Abstract
Background
The genus Amorphophallus (Araceae) contains approximately 250 species, most of which have high ecological and economic significance. The chloroplast genome data and the comprehensive analysis of the chloroplast genome structure of Amorphophallus is limited. In this study, four chloroplast genomes of Amorphophallus were sequenced and assembled. For the first time, comparative analyses of chloroplast genomes were conducted on the 13 Amorphophallus species in conjunction with nine published sequences.
Results
The Amorphophallus chloroplast genomes exhibited typical quadripartite structures with lengths ranging from 164,417 to 177,076 bp. These structures consisted of a large single copy (LSC, 90,705 − 98,561 bp), a small single copy (SSC, 14,172 − 21,575 bp), and a pair of inverted repeats (IRs, 26,225 − 35,204 bp). The genomes contain 108 − 113 unique genes, including 76 − 79 protein-coding genes, 28 − 29 tRNA genes, and 4 rRNA genes. The molecular structure, gene order, content, codon usage, long repeats, and simple sequence repeats (SSRs) within Amorphophallus were generally conserved. However, several variations in intron loss and gene expansion on the IR-SSC boundary regions were found among these 13 genomes. Four mutational hotspot regions, including trnM-atpE, atpB, atpB-rbcL and ycf1 were identified. They could identify and phylogeny future species in the genus Amorphophallus. Positive selection was found for rpl36, ccsA, rpl16, rps4, rps8, rps11, rps12, rps14, clpP, rps3, ycf1, rpl20, rps2, rps18, rps19, atpA, atpF, rpl14, rpoA, rpoC1, rpoC2 and rps15 based on the analyses of Ka/Ks ratios. Phylogenetic inferences based on the complete chloroplast genomes revealed a sister relationship between Amorphophallus and Caladieae. All Amorphophallus species formed a monophyletic evolutionary clade and were divided into three groups, including CA-II, SEA, and CA-I. Amorphophallus albus, A. krausei, A. kachinensis and A. konjac were clustered into the CA-II clade, A. paeoniifolius and A. titanum were clustered into the SEA clade, A. muelleri ‘zhuyajin1’, Amorphophallus sp, A. coaetaneus, A. tonkinensis and A. yunnanensis were clustered into CA- I clade.
Conclusions
The genome structure and gene content of Amorphophallus chloroplast genomes are consistent across various species. In this study, the structural variation and comparative genome of chloroplast genomes of Amorphophallus were comprehensively analyzed for the first time. The results provide important genetic information for species classification, identification, molecular breeding, and evolutionary exploration of the genus Amorphophallus.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11053-z.
Keywords: Amorphophallus, Chloroplast genome, Genome comparison, Phylogenetic analysis
Background
The Amorphophallus genus Blume ex Decne. (Araceae) consists of approximately 200–250 species [1, 2], among which 242 species are accepted by POWO. These species are primarily distributed in tropical or subtropical areas of South Asia and West Africa, including China, Japan, Myanmar, Vietnam, and Indonesia [3]. Southwestern China has been recognized as one of the centers of origin, and there are currently over 21 species recorded for China [3]. Multiple Amorphophallus species have important medicinal, ornamental, edible, and economic values [4]. In China, nine of the 21 species that have been recorded are endemic [3]. Amorphophallus has been cultivated and consumed in China for over 2000 years as an agricultural crop due to its tuber’s abundance of konjac glucomannan (KGM) and starch [5, 6]. KGM is a water-soluble polysaccharide (dietary fiber) that is not only used in the industrial field, food science, nutrition, biotechnology, and pharmacology but also has beneficial health impacts, including weight loss, intestinal health, and the reduction of blood lipids, blood pressure, and blood sugar levels [3, 6–11]. The medicinal properties of Amorphophallus species have been extensively investigated in recent years, including analgesic, neuroprotective, hepatoprotective, anti-inflammatory, anticonvulsant, antibacterial, antioxidant, anticancer, antiobesity, and immunomodulatory effects [7, 11]. Therefore, numerous Amorphophallus species have significant research value due to the combination of industrial, dietary, and medicinal properties. However, research on Amorphophallus primarily focuses on its medical value [11–13], properties of KGM [14, 15], karyotype analysis [5], genetic diversity [16, 17], phylogeny [1, 16, 18, 19], heat production [20, 21] and disease resistance [22–24]. For phylogeny, several chloroplast genome markers (rbcL, matK, trnH, and psbA) and nuclear DNA markers (ribosomal DNA intratranscriptional spacer, ITS) were used to determine relationships and evaluate genetic variation in Amorphophallus genus [1, 19]. However, the current knowledge of genetic relationships and evolution among Amorphophallus species offers merely baseline information [25]. Infrageneric classification and evolution based on intricate morphological traits still have some disagreements [25, 26]. Therefore, developing more effective DNA barcodes is particularly important for Amorphophallus plants.
Chloroplasts are self-replicating organelles with their independent genetic material, playing pivotal roles in photosynthesis, transcription, and translation [27, 28]. The chloroplast genome typically spans a length of 107–218 kb [28]. It maintains a highly conserved quadripartite circular configuration featuring a pair of inverted repeats (IRs), flanking a large single-copy (LSC) region and a small single-copy (SSC) region [29, 30]. Despite the structural conservation of the chloroplast genome, multiple mutational events, including gene rearrangements, single-nucleotide substitutions (SNPs), gene losses, gene duplication, intron loss, and variations in the expansion/contraction of the IR, frequently occur across species and even within individual organisms [27, 29]. These variations can be used for species identification and analysis to improve the current understanding of plant phylogenetic and evolutionary relationships. Compared with variable markers, the complete chloroplast genome sequence is rich in genetic variations, which are valuable tools utilized for various purposes, including phylogenetic analyses, evolutionary studies, comparative genomics, and the development of molecular markers in higher plants [31, 32]. Limited studies are available on the chloroplast genomes of the Amorphophallus, up to now, only nine chloroplast genomes of Amorphophallus have been published [25, 33–41]. These studies indicate that the chloroplasts of Amorphophallus contain 126–131 genes. However, with one exception, Liu et al. suggested that the genus of Amorphophallus contains fewer genes [35]. They reported a loss of some important genes in four Amorphophallus species, including ycf1, accD, psbE, trnL-CAA, and trnG-GCC genes. Deletion of rpl23 and rpl2 was limited to only one IR region [35]. Recent studies have reported the conservation of chloroplast genome structures in Amorphophallus and do not support the gene deletion mentioned above [36, 38]. Meanwhile, most studies on the chloroplast genome of Amorphophallus primarily focus on the basic information description, while comparative studies on the chloroplast genome of Amorphophallus are relatively limited. Additionally, although the chloroplast genome of A. krausei and A. albus have recently been published, there are many local varieties of A. albus. The A. albus in this study is the Xiluodu A. albus, which is the most representative local variety in Yunnan. In addition, A. krausei is a species with extremely rich intraspecific variation. It was collected in Wangya Village, Puer, China in 2019 and has been planted in the Amorphophallus germplasm resource nursery of Kunming University ever since. To develop and utilize local Xiluodu A. albus resources, and to compare the chloroplast of A. albus and A. krausei from different distributed regions, we also sequenced the complete chloroplast genomes of these two species.
In this study, we sequenced and assembled the chloroplast genomes of A. albus Yunnan, A. krausei Yunnan, A. muelleri ‘zhuyajin1’, and Amorphophallus sp (Fig. 1). Furthermore, we compared the chloroplast genome sequences of nine other published Amorphophallus species. Our primary objectives were to (1) compare the genome structures and gene organization of chloroplast genomes within the Amorphophallus genus; (2) identify variations of long repeats, simple sequence repeats (SSRs), and codon usage patterns of these chloroplast genomes in Amorphophallus; (3) identify highly variable regions (hotspots) as potential chloroplast markers for future phylogenetic analyses of the Amorphophallus genus; (4) identify the protein-coding genes under positive selection within the seven plastomes of Amorphophallus and determine the phylogenetic relationships within the Araceae family. These findings can provide valuable genetic resources for further research on the phylogenetic position of Amorphophallus and contribute to the breeding improvement of Amorphophallus.
Fig. 1.

Species reference image of Amorphophallus. A A. krausei; B A. albus Yunnan; C-G A. muelleri ‘zhuyajin1’, C petiole detail, D leaf detail, E flower bud details before flowering, F and G inflorescence; H–L Amorphophallus sp, H petiole detail, I leaf detail, J flower bud details before flowering, K and L inflorescence
Results
Chloroplast genome features of Amorphophallus species
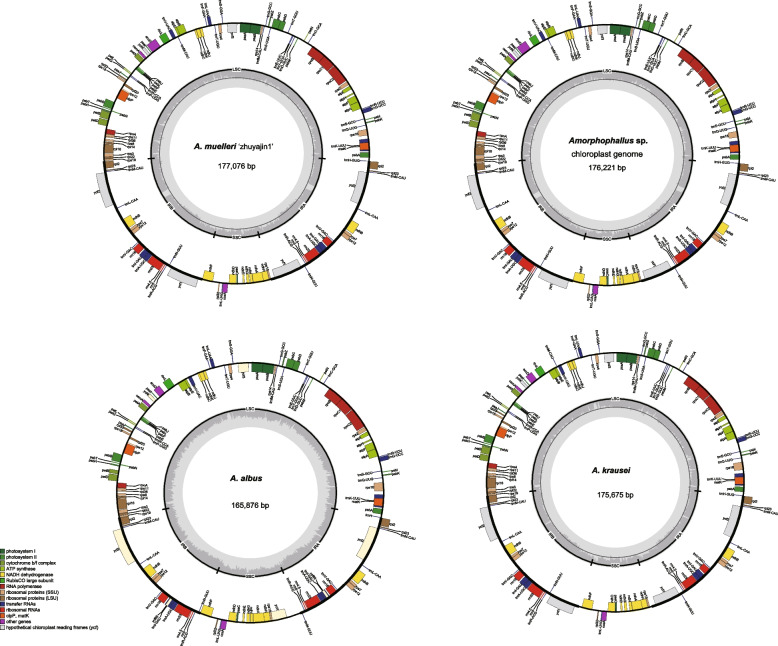
The current study analyzed 13 chloroplast genomes of Amorphophallus species, including four newly sequenced genomes and nine published ones. The four sequenced samples yielded 4.0 to 5.6 GB of raw data (Table S1). After removing adapters and low-quality reads, these samples generated 3.3 to 4.7 GB of clean reads each. De novo assembled chloroplast genomes were deposited in GenBank with accession numbers (A. muelleri ‘zhuyajin1’ OR995733, A. albus Kunming OR438676, A. krausei Kunming PP936071, and Amorphophallus sp. PP936070). Complete chloroplast genomes of 7 species ranged from 164,417 bp (A. yunnanensis) to 177,076 bp (A. muelleri ‘zhuyajin1’) in length, with an overall length variance of approximately 12.66 kb (Table 1). All 13 Amorphophallus chloroplast genomes exhibited typical quadripartite structures with an LSC region (90,705–98,561 bp) and an SSC region (14,172–20,249 bp) separated by two inverted repeat (IR) regions (26,225–35,204 bp) (Fig. 2 and Table 1). The overall GC content in the Amorphophallus chloroplast genomes was 34.5%–36%. The complete chloroplast genomes of Amorphophallus consist of 126–131 genes, including 81–86 protein-coding genes, 36–39 tRNAs, and 8 rRNAs, which were classified into four categories based on their functions (Table 2, Table S2). After removing duplicates, 108–113 unique genes including 76–79 protein-coding, 28–29 tRNAs and 4 rRNAs genes were remained for each genome (Table 1 and Table S3). Specifically, there are 5–7 protein-coding genes, 7 tRNA genes (trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRNA genes duplicate in the IR regions. In 13 of the species, except for A. coaetaneus, A. yunnanensis and A. tonkinensis, there were 7 duplicated protein-coding genes (rps12, ycf1, ndhB, rps7, ycf2, rpl2 and rpl23) in the IR region. Amorphophallus coaetaneus had one copy of rpl23, and A. albus, A. konja and A.yunnanensis had one copy of ycf1 each. Specifically, the rpl23 was annotated within the IR (IRa and IRb) regions of the 10 chloroplast genomes. Nevertheless, in A. coaetaneus, it was only detected in the IRb region and was missing in the IRa region. Additionally, the rpl23 was lost in the chloroplast genomes of A. yunnanensis and A. tonkinensis. The infA gene was only present in A. titanium and was a non-functional gene. Furthermore, A. coaetaneus contained three trnQ-UUG genes, while the remaining six genomes contained one trnQ-UUG.
Table 1.
The basic chloroplast genome information of 13 Amorphophallus species
| Name | Total | IR | LSC | SSC | Total | Total | Protein | rRNAs | tRNAs | Accession | Locations |
|---|---|---|---|---|---|---|---|---|---|---|---|
| length (bp) | length (bp) | length (bp) | length (bp) | GC% | Genes (unique) | coding genes (unique) | (unique) | (unique) | number | ||
| A. krausei | 175,675 | 32,945 | 93,724 | 16,061 | 34.9 | 130 (111) | 85 (78) | 8 (4) | 37 (29) | PP936071 | Kunming, Yunnan |
| Amorphophallus sp | 176,221 | 34,891 | 91,718 | 14,172 | 34.7 | 130 (111) | 85 (78) | 8 (4) | 37 (29) | PP936070 | Kunming, Yunnan |
| A. muelleri ‘zhuyajin1’ | 177,076 | 35,204 | 91,947 | 14,721 | 34.5 | 130 (111) | 85 (78) | 8 (4) | 37 (29) | OR995733 | Kunming, Yunnan |
| A. albus | 165,876 | 26,225 | 93,177 | 20,249 | 35.6 | 130 (113) | 86 (79) | 8 (4) | 36 (30) | OR438676 | Kunming, Yunnan |
| A.konja | 167,470 | 26,226 | 93,443 | 21,575 | 35.4 | 130 (113) | 86 (79) | 8 (4) | 36 (30) | OR438675 | Kunming, Yunnan |
| A. titanum | 176,835 | 32,708 | 95,475 | 15,944 | 34.5 | 130 (111) | 85 (78) | 8 (4) | 37 (29) | MN046883 | Sumatra, Indonesia |
| A. coaetaneus | 175,465 | 30,200 | 98,561 | 16,504 | 34.9 | 131 (111) | 84 (78) | 8 (4) | 39 (29) | OQ404947 | Xishuangbanna, Yunnan |
| A. tonkinensis | 169,341 | 31,498 | 90,705 | 15,640 | 36 | 128 (110) | 83 (77) | 8 (4) | 37 (29) | NC_086855 | Hekou County, Yunnan |
| A. yunnanensis | 164,417 | 28,543 | 92,149 | 15,182 | 36 | 126 (108) | 81 (76) | 8 (4) | 37 (28) | NC_082906 | Wangmo County, Guizhou |
| A. paeoniifolius | 176,258 | 33,647 | 93,951 | 15,013 | 34.8 | 130 (111) | 85 (78) | 8 (4) | 37 (29) | NC_086625 | Kunming, Yunnan |
| A. kachinensis | 173,330 | 33,091 | 92,030 | 15,118 | 35 | 130 (110) | 85 (78) | 8 (4) | 37 (28) | PP072244 | Xishuangbanna, Yunnan |
| A. krausei | 172,418 | 32,422 | 91,983 | 15,591 | 35.23 | 130 (110) | 85 (78) | 8 (4) | 37 (28) | OR416863 | Xishuangbanna, Yunnan |
| A. albus | 175,728 | 33,693 | 93,091 | 15,251 | 34.94 | 129 (111) | 84 (77) | 8 (4) | 37 (29) | OM037675 | - |
Total genes (unique): total number of genes; Protein-coding genes (unique): number of protein encoding genes (unique); rRNAs(unique): Number of rRNA genes(unique); tRNAs: Number of tRNA genes(unique)
Fig. 2.
Chloroplast genome maps of Amorphophallus with annotated genes. Genes within the circle are clockwise, while those beyond the circle are counterclockwise. Different colors indicate functional gene groups. The darker and lighter shades of gray in the inner circle represent the content of GC and AT, respectively
Table 2.
Gene contents of chloroplast genome in 13 Amorphophallus species
| Function | Gene group | Name of genes |
|---|---|---|
| Photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI |
| Subunits of NADH-dehydrogenase | ndhA*, ndhB*(× 2), ndhC, ndhD, ndhE | |
| ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | ||
| Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petN, petL | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE | |
| psbF, psbH, psbI, psbJ, psbK, | ||
| psbL, psbM, psbN, psbT, psbZ | ||
| Subunits of rubisco | rbcL | |
| Self-replication | Small subunit of ribosome | rps11, rps12*(× 2), rps14, rps15, rps16* |
| rps18, rps19, rps2, rps3, rps4, rps7(× 2), rps8 | ||
| Large subunit of ribosome | rpl2* (× 2), rpl14, rpl16*, rpl20, rpl22, | |
| rpl23(× 2), rpl32, rpl33, rpl36 | ||
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| Ribosomal RNA genes | rrn16(× 2), rrn23(× 2), rrn4.5(× 2), rrn5(× 2) | |
| Transfer RNA genes | trnA-UGC(2)*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA | |
| trnG-GCC, trnG-UCC*, trnH, trnI-GAU(×2)*, trnM-CAU(× 3) | ||
| trnK-UUU*, trnL-UAA*, trnL-CAA(× 2), trnL-UAG, trnN-GUU(× 2) | ||
| trnP-UGG, trnQ-UUG (× 3), trnR-UCU, trnR-ACG(× 2), trnS-GCU | ||
| trnS-UGA, trnS-GGA, trnT-UGU, trnT-GGU, trnV-UAC* | ||
| trnV-GAC(× 2), trnW-CCA, trnY-GUA, trnfM-CAU | ||
| other genes | Maturase | matK |
| Protease | clpP** | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD* | |
| c-type cytochrome synthesis | ccsA | |
| translation initiation factor | infA | |
| Genes of unknown function | Conserved open reading frames | ycf1b (c×2), ycf2(× 2), ycf3**, ycf4, ycf68*(× 2)a |
*genes containing one intron
**genes containing two introns; (×2) genes with two copies; (×3) genes with three copies
aycf68 is only present in two chloroplast genomes of A. albus Kunming and A. konja
bgene was one in A. albus Kunming, A. konja and A. yunnanensis
cgenes are two copies four chloroplast genomes of A. muelleri ‘zhuyajin1’, Amorphophallus SP., A. titanum and A. Krause Kunming; ①: ndhA has no intron in two chloroplast genomes of A. albus Kunming and A. konja; ②: rpl23 has only one copy in chloroplast genome of A. coaetaneus, and this gene is absent in the chloroplast genomes of A. yunnanensis and A.tonkinensis; ③: trnA-UGC has no intron in two chloroplast genomes of A. albus Kunming and A. konja; ④: trnK-UUU has no intron in chloroplast genome of A. tonkinensis; ⑤: trnL-UAA has no intron in chloroplast genome of A. coaetaneus; ⑥: rpl16 has no intron in chloroplast genome of A. krausei;⑦: trnQ-UUG has three copies only in chloroplast genome of A. coaetaneus; ⑧: infA is only present in chloroplast genome of A.titanum. ⑨ accD has one intron only in three chloroplast genomes of A. albus, A. konja and A. coaetaneus
Fourteen genes (rps16, atpF, rpoC1, petB, petD, rpl2, ndhB, ndhA, rps12, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) contained one intron in all genomes except in A. albus, A. konja and A. coaetaneus (Table 2). In addition to the 14 genes mentioned above, the accD gene had a single intron in three species (A. albus, A. konja, and A. coaetaneus), trnK-UUU had no intron only in chloroplast genome of A. tonkinensis, rpl16 had no intron only in chloroplast genome of A.kachinensis, while ycf68 contained one in A. albus and A. konja. In all 13 species, two introns were found in ycf3 and clpP. The rps12 gene was identified as a trans-splicing gene with 5' exon located in the LSC region and the 3' exon duplicated and located in the IR (IRa and IRb) regions in all species.
Codon usage
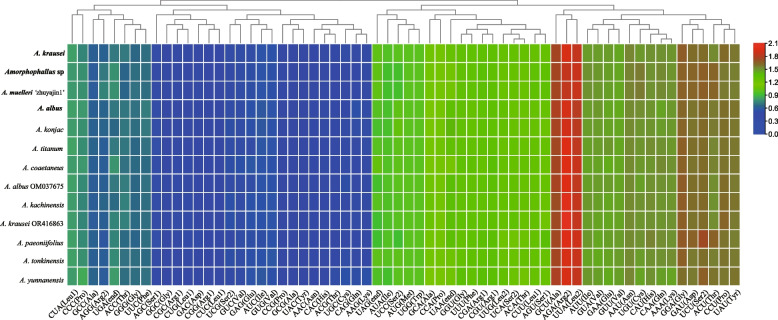
The codon usages of the protein-coding genes in the chloroplast genome from seven Amorphophallus species were analyzed. A total of 64 RSCU were presented in the Amorphophallus plastomes, and the number of codons ranged from 25,520 to 28,798 (Table S4). Within these codons, leucine (Leu) was the most abundant amino acid, comprising 10.01%–10.35% of the total occurrences, followed by isoleucine (Ile) with 8.50% (A. albus) and 8.84% (Amorphophallus sp.). However, cysteine (Cys) was the least prevalent amino acid, accounting for only 1.12% (A. albus OM037675) and 1. 33% (Amorphophallus sp.) (Table S4). The codons ATG and TGG, which encode methionine (Met) and tryptophan (Trp), respectively, showed no codon bias with RSCU values of 1.00 in these Amorphophallus genomes (Fig. 3; Table S4). Thirty-three codons were identified with an RSCU value greater than 1. Among them, except for UUG (Leu), all codons ended with A or U(T) nucleotides (Fig. 3 and Table S4). This observation suggested a preference for A and T as the terminal bases in codons.
Fig. 3.
Heat map analysis for relative synonymous codon usage (RSCU) values of all protein-coding genes of 13 complete chloroplast genomes in Amorphophallus. Red and blue indicates higher and lower RSCU values, respectively. The species in bold are sequenced in this study
Repeat sequence and SSR analyses
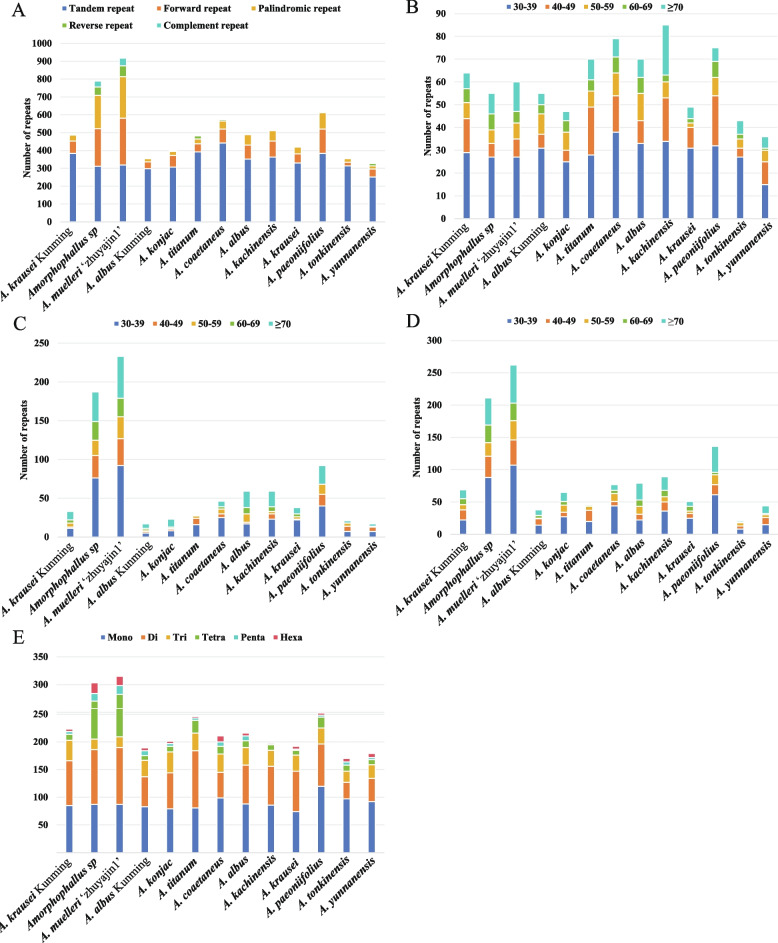
In the chloroplast genomes of Amorphophallus, a comprehensive analysis revealed the presence of 4,446 tandem repeats (Table S5). Amorphophallus yunnanensis (252) had the lowest, and A. coaetaneus (442) had the highest number of tandem repeats (Fig. 4A; Table S5). The length of tandem repeats varied among the 13 chloroplast genomes; however, most tandem repeats existed in the 30–39 bp (Fig. 4B). The 13 Amorphophallus chloroplast genomes had four categories of long repeats, including forward, reverse, complement, and palindromic repeats (Fig. 4A). The long repeats ranged from 48 (A. tonkinensis) to 599 (A. muelleri ‘zhuyajin1’) (Table S6). The maximum number of long repeats were forward repeats, ranging from 21 (A. tonkinensis) to 262 (A. muelleri ‘zhuyajin1’), followed by palindromic repeats, varied from 17 (A. albus) to 233 (A. muelleri ‘zhuyajin1’) (Fig. 4A). The reverse repeats and complement repeats ranged from 3 (A. kachinensis) to 61(A. muelleri ‘zhuyajin1’), and 0 (A. kachinensis A. albus, A. konjac) to 43 (A. muelleri ‘zhuyajin1’), respectively (Fig. 4A-D). The repeat sequence length was 30–204 bp and primarily 30–39 bp among Amorphophallus (Fig. 4B–D).
Fig. 4.
Analysis of repeats and SRRs in seven complete chloroplast genomes of the Amorphophallus. A Different types of repeats in each chloroplast genome. B Numbers of tandem repeats more than 30 bp long in each chloroplast genome. C Numbers of palindromic repeats more than 30 bp long in each chloroplast genome. D Numbers of forward repeats more than 30 bp long in each chloroplast genome. E Total numbers and different types of SSRs detected in each chloroplast genome. Mono: mononucleotide, Di: dinucleotide, Tri: trinucleotides, Tetra: tetranucleotide, Penta: pentanucleotide, Hexa: hexanucleotide. The species in bold are sequenced in this study
In the Amorphophallus chloroplast genomes, a total of 170–315 SSRs were identified, with numbers of mononucleotides, dinucleotides, trinucleotides, tetranucleotides, pentanucleotide, and hexanucleotide SSRs ranging from 74–120, 30–103, 19–38, 8–73, 3–16, and 2–19, respectively (Fig. 4E; Table S7). It was observed that mononucleotide and dinucleotide SSRs were very common in all sequenced genomes. Most mononucleotide repeats consisted of A/T with minimal G/C content, and most of the dinucleotide repeats consisted of AT/TA sequences in all seven species (Table S7).
IR expansion and contraction
We compared the expansion and contraction of IRs regions at the LSC/IRs/SSC boundaries among seven Amorphophallus species (Fig. 5). The complete chloroplast genome structure of seven Amorphophallus species was different and classified into five types based on gene positions at the LSC/IRs/SSC boundaries. Type I consisted of A. krausei, Amorphophallus sp, A. muelleri ‘zhuyajin1’, A. titanum, A. albus OM037675, A. kachinensis, A. krausei PPO72244 and A. tonkinensis. In this type, the JLA (IRa/LSC) and JLB (LSC/IRb) junctions were highly conserved.These boundaries were between rps19 and rpl2 (JLA) or within rpl2 and trnH-GUG (JLB), with varying distances from the border in all species (Fig. 5). The distances between the ends of rpl2 and IRa/LSC borders ranged from 41—46 bp. trnH expanded into the IRa regions with distances ranging from 1—8 bp from the IRa/LSC borders (Fig. 5). At the junction of JLB (LSC/IRb) regions, rps19 was justly located within the LSC region, and a total of 29–162 bp were found between the ends of rps19 and the LSC/IRb borders. rpl2 was present completely in the IR regions with distances ranging from 43–46 bp from the IRb/LSC borders. Regarding the SSC/IRa boundaries regions, the rps15 and ycf1 genes were found in the SSC and IRa regions, respectively. The rps15 expanded into the IRa regions ranging from 1–9 bp in Amorphophallus sp, A. muelleri ‘zhuyajin1’, A. titanum, A. kachinensis and A. albus OM037675 five genomes (Fig. 5). In contrast, the end of the rps15 gene was present completely in SSC region in A. krausei and A. tonkinensis genomes. ycf1 was located in the SSC region in these four genomes, with the lengths ranging from 501—1718 bp from the SSC/IRa boundaries. For IRb/SSC boundaries, the ycf1 and ndhF genes were located at the boundaries in these eight genomes, respectively. The start of ycf1 and the SSC/IRa boundaries ranged from 256–1275 bp (Fig. 5). The ndhF expanded into the IRb regions 2 bp in A. muelleri ‘zhuyajin1’ and Amorphophallus sp. genomes. However, in the remaining species, the ndhF gene was justly located within the IRb/SSC boundaries, with the length ranging from 14–665 bp (Fig. 5). Type II, comprising A. albus (OR438676) Yunnan and A. konjac, was characterized by the presence of ycf1 and trnN at JSA, trnN and ndhF at JSB, and the complete existence of the ycf1 gene in SSC regions. Additionally, the JLB (LSC/IRb) junction regions of this type were between rps19 and rpl2, and JLA (IRa/LSC) were between rpl2 and trnH-GUG, similar to species in type I. Our findings indicated that the IRs in type I were longer, ranging from 32,708–35,204 bp, compared to type II, which had IRs ranging from 26,225–26,226 bp. This expansion of the IR regions may be associated with the replication and length increase of the ycf1 gene in tye I, whereas the ycf1 gene is absent from the IR regions of type II species. Type III, represented by A. coaetaneus, exhibited the IRa/LSC border within rpl2 and trnM, and the LSC/IRb border within rpl23 and trnM. In addition, its IRa/LSC and LSC/IRb boundary genes were similar to type I (Fig. 5). In A. coaetaneus, at the junction of the LSC and IRb regions, rpl23 existed in the LSC region instead of rps19, whereas trnM existed in the IRb region instead of rpl2. At the junction of LSC and IRa, trnM existed in the IRa, and rpl2 existed in the LSC instead of trnH-GUG. Type IV, represented by A. paeoniifolius, the IRb/SSC border in this type was located within ndhB and ndhF, the SSC/IRa was located within rps15 and ndhB (Fig. 5). Type V, represented by A. yunnanensis, the IRb/SSC border in this type was locaed within ycf1, the SSC/IRa was located within rps15 and trnN-GUU.
Fig. 5.
Comparison of the LSC, SSC, and IR boundaries among 13 chloroplast genomes. The light blue, orange, and light green blocks indicate the LSC, IR, and SSC regions. JLB: junction of the LSC and the IRb; JSB: junction of the IRb and the SSC; JSA: connection of the IRa and the SSC; JLA: connection of the SSC and the IRb. The species in bold are sequenced in this study
Comparative chloroplast genomic analysis
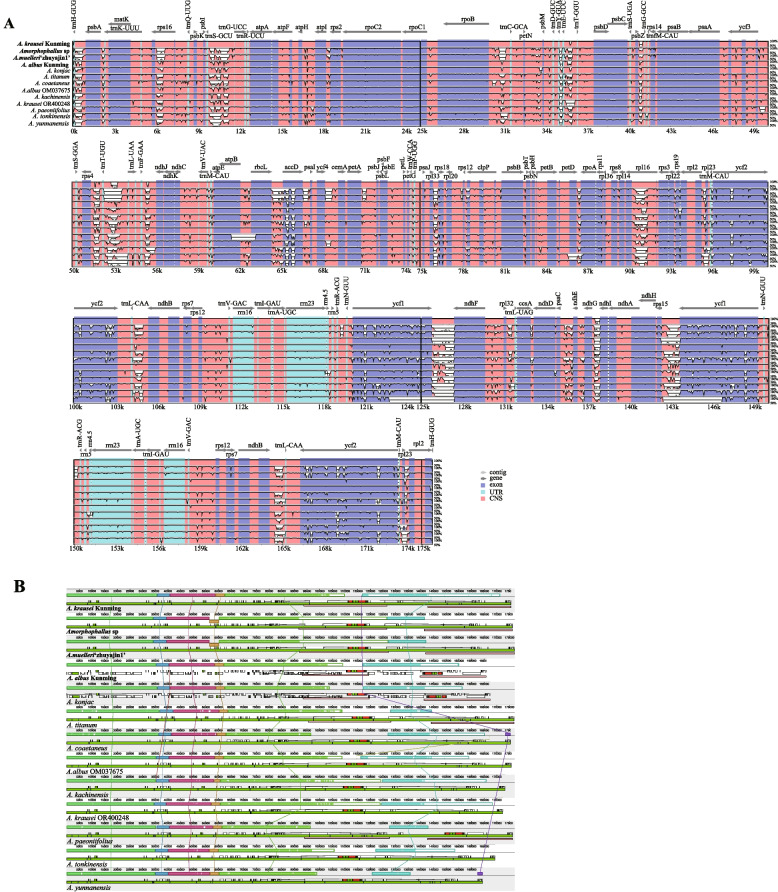
We used the mVISTA tool to compare the divergences among the chloroplast genomes of 13 Amorphophallus species with A. krausei as a reference. The results showed that the seven chloroplast genomes were highly conserved, and the sequences in coding regions were more conserved than in non-coding regions (Fig. 6). The intergenic regions of trnH-GUG-psbA, trnS-GCU-trnG-UCC, rpoB- trnC-GCA, trnY-GUA-trnT-GGU, psbZ-trnG-GCC, rps4-trnT-UGU, trnT-GGU-trnL-UAA, trnF-GAA-ndhJ, rbcL-accD, trnL-CAA-ndhB, ycf1-ndhF, ndhF-rpl32, psaC-ndhE, ndhG-ndhI, rps15-ycf1 and genes of rps16 and rpl16 exhibited high variation (Fig. 6A). They were located in the conserved non-coding regions (CNS) of the chloroplast genomes of these species. Of the exon regions, the greatest divergence was observed in ycf1, ycf2, and accD (Fig. 6A). Mauve collinearity analysis showed that among these seven Amorphophallus species, 6 large conserved regions were observed in the chloroplast genome sequences of 13 species, and their arrangements were similar, indicating that their chloroplast genomes were relatively conserved. However, Amorphophallus sp and A. muelleri 'zhuyajin1' exhibited gene inversions (Fig. 6B).
Fig. 6.
Comparison of the chloroplast genome sequences of 13 Amorphophallus species. A Sequence variation analysis generated with mVISTA. Gray arrows indicated the position and direction of each gene. Purple, blue, pink, and gray bars represent exons, untranslated regions (UTRs), non-coding sequences (CNS), and mRNA, respectively. The scales on the Y-axis represent the average percent identity of sequence similarity ranging from 50 to 100%. B Collinear block analyses of Amorphophallus genome. The white, black, green and colours blue represent protein-coding genes, tRNA genes, intron containing tRNA genes, and rRNA genes, respectively. The species in bold are sequenced in this study
Divergence hotspot region
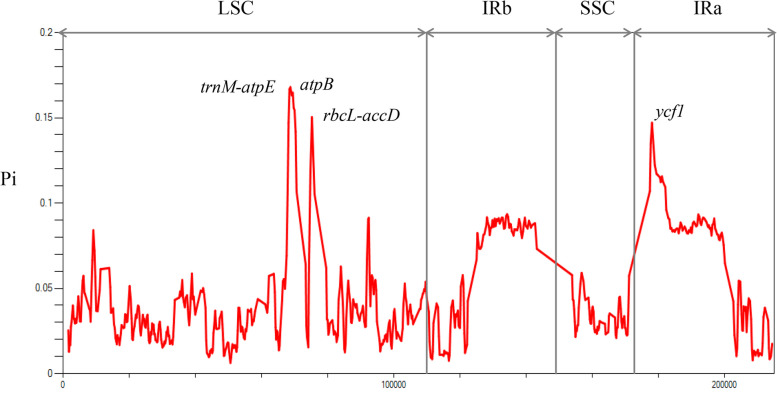
We used the DnaSP v6.0 to perform a sliding window analysis to identify mutation hotspot regions in the 13 chloroplast genomes of Amorphophallus. This analysis enabled us to calculate nucleotide variability (Pi) and identify sequence-level divergences (Fig. 7). The results showed that the Pi values of these 13 Amorphophallus species ranged from 0–0.164, with an average value of 0.029 (Table S8). The LSC region was more divergent than the IR and SSC regions. The trnM-atpE (0.16), atpB (0.155) and atpB-rbcL (0.15) regions were the highest variables among the LSC region (Fig. 7). The ycf1 (0.147) were the most variables in the IR regions. They could be used as specific molecular markers for identifying Amorphophallus species.
Fig. 7.
Sliding window analysis for the nucleotide diversity (Pi) of the whole chloroplast genomes for Amorphophallus species. Window length and step size are 600 bp and 200 bp, respectively. The y-axis represents the nucleotide diversity of each window; the X-axis represents the position of the window’s midpoint. The species in bold are sequenced in this study
Selective pressure analyses
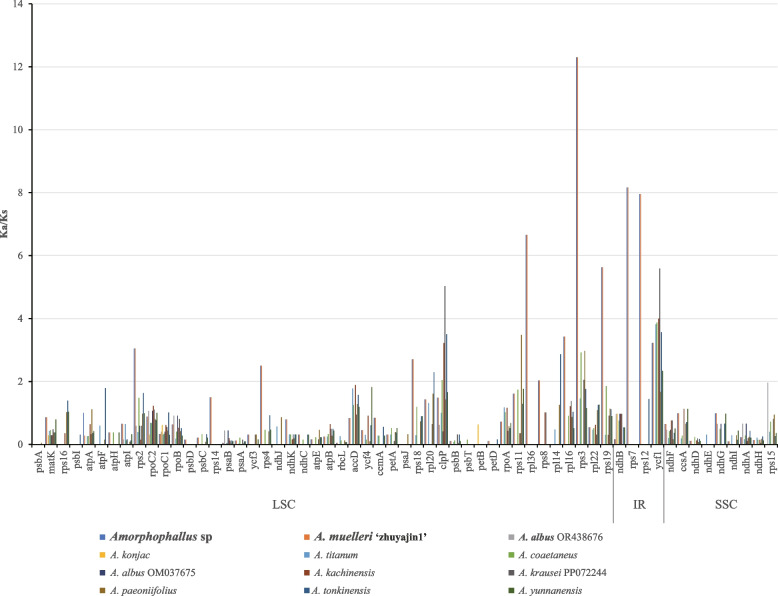
The non-synonymous (Ka)/synonymous (Ka) ratio (Ka/Ks) was calculated for 13 Amorphophallus species (Fig. 8), using the genome of A. krausei as a reference. We found most genes with Ka/Ks < 1 that were supposed to be negatively-selected genes (Table S9). The highest Ka/Ks value was 12.3 for the rps3 gene in Amorphophallus sp. and A. muelleri ‘zhuyajin1’. Furthermore, four genes (rpl36, rps4, rps7, and rps14) with Ka/Ks > 1.00 were identified only in Amorphophallus sp. and A. muelleri ‘zhuyajin1’. The Ka/Ks ratios of ycf1 in Amorphophallus sp, A. muelleri ‘zhuyajin1’, A. titanium, A. coaetaneus, A. krausei, A. paeoniifolius, A. tonkinensis, and A. yunnanensis, rpl20 in Amorphophallus sp, A. muelleri ‘zhuyajin1’, A. titanum, A. paeoniifolius and A. tonkinensis, rps2, rps11, and rps19 in Amorphophallus sp, A. muelleri ‘zhuyajin1’, A. coaetaneus, A. tonkinensis, ccsA in A. kachinensis and A. yunnanensis, and rps12 in Amorphophallus sp, A. muelleri ‘zhuyajin1’ and A. tonkinensis were all > 1.00, indicating that these genes underwent positive selection in different species. Additionally, we also observed accD, atpA, atpF, clpP, rpl14, rpl16, rpl20, rpl22, rpoA, rpoC1, rpoC2, rps8, rps15,rps16, rps18 and ycf4 exhibit Ka/Ks greater than 1 in some species. Overall, there was a more positive selection of genes in triploid Amorphophallus (Amorphophallus sp. and A. muelleri ‘zhuyajin1’).
Fig. 8.
Comparison of non-synonymous (Ka)/synonymous (Ks) substitution ratios among 13 species of Amorphophallus. The species in bold are sequenced in this study
Additionally, using the codon models for estimating gene selection pressure, a small number (34) of protein-coding genes were under positive selection with a posterior probability greater than 0.9 using the BUSTED (Table S10), which was similar to the Ka/Ks method. All the positive—selection genes screened out by the Ka/Ks method were also detected in the codon models, except rpl22, ycf4, rps16 and accD, indicating that these shared genes underwent positive selection. Subsequently, we used FUBAR to detect rare sites that might be under positive selection. The results revealed that the gene ycf1 possesses most positive selective sites, followed by clpP (4) rpoC2 (3), rps11 (8), rps3 (8), rps12 (6) and rps18 (6), whereas one positive selective site was observed in the atpA, atpF, ccsA, rpl36 and rpoA (Table S10).
Phylogenetic relationship analysis
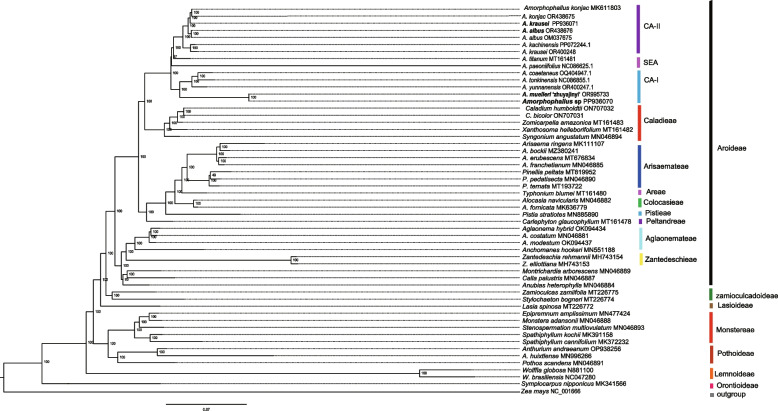
The gene content within chloroplast DNA exhibited high conservation across most land plants. To identify the phylogenetic positions of the A. krausei, Amorphophallus sp, A. muelleri ‘zhuyajin1’, and A. albus within the subfamily Aroideae, we utilized the complete chloroplast genome of 49 species from the seven subfamilies within Araceae, including Aroideae, Lasioideae, Lemnoideae, Monsteroideae, Orontioideae, Pothoideae, and Zamioculcadoideae (Table S11). We constructed a phylogenetic tree using Zea mays as the outgroup. Within the subfamily Aroideae, Amorphophallus species formed a distinct clade with robust bootstrapping values of 100%, constituting a well-supported monophyletic evolutionary branch. The phylogenetic tree indicated that these Amorphophallus species were divided into three clades, including continental Asia II (CA-II), continental Asia I (CA- I), and Southeast Asia clade (SEA) clade (Fig. 9). Within the CA-II clade, A. krausei, A. kachinensis, A. albus, and A. konjac clustered together, with a bootstrap value of 100%, indicating that these species had a close relationship. In terms of CA- I clade, Amorphophallus sp exhibited a particularly close relationship with A. muelleri ‘zhuyajin1’, both members of the same branch, with a bootstrap value of 100%, indicating that A. muelleri ‘zhuyajin1’ and Amorphophallus sp are the most closely related species. Nevertheless, A. coaetaneus and A. tonkinensis cluster into a subclade and form a sister relationship with the subclade of A. yunnanensis, suggesting that they are more closely related. The SEA clade contains two species, A. paeoniifolius and A. titanium. Furthermore, the genus Amorphophallus was found to be a sister to the genera Caladium, Zomicarpella, Xanthosoma, and Syngonium (Fig. 9). The subfamily Aroideae was the crown group, exhibiting a sister relationship with the subfamily Zamioculcadoideae. The subfamily Monsteroideae revealed a sister relationship with the subfamily Pothoideae. The subfamily Orontioideae was the basal group, followed by Lemnoideae.
Fig. 9.
Phylogenetic trees were constructed using the maximum likelihood (ML) based on the complete chloroplast genomes of 54 Araceae species. The numbers above the nodes indicate support values. The species in bold are sequenced in this study
Discussion
In this study, we characterized the complete chloroplast genomes of four Amorphophallus species and compared them with those of nine available species within this genus. The results showed that the chloroplast genome structure in Amorphophallus is highly conserved, comprising IRa and IRb, which separate LSC and SSC regions. Interestingly, these seven species exhibited variation in chloroplast genome size (Table 1), with the largest chloroplast genome size in A. muelleri’ ‘zhuyajin1’ and the smallest in A. yunnanensis, with a difference of 12,659 bp. This phenomenon may be due to IR expansion, contraction, and recombination of the chloroplast genome among these Amorphophallus species [42]. In addition, they shared similar GC content (34.5% − 36%), rRNAs, most of the protein-coding genes, and tRNAs, which also had been found in other plants [42, 43]. Our study identified 126 − 131functional genes, comprising 81 − 86 protein-coding genes, 36 − 39 tRNA genes, and 8 rRNA genes, which are in agreement with previous reports of the species in Araceae, including A. konjac [33, 38], Alocasia fornicate [36], Colocasia gigantea, Caladium bicolor and Xanthosoma sagittifolium [42]. However, a recent study reported reduced gene content in four species of the genus Amorphophallus [35], which is an exception. In particular, A. albus, A. bulbifer, A. konjac, and A. muelleri were found to possess 113 (79 protein-coding genes, 30 tRNAs, and 4 rRNAs), 111 (78 protein-coding genes, 29 tRNAs, and 4 rRNAs), 111 (78 protein-coding genes, 29 tRNAs, and four rRNAs), and 113 (80 protein-coding genes, 29 tRNAs, and 4 rRNAs) genes, respectively [35]. Although these 13 chloroplast genomes were highly conserved, intron loss, gene duplication, and gene loss were observed in this study. For example, the chloroplast genomes of all Amorphophallus species except A. coaetaneus, A.yunnanensis and A. tonkinensis had two copies of rpl23. Amorphophallus coaetaneus had one copy of rpl23, while A.yunnanensis and A. tonkinensis lost rpl23. Moreover, accD had no intron in the genomes of A. krausei, A. muelleri ‘zhuyajin1’, Amorphophallus sp, and A. titanum, while the other three genomes exhibited one intron in this protein-coding gene, indicating that intron loss had occurred during the evolutionary history of A. krausei, A. muelleri ‘zhuyajin1’, Amorphophallus sp and A. titanum. In contrast, Liu et al. [35] reported the deletion of rpl23, rpl2, trnL-CCA, trnG-GCC, accD, and psbE in the genus Amorphophallus. In this study, we sequenced and de novo assembled the chloroplast genomes of A. krausei, A. muelleri ‘zhuyajin1’, Amorphophallus sp, and A. albus. The gene content of these genomes is similar to that of previous reports in aroids, as well as A. titanum [36], A. konjac [38] and other Monsteroideae (Araceae), including Spathiphyllum patulinervum, Stenospermation multiovulatum, Monstera adansonii, and Rhaphidophora amplissima [43]. Nevertheless, the gene deletion mentioned above was not supported. Furthermore, certain events of intron loss, gene duplication, and gene loss were reported within other plants, including Aglaonema cultivars [44], Zingiberoideae species [45], Costaceae species [46]. The gene loss events involved ycf68, trnS-CGA, trnS-GGA, and trnT-GGU, and intron loss events involved trnG-UCC in the Aglaonema cultivars [44].
Previous studies have shown that IR contraction and expansion of the chloroplast genomes were considered significant evolutionary events. These events can result in chloroplast genome size variations, production of pseudogenes, gene duplication, and reduced duplicate genes to one copy [47]. Our results also indicated that genome lengths and boundaries of IR expansion exhibited variations among these 13 genomes. Amorphophallus species in type I and type III showed two functional copies of ycf1 gene due to duplication in IR regions, one each in IRa and IRb, while ycf1 is present completely in the SSC region and hence exists as single copy with in type II. The same phenomenon exists in some species of the family Araceae. For example, A. simorrhinum, C. glaucophyllum, and T. blumei in the Dracunculus clade (Araceae) contained two functional copies of the ycf1 gene, one each in IRa and IRb [36]. In Colocasia gigantean [42], Anthurium huixtlense, Pothos scandens [47], P. pedatisecta, C. esculenta, A. franchetianum, Alocasia fornicate, and Steudnera colocasiifolia [36], this gene existed completely as one functional copy in SSC regions. Moreover, in Caladium bicolor and Xanthosoma sagittifolium, this gene extended into IRa from SSC and existed as a single copy in the plastom genome [42]. However, in some Araceae species, including Anubias heterophylla, Aglaonema costatum, Syngonium angustatum, Xanthosoma helleborifolium, and Zomicarpella amazonica, the functional copy of the ycf1 gene extended into IRa from SSC; however, a truncated copy also existed in IRb [36, 48]. Previous studies of angiosperm chloroplast genomes revealed the complete existence of trnH-GUG in the LSC region or integration of trnH-GUG into the IRa region [42, 44, 48]. In this study, trnH-GUG was found to be at JLA in all 12 Amorphophallus except A. coaetaneus (type II), either starting inside the IRa (6 − 8 bp) in A. krausei, Amorphophallus sp, A. muelleri ‘zhuyajin1’ and A. titanum or starting up to 1 bp (A. albus) and 86 bp (A. konjac) after the start of LSC region. In A. coaetaneus, at the junction of the JLA region, rpl2 exists in the LSC instead of trnH-GUG. In Anchomanes hookeri, at the junction of JLA, psbK exists in the LSC instead of trnH-GUG [48]. As reported by Abdullah et al. the duplication events of ycf1 and/or other genes that present at the junction of single copy and inverted repeats in chloroplast genome are species-specific rather than cladistic synapomorphies [36]. Further genomic resources may provide a better understanding of the phylogenetic level of IR contraction and amplification in the Amorphophallus genus as well as Araceae family.
Chloroplast genomes are rich in SSRs, long repeats, and highly divergent regions, widely used to determine phylogenetic relationships between organisms and identify species and cultivars. Our study indicated that most SSRs were mononucleotide repeats and were short A/T repeats consistent with previous studies [42]. In current study, repeats detection results were the same as most other Araceae plants; for example, four types of oligonucleotide repeats were identified, and forward repeats were the most abundant types of repeats [42, 47]. Previous studies have shown that these repeats may be associated with generating substitutions and InDels [49]. Furthermore, divergent analyses implemented by mVISTA revealed that the non-coding regions were more divergent and variable than the coding regions, indicating that non-coding regions are suitable for molecular marker identification in Amorphophallus, consistent with previous studies in Araceae chloroplast genomes [42, 50]. Twenty-two regions (trnH-GUG-psbA, trnS-GCU-trnG-UCC, rpoB-trnC-GCA, trnY-GUA-trnT-GGU, psbZ-trnG-GCC, rps4-trnT-UGU, trnT-GGU-trnL-UAA, trnF-GAA-ndhJ, rbcL-accD, trnL-CAA-ndhB, ycf1-ndhF, ndhF-rpl32, psaC-ndhE, ndhG-ndhI, rps15-ycf1, ndhB-trnL-CAA, atpB-rbcL, rbcL-accD, rps16, rpl16, ycf1, ycf2, and accD) with high variation were identified from Amorphophallus based on the mVISTA analysis. Similarly, trnH-psbA, rps4-trnT-UGU, trnL-ndhB, psaC-ndhE, rps15-ycf1, rpl16, ycf1, and ycf2 showed divergence in four Zantedeschia (Araceae) [51]. Additionally, divergent analyses implemented by nucleotide diversity revealed 4 highly divergent regions among 13 Amorphophallus chloroplast genomes, including trnM-atpE, atpB, atpB-rbcL and ycf1. In previous studies, 12 regions (trnH-GUG-CDS1, trnH-GUG-CDS1_psbA, trnS-GCU_trnS-CGA-CDS1, psbC-trnS-UGA, rps4-trnT-UGU, trnF-GAA-ndhJ, psbF-psbE, petD-CDS2-rpoA, ycf1-ndhF, rps15-ycf1-D2, ccsA-ndhD, and trnY-GUA-trnE-UUC) showed significantly higher Pi values in seven Aglaonema species [44]. Besides, 8 highly divergent regions (rps16-trnQ-UUG, trnS-GCU-trnG-UCC, atpH-atpI, petA-psbJ, psbE-petL, ndhF, rpl32, and ndhE) were identified in seven Lemnoideae species [52]. Similarly, 14 highly divergent regions (trnS-trnA, psbI-trnS, ndhF, ycf1, trnQ-psbK, rpl32-trnL, trnC-petN, trnT-trnL, rps16-trnQ, trnT-psbD, rpoB-trnC, trnL-ccsA, psbK-psbI, and petA-psbJ) were found in Symplocarpus [50]. Furthermore, 16 regions (trnN-ndhF, trnS-trnG, rpl32-trnL, psaC-ndhE, ndhG-ndhI, accD-psaI, ccsA-ndhD, rps15-ycf1, trnL-ccsA, psbI-trnS, petD-rpoA, rps19-rpl2, atpH-atpI, ccsA, ndhF, and ndhD) with high nucleotide diversity were identified in Aroideae [42]. Six highly divergent regions (trnH-GUG_psbA, rps4-trnT-UGU, trnF-GAA-ndhJ, rps15-ycf1, ccsA-ndhD, and petD-rpoA) were used as DNA barcodes in Araceae species or were in the marker development of DNA barcodes [42, 44, 48]. When these hotspot regions based on the representative lineages within the family Araceae were compared to those from seven congeneric species of Amorphophallus, we found that highly variable regions on the family level were not the same as those within the genus Amorphophallus. However, trnH-GUG-psbA, ndhB-trnL, psaC-ndhE, trnS-trnG, and rps4-trnT regions were consistent and highly variable in most Araceae species. The absence of hotspot regions in other Araceae species in the remaining regions of Amorphophallus suggested that there was no universal "best" region. Additionally, these regions may evolve rapidly within the genus Amorphophallus and can be used as special DNA barcodes for Amorphophallu species. Claudel et al. reported that the rbcL gene had significant potential as a DNA barcoding tool for specific Amorphophallu species [1]. Our results also support the high variability of the atpB-rbcL and rbcL-accD regions in 13 Amorphophall chloroplast genomes. In summary, we identified several highly variable plastid regions in the Amorphophall genus, which may help determine phylogenetic relationships and can serve as markers for barcoding and phylogenetic studies at higher taxonomic levels.
The Ka/Ks ratio is a vital tool for determining genome evolution. In this study, most genes displayed ratios of less than 1.00, which aligns with observations in other high plant chloroplast genomes [53]. However, certain Araceae species have reported higher Ka/Ks values, signifying a positive selection of genes [42, 47]. Our preliminary results indicated the presence of 23 genes (atpA, atpF, rpl14, rpoA, rpoC1, rpoC2,rpl36, ccsA, rpl16, rps4, rps7, rps8, rps11, rps12, rps14, clpP, rps3, ycf1, rpl20, rps2, rps18, rps19 and rps15) undergoing positive selection in the chloroplast genomes of Amorphophallus. Among them, four genes (rpl36, rps4, rps7, rps8and rps14) exhibited positive selection in Amorphophallus sp and A. muelleri ‘zhuyajin1’. atpF and rpoC1 only showed positive selection in A. tonkinensis, atpA in A. paeoniifolius, and ycf4 only in A. yunnanensis. The remaining genes showed positive selection in more than one species of Amorphophallus species, indicating that these genes underwent adaptive evolution in different environments. Previous studies have shown that positive selection of rps3, ycf1, and ycf2 in angiosperms may be very common [44]. rps3, ycf1 and ycf2 showed positive selection in 16 Aglaonema species [44] and four Zingiber species [45]. In addition, ycf exhibited positive selection in Alocasia fornicata, Colocasia esculenta, Steudnera colocasiifolia, Arisaema franchetianum, Arisarum simorrhinum, and Carlephyton Glaucophyllum [36], and ycf1 in four Pinellia [42]. Furthermore, other genes experiencing positive selection have been identified, including rpl2 in Epipremnum aureum [52], rps2 in 16 Araceae species [44] and ndhF, ndhK, rbcL, rpoC1, rpoC2, and matK in Colocasia gigantea, Caladium bicolor, and Xanthosoma sagittifolium [42], ccsA, matK, and ndhF in four Anubias (Araceae) [54], rpl33 in Typhonium blumei, rps8 in Xanthosoma helleborifolium, rps16 in Zomicarpella amazonica [36], and clpP and rpl36 in Stylochaeton bogneri [55]. Aroideae species inhabit diverse habitats, including swamps, river margins, and damp sites [56]. Therefore, various types of positively selected genes in these species may be associated with distinct ecological pressures of their respective niches [47].
Chloroplast genomes containing sufficient variable loci are valuable for determining evolutionary and phylogenetic relationships [57]. In our study, we analyzed the chloroplast genomes of 54 species from seven subfamilies of Araceae, including Aroideae, Lasioideae, Lemnoideae, Monsteroideae, Orontioideae, Pothoideae, and Zamioculcadoideae to gain insights into their evolutionary relationships. The maximum likelihood (ML) phylogeny confirmed the phylogenetic position of Amorphophallus within the subfamily Aroideae, with the Amorphophallus species forming a single monophyletic group with a bootstrap value of 100. Previously, the genus Amorphophallus was also identified as monophyletic. The phylogenetic tree also indicated that these Amorphophallus species were divided into three clades. Amorphophallus albus, A. krausei, A. kachinensis and A. konjac were clustered into the continental Asia II (CA-II) clade, A. coaetaneus, A. tonkinensis, A. yunnanensis, A. muelleri ‘zhuyajin1’ and Amorphophallus sp were clustered into continental Asia I (CA- I) clade, and A. titanium belongs to the Southeast Asia clade (SEA), which was in line with the previous study with nuclear (ITS1) and plastid (rbcL and matK) regions [1]. Moreover, the genus Amorphophallus was most closely related to Caladium, Zomicarpell, Xanthosoma, and Syngonium, consistent with previous studies on the Araceae family [36]. Generally, the phylogenetic inference among the species of seven subfamilies of Araceae was in agreement with previous findings [42, 44]. The subfamily Aroideae showed a sister relationship with the subfamily zamioculcadoideae, and the subfamily Pothoideae was closer to the subfamily Monsteroideae. The subfamily Orontioideae was the basal group, while the subfamily Aroideae was the crown group.
Conclusions
In the current study, we sequenced the chloroplast genomes of four Amorphophallus and compared them with three previously published chloroplast genomes. These seven genomes exhibited a typical quadripartite structure, similar GC content, rRNAs, codon usage, long repeats, and SSRs. However, there were variations in genome lengths, tRNA gene contents, protein-coding genes introns, and IR borders. A previous study reported that ycf1, accD, psbE, trnL-CAA, and trnG-GCC genes were absent in four Amorphophallus species; however, our study does not support the aforementioned gene loss. Comparative analyses of these chloroplast genomes identified 4 divergent hotspots (trnM-atpE, atpB, atpB-rbcL and ycf1) with potential application as molecular markers for future population genetic studies within the Amorphophallus. The Ka/Ks, BUSTED and FUBAR analyses of 13 Amorphophallus species showed that atpA, atpF, rpl14, rpoA, rpoC1, rpoC2, rpl36, ccsA, rpl16, rps4, rps7, rps8, rps11, rps12, rps14, clpP, rps3, ycf1, rpl20, rps2, rps18, rps19, and rps15 were under positive selection, which can be due to adaptation to the environment. Phylogenetic trees based on whole chloroplast genomes revealed that Amorphophallus was a sister to Caladieae and had significant support. All Amorphophallus species formed a monophyletic evolutionary clade and were divided into three groups: CA-II, SEA, and CA-I. These findings provide a valuable reference for studying the phylogeny and conservation of Amorphophallus and lay a solid foundation for conducting phylogenetic analyses, classification efforts, and the exploration of genetic diversity within the broader Araceae family.
Methods
Plant material sampling, DNA extraction, and sequencing
The samples of cultivated plants, including A. albus, A. krausei and A. muelleri ‘zhuyajin1’ were collected from Konjac Genetic Resources Garden of Kunming University, Yunnan Province (24.97406°N, 102.79605°E). The sample of wild plant Amorphophallus sp was collected by Li Yu on July 16, 2018, from Mansai Village (22.131258981521°N, 101.31695415018°E), Xiangming Township, Mengla County, Xishuangbanna, Yunnan province, China. The taxonomic identifcation is authenticated by Professor Lei Yu (the head of Yunnan Key Laboratory of Konjac Biology, Kunming University), the author of the study of areas of the genus Amorphophallus. Now, Amorphophallus sp has been introduced in the konjac germplasm resource garden of Kunming University, China and is growing well. Because the Amorphophallus species we collected from field were currently not protected species, no permission was required during the sampling process. Voucher specimens were placed in the deposited in the Herbarium of Yunnan Urban Agricultural Engineering and Technological Research Center, Kunming University under voucher specimens numbers BMY001(A. albus), A. krausei (XMY001), A. muelleri ‘zhuyajin1’(ZYJ01b) and Amorphophallus sp (ZY010). Total genomic DNA was extracted from each fresh leaf using the Qiagen DNeasy Plant Mini kit (Qiagen Co., Hilden, Germany). Subsequently, DNA quality and quantity were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1% (w/v) agarose gel (Fig. S1), respectively. Paired-end libraries were constructed using the NEBNext UltraTM DNA library prep kit and sequenced on an Illumina HiSeq 2500 platform, resulting in 2 × 150 bp paired-end reads. In addition, nine published plastids of Amorphophallus (Table 1) were also added to determine their inter—generic variation.
Chloroplast genome assembly and annotation
The quality of the raw paired-end reads was assessed using FastQC [58] and trimmed using Trimmomatic software [59]. Then, the trimmed reads were de novo assembled into contigs using SOAPdenovo v.2.04 with the default parameters [60] and GetOrganelle v1.7.8 [49]. To validate the contigs, they were aligned against a reference chloroplast genome of A. konjac (NC_046702) using the Blast program. The aligned contigs were then oriented according to the reference chloroplast genome. Complete chloroplast genomes with the default parameters were annotated using CPGAVAS2 [61] and GeSeq [62]. Subsequently, tRNAs were identified using tRNAscan-SE with the default parameters [63]. The circular map of the genomes was constructed using Organellar Genome DRAW (OGDRAW) version 1.3.1 [64].
Sequence analysis
The relative synonymous codon usage (RSCU) analysis in protein-coding genes was performed using Geneious R8.1 [65]. SSR was predicted using MISA [66] with minimum repeat thresholds set at ten for mononucleotide repeats, five for dinucleotide repeats, four for trinucleotide repeats, three for tetranucleotide repeats, three for pentanucleotide repeats, and three for hexanucleotide repeats. Tandem repeats with default parameters were determined using the tandem repeat finder (http://tandem.bu.edu/trf/trf.submit.options.html).
Comparative genome and sequence divergence analyses
The mVISTA software [67] in Shuffle-LAGAN mode was used to compare the seven complete chloroplast genomes of Amorphophallus, using the A. krausei sequence as the reference. To visualize the contraction and expansion of the IR junction sites, IRScope [68] was used. For interspecific comparisons, MAFFT v.7 [69] was used to align the complete chloroplast genomes of the seven species. Subsequently, DnaSP version 6.0 [70] was employed to perform a sliding window analysis with a window length of 600 bp and a step size of 200 bp to determine the nucleotide diversity (Pi) of the plastome based on the alignment results.
Analysis of synonymous (Ks) and non-synonymous (Ka) substitution rate
We used synonymous substitution rates (Ks) and non-synonymous substitution rates (Ka), along with their ratio Ka/Ks, to determine the role of natural selection in shaping the molecular evolution of the A. albus chloroplast genome. All protein-coding genes were aligned using MAFFT. The Ks, Ka, and Ka/Ks values were calculated using the KaKs_Calculator 2.0 software [71]. Values of Ka/Ks > 1, Ka/Ks = 1, and Ka/Ks < 1 indicate positive, neutral, and purifying selection, respectively.
Phylogenetic analysis
To determine the phylogenetic relationships and verify the phylogenetic placement of Amorphophallus., 49 aroid taxa were considered, including 54 species obtained from NCBI (Table S11) and four species introduced in this study. All chloroplast genome sequences were aligned using MAFFT V7, with Zea mays serving as outgroup. The phylogenetic tree was constructed using IQ-TREE v. 1.4.2 [68]. A bootstrap test was performed with 1000 iterations to calculate the maximum likelihood (ML) bootstrap value. The best-fit model used for this analysis was TVM + F + R9.
Supplementary Information
Acknowledgements
Thank all those who have helped us.
Abbreviations
- IR
Inverted repeat regions
- LSC
Large single copy region
- SSC
Small single copy region
- SSR
Simple sequence repeats
- Ka
Non-synonymous
- Ks
Synonymous
- ML
Maximum Likelihood
Authors’ contributions
L.F.L. conceived and designed the experiments, generated and analyzed the data, wrote the draft of the manuscript and revised it. M.Y., Y.Q., P.H.G. and S.W.Y. conceived the study. Y.T.Z. and J.W.G. collected plant materials. H.Y.W., J.N.L. and J.R.Z .analyzed the data and help to revise the manuscript. F.Y.H. and L.Y. planned and directed the study and revised the manuscript. All authors contributed to the experiments and approved the final draft of the manuscript.
Funding
This study was funded by Yunnan Provincial Science and Technology Department (grant no. 202401AU070020, 202201AU070043, 202101BA070001-174, 202201AT070113, 202101AO070075), Yunnan Education Department Research Project (grant no. 2024J0771), Kunming University Talent Program (grant no. YJL23026, YJL23010, YJL23005, YJL23007).Yunnan Province Yu Lei Expert Grassroots Research Workstation, Yunnan Province Youth Talent Support Program (Grant No. YNWR-QNBJ-2018–324). Yunnan Provincial Science and Technology Department (grant no. 202449CE340009).
Data availability
The genome raw reads have been deposited in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under a Project accession: PRJNA1126114. The four newly sequenced complete chloroplast genomes in this study have been submitted to GenBank (https://www.ncbi.nlm.nih.gov) with accession numbers OR995733, OR438676, PP936070 and PP936071, and available in NCBI (https://www.ncbi.nlm.nih.gov/) (see Table S1). The materials are available from the corresponding author on reasonable request after the publication of the work.
Declarations
Ethics approval and consent to participate
The materials involved in the article does not an endangered or protected species; therefore, permission is not required to collect this species. Research on these species, including the collection of plant materials has been carried out in accordance with guidelines provided by Kunming University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fei-Yan Huang, Email: 125593879@qq.com.
Lei Yu, Email: yulei0425@163.com.
References
- 1.Claudel C, Buerki S, Chatro LW, Antonelli A, Alvarez N, Hetterscheid WLA. Large-scale phylogenetic analysis of Amorphophallus (Araceae) derived from nuclear and plastid sequences reveals new subgeneric delineation. J Linn Soc Bot. 2017;184:32–45. [Google Scholar]
- 2.Promprom W, Chatan W, Pasorn P, Prasertsri N, Angkahad T. A new species of Amorphophallus (Araceae) from northeastern Thailand. PhytoKeys. 2023;229:131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi HD, Zhang WQ, Lu HY, Zhang WQ, Ye H, Liu DD. Functional characterization of a starch synthesis-related gene AmAGP in Amorphophallus muelleri. Plant Signal Behav. 2020;15(11):1805903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islam F, Labib RK, Zehravi M, Lami MS, Das R, Singh LP, Mandhadi JR, Balan P, Khan J, Khan SL, Nainu F, Nafady MH, Rab SO, Emran TB, Wilairatana P. Genus Amorphophallus: a comprehensive overview on phytochemistry, ethnomedicinal uses, and pharmacological activities. Plants (Basel). 2023;12(23):3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao C, She X, Liu E, et al. A mixed ploidy natural population of Amorphophallus muelleri provides an opportunity to trace the evolution of Amorphophallus karyotype. J Genet. 2021;100:10. [PubMed] [Google Scholar]
- 6.Zhao C, Harijati N, Liu E, Jin S, Diao Y, Hu Z. First report on DNA content of three species of Amorphophallus. J Genet. 2020;99:36. [PubMed] [Google Scholar]
- 7.Dai J, Chen J, Qi J, Ding M, Liu W, Shao T, Han J, Wang G. Konjac Glucomannan from Amorphophallus konjac enhances immunocompetence of the cyclophosphamide-induced immunosuppressed mice. Food Sci Nutr. 2020;9(2):728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zalewski BM, Chmielewska A, Szajewska H. The effect of glucomannan on body weight in overweight or obese children and adults: a systematic review of randomized controlled trials. Nutrition. 2015;31(3):437-42.e2. [DOI] [PubMed] [Google Scholar]
- 9.Harmayani E, Aprilia V, Marsono Y. Characterization of glucomannan from Amorphophallus oncophyllus and its prebiotic activity in vivo. Carbohydr Polym. 2014;112:475–9. [DOI] [PubMed] [Google Scholar]
- 10.Cui H, Zhu X, Wang Z, Fang J, Yuan T. A purified glucomannan oligosaccharide from Amorphophallus konjac improves colonic mucosal barrier function via enhancing butyrate production and histone protein H3 and H4 acetylation. J Nat Prod. 2021;84(2):427–35. [DOI] [PubMed] [Google Scholar]
- 11.Devaraj RD, Reddy CK, Xu B. Health-promoting effects of konjac glucomannan and its practical applications: a critical review. Int J Biol Macromol. 2019;126:273–81. [DOI] [PubMed] [Google Scholar]
- 12.Majumder M, Sharma M, Maiti S, Mukhopadhyay R. Edible Tuber Amorphophallus paeoniifolius (Dennst.) Extract induces apoptosis and suppresses migration of breast cancer cells. Nutr Cancer. 2021;73(11–12):2477–90. [DOI] [PubMed] [Google Scholar]
- 13.Dwiputri E, Lestari KD, Tan GHK, Sulijaya B, Soeroso Y, Masulili SLC, Takahashi N, Tabeta K, Tadjoedin FM. Osteoclastogenesis inhibitor and antioxidant properties of Konjac Glucomannan in a periodontitis mice model: an in vivo study. Int J Dent. 2023;2023:7400421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jian W, Siu KC, Wu JY. Effects of pH and temperature on colloidal properties and molecular characteristics of Konjac glucomannan. Carbohydr Polym. 2015;134:285–92. [DOI] [PubMed] [Google Scholar]
- 15.Zhu F. Modifications of konjac glucomannan for diverse applications. Food Chem. 2018;256:419–26. [DOI] [PubMed] [Google Scholar]
- 16.Gao Y, Yin S, Yang H, Wu L, Yan Y. Genetic diversity and phylogenetic relationships of seven Amorphophallus species in southwestern China revealed by chloroplast DNA sequences. Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29(5):679–86. [DOI] [PubMed] [Google Scholar]
- 17.Tang R, Liu E, Zhang Y, Schinnerl J, Sun W, Chen G. Genetic diversity and population structure of Amorphophallus albus, a plant species with extremely small populations (PSESP) endemic to dry-hot valley of Jinsha River. BMC Genet. 2020;21(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gholave AR, Pawar KD, Yadav SR, Bapat VA, Jadhav JP. Reconstruction of molecular phylogeny of closely related Amorphophallus species of India using plastid DNA marker and fingerprinting approaches. Physiol Mol Biol Plants. 2017;23(1):155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grob GB, Gravendeel B, Eurlings MC. Potential phylogenetic utility of the nuclear FLORICAULA/LEAFY second intron: comparison with three chloroplast DNA regions in Amorphophallus (Araceae). Mol Phylogenet Evol. 2004;30(1):13–23. [DOI] [PubMed] [Google Scholar]
- 20.Kirschner GK. A hot topic: thermogenesis in Amorphophallus. Plant J. 2023;115(4):872–3. [DOI] [PubMed] [Google Scholar]
- 21.Claudel C, Loiseau O, Silvestro D, Lev-Yadun S, Antonelli A. Patterns and drivers of heat production in the plant genus Amorphophallus. Plant J. 2023;115(4):874–94. [DOI] [PubMed] [Google Scholar]
- 22.Gao P, Qi Y, Li L, Yang S, Guo J, Liu J, Wei H, Huang F, Yu L. Phenylpropane biosynthesis and alkaloid metabolism pathways involved in resistance of Amorphophallus spp. against soft rot disease. Front Plant Sci. 2024;15:1334996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P, Qi Y, Li L, Yang S, Liu J, Wei H, Huang F, Yu L. Amorphophallus muelleri activates ferulic acid and phenylpropane biosynthesis pathways to defend against Fusarium solani infection. Front Plant Sci. 2023;14:1207970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M, Qi Y, Liu J, Gao P, Huang F, Yu L, Chen H. Different response mechanisms of rhizosphere microbial communities in two species of Amorphophallus to Pectobacterium carotovorum subsp. carotovorum infection. Plant Pathol J. 2023;39(2):207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin S, Chen H, Wu W, Gao Y. The complete chloroplast genome assembly of Amorphophallus tonkinensis Engler and Gehrmann 1911 from southwestern China. Mitochondrial DNA B Resour. 2024;9(5):592–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srzednicki G, Borompichaichartkul C. Konjac glucomannan-production, processing, and functional applications. Boca Raton: CRC Press; 2020. [Google Scholar]
- 27.Chen Y, Hu N, Wu H. Analyzing and characterizing the chloroplast genome of Salix wilsonii. BioMed Res Int. 2019;2019:5190425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y, Zhao W, Xu J, Li M, Zhang Y. Chloroplast genome evolution and species identification of Styrax (Styracaceae). BioMed Res Int. 2022;2022:5364094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Qu D, Ma Y. Characterization of the chloroplast genome of Argyranthemum frutescens and a comparison with other species in Anthemideae. Genes. 2022;13(10):1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Wang S, Liu Y, Yuan Q, Sun J, Guo L. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics. 2021;22(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui N, Chen W, Li X, Wang P. Comparative chloroplast genomes and phylogenetic analyses of Pinellia. Mol Biol Rep. 2022;49(8):7873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu H, Liua J, Wang B, An J, Wang Q. Characterization of the complete chloroplast genome of Amorphophallus konjac (Araceae) and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 2019;4:1658–9. [Google Scholar]
- 34.Yin S, Gao Y. Characterization of the complete chloroplast genome assembly of Amorphophallus yunnanensis Engler, Pflanzenr (Araceae) from southwestern China. Mitochondrial DNA B Resour. 2023;8(12):1445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu E, Yang C, Liu J, Jin S, Harijati N, Hu Z, Diao Y, Zhao L. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci Rep. 2019;9(1):809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdullah Henriquez CL, Mehmood F, Hayat A, Sammad A, Waseem S, Waheed MT, Matthews PJ, Croat TB, Poczai P, Ahmed I. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics. 2021;113(1 Pt 1):183–92. [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Dong K, Xiao P, Wu W, Yin S. Complete assembly of the chloroplast genome of Amorphophallus coaetaneus SY Liu & SJ Wei 1986 (Araceae) from southwestern China. Mitochondrial DNA B Resour. 2023;8(7):766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Qi Y, Gao P, Yang S, Zhao Y, Guo J, Liu J, Huang F, Yu L. The complete chloroplast genome sequence of Amorphophallus konjac (Araceae) from Yunnan, China and its phylogenetic analysis in the family Araceae. Mitochondrial DNA B Resour. 2024;9(1):41–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Yin S. The complete chloroplast genome assembly of Amorphophallus kiusianus makino 1913 (araceae) from Southern China. Mitochondrial DNA B Resour. 2024;9(4):522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y, Yin S. Assembly and analysis of the chloroplast genome of Amorphophallus kachinensis Engler & Gehrmann (Araceae) from Southwestern China: implications for conservation and utilization. Mitochondrial DNA B Resour. 2024;9(4):452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin S, Gao Y. The complete chloroplast genome assembly of Amorphophallus krausei Engler, Pflanzenr 1911 (Araceae) from southwestern China. Mitochondrial DNA B Resour. 2023;8(12):1339–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Liu T, Ali A, Xiao Y, Shan N, Sun J, Huang Y, Zhou Q, Zhu Q. Complete chloroplast genome sequences of three aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics. 2022;23(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henriquez CL, Abdullah, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae). Planta. 2020;251(3):72. [DOI] [PubMed] [Google Scholar]
- 44.Li DM, Zhu GF, Yu B, Huang D. Comparative chloroplast genomes and phylogenetic relationships of Aglaonema modestum and five variegated cultivars of Aglaonema. PLoS One. 2022;17(9):e0274067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li DM, Ye YJ, Xu YC, Liu JM, Zhu GF. Complete chloroplast genomes of Zingiber montanum and Zingiber zerumbet: genome structure, comparative and phylogenetic analyses. PLoS One. 2020;15:e0236590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li DM, Pan YG, Liu HL, Yu B, Huang D, Zhu GF. Thirteen complete chloroplast genomes of the costaceae family: insights into genome structure, selective pressure and phylogenetic relationships. BMC Genomics. 2024;25(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdullah, Henriquez CL, Mehmood F, Carlsen MM, Islam M, Waheed MT, Poczai P, Croat TB, Ahmed I. Complete Chloroplast Genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): unique inverted repeat expansion and contraction affect rate of evolution. J Mol Evol. 2020;88(7):562–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henriquez CL, Abdullah, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics. 2020;112(3):2349–60. [DOI] [PubMed] [Google Scholar]
- 49.Abdullah, Henriquez CL, Croat TB, Poczai P, Ahmed I. Mutational dynamics of aroid chloroplast genomes II. Front Genet. 2021;11:610838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SH, Yang J, Park J, Yamada T, Maki M, Kim SC. Comparison of whole plastome sequences between thermogenic skunk cabbage Symplocarpus renifolius and nonthermogenic S. nipponicus (Orontioideae; Araceae) in East Asia. Int J Mol Sci. 2019;20(19):4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He S, Yang Y, Li Z, Wang X, Guo Y, Wu H. Comparative analysis of four Zantedeschia chloroplast genomes: expansion and contraction of the IR region, phylogenetic analyses and SSR genetic diversity assessment. PeerJ. 2020;8:e9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park H, Park JH, Kang YJ. Characterization of the complete chloroplast genome of Wolffia arrhiza and comparative genomic analysis with relative Wolffia species. Sci Rep. 2024;14(1):5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian N, Han L, Chen C, Wang Z. The complete chloroplast genome sequence of Epipremnum aureum and its comparative analysis among eight Araceae species. PLoS One. 2018;13(3):e0192956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Liu C, Hou K, Liu W. Comparative analyses of plastomes of four anubias (Araceae) taxa, tropical aquatic plants endemic to africa. Genes (Basel). 2022;13(11):2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdullah, Henriquez CL, Mehmood F, Shahzadi I, Ali Z, Waheed MT, Croat TB, Poczai P, Ahmed I. Comparison of chloroplast genomes among species of unisexual and bisexual clades of the monocot family araceae. Plants (Basel). 2020;9(6):737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomlinson P, Mayo S, Bogner J, Boyce P, Catherine E. The Genera of Araceae, 199; vol. 53
- 57.Peng JY, Zhang XS, Zhang DG, Wang Y, Deng T, Huang XH, Kuang TH, Zhou Q. Newly reported chloroplast genome of Sinosenecio albonervius Y Liu & QE Yang and comparative analyses with other Sinosenecio species. BMC Genomics. 2022;23(1):639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Sena Brandine G, Smith AD. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 2019;8:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;47(W1):W65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan PP, Lin BY, Mak AJ, Lowe TM. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49(16):9077–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52(5–6):267–74. [DOI] [PubMed] [Google Scholar]
- 65.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1. [DOI] [PubMed] [Google Scholar]
- 69.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–302. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4(4):259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome raw reads have been deposited in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under a Project accession: PRJNA1126114. The four newly sequenced complete chloroplast genomes in this study have been submitted to GenBank (https://www.ncbi.nlm.nih.gov) with accession numbers OR995733, OR438676, PP936070 and PP936071, and available in NCBI (https://www.ncbi.nlm.nih.gov/) (see Table S1). The materials are available from the corresponding author on reasonable request after the publication of the work.