Abstract
Fungal communities inhabiting plant tissues are complex systems of inter-species interactions, consisting of both the “abundant biosphere” and “rare biosphere”. However, the composition, assembly, and stability of these subcommunities, as well as their contributions to productivity remain unclear. In this study, the taxonomic and functional composition, co-occurrence, and ecological assembly of abundant and rare fungal subcommunities in different tissues of three Panax species were investigated. Abundant subcommunities were dominated by potential plant pathogens belonging to Microbotryomycetes, while saprotrophic fungi like Agaricomycetes and Mortierellomycetes were more prevalent in rare subcommunities. The rare taxa played a central role in upholding the stability of the fungal networks as driven by Dothideomycetes and Sordariomycetes. Homogeneous selection played a larger role in the assembly of abundant fungal subcommunities compared to the rare counterparts, which was more dominated by stochastically ecological drift in all plant species. Rare biospheres played a larger role in the accumulation of saponin compared to their abundant counterparts, especially in the leaf endosphere, which was mainly affected by environmental factors (Mg, pH, OC, and etc.). Furthermore, we found that rare species belonging to unidentified saprotrophs were associated with saponin formation. This study provides hypotheses for future experiments to understand mechanisms accounting for the variations in the composition and function of rare fungal subcommunities across different Panax species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00645-7.
Introduction
Plants form complex symbiotic relationships with a wide variety of endophytic fungi, which inhabit their tissues (e.g., leaves and roots) across diverse plant lineages [71, 76]. These symbiotic associations are essential for plant fitness by affecting nutrient absorption and stress tolerance of the host species [59]. Although there has been significant research on the mechanisms driving the assembly and stability of plant-associated mycobiota and their effects on plant immune responses and productivity, most of these studies primarily focused on rhizosphere habitats [17, 56]. Given that endophytes play direct roles in leaf and root metabolisms, a deeper understanding of endophytic fungal communities is critical for plant health and ecology [4, 9, 81]. However, the assembly of fungal communities within the complex endophytic ecosystem remains underexplored, particularly in medicinal plants that are rich in a range of secondary metabolites [82].
In various ecosystems, including plant tissues, fungal communities typically comprise a few dominant species and a multitude of low-abundance species, referred to as the “rare biosphere” [41]. Differences in distribution patterns and functional traits have been observed between abundant and rare microbial taxa [55]. Past research indicates that both abundant and rare fungal subcommunities in plant-associated niches can significantly influence host-related ecological processes and functions. Generally, abundant fungi have effects on plant reproductive output [21] and influence the temporal dynamics of vegetation successional patterns [1], while rare species are important for maintaining the stability of crop mycobiota and ecosystem functions [34, 77]. For instance, studies have found that Basidiomycota was always classified into rare biosphere within plants, which have the potential to produce antimicrobials [60, 69]. A recent study demonstrated that a collection of rare species interacts with prevalent, inheritable fungal endophytes to affect plant performance [25]. Researchers also found that the relative contribution of deterministic processes to rare abundant species was larger in the root endosphere than the leaf endosphere, which might be caused by a complex environment underground [77]. It is, therefore, crucial to understand the underlying mechanisms supporting the taxonomic and functional compositions, host-microbe-environment interactions, as well as microbiome assembly of abundant and rare mycobiota in plant habitats.
Understanding the relative contributions of deterministic and stochastic processes to microbial community assembly remains a fundamental challenge in microbial ecology [52]. Deterministic processes drive community assembly through directed ecological selection, such as environmental filtering and species interactions. Conversely, stochastic processes involve random community changes resulting from birth, death, and dispersal [66]. Some studies have found that rare subcommunities could be more influenced by stochastic processes compared to abundant subcommunities, possibly potentially attributed to their vulnerability to ecological drift as a result of low abundance [31, 46, 82]. Other research suggests that stochastic processes may play a larger role in the assembly of abundant subcommunities, potentially due to their extensive environmental adaptability [26, 32]. Recently, a new framework called iCAMP (Infer Community Assembly Mechanisms by Phylogenetic-bin-based null model) was introduced to assess the relative importance of assembly processes in shaping microbial diversity and dynamics, focusing on specific groups defined by phylogenetic distance [52]. This framework is essential for exploring assembly mechanisms in microbial communities subjected to environmental variations. It may be worthwhile to mention that the results of these models can be used to design experiments testing these assembly hypotheses. Additionally, the co-occurrence of taxa, analyzed through ecological network construction, are crucial deterministic drivers of community assembly, offering insights into community interactions and coexistence [5, 17, 36]. By scrutinizing the topologies of networks, a more profound comprehension of community interactions can be attained [42]. However, studies focusing on the community assembly and network structures of abundant and rare fungal subcommunities in plant tissues are still limited [77].
The cultivated Panax ginseng (PG), P. quinquefolium (PQ), and P. notoginseng (PN) are representative perennials within the important medical Panax genus. The main medicinal feature of Panax is ginsenosides (tetracyclic triterpenoid saponins), which can play a great role in treating cardiovascular disease, diabetes mellitus, cancers, stress, and immunostimulation [37]. As the quintessential secondary metabolites of the esteemed Panax genus, saponins have garnered extensive acclaim for their pivotal role as the principal bioactive constituents underpinning the pharmacological prowess of Panax species. These saponins are widely regarded as a cornerstone of the plant’s defense arsenal against pathogens and herbivorous threats, whose lineage-specific nature suggests a remarkable evolutionary trajectory, hinting at a period of rapid biosynthetic adaptation amidst the ancient “conflict” between flora and potential interlopers [3, 18]. The relationships between Panax-associated mycobiota and saponin diversification have been explored in previous studies. For example, saponins may play a key role in the co-evolution of mycobiota and Panax genus [81], and low-efficiency phyllosphere fungal networks could drive saponin accumulation in leaves as feedback [80].
Above all, there exhibit complex and intimate relationships between saponins and fungal endophytes, while responses of abundant and rare endophytic fungal subcommunities to environmental selection, and their potential contributions to saponin biosynthesis, remain largely unexplored. In this study, we analyze the abundant and rare fungal subcommunities within the root endosphere and leaf endosphere of these three Panax species, focusing on their diversity, coexistence, ecological assembly, as well as links to saponin accumulation. Our goals are to (1) identify variations in the taxonomic and functional compositions of abundant and rare fungal subcommunities; (2) reveal the distinct roles of these subcommunities in maintaining potential community interaction structures; (3) quantify the contributions of specific ecological processes to the assembly of abundant and rare subcommunities; and (4) uncover the potential contributions of these subcommunities to saponin accumulation.
Materials and methods
Sampling
Plant and soil samples were collected in PG, PQ, and PN fields at the main producing regions. The locations of PG and PQ farmlands were in Baishan City, Jilin Province, northeast China, and the locations of PN were in Wenshan Prefecture, Yunnan Province, southeast China (Table S1). These sampling fields are sited in the Daodi areas, which are regarded as the most suitable areas for the growing and saponin accumulation of the three Panax species. The sampling time of the three Panax species ranged from September 2019 to October 2019, which was at the end of the root growth stage of all plants [81]. To prevent the effect of alien microbes, plant individuals were all planted in fields as seeds instead of transplanting, and were managed according to Good Agricultural Practice [78, 79].
Three fields were chosen for the three plant species, each of which was cultivated for 2 years, 3 years, and 4 years, since plant growth stage plays critical roles in shaping plant–microbe interactions [79]. A total of nine 1.5 × 2 m2 plots across each field were selected as replicates. Unlike the genotypes of model plants or crops, PG, PQ, and PN are heterozygous germplasm. There are significant genetic differences among individuals, which may hinder the discovery of regularities. Ten healthy plants in each plot were randomly selected and mixed as one sample to reduce the impact of genetic heterogeneity of plants on the results. Plants were dug out gently with a sterilized shovel, and the soil loosely attached to the root was removed as the rhizosphere soil (RS). Sterilized 2-mm meshes were used to remove plant roots and other plant materials in RS samples, and the rest of the small plant residues were picked out manually using sterilized gloves. To avoid environmental interference, the RS samples were shifted to the laboratory within 2 h after homogenized. The samples were used to determine physicochemical properties.
After the plants were separated and thoroughly washed to eliminate surface residues, the root samples were subjected to surface disinfection by immersion in 70% ethanol for 1 min, 5% sodium hypochlorite for 5 min, and 70% ethanol for 1 min [73]. Then, the samples were washed four times with sterile ultrapure water. After cutting into small pieces using sterile scissors and grounded into powder with liquid nitrogen to avoid metabolic conversions, root samples were stored at − 80 °C to determine endophytic spectrum (RE) and saponin content [79]. The method described in Zhang et al. [77–83] was followed to eliminate the phylloplane fungi from leaf surfaces. Afterward, leaves were immersed in 75% ethanol for 1 min initially; then treated with 3.25% sodium hypochlorite for 3 min; further immersed in 75% ethanol for 30 s; finally rinsed four times using sterile ultrapure water. To abolish metabolic conversions in tissues, the sterilized leaves were pulverized using liquid nitrogen and stored at − 80 °C as endophyte profiling (LE) for saponin measurement. A total of 162 samples were acquired from 324 plants: three plant species (PG, PQ, and PN) × 3 growth years (2 y, 3 y, and 4 y) × two compartments (RE and LE) × nine plots (replicates) = 162 samples. It is important to acknowledge that the plant specimens examined in this research were sourced from various fields, and thus, the potential impact of these field-specific variations on species distinctions cannot be entirely discounted.
Measurement of edaphic factors and plant saponins
The measurement of twelve edaphic factors was conducted on all rhizosphere samples following previous studies [63, 86], including pH, organic carbon (OC), total nitrogen (TN), available phosphorus (AP), available potassium (AK), available sulfur (S), exchangeable calcium (Ca), magnesium (Mg), and particle size distribution (PSD). According to clay proportion, the PSD was clay (< 0.002 mm), silt (0.002–0.02 mm), fine sand (Fsand, 0.02–0.2 mm), and coarse sand (Csand, 0.2–2 mm).
The ginsenosides found in the Panax species are various, but most of them are not abundant in nature. Here, a total of eight major types of saponins were measured with HPLC (Waters, USA) using a Zorbax SB-AQ C18 column (Agilent Technologies, United States) [73], including ginsenosides Rb1, Rb2, Rc, Rd, Re, Rg1, and F1 as well as notoginsenoside R1 [35, 73]. In brief, 0.2 g of plant tissue powder that had been dried was combined with 15 ml of pure ethanol. The mixture was homogenized using a vortex mixer and then sonicated for half an hour at 25 °C. After frozen at − 20 °C for one hour, the mixture was centrifugated at 10,000 × g for 10 min. The supernatant was collected and filtered through a membrane with a pore size of 0.22 μm. An Agilent C18 column was employed to separate various types of saponins. The column temperature was set at 35 °C and the flow rate was maintained at 1 ml min−1. Elution was performed using two mobile phases: acetonitrile (A) and ultrapure water (B). The elution process followed a gradient pattern as described below: at the beginning of 12 min, B concentration decreased by 81%; from 12 to 70 min, B concentration further reduced from 81 to 64%; from 71 to 76 min, B concentration increased back from 64 to 81%; finally, until completion at the 76-min mark, B concentration remained constant at 81%. The retention time and standard curve for each saponin were determined by analyzing their respective standard substances.
DNA extraction and amplicon sequencing
Genomic DNA was extracted from samples using the FastDNA SPIN Kit for Soil (MoBio Laboratories, Inc., USA). For ITS gene amplification, each 25 μl PCR reaction containing 12.5 μl Premix Taq DNA polymerase (Takara, China), 0.5 μl (200 nM) each primer, 2 μl template DNA (~ 5 ng μl−1), and 9.5 μl PCR-grade water. The PCR amplifications (performed in triplicate for each sample) were carried out using the following program: 2 min initial denaturation at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 55 °C, and 45 s at 72 °C, with a final 10-min elongation at 72 °C. Primer pair ITS1F/ITS2R was utilized for conducting polymerase chain reaction [32], followed by sequencing on the Illumina MiSeq PE300 platform (Shanghai Biozeron Co., Ltd., China). This primer pair can effectively avoid host contamination and has been widely used in plant-associated mycobiota profiling. Then, we completed the whole process of bioinformatics analysis independently as follows: Low-quality reads and adaptors in raw sequences were removed using the fastp software (default parameters) [10]. The meticulously cleaned data were subsequently processed through the cutting-edge cutadapt software, which meticulously excised primer sequences andused stringent parameters: “–errors 0.13 || –overlap 5” [44]. These refined sequences were then seamlessly imported into QIIME2 through the dedicated import plugin. The dada2 denoise-paired command was executed with precise parameters: “-p-trunc-len-f 220 || -p-trunc-len-r 220” to generate amplicon sequence variants (ASVs) and remove singletons and chimeras [7]. For taxonomy annotation, the QIIME2 platform’s classify-sklearn method was utilized, leveraging a Bayes-trained classifier based on the comprehensive UNITE database (version V16.10.2022) with its default settings [49]. Our dataset was meticulously pruned to exclude unclassified ASVs. Additionally, to ensure purity, the remaining ASVs were rigorously inspected for potential plant DNA contamination through blast analysis.
Data analysis
All samples were rarefied based on the lowest sequence depth 12,412 sequences for fungi to minimize the impact of read-count variation from the different samples. ASVs were classified as abundant or rare according to their relative abundances in each treatment. In the present research, we established relative abundance thresholds at 0.01% for infrequent taxa and 1% for frequent taxa, categorizing all ASVs into six distinct groups (AAT, CAT, MT, ART, CRT, CRAT) based on recent literature [39]. The specific categories are delineated as follows: (1) consistently frequent taxa (AAT) are those OTUs with a prevalence of 1% or more across all samples; (2) consistently infrequent taxa (ART) are those OTUs with a prevalence less than 0.01% across all samples; (3) moderately frequent taxa (MT) are those OTUs with a prevalence ranging between 0.01 and 1% across all samples; (4) conditionally infrequent taxa (CRT) are those OTUs with a prevalence below 1% overall but less than 0.01% in certain samples; (5) conditionally frequent taxa (CAT) are those taxa with a prevalence of 0.01% or more across all samples and 1% or more in some samples, but never less than 0.01%; and (6) conditionally infrequent and frequent taxa (CRAT) are those OTUs whose prevalence varies from rare (< 0.01%) to frequent (≥ 1%). For further analysis, we artificially grouped the AAT, CAT, and CRAT as abundant taxa. This amalgamation of the three categories (AAT, CAT, and CRAT) was termed “Abundant subcommunity” to prevent confusion. We also grouped the CRT and CRAT as “Rare subcommunities”. These thresholds for the definition of abundant and rare ASVs were specified in previous studies [11, 68, 74].
Fungal biodiversity
Alpha diversities of the fungal subcommunities were estimated by calculating the Richness and Shannon index using the “vegan” package in R [52]. The β-diversities of the fungal abundant and rare subcommunities were estimated based on the Bray–Curtis distance matric, which represents the taxonomic dissimilarities, and were calculated using the vegan package in R. To compare the variations between subcommunities, the β dispersion of both abundant and rare communities were also calculated using vegan [52]. The description of environmental factors on β-diversity based on Bray–Curtis was evaluated using a distanced-based linear model and forward selection procedure and shown using the Constrained Analysis of Principal Coordinates (CAP) since constrained analysis considers not only the response variables but also the explanatory variables during the dimensionality reduction process. Variables with strong collinearity (Spearman ρ2 > 0.9) were ignored before the evaluation. Principal Coordinate analysis (PCoA) was used to represent the community variation in subcommunities. The Pearson's correlation coefficients relationships of microbial diversities and phyla with environmental factors were assessed using Pearson correlation method.
Network analysis
Network analysis was used to depict the co-occurrence pattern within a complex community. The co-occurrence network analysis was carried out on RE and LE samples after combining the abundant and rare subcommunities. A valid co-occurrence was defined as a statistically significant correlation between ASVs, with a spearman’s correlation coefficient |r|> 0.6 and P < 0.01 [76]. The P values have been adjusted for multiple testing using the Benjamini-Hochberg’s FDR method. Topological properties of the network, including the graph density, shortest path length, and clustering coefficient, as well as properties of nodes, were calculated, such as the node degree, betweenness centrality, closeness centrality, and eigenvector centrality to infer the ability of each node to alter the abundance of other species and the structure of the community. Closeness centrality is a measure of the average shortest distance from each vertex to each other vertex [24]. Nodes with high closeness centrality values were identified as key species in co-occurrence networks (van der Heijden and Hartmann, 2016). Eigenvector centrality was also taken into consideration since it could take into account both the number of connections of each node and its importance in terms of influence within the network (Ruhnau, 2000). Statistical analysis and display of the networks were conducted with the “gephi 0.9.2” software [87]. The module is defined as a consortium comprising a group of microbes with relatively high levels of interaction, through which they can actively execute specific dominant functions [15]. The role of each node was determined based on its Zi degree (connection to other nodes within the same module) and Pi degree (connection to nodes in other modules) [23, 80]. Following the suggested thresholds for Zi and Pi degrees, nodes were classified into four subcategories: peripherals (Zi ≤ 2.5 and 0 ≤ Pi ≤ 0.62), connectors (Zi ≤ 2.5 and Pi > 0.62), module hubs (Zi > 2.5 and Pi ≤ 0.62), and network hubs(Zi > 2.5 and Pi > 0.62). Networks with more connectors than module hubs were regarded to have strong stability [17].
Assembly
There exist two crucial and complementary mechanisms that regulate the formation of microbial communities: deterministic mechanisms (niche-based theory), where environmental filtering and diverse biological interactions govern the patterns of microbial communities; and stochastic mechanisms (neutral theory), which highlight the significance of probabilistic dispersal and ecological drift [8, 13, 70]. To ensure the result of ecological patterns was robust, three models were conducted in community assembly: the phylogenetic model—iCAMP calculation, as well as a taxonomic model—the Sloan NCM, and the normalized stochasticity ratio (NST). Considering that the iCAMP calculation shows higher accuracy compared to those from the entire community-based approach, it was conducted using the iCAMP package in R to explore the assembly of subcommunities [52]. A total of five assembly mechanisms were identified according to the iCAMP, which can be classified into stochastic processes (e.g., dispersal limitation: DL, homogenizing dispersal: HD, and drift: DR) and deterministic processes (e.g., homogeneous selection: HoS and heterogeneous selection: HeS). To quantify the contributions of these ecological processes, the iCAMP first separated the observed taxa into various groups (i.e., bins) based on their phylogenetic relationships. Afterward, the process controlling each bin was identified according to the null model analysis of the phylogenetic diversity characterized by beta Net Relatedness Index (βNRI) and taxonomic β-diversities using modified Raup–Crick metric (RC). The high variability of fungal ITS would affect the accuracy of the beta nearest taxon index (βNTI) by preventing the alignment of distant taxa, but not affecting the accuracy of βNRI [22]. The quantification of the contributions of ecological processes includes two steps: (i) Taxa were classified into different groups (called bins according to their phylogenetic relationships, (ii) The null model analysis of the phylogenetic diversity marked by beta Net Relatedness Index (βNRI) and taxonomic β-diversities with modified Raup–Crick metric (RC) was used to identify the assembly process of each bin. In detail, the relative contribution of HoS process and HeS process was categorized according to βNRI < − 1.96 and βNRI > 1.96, separately. For the left pairwise comparisons, pairs |βNRI|≤ 1.96 and RC < 0.95 were classified into HD process, and those with βNRI ≤ 1.96 and RC > 0.95 were defined as DL process. Finally, the remaining pairs with βNRI ≤ 1.96 and RC ≤ 0.95 were classified into the DR process.
The Sloan NCM was subsequently used for subcommunities to identify the relative contribution of stochastic processes to the assembly with the proportion of predicted relationships between ASVs in the local community. The NST was further applied to evaluate the microbial community assembly [51]. The relative importance of deterministic and stochastic processes can be quantified by the index NST with 50% as the boundary point between more deterministic (NST < 50%) and more stochastic (NST > 50%). Moreover, the normalized stochasticity ratio (NST) was employed to assess the assembly of microbial communities. This index (NST) could effectively quantify the relative contribution of deterministic and stochastic processes, with 50% serving as a critical threshold. Values below 50% indicate a greater deterministic process, whereas those exceeding 50% suggest a more stochastic process in the community assembly.
Functional prediction
The putative function of the fungal ASVs was estimated by FUNGuild [48], a database linking the fungal community with function at the ecological guild level. Main trophic types and guilds were classified by FUNGuild. Only highly probable and probable guilds with identified trophic modes were used in further analysis.
Results
Different taxonomic and functional compositions between rare and abundant subcommunities
A total of 1239 (PG), 954 (PQ), and 1528 (PN) fungal ASVs were detected in the RE, and 1666 (PG), 2946 (PQ), 2555 (PN) fungal ASVs were detected in the LE, respectively, and there were more rare ASVs than abundant ASVs detected in all the three Panax species (Table 1). On the local level, the relative abundance of abundant fungal ASVs ranges from 90.71 to 95.99% for all samples of PG and PQ, 14 folds higher than the abundance of rare ASVs (4.01–9.29%) (Table S2). For PN, the abundance of rare ASVs in the RE and LE were 45.22% and 45.68%, respectively (Table S2). However, the abundant ASVs in the RE and LE of PN accounted for only 54.78% and 8.87% of the whole communities, much lower than those in PG and PQ (Table S2). The richness index showed that in the RE, the number of rare ASVs was only a few more than abundant ASVs (6.67–66.26% higher), while in the LE, the ratios of rare/abundant ASV numbers were larger than 2 folds (Table S3).
Table 1.
Fungal ASVs detected in abundant and rare subcommunities in the leaf endophyte (LE) and root endophyte (RE) in the cultivated P. ginseng (PG), P. quinquefolium (PQ) and P. notoginseng (PN)
| PG | PQ | PN | ||
|---|---|---|---|---|
| RE | Abundant | 98 | 74 | 92 |
| Rare | 1141 | 880 | 1436 | |
| Total | 1239 | 954 | 1528 | |
| LE | Abundant | 31 | 46 | 20 |
| Rare | 1635 | 2900 | 2535 | |
| Total | 1666 | 2946 | 2555 |
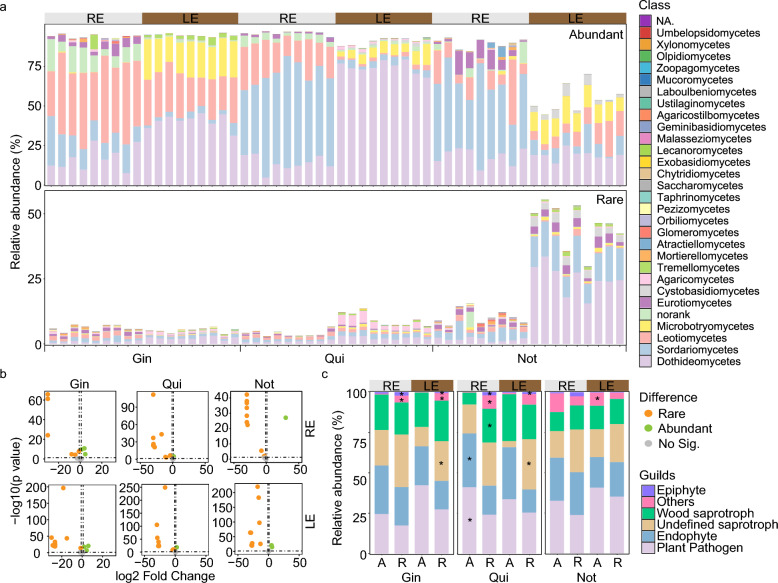
For the community composition, Leotiomycetes and Sordariomycetes were more abundant in the RE than the LE (significant within the PQ and PN species), while more depleted of Dothideomycetes and Themellomycetes (Fig. 1a, b; Table S4; P < 0.05). Differential analysis showed that relative abundances of ASVs in rare and abundant subcommunities differed a lot on class level (Fig. 1a, b; Table S4). In the LE, Agaricomycetes, Eurotiomycetes and Sordariomycetes were more abundant in rare than in abundant subcommunities, while Microbotryomycetes and Dothideomycetes were the opposite. In the RE, Agaricomycetes, Eurotiomycetes and Mortierellomycetes were more abundant in the rare subcommunities (Fig. 1a, b; Table S5). According to the composition of guild abundance, the trophic types and functional potential were similar in the LE and the RE (Fig. 1c). Differential analysis indicated that the relative abundance of undefined saprotrophs was higher in the rare than the abundant counterparts, and the difference was significant in the LE of PG and PQ (Fig. 1c; P < 0.05). Potential plant pathogens were more dominant in abundant than the rare counterparts in all groups, though it was only significant in the RE of PQ (Fig. 1c; P < 0.05). Based on the top 5 abundant species in each subcommunity, some species were found to be dominant generally in specific habitats. For example, in the RE, Plectosphaerella_cucumerina and Leptosphaeria_sp were dominant in the abundant subcommunity, while Exophiala_sp._Ppf18 was abundant in the rare subcommunity (Fig. S1). In the LE, Rhodotorula_glutinis and Sporidiobolus_sp. were dominant in abundant and rare subcommunities, separately (Fig. S1). These results indicated that the abundant and rare subcommunities exhibited significant differences in terms of both community compositions and trophic types.
Fig. 1.
Taxonomic composition and functional guilds of abundant and rare fungal taxa in the leaf endophyte (LE) and root endophyte (RE) of the cultivated Panax ginseng (PG), P. quinquefolium (PQ) and P. notoginseng (PN). a Relative abundance of fungi on class level in abundant and rare subcommunities. b Enrichment of classes in abundant and rare subcommunities based on the DeSeq2 analysis (|log2FoldChange|> 1 and P < 0.05). c Richness of guilds in abundant and rare subcommunities and their enrichment in abundant and rare subcommunities based on the DeSeq2 analysis (|log2FoldChange|> 1 and P < 0.05). Significant differential guilds were marked with *
Rare taxa drive fungal co-occurrence patterns in Panax species
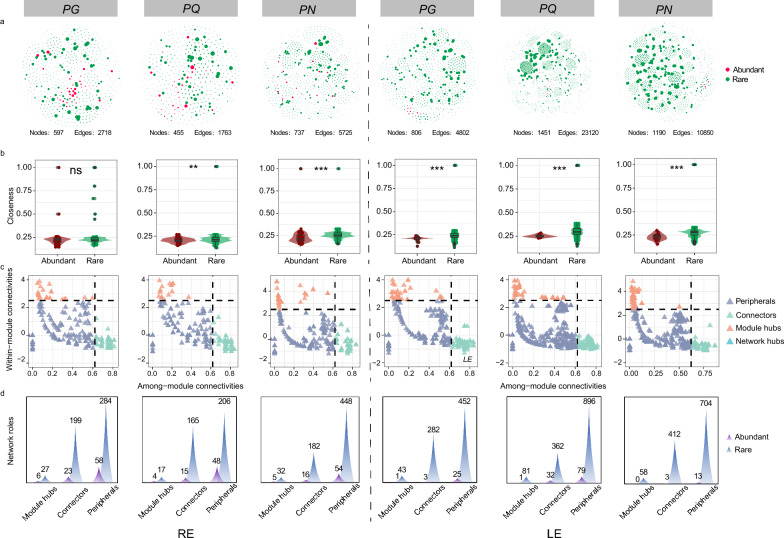
The networks for mycobiotas in the RE and LE of three Panax species were individually constructed and their topological properties were examined (Fig. 2). The network sizes showed that the LE networks (nodes: 806–1451; edges:4802–23,120) were larger than those in the RE (nodes: 455–737; edges:1763–5725) with more nodes and edges, and the occurrence of rare species was larger than abundant species, especially in the LE (Fig. 2a, b; Table S6). The closeness and eigenvector of rare species were larger than those of abundant species in all networks except for the RE network of PG (P < 0.001) (Fig. S2), which should be caused by taxa that occur in few samples. Based on the connectivity within and among modules (Zi and Pi, respectively), nodes can be categorized into four topological roles: peripherals, connectors, module hubs, and network hubs (Fig. 2c, d). No network hubs were observed in any of the networks in the RE and LE. The majority of nodes (61.31%) belonged to the peripheral category in all networks followed by connectors (33.50%), while only 5.19% of nodes belonged to module hubs (Fig. 2c, d; Table S7). The proportion of abundant species classified as peripherals exceeded those categorized as connectors and module hubs in the LE. Furthermore, The large proportion of connectors compared to module hubs indicates that these networks were strongly stable. It was also found that Sordariomycetes and Dothideomycetes were dominant in connectors and module hubs in all networks, most of which belonged to rare species (Table S8). To emphasize the interaction between key taxa, we conducted an overall correlation analysis and the results showed that rare key taxa were more tightly linked to others compared to abundant taxa, and a majority of them were negative (Fig. S3; Table S9). Overall, rare taxa played an important role in maintaining the stability of networks, particularly in the LE.
Fig. 2.
The co-occurrence networks and topological characteristics of abundant and rare subcommunities in the RE and LE of three Panax species. a The co-occurrence networks of abundant and rare subcommunities. Nodes belonging to abundant and rare subcommunities in networks were represented with red and green colors, respectively. b The clossness of abundant and rare taxa within the network, the outliers above of some rare taxa was because they were found in more samples than others. Wilcoxon rank sum test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001. c Among-module (Pi) and within-module (Zi) of taxa within the networks. d numbers of abundant taxa and rare belonging to different subcategories
Rare and abundant subcommunities respond similarly to environmental changes
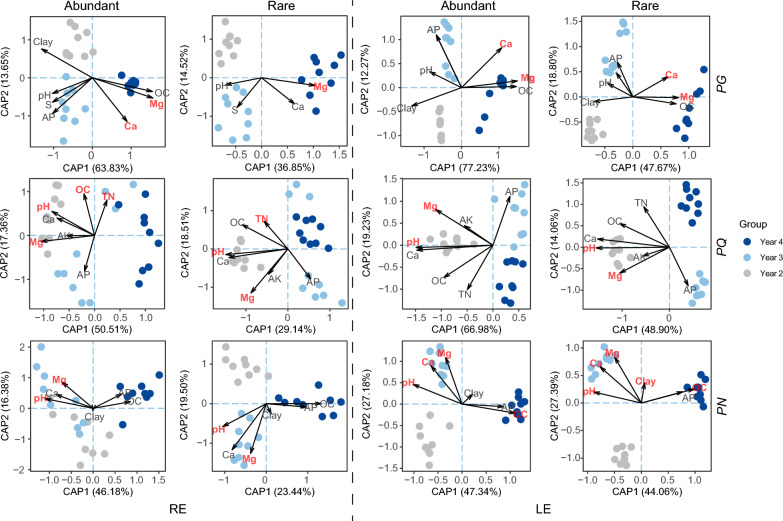
The Bray–Curtis distances were calculated to represent the community variations of abundant and rare subcommunities. According to the β dispersion, the rare subcommunities exhibited more obvious variation compared to the abundant counterparts in all habitats (P < 0.001) except the RE in PN (Fig. S4; Table S10). CAP analysis was used to depict the contributions of soil properties to the variations of fungal subcommunities (Fig. 3; Table S11). According to the CAP analysis, abundant and rare subcommunities showed similar responses to environmental changes, and Mg was an important factor in both abundant (explained variation: 8.72–49.71%) and rare (explained variation: 4.62–21.08%) subcommunities (Table S10). For the PG species, Ca is the second unignorable explanatory factor, while for PQ and PN species, pH, OC, TN, as well as Clay, were additional key factors for explaining the subcommunities (Table S10). The results revealed that responses of rare and abundant subcommunities to the indirect influence of physicochemical properties in the soil were parallel.
Fig. 3.
CAP of the abundant and rare subcommunities based on Bray–Curtis. Arrows represent important variables identified by linear models and forward selection. Points in coordinate spaces represent samples, and point color represented different sampling fields cultivated with plant with different ages. Significant explanatory factors (P < 0.05) are displayed in red font
Assembly processes of rare and abundant subcommunities
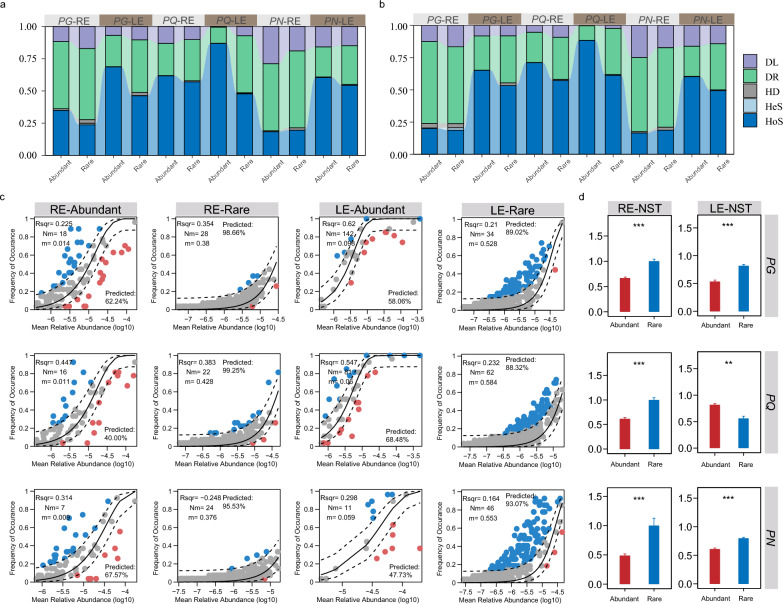
The ecological assembly mechanisms of different fungal subcommunities were analyzed based on the iCAMP framework, as well as the null model and the neutral model in each compartment. The relative importance of homogeneous selection in the assembly of rare subcommunities was weaker compared to the abundant counterparts (Fig. 4a), which was similar to differences between ecological processes of plant pathogens in abundant and rare subcommunities, and the differences were more pronounced in the LE than in the RE (Fig. 4b). Similar difference, however, was not detected in ecological processes of undefined saprotrophs within abundant and rare subcommunities (Fig. S5). Moreover, proportions of assembly explained by homogenizing selection in LE were higher compared to RE within each plant species. The neutral model fitting results showed that the relative abundances and occupancy of both abundant and rare fungal taxa exhibited positive correlations, indicating the important role of neutral processes (Fig. 4c). Based on the ratio of ASVs within the model predictions, the results indicated that stochastic processes played a more prominent role in shaping rare subcommunities in contrast to their influence on abundant counterparts. The NST indexes of rare subcommunities in the RE were significantly higher than abundant counterparts, indicating that the assembly of rare subcommunities was more dominated by stochastic processes than abundant subcommunities in this habitat (Fig. 4d). In the LE, the deterministic effects in the assembly of abundant subcommunities were also larger compared to the rare counterparts. In summary, these results suggested the important role of stochastic processes in the assembly of rare and abundant subcommunities.
Fig. 4.
The quantification of different ecological processes on the assembly of abundant and rare subcommunities. a The relative importance of different ecological processes based on the iCAMP framework of abundant and rare subcommunities. b The relative importance of different ecological processes based on the iCAMP framework of abundant and rare plant pathogens.. Dispersal limitation: DL, homogenizing dispersal: HD, drift: DR, homogeneous selection: HoS, heterogeneous selection: HeS. The neutral community model (NCM) of fungi in the abundant and rare subcommunities of RE and LE and the normalized stochasticity ratio (NST) index of fungi in the abundant and rare subcommunities were displayed in (c) and (d). Wilcoxon rank sum test. *, P < 0.05; **, P < 0.01; ***, P < 0.001
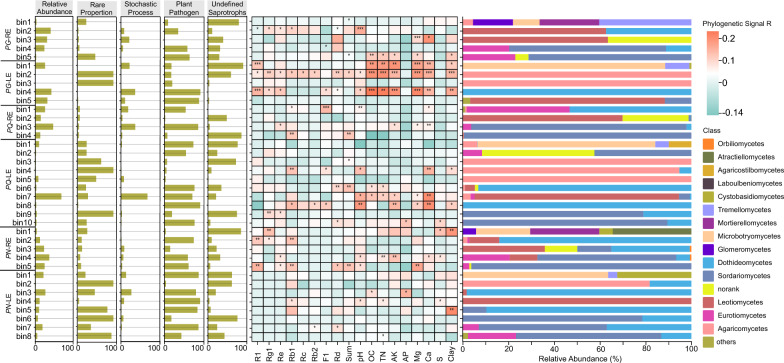
Considering that various phylogenetic groups of fungi differ greatly in their responses to environmental variations, the assembly mechanisms, relative abundance and proportion of abundant species, plant pathogens and undefined saprotrophs, as well as community compositions of the identified 37 phylogenetic bins were quantified as well, and the results are shown in Fig. 5. For most phylogenetic bins clustered by iCAMP, the proportion of abundant species was larger than rare species, and the ecological processes were dominated by a deterministic assembly process. Besides, plant pathogens and undefined saprotrophs were abundant in the bins, which are widely present across the fungi kingdom. The relative abundance of them was found to be higher than 50% in 20 and 11 bins, separately. Bins in the LE of PG and PQ were more sensitive to environmental changes than in the RE, which was also more closely linked to the concentrations of saponin, especially those mainly belonging to Agaricomycetes, Leotiomycetes, and Sordariomycetes.
Fig. 5.
Assembly mechanisms across different phylogenetic groups (bins), and their links to saponin accumulation and environmental factors. The relative abundance, relative importance of stochastic process, as well as relative abundances of abundant species, plant pathogens and undefined saprothophs were displayed with barplot. Phylogenetic signals for each groups were showed using heatmap. Community compositions of each groups were also displayed in our figure. Wilcoxon rank sum test. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Effects of fungal indicators on saponin concentrations
Pearson’s correlation analysis was conducted to explore the interactions between fungal subcommunities and saponins (Fig. S6). Our results showed that only a few relationships were exhibited between saponins and the properties of communities in the RE, especially rare subcommunities. However, more correlations were detected in the LE. For the PG species, some saponins (eg., R1 and Re) showed highly significant positive relationships with Shannon diversities and Richness of rare and abundant subcommunities, while other saponins (F1 and Rd) showed negative relationships. The effects of Shannon diversities of abundant and rare subcommunities on saponins were the opposite within the PQ species. For example, the diversity of abundant subcommunity was negatively linked to Rg1 and positively linked to Rb1, while the rare subcommunity was the opposite. Unlike the PG and PQ species, nearly no saponins in the LE of PN were associated with the diversities of abundant and rare subcommunities. Overall, rare subcommunities were more tightly linked to the accumulation of saponins in all three Panax species compared to the abundant counterparts, especially in the LE.
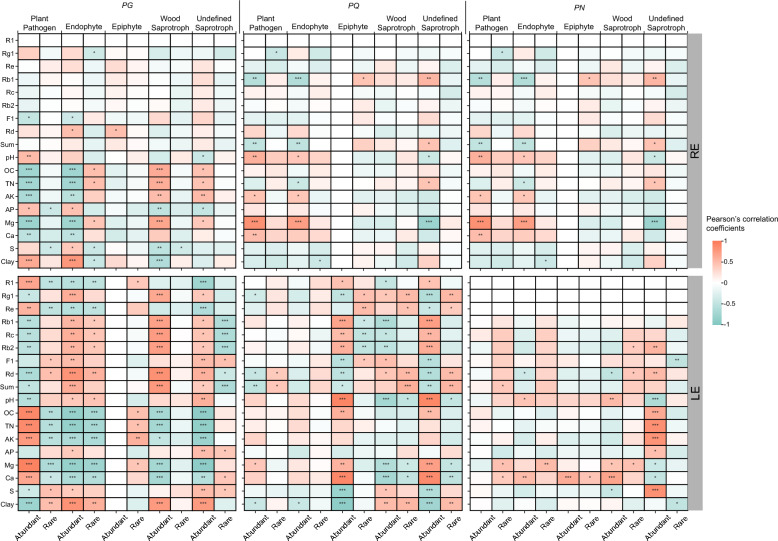
By relating abundant and rare taxa on class level and functional guilds to saponin contents and soil properties, we explored potential microbiological mechanisms of saponin production influenced by the environment (Figs. 6, S7). Significant correlations between saponin contents and fungal taxa, as well as functional guilds, were much more in the LE compared to the RE. For the LE, relative abundances of Agaricomycetes, Eurotiomycetes and Microbotryomycetes in rare subcommunities were sensitive to environmental factors and were tightly associated with saponin contents. Differently, in the RE, rare taxa showed few associations with saponin contents, though some of them (Agaricomycetes, Dothideomycetes, Eurotiomycetes, and Leotiomycetes) were strongly influenced by soil properties. The plant-associated guilds (plant pathogens and endophytes) and undefined saprotrophs were linked to saponin accumulation in abundant but not rare subcommunities. We also observed that the majority of positive relationships were evident in undefined saprotrophs (PG: 29.17%; PQ: 46.15%; PN: 100%) (Fig. 6). In summary, our results indicated the tight relationships between saponin accumulation and specific rare and abundant taxa.
Fig. 6.
Relationships between functional guilds predicted by FUNGuild and environmental variables and saponin contents in each subcommunity based on Pearson correlation analysis. Statistical p-value. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Discussion
Understanding the factors that shape the taxonomic and functional diversity of abundant and rare fungal subcommunities in different tissues of Panax species is crucial for predicting and managing fungal influences on plant growth and saponin biosynthesis. In this study, we explored the diversities, functional traits, co-occurrence structures, and ecological assembly patterns of both abundant and rare fungal subcommunities. Our findings reveal several key insights: (i) abundant and rare subcommunities exhibit distinct community compositions and guilds, with more pronounced differences in the leaf endosphere (LE); (ii) rare species assume more critical roles within the co-occurrence networks than abundant species, highlighting their importance in maintaining community stability; (iii) stochastic processes have a more significant impact on the assembly of the rare subcommunities compared to the abundant counterparts, and (iv) the microbial diversities of rare subcommunities are closely associated with variations in saponin concentrations.
Variations in community composition and functional guilds
Fungal communities have been shown to inhabit various tissues of terrestrial plants and display tissue-specific distribution patterns. Despite the dominance of a limited subset of endophytic fungi across different systems [57], the vast majority of uncultured fungi could be detected with advances advancements in next-generation sequencing (NGS) technologies [50]. Prior studies in diverse ecosystems such as rivers, grasslands, and agricultural areas have demonstrated that abundant and rare fungal taxa often exhibit distinct distribution patterns and functional traits [32, 84]. However, few studies have compared the distribution patterns of abundant and rare endophytic subcommunities. It has been observed that the α-diversity of endophytic fungal communities in plants experiences a reduction compared to the external environment due to host selection pressures such as host immune and plant exudates, with this selection being more pronounced in leaves than in roots [76]. We observed that the Shannon and Richness indices for rare subcommunities in the leaf endosphere (LE) were higher than those in the root endosphere (RE), contrary to indices in abundant subcommunities, suggesting that rare subcommunities are less responsive to host selection than abundant ones [77]. Another reason for the higher diversity in the LE might be that fungi can potentially spread much further and faster in the air as spores than through the soil as mycelium [62].
The evolutionary symbiosis of dominant taxa with the host can have significant ecological impacts, yet understanding the role of rare species in ecosystems remains challenging. In our study, abundant and rare fungal subcommunities exhibited substantial differences in taxonomic and functional compositions. For instance, abundant taxa in the leaf endophyte predominantly belong to Microbotryomycetes, which are generally identified as plant pathogens [47], whereas rare taxa from Agaricomycetes and Mortierellomycetes demonstrated a saprobic lifestyle, coinciding with an enrichment of undefined saprotrophs in rare subcommunities. Moreover, we found that abundant subcommunity dominant species Plectosphaerella_cucumerina, Leptosphaeria_sp and Rhodotorula_glutinis were specific plant pathogens, while rare subcommunity dominant species Exophiala_sp._Ppf18 and Sporidiobolus_sp. were specific undifined saprotrophs. Therefore, it is clear that differences in the taxonomic and functional composition of abundant and rare subcommunities are consistent across various Panax species. However, fungal function (and microbial function in general) is highly context-dependent and FunGUILD, further studies are demanded to classify functional fungi by using metagenomics and sequence similarity analysis and metabolomics. Constrained analysis of principal coordinates (CAP) demonstrated that magnesium (Mg) was a key factor influencing all subcommunities. Previous research identified Mg as a crucial role in driving the root fungal community composition in foxtail millet (Setaria italica) [33]. Mg is also involved in several vital plant functions, such as chlorophyll synthesis, enzyme activity, and root development, which may be closely linked to the endophytic fungal community composition of Panax species [67]. In return, the endophytic fungal community might enhance Mg acquisition, thereby promoting plant growth [6].
More central topological role of rare taxa in co-occurrence networks
There is growing evidence that the characteristics of ecological networks, which depict the potential interrelationships among coexisting organisms, can significantly influence community responses to environmental changes [17, 64]. In our study, we discovered that nodes associated with rare subcommunities based on covariation in relative abundances possessed greater closeness and eigenvector centrality than those of abundant amplicon sequence variants (ASVs), indicating that these rare taxa are connected to a large number of nodes and could be considered hubs or mediators within ecological niches [83]. This suggests that the presence of rare species might be crucial for maintaining the stability of the fungal network in plants. However, further experimental studies were demanded to verification the key role of rare species. Typically, networks characterized by fewer module hubs and more connectors are better equipped to resist environmental disturbances and thus maintain community stability [17]. In all examined networks, the proportion of module hubs was significantly smaller than that of connectors, and most module hubs were represented by rare ASVs, suggesting that the network structure within the root endosphere (RE) and leaf endosphere (LE) driven by rare subcommunities is not hub-based.
The positive impact of the rare biosphere on community stability can be attributed to its ability to finely balance host growth and defense mechanisms [34]. Additionally, we observed a decrease in connectors and an increase in module hubs in the RE compared to the LE, implying lower ecological stability of fungal communities in the RE. This may be driven by the more diverse resource availability and broader niches in the root environment for microorganisms [16, 72]. We also found that nodes belonging to the Dothideomycetes and Sordariomycetes were predominant in both module hubs and connectors, corroborating findings from a previous study [77]. Furthermore, the overall network of key species showed that ASV1, an abundant key taxon belonging to plant pathogenic Cladosporium_sp., was associated with many rare taxa, indicating the complex interplay between dominant and understudied species in shaping the ecosystem dynamics. These groups exhibit a high functional diversity, including roles as plant pathogens, endophytes, and epiphytes [30, 43]. These results further underscore the importance of rare taxa in plant mycobiota, particularly regarding their significant contributions to the ecological networks associated with plants.
Dominant host selection in shaping the abundant fungal subcommunities
In our research, abundant subcommunities were predominantly shaped by deterministic assembly processes, as evidenced by the application of Sloan’s Neutral Community Model (NCM), Neutral Species Turnover (NST) index, and the iCMAP framework. These findings align with previous research in reservoirs and oil-contaminated soils, which demonstrated that stochastic processes predominantly influence the assembly of rare communities [19, 31, 75]. Ecological drift has been identified to be positively correlated with microbial richness, particularly when community sizes are small since a larger species pool relative to the local community size increases the likelihood of species extinction [19]. Therefore, the higher richness of the rare biosphere compared to the abundant biosphere could explain the greater influence of ecological drift on rare subcommunity assembly.
Additionally, our results suggested that stochastic processes play a larger role in shaping rare plant pathogenic communities compared to abundant communities. This distinction between the abundant and rare biospheres, alongside overall community dynamics, indicates a significant participation of plant pathogens in community assembly (eg., Rhodotorula_glutinis and Botrytis_cinerea). The prominent position of pathogens in our findings could be attributed to two factors: balanced antagonism and saponin selection. According to the balanced antagonism hypothesis, fungal pathogens can thrive within the plant by evading the host's defense mechanisms without inducing symptoms [61], allowing for their asymptomatic coexistence with the host, as observed in our study [54]. Furthermore, as primary secondary metabolites in Panax tissues, saponins may influence the endophytic fungal community by disrupting the cell membranes of certain fungi and promoting the growth of others by providing carbon sources [40, 45]. Pathogens have been shown to degrade saponins into less toxic products, thereby avoiding cell membrane damage, as evidenced in studies on wheat, oats, and the P. notoginseng rhizosphere [40, 53]. Although iCAMP performed well in predicting the ecological processes, it still has limitations in dealing with the ‘drift’ part for the the lack of consideration for diversification. This is particularly important for rare species, as their distribution may be more heavily influenced by stochastic processes [52].
Potential microbial drivers of saponin accumulation
Saponins may act as chemical defenses against fungal infections at high concentrations, and could be induced by pathogen invasion attempts, serving as a feedback mechanism [58]. Thus, endophytic fungi can influence saponin composition, potentially promoting the rapid evolution of saponins in the ongoing conflict between plants and fungi [3, 18, 81]. For P. ginseng and P. quinquefolium, links between microbial diversity and saponin concentrations were more pronounced in the leaf endosphere (LE) than in the root endosphere (RE), highlighting a substantial contribution of fungal subcommunities in the LE. Microbial α-diversity may influence saponin concentrations by affecting the functional diversity of the microbiome [20]. In contrast, only a few correlations were observed in P. notoginseng, which could be attributed to the distinct evolutionary symbioses between endophytic taxa and the host across different Panax species, noting that PG and PQ are closely related allotetraploid species [65, 81]. Another possible explanation is the environmental adaptation of these species; PN primarily inhabits the warmer mountain areas of Southwest China, while PG and PQ are found in the colder winter environments of Northeast Asia and North America [81]. Moreover, diversities within rare subcommunities were linked to saponin accumulation in the LE of all examined Panax species, suggesting that the rare biosphere might play a more significant role in saponin accumulation compared to the abundant biosphere. Earlier studies have also highlighted that rare taxa contribute importantly to plant protection and productivity by sustaining ecological multifunctionality [12, 28, 29, 77].
Our findings also underscore the vital roles of rare subcommunity-enriched taxa (Agaricomycetes, Eurotiomycetes, Sordariomycetes, and Mortierellomycetes) and their saprobic lifestyle in saponin accumulation. We propose that undefined saprotrophs might contribute to the form of saponins, and saponin production could also benefit for the survival of undefined saprotrophs in reverse in the Panax genus. Previous research has shown that saprophytes have a strong capacity to utilize small molecular compounds and produce diverse secondary metabolites [14], which presents significant prospects for saponin production [2]. Agaricomycetes and Mortierellomycetes were beneficial for plant growth by enhancing nutrient uptake or suppressing soil-borne pathogens [27, 45]. Although most positive links between undefined saprotrophs and saponin accumulation were observed in the abundant biosphere, our results demonstrated that undefined saprotrophs in the rare biosphere were more diverse and phylogenetic bins consisting of these rare undefined saprotrophs were positively correlated with saponin variation. From this, we conclude that the rare biosphere may influence saponin accumulation as a “regional species pool” for taxa, such as undefined saprotrophs, to enhance saponin production. Our results align with the role of rare communities in sustaining microbiome function, as they are presumed to be highly resistant to stress and functionally redundant [38, 85]. Overall, this study demonstrates the crucial role of rare species in saponin generation from both taxonomic and functional perspectives.
Conclusion
The taxonomic and functional compositions, co-occurrence patterns, and ecological assembly processes of fungal subcommunities exhibit distinct differences between abundant and rare taxa in the root and leaf tissues of three cultivated Panax species. Undefined saprotrophs, which were predominantly enriched in rare subcommunities, exhibited a strong association with the accumulation of saponins. Furthermore, positive association-dominated inter-species interactions were predominantly maintained by rare species, which might lead to no hub-based patterns, particularly noticeable in the leaf endosphere (LE). The assembly of rare subcommunities could be more influenced by stochastic processes compared to abundant subcommunities for its ability to coexist with saponins effectively. This study provides insights into the differential distributions between the abundant and rare biospheres of fungi within plants, emphasizing the potentially essential role of rare species in sustaining ecological stability and enhancing saponin accumulation. Additionally, our findings offer valuable directions for future experimental studies to unravel the life strategies of endophytic fungi within Panax species. In the future, SynComs and metagenomics methods could be used to further validate our viewpoint.
Supplementary Information
Acknowledgements
We thank the Shanghai Biozeron Co., Ltd. for their help in high-throughput sequencing.
Author contributions
LLD, GZZ and YL developed the concept. YL analysed the data and wrote the manuscript. LPS, FH, GFW, ZZJ, XYW and JJP collected the data and discussed the content. LPS, FH and JJP revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Key Research and Development Program (2022YFC3501802, 2022YFC3501803, 2022YFC3501804), Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences (CI2023E002), Specific research fund of The Innovation Platform for Academicians of Hainan Province (YSPTZX202137), National Natural Science Foundation of China (82304663), and Fundamental Research Funds for the Central Public Welfare Research Institutes (ZXKT22001, ZXKT22052, ZXKT22060, ZXKT23018, ZZ13-YQ-049, ZZ16-XRZ-072, ZZ17-YQ-025).
Data availability
The datasets generated and/or analyzed during the current study are available in the National Genomics Data Center under the BioProject PRJCA007643. R codes used for statistical analyses are available at “https://github.com/githubzgz”. No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guozhuang Zhang, Email: gzzhang@icmm.ac.cn.
Linlin Dong, Email: lldong@icmm.ac.cn.
References
- 1.Afkhami ME, Strauss SY. Native fungal endophytes suppress an exotic dominant and increase plant diversity over small and large spatial scales. Ecology. 2016;97:1159–69. [DOI] [PubMed] [Google Scholar]
- 2.An C, Ma S, Shi X, Liu C, Ding H, Xue W. Diversity and ginsenoside biotransformation potential of cultivable endophytic fungi associated with Panax bipinnatifidus var. bipinnatifidus in Qinling Mountains, China. Front Pharmacol. 2022;13:762862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Augustin JM, Kuzina V, Andersen SB, Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72:435–57. [DOI] [PubMed] [Google Scholar]
- 4.Bahram M, Netherway T, Hildebrand F, Pritsch K, Drenkhan R, Loit K, Anslan S, Bork P, Tedersoo L. Plant nutrient-acquisition strategies drive topsoil microbiome structure and function. New Phytol. 2020;227:1189–99. [DOI] [PubMed] [Google Scholar]
- 5.Barberan A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron NC, Rigobelo EC. Endophytic fungi: a tool for plant growth promotion and sustainable agriculture. Mycol Int J Fungal Biol. 2022;13:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory CJ. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chave J. Neutral theory and community ecology. Ecol Lett. 2004;7:241–53. [Google Scholar]
- 9.Chen Q, Ding J, Zhu D, Hu H, Delgado-Baquerizo M, Ma Y, He J, Zhu Y. Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol Biochem. 2020;141:107686. [Google Scholar]
- 10.Chen S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta. 2023;2:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Ren K, Isabwe A, Chen H, Liu M, Yang J. Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome. 2019;7:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Nomura K, Wang X, Sohrabi R, Xu J, Yao L, Paasch BC, Ma L, Kremer J, Cheng Y, Zhang L, Wang N, Wang E, Xin X, He SY. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature. 2020;580:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–66. [Google Scholar]
- 14.Crowther TW, Boddy L, Jones TH. Functional and ecological consequences of saprotrophic fungus-grazer interactions. ISME J. 2012;6:1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Ye, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinform. 2012;13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S. Microbial interkingdom interactions in roots promote arabidopsis survival. Cell. 2018;175:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan K, Weisenhorn P, Gilbert JA, Chu H. Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol Biochem. 2018;125:251–60. [Google Scholar]
- 18.Frantzeskakis L, Di Pietro A, Rep M, Schirawski J, Wu C, Panstruga R. Rapid evolution in plant-microbe interactions—a molecular genomics perspective. New Phytol. 2020;225:1134–42. [DOI] [PubMed] [Google Scholar]
- 19.Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Singan V, Vogel J, Hutmacher RB, Dahlberg JA, Coleman-Derr D, Lemaux PG, Taylor JW. Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nat Commun. 2020;11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Garcia N, Tamames J, Linz AM, Pedros-Alio C, Puente-Sanchez F. Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. ISME J. 2019;13:2969–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorischek AM, Afkhami ME, Seifert EK, Rudgers JA. Fungal symbionts as manipulators of plant reproductive biology. Am Nat. 2013;181:562–70. [DOI] [PubMed] [Google Scholar]
- 22.Goss-Souza D, Tsai SM, Rodrigues JLM, Klauberg-Filho O, Sousa JP, Baretta D, Mendes LW. Biogeographic responses and niche occupancy of microbial communities following long-term land-use change. Antonie Van Leeuwenhoek. 2022;115:1129–50. [DOI] [PubMed] [Google Scholar]
- 23.Guimera R, Amaral L. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen D, Shneiderman B, Smith M. Calculating and visualizing network metrics. Amsterdam: Elsevier; 2011. p. 69–78. [Google Scholar]
- 25.Harrison JG, Beltran LP, Buerkle CA, Cook D, Gardner DR, Parchman TL, Poulson SR, Forister ML. A suite of rare microbes interacts with a dominant, heritable, fungal endophyte to influence plant trait expression. ISME J. 2021;15:2763–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Z, Liu D, Shi Y, Wu X, Dai Y, Shang Y, Peng J, Cui Z. Broader environmental adaptation of rare rather than abundant bacteria in reforestation succession soil. Sci Total Environ. 2022;828:154364. [DOI] [PubMed] [Google Scholar]
- 27.Hibbett DS, Bauer R, Binder M, Giachini AJ, Hosaka K, Justo A, Larsson E, Larsson KH, Lawrey JD, Miettinen O, Nagy LG, Nilsson RH, Weiss M, Thorn RG. 14 Agaricomycetes. In: McLaughlin DJ, Spatafora JW, editors. Systematics and evolution: part A. Berlin: Springer; 2014. p. 373–429. [Google Scholar]
- 28.Hol WHG, de Boer W, de Hollander M, Kuramae EE, Meisner A, van der Putten WH. Context dependency and saturating effects of loss of rare soil microbes on plant productivity. Front Plant Sci. 2015;6:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hol WHG, de Boer W, Termorshuizen AJ, Meyer KM, Schneider JHM, van Dam NM, van Veen JA, van der Putten WH. Reduction of rare soil microbes modifies plant-herbivore interactions. Ecol Lett. 2010;13:292–301. [DOI] [PubMed] [Google Scholar]
- 30.Hyde KD, Jones EBG, Liu J, Ariyawansa H, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai D, Diederich P, Dissanayake A, Doilom M, Doveri F, Hongsanan S, Jayawardena R, Lawrey JD, Li Y, Liu Y, Lucking R, Monkai J, Muggia L, Nelsen MP, Pang K, Phookamsak R, Senanayake IC, Shearer CA, Suetrong S, Tanaka K, Thambugala KM, Wijayawardene NN, Wikee S, Wu H, Zhang Y, Aguirre-Hudson B, Alias SA, Aptroot A, Bahkali AH, Bezerra JL, Bhat DJ, Camporesi E, Chukeatirote E, Gueidan C, Hawksworth DL, Hirayama K, De Hoog S, Kang J, Knudsen K, Li W, Li X, Liu Z, Mapook A, McKenzie EHC, Miller AN, Mortimer PE, Phillips AJL, Raja HA, Scheuer C, Schumm F, Taylor JE, Tian Q, Tibpromma S, Wanasinghe DN, Wang Y, Xu J, Yacharoen S, Yan J, Zhang M. Families of dothideomycetes. Fungal Divers. 2013;63:1–313. [Google Scholar]
- 31.Jiao S, Chen W, Wei G. Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol Ecol. 2017;26:5305–17. [DOI] [PubMed] [Google Scholar]
- 32.Jiao S, Lu Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob Change Biol. 2020;26:4506–20. [DOI] [PubMed] [Google Scholar]
- 33.Jin T, Wang Y, Huang Y, Xu J, Zhang P, Wang N, Liu X, Chu H, Liu G, Jiang H, Li Y, Xu J, Kristiansen K, Xiao L, Zhang Y, Zhang G, Du G, Zhang H, Zou H, Zhang H, Jie Z, Liang S, Jia H, Wan J, Lin D, Li J, Fan G, Yang H, Wang J, Bai Y, Xu X. Taxonomic structure and functional association of foxtail millet root microbiome (vol 6, pg 1, 2017). GigaScience. 2018;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Kuesel K, Rillig MC, Rivett DW, Salles JF, van der Heijden MGA, Youssef NH, Zhang X, Wei Z, Hol WHG. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D. Chemical diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng. J Ginseng Res. 2012;36:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Kim C, Lee Y. The single-seed microbiota reveals rare taxa-associated community robustness. Phytobiomes J. 2023;7:324–38. [Google Scholar]
- 37.Kim JH, Yi Y, Kim M, Cho JY. Role of ginsenosides, the main active components of Panax ginseng, in inflammatory responses and diseases. J Ginseng Res. 2017;41:435–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Yang J, Yu Z, Wilkinson DM. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 2015;9:2068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo L, Yang L, Yan Z, Jiang B, Li S, Huang H, Liu Y, Zhu S, Yang M. Ginsenosides in root exudates of Panax notoginseng drive the change of soil microbiota through carbon source different utilization. Plant Soil. 2020;455:139–53. [Google Scholar]
- 41.Lynch MD, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13(4):217–29. [DOI] [PubMed] [Google Scholar]
- 42.Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, Brookes PC, Xu J, Gilbert JA. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016;10:1891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat JD, Dayarathne MC, Huang S, Norphanphoun C, Senanayake IC, Perera RH, Shang Q, Xiao Y, D’Souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang W, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang J, Li Q, Liu JK, Liu XZ, Liu Z, Luangsa-Ard JJ, Pang K, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen T, Wijayawardene NN. Families of sordariomycetes. Fungal Divers. 2016;79:1–317. [Google Scholar]
- 44.Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. [Google Scholar]
- 45.Miao C, Mi Q, Qiao X, Zheng Y, Chen Y, Xu L, Guan H, Zhao L. Rhizospheric fungi of Panax notoginseng: diversity and antagonism to host phytopathogens. J Ginseng Res. 2016;40:127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mo Y, Zhang W, Yang J, Lin Y, Yu Z, Lin S. Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J. 2018;12:2198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naranjo-Ortiz MA, Gabaldón T. Fungal evolution: diversity, taxonomy and phylogeny of the Fungi. Biol Rev. 2019;94:2101–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–8. [Google Scholar]
- 49.Nilsson RH, Larsson K, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Gloeckner FO, Tedersoo L, Saar I, Koljalg U, Abarenkov K. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47:259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson RH, Tedersoo L, Lindahl BD, Kjoller R, Carlsen T, Quince C, Abarenkov K, Pennanen T, Stenlid J, Bruns T, Larsson K, Koljalg U, Kauserud H. Towards standardization of the description and publication of next-generation sequencing datasets of fungal communities. New Phytol. 2011;191:314–8. [DOI] [PubMed] [Google Scholar]
- 51.Ning D, Deng Y, Tiedje JM, Zhou J. A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci USA. 2019;116:16892–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ning D, Yuan M, Wu L, Zhang Y, Guo X, Zhou X, Yang Y, Arkin AP, Firestone MK, Oksanen J, Simpson G, Blanchet FG, Kindt R, Legendre P, Minchin P, Hara R, Solymos P, Stevens H, Szöcs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, De Cáceres M, Durand S, Weedon J. Vegan community ecology package version 2.6. 2022.
- 53.Osbourn A. Saponins and plant defence—A soap story. Trends Plant Sci. 1996;1:4–9. [Google Scholar]
- 54.Palencia ER, Hinton DM, Bacon CW. The black aspergillus species of maize and peanuts and their potential for mycotoxin production. Toxins. 2010;2:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedros-Alio C. The rare bacterial biosphere. Ann Rev Mar Sci. 2012;44:449–66. [DOI] [PubMed] [Google Scholar]
- 56.Pellitier PT, Zak DR. Ectomycorrhizal root tips harbor distinctive fungal associates along a soil nitrogen gradient. Fungal Ecol. 2021;54:101111. [Google Scholar]
- 57.Petrini O, Sieber TN, Toti L, Viret O. Ecology, metabolite production, and substrate utilization in endophytic fungi. Nat Toxins. 1993;1:185–96. [DOI] [PubMed] [Google Scholar]
- 58.Piasecka A, Jedrzejczak-Rey N, Bednarek P. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 2015;206:948–64. [DOI] [PubMed] [Google Scholar]
- 59.Qiao Y, Liu L, Miao C, Zhu G, Miao L, She W, Qin S, Zhang Y. Coupling of leaf elemental traits with root fungal community composition reveals a plant resource acquisition strategy in a desert ecosystem. Plant Soil. 2023;484:115–31. [Google Scholar]
- 60.Rungjindamai N, Jones EBG. Why are there so few basidiomycota and basal fungi as endophytes? A review. J Fungi. 2024;10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulz B, Römmert AK, Dammann U, Aust HJ, Strack D. The endophyte-host interaction: a balanced antagonism? Mycol Res. 1999;103:1275–83. [Google Scholar]
- 62.Segers FJJ, Dijksterhuis J, Giesbers M, Debets AJM. Natural folding of airborne fungal spores: a mechanism for dispersal and long-term survival? Fungal Biol Rev. 2023;44:100292. [Google Scholar]
- 63.Shi Y, Li Y, Xiang X, Sun R, Yang T, He D, Zhang K, Ni Y, Zhu Y, Adams JM, Chu H. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome. 2018;6:27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi S, Nuccio EE, Shi ZJ, He Z, Zhou J, Firestone MK. The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol Lett. 2016;19:926–36. [DOI] [PubMed] [Google Scholar]
- 65.Shim H, Waminal NE, Kim HH, Yang T. Dynamic evolution of Panax species. Genes Genomics. 2021;43:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6:1653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su J, Wang Y, Bai M, Peng T, Li H, Xu H, Guo G, Bai H, Rong N, Sahu SK, He H, Liang X, Jin C, Liu W, Strube ML, Gram L, Li Y, Wang E, Liu H, Wu H. Soil conditions and the plant microbiome boost the accumulation of monoterpenes in the fruit of Citrus reticulata ‘Chachi.’ Microbiome. 2023;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Y, Deng X, Tao C, Liu H, Shen Z, Liu Y, Li R, Shen Q, Schadt CW. Temporal dynamics of rare and abundant soil bacterial taxa from different fertilization regimes under various environmental disturbances. mSystems. 2022. 10.1128/msystems.00559-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang L, Xue K, Pang Z, Jiang L, Zhang B, Wang W, Wang S, Xu Z, Rui Y, Zhong L, Che R, Li T, Zhou S, Wang K, Du J, Wang Z, Cui X, Hao Y, Wang Y. Plant community associates with rare rather than abundant fungal taxa in alpine grassland soils. Appl Environ Microbiol. 2023. 10.1128/aem.01862-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tilman D. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci USA. 2004;101:10854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant—microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21. [DOI] [PubMed] [Google Scholar]
- 72.van der Heijden MGA, Dombrowski N, Schlaeppi K. Continuum of root—fungal symbioses for plant nutrition. Proc Natl Acad Sci USA. 2017;114:11574–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei G, Chen Z, Wang B, Wei F, Zhang G, Wang Y, Zhu G, Zhou Y, Zhao Q, He M, Dong L, Chen S. Endophytes isolated from Panax notoginseng converted ginsenosides. Microb Biotechnol. 2021;14:1730–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu M, Huang Q, Xiong Z, Liao H, Lv Z, Chen W, Luo X, Hao X. Distinct responses of rare and abundant microbial taxa to in situ chemical stabilization of cadmium-contaminated soil. mSystems. 2021;6:10–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xue Y, Chen H, Yang JR, Liu M, Huang B, Yang J. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 2018;12:2263–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiong C, He J, Singh BK, Zhu Y, Wang J, Li P, Zhang Q, Han L, Shen J, Ge A, Wu C, Zhang L. Rare taxa maintain the stability of crop mycobiotas and ecosystem functions. Environ Microbiol. 2021;23:1907–24. [DOI] [PubMed] [Google Scholar]
- 77.Xiong C, Zhu Y, Wang J, Singh B, Han L, Shen J, Li P, Wang G, Wu C, Ge A, Zhang L, He J. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021;229:1091–104. [DOI] [PubMed] [Google Scholar]
- 78.Zhang B, Peng Y, Zhang Z, Liu H, Qi Y, Liu S, Xiao P. GAP production of TCM herbs in China. Planta Med. 2010;76:1948–55. [DOI] [PubMed] [Google Scholar]
- 79.Zhang G, Shi L, Liu C, Huang Z, Zheng Y, Dong L. Rhizosphere effects on the microbial community: specificity and conservatism across geographically disjunct Panax species. Appl Soil Ecol. 2023;192:105075. [Google Scholar]
- 80.Zhang G, Shi L, Liu C, Mao R, Xia B, Huang Z, Wei X, Wu L, Zheng Y, Wei G, Xu J, Gao S, Chen S, Dong L. Modules in robust but low-efficiency phyllosphere fungal networks drive saponin accumulation in leaves of different Panax species. Environ Microbiome. 2023;18:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang G, Wei F, Chen Z, Wang Y, Jiao S, Yang J, Chen Y, Liu C, Huang Z, Dong L, Chen S. Evidence for saponin diversity—mycobiota links and conservatism of plant—fungi interaction patterns across holarctic disjunct Panax species. Sci Total Environ. 2022;830:154583. [DOI] [PubMed] [Google Scholar]
- 82.Zhang G, Wei F, Chen Z, Wang Y, Zheng Y, Wu L, Chen S, Dong L. Rare biosphere in cultivated Panax rhizosphere shows deterministic assembly and cross-plant similarity. Ecol Ind. 2022;142:109215. [Google Scholar]
- 83.Zhang S, Li K, Hu J, Wang F, Chen D, Zhang Z, Li T, Li L, Tao J, Liu D, Che R. Distinct assembly mechanisms of microbial subcommunities with different rarity along the Nu River. J Soils Sediments. 2022;22:1530–45. [Google Scholar]
- 84.Zhang Y, Dong S, Gao Q, Ganjurjav H, Wang X, Geng W. “Rare biosphere” plays important roles in regulating soil available nitrogen and plant biomass in alpine grassland ecosystems under climate changes. Agr Ecosyst Environ. 2019;279:187–93. [Google Scholar]
- 85.Zhang Z, Lu Y, Wei G, Jiao S. Rare species-driven diversity - ecosystem multifunctionality relationships are promoted by stochastic community assembly. MBio. 2022;13:e00449-e1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang G, Wei G, Wei F, Chen Z, He M, Jiao S, Wang Y, Dong L, Chen S. Dispersal limitation plays stronger role in the community assembly of fungi relative to bacteria in rhizosphere across the arable area of medicinal plant. Front Microbiol. 2021;12 [DOI] [PMC free article] [PubMed]
- 87.Zhong Y, Hu J, Xia Q, Zhang S, Li X, Pan X, Zhao R, Wang R, Yan W, Shangguan Z, Hu F, Yang C, Wang W. Soil microbial mechanisms promoting ultrahigh rice yield. Soil Biol Biochem. 2020;143:107741. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the National Genomics Data Center under the BioProject PRJCA007643. R codes used for statistical analyses are available at “https://github.com/githubzgz”. No datasets were generated or analysed during the current study.