Abstract
This study elucidates the in vivo genetic mechanisms contributing to the emerging resistance to carbapenem in Shewanella algae through a lens of adaptive microbial evolution. Leveraging PacBio amplification-free sequencing, we tracked the evolution of β-lactam resistance in clinical isolates from a persistent S. algae bacteremia case amidst antimicrobial therapy. Our investigation spotlighted a recurrent G547W mutation in the sensor histidine kinase (pdsS), which was associated with the overexpression of an OmpA-like protein (pdsO) within a proteobacteria-specific sortase system. Intriguingly, we observed a recurrent switch between wild-type and G547W alleles, revealing an adaptive expansion and contraction of underlying cell subpopulations in response to β-lactam exposure. Comparative transcriptome analyses further demonstrated the overexpression of genes pivotal for membrane integrity, biofilm formation, immune evasion, and β-lactamase activation in resistant samples. This underscores the pre-existence of resistant cells at minuscule frequencies even without antibiotic pressure, potentially explaining the within-host emergence of resistance during antibiotic treatments. Our findings provide pivotal insights into the dynamic genetic adaptations of S. algae under therapeutic pressures, unmasking intricate resistance mechanisms and highlighting the critical role of subpopulation dynamics in treatment outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12941-024-00759-3.
Keywords: Shewanella algae, Carbapenem resistance, Adaptive microbial evolution, β-lactam antibiotics
Background
Bacterial populations often exhibit remarkable heterogeneity, enabling survival and adaptation in harsh and dynamically changing environments. This phenotypic diversity, particularly the manifestation of heteroresistance—wherein a single bacterial population harbors both antibiotic-resistant and susceptible phenotypes—can pose significant challenges in clinical settings [1, 2]. This phenotypic variability, specifically the phenomenon of heteroresistance, in which a singular bacterial population exhibits both antibiotic-resistant and susceptible phenotypes, poses substantial challenges in clinical contexts [3]. Such population heterogeneity can be pivotal, enabling the sustenance of resistant subpopulations even at low frequencies, thereby providing a reservoir from which resistance can emerge under the selective pressure of antibiotic treatment [4].
Shewanella algae a prominent human pathogen within the genus Shewanella, is implicated in various infections, including bacteremia, intraabdominal infections, and skin and soft tissue infections, making it a significant concern for public health [5]. The incidence of multidrug-resistant isolates and treatment failures, particularly in response to carbapenem therapies, have been documented over the years [6]. However, the mechanisms underlying this emerging resistance in S. algae are yet to be fully elucidated. Limited knowledge regarding the dynamics and mechanisms of carbapenem resistance in S. algae underscores the urgency of advancing research in this domain to develop effective therapeutic strategies.
Next-generation sequencing has illuminated our understanding of within-host heteroresistant evolution, revealing intricate genetic landscapes and adaptive trajectories of bacteria under antibiotic pressure [7, 8]. For instance, genome comparisons in vancomycin-resistant Staphylococcus aureus have identified mutations associated with reduced susceptibility [9]. Under low-dose colistin treatment, a nonsynonymous mutation in PmrB emerged and remained stable for 50 generations [10]. Deep sequencing of pneumococcal samples identified enriched alleles during antimicrobial therapy, and in the absence of antibiotics, susceptible lineages outcompeted resistant lineages within the host [11]. However, the detection of within-host diversity, especially involving subpopulations with low minor allele frequency (MAF), remains challenging with existing methodologies. Traditional antibiotic susceptibility tests, like Minimum Inhibitory Concentration (MIC), are geared towards assessing population-wide resistance and often fall short in detecting low-frequency resistant cells [12, 13]. The most reliable method is the population analysis profile (PAP) assay [4]. Combining PAP with whole-genome sequencing found most methicillin-resistant Staphylococcus aureus is heteroresistant, and the resistance cells are present at very low frequencies [14]. Unfortunately, PAP assays are labor-intensive and not applicable in clinical practice [3]. Existing PAP-free sequencing methods are prone to biases such as strand bias, GC bias, and coverage bias [15–17].
Emerging third-generation sequencing platforms, such as PacBio, enable single-molecule, amplification-free sequencing, which can minimize biases and provide uniform coverage, offering a novel window into investigating within-host heteroresistant evolution [18, 19]. In this study, we employ PacBio sequencing to trace the within-host evolutionary trajectory of S. algae during antibiotic treatments. By illuminating the competitive interplay between sensitive and resistant subpopulations and unearthing novel insights into the mechanisms of emerging resistance, this work seeks to advance our understanding of bacterial adaptive strategies and inform the development of enhanced therapeutic interventions.
Methods
Bacterial strain isolation and antimicrobial susceptibility testing
Four S. algae isolates were obtained from the blood of a patient with persistent bacteremia. The isolates were inoculated on trypticase soy agar with 5% sheep blood (Becton–Dickinson, Franklin Lakes, NJ, United States) and incubated aerobically at 37 °C overnight. MALDI-TOF mass spectrometry was used for preliminary identification. The MICs of each sample were measured by VITEK 2 Automated System (BioMerieux, Marcy l'Etoile, France), with susceptibility interpreted according to Clinical and Laboratory Standards Institute-established criteria (CLSI M100-S29).
Library preparation and genome sequencing
Overnight cultures of the S. algae isolates were grown in Luria–Bertani broth overnight at 37℃. Genomic DNA was extracted from these cultures using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA), and high-molecular-weight gDNA was sheared to approximately 10-kb lengths using g-TUBES (Covaris, Woburn, MA, USA). PacBio sequencing libraries were prepared from this sheared DNA using standard protocols (Pacific Biosciences, Menlo Park, CA, USA), and whole genome sequencing was performed on the PacBio RS II sequencer (Pacific Biosciences, Menlo Park, CA, USA). Sequence runs of 12 single-molecule real-time (SMRT) cells were performed, with a movie time of 120-min per SMRT cell. SMRT Analysis portal version 2.1 was used to filter and trim the reads, and post-filtered data (1.479 Gb) with approximately 404X coverage and an average read length of approximately 6.2 kb were used for subsequent assembly (Supplementary Table S1).
Genome assembly and gene annotation
The post-filtered genome reads were de novo assembled by Canu (v1.4) [20], which produced one single large chromosome (~ 4.9 Mb) and one plasmid (~ 132kbp) (Supplementary Table S2). Circlator was used to circularize the genome and plasmid [21]. Protein-coding and non-coding genes in the genomes were annotated using National Center for Biotechnology Information (NCBI) Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP). Identification of nonsynonymous mutations and measurement of allele frequency was carried out by aligning reads from all samples (S1, S2, S3, S4) onto the S1 genome by minimap2 followed by codon translation (Supplementary Table S6). The function of the pdsO protein was inferred as an OmpA-like protein using InterPro domain analysis and the STRING database [22].
Annotation of antibiotic-resistance genes
The resistome was annotated using the Resistance Gene Identifier from the Comprehensive Antibiotic Resistance Database [23]. The strict criteria were chosen for prediction using the homology model, in which BLAST is used to detect functional homologs of antimicrobial resistance genes, which are then confirmed by alignment against the Integrated Microbial Genomes (IMG) database using BLASTN with a 95% sequence identity threshold [24].
Validation of pdsS G547W mutation by PCR and Sanger sequencing
Two pairs of primers of specific sequences spanning the upstream and downstream regions of the pdsS G547W mutation were designed (Supplementary Figure S1). The primer sequences, temperatures, and sizes of the PCR products are shown in Supplementary Table S3. The PCR products were amplified using these primer pairs, purified using the GenElute PCR Clean-Up Kit (Sigma-Aldrich, Dorset, United Kingdom), and diluted to a concentration of 1–3 ng/µl following quantification in a NanoDrop 2000 UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The purified PCR products were sequenced using the dye terminator chemistry (BigDye, version 3.1) on the 3130xl ABI PRISM Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA). The forward and reverse sequences of the four samples were shown in Supplementary Figure S2.
Transcriptome analysis
The transcriptomes of carbapenem-susceptible (S1) and carbapenem-resistant (S2) S. algae isolates were sequenced using the Illumina platform. The RNA-seq reads were mapped to the PGAAP-annotated transcripts by kalisto [25], and the expression abundance of each gene between the samples was normalized by Transcripts Per Million (TPM) (Supplementary Table S7). The fold change in expression levels of each gene between S1 and S2 was then calculated and sorted. Differential expression analysis was carried by the NOIseq package with three simulated replicates to assess the significance of fold changes. The protein functions of over-expressed genes were annotated according to PGAAP.
Results
Longitudinal sequencing during antibiotic treatments
Four S. algae isolates (S1-S4) were isolated from blood of a patient with persistent S. algae bacteremia (Fig. 1). These samples were subjected to deep sequencing (~ 300x) using the PacBio platform and assembled into four complete circular genomes (~ 4.7 Mb) and one circular plasmid (~ 132 kb) (Supplementary Table S1 and S2). The minimum inhibitory concentrations (MICs) of these samples indicated that the initial S1 isolate was susceptible to imipenem, while the others (S2, S3, S4) conferred resistance to imipenem, ampicillin/sulbactam, and trimethoprim-sulfamethoxazole.
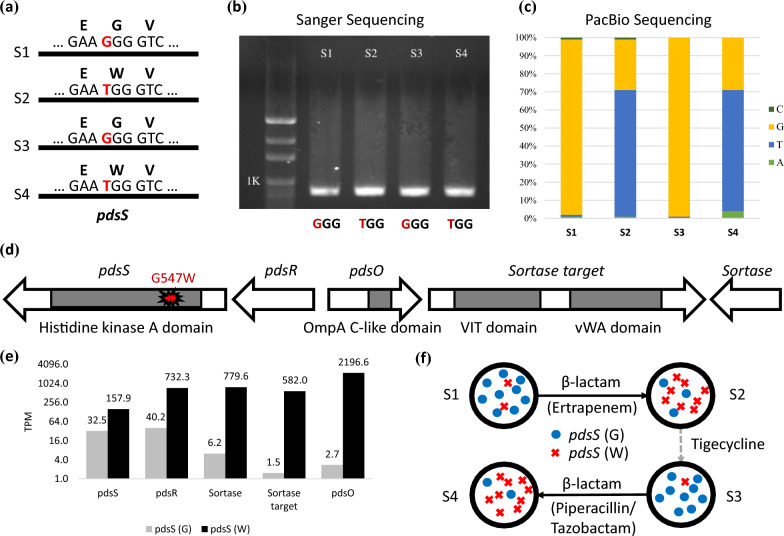
Fig. 1.
Overview of the study that examines the time course of antibiotic treatments and the development of antibiotic resistance in a series of bacterial isolates. The study involves four bacteremic isolates, labeled as S1, S2, S3, and S4, which were collected at different points during antimicrobial therapy. S1 represents the first isolate. S2, S3, and S4 represent subsequent isolates during antimicrobial therapy. The four samples were subject to complete genome sequencing
Recurrent pdsS G547W mutations upon β-lactam delivery
A comparison of the four genomes identified a recurrent missense mutation in the proteobacterial dedicated sortase system histidine kinase (pdsS) (Fig. 2a, Supplementary Figures S3). This G-to-T substitution resulted in a change from Glycine (G) to Tryptophan (W) at position 547 of pdsS. The G547W mutation first appeared in the S2 genome after the delivery of ertapenem, which was restored to G in the S3 genome after switching to tigecycline. The mutation reoccurred in the S4 genome after the addition of piperacillin/tazobactam. We validated these nucleotide switches by PCR and Sanger sequencing (Fig. 2b, Supplementary Figures S1 and S2).
Fig. 2.
Heteroresistance to carbapenem in S. algae. a A non-synonymous SNP (G to T) leading to amino acid change (G to W) in pdsS was recurrently detected after delivering β-lactams (S2 and S4); b and c allele frequencies of the Sanger sequencing and PacBio sequencing; d the genes surrounding pdsS disclose the other members of the proteobacterial dedicated sortase system, including pdsR, pdsO, sortase target, and sortase; e comparison of expression levels between the wild type pdsS (G) and mutant pdsS (W); f illustration of the adaptive expansion and reduction of the pdsS G547W subpopulations with or without β-lactams
The amplification-free sequencing of PacBio allowed investigation of underlying cell population. We found that a heterogeneous cell population existed before antibiotic treatment and the subpopulation carrying the G547W mutation expands and reduces depending on the exposure to beta-lactams. Analysis of the PacBio reads revealed diversifying cell populations in all four samples (Fig. 2c), with the G547W mutation expanding (in S2 and S4) and reducing (in S1 and S3) depending on the exposure to beta-lactams (Fig. 2c). In sample S1, although the amino acid G was the dominant allele (~ 99%), W was also detected at low MAF (< 1%). After the delivery of ertapenem (S2), the allele frequency of W surged to ~ 70%, while G reduced to 30%. The frequencies were restored after switching to tigecycline in S3 (i.e., G = ~ 99%, W < 1%). Finally, after starting piperacillin/tazobactam (S4), the frequency of W surged again to 70%. Consequently, a heterogeneous cell population existed before antibiotic treatments. The subpopulation carrying the G547W mutation expands (i.e., S2 and S4) and reduces (i.e., S1 and S3) depending on the exposure to β-lactams.
BLAST search of the NCBI database showed that all public available pdsS sequences carry the wild type (G) allele, implying that the pdsS (G) may have better fitness when free off antibiotic pressure. Therefore, the co-existence of the alternative allele (W) with low MAF might be likely overlooked by conventional amplification-based PCR and sequencing.
Up-regulation of an OmpA-like protein by the pdsS G547W mutation
We found that the G547W mutation in the sensor histidine kinase pdsS associates with a nearby OmpA-like protein called pdsO, which may help maintain membrane integrity and increase resistance to beta-lactam antibiotics. Analysis of genes surrounding pdsS revealed five members of the proteobacterial dedicated sortase system (Fig. 2d). The response regulator (pdsR) is located next to pdsS, followed by a sortase-associated OmpA-like protein (pdsO), sortase target protein, and sortase. The G547W mutation occurs within the histidine kinase A domain in pdsS, and its regulatory role in this sortase system was assessed. The expression levels of all five sortase system members were significantly up-regulated in the pdsS (W) sample (Fig. 2d, p < 0.05). When compared with the wild type, the expression levels of pdsS and pdsR with the G547W mutation increase by 4.8 and 18.2 folds, respectively. The expression levels of the three downstream members (sortase, sortase target, and pdsO) were significantly elevated by 126, 388, and 799 folds, respectively (Fig. 2e, p < 0.01). qPCR with three replicates also confirmed the overexpression of this sortase system (Supplementary Tables S4 and S5). The C-terminal domain of OmpA-like protein is capable of binding to peptidoglycan, which effectively links the outer membrane to the cell wall and stabilizes the outer membrane. Together, these results suggest that the G547W mutation in pdsS is associated with the overexpression of pdsO, which may help to maintain the membrane integrity and increase resistance to β-lactams [26]. The recurrent expansion and reduction of pdsS (W) suggest a substantial fitness cost of over-expressing pdsO without β-lactams pressure. These findings suggest that the presence of pdsS diversity allows the isolate to adapt to different antimicrobial agents during the treatment course, potentially contributing to the persistence of the infection (Fig. 2f).
Synergistic resistance mechanisms revealed by transcriptome sequencing
The study found that over-expression of pdsO was correlated with resistance to carbapenem. Transcriptomes of the S1 (carbapenem-susceptible) and S2 (carbapenem-resistant) samples were sequenced and compared. The RNA-seq reads were mapped to the PGAAP annotated transcripts, and the expression abundance of each gene was normalized by Transcripts Per Million (TPM). The fold change in expression levels of each gene between S1 and S2 was then calculated and sorted. The transcriptomes also revealed that genes related to membrane integrity, biofilm formation, immune evasion, and beta-lactamase activation were over-expressed in the resistant sample.
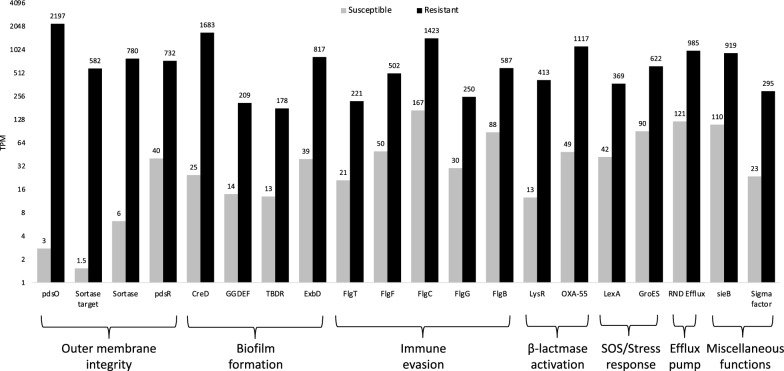
Figure 3 shows the top 20 over-expressed genes in the resistant sample (p < 0.01, see method). Four pdsS/pdsR sortase system members are included, and the fold change of pdsO being the largest of all genes. Four biofilm-associated genes (creD, GGDEF gene, TonB-dependent receptor [TBDR] gene, and exbD) are also significantly up-regulated. CreD is involved in biofilm development in the presence of β-lactams [27], and over-expression of GGDEF-containing proteins has been shown to increase biofilm formation [28]. The exbB-exbD-TonB system contributes to biofilm formation by influencing the production of N-acyl homoserine lactones [29]. In addition, five flagella-related genes (e.g., flgT and flgF) are significantly over-expressed, which may increase motility or evade immune recognition [30]. Finally, over-expression of LysR/OXA-55, RND efflux, and LexA/GroES confirms the activation of β-lactamase, efflux pumps, and SOS/stress responses [31]. These findings suggest that the in vivo resistance of this isolate to multiple antibiotic agents is likely due to a synergistic effect of outer membrane alteration, biofilm formation, immune evasion, and activation of β-lactamase/efflux pumps.
Fig. 3.
Functional classification of the top 20 genes over-expressed in the resistant sample. Eight genes contribute to biofilm formation. Five flagellar genes are possibly involved with immune evasion. OXA-55 and LysR are associated with β-lactamase activation. LexA and GroES are related to SOS/stress response. RND efflux is associated with the efflux pump
Discussion
In the study, we identified a mutation in a sensor histidine kinase called pdsS, which leads to the over-expression of an OmpA-like protein pdsO via a proteobacteria-dedicated sortase system. Our findings suggest that this mutation in pdsS is associated with the overpexression of an OmpA-like protein pdsO, which can contribute to resistance to β-lactam antibiotics by maintaining membrane integrity and potentially through its involvement in biofilm formation [26]. Previous research has shown that OmpA can decrease susceptibility to several antibiotics, including nalidixic acid, chloramphenicol, aztreonam, imipenem, and meropenem. Over-expression of OmpA decreased the susceptibility to nalidixic acid, chloramphenicol, aztreonam, imipenem, and meropenem [32]. The deletion of OmpA has also been shown to increase susceptibility to β-lactams in Acinetobacter baumannii [33]. Additionally, over-expression of OmpA has been linked to biofilm formation in E coli [34]. Thus, the up-regulation of pdsO is likely a key driving force contributing to carbapenem resistance in S. algae.
In the study, we present novel findings that shed light on the intricate mechanisms underlying the emergence of carbapenem resistance in S. algae. For many years, it has been observed that carbapenem resistance emerges in S. algae during antimicrobial therapy, leading to treatment failure [6]. This phenomenon has been has been linked to the presence of a chromosome-encoded carbapenem-hydrolyzing Ambler class D OXA-55 β-lactamase, a gene that is conserved in S. algae regardless of its susceptibility to carbapenem [35]. Recent study have shown that there is an increasing level of blaOXA-55 transcription and β-lactamase activity in carbapenem-resistant S. algae isolate compared with the carbapenem-susceptible isolates [31]. Our research has revealed an addition mechanism of resistance, involving a mutation in a sensor histidine kinase (pdsS) and subsequent over-expression of an OmpA-like protein pdsO through a proteobacteria-dedicated sortase system. Furthermore, our transcriptome analysis revealed that not only is the beta-lactamase OXA-55 activated in carbapenem resistant S. algae isolate, but also genes related to membrane integrity, biofilm formation, immune evasion are over-expressed in the carbapenem-resistant isolate. The identification of a novel mechanism of resistance involving mutation in pdsS and multiple overexpressing genes, in addition to the well-known role of OXA-55, underscores the complexity of carbapenem resistance in S. algae and the need for multi-faceted approaches to tackle this problem.
We found that the G547W mutation existed at low frequency before antibiotic treatments. The resistant G547W subpopulation adaptively expanded and reduced in response to exposure to β-lactams. This phenomenon, known as heteroresistance, has been previously observed in the PhoP–PhoQ two-component system in E. cloacae. In that case, heteroresistant E. cloacae cells were able to withstand colistin treatment by modifying the lipopolysaccharide component of their outer membrane [36] which is dependent on the histidine kinase PhoQ [37]. This suggests that heteroresistance, mediated by distinct subpopulations with different outer membrane compositions, may be a common phenomenon in bacteria. In our study, the G547W mutation is located in the sensor histidine kinase pdsS, which is a TCS-like system. Thus, it is possible that similar bimodal responses mediated by heterogeneous subpopulations may exist in other TCS systems in bacteria [38].
We note that the pdsS G547W is not the only reason driving the multidrug resistance. The analysis of RNA-seq showed that the up-regulated pathways also include other resistant pathways (e.g., over-expression of flagella-related genes, efflux pumps, and OXA-55). High-level expression of OXA-55 alone has been shown to confer carbapenem, but our study can’t link the G547W mutation to the over-expression of OXA-55. We hypothesize the multi-drug resistance may be attributed to a synergistic effect of several resistant mechanisms, although each has its unique fitness cost. For instance, the G547W mutation may have better fitness than over-expression of OXA-55/efflux pumps under beta lactam pressure, albeit each of them can confer resistance to carbapenem. Consequently, this coordination reduced the treatment efficacy no matter how we changed the antibiotics or used multiple antibiotics, which finally led to the death of the patient.
Resistant mutation often comes at fitness costs. For instance, mutations reducing the influx or enhancing efflux of antibiotics can also decrease the absorption of nutrient compounds [39]. As a result, compensatory mutations may arise to compensate for the fitness of the resistant allele [40, 41]. Our findings suggest that the heterogeneous subpopulations in this isolate are able to balance the fitness costs by retaining the resistant subpopulations at low frequency and the wild type at high frequency without β-lactam exposure. When coupled with other resistant mechanisms, this within-host diversity and adaptiveness greatly reduce the drug efficacy during treatments.
As we only focused on the transcriptomic profiles of S1 (susceptible) and S2 (resistant), the lack of S3 and S4 transcriptomes for understanding the dynamics of all resistant pathways is a limitation of this study. It is possible that the wild type subpopulation may benefit from the over-expression of OXA-55 from other subpopulations to combat the beta-lactam treatments without the need of G547W mutation. The allele frequency analysis implies the G547W subpopulation might have better fitness than others with over-expressed OXA-55/efflux pumps, albeit all of them may confer substantial resistance to beta lactams. Further in vitro experiments are required to justify this hypothesis.
In conclusion, we identified a β-lactam-resistant mutation in a sensor histidine kinase called pdsS, which leads to the over-expression of an OmpA-like protein pdsO via a proteobacteria-dedicated sortase system. We found that the G547W mutation in pdsS is associated with overexpressed pdsO, which can contribute to β-lactam resistance by maintaining membrane integrity and potentially through its involvement in biofilm formation. The G547W-encoding cells existed at low frequency before antibiotic treatment, but adaptively expanded and reduced in response to exposure to β-lactams. This within-host diversity and adaptiveness greatly reduce the efficacy of antibiotic treatment, especially when coupled with other resistant mechanisms [42–44]. These findings provide insights into the mechanisms of antibiotic resistance and highlight the importance of considering heteroresistant subpopulations in the treatment of bacterial infections.
Conclusion
The study provides new insights into the within-host evolution of microbial populations during antibiotic treatments and the dynamics of allele switch in the development of emerging resistance to carbapenem in Shewanella algae. The identification of a recurrent G547W mutation in the sensor histidine kinase gene and its correlation with the an OmpA-like protein, as well as the adaptive expansion and reduction of the cell subpopulations in response to β-lactam exposure, offers a deeper understanding of the mechanisms of antibiotic resistance and the potential for resistant subpopulations to exist at very low frequencies even in the absence of antibiotic pressure. This information can inform the development of more effective treatment strategies to combat antibiotic resistance.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- IMG
Integrated Microbial Genomes
- MAF
Minor allele frequency
- MIC
Minimum Inhibitory Concentration
- NCBI
National Center for Biotechnology Information
- PAP
Population analysis profile
- pdsS
Proteobacterial dedicated sortase system histidine kinase
- PGAAP
Prokaryotic Genomes Automatic Annotation Pipeline
- SMRT
Single-molecule real-time
- TPM
Transcripts Per Million
Author contributions
Conceptualization: Yao-Ting Huang, Po-Yu Liu; methodology: Yao-Ting Huang, Po-Yu Liu; formal analysis: Yao-Ting Huang, Po-Yu Liu; investigation: Yao-Ting Huang, Po-Yu Liu; data curation: Yao-Ting Huang, Po-Yu Liu; writing—original draft: Yao-Ting Huang, Po-Yu Liu; writing—review and editing: Yao-Ting Huang, Po-Yu Liu; project administration: Yao-Ting Huang, Po-Yu Liu; funding acquisition: Yao-Ting Huang, Po-Yu Liu. All authors had final approval of the version to be published and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by the National Science and Technology Council [109-2221-E-194-038-MY3, 111-2221-E-194-031-MY3, and 112-2314-B-075A-006] and Taichung Veterans General Hospital [TCVGH-1133901C, TCVGH-1133901D and TCVGH-NCHU1137608].
Availability of data and materials
The assembled genome and gene annotations have been deposited at NCBI/GenBank as BioProject PRJNA356098 (with accession number CP018456).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508. [DOI] [PubMed] [Google Scholar]
- 2.Norman TM, Lord ND, Paulsson J, Losick R. Stochastic switching of cell fate in microbes. Annu Rev Microbiol. 2015;69:381–403. [DOI] [PubMed] [Google Scholar]
- 3.Dewachter L, Fauvart M, Michiels J. Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol Cell. 2019;76:255–67. [DOI] [PubMed] [Google Scholar]
- 4.Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17:479–96. [DOI] [PubMed] [Google Scholar]
- 5.Tellgren-Roth C, Thorell K, Galperin MY, Krell T, Römling U, Sjöling Å, et al. Complete genome sequence and methylome of the type strain of Shewanella algae. Microbiol Resour Announc. 2021;10: e0055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D-M, Kang C-I, Lee CS, Kim H-B, Kim E-C, Kim NJ, et al. Treatment failure due to emergence of resistance to carbapenem during therapy for Shewanella algae bacteremia. J Clin Microbiol. 2006;44:1172–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014;30:401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 2016;14:150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci USA. 2007;104:9451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, et al. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother. 2014;58:4399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tonkin-Hill G, Ling C, Chaguza C, Salter SJ, Hinfonthong P, Nikolaou E, et al. Pneumococcal within-host diversity during colonization, transmission and treatment. Nat Microbiol. 2022;7:1791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. MBio. 2018. 10.1128/mbio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicoloff H, Hjort K, Levin BR, Andersson DI. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol. 2019;4:504–14. [DOI] [PubMed] [Google Scholar]
- 14.Dordel J, Kim C, Chung M, de la Gándara MP, Holden MTJ, Parkhill J, et al. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio. 2014;5: e01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato MP, Ogura Y, Nakamura K, Nishida R, Gotoh Y, Hayashi M, et al. Comparison of the sequencing bias of currently available library preparation kits for Illumina sequencing of bacterial genomes and metagenomes. DNA Res. 2019;26:391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y, Li J, Li C-I, Long J, Samuels DC, Shyr Y. The effect of strand bias in Illumina short-read sequencing data. BMC Genomics. 2012;13:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browne PD, Nielsen TK, Kot W, Aggerholm A, Gilbert MTP, Puetz L, et al. GC bias affects genomic and metagenomic reconstructions, underrepresenting GC-poor organisms. Gigascience. 2020. 10.1093/gigascience/giaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, et al. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genom. 2013;14:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucl Acids Res. 2020;49:D605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucl Acids Res. 2019;48:D517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, et al. The IMG/M data management and analysis system v.7: content updates and new features. Nucl Acids Res. 2023;51:D723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 26.Choi U, Lee C-R. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front Microbiol. 2019;10:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamorano L, Moyà B, Juan C, Mulet X, Blázquez J, Oliver A. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother. 2014;58:5084–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gjermansen M, Ragas P, Tolker-Nielsen T. Proteins with GGDEF and EAL domains regulate Pseudomonas putida biofilm formation and dispersal. FEMS Microbiol Lett. 2006;265:215–24. [DOI] [PubMed] [Google Scholar]
- 29.Abbas A, Adams C, Scully N, Glennon J, O’Gara F. A role for TonB1 in biofilm formation and quorum sensing in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2007;274:269–78. [DOI] [PubMed] [Google Scholar]
- 30.Rossez Y, Wolfson EB, Holmes A, Gally DL, Holden NJ. Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog. 2015;11: e1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohama Y, Aoki K, Harada S, Nagasawa T, Sawabe T, Nonaka L, et al. Genetic environment surrounding blaOXA-55-like in clinical isolates of Shewanella algae clade and enhanced expression of blaOXA-55-like in a carbapenem-resistant isolate. MSphere. 2021;6: e0059321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon HI, Kim S, Oh MH, Na SH, Kim YJ, Jeon YH, et al. Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J Antimicrob Chemother. 2017;72:3012–5. [DOI] [PubMed] [Google Scholar]
- 33.Smani Y, Fàbrega A, Roca I, Sánchez-Encinales V, Vila J, Pachón J. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58:1806–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orme R, Douglas CWI, Rimmer S, Webb M. Proteomic analysis of Escherichia coli biofilms reveals the overexpression of the outer membrane protein OmpA. Proteomics. 2006;6:4269–77. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z-Y, Huang Y-T, Chao W-C, Ho S-P, Cheng J-F, Liu P-Y. Reversal of carbapenem-resistance in Shewanella algae by CRISPR/Cas9 genome editing. J Advert Res. 2019;18:61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang KN, Klein DR, Kazi MI, Guérin F, Cattoir V, Brodbelt JS, et al. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol Microbiol. 2019;111:1604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol. 2016;1:16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ram S, Goulian M. The architecture of a prototypical bacterial signaling circuit enables a single point mutation to confer novel network properties. PLoS Genet. 2013;9: e1003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794:808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jochumsen N, Marvig RL, Damkiær S, Jensen RL, Paulander W, Molin S, et al. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat Commun. 2016;7:13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dößelmann B, Willmann M, Steglich M, Bunk B, Nübel U, Peter S, et al. Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob Agents Chemother. 2017. 10.1128/AAC.00043-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jean S-S, Lee Y-L, Liu P-Y, Lu M-C, Ko W-C, Hsueh P-R. Multicenter surveillance of antimicrobial susceptibilities and resistance mechanisms among Enterobacterales species and non-fermenting Gram-negative bacteria from different infection sources in Taiwan from 2016 to 2018. J Microbiol Immunol Infect. 2022;55:463–73. [DOI] [PubMed] [Google Scholar]
- 43.Liu P-Y, Ko W-C, Lee W-S, Lu P-L, Chen Y-H, Cheng S-H, et al. In vitro activity of cefiderocol, cefepime/enmetazobactam, cefepime/zidebactam, eravacycline, omadacycline, and other comparative agents against carbapenem-non-susceptible Pseudomonas aeruginosa and Acinetobacter baumannii isolates associated from bloodstream infection in Taiwan between 2018–2020. J Microbiol Immunol Infect. 2022;55:888–95. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y-L, Ko W-C, Hsueh P-R. In vitro activity of imipenem/relebactam, meropenem/vaborbactam and comparators against Enterobacterales from patients with intra-abdominal infections: results of the study for monitoring antimicrobial resistance trends (SMART) in Taiwan, 2020. J Microbiol Immunol Infect. 2023;56:75–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled genome and gene annotations have been deposited at NCBI/GenBank as BioProject PRJNA356098 (with accession number CP018456).