SUMMARY
Whole salivary gland generation and transplantation offer potential therapies for salivary gland dysfunction. However, the specific lineage required to engineer complete salivary glands has remained elusive. In this study, we identify the Foxa2 lineage as a critical lineage for salivary gland development through conditional blastocyst complementation (CBC). Foxa2 lineage marking begins at the boundary between the endodermal and ectodermal regions of the oral epithelium before the formation of the primordial salivary gland, thereby labeling the entire gland. Ablation of Fgfr2 within the Foxa2 lineage in mice leads to salivary gland agenesis. We reversed this phenotype by injecting donor pluripotent stem cells into the mouse blastocysts, resulting in mice that survived to adulthood with salivary glands of normal size, comparable to those of their littermate controls. These findings demonstrate that CBC-based salivary gland regeneration serves as a foundational experimental approach for future advanced cell-based therapies.
In brief
Tanaka et al. demonstrated that the Foxa2 lineage labels the boundary region of the endoderm and surface ectoderm on the oral epithelium, crucial for Fgfr2-mediated salivary gland primordial formation. Foxa2-driven Fgfr2 knockout mice exhibit a salivary gland agenesis phenotype, restored by blastocyst injection of mouse PSCs, which leads to functional salivary gland generation.
Graphical Abstract
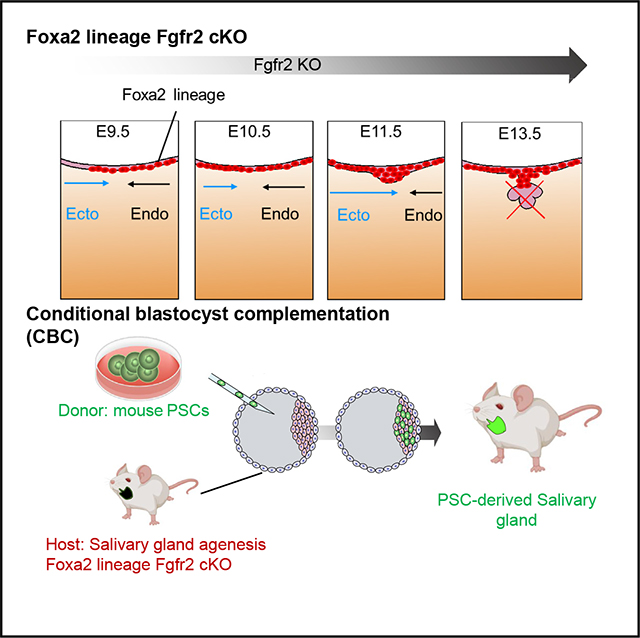
INTRODUCTION
Cell-based therapy for generating salivary glands is a promising next-generation therapy for patients suffering from dry mouth, rampant caries, and fungal infections due to Sjögren’s syndrome or the side effects of radiotherapy for head and neck cancers.1 The strategies of isolating and expanding tissue-specific stem cells in the salivary gland tissues have been proposed,2–5 although the extent to which endogenous tissue stem cells play regenerative/restorative roles in damaged salivary gland function remains controversial. To fill this knowledge gap, we reported a strategy to transplant exogenous salivary gland progenitor organoids induced to differentiate from pluripotent stem cells (PSCs).6,7 These PSC-derived salivary gland organoids were engrafted orthotopically into salivary-gland-resected mice. However, post-transplantation, the PSC-derived salivary gland organoids were smaller than usual, and the recovery effect on salivary secretion was limited.6 A better strategy that produces whole salivary glands derived from PSCs to overcome size issues was needed.
Blastocyst complementation (BC) has been proposed as a promising option for creating fully functional organs derived from PSCs in vivo.8 This unique technique has further evolved into generating intra- and interspecies organs such as kidneys, pancreas, and blood vessels.9–12 Previously, we refined the BC approach to a conditional BC (CBC) strategy, which, by targeting a specific lineage to be rescued via PSC timed injections into the blastocysts, allowed us to generate fully functional organs.13 Developing an empty niche in a lineage is the key to successful CBC; however, the lineage and the genes that vacate the salivary gland niche have been unknown.
It is well known that a fibroblast growth factor 10 (Fgf10) ligand, expressed in mesenchymal cells during mouse and human development, is crucial for salivary gland organogenesis through interactions with its receptor, fibroblast growth factor receptor 2 (Fgfr2), which is expressed in epithelial cells.14–17 In the systemic knockout (KO) mice of Fgfr2, various organs, including salivary glands, lungs, and limbs, show aplasia or hypoplasia phenotypes,18–21 though the timing and lineage that require Fgfr2 for salivary gland formation remain unclear.
The origin of the salivary glands before primordial salivary gland initiation has been debated. Based on previous Sox17-LacZ lineage-tracing mouse analyses, most endodermal SRY-box transcription factor 17 (Sox17) lineages do not label salivary gland epithelium.22 Thus, the salivary gland has been believed to originate from the ectodermal oral mucosa and not the endoderm. Contradictorily, Sonic hedgehog (Shh), primarily known as a marker of the endodermal epithelium, has also been reported to label the salivary gland epithelial lineage.23 This confusion arises because the developmental origins of the submandibular and sublingual glands anatomically arise around the boundary region between the ectodermal and the endodermal-derived oral epithelium (OE).24,25 Based on this, for generating PSC-derived salivary glands by CBC, we investigated the lineage to label the boundary region, which is potentially critical for salivary gland formation.
RESULTS
Foxa2 lineage contributes to salivary gland development
Shh lineage has been reported to contribute to the endoderm, pharyngeal pouch,26 and salivary gland epithelium at the later stage of development at embryonic day (E)15.5, while Shh regulates the branching morphogenesis of salivary glands.27 On the other hand, the paired like homeodomain 2 (Pitx2) lineage is another potential lineage of the salivary gland because its protein is known to be expressed in the embryonic OE and dental epithelium before salivary gland formation, and the Pitx2 defect results in a range of developmental deficits, including defective body-wall closure, right pulmonary isomerism, and altered cardiac position.28–30 Using ShhCre/+; RosaLSL-tdTomato/+ or Pitx2Cre/+; RosaLSL-tdTomato/+ lineage-tracing mice, we examined whether Shh or Pitx2 lineage could contribute to salivary gland epithelium. We found that 97.2% ± 1.59% in Shh lineage-tracing mice and 98.7% ± 0.66% in Pitx2 lineage-tracing mice of the salivary gland epithelium were tdTomato+ at E18.5 in both lineage-tracing analyses (Figures S1A–S1C), which is supported by previous studies.23 Surprisingly, we found that the salivary gland was decently formed in both lineage-specific Fgfr2 KO mice (Figures S1A and S1B). These results indicate that the Pitx2 or Shh lineage-based strategy is insufficient for inducing the phenotype of salivary gland agenesis, though it is unclear why Fgfr2 deficiency did not cause a salivary gland defective phenotype in vivo despite the evidence that Fgfr2 is known to be critical for salivary gland branching morphogenesis in ex vivo culture and KO study.14,31 We reasoned that this resulted from using conditional KO (cKO) alleles and that in these models, Pitx2 and Shh lines did not target all salivary gland precursors to deplete Fgfr2. The time lag between Cre driver expression and Fgfr2 KO was expected. Therefore, it is necessary to use driver genes that are expressed early enough before the onset of salivary gland development, which begins with the thickening of the OE at E11.5. In organ formation by CBC, it is important to form an empty niche before the emergence of the desired organ primordium.13,32 To address this issue, we further investigated another potential lineage involved in the salivary gland precursor niche around the boundary region of the endoderm and ectoderm, potential salivary gland precursor niches by analyzing deposited single-cell RNA sequencing (scRNA-seq) data from E8.5 mouse33 and E12.0 OE.34 In the E12.0 OE scRNA-seq analysis, we identified specific genes in the posterior-lateral epithelium (PL) and tongue epithelium (T) adjacent to the salivary gland primordium, which are known to give rise to salivary gland primordium (Figures S1D–S1F). The endodermal markers Foxa2 and Pitx2 were expressed in T, while Sox9 and Shh did not express T, respectively (Figures S1D and S1E).22 Because Pitx2 lineage-driven Fgfr2 KO analyses did not show the salivary gland agenesis phenotype, we speculate that targeting both the PL and T is crucial for causing the agenesis phenotype. Based on this, we checked the earlier developmental time of OE formation.
Using E8.5 mouse scRNA-seq, we identified Foxa2 as the potential gene expressed around the boundary region of the E8.5 OE between the ectoderm and endoderm, indicating the salivary gland precursors of the growing boundary region (black dotted area) for future salivary gland primordia formation (Figures S1G and S1H). Conversely, neither Shh nor Pitx2 was expressed on the precursor of the E8.5 OE (Figure S1H). These data suggested that the Foxa2 may label the boundary of the E10.5 OE as a salivary gland precursor niche before salivary gland primordia formation.
To confirm this observation, we performed lineage-tracing studies utilizing Foxa2Cre/+; RosatdTomato/+ mice. Before the boundary region formation by connecting the ectodermal OE and the endodermal epithelium (Figure 1A), we found that a few Foxa2 lineage cells appeared in the ectodermal OE (Figure 1B). Significantly, the boundary region of the E9.5 epithelial cells on the oral floor was entirely labeled by the tdTomato+ Foxa2 lineage (Figure 1C, arrowhead). In contrast, the Shh or Pitx2 lineage did not mark tdTomato on the E9.5 OE of the boundary region (Figure S1I, arrows). At E10.5, the Foxa2 lineage labeling had extended to the anterior region of the OE, while it did not contribute to the ventral surface epidermis (Figure 1D). Interestingly, at this point, the expression of the Foxa2 protein itself was limited to the posterior OE (Figure 1E). As evidenced by the tdTomato signal and histological analysis (Figures 1F and 1G), the Foxa2 lineage in the OE contributed to the invaginating OE (Figure 1G, arrow) and primordium formation (Figure 1G, asterisk) at E13.5 and occupied the adjacent epithelium. Remarkably, we observed that the Foxa2 lineage significantly labeled 99.3% ± 0.64% of the salivary E15.5 gland epithelium compared to mesenchyme labeling (2.9% ± 2.11%) (Figures 1H and 1I).
Figure 1. Foxa2 lineage labeled the boundary region between the endoderm and ectoderm, leading to marking the whole salivary gland epithelium.
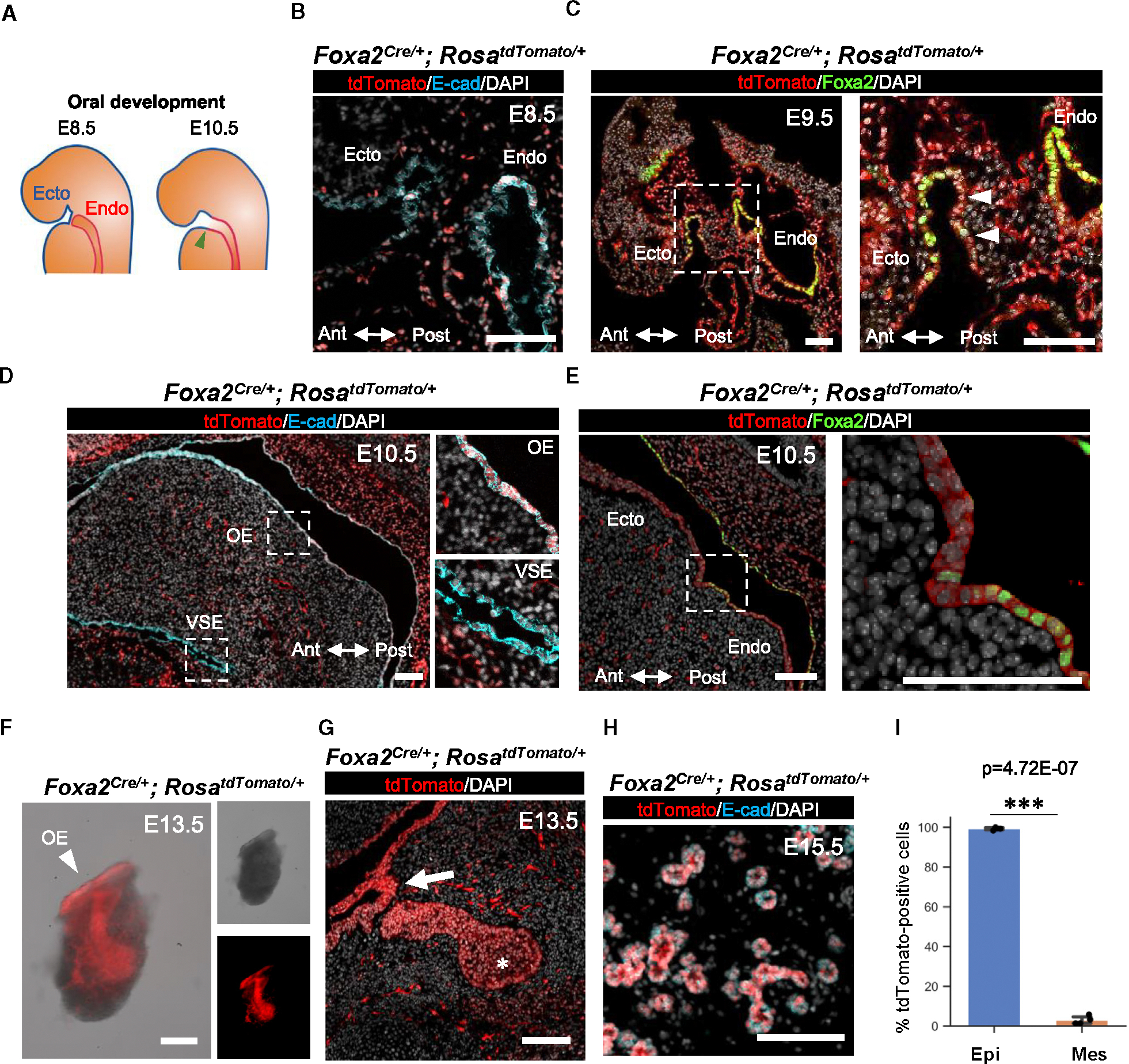
(A) Schematics of the boundary region development located between endodermal (a red line) and ectodermal oral (a blue line) epithelial cells at embryonic day (E) 8.5 and E10.5 before salivary gland primordial formation. The green arrowhead indicates the boundary region of salivary gland development. Ecto, ectoderm; Endo, endoderm.
(B–E) Representative immunofluorescence (IF)-confocal imaging of the ectodermal (Ecto) oral mucosal and endodermal (Endo) epithelial boundary in E8.5 (B), E9.5 (C), and E10.5 (D and E) Foxa2Cre/+; RosaLSL-tdTomato/+ lineage-tracing mice. Immunostaining of tdTomato: red, E-cadherin (E-cad): cyan, DAPI: gray, and Foxa2: green. Each right image of (C)–(E) is an enlarged image of a white dotted box. Ant, arteriolar; Post, posterior axis; VSE, ventral surface epidermis; OE, oral epithelium (n = 3 at each time point, biological replicates).
(F) Representative merged image (left) of bright-field (top right) and tdTomato fluorescent signal (bottom right) of the isolated salivary gland from E13.5 Foxa2Cre/+; RosaLSL-tdTomato/+ lineage-tracing mice (n = 3).
(G and H) Representative confocal imaging of E13.5 salivary gland primordium (G) and E15.5 salivary glands (H) of Foxa2Cre/+; RosaLSL-tdTomato/+ lineage-tracing mouse (n = 4).
(I) Graphs: the morphometric lineage-tracing analysis: percentage of Foxa2 lineage labeling in E-cad+ epithelial (blue bar) and E-cad− mesenchymal (orange bar) cells from E15.5 Foxa2Cre/+; RosaLSL-tdTomato/+ lineage-tracing mouse salivary glands (n = 4). Statistical analyses: unpaired Student’s t test, significant: p < 0.05.
***p < 0.001. Error bars represent mean ± SD. Scale bars: 100 μm.
Foxa2-driven Fgfr2 cKO caused salivary gland agenesis phenotype
Since the Foxa2 lineage contributes to the E8.5–E9.5 boundary region between the endodermal and ectodermal OE, leading to the entire salivary gland labeling, we investigated whether mice with Fgfr2 depletion in the Foxa2 lineage would show the salivary gland agenesis phenotype. We utilized the Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+ mice. In this mouse model that we previously reported,32 formation of the lungs and thymus is absent. However, other major endodermal organs such as the liver, esophagus, pancreas, and intestines do not show an agenesis phenotype. This is because, although these organs were marked as the Foxa2 lineage, Fgfr2 is not critical for their formation. Additionally, the phenotypes of organ agenesis known to form in systemic Fgfr2 KO, such as the kidneys, hair follicles, and limbs, are preserved, which indicates that the Foxa2 lineage does not target these organs enough to cause these agenesis phenotypes. In the Fgfr2 heterozygous KO mice (Foxa2Cre/+; Fgfr2flox/+; RosaLSL-tdTomato/+, hereafter called Fgfr2hetero), tdTomato+ salivary gland primordia were detected at the base of the oral cavity at E13.5 (Figure 2A). Conversely, tdTomato+ tissue was absent in Fgfr2cKO mice (Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+), suggesting the salivary gland agenesis phenotype (Figure 2B). In contrast to the lineage-tracing results (Figure 1G), histological analysis showed tdTomato+ OE invagination into mesenchyme (Figure 2B, arrow), but it lost salivary gland primordia formation in the Fgfr2cKO mice at 13.5 (Figure 2B, asterisk). This result indicates that Fgfr2 is critical for initiating salivary gland primordia formation after the invagination of OE but is not essential for invagination.
Figure 2. Foxa2-driven Fgfr2 conditional knockout (Fgfr2cKO) caused the salivary gland agenesis phenotype.
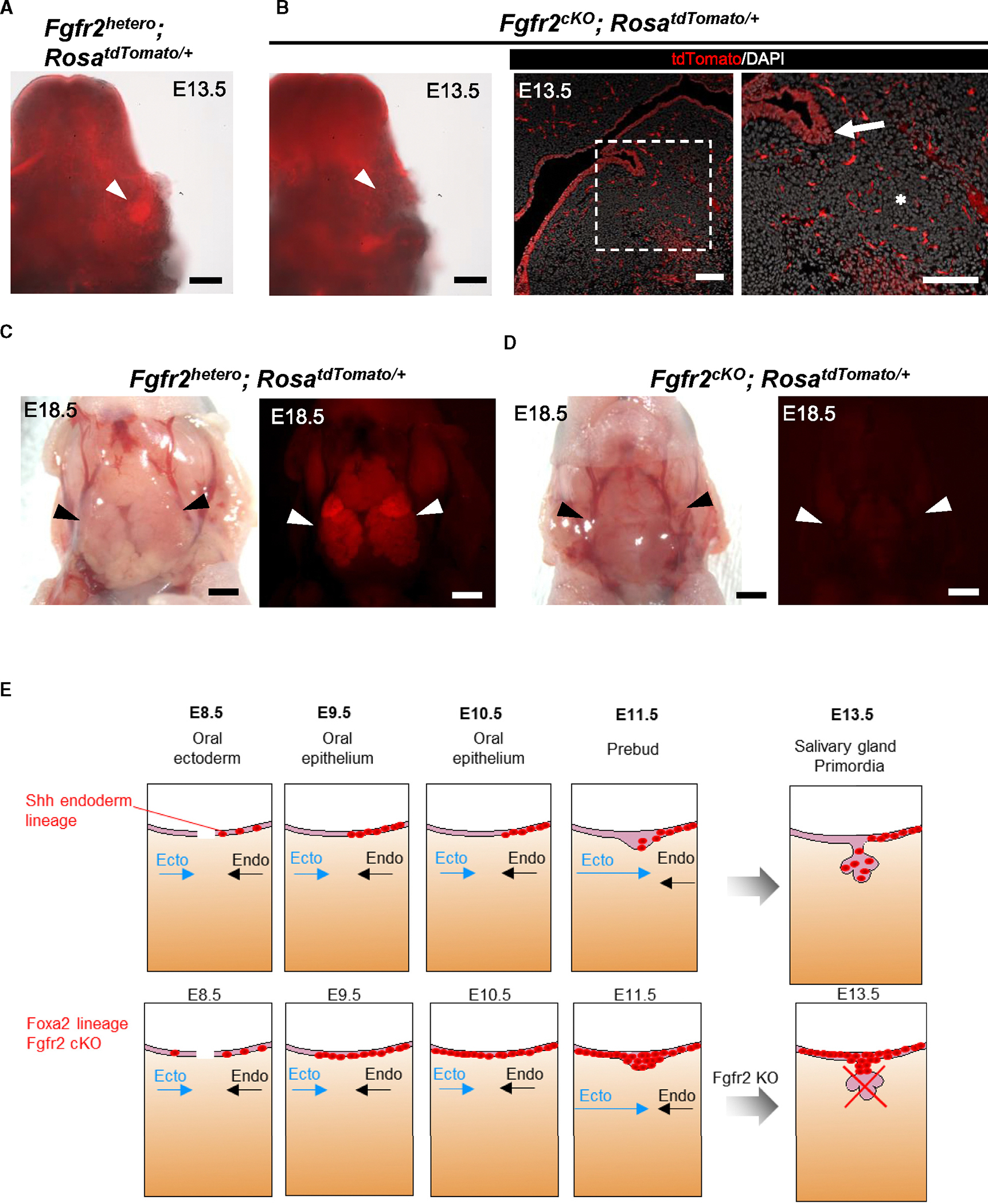
(A and B) Representative macroscopic merged images of bright-field and tdTomato fluorescent signals of salivary gland primordium formation (white arrowheads) from E13.5 Fgfr2hetero (Foxa2Cre/+; Fgfr2flox/+; RosaLSL-tdTomato/+) mice (A) and E13.5 Fgfr2cKO (Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+) mice: no salivary gland primordia formation (B, left). Representative IF-confocal imaging of the salivary gland rudiment in E13.5 Fgfr2cKO (Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+) mice (n = 3): no salivary gland primordium formation (asterisk), but invaginating of the OE occurs (arrow), indicating the salivary gland rudiment (B, middle and right).
(C and D) Representative macroscopic bright-field images (left) and tdTomato fluorescent signals (right) of salivary glands (arrowhead) from E18.5 Fgfr2hetero (Foxa2Cre/+; Fgfr2flox/+; RosaLSL-tdTomato/+) (C) and E18.5 Fgfr2cKO (Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+) (D) mice (n = 3): salivary gland agenesis phenotype in the E18.5 Fgfr2cKO (Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+). tdTomato: red.
(E) Proposed schematic models for the initial salivary gland development from the junction of Ecto oral mucosa and Endo epithelium based on Foxa2 or Shh lineage-tracing analysis: when Ecto oral mucosa (blue arrows) and Endo (arrows) epithelium are closer and connecting, Shh lineage (red)-labeled cells appeared only on the oral Endo side (black arrows) of the E8.5 and E9.5 boundary region but not the oral Ecto side of salivary gland precursors (SGPs) before the boundary formation. Later, the Shh lineage labeled nearly the entire E15.5 epithelium, supported by a previous study.23 In contrast, the Foxa2 lineage initiated labeling of the E8.5 SGP and the entire E9.5 SGP on the boundary region before the invagination of the SGPs. Therefore, the KO of Fgfr2 in the Foxa2 lineage resulted in the invagination of SGPs but failed to form salivary gland primordia, leading to the salivary gland agenesis phenotype.
Scale bars: (B, left) 200 μm, (B, right) 100 μm, and (C and D) 1 mm.
To ensure that these observations were not due to developmental delays by the Fgfr2 deficiency in the Foxa2 lineage, we performed a macroscopic analysis of E18.5 Fgfr2cKO mice. While tdTomato+ salivary gland tissues were formed in the E18.5 Fgfr2hetero mice (Figure 2C, arrow), tdTomato+ salivary glands were not observed in the Fgfr2cKO mice (Figure 2D, arrow). These results indicate the salivary gland agenesis phenotype rather than a developmental delay. Taken together, Fgfr2 depletion in the Foxa2 lineage-based niche is efficient for vacating the niche critical for salivary gland formation, which can be utilized for CBC (Figure 2E).
CBC using Foxa2-driven Fgfr2 cKO mice rescued salivary gland formation during development
To address whether CBC would rescue the salivary gland agenesis phenotype, we injected donor mouse induced pluripotent stem cells (iPSCs) expressing nuclear EGFP (nEGFP) into host Fgfr2cKO mice. In this CBC approach, donor iPSC-derived cells were GFP+, while host cells from the Foxa2 lineage were tdTomato+ and non-Foxa2 lineage cells were tdTomato−, allowing us to distinguish them visually (Figure 3A). Analysis at E17.5 revealed the formation of chimeric mice expressing both GFP and tdTomato signals. Furthermore, no significant external abnormalities were observed in the Fgfr2cKO, nEGFP+ iPSC chimeric mice or littermate Fgfr2hetero, nEGFP+ iPSC chimeric mice (Figures 3B and 3C). Macroscopic analysis showed sporadic detection of GFP and tdTomato signals in the Fgfr2hetero chimeric mice’s submandibular and sublingual glands (Figure 3B). In contrast, in the salivary glands of Fgfr2cKO, nEGFP+ iPSC chimeric mice, a strong GFP signal was observed, while the tdTomato signal was low (Figure 3C). To quantify this observation, we performed a histological analysis. In the Fgfr2hetero chimeric mice, the E-cadherin (E-cad)+ salivary gland epithelium was formed as a chimera of host tdTomato+ cells and donor nEGFP+ cells (Figure 3D). In the Fgfr2cKO, nEGFP+ iPSC chimeric mice, all the E-cad+ salivary gland epithelium and smooth muscle actin (αSMA)+ myoepithelial cells were rescued exclusively by donor nEGFP+ cells without tdTomato+ host cells (Figure S2). These chimerism rates in the Fgfr2cKO, nEGFP+ iPSC chimeric mice were significantly higher than in the heterozygous, nEGFP+ iPSC chimeric mice (Fgfr2cKO, nEGFP+ iPSCs vs. Fgfr2hetero, nEGFP+ iPSCs: 100.0% ± 0% vs. 74.7% ± 13.44%) (Figure 3F). In contrast, the chimerism in mesenchymal cells did not show a significant difference between the Fgfr2hetero and Fgfr2cKO chimeric mice (Fgfr2cKO, nEGFP+ iPSCs vs. Fgfr2hetero, nEGFP+ iPSCs: 62.8% ± 16.35% vs. 64.3% ± 19.25%) (Figure 3G).
Figure 3. Rescue of salivary gland agenesis phenotype of Fgfr2cKO mice by Foxa2 lineage-based CBC.
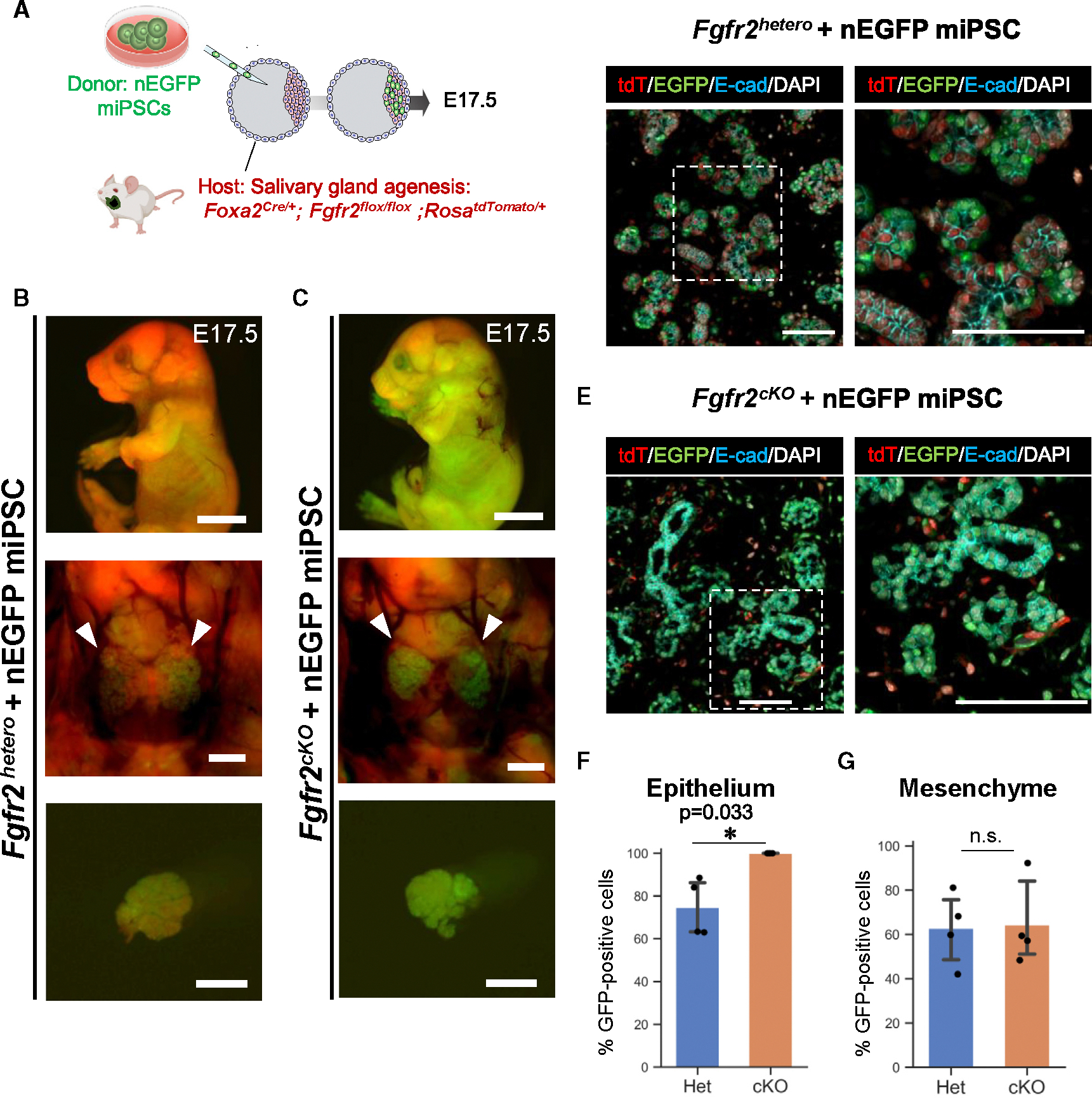
(A) Schema of CBC experiment: we injected nuclear EGFP (nEGFP)+ mouse iPSCs (nEGFP miPSCs) into the blastocysts of Foxa2-driven Fgfr2cKO (Foxa2Cre/+; Fgfr2flox/flox; RosaLSL-tdTomato/+) mice or littermate controls: Fgfr2hetero mice (Foxa2Cre/+; Fgfr2flox/+; RosaLSL-tdTomato/+).
(B and C) Representative images of fluorescent signals of the host (tdTomato: red) and donor nEGFP signal of nEGFP miPSCs (green) from embryos (top), salivary glands (middle: arrows), and isolated salivary glands (bottom) from the E17.5 chimeric mice of Fgfr2cKO+nEGFP miPSCs (C) or littermate controls: Fgfr2hetero+nEGFP miPSCs (B).
(D and E) Representative IF-confocal imaging of the salivary glands in the E17.5 chimeric embryos of Fgfr2cKO+nEGFP miPSCs (E) or littermate controls: Fgfr2hetero+nEGFP miPSCs (D). Immunostaining of tdTomato: red, EGFP: green, E-cad: cyan, and DAPI: gray. Each right image of (D) and (E) is an enlarged image of a white dotted box.
(F) Graphs: the morphometric analysis: percentage of nGFP+ donor cells in E-cad+ epithelial cells from E17.5 chimeric embryos of Fgfr2hetero+nEGFP miPSCs (n = 3 per biological replicates, 5 fields per group).
(F and G) The morphometric analysis: percentage of nEGFP+ donor cells in E-cad+ epithelium (F) or E-cad− mesenchyme (G) from E17.5 chimeric embryos of Fgfr2cKO+nEGFP miPSCs or littermate controls: Fgfr2hetero+nEGFP miPSCs (n = 4 per biological replicates, 5 fields per group). Statistical analyses: unpaired Student’s t test, significant: *p < 0.05. No significant change, n.s. Error bars represent mean ± SD.
Scale bars: (B and C) 5 mm (top) and 1 mm (middle and bottom) and (D and E) 100 μm.
To confirm whether the fully complemented salivary glands in the Fgfr2cKO, nEGFP+ iPSC chimeric mice differentiate well, we examined the expression of various progenitor and differentiation markers in the complemented salivary glands(Figure S2). Indeed, E17.5 nEGFP+ intercalated duct cells and acinar cells expressed Sox9, a marker for salivary gland progenitors. nEGFP+ rescued ductal cells expressed keratin 5 (K5), a marker for basal cells, and myoepithelial marker α-Sma around the acinar cells. E17.5 acinar cells expressed Mist1, a mature acinar marker, and Aqp5, a marker of mature acinar aggregated on the luminal surface of the Mist1+ acinar cells. These results indicate that the salivary gland epithelial cells complemented with mouse iPSCs develop and differentiate well at E17.5 without any developmental delays. In addition, we observed salivary gland generation in all cKO mice, regardless of the general chimerism (55.1% ± 22.92%) represented in hematopoietic cells (Tables S1 and S2).
Complemented salivary glands develop well enough in size and differentiation until adulthood
Based on this, we further examined whether the CBC-mediated rescued salivary glands would develop well until adulthood. To address this question, we injected donor mouse PSCs13 expressing EGFP under a CAG promoter (PSCCAG-EGFP) into the host Foxa2-driven Fgfr2 cKO mice. Based on the macroscopic fluorescent analysis of 4-week-old mice, the salivary glands of Fgfr2 heterozygous, PSCCAG-EGFP chimeric mice formed a chimera consisting of tdTomato+ host cells and EGFP+ donor cells (Figure 4A). In contrast, the Fgfr2cKO, PSCCAG-EGFP chimeric mice exhibited few tdTomato+ signals (Figure 4B). In histological analysis, the salivary glands of the Fgfr2hetero, PSCCAG-EGFP chimeric mice showed both tdTomato and EGFP signals in the epithelial cells (Figure 4C). Conversely, in the rescued Fgfr2cKO, PSCCAG-EGFP chimeric mice, the epithelial compartment was entirely labeled by EGFP, while the E-cad− mesenchymal cells showed a few tdTomato signals (Figure 4D). Furthermore, there was no significant difference in the weight of the complemented submandibular and sublingual glands between the chimeric heterozygous mice and the Fgfr2cKO mice (Figure 4E). The histological analysis showed that the complemented salivary glands formed periodic-acid-Schiff (PAS)+ mucin-producing acinar cells, similar to those in heterozygous mice, and the body weight of the mice was also comparable to that of heterozygous mice (Figures S3A and S3B). These findings indicate that a sufficiently sized and functional salivary gland tissue was generated from donor PSCs via the Foxa2 lineage-based CBC approach. Additionally, we compared the EGFP+ donor cell chimerism with differentiation markers (the proportion of EGFP+ cells with each marker) between the Fgfr2cKO, PSCCAG-EGFP and Fgfr2hetero, PSCCAG-EGFP chimeric mice (Figures 4F–4H). In the chimeric salivary glands, GFP was labeled with the markers of Aqp5+ acinar cells (Fgfr2cKO, PSCCAG-EGFP vs. Fgfr2hetero, PSCCAG-EGFP: 100.0% ± 0% vs. 82.1% ± 7.05%), which aggregates on the luminal side with abundant cytoplasm, and α-Sma+ spindle-shaped myoepithelial cells (Fgfr2cKO, PSCCAG-EGFP vs. Fgfr2hetero, PSCCAG-EGFP: 100.0% ± 0% vs. 59.9% ± 9.75%) surrounding the acini reflect the tissue structure of adult salivary glands as well as K5+ basal cells (Fgfr2cKO, PSCCAG-EGFP vs. Fgfr2hetero, PSCCAG-EGFP: 100.0% ± 0% vs. 63.3% ± 13.39%) (Figure 4H). Overall, in the complemented salivary glands in the Fgfr2cKO, PSCCAG-EGFP mice, the Aqp5+ acinar cells, K5+ basal cells, and α-Sma+ myoepithelial cells all showed a GFP+ rate of 100%, significantly higher than that in Fgfr2hetero, PSCCAG-EGFP mice (Figure 4H). These results demonstrated that depleting Fgfr2 in the Foxa2 lineage in the empty niche is critical for generating a fully sized, well-differentiated adult salivary gland, followed by PSC injection into the blastocysts.
Figure 4. Fully mature adult salivary gland generation via Foxa2 lineage-based CBC.
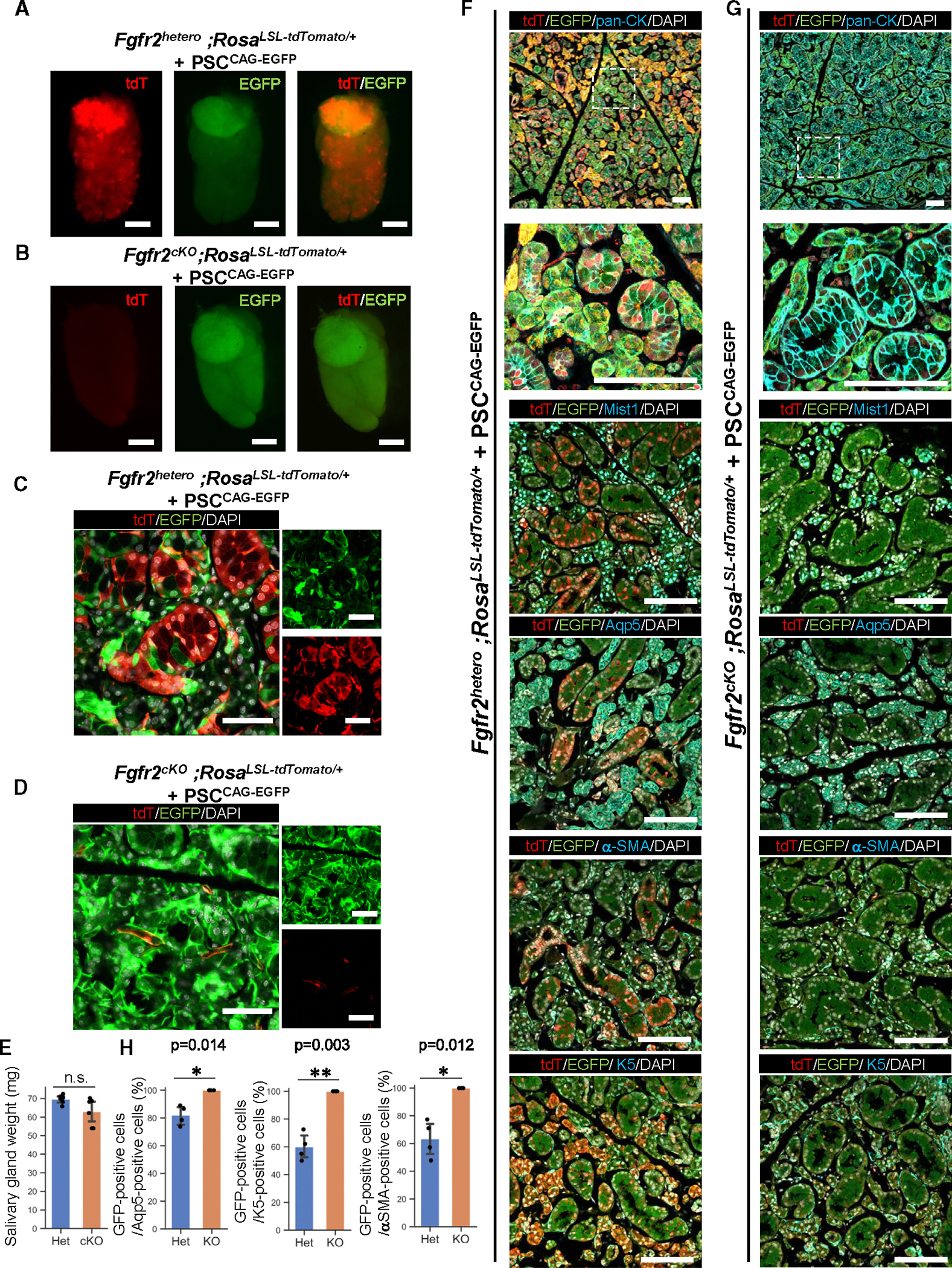
(A and B) Representative images of fluorescent signals of the host (tdTomato: red) and donor EGFP signal of PSCCAG-EGFP (green) from isolated salivary glands from the 4-week-old chimeric mice of littermate controls: Fgfr2hetero, PSCCAG-EGFP (A) or of Fgfr2cKO, PSCCAG-EGFP (B).
(C and D) Representative confocal imaging for native fluorescence signals of the salivary glands in the 4-week-old chimeric embryos of littermate controls:</p/>Fgfr2hetero, PSCCAG-EGFP (C) or Fgfr2cKO, PSCCAG-EGFP (D).
(E) Analysis of salivary gland weight. The combined weight of the submandibular gland and the sublingual gland was measured. Het: Fgfr2hetero, PSCCAG-EGFP, cKO: Fgfr2cK, PSCCAG-EGFP. Statistical analyses: unpaired Student’s t test, significant: p < 0.05. No significant change, n.s. Error bars represent mean ± SD.
(F and G) Representative IF-confocal imaging of the salivary gland from the 4-week-old chimeric mice of littermate controls: Fgfr2hetero, PSCCAG-EGFP (F) or Fgfr2cKO, PSCCAG-EGFP (G). The second image from the top is an enlarged image of a white dotted box. Scale bars: 100 μm.
(H) The morphometric analysis: percentage of EGFP+ donor cells in Aqp5+ acinar cells (left), percentage of EGFP+ donor cells in K5+ basal cells (middle), percentage of EGFP+ donor cells in α-Sma+ myoepithelial cells (right) from the 4-week-old chimeric mice of littermate controls: Fgfr2hetero, PSCCAG-EGFP (Het) or Fgfr2cKO, PSCCAG-EGFP (KO) (n = 4 per biological replicates, 5 fields per group). Statistical analyses: unpaired Student’s t test, significant: p < 0.05. No significant change, n.s. *p < 0.05 and **p < 0.01. Error bars represent mean ± SD.
Scale bars: (A and B) 2 mm and (D) 50 μm.
DISCUSSION
Radiotherapy for head and neck cancers35–37 or Sjögren’s syndrome38,39 can result in fibrosis and the disappearance of salivary gland acinar cells, resulting in hyposalivation. Hyposalivation can cause various symptoms, including dry mouth, rampant caries, and fungal infections, but current treatment options for these symptomatic diseases are limited.40 In this regard, cell-based therapy is one of the most promising approaches. We harnessed a CBC approach to generate a fully functional salivary gland. The salivary glands produced by CBC showed weight equivalent to littermate controls and good differentiation, indicating that our approach generated fully sized, PAS+ mucin-producing salivary glands derived from PSCs.
Fgf7 and Fgf10, which are ligands of Fgfr2, have been proven to be crucial for the branching formation and duct elongation of salivary glands using ex vivo organ culture.14,17 In this study, we provided a piece of evidence that Foxa2 lineage-driven Fgfr2 knockout mice exhibit salivary gland agenesis phenotype but salivary gland rudiment formation still occurs and invaginates the OE. Our results indicate that Fgfr2 is critical for salivary gland primordia formation. We rescued this phenotype in the CBC experiment, and the salivary gland epithelium was entirely replaced by donor cells derived from PSCs. These results suggest that generating the vacant niche by Fgfr2 depletion in the invaginating OE was sufficient to rescue this phenotype via CBC, leading to complementing the entire salivary gland epithelium.
Salivary gland primordia arise from the OE at E11.5. The ectodermal OE arises from the boundary region after the junction of the ectoderm and the endoderm around E8.5–E9.5, but its exact origin has been unknown. Intriguingly, Shh lineage-tracing mice are known to label endoderm and also all salivary gland epithelial cells using ShhCre/+; RosaLSL-YFP/+ mice, while the Sox17 Cre/+; RoaLSL-LacZ/+ mice, which are another endodermal lineage-tracing model, do not label salivary gland epithelial cells.22,23
We revealed that Foxa2 is the lineage marking around the E9.5 boundary region critical for salivary gland primordial formation. In the previous study using Krt14 lineage-driven Fgfr2 cKO, a small salivary gland primordium was formed,41 while our lineage-tracing analysis using Shh or Pitx2 did not label the E9.5 boundary region of the oral mucosa. Based on these data, scRNA-seq data allowed us to draw the area most likely indicating the boundary region (black dotted lines in Figure S1H). These scRNA-seq results indicated that targeting the boundary region using the Foxa2 lineage mice is critical for controlling the entire salivary gland’s epithelial precursor behaviors before primordial salivary gland formation, while further analyses of the boundary region at a single-cell level are required in the near future.
Systemic Fgfr2 KO17 and our Foxa2-driven Fgfr2 KO phenotype support the requirement of Fgfr2 for salivary gland primordia formation. Our lineage-tracing analysis using ShhCre/+; RosatdTomato/+ mice faithfully labeled the endoderm but not the OE right before the E8.5 boundary regions, while the previous study at E15.5 showed that the lineage labels the entire salivary gland,23 and we showed the entire salivary gland at E18.5, suggesting that the Shh lineage spontaneously increased its labeling from the boundary region formation at E15.5 to the entire salivary gland.
Since the Shh-driven Fgfr2 KO phenotype showed the lung agenesis phenotype, the Cre driver activity and genotyping cannot be mistaken in our experiments.13,32 Interestingly, the Shh-driven Fgfr2 KO phenotype did not show gross morphological changes in E18.5 salivary glands compared to the littermate controls, which is contradictory evidence of Fgfr2’s effect in ex vivo culture.14 Based on our data, we speculate that Fgfr2 depletion requires specific time windows to induce the salivary gland agenesis phenotype, most likely in the salivary gland precursors around the boundary regions, as the lung agenesis model requires the depletion of the genes before the lung primordia formation.13,42 For the exact requirement of Fgfr2 during salivary gland branching morphogenesis in vivo, spatiotemporal Fgfr2 depletion using ShhCreERT2/+; Fgfr2flox/flox or Foxa2 CreERT2/+; Fgfr2flox/flox during salivary gland development is required in future experiments.
Our study results collectively demonstrate that the Foxa2 lineage is essential for the formation of salivary gland primordia and for generating fully matured salivary glands via CBC, which provides a critical experimental foundation for future interspecies salivary gland generation using human iPSCs.43–45
Limitations of the study
Although sex-hormone-mediated salivary gland size dimorphism was reported, it is unclear which cell types in the salivary gland cause the dimorphism phenotype.46 In our study, we determined the sex based on the appearance of the sex gland but not at the cellular level, and we used donor male PSCs for adult analysis. Interestingly, all of the chimeric adult animals showed a penis, a male phenotype appearance, most likely caused by the occupation of donor male PSCs into germ lines. Therefore, we did not expect to consider the dimorphism in our studies. Further studies are needed to determine the extent to which the sex of the germ line would affect the size of the complement salivary gland when the donor cell’s sex is different, which is an interesting question.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Munemasa Mori (mm4452@cumc.columbia.edu).
Materials availability
This study did not generate unique reagents.
Data and code availability
This study did not generate any unique datasets
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mouse
ShhCre/+ mice (cat. 05622), Rosa26tdTomato/tdTomato mice (cat. 07914) were obtained from the Jackson Lab. X. Zhang kindly gifted Fgfr2flox/flox mice. We further backcrossed these mice for over three generations with CD-1 mice (cat. 022) from the Charles River. Dr. Nicole C Dubois kindly provided Foxa2Cre/Cre mice (129xB6 mixed background).47 For conditional deletion of Fgfr2 (Fgfr2cKO), we crossed Fgfr2flox/flox; Rosa26tdTomato/tdTomato females with Foxa2Cre/Cre; Fgfr2flox/+, Foxa2Cre/+; Fgfr2flox/+ or ShhCre/+; Fgfr2flox/+ males, respectively. PCR performed genotyping of the Shh-Cre and Rosa26-tdTomato alleles according to the protocol provided by the vendor. All animals in this study were housed under specific pathogen-free conditions in a 12h light/dark cycle. All animal experiments were approved by Columbia University Institutional Animal Care and Use Committee in accordance with US National Institutes of Health guidelines.
Murine embryonic stem cells and induced pluripotent stem cells (iPSC)
nGFP+iPSC derived from Rosa nT-nG/+ mouse embryonic fibroblasts (129S6/SvEvTac x C57BL/6NCr background, male and female iPSC expressing nuclear EGFP under Rosa Locus)32 and PSC CAGGFP (C57BL/6 N × 129 S6 background, male mouse embryonic stem cells, MTI-GlobalStem: cat. no. GSC-5003)13 were cultured in a2i/VPA/LIF medium on a feeder, as previously reported.13 These pluripotent stem cells (PSCs) were passaged and seeded in 105 cells in 6-well plate every 2–3 days. All cell cultures were grown at 37°C and 5% CO2.
METHOD DETAILS
Culture of mouse iPSCs and PSC
We cultured iPSCs and male PSCs in a2i/VPA/LIF medium on a feeder, as previously reported.13 These PSC cells were passaged at a split ratio 1:10 every 2–3 d. For the CBC donor cell preparation, nGFP+iPSCs and PSCCAGEGFP, cultured in a2i/VPA/LIF, were trypsinized and resuspended in 4 mL cold DMEM +10% FBS immediately, and filtering the cells with a 40-μm filter. Cells were centrifuged at 350 rcf, 4°C, for 3 min, and the supernatant was removed. After being washed with flow buffer containing 0.2% BSA, 1% Glutamax, and 1μM Y27632, 1 million cells were resuspended in 100 μL of flow buffer. The following antibodies were added: Epcam-BV421 (1:50), SSEA1-PE (1:50), CD31-APC (1:50), Zombie Aqua Fixable Viability Kit (1:100). EpcamhighSSEA1highCD31high cells were sorted by FACS (SONY MA900) and subsequently prepared for the injection.
Immunofluorescence
Before the immunostaining, antigen retrieval was performed using Unmasking Solution (Vector Laboratories, H-3300) for 10 min at around 100°C by microwave. 4-μm tissue sections were incubated with primary antibodies in the buffer of M.O.M. kit (Vector Laboratories, MKB-2213–1) overnight at 4°C, washed in PBS, and incubated with secondary antibodies conjugated with Alexa 488, 567, or 647 (ThermoScientific, 1:400) with DAPI for 1.5 h, and mounted with ProLong Gold antifade reagent (Invitrogen, P36930). The images were captured by a Zeiss confocal 710 microscopy.
Flow cytometry of chimeric mouse lung and liver for genotyping
Lung and liver tissues were minced with micro scissors, and 1 mL of pre-warmed dissociation buffer (1 mg/mL DNase (Sigma, DN25), 5 mg/mL collagen (Roche, 10103578001), and 15 U/ml Dispase II (Stemcell Technologies, 7913) in HBSS), incubated at 37°C on the rocker with 50 r.p.m. speed, and neutralized with the dissociation buffer by FACS buffer containing 2% FBS, Glutamax, 2mM EDTA and 10mM HEPES in HBSS after the 30 min incubation. After filtrating the cells with a 40-μm filter (FALCON, 352235), cell pellets were resuspended with 1 mL of cold RBC lysis buffer (Biolegend, 420301) to lyse the remaining erythrocytes for 5 min on ice and neutralized by 1 mL cold FACS buffer. After that, it was centrifuged them at 350 rcf, 4°C, for 3 min to remove the lysed blood cells. Cells were resuspended in 500 μL FACS buffer with PI for the subsequent analyses using SONY MA900. For the CBC, the genotyping of chimeric animals was confirmed by GFP-negative sorted liver cells and lung cells. For detecting the Fgfr2 floxed allele, we performed PCR using the primer sets: FR2-F1, 5′-ATAGGAGCAACAGGCGG-3′, and FR2-F2, 5′-CAAGAGGCGACCAGTCA-3’.
Blastocyst preparation and embryo transfer
Blastocysts were prepared by mating Foxa2Cre/Cre; Fgfr2flox/+, Foxa2Cre/+; Fgfr2flox/+ or ShhCre/+; Fgfr2flox/+ males (all 129 × B6 × CD-1 background) with superovulated Fgfr2flox/flox; Rosa26tdTomato/tdTomato females (129 × B6 × CD-1 background). Blastocysts were harvested at E3.5 after superovulation.13 Twenty sorted nGFP+ iPSCs were injected into each blastocyst. After the iPSC injection, blastocysts were cultured in an M2 medium (Cosmobio) for a few hours in a 37°C, 5% CO2 incubator for recovery. Then, blastocysts were transferred to the uterus of the pseudopregnant foster mother.
Quantification and statistical analysis
Data analysis was performed using R-studio. Data acquired by performing biological replicas of two or three independent experiments are presented as the mean ± SD. Statistical significance was determined using a student t-test. *p < 0.05, ns: non-significant.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-E-cadherin | Invitrogen | 131900 |
| Anti-tdTomato | Abcam | Orb182397 |
| Anti-aSMA | Sigma-Aldrich | Ab5694 |
| Anti-Foxa2 | Cell Signaling Technology | 8186S |
| Anti-Sox9 | Sigma-Aldrich | AB5535 |
| Anti-GFP | Aves lab | GFP1020 |
| Anti-Pan-CK | Sigma-Aldrich | C2562 |
| Anti-Mist1 | Cell Signaling Technology | 14896 |
| Anti-CK5 | Abcam | Ab52635 |
| Anti-CK14 | Abcam | Ab7800 |
| Anti-AQP5 | Alomone labs | AQP-005 |
| Donley anti Rabbit IgG (H + L), Alexa Fluor 488 | Invitrogen | A21206 |
| Donley anti Mouse IgG (H + L), Alexa Fluor 488 | Invitrogen | A21202 |
| Donley anti Chicken IgG (H + L), Alexa Fluor 488 | Jackson Immunoresearch | 703545155 |
| Donley anti Rabbit IgG (H + L), Alexa Fluor 568 | Invitrogen | A10042 |
| Donley anti Goat IgG (H + L), Alexa Fluor 568 | Invitrogen | A11057 |
| Donley anti Rabbit IgG (H + L), Alexa Fluor 647 | Invitrogen | A31572 |
| Donley anti Goat IgG (H + L), Alexa Fluor 647 | Invitrogen | A21447 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| nGFP+ iPSC | Miura et al.32 | N/A |
| PSC CAG-GFP | MTI-GlobalStem | GSC-5003 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Rosa26tdTomato/tdTomato mice | The Jackson Laboratory | 07914 |
| Rosa26nT-nG/nT-nG mice | The Jackson Laboratory | 023035 |
| Foxa2Cre/Cre mice | Horn et al.47 | From Dr. Nicole C Duboi |
| Fgfr2 flox/flox | Miura et al.32 | From Dr. X. Zhang |
| CD1 | Charles River | 022 |
| Shh Cre/+ mice | The Jackson Laboratory | 05622 |
Highlights.
Foxa2 lineage labels endoderm and ectoderm boundary region in oral epithelium (OE)
Fgfr2 is critical for salivary gland primordia formation but not OE invagination
Foxa2-driven Fgfr2 knockout mice display a salivary gland agenesis phenotype
PSC generates functional salivary glands via conditional blastocyst complementation
ACKNOWLEDGMENTS
We sincerely appreciate the considerate support and scientific input from Dr. Wellington Cardoso at the Columbia Center for Human Development (CCHD) and the members of Cardoso’s lab and CCHD. We acknowledge the support from the CCHD Medicine Microscopy core (MMC), the Columbia Stem Cell Initiative (CSCI) Flow Cytometry core (SONY MA900), and the Genetically Modified Mouse Model Shared Resource (GMMMSR) for blastocyst injection. We thank Kathryn K. Kennedy for checking the manuscript grammatically. We thank Dr. Heiko Lickert of the Technical University of Munich (TUM) for sharing Foxa2-Cre mice. This work was financially supported by NIH-NHLBI 1R01 HL148223-01, DoD PR190557, and PR191133, USA to M.M., JSPS21KK0290, and The Uehara Memorial Foundation, Japan to J.T.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114340.
REFERENCES
- 1.Atkinson JC, Grisius M, and Massey W (2005). Salivary hypofunction and xerostomia: diagnosis and treatment. Dent. Clin. North Am. 49, 309–326. [DOI] [PubMed] [Google Scholar]
- 2.Zhang C, Li Y, Zhang X-Y, Liu L, Tong H-Z, Han T-L, Li W-D, Jin X-L, Yin N-B, Song T, et al. (2017). Therapeutic potential of human minor salivary gland epithelial progenitor cells in liver regeneration. Sci. Rep. 7, 12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng J, van der Zwaag M, Stokman MA, van Os R, and Coppes RP (2009). Isolation and characterization of human salivary gland cells for stem cell transplantation to reduce radiation-induced hyposalivation. Radiother. Oncol. 92, 466–471. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan PP, Patel VN, Liu S, Harrington DA, Hoffman MP, Jia X, Witt RL, Farach-Carson MC, and Pradhan-Bhatt S (2017). Primary Salivary Human Stem/Progenitor Cells Undergo Microenvironment-Driven Acinar-Like Differentiation in Hyaluronate Hydrogel Culture. Stem Cells Transl. Med. 6, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pringle S, Maimets M, Van Der Zwaag M, Stokman MA, Van Gosliga D, Zwart E, Witjes MJH, De Haan G, Van Os R, and Coppes RP (2016). Human salivary gland stem cells functionally restore radiation damaged salivary glands. Stem Cell. 34, 640–652. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka J, Ogawa M, Hojo H, Kawashima Y, Mabuchi Y, Hata K, Nakamura S, Yasuhara R, Takamatsu K, Irié T, et al. (2018). Generation of orthotopically functional salivary gland from embryonic stem cells. Nat. Commun. 9, 4216. 10.1038/S41467-018-06469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka J, Senpuku H, Ogawa M, Yasuhara R, Ohnuma S, Takamatsu K, Watanabe T, Mabuchi Y, Nakamura S, Ishida S, et al. (2022). Human induced pluripotent stem cell-derived salivary gland organoids model SARS-CoV-2 infection and replication. Nat. Cell Biol. 24, 1595–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Lansford R, Stewart V, Young F, and Alt FW (1993). RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. USA 90, 4528–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamanaka S, Umino A, Sato H, Hayama T, Yanagida A, Mizuno N, Kobayashi T, Kasai M, Suchy FP, Yamazaki S, et al. (2018). Generation of Vascular Endothelial Cells and Hematopoietic Cells by Blastocyst Complementation. Stem Cell Rep. 11, 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee Y-S, Usui J-I, Knisely AS, et al. (2010). Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799. [DOI] [PubMed] [Google Scholar]
- 11.Usui J-I, Kobayashi T, Yamaguchi T, Knisely AS, Nishinakamura R, and Nakauchi H (2012). Generation of Kidney from Pluripotent Stem Cells via Blastocyst Complementation. Am. J. Pathol. 180, 2417–2426. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi T, Sato H, Kato-Itoh M, Goto T, Hara H, Sanbo M, Mizuno N, Kobayashi T, Yanagida A, Umino A, et al. (2017). Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196. [DOI] [PubMed] [Google Scholar]
- 13.Mori M, Furuhashi K, Danielsson JA, Hirata Y, Kakiuchi M, Lin C-S, Ohta M, Riccio P, Takahashi Y, Xu X, et al. (2019). Generation of functional lungs via conditional blastocyst complementation using pluripotent stem cells. Nat. Med. 25, 1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, and Hoffman MP (2005). FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development 132, 1223–1234. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman MP, Kidder BL, Steinberg ZL, Lakhani S, Ho S, Kleinman HK, and Larsen M (2002). Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF- dependent mechanisms. Development 129, 5767–5778. 10.1242/dev.00172. [DOI] [PubMed] [Google Scholar]
- 16.Chatzeli L, Gaete M, and Tucker AS (2017). Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development 144, 2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaskoll T, Abichaker G, Witcher D, Sala FG, Bellusci S, Hajihosseini MK, and Melnick M (2005). FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev. Biol. 5, 11. 10.1186/1471-213x-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arman E, Haffner-Krausz R, Gorivodsky M, and Lonai P (1999). Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc. Natl. Acad. Sci. USA 96, 11895–11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Langhe SP, Carraro G, Warburton D, Hajihosseini MK, and Bellusci S (2006). Levels of mesenchymal FGFR2 signaling modulate smooth muscle progenitor cell commitment in the lung. Dev. Biol. 299, 52–62. [DOI] [PubMed] [Google Scholar]
- 20.De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, and Dickson C (2000). An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483–492. [DOI] [PubMed] [Google Scholar]
- 21.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, and Kato S (1999). Fgf10 is essential for limb and lung formation. Nat. Genet. 21, 138–141. [DOI] [PubMed] [Google Scholar]
- 22.Rothova M, Thompson H, Lickert H, and Tucker AS (2012). Lineage tracing of the endoderm during oral development. Dev. Dyn. 241, 1183–1191. [DOI] [PubMed] [Google Scholar]
- 23.Szymaniak AD, Mi R, McCarthy SE, Gower AC, Reynolds TL, Mingueneau M, Kukuruzinska M, and Varelas X (2017). The Hippo pathway effector YAP is an essential regulator of ductal progenitor patterning in the mouse submandibular gland. Elife 6, e23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono-Minagi H, Nohno T, Serizawa T, Usami Y, Sakai T, Okano H, and Ohuchi H (2023). The Germinal Origin of Salivary and Lacrimal Glands and the Contributions of Neural Crest Cell-Derived Epithelium to Tissue Regeneration. Int. J. Mol. Sci. 24, 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.We R (1982). Evolution of dermal skeleton and dentition in vertebrates. Evol. Biol. 15, 287–368. [Google Scholar]
- 26.Yamagishi C, Yamagishi H, Maeda J, Tsuchihashi T, Ivey K, Hu T, and Srivastava D (2006). Sonic Hedgehog Is Essential for First Pharyngeal Arch Development. Pediatr. Res. 59, 349–354. [DOI] [PubMed] [Google Scholar]
- 27.Jaskoll T, Leo T, Witcher D, Ormestad M, Astorga J, Bringas P Jr., Carlsson P, and Melnick M (2004). Sonic hedgehog signaling plays an essential role during embryonic salivary gland epithelial branching morphogenesis. Dev. Dyn. 229, 722–732. [DOI] [PubMed] [Google Scholar]
- 28.Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisúa-Belmonte JC, and Rosenfeld MG (1999). Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282. [DOI] [PubMed] [Google Scholar]
- 29.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, et al. (1999). Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 126, 5749–5758. [DOI] [PubMed] [Google Scholar]
- 30.Gage PJ, Suh H, and Camper SA (1999). Dosage requirement of Pitx2 for development of multiple organs. Development 126, 4643–4651. [DOI] [PubMed] [Google Scholar]
- 31.Aure MH, Symonds JM, Villapudua CU, Dodge JT, Werner S, Knosp WM, and Hoffman MP (2023). FGFR2 is essential for salivary gland duct homeostasis and MAPK-dependent seromucous acinar cell differentiation. Nat. Commun. 14, 6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura A, Sarmah H, Tanaka J, Hwang Y, Sawada A, Shimamura Y, Otoshi T, Kondo Y, Fang Y, Shimizu D, et al. (2023). Conditional blastocyst complementation of a defective Foxa2 lineage efficiently promotes the generation of the whole lung. Elife 12, e86105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye Q, Bhojwani A, and Hu JK (2022). Understanding the development of oral epithelial organs through single cell transcriptomic analysis. Development 149, dev200539. 10.1242/dev.200539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisbruch A, Kim HM, Terrell JE, Marsh LH, Dawson LA, and Ship JA (2001). Xerostomia and its predictors following parotid-sparing irradiation of head-and-neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 50, 695–704. [DOI] [PubMed] [Google Scholar]
- 36.Cheng SCH, Wu VWC, Kwong DLW, and Ying MTC (2011). Assessment of post-radiotherapy salivary glands. Br. J. Radiol. 84, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dirix P, Nuyts S, and Van den Bogaert W (2006). Radiation-induced xerostomia in patients with head and neck cancer: a literature review. Cancer 107, 2525–2534. [DOI] [PubMed] [Google Scholar]
- 38.Jensen SB, and Vissink A (2014). Salivary gland dysfunction and xerostomia in Sjögren’s syndrome. Oral Maxillofac. Surg. Clin. North Am. 26, 35–53. [DOI] [PubMed] [Google Scholar]
- 39.Baer AN, and Walitt B (2017). Sjögren Syndrome and Other Causes of Sicca in Older Adults. Clin. Geriatr. Med. 33, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibson B, Periyakaruppiah K, Thornhill MH, Baker SR, and Robinson PG (2020). Measuring the symptomatic, physical, emotional and social impacts of dry mouth: A qualitative study. Gerodontology 37, 132–142. [DOI] [PubMed] [Google Scholar]
- 41.Ray AT, and Soriano P (2023). FGF signaling regulates salivary gland branching morphogenesis by modulating cell adhesion. Development 150, dev201293. 10.1242/dev.201293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura A, Sarmah H, Tanaka J, Hwang Y, Sawada A, Shimamura Y, Fang Y, Shimizu D, Ninish Z, Suer JL, et al. (2023). Conditional blastocyst complementation of a defective Foxa2 lineage efficiently promotes generation of the whole lung. Preprint at bioRxiv. 10.1101/2022.10.31.514628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kano M, Mizutani E, Homma S, Masaki H, and Nakauchi H (2022). Xenotransplantation and interspecies organogenesis: current status and issues. Front. Endocrinol. 13, 963282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng C, Ballard EB, and Wu J (2021). The road to generating transplantable organs: from blastocyst complementation to interspecies chimeras. Development 148, dev195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarmah H, Sawada A, Hwang Y, Miura A, Shimamura Y, Tanaka J, Yamada K, and Mori M (2023). Towards human organ generation using interspecies blastocyst complementation: Challenges and perspectives for therapy. Front. Cell Dev. Biol. 11, 1070560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukaibo T, Gao X, Yang N-Y, Oei MS, Nakamoto T, and Melvin JE (2019). Sexual dimorphisms in the transcriptomes of murine salivary glands. FEBS Open Bio 9, 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horn S, Kobberup S, Jørgensen MC, Kalisz M, Klein T, Kageyama R, Gegg M, Lickert H, Lindner J, Magnuson MA, et al. (2012). Mind bomb 1 is required for pancreatic β-cell formation. Proc. Natl. Acad. Sci. USA 109, 7356–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


