differential protein expression. With IHC for COX-2, there was a trend towards increased expression in EBV(−) compared with EBV+ tumors, with means of 56% versus 25% of tumor cells expressing COX-2, respectively, (p=0.068, one-sided) (figure 5A). Of interest, we also observed uniform COX-2 expression in the normal gastric epithelium overlying GC specimens, where PGE2 plays a normal homeostatic role. COX-2 was expressed by multiple cell types in GC, including tumor cells, myeloid cells, and non-myeloid stromal cells (figure 5B).
Figure 5. COX-2 protein expression in EBV(−) versus EBV+GCs, detected with IHC. (A) Left, COX-2 expression among tumor cells was quantified in 25% increments. There was a trend towards overexpression of COX-2 among tumor cells in EBV(−) versus EBV+ GC. P value from Wilcoxon rank-sum test (one-sided test, based on prior differential gene expression result). Right, representative H&E and COX-2 immunohistochemistry images from an EBV(−) GC. COX-2 was expressed by tumor cells (yellow star) and normal gastric epithelium (red arrow). Scale bar, 100 um. H&E, hematoxylin and eosin. (B) COX-2 and CD68 expression in serial sections of an EBV(−) GC. Expression was observed in adenocarcinoma cells (red arrow), CD68+ myeloid cells (blue arrow), and non-myeloid stromal cells. This specimen is devoid of normal gastric epithelium. Scale bar, 100 microns. COX-2, cyclooxygenase 2; EBV, Epstein-Barr virus; GC, gastric cancer.
COX-2 analysis in TCGA data
To further explore the implications of COX-2 expression in GC, we conducted an analysis of published TCGA data from 415 GCs, including 30 EBV+ and 385 EBV(−) specimens. Examining PTGS2 gene expression, there was highly significant upregulation in EBV(−) compared with EBV+ GCs, consistent with findings from our current study (figure 6A). Of interest, EBV(−) GCs also showed significantly lower expression of a 13-gene inflammation score,35 consistent with the known immunosuppressive effects of the COX-2/PGE2 pathway (figure 6B). Furthermore, in an analysis of TCGA data across 25 different cancer types having transcriptional data from at least 100 specimens, GC was among the highest PTGS2-expressing tumor types (online supplemental figure S4). Analyzing OS among 407 patients with GC as a function of the level of tumor PTGS2 expression, we found significantly lower OS in the highest PTGS2-expressing quartile compared with the lowest expressing quartile (p=0.032; figure 6C). Thus, increased COX-2 gene expression was associated with EBV(−) tumor status and reduced OS in patients with GC.
Figure 6. PTGS2 (COX-2) expression in EBV(−) versus EBV+ GC and association with overall survival in TCGA. Analysis of GC transcriptional profiling from TCGA shows that EBV(−) tumors have (A) significantly higher PTGS2 expression and (B) significantly lower expression of a 13-gene inflammation score. In (A), gene expression is measured as log2TPM (transcripts per million). Standard box and whisker plot notation are used to represent the data distribution: thick horizontal line in the middle of the box for median value, box for IQR (25%–75%), and whiskers for non-outlier data range (1.5×IQR). (C) Patients whose GCs have the highest quartile of PTGS2 expression have significantly worse overall survival (OS; red line) than those whose tumors have the lowest quartile of PTGS2 expression (blue line; p=0.032). EBV, Epstein-Barr virus; GC, gastric cancer; IQR, interquartile range; OS, overall survival; TCGA, The Cancer Genome Atlas.
Discussion
This comparative study of the TiME of EBV-associated versus non-EBV-associated GC is immediately relevant to the striking clinical observation that responsiveness to PD-1 pathway blockade is substantially enhanced in the EBV+ GC subtype. While the TiMEs of both GC subsets harbor a variety of immunosuppressive features, our findings of heightened inflammation in EBV+ compared with EBV(−) GC, with higher densities of infiltrating CD8+ T cells and upregulation of IFN-gamma-inducible genes such as IDO and CXCL9, are consistent with previously published observations.2639,41 EBV(−) GCs are heterogeneous, comprising chromosome instable, genome instable, and microsatellite instability-high (MSI-H) molecular subtypes identified by TCGA.15 Among these EBV(−) subtypes, only MSI-H GC responds favorably to PD-1 blockade, highlighting the need for improved therapeutic options for the majority of GCs.
Our GEP analysis revealed that, compared with EBV+ GCs, EBV(−) GCs differentially overexpressed certain genes associated with immunosuppression that could potentially inhibit local antitumor immunity in the TiME. Notably, PTGS2, encoding the COX-2 enzyme that catalyzes the production of the lipid-derived prostaglandin PGE2, was the most highly upregulated gene in our EBV(−) GC cohort. PGE2 mediates protumorigenic signals through the G-protein coupled EP receptors EP2 and EP4 (encoded by PTGER2 and PTGER4); importantly, PTGER4 was upregulated in EBV(−) tumors alongside PTGS2 in this study. Of interest, transient transfection of the EBV latent membrane proteins LMP1 and LMP2A into a COX-2-expressing EBV(−) cultured GC line was reported to significantly downregulate COX-2 expression, accompanied by downregulation of the anti-apoptotic signaling molecule TRAF2; these findings highlight a biological interaction between EBV infection and COX-2 expression that may potentially render EBV+ GC more susceptible than EBV(−) GC to immune attack.42
Additional upregulated genes that we identified in EBV(−) GCs, CXCL2 and IL1B, have been associated with high PTGS2 expression in human melanomas.43 We previously showed that the COX-2/PGE2 pathway is upregulated in human cancer cell lines and myeloid cells following in vitro exposure to interleukin-1 beta (IL-1B) and certain other tumor microenvironment (TME)-resident cytokines, and that PGE2 can functionally suppress several immune cell subsets including T and myeloid cells.29 44 45 Results from our current analysis of PTGS2 gene expression as well as a multigene inflammation score in TCGA data including 415 GC specimens are consistent with findings from our in-depth analysis of 25 GCs revealing increased PTGS2 expression and decreased inflammation in EBV(−) versus EBV+ GC.
Our laboratory’s previous studies of the TiMEs of two different EBV-associated cancers, classical Hodgkin’s lymphoma (CHL)28 and nasopharyngeal carcinoma (NPC),46 demonstrated that the presence of tumor-associated EBV is not in itself sufficient to generate a vigorous tumor-reactive immune milieu. While CHL is highly responsive to anti-PD-1 monotherapy,47 the NPC response rate to anti-PD-1 monotherapy is only ~20%.48 Comparing EBV+ versus EBV(−) CHLs, we found a Th1-associated IFN-driven pro-inflammatory TiME in EBV+ CHL versus a pathogenic Th17-driven TiME in EBV(−) CHL. Strikingly, NPCs, of which >95% are EBV+, were characterized by a more immunosuppressive TiME when compared with EBV+ CHL. In particular, COX-2 was highly upregulated in NPC compared with EBV+ CHL. Thus, cancers associated with the same virus may have distinct immune biologies based on tissue context,49 suggesting the need to customize anti-PD-1-based treatment strategies according to disease application.
In summary, the COX-2/PGE2 pathway may interact through multiple mechanisms to down-modulate antitumor immunity in the TiME and mitigate the therapeutic effects of immunotherapies that are currently in standard use for treating GC. Whether COX-2/PGE2 pathway inhibition as a treatment for established cancers will mediate similar beneficial effects to those observed in preventing the occurrence of new cancers is an open question.50 51 Combining PD-1 pathway blockade with inhibition of the COX-2/PGE2 pathway, by inhibiting COX-2-inducing cytokines such as IL-1B, or by blocking the PGE2 receptors EP2 and/or EP4, are strategies that are now in clinical testing across a wide array of cancer types and may offer new opportunities to improve therapeutic outcomes for patients with EBV(−) GCs.
supplementary material
Acknowledgements
This work was presented in part at the 33rd Annual Meeting of the Society for Immunotherapy of Cancer, November 9, 2018, Washington, DC, USA. The authors would like to thank Dr Shuming Chen (Johns Hopkins University School of Medicine) for helpful discussions.
Footnotes
Funding: This study was supported by NCI R01CA142779 (to SLT and JT), Bristol Myers Squibb (to SLT), The Mark Foundation (to JT), and the Bloomberg-Kimmel Institute for Cancer Immunotherapy.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: The study was approved by the Johns Hopkins Medicine Institutional Review Board.
Data availability free text: Data are available from the corresponding authors upon reasonable request.
Contributor Information
Tracee L McMiller, Email: tmcmill3@jhmi.edu.
Sepideh Besharati, Email: sb4433@cumc.columbia.edu.
Mark Yarchoan, Email: mark.yarchoan@jhmi.edu.
Qingfeng Zhu, Email: qzhu6@jhmi.edu.
Keziban Ünsal-Kaçmaz, Email: Kezi.UnsalKacmaz@biontech.us.
Ke Xu, Email: kxu7865@gmail.com.
Junghwa Lee, Email: junghwa.lee@ip-korea.org.
Feriyl Bhaijee, Email: fbhaijee@gmail.com.
Logan L Engle, Email: eengle6@jhmi.edu.
Janis M Taube, Email: jtaube1@jhmi.edu.
Alan E Berger, Email: aberger9@jhmi.edu.
Robert A Anders, Email: rander54@jhmi.edu.
Suzanne L Topalian, Email: stopali1@jhmi.edu.
Data availability statement
Data are available upon reasonable request.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Wagner AD, Unverzagt S, Grothe W, et al. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010;3:CD004064. doi: 10.1002/14651858.CD004064.pub3. [DOI] [PubMed] [Google Scholar]
- 3.Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–71. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs CS, Doi T, Jang RW, et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer. JAMA Oncol. 2018;4:e180013. doi: 10.1001/jamaoncol.2018.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada K. Epstein-Barr virus and gastric carcinoma. Mol Pathol . 2000;53:255–61. doi: 10.1136/mp.53.5.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boysen T, Mohammadi M, Melbye M, et al. EBV-associated gastric carcinoma in high- and low-incidence areas for nasopharyngeal carcinoma. Br J Cancer. 2009;101:530–3. doi: 10.1038/sj.bjc.6605168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449–58. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 8.Bai Y, Xie T, Wang Z, et al. Efficacy and predictive biomarkers of immunotherapy in Epstein-Barr virus-associated gastric cancer. J Immunother Cancer. 2022;10:e004080. doi: 10.1136/jitc-2021-004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy G, Pfeiffer R, Camargo MC, et al. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009;137:824–33. doi: 10.1053/j.gastro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsao S-W, Tsang CM, To K-F, et al. The role of Epstein-Barr virus in epithelial malignancies. J Pathol. 2015;235:323–33. doi: 10.1002/path.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Jin H, Cheung KF, et al. Zinc finger E-box binding factor 1 plays a central role in regulating Epstein-Barr virus (EBV) latent-lytic switch and acts as a therapeutic target in EBV-associated gastric cancer. Cancer. 2012;118:924–36. doi: 10.1002/cncr.26184. [DOI] [PubMed] [Google Scholar]
- 12.Chong J-M, Sakuma K, Sudo M, et al. Global and non-random CpG-island methylation in gastric carcinoma associated with Epstein-Barr virus. Cancer Sci. 2003;94:76–80. doi: 10.1111/j.1349-7006.2003.tb01355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 14.Zong L, Seto Y. CpG Island Methylator Phenotype, Helicobacter pylori, Epstein-Barr Virus, and Microsatellite Instability and Prognosis in Gastric Cancer: A Systematic Review and Meta-Analysis. PLoS ONE. 2014;9:e86097. doi: 10.1371/journal.pone.0086097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarchoan M, Johnson BA, 3rd, Lutz ER, et al. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17:209–22. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal R, Şenbabaoğlu Y, Desrichard A, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829. doi: 10.1172/jci.insight.89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Succaria F, Kvistborg P, Stein JE, et al. Characterization of the tumor immune microenvironment in human papillomavirus-positive and -negative head and neck squamous cell carcinomas. Cancer Immunol Immunother. 2021;70:1227–37. doi: 10.1007/s00262-020-02747-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipson EJ, Vincent JG, Loyo M, et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus and overall survival. Cancer Immunol Res. 2013;1:54–63. doi: 10.1158/2326-6066.CIR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nghiem P, Bhatia S, Lipson EJ, et al. Durable Tumor Regression and Overall Survival in Patients With Advanced Merkel Cell Carcinoma Receiving Pembrolizumab as First-Line Therapy. J Clin Oncol. 2019;37:693–702. doi: 10.1200/JCO.18.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang MS, Lee HS, Kim CW, et al. Clinicopathologic characteristics of Epstein-Barr virus-incorporated gastric cancers in Korea. Pathol Res Pract. 2001;197:395–400. doi: 10.1078/0344-0338-00052. [DOI] [PubMed] [Google Scholar]
- 22.Gulley ML, Pulitzer DR, Eagan PA, et al. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol. 1996;27:20–7. doi: 10.1016/s0046-8177(96)90133-1. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Li J, Hao Y, et al. Differentiated tumor immune microenvironment of Epstein-Barr virus-associated and negative gastric cancer: implication in prognosis and immunotherapy. Oncotarget. 2017;8:67094–103. doi: 10.18632/oncotarget.17945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokunaga M, Land CE, Uemura Y, et al. Epstein-Barr virus in gastric carcinoma. Am J Pathol. 1993;143:1250–4. [PMC free article] [PubMed] [Google Scholar]
- 25.van Beek J, zur Hausen A, Snel SN, et al. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. Am J Surg Pathol. 2006;30:59–65. doi: 10.1097/01.pas.0000176428.06629.1e. [DOI] [PubMed] [Google Scholar]
- 26.Derks S, de Klerk LK, Xu X, et al. Characterizing diversity in the tumor-immune microenvironment of distinct subclasses of gastroesophageal adenocarcinomas. Ann Oncol. 2020;31:1011–20. doi: 10.1016/j.annonc.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim AK, Gani F, Layman AJ, et al. Multiple Immune-Suppressive Mechanisms in Fibrolamellar Carcinoma. Cancer Immunol Res. 2019;7:805–12. doi: 10.1158/2326-6066.CIR-18-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duffield AS, Ascierto ML, Anders RA, et al. Th17 immune microenvironment in Epstein-Barr virus-negative Hodgkin lymphoma: implications for immunotherapy. Blood Adv. 2017;1:1324–34. doi: 10.1182/bloodadvances.2017007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, McMiller TL, Soni A, et al. Comparing anti-tumor and anti-self immunity in a patient with melanoma receiving immune checkpoint blockade. J Transl Med. 2024;22:241. doi: 10.1186/s12967-024-04973-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan JS, Reed A, Chen F, et al. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisen M, Hoon M. Cluster 3.0 manual. 2002. http://bonsai.hgc.jp/~mdehoon/software/cluster/cluster3.pdf Available.
- 32.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saldanha AJ. Java treeview user’s manual revision: 1.26. 2004. http://jtreeview.sourceforge.net/docs/JTVUserManual/single.html Available.
- 34.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 35.Harlin H, Meng Y, Peterson AC, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–85. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hothorn T, Hornik K, van de Wiel MA, et al. Implementing a Class of Permutation Tests: The coin Package. J Stat Soft. 2008;28:1–23. doi: 10.18637/jss.v028.i08. [DOI] [Google Scholar]
- 37.Thompson ED, Zahurak M, Murphy A, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2017;66:794–801. doi: 10.1136/gutjnl-2015-310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Derks S, Liao X, Chiaravalli AM, et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget. 2016;7:32925–32. doi: 10.18632/oncotarget.9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gullo I, Oliveira P, Athelogou M, et al. New insights into the inflamed tumor immune microenvironment of gastric cancer with lymphoid stroma: from morphology and digital analysis to gene expression. Gastric Cancer. 2019;22:77–90. doi: 10.1007/s10120-018-0836-8. [DOI] [PubMed] [Google Scholar]
- 41.Kim SY, Park C, Kim HJ, et al. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology. 2015;148:137–47. doi: 10.1053/j.gastro.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 42.Qi YF, Liu M, Zhang Y, et al. EBV down-regulates COX-2 expression via TRAF2 and ERK signal pathway in EBV-associated gastric cancer. Virus Res. 2019;272:197735. doi: 10.1016/j.virusres.2019.197735. [DOI] [PubMed] [Google Scholar]
- 43.Zelenay S, van der Veen AG, Böttcher JP, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257–70. doi: 10.1016/j.cell.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, McMiller T, Sankaran P, et al. 288 The COX-2 pathway as a mediator of resistance to anti-PD-1 therapy. J Immunother Cancer. 2021;9:A312. doi: 10.1136/jitc-2021-SITC2021.288. [DOI] [Google Scholar]
- 45.Chen S, Lee S, McMiller TL, et al. Abstract 4159: The COX-2/PGE2 pathway as a mediator of resistance to anti-PD-1 therapy. Cancer Res. 2023;83:4159. doi: 10.1158/1538-7445.AM2023-4159. [DOI] [Google Scholar]
- 46.Duffield AS, Ascierto ML, Anders RA, et al. Abstract 4750: The immunosuppressive tumor microenvironment (TME) in nasopharyngeal carcinoma: implications for immunotherapy. Cancer Res. 2018;78:4750. doi: 10.1158/1538-7445.AM2018-4750. [DOI] [Google Scholar]
- 47.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma BBY, Lim W-T, Goh B-C, et al. Antitumor Activity of Nivolumab in Recurrent and Metastatic Nasopharyngeal Carcinoma: An International, Multicenter Study of the Mayo Clinic Phase 2 Consortium (NCI-9742) J Clin Oncol. 2018;36:1412–8. doi: 10.1200/JCO.2017.77.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pao W, Ooi C-H, Birzele F, et al. Tissue-Specific Immunoregulation: A Call for Better Understanding of the “Immunostat” in the Context of Cancer. Cancer Discov. 2018;8:395–402. doi: 10.1158/2159-8290.CD-17-1320. [DOI] [PubMed] [Google Scholar]
- 50.Brown JR, DuBois RN. COX-2: A Molecular Target for Colorectal Cancer Prevention. JCO. 2005;23:2840–55. doi: 10.1200/JCO.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 51.Ridker PM, MacFadyen JG, Thuren T, et al. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390:1833–42. doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.