Abstract
Transposable elements (TEs) are major components of plant genomes, profoundly impacting the fitness of their hosts. However, technical bottlenecks have long hindered our mechanistic understanding of TEs. Using RNA-Seq and long-read sequencing with Oxford Nanopore Technologies’ (ONT) direct cDNA sequencing, we analyzed the heat-induced transcription of TEs in three natural accessions of Arabidopsis thaliana (Cvi-0, Col-0, and Ler-1). In addition to the well-studied ONSEN retrotransposon family, we confirmed Copia-35 as a second heat-responsive retrotransposon family with particularly high activity in the relict accession Cvi-0. Our analysis revealed distinct expression patterns of individual TE copies and suggest different mechanisms regulating the GAG protein production in the ONSEN versus Copia-35 families. In addition, analogously to ONSEN, Copia-35 activation led to the upregulation of flanking genes such as APUM9 and potentially to the quantitative modulation of flowering time. ONT data allowed us to test the extent to which read-through formation is important in the regulation of adjacent genes. Unexpectedly, our results indicate that for both families, the upregulation of flanking genes is not predominantly directly initiated by transcription from their 3′ long terminal repeats. These findings highlight the intraspecific expressional diversity linked to retrotransposon activation under stress.
Keywords: Arabidopsis thaliana, heat stress, Oxford Nanopore Sequencing, retrotransposon, ONSEN
Significance.
Transposable elements (TEs) play a crucial role in genome evolution, but their stress-induced transcriptional behavior across natural populations remains underexplored. Our study reveals significant variation in heat-responsive TEs, including the activation of a less-studied retrotransposon family, Copia-35, alongside the well-known ONSEN. These findings show how stress-responsive TEs can impact neighboring gene expression, potentially influencing important traits like flowering time. This research provides new insights into how TEs contribute to natural genetic diversity and potentially to plant adaptation under environmental stress.
Introduction
Transposable elements (TEs) have a profound impact on genome architectures of plants. In crops such as maize, wheat, and barley, TEs account for a majority of the genome, ranging from 64% to more than 80% (Jiao et al. 2017; Wicker et al. 2017, 2018). Besides their impact on genome size, TEs are well known to alter the expression of nearby genes (e.g. Wang et al. 2013; Domingez et al. 2020; Quadrana et al. 2020) and in fine the phenotype of their host (for review Lisch 2013; Vitte et al. 2014; Baduel and Quadrana 2021). Yet, due to their potentially deleterious effects, most TEs are silenced by DNA methylation and through packaging into a heterochromatin state. In particular, one of the most studied plant-specific TE silencing mechanisms is the RNA-directed DNA methylation (RdDM) pathway (Matzke and Mosher 2014). The canonical RdDM pathway features two plant-specific RNA polymerases (Pol IV and Pol V), which, via complex processes, facilitate DNA methylation and, ultimately, the silencing of TEs. Despite widespread silencing, some TEs are still able to transpose in the wild, hereby creating genetic diversity among populations of a given species. For example, a recent study identified ∼23,000 TE insertion polymorphisms (TIPs) across 1,047 natural accessions (Baduel et al. 2021) in Arabidopsis thaliana, in which TEs account for ∼21% of the genome (Berardini et al. 2015).
Abiotic as well as biotic stresses can provide the conditions that allow specific TE families to evade the host’s silencing mechanisms (Negi et al. 2016). One of the best characterized stress-responsive plant TEs is the retrotransposon (RT) ONSEN (or ATCOPIA78) in A. thaliana (Pecinka et al. 2010; Tittel-Elmer et al. 2010; Ito et al. 2011, 2013). ONSEN contains long terminal repeats (LTRs) on both ends, as well as coding sequences for gag, the reverse transcriptase and other enzymes, which are essential for its transposition process and typical of Copia elements (Wicker et al. 2007). When A. thaliana seedlings are treated with heat, ONSEN becomes transcriptionally active, and, upon loss of major epigenetic regulators (Ito et al. 2011) or a transient chemical demethylation (Thieme et al. 2017), it transposes at high frequency, resulting in the stable inheritance of novel ONSEN copies.
A particularly interesting feature of ONSEN is the fact that its insertions can also confer neighboring genes with heat responsiveness (Ito et al. 2011; Baduel et al. 2021; Roquis et al. 2021), leading to a reshuffling of transcriptional networks. The heat-induced transcription of ONSEN flanking genes is attributed to heat-responsive elements in ONSEN’s LTRs. These elements recruit heat shock factors that engage the transcription machinery as trimers, resulting in an upregulation of downstream genes (Wu 1995; Cavrak et al. 2014). The finding that ONSEN can mediate the expression of flanking regions under heat stress has evolutionary implications since numerous studies have confirmed insertion polymorphisms of ONSEN among natural populations (Cavrak et al. 2014; Masuda et al. 2016; Quadrana et al. 2016; Baduel et al. 2021) as well as an insertion bias toward exons and H2A.Z enriched regions (Quadrana et al. 2019; Roquis et al. 2021).
Since the initial discovery of ONSEN (Ito et al. 2011), additional heat-responsive TEs have been identified in A. thaliana. Two comprehensive experiments using RNA-Seq revealed that in Col-0, both ONSEN and ROMANIAT5 (referred to as Copia-35 in Repbase) (Pietzenuk et al. 2016; Sun et al. 2020) display heat-dependent transcription. However, while ONSEN has been studied in detail, our understanding of Copia-35 remains limited. A few studies have focused on a particular copy of Copia-35, AT1TE43225, owing to its role in modulating the expression of its 3′ flanking gene APUM9, which encodes the RNA-binding protein Arabidopsis PUMILIO9 that triggers the decay of target mRNA (Sanchez and Paszkowski 2014; Hristova et al. 2015). However, the natural diversity of the APUM9 locus, and more specifically the role of Copia-35 in driving its expression under heat stress, have not been examined across multiple natural accessions, meaning that our current understanding of the TE contribution to heat responsiveness is superficial at best.
While technical bottlenecks have been largely responsible for this knowledge gap, the advent of next-generation sequencing now allows to decipher the natural genetic diversity linked to TEs. The availability of polished genome assemblies, produced by long-read sequencing, provides access to the complete sequences of insertions, thereby facilitating a more comprehensive analysis of the genetic features of these insertions. In terms of characterizing the effects of TEs, RNA-Seq has allowed us to survey the entire transcriptome at once, irrespective of the limitations to perceptible phenotypic traits. Technical hurdles persist, however, as the task of aligning short reads from RNA-Seq to multi-copy TEs remains challenging (Lanciano and Cristofari 2020), particularly when the TE copies exhibit a high degree of identity. As a result, transcriptional studies of TEs using RNA-Seq are either based on consensus sequences such as SalmonTE (Jeong et al. 2018) or distribute reads evenly to all copies (Jin et al. 2015). In this context, the breakthrough recently brought by Oxford Nanopore Technologies’ (ONT) direct cDNA sequencing, which generates longer reads, has begun to drastically reduce alignment ambiguities, hereby facilitating the detection of TE expression at the single insertion level. As such, ONT has recently succeeded in improving existing TE annotations. For example, ONT’s cDNA sequencing on an A. thaliana mutant with transcriptionally reactivated TEs has allowed to identify and annotate the active TE loci (Panda and Slotkin 2020). Similarly, long reads generated by ONT recently enabled the identification of chimeric gene-transposon transcripts in A. thaliana (Berthelier et al. 2023), further highlighting the advantage of this powerful sequencing technique.
In this study, we examined the patterns of TE expression among natural accessions of heat-stressed A. thaliana (particularly between individual TE insertions) and the subsequent effects of TE activation on neighboring genes, by combining the powers of RNA-Seq and Oxford Nanopore Technologies’ (ONT) direct cDNA sequencing. Arabidopsis thaliana accessions group into ten genetic clusters spanning from the United States of America to Asia (The 1001 Genomes Consortium 2016). To optimize genetic diversity, we chose accessions from three distinct groups: a relict accession from Cape Verde (Cvi-0), a nonrelict accession from the United States of America (Col-0) and an accession from the admixture group (Ler-1) originating from Germany (The 1001 Genomes Consortium 2016). Importantly, each of these accessions previously had polished chromosomal-level PacBio assemblies and annotated genes. Using ONT direct cDNA, we were also able to precisely profile the transcription of heat-activated TEs as well as their impact on adjacent gene expression. The regulation of genes by TEs could occur through the formation of read-through from the TE to the gene (e.g. like for the iconic blood orange; for review Lisch 2013) or, alternatively, via cis-regulatory effects mediated by the recruitment of the TE transcription machinery (Zhao et al. 2018; Fagny et al. 2020; Deneweth et al. 2022). ONT allowed us to discriminate between the two mechanisms and as such, our work speak against a major role of read-through in transcriptional novelty.
Results
Global Comparison of ONT and RNA-Seq Datasets
We grew Col-0, Ler-1, and Cvi-0 plants under controlled or heat stress conditions and performed RNA-sequencing with classical Illumina short-read RNA-Seq and ONT. We first assessed the data quality of our RNA- and ONT-Seq runs (supplementary table S1, Supplementary Material online). To verify the effectiveness of the heat stress treatment, we performed a Principal Coordinate Analysis (PCoA) on gene expression using all samples. We found a clear separation of samples based on their treatment and genotype (Fig. 1a), indicating that the applied heat stress induced an accession-specific stress response. Most importantly, this showed that our ONT data were reproducible, and that differences between sequencing technologies did not overshadow global gene expression estimates.
Fig. 1.
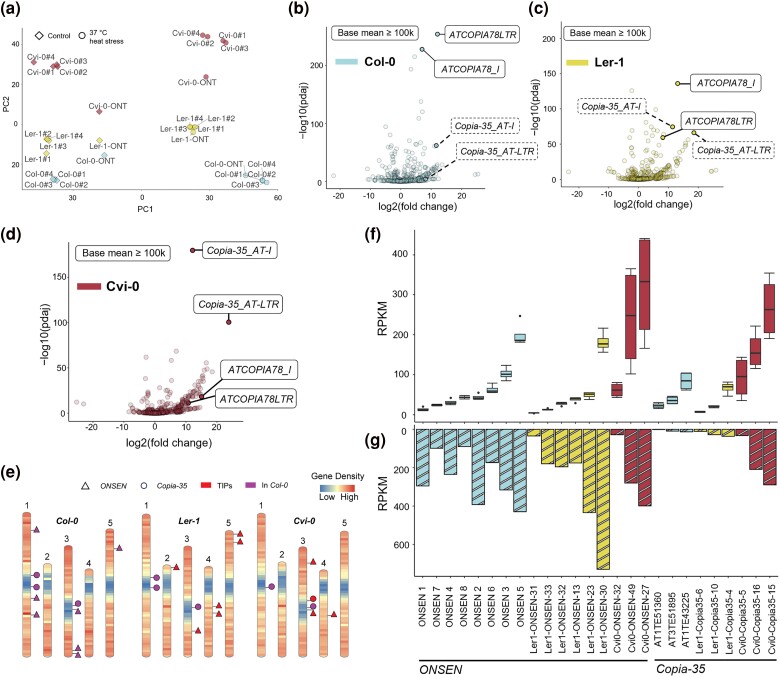
Expression of ONSEN and Copia-35. a) PCoA on gene expression in all sequenced samples. b to d) SalmonTE analysis with RNA-Seq data. Labeled consensus sequences in solid outlines represent candidates that have a base mean value that is greater than 100,000. Copia-35 consensus sequences in b) and c) are labeled in dashed boxes due to below-cutoff base mean. e) Annotation of ONSEN and Copia-35 full-length copies in the three accessions. Reference insertions (in Col-0) and TE insertion polymorphisms (TIPs) are marked. f and g) Expression of TE per copy measured using RNA-Seq (four replicates) and ONT direct cDNA sequencing, respectively. As seedling was pooled for mRNA extraction, replicates cannot be plotted for ONT data and barplots display expression level instead.
Activity of Heat-Responsive TEs Differs Across Accessions
We first aimed to identify TE candidates responsive to heat stress in each of the accessions. For this purpose, we used a consensus sequence-guided approach. Based on the library from Repbase (Bao et al. 2015), which contains 1,136 A. thaliana specific TE consensus sequences, we measured the transcriptional abundance of TEs in our RNA-Seq data using SalmonTE. Notably, in the Repbase library, the LTR and the internal (i.e. coding domain; Wicker et al. 2007) consensus of LTR retrotransposons were constructed separately, enabling us to distinguish the expression of LTR versus internal sequences. To reduce noise and to only focus on high-confidence TEs that would react to heat stress, we applied a stringent filter of log2fold change ≥ 2 and Padj ≤ 10−10, and a baseMean exceeding 100,000. We found that in all three accessions, the internal (ATCOPIA78_I) and the LTR (ATCOPIA78LTR) segments of ONSEN were consistently and significantly upregulated and with a high baseMean (Fig. 1b and d), confirming the robustness of ONSEN’s activation under heat stress. Importantly, in addition to the well-known case of ONSEN, we also found Copia-35 in Cvi-0 that emerged as a top candidate, passing the same stringent filters as ONSEN (Fig. 1d). In Cvi-0, both Copia-35_AT-I and Copia-35_AT-LTR showed a high level of expression and even greater statistical significance when compared to the activation of the ONSEN family.
Variations of Expression of Individual TE Insertions
After assessing the global expression of ONSEN and Copia-35 based on consensus sequences and RNA-Seq data, we combined the ONT direct cDNA- (ONT in short) and RNA-Seq data to explore variations in expression among individual full-length TE copies of the same family. We first generated high-confidence annotations of the two identified heat-responsive retrotransposon families ONSEN and Copia-35 in all three accessions. In total, we identified six full-length ONSEN copies in Ler-1 and three in Cvi-0, as well as three full-length Copia-35 copies in both accessions (Fig. 1e, Tables 1 and 2). For Col-0, we adopted the TAIR10 annotation IDs for the full-length ONSEN and three Copia-35 elements. However, we refined their annotations to include both LTRs. Interestingly, we found all full-length ONSEN insertions in Ler-1 and Cvi-0 to be polymorphic, representing TIPs (Fig. 1e). For Copia-35, one TIP was identified on chromosome 3 of Cvi-0, whereas all other full-length Copia-35 insertions in Ler-1 and Cvi-0 were shared with Col-0 (Fig. 1e).
Table 1.
Full-length ONSEN elements and their S1 strength
| TE ID | Chr | Start | End | S1 reads | S2 reads | S1 strength S1/(S1 + S2) ∗ 100% | S1/S2 | 3′ gene | Distance TE/gene | 3′ gene reached by S2 reads |
|---|---|---|---|---|---|---|---|---|---|---|
| AT1G11265/ONSEN1 | Chr1 | 3780765 | 3785721 | 138 | 19 | 87.90% | 7.26 | AT1G11280 | 1613 | False |
| AT1G21945/ONSEN7 | Chr1 | 7717255 | 7722647 | NA | NA | NA | NA | AT1G21950 | 612 | False |
| AT1G48710/ONSEN5 | Chr1 | 18013162 | 18018435 | 321 | 18 | 94.70% | 17.83 | AT1G48730 | 1902 | True |
| AT1G58140/ONSEN4 | Chr1 | 21524995 | 21529851 | 60 | 148 | 28.80% | 0.41 | AT1G58130 | 939 | True |
| AT3G32415/ONSEN8 | Chr3 | 13369174 | 13374108 | NA | NA | NA | NA | AT3G32425 | 6636 | False |
| AT3G59720/ONSEN6 | Chr3 | 22059535 | 22064329 | 102 | 41 | 71.30% | 2.49 | AT3G59740 | 2658 | False |
| AT3G61330/ONSEN2 | Chr3 | 22695566 | 22700522 | 291 | 11 | 96.40% | 26.45 | AT3G61310 | 2732 | False |
| AT5G13205/ONSEN3 | Chr5 | 4208083 | 4213084 | 230 | 39 | 85.50% | 5.9 | AT5G13210 | 1420 | False |
| Ler1-ONSEN-13 | Chr2 | 586740 | 591695 | 397 | 18 | 95.70% | 22.06 | ATLER-2G11780 | 1572 | False |
| Ler1-ONSEN-23 | Chr3 | 17904360 | 17909316 | 146 | 22 | 86.90% | 6.64 | ATLER-3G66970 | 142 | True |
| Ler1-ONSEN-30 | Chr4 | 8394474 | 8399430 | 699 | 0 | 100% | inf | ATLER-4G38650 | 5355 | False |
| Ler1-ONSEN-31 | Chr4 | 9796312 | 9801243 | NA | NA | NA | NA | ATLER-4G42665 | 3975 | False |
| Ler1-ONSEN-32 | Chr5 | 751500 | 756456 | 179 | 8 | 95.70% | 22.38 | ATLER-5G12430 | 550 | True |
| Ler1-ONSEN-33 | Chr5 | 2476353 | 2481309 | 173 | 1 | 99.40% | 173 | ATLER-5G17680 | 1923 | False |
| Cvi0-ONSEN-27 | Chr3 | 3410998 | 3415955 | 640 | 141 | 81.90% | 4.54 | ATCVI-3G20890 | 2067 | True |
| Cvi0-ONSEN-32 | Chr3 | 13231539 | 13236316 | 51 | 3 | 94.40% | 17 | ATCVI-3G50870 | 294 | True |
| Cvi0-ONSEN-49 | Chr4 | 9597983 | 9602941 | 473 | 76 | 86.20% | 6.22 | ATCVI-4G40910 | 2029 | True |
Table 2.
Full-length Copia-35 elements and their S1 strength
| TE ID | Chr | Start | End | S1 reads | S2 reads | S1 strength S1/(S1 + S2) ∗ 100% | S1/S2 | 3′ gene | Distance TE/gene | 3′ gene reached by S2 reads |
|---|---|---|---|---|---|---|---|---|---|---|
| AT1TE43225 | Chr1 | 13230575 | 13236255 | 39 | 2 | 95% | 19.5 | AT1G35730 | 779 | False |
| AT1TE51360 | Chr1 | 15610250 | 15615952 | 3 | 1 | 75% | 3 | AT1G41830 | 2284 | False |
| AT3TE51895 | Chr3 | 12602886 | 12608833 | 28 | 0 | 100% | 28 | AT3G30842 | 2454 | False |
| Ler1-Copia35-4 | Chr1 | 13303453 | 13309151 | 42 | 2 | 95% | 21 | ATLER-1G48170 | 760 | False |
| Ler1-Copia35-6 | Chr1 | 15445886 | 15451583 | 10 | 2 | 83% | 5 | ATLER-1G55480 | 2426 | False |
| Ler1-Copia35-10 | Chr3 | 12933534 | 12939434 | 34 | 0 | 100% | inf | ATLER-3G49220 | 2453 | False |
| Cvi0-Copia35-5 | Chr1 | 15536790 | 15542484 | 76 | 0 | 100% | inf | ATCVI-1G55690 | 2915 | False |
| Cvi0-Copia35-15 | Chr3 | 11316521 | 11322293 | 610 | 66 | 76% | 3.24 | ATCVI-3G43580 | 1082 | False |
| Cvi0-Copia35-16 | Chr3 | 12762880 | 12768764 | 483 | 6 | 99% | 80.5 | ATCVI-3G48960 | 2484 | False |
Subsequently, we aligned the RNA-Seq and ONT reads to their respective genomic assemblies. Because ONSEN and Copia-35 are still active, copies can show high sequence similarity among each other. We thus allowed multi-mapping reads (see Materials and methods) for downstream analysis (Teissandier et al. 2019). Overall, the pattern of expression levels was generally highly consistent between the RNA-Seq and ONT for a given accession (e.g. ONSEN 5 was the most expressed copy in Col-0, as was ONSEN 30 in Ler-1, according to both datasets) (Fig. 1f and g). Both RNA-Seq and ONT revealed a significant variation of expression levels between individual ONSEN and Copia-35 copies (Fig. 1f and g). In accordance with our consensus-based analysis, we found a specifically high activity of Copia-35 in Cvi-0 compared to the other two accessions. Indeed, the least transcribed copy in Cvi-0, Cvi0-Copia35-5, reached expression levels resembling those of the most expressed Copia-35 copies in the other two accessions. In addition, both RNA-Seq and ONT datasets revealed similar expression levels of both TE families in Cvi-0, with the highest expression level approximating 400 RPKM. Note that ONSEN 7 was not included in further analyses as it harbors a large insertion, which together with its low expression level (Fig. 1e), suggests that this copy is not functional.
ONT Allows for a High-Resolution Profiling of ONSEN and Copia-35
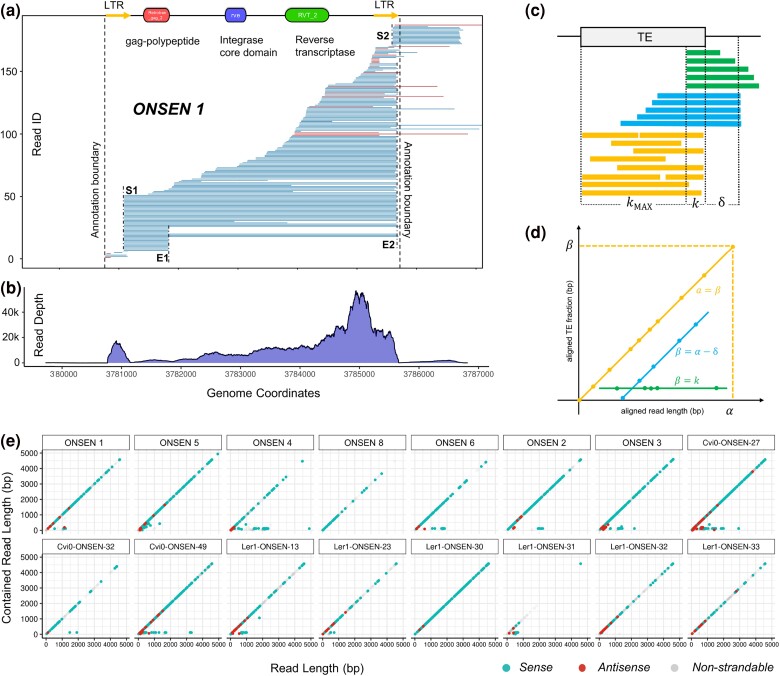
Given the substantial differences in abundance of per-copy expression of ONSEN and Copia-35, we investigated the expression of individual copies in detail with ONT. Using the alignment of one of the most active and autonomous ONSEN copies (ONSEN 1) (Cavrak et al. 2014; Roquis et al. 2021), we found that, under heat stress, active full-length ONSEN copies have two transcription starting sites (TSS), namely S1 and S2, one within each of their LTRs (Fig. 2a, supplementary figs. S2 to S4, Supplementary Material online). Moreover, we identified two transcription termination sites, E1 and E2. E1 is located just after the detected gag domain and E2 is situated at the 3′ LTR. A read from S1 to E2 thus represents a full-length mRNA that serves as a precursor for subsequent reverse transcription to ONSEN. Importantly, the read depth peak observed around 37,810 kb shows that RNA-Seq data failed to resolve the transcription starts and ends (Fig. 2b).
Fig. 2.
Transcriptional profile of ONSEN. a) Long-read alignment of ONSEN 1. Blue reads indicate matching orientation to the TE annotation (sense), while red reads indicate opposite orientation (antisense). b) Read depth of ONSEN 1 in RNA-Seq data. c and d) Principles of an aligned TE fraction versus read length plot. c) Read-through reads of a TE annotation can be divided into three groups: alignments that cover the entire TE annotation or contained in the TE annotation (yellow); alignments that start outside of a TE annotation (blue); and alignments that start within the TE annotation (green). Aligned read length and aligned TE fraction length are denoted as α and β, respectively. The symbol δ indicates the distance between a transcription starting site outside of a TE annotation and the TE. The symbol k indicates the distance between the transcription starting site to one end of the TE. d) Theoretical example of a Transposon-Read Alignment Length Analysis (TRALA) plot, in which α is plotted against β. e) TRALA plot of 16 full-length ONSEN copies.
We also found that the 5′ LTR acts as a more dominant promoter than the 3′ LTR driving the selective expression of the gag-polypeptide or of the entire element, respectively. To quantify the difference in strength, we counted the number of reads from S1 and S2 for active ONSEN copies of all accessions (Table 1). We assumed that reads with starting sites between S1 and S2 were also transcribed from S1. This assumption was based on the rationale that many mRNA molecules were not fully sequenced to their 5′ ends, as suggested from the continuous distribution of reads across the entire elements (Fig. 2a), likely due to limitations of the reverse transcriptase during ONT library preparation. We found that the 5′ LTR accounts for 71.3% to 100% of the ONSEN transcripts, except for ONSEN 4, where the 3′ LTR accounts for 71.2% of the total transcription.
To assess the global variations of full-length ONSEN copies, we implemented a graphical analysis by plotting the aligned read length of an ONT read against the length covered by a TE annotation (Fig. 2c and d), which we refer to as Transposon-Read Alignment Length Analysis (TRALA) plot (Fig. 2d). As aforementioned, for most ONSEN copies, reads were initiated from S1 and therefore contained in the annotation, appearing as dots on the diagonal line. However, ONSEN 4, Cvi0-ONSEN-27, and Cvi0-ONSEN-49 form a horizontal line at the bottom due to substantial amounts of reads initiated from S2, hence directly driving the expression of their flanking regions. Moreover, the TRALA plots revealed differences in the abundance of antisense transcription substantiating the expressional diversity among individual ONSEN copies (Fig. 2e).
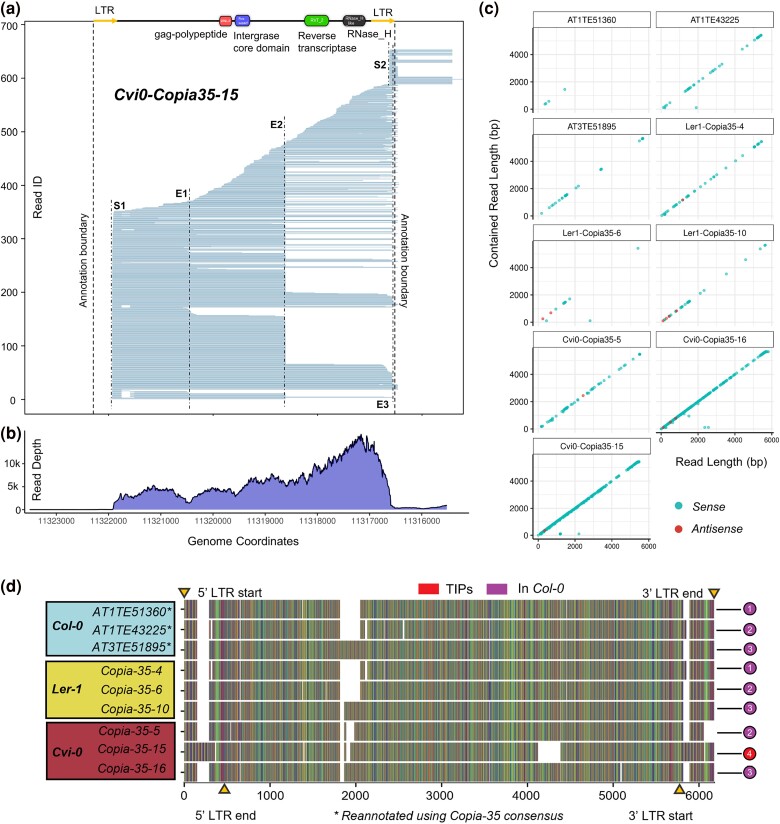
We found that, like ONSEN, when exposed to heat stress, full-length copies of Copia-35 show a continuous distribution of reads and have TSS S1 and S2 within each of their LTRs (Fig. 3a, supplementary figs. S5 to S7, Supplementary Material online). In contrast to ONSEN, we identified three termination sites: E1, E2, and E3 in Copia-35. E1 is located between the 5′ LTR and the gag-polypeptide, E2 is between the integrase and reverse transcriptase domain, while E3 lies at the 3′ LTR. Hence, a read from S1 to E3 represents a full-length mRNA that serves as a precursor for subsequent reverse transcription to Copia-35 cDNA. As shown for the most active Copia-35 copy (Cvi0-Copia35-15) and in contrast to the ONT data, RNA-Seq again failed to identify the transcription start and end points (Fig. 3b). Notably, the high resolution provided by the ONT data also revealed that some of the reads aligning to Copia-35 were spliced between S1 and E1 (Fig. 3a, supplementary figs. S4 to S6, Supplementary Material online).
Fig. 3.
Expression of Copia-35 profiled with long reads. a) Long reads alignment of Cvi0-Copia35-15. Blue reads indicate matching orientation to the TE annotation (sense). b) Read depth of Cvi0-Copia35-15 in RNA-Seq data. c) TRALA plot of nine full-length Copia-35 copies. d) Alignment of full-length Copia-35 copies in Col-0, Ler-1, and Cvi-0. LTR boundaries are marked by yellow triangles. Col-0 copies are numbered 1 to 3; numbers on sequences of other accessions correspond to these Col-0 copies conserved among accessions. The Cvi-0 TIP is labeled as copy no. 4.
The TRALA plot of all nine Copia-35 copies revealed that most reads are contained within the Copia-35 annotations (Fig. 3c), with the exception of Cvi0-Copia35-15 and Cvi0-Copia35-16, which both show the existence of read-through transcripts. In addition to substantial differences between the number of transcripts per copy, the dots on the diagonal line in the TRALA plots of most Copia-35 copies in Col-0 and Ler-1 contained large gaps, suggesting that not the entire length of the element is transcribed. To investigate whether obvious structural differences were responsible for this discrepancy between copies, we aligned all full-length Copia-35 elements. We found that despite having greater expression, the full-length copies in Cvi-0 exhibited no major structural differences compared to copies in Ler-1 and Col-0 (Fig. 3d). For example, Cvi0-Copia35-16 and Ler1-Copia35-10 showed different expression levels under heat stress, but were identical in terms of structure, except for a small deletion in Cvi0-Copia35-16 at around 5,000 bp. Notably, we observed that the most active copy Cvi0-Copia-35-15, that is also a TIP, carried an insertion in both its LTRs.
Both ONSEN and Copia-35 Confer Heat Responsiveness to Their Flanks
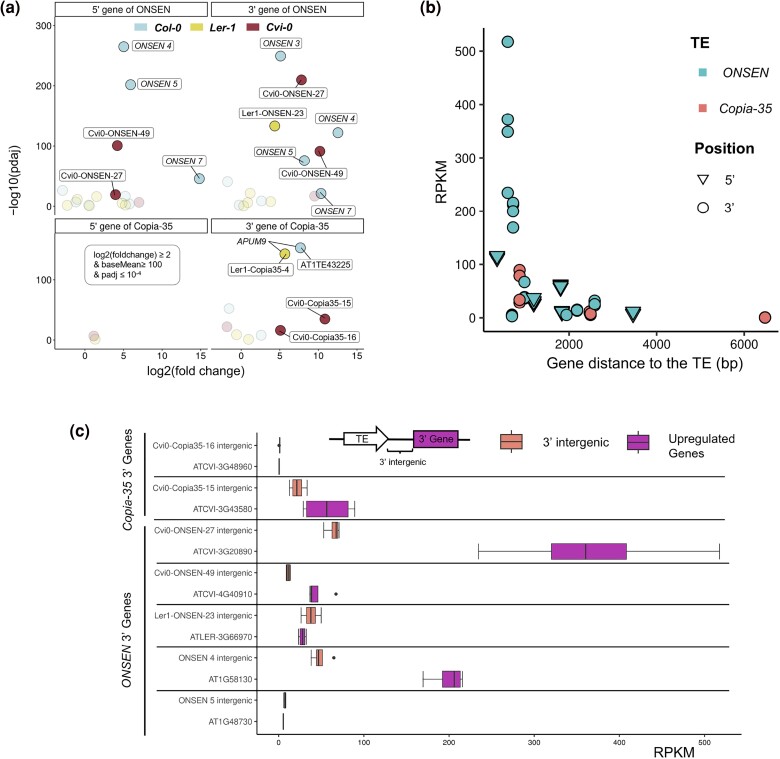
It is well established that full-length ONSEN elements can trigger the expression of adjacent genes under heat stress (Ito et al. 2011; Roquis et al. 2021), a pattern we confirmed in our RNA-Seq data. Among the seven full-length ONSEN copies in Col-0, three were associated with the upregulation of both their 5′ and 3′ flanking genes, and a fourth with the upregulation of the 3′ genes only (Fig. 4a). This pattern was also observed with two copies in Cvi-0 with the upregulation of flanking genes on both sides of Cvi-0-ONSEN-27 and Cvi-0-ONSEN-49 showing a log2fold change > 2 and Padj < 10−4 (Fig. 4a), while in Ler-1, this was only observed for Ler1-ONSEN-23 in the 3′ direction. Similarly, our data confirmed that the expression of Cvi0-Copia35-15 and Cvi0-Copia35-16, two predominantly expressed copies in Cvi-0, was associated with an upregulation of their 3′ flanking genes (Fig. 4a). Notably, while Cvi0-Copia35-16 was shared between the three accessions (Fig. 3d), the upregulation of its 3′ gene was only observed in Cvi-0.
Fig. 4.
Upregulation of ONSEN and Copia-35 flanking genes. a) Volcano plots (RNA-Seq) highlighting genes adjacent to ONSEN and Copia-35 with the following criteria: log2(fold change) ≥ 2, baseMean ≥ 100, and Padj ≤ 10−4. Highlighted genes are labeled with the names of their corresponding TE. b) Relationship between a gene’s distance to its corresponding TE and RPKM (RNA-Seq). ONSEN genes are depicted in cyan, and Copia-35 genes in coral. 5′ flanking genes (triangles) and 3′ flanking genes (circles) are denoted. c) A comparison of the RPKM of the 3′ intergenic regions (located between the TE and its 3′ gene) against the RPKM of the corresponding upregulated 3′ gene (ONT), as described in a). Only upregulated 3′ genes reached by reads originating in the S2 of the TE are included in the analysis.
Two mechanisms could lead to the TE-drive upregulation of a gene. The upregulation could occur through the formation of read-through from the TE to the gene. Alternatively, TEs can lead to indirect upregulation of genes via cis-regulatory effect mediated by their recruitment of the transcription machinery. To disentangle the two scenarios, we first tested whether the distance between the TE and the flanking genes could explain the observed patterns in Fig. 4a. We further plotted the distance between each gene and its associated TE against the gene’s RPKM. As a clear indication of cis-regulation, we uncovered a localized effect of TE-mediated gene activation under heat stress with closer genes showing a stronger heat response (Fig. 4b).
To test whether the upregulation of flanking genes could also be explained by the detected read-through transcription from the 3′-LTR of some TE copies (Figs. 2a and 3a, supplementary figs. S1 to S6, Supplementary Material online and Tables 1 and 2), we then used our ONT reads and plotted the length of all S2 reads of TE copies that exhibit transcription from their 3′ LTR (Fig. 4c). For most copies, the length of S2 reads ranged between 0 and 2 kb. However, for some insertions, we found that S2 reads were spanning up to 4.5 kb of the flanking region, even reaching the 3′ gene in seven cases (Fig. 4d). To assess the importance of those reads in driving gene expression, we quantified the relative transcription level of the intergenic region between the TE and the 3′ flanking gene (Tables 1 and 2). This analysis showed that the expression of the intergenic region was either similar or lower than the actual gene expression. We further noted that the transcription of highly expressed flanking genes such as AT1G58130 and ATCVI-3G20890 was independent from the abundance of reads aligning to the flanking region (Fig. 4d), suggesting that the cis-regulatory effect of the TE is the main driver of their heat response.
Among the genes that were solely upregulated by the cis-regulatory effect of the TE (Fig. 4a, Tables 1 and 2), we detected APUM9, a well-characterized gene that plays an important role in development (Xiang et al. 2014; Hristova et al. 2015). By using Silex reporter (a construct that contains APUM9 upstream region and the Copia-35/ROMANIAT5-2 3′ LTR upstream of a GFP reporter) in Col-0 plants under controlled conditions with and without HS, Pietzenuk et al. (2016) demonstrated at the molecular level that Copia-35 controls the expression of APUM9. In our experiment, APUM9 was accordingly highly expressed in response to heat in Col-0 and Ler-1 but not in Cvi-0, where the Copia-35 insertion was missing (Figs. 3d and 4a). Because this insertion is present in the reference genome Col-0 but absent from Cvi-0, we define this insertion polymorphism as a TAP in the rest of the manuscript (TE absence polymorphism).
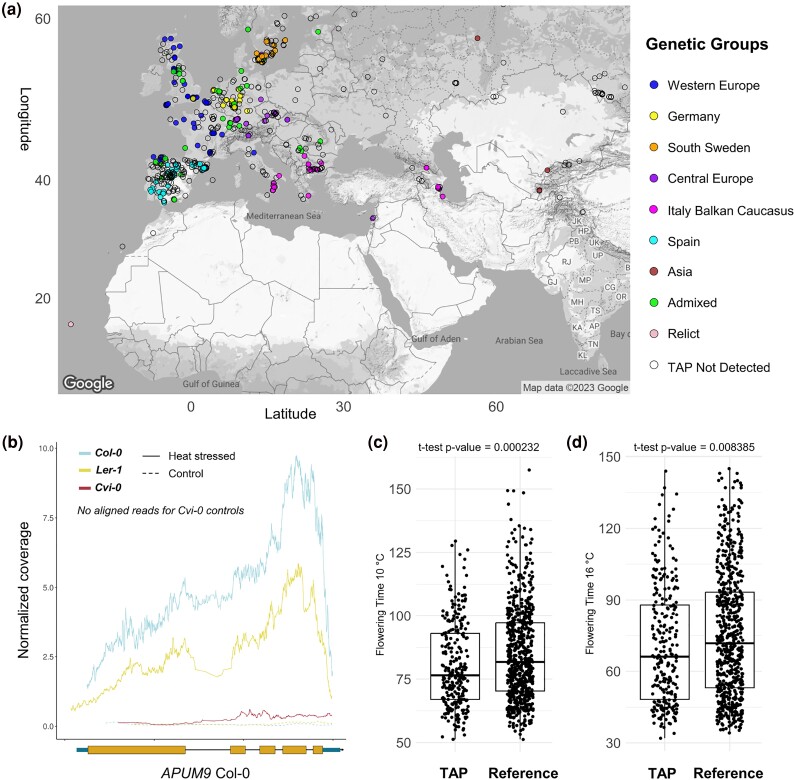
Because the transcriptional changes of APUM9 under heat stress may have phenotypic consequences and thus play a role in adaption, we further determined how frequently this TAP of Copia-35 in the flanking region of APUM9 occurred in natural accessions. After validating our approach using the available PacBio assemblies (supplementary fig. S7, Supplementary Material online), we screened genomic reads of 1,030 available accessions for the presence of this copy. Overall, we detected TAPs in 340 accessions, belonging to all genetic groups of A. thaliana (Fig. 5a). Surprisingly, TAPs were found in accessions geographically close to those carrying the Copia-35 insertion at the APUM9 locus.
Fig. 5.
APUM9 locus and flowering time of natural accessions of A. thaliana. a) Distribution map of the Copia-35 TAP at the APUM9 locus, with accessions color-coded by genetic group. b) Normalized RNA-Seq coverage for the APUM9 gene across three accessions. Solid lines represent heat-stressed samples, while dashed lines represent controls. Normalized coverage is averaged over four replicates. Average flowering time at 10 °C c) and 16 °C d) depending on the detection of a Copia-35 TAP at the APUM9 locus. Reference indicates that no TAP was detected.
Since our analysis showed that the expression of APUM9 under heat stress was potentially associated with the presence of Copia-35 (Figs. 4a and 5b) and knowing that APUM9 is involved in regulating flowering time (Nyikó et al. 2019), we tested the possibility that the presence of Copia-35 may affect this important trait when plants are exposed to different temperatures. By analyzing publicly available data, we found an association between flowering time at 10 (FT10, P < 0.001) and 16 °C (FT16, P < 0.01) and the presence of Copia-35 in the flanking region of APUM9 (Fig. 5c and d).
Discussion
TE activity is an important source of transcriptional novelty (Rebollo et al. 2012) and a major driver of genome evolution. The genetic diversity arising from TE mobility has been documented in wild plants, including A. thaliana (Quadrana et al. 2016; Baduel et al. 2021) and Brachypodium distachyon (Stritt et al. 2020), as well as in crops like rice (Huang et al. 2008; Carpentier et al. 2019; Castanera et al. 2021), maize (Stitzer et al. 2021), and wheat (Wicker et al. 2022). While ONT long-read sequencing has recently been shown to be effective to study TE expression in Arabidopsis mutants impaired for TE silencing (Panda and Slotkin 2020; Berthelier et al. 2023), the availability of high-quality assemblies now makes it possible to investigate the diversity of individual, highly similar TEs in multiple natural accessions of the same species. Using heat as an abiotic stress, our analysis revealed multiple layers of significant expressional diversity linked to stress-inducible TEs in A. thaliana.
Besides confirming the heat responsiveness of the well-studied ONSEN family, the use of three different natural genetic backgrounds allowed for the in-depth characterization of Copia-35, a second retrotransposon family with an increased activity under heat stress. Despite sharing heat as environmental trigger, our data revealed striking differences between both families. Indeed, while none of the ONSEN copies is conserved between all three accessions, we only detected one TIP of Copia-35 in the relict accession Cvi-0. These findings support the view that ONSEN is highly dynamic (Baduel et al. 2021), and could indicate a reduced mobility of Copia-35 in Ler-1 and Col-0 compared to Cvi-0. This argument is further strengthened by the fact that Copia-35 elements in Col-0 are lacking the ability to transpose, pointing toward a nonautonomous nature in this accession (Pietzenuk et al. 2016).
In response to heat treatment, both ONT and RNA-Seq data showed that the transcription of Copia-35 was relatively low in Col-0 and Ler-1 but reached high expression levels, similar to those of ONSEN, in Cvi-0. Our ONT data further confirmed the presence of full-length transcripts that could serve as a template for the reverse transcription resulting in the transposition of Copia-35 in Cvi-0. These results show that the genome of Cvi-0 harbors two independent and potentially mobile TE families, synchronically activated by the same environmental trigger. Whether additional factors, such as specific insertion preferences as observed for ONSEN (Quadrana et al. 2019; Roquis et al. 2021) or their epigenetic regulation by different pathways, are defining separate “niches” (Kidwell and Lisch 1997; Venner et al. 2009) allowing for a coexistence of both families, remains to be elucidated.
The strong variation in the activity of Copia-35, which is equally abundant in all three accessions but differentially expressed, is in line with previous work (Marí-Ordóñez et al. 2013; Thieme et al. 2017; Nozawa et al. 2022), and suggests that factors other than copy number determine the overall activity of a TE family. For instance, Copia-35 expression increases in mutants deficient in epigenetic silencing (Yokthongwattana et al. 2010) while the loss of RdDM alone (i.e. without abiotic stress) does not activate ONSEN (Ito et al. 2011), highlighting differences in the factors governing the activities of both families. Notably, recent work showed that natural variations in the strength of epigenetic silencing under heat stress lead to increased activation of ONSEN in the Kyoto accession that displays reduced methylation in the CHH context (Nozawa et al. 2022). In this regard, it is noteworthy that the relic accession Cvi-0 that displayed a high activity of both TEs in our study is globally hypomethylated compared to Col-0 (Kawakatsu et al. 2016).
The high resolution of the ONT data also revealed striking qualitative expressional differences between both families. Most importantly, we revealed the presence of an additional transcription termination site for Copia-35 compared to ONSEN. This could imply mechanistic variations in the lifecycle of the two families. Analogous to retroviruses, LTR-RT require specific amounts of the structural GAG nucleocapsid, the catalytic polyprotein and the full-length transcript that serves as a template for reverse transcription to complete their lifecycle (Schulman 2013). Besides mechanisms affecting translation (Clare et al. 1988; Matthews et al. 1997; Havecker and Voytas 2003), subgenomic TE expression and splicing resulting in different transcript pools underly the fine-tuning of retrotransposon protein abundances (Chang et al. 2013). The role of alternative splicing is perfectly illustrated by its importance for regulating protein abundances of the Arabidopsis Copia-type retrotransposon EVADÉ (Oberlin et al. 2017). Our work, however, paints a more nuanced picture. While we detected the presence of a few spliced transcripts produced by Copia-35, our ONT analysis suggests the presence of short subgenomic transcripts that may indicate that the diverse RNA pools needed to complete the TE-lifecycle are obtained using a splicing-independent mechanism. These findings therefore open new avenues for elucidating the fundamental processes of plant retrotransposon mobility. This is particular crucial, because while ONSEN has been studied in detail (Ito et al. 2011; Cavrak et al. 2014; Thieme et al. 2017; Baduel et al. 2021), our current mechanistic understanding of plant TEs is overwhelmingly based on studies using few genetic backgrounds, and in the case of heat-responsive TEs, mainly on Col-0.
The influence of TEs on the expression of their flanking regions is well-documented (Butelli et al. 2012; Makarevitch et al. 2015; Rech et al. 2022). Here, we confirmed that ONSEN mediates a heat-dependent upregulation of flanking regions (Ito et al. 2011; Roquis et al. 2021) and further revealed that Copia-35 can also confer heat responsiveness to its neighboring genes, in addition to the previously reported APUM9 locus in Col-0 (Pietzenuk et al. 2016), in multiple accessions. The ONT data further allowed us to unambiguously discriminate between read-through transcription and the indirect upregulation of genes via the cis-regulatory effect mediated by the recruitment of the transcription machinery to the TE (Zhao et al. 2018; Fagny et al. 2020; Deneweth et al. 2022). The formation of TE–gene fusion transcripts is a common phenomenon in Arabidopsis (Lockton and Gaut 2009; Berthelier et al. 2023) and we indeed detected read-through transcription originating from the 3′ LTRs of both ONSEN and Copia-35 TE families under heat stress. However, the formation of read-through is not a predominant phenomenon (Tables 1 and 2) and our data suggest that the cis-regulatory effect is the main driver of TE-mediated expression of the flanking genes.
Interestingly, one of the genes that has previously been shown to be cis-regulated by Copia-35 (Pietzenuk et al. 2016) is APUM9, which is involved in early embryonic development, with a putative role in basal heat tolerance (Nyikó et al. 2019). In addition, an overexpression of APUM9 results in abnormal leaf morphology and a delayed flowering phenotype (Nyikó et al. 2019). Despite its importance in development, the natural diversity of the APUM9 locus and more specifically the role of Copia-35 in driving its expression under heat stress had not been studied across multiple natural accessions. Our data revealed that on a population scale, accessions with the Copia-35 insertion at the APUM9 locus (i.e. putatively heat-responsive based on our expression analyses in Col-0, Ler-1, and CVI-0) tend to flower later, which support the overexpression analysis of Nyikó et al. (2019). The timing of flowering is crucial for a population to survive. Despite their selfish nature, major (epi)genetic effects linked to transposition events are generally viewed as a driving force of plant evolution (for review Lisch 2013), capable of facilitating rapid adaptation (Van't Hof et al. 2016; Thieme et al. 2022), and the link between transposition and modulation of flowering time in A. thaliana has been suggested previously (Thieme et al. 2017; Quadrana et al. 2019; Baduel et al. 2021). Flowering time is a complex trait driven by multiple loci with small quantitative effects (Kinoshita and Richter 2020). The fact that heat triggers the upregulation of Copia-35, resulting in an activation of APUM9, and that the experimentally induced overexpression of APUM9 in Col-0 results in delayed flowering (Nyikó et al. 2019), indeed indicates a quantitative effect of this insertion on flowering time.
Overall, our study revealed a great expressional diversity linked to heat-responsive LTR retrotransposons in A. thaliana. These findings strongly advocate for the use of ONT in studies aiming at understanding both the fundamental mechanisms of LTR-retrotransposon mobility and their adaptive consequences across multiple natural accessions. With the increasing availability of high-quality genomes, similar studies should soon allow us to drastically improve our understanding of the role of TEs in plants that are densely packed with TEs.
Materials and Methods
Heat Stress Experiments, RNA Extractions, and Sequencing
Seeds of Col-0, Ler-1, and Cvi-0 were first stratified on ½ Murashige and Skoog plates for 7 d at 4 °C and then grown under controlled conditions (16 h light at 24 °C, 8 h dark at 22 °C) in a Aralab 600 growth chamber (Rio de Mouro, Portugal). After 7 d of growth, plants were stressed at 37 °C for 24 and 16 h light in a second Aralab 600 growth chamber. Seedlings from control and heat treatment were sampled simultaneously at the end of the stress period. For the ONT direct cDNA sequencing, 20 seedlings per accession per treatment were pooled together for mRNA extraction using oligo-dT beads (#61011) (Thermo Fisher Scientific, Waltham, USA). The Functional Genomic Centre at Zürich performed library preparation and sequencing. Final cDNA libraries were sequenced on ONT Flow Cells (R 9.4.1) (Oxford, UK).
For the Illumina RNA-Seq samples, plants were grown and stressed under the same conditions. Four biological replicates (pools of at least nine seedlings) per condition for each accession were extracted using the QIAGEN RNeasy plant mini kit (#74904) (Venlo, Netherlands). Novogene UK performed the library prep and sequencing.
TE Annotation
For ONSEN, full-length copies (Cavrak et al. 2014) were used to generate annotations using RepeatMasker (version 4.1.1) (repeatmasker.org) with the following options: -a -xsmall -gccalc -nolow. We only conducted the rest of the analysis on the remaining seven functional copies. In addition, TE consensus sequences of A. thaliana from RepBase28.03 (Bao et al. 2015) were used to annotate all other TEs using the same command. ROMANIAT5 consensus sequence was reconstructed by Repbase in 2018, and its name was reverted to Copia-35 (girinst.org/2018/vol18/issue9/Copia-35_AT-I.html). For clarity, this article abandoned the legacy name of ROMANIAT5 and refers to the family as Copia-35. In the case of full-length copies of Copia-35 in Col-0, we adopted their TAIR10 names, AT1E51360, AT1E43225, and AT3TE51895, even after reannotation. For the remaining accessions, the elements were named based on the format: Accession-TE family-Annotation ID. NCBI conserved domain search (CDD v3.20) (Lu et al. 2019) was used to annotate protein domains in TE sequences.
RNA-Seq Analysis
Fastp (version 0.23.2) (Chen 2023) was used to trim adapters and remove low complexity reads using the following options: --qualified_quality_phred 15 --unqualified_percent_limit 40 --n_base_limit 10 --low_complexity_filter --correction --detect_adapter_for_pe --overrepresentation_analysis --dedup --dup_calc_accuracy 6. Ribosomal RNA was then removed using bbduk.sh (version 39.01) from the BBTools suite (sourceforge.net/projects/bbmap/) with the options k = 31 hdist = 1.
Cleaned reads were then mapped to their respective genome assemblies using STAR (version 2.7.10b) (Dobin et al. 2012) with options: --alignIntronMax 5000 –outFilterMultimapNmax 100 –winAnchorMultimapNmax 100. The genome assembly and gene annotation of Col-0 (release 10) were downloaded from the Arabidopsis Information Resource (TAIR) (Berardini et al. 2015). The genome assemblies and gene annotation of Ler-1 and Cvi-0 were downloaded from the 1001 genomes webpage (Jiao and Schneeberger 2020).
We employed RPKM (Reads Per Kilobase of transcript, per Million mapped reads), a commonly used unit of measurement to quantify gene and single TE copy expression levels and normalize the expression levels across replicates. Pair-ended fragments were counted using featureCounts (Liao et al. 2013) against the TE or gene annotations, with the following options: -B -p -P -O. To quantify expression at the TE family level; cleaned RNA-Seq data were also analyzed by SalmonTE (version 0.4) (Jeong et al. 2018). The A. thaliana TE consensus library was downloaded from Repbase (version 28.03.2023) (Bao et al. 2015) and used as the custom library for SalmonTE. Default options of SalmonTE’s “quant” and “test” program were used to quantify expression and perform statistical analyses.
Basecalling and Mapping of ONT Data
Basecalling was performed on the passed fast5 files using Guppy (version 6.1.2) with default options. Guppy is developed by ONT and available via their community website (community.nanoporetech.com). Stranding was then directly performed on the passed output from basecalling using Pychopper (version 2.5.0) (github.com/epi2me-labs/pychopper). Primer configuration for stranding was set to “+:SSP, -VNP|-:VNP, -SSP”, and rescued reads were not used. Porechop (version 0.2.4) (github.com/rrwick/Porechop) was then used to remove sequencing adapters from ONT reads. Finally, ONT reads were mapped to their respective genome assemblies using minimap2 (version 2.24) (Li 2018) with options -ax splice -uf -k14.
Mapping of Whole-Genome Sequencing (WGS) Data
The whole-genome sequencing (WGS) data of 1,135 A. thaliana accessions were downloaded from the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under project PRJNA273563 (The 1001 Genomes Consortium 2016). Fastp (version 0.23.2) (Chen 2023) was used to trim adapters and remove low complexity reads using the following options: --qualified_quality_phred 15 --unqualified_percent_limit 40 --n_base_limit 10 --low_complexity_filter --correction --detect_adapter_for_pe --overrepresentation_analysis --dedup --dup_calc_accuracy 6. BWA-MEM (version 0.7.17) (Li 2013) was used to map the genomic reads to the APUM9 locus of Col-0.
TAP Detection at the APUM9 Locus
Data retrieved from the 1001 Genomes Project (The 1001 Genomes Consortium 2016) were used to screen for TE Absence Polymorphisms (TAPs) at the APUM9 locus. BWA-mem (version 0.7.17) (Li 2013) and detettore (version 2.0.3) (github.com/cstritt/detettore) were used in tandem to first map the reads, and then perform TAP calling using default options.
Flowering Time Analysis
Flowering time at 16 °C (FT16) and 10 °C (FT10) recorded by the 1001 Genomes Project (The 1001 Genomes Consortium 2016) was used to test the association between the number of TAPs and flowering time.
Supplementary Material
Acknowledgments
The author would like to thank the Functional Genomic Center Zurich for processing the samples and Dr. Emmanuelle Botté (https://manuscribe.com.au) for professional editing of the manuscript. The authors would like to thank Angela Hancock for handling our manuscript and the two anonymous reviewers for their comments on the manuscripts. This work was supported by the University of Zurich Research Priority Programs (URPP) Evolution in Action (M.T. and A.C.R.) and the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung. Grant number: 31003A_182785 (A.C.R. and W.X.).
Contributor Information
Wenbo Xu, Department of Plant and Microbial Biology, University of Zürich, 8008 Zürich, Switzerland.
Michael Thieme, Department of Plant and Microbial Biology, University of Zürich, 8008 Zürich, Switzerland.
Anne C Roulin, Department of Plant and Microbial Biology, University of Zürich, 8008 Zürich, Switzerland; Agroscope, 8820 Wädenswil, Switzerland.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
M.T. and A.C.R. conceived the study; W.X. and M.T. conducted experiments; W.X. analyzed the data; W.X. and M.T. wrote the paper with contributions from A.C.R. A.C.R. secured funding. A.C.R. revised the manuscript. All authors approve the paper.
Data Availability
Raw RNA-Seq and base-called ONT data were uploaded to the European Nucleotide Archive (ENA) under project PRJEB64476. The scripts used for the statistics and figure generation were deposited into https://github.com/GroundB/Natural-diversity-of-heat-induced-transcription-of-retrotransposons-in-Arabidopsis-thaliana. The genome assemblies and annotations used in this study are available on TAIR: https://1001genomes.org/data/MPIPZ/MPIPZJiao2020/releases/current/.
Literature Cited
- Baduel P, Leduque B, Ignace A, Gy I, Gil J Jr, Loudet O, Colot V, Quadrana L. Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana. Genome Biol. 2021:22(1):138. 10.1186/s13059-021-02348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Quadrana L. Jumpstarting evolution: how transposition can facilitate adaptation to rapid environmental changes. Curr Opin Plant Biol. 2021:61:102043. 10.1016/j.pbi.2021.102043. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015:6:11. 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015:53(8):474–485. 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelier J, Furci L, Asai S, Sadykova M, Shimazaki T, Shirasu K, Saze H. Long-read direct RNA sequencing reveals epigenetic regulation of chimeric gene-transposon transcripts in Arabidopsis thaliana. Nat Commun. 2023:14(1):3248. 10.1038/s41467-023-38954-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012:24(3):1242–1255. 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier MC, Manfroi E, Wei F-J, Wu H-P, Lasserre E, Llauro C, Debladis E, Akakpo R, Hsing Y-I, Panaud O. Retrotranspositional landscape of Asian rice revealed by 3000 genomes. Nat Commun. 2019:10(1):24. 10.1038/s41467-018-07974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanera R, Vendrell-Mir P, Bardil A, Carpentier M-C, Panaud O, Casacuberta JM. Amplification dynamics of miniature inverted-repeat transposable elements and their impact on rice trait variability. Plant J. 2021:107(1):118–135. 10.1111/tpj.15277. [DOI] [PubMed] [Google Scholar]
- Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014:10(1):e1004115. 10.1371/journal.pgen.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Jääskeläinen M, Li SP, Schulman AH. BARE retrotransposons are translated and replicated via distinct RNA pools. PLoS One. 2013:8(8):e72270. 10.1371/journal.pone.0072270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta. 2023:2(2):e107. 10.1002/imt2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare JJ, Belcourt M, Farabaugh PJ. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci U S A. 1988:85(18):6816–6820. 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneweth J, Van de Peer Y, Vermeirssen V. Nearby transposable elements impact plant stress gene regulatory networks: a meta-analysis in A. thaliana and S. lycopersicum. BMC Genomics. 2022:23(1):18. 10.1186/s12864-021-08215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012:29(1):15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez M, Dugas E, Benchouaia M, Leduque B, Jiménez-Gómez JM, Colot V, Quadrana L. The impact of transposable elements on tomato diversity. Nat Commun. 2020:11(1):4058. 10.1038/s41467-020-17874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagny M, Kuijjer ML, Stam M, Joets J, Turc O, Rozière J, Pateyron S, Venon A, Vitte C. Identification of key tissue-specific, biological processes by integrating enhancer information in maize gene regulatory networks. Front Genet. 2020:11:606285. 10.3389/fgene.2020.606285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Voytas DF. The soybean retroelement SIRE1 uses stop codon suppression to express its envelope-like protein. EMBO Rep. 2003:4(3):274–277. 10.1038/sj.embor.embor773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova E, Fal K, Klemme L, Windels D, Bucher E. HISTONE DEACETYLASE6 controls gene expression patterning and DNA methylation-independent euchromatic silencing. Plant Physiol. 2015:168(4):1298–1308. 10.1104/pp.15.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Lu G, Zhao Q, Liu X, Han B. Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol. 2008:148(1):25–40. 10.1104/pp.108.121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011:472(7341):115–119. 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- Ito H, Yoshida T, Tsukahara S, Kawabe A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene. 2013:518(2):256–261. 10.1016/j.gene.2013.01.034. [DOI] [PubMed] [Google Scholar]
- Jeong H-H, Yalamanchili HK, Guo C, Shulman JM, Liu Z. An ultra-fast and scalable quantification pipeline for transposable elements from next generation sequencing data. Pac Symp Biocomput. 2018:23:168–179. [PubMed] [Google Scholar]
- Jiao W-B, Schneeberger K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat Commun. 2020:11(1):989. 10.1038/s41467-020-14779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin C-S, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017:546(7659):524–527. 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015:31(22):3593–3599. 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang S-SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016:166(2):492–505. 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A. 1997:94(15):7704–7711. 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Richter R. Genetic and molecular basis of floral induction in Arabidopsis thaliana. J Exp Bot. 2020:71(9):2490–2504. 10.1093/jxb/eraa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciano S, Cristofari G. Measuring and interpreting transposable element expression. Nat Rev Genet. 2020:21(12):721–736. 10.1038/s41576-020-0251-y. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, arXiv, arXiv:13033997, preprint: not peer reviewed.
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018:34(18):3094–3100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013:30(7):923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013:14(1):49–61. 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- Lockton S, Gaut BS. The contribution of transposable elements to expressed coding sequence in Arabidopsis thaliana. J Mol Evol. 2009:68(1):80–89. 10.1007/s00239-008-9190-5. [DOI] [PubMed] [Google Scholar]
- Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2019:48(D1):D265–D268. 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015:11(1):e1004915. 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013:45(9):1029–1039. 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- Masuda S, Nozawa K, Matsunaga W, Masuta Y, Kawabe A, Kato A, Ito H. Characterization of a heat-activated retrotransposon in natural accessions of Arabidopsis thaliana. Genes Genet Syst. 2016:91(6):293–299. 10.1266/ggs.16-00045. [DOI] [PubMed] [Google Scholar]
- Matthews GD, Goodwin TJ, Butler MI, Berryman TA, Poulter RT. pCal, a highly unusual Ty1/copia retrotransposon from the pathogenic yeast Candida albicans. J Bacteriol. 1997:179(22):7118–7128. 10.1128/jb.179.22.7118-7128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014:15(6):394–408. 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Negi P, Rai AN, Suprasanna P. Moving through the stressed genome: emerging regulatory roles for transposons in plant stress response. Front Plant Sci. 2016:7:1448. 10.3389/fpls.2016.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Masuda S, Saze H, Ikeda Y, Suzuki T, Takagi H, Tanaka K, Ohama N, Niu X, Kato A, et al. Epigenetic regulation of ecotype-specific expression of the heat-activated transposon ONSEN. Front Plant Sci. 2022:13:899105. 10.3389/fpls.2022.899105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyikó T, Auber A, Bucher E. Functional and molecular characterization of the conserved Arabidopsis PUMILIO protein, APUM9. Plant Mol Biol. 2019:100(1-2):199–214. 10.1007/s11103-019-00853-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin S, Sarazin A, Chevalier C, Voinnet O, Marí-Ordóñez A. A genome-wide transcriptome and translatome analysis of Arabidopsis transposons identifies a unique and conserved genome expression strategy for Ty1/Copia retroelements. Genome Res. 2017:27(9):1549–1562. 10.1101/gr.220723.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K, Slotkin RK. Long-read cDNA sequencing enables a “gene-like” transcript annotation of transposable elements. Plant Cell. 2020:32(9):2687–2698. 10.1105/tpc.20.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Scheid OM. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010:22(9):3118–3129. 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzenuk B, Markus C, Gaubert H, Bagwan N, Merotto A, Bucher E, Pecinka A. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements. Genome Biol. 2016:17(1):209. 10.1186/s13059-016-1072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L. The contribution of transposable elements to transcriptional novelty in plants: the FLC affair. Transcription. 2020:11(3-4):192–198. 10.1080/21541264.2020.1803031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Bortolini Silveira A, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V. The Arabidopsis thaliana mobilome and its impact at the species level. eLife. 2016:5:e15716. 10.7554/eLife.15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui M-A, Guy J, Bortolini Silveira A, Engelen S, Baillet V, Wincker P, et al. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat Commun. 2019:10(1):3421. 10.1038/s41467-019-11385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012:46:21–42. 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Rech GE, Radío S, Guirao-Rico S, Aguilera L, Horvath V, Green L, Lindstadt H, Jamilloux V, Quesneville H, González J. Population-scale long-read sequencing uncovers transposable elements associated with gene expression variation and adaptive signatures in Drosophila. Nat Commun. 2022:13(1):1948. 10.1038/s41467-022-29518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquis D, Robertson M, Yu L, Thieme M, Julkowska M, Bucher E. Genomic impact of stress-induced transposable element mobility in Arabidopsis. Nucleic Acids Res. 2021:49(18):10431–10447. 10.1093/nar/gkab828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DH, Paszkowski J. Heat-induced release of epigenetic silencing reveals the concealed role of an imprinted plant gene. PLoS Genet. 2014:10(11):e1004806. 10.1371/journal.pgen.1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman AH. Retrotransposon replication in plants. Curr Opin Virol. 2013:3(6):604–614. 10.1016/j.coviro.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Stitzer MC, Anderson SN, Springer NM, Ross-Ibarra J. The genomic ecosystem of transposable elements in maize. PLoS Genet. 2021:17(10):e1009768. 10.1371/journal.pgen.1009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Wyler M, Gimmi EL, Pippel M, Roulin AC. Diversity, dynamics and effects of long terminal repeat retrotransposons in the model grass Brachypodium distachyon. New Phytol. 2020:227(6):1736–1748. 10.1111/nph.16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Jing Y, Liu X, Li Q, Xue Z, Cheng Z, Wang D, He H, Qian W. Heat stress-induced transposon activation correlates with 3D chromatin organization rearrangement in Arabidopsis. Nat Commun. 2020:11(1):1886. 10.1038/s41467-020-15809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissandier A, Servant N, Barillot E, Bourc’his D. Tools and best practices for retrotransposon analysis using high-throughput sequencing data. Mob DNA. 2019:10:52. 10.1186/s13100-019-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1001 Genomes Consortium . 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016:166(2):481–491. 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme M, Brêchet A, Bourgeois Y, Keller B, Bucher E, Roulin AC. Experimentally heat-induced transposition increases drought tolerance in Arabidopsis thaliana. New Phytol. 2022:236(1):182–194. 10.1111/nph.18322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme M, Lanciano S, Balzergue S, Daccord N, Mirouze M, Bucher E. Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol. 2017:18(1):134. 10.1186/s13059-017-1265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010:6(10):e1001175. 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Hof AE, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, Saccheri IJ. The industrial melanism mutation in British peppered moths is a transposable element. Nature. 2016:534(7605):102–105. 10.1038/nature17951. [DOI] [PubMed] [Google Scholar]
- Venner S, Feschotte C, Biémont C. Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet. 2009:25(7):317–323. 10.1016/j.tig.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Fustier M-A, Alix K, Tenaillon MI. The bright side of transposons in crop evolution. Brief Funct Genomics. 2014:13(4):276–295. 10.1093/bfgp/elu002. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 2013:9(2):e1003255. 10.1371/journal.pgen.1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Gundlach H, Spannagl M, Uauy C, Borrill P, Ramírez-González RH, De Oliveira R; International Wheat Genome Sequencing Consortium; Mayer KFX, Paux E, et al. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018:19(1):103. 10.1186/s13059-018-1479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007:8(12):973–982. 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wicker T, Schulman AH, Tanskanen J, Spannagl M, Twardziok S, Mascher M, Springer NM, Li Q, Waugh R, Li C, et al. The repetitive landscape of the 5100 Mbp barley genome. Mob DNA. 2017:8:22. 10.1186/s13100-017-0102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Stritt C, Sotiropoulos AG, Poretti M, Pozniak C, Walkowiak S, Gundlach H, Stein N. Transposable element populations shed light on the evolutionary history of wheat and the complex co-evolution of autonomous and non-autonomous retrotransposons. Adv Genet. 2022:3(1):2100022. 10.1002/ggn2.202100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995:11:441–469. 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell. 2014:26(11):4362–4375. 10.1105/tpc.114.132811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokthongwattana C, Bucher E, Caikovski M, Vaillant I, Nicolet J, Mittelsten Scheid O, Paszkowski J. MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 2010:29(2):340–351. 10.1038/emboj.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang W, Chen L, Wang L, Marand AP, Wu Y, Jiang J. Proliferation of regulatory DNA elements derived from transposable elements in the maize genome. Plant Physiol. 2018:176(4):2789–2803. 10.1104/pp.17.01467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-Seq and base-called ONT data were uploaded to the European Nucleotide Archive (ENA) under project PRJEB64476. The scripts used for the statistics and figure generation were deposited into https://github.com/GroundB/Natural-diversity-of-heat-induced-transcription-of-retrotransposons-in-Arabidopsis-thaliana. The genome assemblies and annotations used in this study are available on TAIR: https://1001genomes.org/data/MPIPZ/MPIPZJiao2020/releases/current/.