Abstract
Multiple accidents in nuclear power plants and the growing concerns about the misuse of radiation exposure in warfare have called for the rapid determination of absorbed radiation doses (RDs). The latest findings about circulating microRNA (miRNAs) using several animal models revealed considerable promises, although translating this knowledge to clinics remains a major challenge. To address this issue, we randomly divided 36 nonhuman primates (NHPs) into six groups and exposed these groups to six different radiation doses ranging from 6.0–8.5 Gy in increments of 0.5 Gy. Serum samples were collected pre-irradiation as well as three post-irradiation timepoints, namely 1, 2 and 6 days post-total body irradiation (TBI). Generated from a deep sequencing platform, the miRNA reads were multi-variate analyzed to find the differentially expressed putative biomarkers that were linked to RDs, time since irradiation (TSI) and sex. To increase these biomarkers’ translational potential, we aligned the NHP-miRNAs’ sequences and their functional responses to humans following an in-silico routine. Those miRNAs, which were sequentially and functionally conserved between NHPs and humans, were down selected for further analysis. A linear regression model identified miRNA markers that were consistently regulated with increasing RD but independent TSI. Likewise, a set of potential TSI-markers were identified that consistently shifted with increasing TSI, but independent of RD. Additional molecular analysis found a considerable gender bias in the low-ranges of doses when the risk to radiation-induced fatality was low. Bionetworks linked to cell quantity and cell invasion were significantly altered between the survivors and decedents. Using these biomarkers, an assay could be developed to retrospectively determine the RD and TSI with high translational potential. Ultimately, this knowledge can lead to precise and personalized medicine.
Introduction
Radiological or nuclear events caused by the deliberate or inadvertent release of radiation can have life-threatening consequences. Identifying solutions for nuclear or radiological threats due to terrorist action, accident or military conflict remain a priority for governments around the globe, since such wide radiation exposure can have mass-scale catastrophic consequences to the civilian and military communities [1, 2]. Retrospective physical and biological dosimetry techniques that are currently available include electron paramagnetic resonance, DNA damage profiling, thermoluminescence, dicentrics, chromosomal translocations, premature chromosomal condensation, hematology as well as metabolite and protein biomarkers [3–9]. However, these assays do not provide the required high throughput capability needed to screen a large number of individuals during any mass casualty scenario.
In this context, the molecular phenotyping technologies may be highly useful for the development of minimally invasive biomarker assays for rapid assessment of radiation exposure victims. Biomarkers could be instrumental to inform the impact of specific doses of radiation on biological systems. Such strategy may lead to precise and effective treatments for exposed individuals at risk. Hence, there is a significant need for biomarker identification, validation, and qualification that could assist in the rapid evaluation of absorbed radiation doses and the related health risks [6, 10]. Additionally, radiation biomarkers are also helpful for investigating countermeasure efficacy which can be used to rescale the drug doses from animal models to humans [11]. Using various omics platforms, several metabolites, lipids, proteins, and miRNA molecules have been studied as biomarkers for radiation exposure during the last two decades. These biomarkers have been mostly studied in murine models and have used a small range of radiation doses (RDs) [6, 12, 13], and these findings have been validated using large animal models, albeit to a limited extent [5, 14–16].
MicroRNAs (miRNAs), which are 18–30 nucleotides-long noncoding moieties, control gene regulations via degenerating mRNA and suppression of protein translation [17]. By current estimation, miRNA regulates 30% of the entire human genome [18] and thereby controls a vast spectrum of biofunctions. Recent discoveries threw light on miRNAs’ role in epigenetic activities [19] that could have chronic implications, even heritable consequences. There is potentially a feedback loop between miRNA and other epigenetic mechanisms, including DNA methylation and histone modifications [19, 20]. Such widespread regulatory activities of miRNA underlines its value as diagnostic, therapeutic, and prognostic biomarkers [21]. Measuring the expression levels of cell free circulating miRNA from plasma or serum has several logistic advantages [22]. The miRNA detection process could be rapid, namely a few hours at maximum; thus, miRNA assays could provide a significant time advantage in comparison to current technologies to detect radiation exposure [3–9, 23]. Indeed, promoting miRNAs as potential biomarkers of radiation injury has been gaining significant deliberations in recent years [24–29]. Focusing on animal model-based miRNA studies, a meta-analysis concluded that miRNA could be a promising species-independent candidate to triage irradiated subjects based on the extent of injury [30].
Recent advancements in the high throughput omics approach [31] coupled with increased application of Artificial Intelligence (AI) and Machine Learning (ML)-empowered algorithm has given an unprecedented boost towards biomarker discovery [32]. Using a similar approach, a clinical study probed 16 patients and presented a panel of three serum miRNAs to determine radiation exposure [33]. Similarly, a panel of two serum miRNAs was constructed to diagnose radiotoxicity during the course of radiotherapy [34]; in addition, a fourteen-serum miRNA panel was suggested by an independent study to aid in determining RDs for cancer patients [35]. By fitting the miRNA expression values into a logistic regression curve, the variable health risks of radiation exposure from three different sources was estimated in a mouse model [36]. Clearly, the cell free circulating miRNAs have emerged as great candidates for dose assessment. However, a general concern remains about the translational potential of in vivo knowledge [37].
To address this concern, the current study has been built upon two of our previous publications. In the first of the two previous publications [38], we developed an in silico pipeline to identify phylogenetically conserved miRNAs linked to radiation exposure [38] and this pipeline was reused in the current work. The delivered miRNAs are not only sequentially conserved between NHPs and humans, but these miRNAs would likely to display similar responses to irradiation in both NHPs and humans. The second publication of interest reported a combination of mouse [27] and NHP [39] models and a cross-species interrogation identified seven evolutionary conserved miRNAs, of which a five-miRNA panel was selected as the best candidate to predict radiation-induced fatality [28]. This result essentially justified the current approach, where we used a wide range of radiation doses and a novel in silico approach to find conserved miRNA markers.
Taking advantages from these two studies, we explored a wide range of radiation doses to determine the miRNA markers linked to total-body irradiation (TBI). We present putative markers linked to radiation dose, time since irradiation (TSI), and a panel of miRNAs that could potentially predict the risk to radiation-induced fatality (RRiF), respectively. These biomarkers are expected to be species independent since the seeding pool of miRNAs was sequentially and functionally conserved between NHPs and humans.
Materials and methods
Experimental design
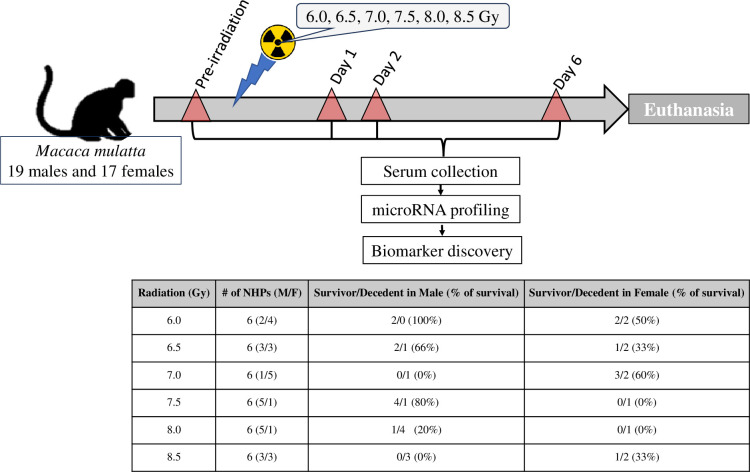
For this study, a total of 36 animals of both sexes were evenly distributed in six different groups. Animals in each group were exposed to one of six different 60Co γ-radiation doses (6.0, 6.5, 7.0, 7.5, 8.0 or 8.5 Gy) [40]. Though these animals were observed for 60 days post-irradiation to score survivors, blood samples for microRNA analysis were collected pre-irradiation and on days 1, 2 and 6 post-irradiation (Fig 1). All 21 animals that moribundity criteria for early euthanasia displayed radiation-exposure related injury.
Fig 1. Study design.
The flow diagram shows the longitudinal plan for collecting blood samples for circulating miRNA pre- and post-irradiation from NHPs. The table displays the sample size (M/F) and the survival outcome.
Animals
Naïve rhesus macaques (Macaca mulatta, Chinese sub-strain, 19 males and 17 females) 2.9–7.3 years of age weighing 3.6–7.4 kg, were acquired from the National Institutes of Health Animal Center (NIHAC, Poolesville, MD, USA) and maintained in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-International. Animals were quarantined for six weeks prior to the initiation of the study. Animal quarantine, exclusion criteria, housing, health monitoring, care, and enrichment during the experimental period have been described in detail earlier [39, 41]. In brief, all NHPs were individually housed in stainless steel cages in environmentally controlled rooms maintained at 22°C ± 2°C, 30–70% relative humidity, 10–15 air change cycles per h, and 12 h light:12 h dark cycle. Animals were fed primate diet (Harlan Teklad T.2050 diet, Madison, WI, USA) twice daily and they received drinking water ad libitum. All the animals received enrichment food once a day Monday–Friday. They received mirrors, toys, and challenge balls for enrichment. TVs were used for sensory enrichment for 4–5 h at least 3 times a week. All animals were able to see, hear and/or touch the conspecifics through the cages. All animals were randomly assigned to one of six different irradiation groups. All procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the Armed Forces Radiobiology Research Institute (AFRRI) Protocol #2015-12-011 and University of Maryland Baltimore Protocol #0581005, as well as the Department of Defense Animal Care and Use Review Office (ACURO). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals [42]. This study used full supportive care including whole blood transfusion, although all samples for this study were collected prior to any blood transfusion to animals [16]. This study was carried out in compliance with the ARRIVE guidelines.
Irradiation
The irradiation procedure and dosimetry have been previously described [43]. In brief, two NHPs were placed on the irradiation platform facing away from each other and were exposed to a specific midline dose of 60Co gamma-radiation at a dose rate of 0.6 Gy/min (bilateral, simultaneous exposure). The radiation field in the area of the NHP location was uniform within ± 1.5%. The dosimetry for photons was based on the alanine/EPR (electron paramagnetic resonance) dosimetry system [44]. Dose measurements and calibrations (EMXmicro spectrometer, Bruker Corp., Billerica, MA, USA) were based on the alanine/electron paramagnetic resonance system [44, 45]. The alanine dosimeters obtained from the National Institute of Standards and Technology (NIST, Gaithersburg, MD, USA) were calibrated using methods previously discussed [41]. Animals were mildly sedated for the procedure. To deliver the precise radiation dose, NHPs’ abdominal widths were measured with digital calipers. Animals were observed throughout the irradiation procedure via in-room cameras [46]. Following irradiation, animals were returned to the transport cart and brought to their cages in the housing area and were closely monitored until they were able to perch within their cage.
Cage-side animal observations
All NHPs were observed pre-irradiation and for 60 d post-irradiation with survival as the primary endpoint. Daily observations for signs of pain and distress were made no less than twice a day by trained staff for executing NHP procedures and observation. During the critical period (approximately 10 to 20 d post-irradiation), animals were observed three times a day for signs of radiation sickness or need for further medical intervention. The animals were evaluated for the following parameters at least three times a week: weight, body temperature, fecal consistency, respiratory rate, heart rate, and overall health assessment [39].
Euthanasia
Euthanasia was conducted in accordance with the most recently approved versions of the IACUC protocol, the Guide, and American Veterinary Medical Association (AVMA) guidelines [42, 47]. When an animal reached a state of moribundity, the animal was immediately euthanized. Moribundity was used as a surrogate for mortality, and animals were euthanized in order to minimize pain and distress [39]. Various parameters were used as guidelines for moribundity, which have been previously listed [39]. Moribundity status of each animal was determined by a joint effort between the institutional veterinarian, principal investigator, research staff, veterinary technicians, and husbandry staff based on the combination of criteria described above. To initiate euthanasia, the animals were sedated using ketamine hydrochloride injection (Mylan Institutional LLC, Rockford, IL) (5–15 mg/kg, im), and then administered pentobarbital sodium iv (Virbac AH Inc., Fort Worth, TX) using either saphenous or cephalic veins, needle size 20–25 gauge (100 mg/kg, 1–5 ml). Intra-cardiac administration was performed if unable to administer pentobarbital sodium through peripheral veins. The animals were deeply anesthetized by Isoflurane (Baxter Healthcare Corporation, Deerfield, IL) (1–5%) with oxygen at 1–4 liters per minute via mask before administering the intra-cardiac injection. The animals were euthanized only under the guidance of a staff veterinarian or a trained technician in consultation with the veterinarian. After pentobarbital sodium administration, the animals were examined by assessing the heart auscultation and pulse to confirm death. At the end of the study (60 d post-irradiation), all surviving animals were sent to another facility for studying late and delayed effects of radiation exposure.
Blood collection and serum separation
On the day of a scheduled sample collection, animals were restrained using the pole-and-collar method and were guided to a restraint chair for blood collection. The blood draw was conducted between 08:00 AM and 10:00 AM, 1–3 h after animals were fed. Blood samples were collected from the saphenous vein of the lower leg after the site was cleaned using a 70% isopropyl alcohol wipe and dried with sterile gauze [48]. A 3 ml disposable luer-lock syringe with a 25-gauge needle was used to collect the desired volume of blood. For serum collection, blood samples were transferred to vacutainer serum separator tubes (Becton, Dickinson Corp, Franklin Lakes, NJ, USA), allowed to clot for 30 min, and then centrifuged for 10 minutes at 400 x g. Serum samples were then stored at -70°C for analysis at a later date [49]. As stated above, for miRNA analysis, samples were collected on days -4 pre-irradiation and days 1, 2, and 6 post-irradiation.
miRNA assay
The assay protocol that we have reported elsewhere [50] was adapted herein with necessary modifications. We used TruSeq small RNA sample preparation kit to construct the sequencing library from 5 μL of NHP serum samples, as the 3′ and 5′ adapters were ligated to small RNA molecules, and the ligated products were reverse transcribed and amplified. The sample library was gel purified and size selected to curate small RNAs that were less than 30 bases; subsequently, the library was quantified by TapeStation (QIAGEN, Inc.). Based on the sample concentrations, equimolar amounts of small RNA-derived libraries were processed in Illumina NextSeq 500 platform to generate 5 million reads for miRNA profiling [29]. Illumina-generated Base Call (BCL) files were converted to FastQ files and electronically demultiplexed using bcl2fastq2 v2.20 software (Illumina Inc., San Diego, CA, USA). Genomics Workbench 20.0.4 software (QIAGEN, Germany) was used for analysis of the samples and to enumerate the miRNA expression values. FastQ file reads were first filtered to reject the poor reads then the screened reads from different lanes were conjoined. The trimming was performed using a score of 0.05 with the maximum number of ambiguities equaled to 2. We used a custom generated trim adaptor list containing an Illumina small adapter, as the allowed length of sequences were between 19 and 25 with a maximum mismatch of 2. Then, miRNA expression counts were determined using miRbase Release 22.1 with reference for NHP gene assembly. Read counts of mature miRNA were then normalized to CPM values using the trimmed mean of the M values (TMM) normalization method. Mature miRNA expression values were then subjected to differential analysis.
The TMM normalized data was first prefiltered to remove miRNAs with < 40% non-zero reads across all samples. Differential expression (DE) analysis was then performed by calculating the logfold2 change values using the pre-irradiation samples as the baseline. Preliminary analysis used four major co-factors of this study: (i) RDs (6.0 Gy, 6.5 Gy, 7.0 Gy, 7.5 Gy, 8.0 Gy and 8.5 Gy); (ii) TSI, namely pre-irradiation, day 1, day 2 and day 6 post-irradiation, respectively; (iii) male and female sex; and (iv) RRiF, or animals that survived until 45 days post-irradiation were reported as survived.
Four-way ANOVA was computed to find differentially expressed miRNAs linked to 15 dependent and independent variables at the cut-off of p<0.05. The list included four independent variables, namely RD, TSI, Sex and RRiF, and the corresponding 11 dependent variables, where multiple variables were connected via *; the list was the following: RD*TSI, RD*Sex, RD*RRiF, TSI*Sex, TSI*RRiF, Sex* RRiF, RD*TSI* RRiF, RD*Sex* RRiF, RD*TSI* RRiF, TSI*Sex* RRiF and RD*TSI*Sex* RRiF.
We used the RNASeqPower Bioconductor package [51] to determine whether the sample size of 36 was sufficient to detect the differences corresponding to all of the multiple variables listed above. The current assay is expected to generate 400 million reads (length 50 bases per read) on a NextEeq500 platform. Given that NHP have nearly 3,100 million bps in its whole reference genome, we assumed to meet a 6.45x coverage in our assay. To measure the statistical power for the ANOVA analysis, the effect sizes (η2) of all combinations of variables were computed, and the mean effect size and its coefficient of variation (CV) was determined as 0.21, and 1.7, respectively. With an alpha value at 0.05, the power of the study was expected to be greater than 80%, which was sufficient to proceed with further analysis.
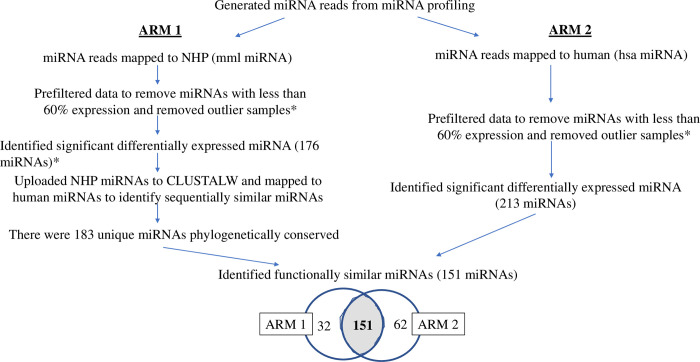
Subsequent analysis was focused on curating those DE mml-miRNAs, which would be sequentially conserved and functionally similar between humans and NHPs exposed to TBI (Fig 2). We described this process elsewhere in the context of a minipig model [38]. Briefly, we pooled the DE mml-miRNAs that were selected by 4-way ANOVA, namely 176 miRNAs after removing the duplicates. These miRNAs were seeded into the multiple-sequence alignment tool, ClustlW [52], to curate sequentially conserved miRNAs between humans and NHPs. FASTA sequences of all mature miRNA sequences were retrieved from miRBase. A pairwise sequence alignment between DE mml-miRNAs and the retrieved human miRNAs was conducted with specified loci positions to meet the following parameters: the k-tuple word size 1 and window size 5 with top diagonals of 5 with percent method. Gap penalty of 10 and penalty of extension 0.1 with a BLOSUM scoring weight matrix were applied. Scores were calculated using the absolute and percent method and the top match(es) for each NHP miRNA was determined. As a result, we identified a set of mml-miRNAs, which were sequential homologues to the known human miRNAs. We named this set of miRNAs as sequentially conserved-miRNAor sc-miRNA.
Fig 2. A flow chart to depict the process to deliver miRNAs that were not only sequential homologues between humans and NHPs, but were also likely to respond similarly to irradiation across these two species.
In parallel, we employed miRBase to align the quality filtered and adaptor-trimmed reads to the human genome assembly, hg38. Thereby, we followed the previously described analysis pipeline with one important difference, namely the miRDeep2 core algorithm in miRBase curated known hsa-miRNAs, not mml-miRNA. The resultant hsa-miRNA matrix was quality filtered and the counts were normalized to generate hsa-miRNA sets. Four-way ANOVA with the cut-off set at p < 0.05 found a set of miRNAs that met the cut-off in at least one of the 15 feature selection criteria as mentioned earlier. We named this set of hsa-miRNAs as functionally similar-miRNA or fs-miRNA. A subsequent Venn approach identified those miRNAs that were conserved between sc-miRNA and fs-miRNA. We named these overlapping features as conserved-miRNA (cnvd-miRNA). Henceforth, all statistical analysis was computed using this cnvd-miRNAs. The raw data is saved in GEO database, GSE247225.
Statistical analysis
R Studio was used to generate principal component analysis (PCA) plots and carry out hierarchical clustering computed by Euclidian distance analysis using heatmap2 function from the gplots. The time and dose dependent miRNA discovery was computed using GraphPad v8 (Prism, Inc.). A linear regression plot was drawn to measure if the slope significantly deviates from zero and the cut-off was set at p < 0.05.
Ingenuity Pathway Analysis (IPA, QIAGEN, Inc., version 01–13) was used for functional analysis and bionetwork building. Differentially expressed miRNAs of the following five groups were of particular interest: (i) RD*TSI, (ii) Sex*RD*TSI, (iii) RRiF, (iv) RRiF*RD, and (v) Sex*RRiF*RD. DE miRNAs from these groups were transferred to the IPA analysis portal, and the canonical and non-canonical bionetworks that were significantly enriched at each of the five groups were curated (hypergeometric test, p <0.05). Henceforth, all the canonical and noncanonical bionetworks were together termed as bionetworks. Those bionetworks that displayed a Z score of greater than 1 (≥1) were considered activated bionetworks. In contrast, those bionetworks, which displayed Z scores of less than 1 (≤ -1) were considered inhibited bionetworks. In any given group, if a bionetwork’s Z score was greater than |1| in at least one instance, that bionetwork was curated for further interrogation. The correlation matrix was computed using the Pearson correlation formula.
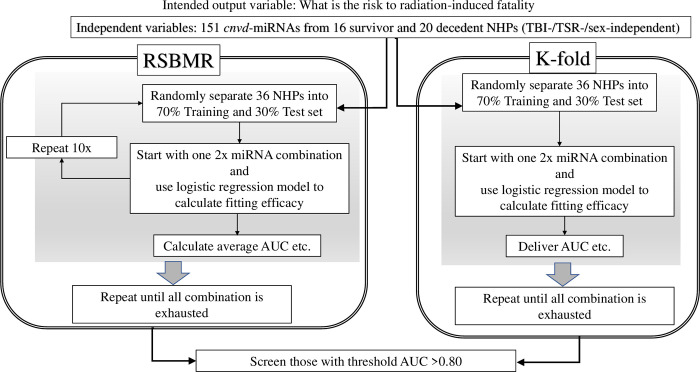
To discover the panel of miRNAs that can potentially predict RRiF, we used our proprietary machine learning routine named the Biomarker Discovery Process at Binomial Decision Point (2BDP) [53], which is trained to systematically incorporate independent variables (e.g., multiple DE miRNAs) to explain an output variable (e.g., RRiF) that is binary in nature. As depicted in Fig 3, 2BDP operated on the cnvd-miRNAs via an exhaustive search to generate the best fitting logistic regression model as described in Eq 1.
Fig 3. A flow diagram to explain the algorithm named 2BDP (Biomarker Discovery Process at Binomial Decision Point) that we used to find the best fitting model to predict RRiF.
| (Eq 1) |
Where logit() is the log odds function of a value, P is the probability of successful determination of the outcome variable, a is the intercept of the equation, b through n are coefficient estimates of the independent variables, and X1 through Xn are the expression values of the independent variables 1 to n, respectively. The fitting criteria of these independent variables were measured by multiple R2, adjusted R2, confidence intervals (CIs) and p values (Chi-square). For the present purpose, the fitting models allowed the independent variables to change from 2 to 10 (2≤ n ≤ 10) per panel.
Thereby, 2BDP algorithm measured the efficacy of the individual biomarker panel in determining the outcome variables or RRiF. The panel contained a group of independent features that were linearly associated via unique weight factors or coefficients. The efficacy of the individual panel was quantitively measured by the area under the curve (AUC) of the receiver operating characteristic (ROC) curve. We used two methods to measure the ROC curve.
Method 1. Random Single Bin Multiple Repeats (RSBMR): The independent features from cnvd-miRNAs were used in a random forest 2000 times, where the top 200 panels of most frequent genes were selected. For each panel, every combination of genes, from 2 to 10 feature sized panels, constructed a series of unique subpanels. Hence, the maximum count of features in one panel should not exceed 10. Next, the whole NHP cohort all-inclusive on sex, RD and TSI was randomly sorted into Training and Test set by the 70:30 ratio. A unique panel of independent features was fitted onto the Training set to construct a linear regression model as shown in Eq 1. Afterwards, this linear regression model was operated on the Test set to calculate the AUC and sensitivity/specificity. These deliverables were retained while the exhausted Training and Test sets were replaced by new Test and Training sets. This cycle was repeated 10 times for each subpanel and finally, the mean values of AUCs and sensitivity/specificity were calculated over these 10 iterations and were then reported. This iterative process continued until all features in cnvd-miRNAs were exhausted.
Method 2. k-fold method: The independent features from cnvd-miRNAs were used in a random forest 2000 times, where the top 200 panels of most frequent genes were selected. For each panel, every combination of genes, from 2 to 10 feature sized panels, constructed a series of unique subpanels. For any given unique panel, the entire cohort was segregated into 10 (k = 10) groups, and one randomly selected group was chosen as the Test set. The remaining groups were taken as Training sets, where the unique panel was fitted to construct a linear regression model as shown in Eq 1. Next, the AUC was calculated using the Test set and was reported. This iterative process continued until all features in cnvd-miRNAs were exhausted. Finally, all panels curated by RSBMR and k-fold methods were screened to find those which had a significantly high fitting score, p < 0.05 and (b) AUC > 0.80, respectively.
Results
Thirty-six NHPs were randomly assigned to six groups and each group was exposed to different RDs starting from 6.0 Gy to 8.5 Gy in 0.5 Gy increments (Fig 1). Each group consisted of 6 animals with variable male to female ratios. A table attached to Fig 1 displays the number of animals that survived from each radiation dose.
Cross-species conserved miRNA
Sequencing reads were mapped to 3,127 known miRNAs in the NHP genome assembly, and a total of 1,327 known miRNAs were detected in at least one of the 144 samples. On average, 364.6 ± 0.46 miRNAs were detected per sample, where the error was calculated by standard error of the mean (SEM). After filtering out the features with more than 60% samples, 1,040 miRNAs became eligible for multivariance analysis. S1 Fig depicts the PCA plots of these miRNAs. The plot explained 22.09% of total variance, but no apparent trend emerged in this plot. S1A Table lists the number of DE miRNAs curated by 15 dependent and independent variables. We combined all these miRNAs and removed the duplicates down to select 176 miRNAs (S1B Table), which were significantly altered in at least one of the 15 comparative analyses computed by the 4-way ANOVA model. These 176 miRNAs were further processed through multiple sequence alignments between human and NHP miRNA assembly to deliver miRNAs of high translational potential (Fig 2). Briefly, we first identified 183 miRNAs that are sequential homologues between humans and NHPs using a multiple sequence alignment routine [52] and named this miRNA panel as sequentially conserved-miRNA (sc-miRNA). In a parallel pipeline, the quality controlled NextSeq reads were directly blasted to human genome assembly, and differential expression analysis was performed to curate 213 significantly different hsa-miRNAs from the pre-TBI baseline. We labeled this second set as fs-miRNA. Ultimately, 183 sc-miRNAs and 213 fs-miRNAs were overlapped to find 151 miRNAs that were common between these two sets, which was named cnvd-miRNA (S1C Table). Subsequent analysis of this study used this cnvd-miRNA panel, since we posit that these miRNAs are not only sequentially conserved between humans and NHPs, but also that their functional response to TBI and TSI are expected to be similar. Hence, these cnvd-miRNAs had considerable translational potential.
miRNAs markers showing linear response to dose and time
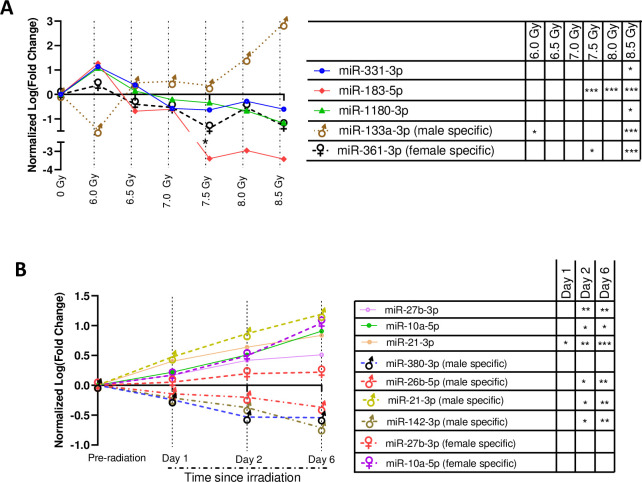
A linear regression model-based screening of 151 cnvd-miRNAs curated five miRNAs showing consistent and significant shifts across doses, but independent of time (Fig 4A). The features of Fig 4A was curated from the sex*RD ANOVA analysis, where TSI was precluded as a dependent variable. Three of these 5 miRNAs, namely miR-331-3p (F = 7.26, p = 0.04), miR-183-5p (F = 22.13, p = 0.009) and miR-1180-3p (F = 52.7, p = 0.001) emerged as sex-independent markers, as they gradually decreased with increasing dose. Here, the F values and p-values characterized the deviations of their slopes across doses from baseline. One miRNA, specifically miR-183-5p, was significantly inhibited from the baseline at the three highest doses of radiation (namely 7.5 Gy, 8.0 Gy and 8.5 Gy), and displayed significant separation of the entire dose range with an AUC ROC curve of 0.95. Its 95% confidence interval (CI) ranged from 0.84 to 1.00. Another miRNA, miR-133a (F = 11.42, p = 0.002), was a male-specific marker that significantly increased with increasing RD; contrastingly, miR-361-3p (F = 9.52, p = 0.027) was a female-specific marker that significantly decreased with increasing RD. All 5 miRNAs were significantly altered from the pre-irradiation baseline at 8.5 Gy, irrespective of TSI. However, none of these miRNAs showed a significant shift from the baseline at the lowest dose, namely 6.0 Gy.
Fig 4. Putative miRNA markers of irradiation.
All miRNAs sequentially and functionally conserved between humans and NHPs. (A) Time since irradiation (TSI)-independent, radiation dose (RD)-miRNA markers. Here, the x-axis and y-axis represented the radiation dose and normalized log2 (Fold change), respectively. The fold changes were computed on the pre-irradiation baseline controls. All these miRNAs displayed a consistent shift (p < 0.05 in linear regression curve) across dosimetry independent of TSI. * p < 0.05; *** p < 0.001. (B) RD-independent, TSI-miRNA markers. Here, the x-axis and y-axis represented the TSI and normalized log2 (Fold change), respectively. The fold changes were computed on the pre-irradiation baseline controls. All these miRNAs displayed consistent shift (p < 0.05 in linear regression curve) across TSI. * p<0.05; ** p<0.05; *** p<0.001.
Fig 4B displays 11 miRNA that consistently shifted across time or TSI. These features of Fig 4B was curated from the sex*TSYI ANOVA analysis, where RD was precluded as a dependent variable. There were 3 TSI-markers, namely miR-27b-3p (F = 74.62, p = 0.013), miR-21-3p (F = 66.84, p = 0.015) and miR-10a-3p (F = 11.9, p = 0.009), that were significantly upregulated with increasing time. One miRNA, miR-21-3p, was significantly altered from the baseline across the entire TSI profile and displayed a separation of the group with an AUC of 0.72, and its 95% CI ranged from 0.33 to 1.00. In addition, there were 5 male specific and 3 female specific miRNAs that consistently shifted their regulations with increasing time. To note, miR-21-3p emerged as both a gender-inclusive and male-exclusive marker. Indeed, miR-21-3p (F = 281.6, p = 0.003) was the only candidate that was significantly upregulated starting from day 1 post-TBI. Three candidates including miR-26b-5p (F = 68.38, p = 0.014), miR-125b-2-3p (F = 69.21, p = 0.014) and miR-142-3p (F = 80.68, p = 0.122) were consistently downregulated with increasing time, while the remaining were increasingly upregulated with time.
RD- and TSI-specific bionetworks
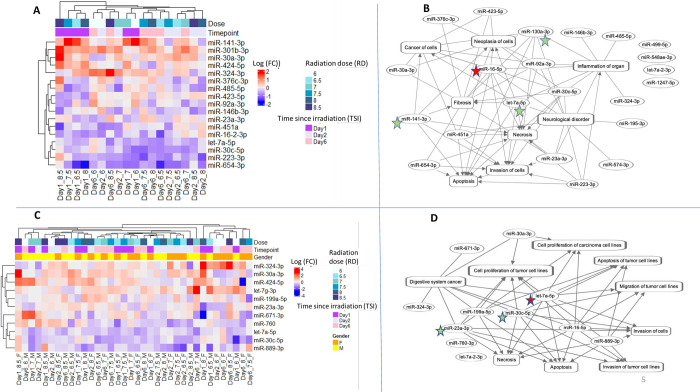
There were 17 miRNAs that were significantly altered by RD*TSI; furthermore, these 17 miRNAs were a subset of cnvd-miRNAs as described in Fig 2. Fig 5A depicted a hierarchical cluster of these 17 miRNAs, and Fig 5B displayed a functional network using these 17 miRNAs to reveal their biological interrelationships. One miRNA, miR-165p, emerged as the most interconnected candidate, followed by miR-130a-3p, miR-7a-3p, and miR-141-3p. Together, these four miRNAs could be a set hub of molecules or key regulators of the functions associated with RD*TSI.
Fig 5. Functional association among the differentially expressed miRNA cluster linked to RD and TSI.
The color keys of log2 (Fold change) were added at top left corners. In the networks, the oval and rectangular shaped nodes represent miRNA and biofunctions, respectively. The edges represent the relationship between two nodes: the solid lines represent their association, and pointed arrowheads denote the activating relationship between the two nodes. Of these, miRNAs that appeared most connected to the neighbors were annotated in the red colored stars for > 90% percentile, and green colored stars for > 75% percentile. (A) Hierarchical cluster of 17 miRNAs that were differentially expressed by the cumulative effects of RD and TSI. These miRNAs are sex-independent markers. (B) Network cluster of the same 17 miRNAs that were differentially expressed by RD*TSI. (C) Hierarchical cluster of 11 miRNAs that were differentially expressed by the cumulative effects of Sex, RD and TSI. (D) Network cluster of the same 11 miRNAs that were differentially expressed by Sex*RD*TSI.
Functional enrichment analysis (Tables 1 and 2) found 11 bionetworks that were significantly regulated due to the dependent effects of RD*TSI; of which the bionetwork linked to invasion of tumor cells was the most enriched function. Bionetworks linked to apoptosis and cell migration emerged as other key candidates that were perturbed by RD*TSI. S2 Table lists the miRNAs linked to all 3 of these bionetworks.
Table 1. List of bionetworks enriched by a subset of cnvd-miRNA: Bionetworks significantly altered by the cumulative effects of RD and TSI (RD*TSI).
| Day 1 | Day 2 | Day 6 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diseases and Bio Functions | 6.0 Gy | 6.5 Gy | 7.0 Gy | 7.5 Gy | 8.0 Gy | 8.5 Gy | 6.0 Gy | 6.5 Gy | 7.0 Gy | 7.5 Gy | 8.0 Gy | 8.5 Gy | 6.0 Gy | 6.5 Gy | 7.0 Gy | 7.5 Gy | 8.0 Gy | 8.5 Gy |
| Invasion of tumor cell lines | -1.04 | -0.47 | -1.04 | 1.5 | 0.39 | -0.33 | 1.16 | 0.64 | 2.65 | 0.28 | -1.04 | -0.81 | -1.96 | -1.16 | 0.64 | 2.65 | 1.79 | 1.88 |
| Invasion of cells | -0.56 | -0.83 | -1.35 | 1.77 | 0.75 | -0.7 | 1.46 | 0.98 | 2.83 | -0.14 | -0.56 | -1.14 | -1.4 | -0.67 | 0.19 | 2.04 | 1.25 | 2.12 |
| Apoptosis of tumor cell lines | 1.43 | 0.66 | 0.66 | -0.2 | 1.14 | 0.18 | -0.09 | 0.66 | -1.24 | -1.82 | 1.43 | 0.66 | 2.01 | 1.43 | -0.11 | -2.01 | -1.14 | -0.47 |
| Migration of tumor cell lines | 0.22 | 1.21 | 1.12 | 1.03 | -0.4 | -0.76 | -1.39 | 1.03 | 0.05 | 1.66 | 0.22 | 2.01 | 0.31 | 0.58 | 1.93 | 0.4 | 0.4 | 0.05 |
| Cell proliferation of tumor cell lines | -1.12 | 0.47 | -0.81 | 0.13 | 0.22 | -0.73 | 0.2 | -0.49 | 1.4 | 1.14 | -1.12 | 0.5 | -1.04 | -0.73 | 0.23 | 1.71 | 1.09 | 1.61 |
| Apoptosis | 0.94 | 0.94 | 0.2 | 0.67 | 1.4 | -0.07 | 0.35 | 1.5 | -0.2 | -2.05 | 0.94 | 1.5 | 1.68 | 1.68 | 0.76 | -0.94 | -0.11 | 0.53 |
| Migration of cells | 0.37 | 1.34 | 1.25 | 0.63 | -0.82 | -1.14 | -0.99 | 0.63 | -0.06 | 2.02 | 0.37 | 2.1 | 0.37 | 0.63 | 2.02 | 0.82 | 0.31 | 0.46 |
| Cell viability of tumor cell lines | -1.39 | -1.39 | -1.39 | -0.28 | -1.94 | -0.83 | 0.28 | -0.28 | 0.83 | 0.83 | -1.39 | -0.28 | -1.94 | -0.83 | -0.28 | 0.83 | 0.83 | -0.28 |
| Cell proliferation of colorectal cancer cell lines | -0.18 | 0.43 | -0.93 | 0.43 | -0.58 | -0.58 | 0.18 | 0.43 | 1.18 | -0.18 | -0.18 | 0.43 | 0.18 | -0.58 | 0.43 | 1.18 | 1.18 | 1.18 |
| Metastasis | -0.19 | 0.75 | 0.75 | -0.19 | -0.19 | 0.75 | 0.75 | -0.19 | 0.75 | 0.19 | -0.19 | -0.75 | -0.19 | -1.69 | 0.75 | 1.69 | 1.69 | 0.75 |
| Cell proliferation of carcinoma cell lines | -0.63 | -0.63 | -0.63 | -0.16 | -0.16 | -1.41 | 0.63 | -0.16 | 0.63 | 0.16 | -0.63 | 0.63 | -0.63 | -0.94 | 0.63 | 0.63 | 0.63 | 0.63 |
Bionetworks with a Z score of ≥ 1 and ≤ -1 were considered activated and inhibited networks, respectively. The tables included those bionetworks, which scored ≥ |1| in at least one of all tested conditions.
Table 2. List of bionetworks enriched by a subset of cnvd-miRNA: Bionetworks significantly altered by the cumulative effects of sex, RD and TSI (sex*RD*TSI).
| Male | ||||||||||||||||||
| 6.0 Gy | 6.5 Gy | 7.0 Gy | 7.5 Gy | 8.0 Gy | 8.5 Gy | |||||||||||||
| Diseases and Bio Functions | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 |
| Apoptosis | 1.20 | -0.57 | 0.57 | -0.27 | 1.27 | -0.59 | -0.88 | 0.37 | 0.35 | 1.37 | -1.26 | 0.21 | 0.25 | -0.25 | -0.63 | 0.74 | -0.74 | -0.59 |
| Apoptosis of tumor cell lines | 0.62 | -0.62 | 0.62 | 0.41 | 0.68 | -1.10 | -0.77 | 0.30 | -0.19 | 0.78 | -1.29 | -0.48 | 0.31 | -0.31 | -1.29 | 0.78 | -1.39 | -1.10 |
| Cell proliferation of carcinoma cell lines | -0.76 | 1.07 | -1.07 | -1.07 | -0.76 | 0.15 | N/A | 1.99 | 0.52 | 0.15 | 0.76 | 0.15 | 0.15 | 0.76 | 1.07 | -1.07 | 0.15 | 0.15 |
| Cell proliferation of tumor cell lines | -0.43 | 0.55 | -0.55 | -0.97 | -0.39 | 1.25 | 0.52 | 0.22 | 0.42 | -0.35 | 1.18 | 0.44 | 0.10 | 0.39 | 1.30 | -0.97 | 1.26 | 1.25 |
| Invasion of cells | 1.60 | -0.64 | 0.64 | -1.24 | 0.64 | 0.70 | N/A | -0.63 | 0.90 | -0.27 | 0.33 | 2.21 | 0.64 | -0.64 | 1.30 | -1.24 | 2.21 | 0.70 |
| Invasion of tumor cell lines | 1.29 | -0.20 | 0.20 | -0.88 | 0.20 | 1.30 | 0.20 | -0.20 | 1.29 | 0.20 | 0.88 | 1.97 | 0.20 | -0.20 | 1.97 | -0.88 | 1.97 | 1.29 |
| Migration of tumor cell lines | 0.65 | -0.65 | 0.65 | 0.90 | -0.53 | 1.88 | N/A | -0.65 | 1.76 | 0.70 | 1.51 | 0.28 | -0.33 | 0.33 | 1.51 | 0.70 | 0.28 | 1.88 |
| Female | ||||||||||||||||||
| 6.0 Gy | 6.5 Gy | 7.0 Gy | 7.5 Gy | 8.0 Gy | 8.5 Gy | |||||||||||||
| Diseases and Bio Functions | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 | Day 1 | Day 2 | Day 6 |
| Apoptosis | 0.42 | 0.21 | -1.37 | -0.38 | 0.21 | 0.57 | -0.38 | -0.74 | 0.36 | -0.59 | -1.37 | 1.00 | 0.83 | -0.63 | -0.25 | -0.27 | -1.58 | 0.21 |
| Apoptosis of tumor cell lines | -0.13 | 0.13 | -0.78 | 0.31 | -0.48 | 1.23 | -0.29 | -0.78 | -0.19 | -0.50 | -1.39 | 0.28 | N/A | -1.29 | -0.31 | 0.42 | -2.20 | -0.48 |
| Cell proliferation of carcinoma cell lines | 1.09 | 1.07 | -0.15 | -1.98 | 1.07 | -1.98 | -1.98 | 0.15 | 0.15 | 1.07 | -1.07 | N/A | -1.98 | 1.98 | -0.15 | -1.07 | 1.98 | 1.07 |
| Cell proliferation of tumor cell lines | 0.95 | 0.15 | 0.35 | -1.01 | 0.93 | -1.82 | -0.22 | 0.48 | 0.43 | 0.95 | 0.65 | 0.64 | -2.19 | 1.79 | -0.10 | -0.97 | 2.61 | 0.93 |
| Invasion of cells | -0.27 | 1.24 | 0.27 | -0.33 | 2.21 | -0.33 | 0.64 | 1.24 | 0.70 | -0.27 | 1.24 | 0.03 | -0.93 | 1.30 | -0.64 | -1.24 | 1.30 | 2.21 |
| Invasion of tumor cell lines | 0.20 | 0.88 | -0.20 | -0.88 | 1.97 | -0.88 | 0.20 | 0.88 | 1.29 | 0.20 | 0.88 | N/A | N/A | 1.97 | -0.20 | -0.88 | 1.97 | 1.97 |
| Migration of tumor cell lines | 0.70 | -0.70 | -0.70 | -0.33 | 0.28 | -0.33 | 0.65 | -0.70 | 1.88 | 0.90 | 0.28 | N/A | N/A | 1.51 | 0.33 | 0.90 | 1.51 | 0.28 |
Bionetworks with a Z score of ≥ 1 and ≤ -1 were considered activated and inhibited networks, respectively. The tables included those bionetworks, which scored ≥ |1| in at least one of all tested conditions.
A correlation matrix was computed to comprehend the degree of coregulation among the 11 bionetworks that were perturbed by RD*TSI (S3A Table). In general, the cell invasion-related bionetworks were positively correlated with the bionetworks linked to proliferation and metastasis, while negatively correlated with bionetworks related to apoptosis. Bionetworks linked to invasion and proliferation were activated at 6-day after the exposure to three highest doses of this study including 7.5–8.5 Gy. In contrast, these bionetworks remained inactivated 6 days post-irradiation to the two lowest doses of this study including 6.0–6.5 Gy. A completely opposite picture emerged for the bionetworks linked to apoptosis. In general, the correlation profile among the networks was most significant at 6 days post-irradiation (S3B Table).
Sex-, RD- and TSI-sensitive bionetworks
We further probed a list of 11 miRNAs that emerged as significantly altered due to Sex*RD*TSI. As noted earlier, these 11 miRNAs were a subset of cnvd-miRNAs. Fig 5C depicted a hierarchical cluster of these 11 miRNAs, and Fig 5D displayed a functional network using these 11 miRNAs to reveal their biological interrelationships. Six of these 11 miRNAs perturbed by Sex*RD*TSI overlapped with the 17 miRNAs perturbed by RD*TSI (S2A Fig). Of these miRNAs, let-7a-5p emerged as the most interconnected feature as shown in Fig 5D, followed by miR-30c-5p and miR-23a-3p. Together, these three miRNAs could be the hub molecules or key regulators of the functions associated with Sex*RD*TSI.
Table 2 lists 7 bionetworks that were differentially enriched by male and female cohorts across the RD*TSI profile. Bionetworks linked to apoptosis, cell invasion and migration again emerged as the most perturbed bionetwork following the trend associated with RD*TSI. Nevertheless, a correlation matrix (S1C Table) suggested sex-dependent bias in these bionetworks’ regulation profile. For instance, the negative correlation profile at 6.0 Gy (-0.24) between males and females became positive (+0.54) at 8.5 Gy. Maximum sex-biased differences in bionetworks’ regulation profile were observed at the lower doses, while a majority of the bionetworks were positively co-regulated between males and females at 8.5 Gy TBI.
miRNAs and corresponding bionetworks linked to risk to radiation-induced fatality (RRiF)
As expected RRiF decreased with increasing radiation doses (Fig 1). Among the cohort of 18 NHPs that were exposed to 7.0 Gy or lower, 10 NHPs survived with a 56% survival rate. On the other hand, only 6 among the 18 NHPs survived from 7.5 Gy or higher.
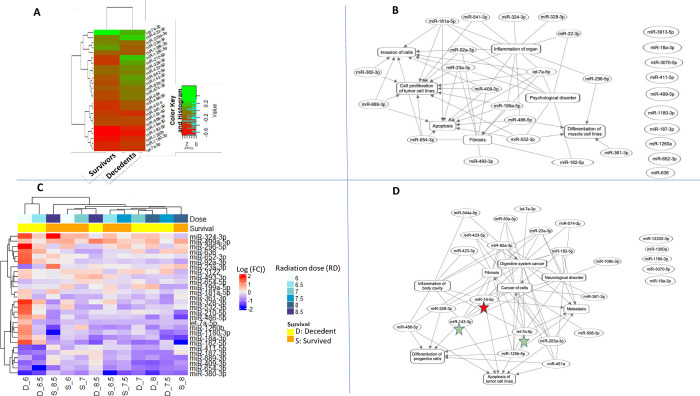
There were 25 miRNAs that were significantly altered due to the independent variable RRiF. As previously mentioned, these 25 miRNAs were a subset of cnvd-miRNAs (Fig 2). Fig 6A depicted a hierarchical cluster of these 25 miRNAs, and Fig 6B displayed a functional network using these 25 miRNAs to reveal their biological interrelationships. Of these, miR-23a-3p and let-7a-5p emerged as the most interconnected features, followed by miR-92a-3p, miR-409-3p and miR-141-3p. Together, these four miRNAs could be the hub molecules or key regulators of the functions associated with RRiF. Functional enrichment analysis (Table 3) found that the activated bionetworks linked to cell quantity, particularly in muscle cells, were associated with the survivors, whereas the bionetwork linked to cell invasion was inhibited in the survivors. S2 Table lists the miRNAs linked to these bionetworks.
Fig 6. Functional association among the differentially expressed miRNA cluster linked to RRiF.
The color keys of log2 (Fold change) were added at the top left corners. In these networks, the oval and rectangular shaped nodes represented miRNA and biofunctions, respectively. The edges represented the relationship between two nodes, the solid lines represented their association, and pointed arrowheads denoted the activating relationship between the two nodes. Red colored stars annotate miRNAs that were most connected to the neighbors for > 90% percentile, and green colored stars for > 75% percentile. (A) Hierarchical cluster of 21 differentially expressed miRNAs linked to RRiF. (B) Network cluster of the same 21 differentially expressed miRNAs linked to RRiF. (C) Hierarchical cluster of 28 differentially expressed miRNAs linked to the cumulative effects of radiation dose and RRiF. (D) Network cluster of the same 28 differentially expressed miRNAs linked to RD*RRiF.
Table 3. List of bionetworks enriched by a subset of cnvd-miRNA: Bionetworks significantly altered by the RRiF.
| Diseases and Bio Functions | Survivors | Decedents |
|---|---|---|
| Quantity of cells | 1.96 | -0.44 |
| Quantity of muscle cell lines | 1 | -1 |
| Differentiation of muscle cell lines | 1.07 | -0.76 |
| Metastasis | 1.09 | -0.22 |
| Differentiation of hematopoietic progenitor cells | 1.15 | 0.07 |
| Differentiation of progenitor cells | 1.74 | 0.84 |
| Cell viability | -0.34 | -1.03 |
| Neoplasia of tumor cell lines | 0 | 1 |
| Invasion of cells | -1.46 | 0.33 |
| Invasion of tumor cell lines | -1.13 | 0.86 |
Bionetworks with a Z score of ≥ 1 and ≤ -1 were considered activated and inhibited networks, respectively. The tables included those bionetworks, which scored ≥ |1| in at least one of all tested conditions.
To understand the dose response in RRiF, 28 miRNAs were identified that were significantly altered by RD*RRiF. To note, there were 10 miRNAs that were common between the two sets that were linked to RRiF and TBI*RRiF, respectively (S2B Fig). Fig 6C depicted a hierarchical cluster of these 28 miRNAs, and Fig 6D displayed a functional network using these 28 miRNAs to reveal their biological interrelationships. One miRNA, miR-16p-5p, emerged as the most interconnected features, followed by miR-143-3p and miR-7a-5p. Together, these three miRNAs could be the hub molecules or key regulators of the functions associated with RD*RRiF. Functional enrichment analysis (Table 4) found that the inhibited bionetwork linked to the quantity of muscle cells emerged as a potential signature of fatality due to the low dose of radiation. S2 Table lists the miRNAs linked to this bionetwork.
Table 4. List of bionetworks enriched by a subset of cnvd-miRNA: Bionetworks significantly altered by the cumulative effects of RRiF and RD (RRiF *RD).
| Dose | S/F | Invasion of cells | Cell proliferation of tumor cell lines | Migration of tumor cell lines | Apoptosis of tumor cell lines | Neoplasia of tumor cell lines | Quantity of muscle cell lines |
|---|---|---|---|---|---|---|---|
| 6.0 Gy | S | 0.26 | -0.47 | -0.17 | -0.06 | -1.09 | 1.00 |
| F | 0.69 | -0.31 | 0.74 | 0.06 | 0.22 | 2.00 | |
| 6.5 Gy | S | 0.69 | 1.29 | -0.02 | 0.06 | 0.22 | 2.00 |
| F | 2.02 | -0.31 | 0.74 | 0.06 | 0.22 | -2.00 | |
| 7.0 Gy | S | 1.20 | 1.51 | 1.68 | -1.41 | 0.66 | -1.00 |
| F | 2.02 | 2.52 | 1.68 | -1.51 | -0.22 | -2.00 | |
| 7.5 Gy | S | 2.79 | 1.73 | 0.74 | -0.22 | -1.09 | 0.00 |
| F | 2.99 | 1.73 | 0.74 | -0.67 | -0.22 | -2.00 | |
| 8.0 Gy | S | 1.25 | 2.07 | 0.93 | -0.67 | -1.09 | -1.00 |
| F | 2.79 | 1.73 | 0.74 | -0.77 | 1.09 | -2.00 | |
| 8.5 Gy | S | -0.82 | -0.35 | 0.74 | 0.62 | -0.22 | -1.00 |
| F | -0.69 | -0.43 | -0.74 | 0.21 | 0.22 | -1.00 |
Bionetworks with a Z score of ≥ 1 and ≤ -1 were considered activated and inhibited networks, respectively. The tables included those bionetworks, which scored ≥ |1| in at least one of all tested conditions.
S: survived; F: fatal.
Prognostic biomarkers to score the risk of radiation-induced fatality or RRiF
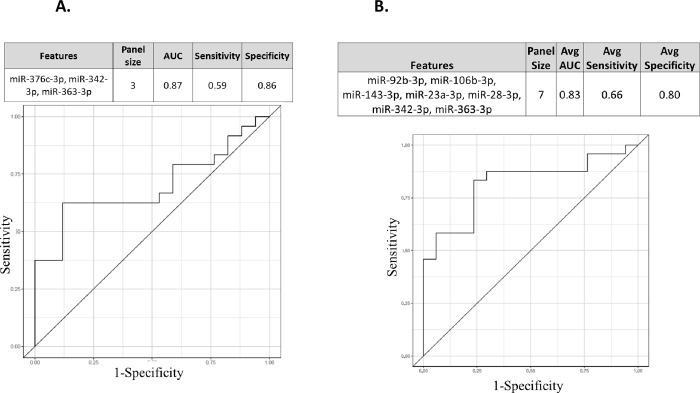
The proprietary algorithm 2BDP curated miRNA panels that can predict RRiF. As described before, 2BDP used two methods to calculate the AUC of a given panel of miRNA biomarkers (Fig 3). Method 1, namely k-fold algorithm, identified 7 panels of miRNA biomarkers that could predict the RRiF with AUC > 0.85 (S4A Table). The top ranked candidate panel is presented in Fig 7A that conjoined three miRNAs (miR-376c-3p, miR-342-3p and miR-363-3p) through the following Eq 2 to achieve AUC = 0.87 with 95% CI ranging from 0.53 to 0.86.
Fig 7. An miRNA panel to predict RRiF.
Two putative top performing markers were listed herein. (A) This three-miRNA enriched panel was determined by k-fold routine of the 2BDP algorithm. AUC ROC plot is included herewith. (B) A seven-miRNA enriched panel was determined by RSBMR routine of the 2BDP algorithm. AUC ROC plot is included herewith.
| (Eq 2) |
Method 2, namely RSBMR, failed to identify any panel that scored greater than 0.85 AUC. Herein, we listed 14 panels of miRNA biomarkers that could predict the RRiF with AUC >0.82 (S4B Table). The top ranked candidate panel is presented in Fig 7B that conjoined seven miRNAs namely miR-92b-3p, miR-106b-3p, miR-143-3p, miR-23a-3p, miR-28-3p, miR-342-3p, and miR-363-3p through the following Eq 3 to achieve AUC = 0.83 with 95% CI ranging from 0.65 to 0.94.
| (Eq 3) |
Discussion
A significantly low success rate of human clinical studies has long been attributed to the ineffective translation of animal data to the clinical space [37]. This challenge is compounded in radiation studies due to the scarcity of human data with ‘true’ signal linked to radiation without any confounding factors, such as comorbidities like cancer. NHPs present the most viable platform to test radiation biomarkers since the gene heterogeneity between humans and NHPs is around 2%, although the scale of heterogeneity in small non-coding RNA regions is potentially higher [54, 55]. Hence, we took care in curating those miRNAs that registered sequential homology between humans and NHPs and would potentially display similar functionality in responding to radiation exposure. We identified a pool of 151 miRNAs and named it cnvd-miRNAs following an in silico routine that we have used earlier to identify miRNAs that were potential homologues between minipigs and humans [38]. In this previous study, male adult minipigs were exposed to LD70/45 doses and 92 miRNAs were found to be functionally and sequentially conserved between minipigs and humans. Of these 92 miRNAs, 27 were common with the present pool of 151 cnvd-miRNAs, potentially suggesting a phylogenetically conserved pool of miRNAs linked to radiation response.
In this respect, we have reported 7 miRNAs that were conserved among mice, humans and NHPs with radiation doses ranging from LD30/60, LD50/60, and LD70/60 [28]. A Venn analysis (S3 Table) found 5 of these seven miRNAs, namely miR-133a-3p, miR-215-5p, miR-150-5p, miR-30a-5p and miR-375-3p overlapped with 151 cnvd-miRNAs. The divergence between our current result and that of our earlier study [28] could be attributed to differences in the approaches to identify conserved miRNAs. For instance, our earlier study [28] established a cross-species homology by curating those miRNAs that target genes with overlapping sequences across mice, NHPs and humans. Furthermore, the range of radiation doses were mostly different between these two studies; namely, our earlier study [28] exposed NHPs to the following radiation doses: 5.8, 6.5, or 7.2 Gy, while the current study exposed NHPs to a wider range of radiation doses, namely 6.0, 6.5, 7.0. 7.5, 8.0 and 8.5 Gy. Regardless, the outcomes of these studies overlapped as described in S5 Table.
S5 Table lists the miRNAs distributed among the three pools, namely (i) 151 cnvd-miRNAs or the conserved miRNAs between humans and NHPs, which is the deliverable of the current study, (ii) 92 cnvd-miRNAs or the conserved miRNAs between humans and minipigs as reported here [38], and (iii) seven evolutionary conserved miRNAs among mice, NHPs and humans, which was published here [28]. S5 Table listed the potentially conserved miRNAs with high translational potential. Furthermore, we mapped our current data with a systematic review of more than 450 articles [30], which reported eight miRNAs that were significantly linked to radiation irrespective of the species model. The 151 cnvd-miRNA pool included 6 of these 8 miRNAs (miR-150, miR-29a, miR-30c, miR-200b, miR-30a and miR-150), emphasizing a cross-species applicability of these miRNAs in radiation biology.
We employed this pool of 151 cnvd-miRNAs to meet two objectives. The first objective was to determine the RD- and TSI-sensitive miRNA signatures and corresponding molecular bionetworks. Thereby, we elucidated the post-irradiation biomechanism dynamics with gender-specific resolution. The second objective was to determine the bionetworks leading to the survival of the subjects and pertinent dose-specific information was presented. Furthermore, we used our proprietary ML-algorithm named 2BDP [53] to deliver the putative prognostic markers linked to the risk of radiation-induced fatality or RRiF.
A multivariant analysis followed by a linear regression model delivered a set of miRNAs that consistently and significantly shifted with either increasing radiation dose or time. Of this set of miRNAs, miR-183-5p significantly distinguished the irradiated subjects from the non-TBI baseline, irrespective of the radiation doses; thereby, miR-183-5p was promoted as a potential radiation marker. In fact, multiple studies endorsed miR-183-5p as a potential signature of various carcinomas like prostate cancer [56], renal cancer [57] and gastric cancer [58], and additional reports found miR-183-5p’s radioresistant ability [59]. Our model found miR-183-5p was inhibited by high RDs, which could be linked to the role of down-regulated miR-183-5p in modulating potassium channels [60] and to escalated neuropathic pain disorders [61].
Subsequent multivariate analysis identified a group of miRNAs and their corresponding networks that could explain the dependent variable namely RD*TSI, and its gender bias, namely Sex*RD*TSI. Of this group of miRNAs, miR-424-5p, miR-30c-5p, and miR-30a-3p were functionally and sequentially conserved across minipigs, NHPs and humans and biologically both miRNAs are linked to the invasion and proliferation of carcinoma [62, 63].
Enumeration of the interconnecting nodes and edges of the miRNA networks linked to RD*TSI and Sex*RD*TSI revealed the hub molecules, which can contribute to multiple biofunctions in pleiotropic fashions [64, 65]. Indeed, one hub molecule, namely let-7a-5p was promoted as a potential radiation marker using a hematopoietic humanized mouse model [66]; furthermore, multiple studies reported the carcinogenic attributes of let-7a-5p [67]. In fact, all the hub miRNAs that were reported in the current study were found to be oncogenic in character highlighting the carcinogenic consequences of radiation [68].
Functional analysis of the miRNA profiles associated with RD*TSI revealed a dose-sensitive trend, particularly at 6 days post-irradiation. For instance, the bionetwork linked to apoptosis was activated and in conjunction with the cell viability network, was inhibited at low doses, namely 6.0–7 Gy [69]. Beyond 7 Gy, bionetworks linked to tumor cell invasion became activated, re-emphasizing the direct association between cancer and radiation [68]. A sex-biased trend dominated at the lower range of RDs, but both sexes responded similarly to the higher doses of radiation. This result might help in developing precision radiation therapy since a sex-based customization of radiation therapy seemed more warranted at the low range of radiation doses rather that its high range of doses.
The determination of prognostic markers linked to radiation-induced fatality and the comprehension of pertinent molecular attributes remain of high importance in radiation epidemiology studies. Since the survival tests depend on the animal model, its transition potentials are often questioned. Hence, we not only assayed an NHP model, which is one of the closest neighbors of humans in the phylogenetic tree, but also applied an in silico pipeline to compile those miRNAs that displayed sequential and potentially functional homology between humans and NHPs. Let-7a-5p re-emerged as a major hub molecule among the group linked to RRiF highlighting this marker’s diagnostic and therapeutic values in the context of radiation [66]. There were three additional miRNAs that were repeatedly featured as hub molecules in the networks linked to RD*TSI and RRiF; among them, miR-23a-3p [70] and miR-16-5p [71] are established oncogenic markers, and miR-7a-5p was identified as a potential marker of radiation exposure in a humanized mouse model [66].
Functional analysis found inhibited bionetworks linked to cell quantity, particularly muscle cell quantity, as the distinct features leading to radiation-induced fatality. These bionetworks were typically suppressed from 6.0 to 8.5 Gy suggesting a link between muscle wasting and radiation-induced fatality. Muscle wasting is a typical comorbidity of cachexia [72], a signal of the terminal phase of several diseases including cancer [73]. In fact, a recently published review article presented a panel of six miRNAs that were potentially linked to cachexia development [74] and one of them, namely miR-92a-3p, overlapped with our list that is linked to RRiF.
The current hypothesis states that a systematic integration of a group of biomarkers can potentially demonstrate higher efficacy than a single candidate biomarker [75]. Indeed, there is a clear trend in finding biomarker panels of different disease pathogenesis by taking the advantages from high throughput and high-resolution multi-omics readouts [75–77]. One of the early breakthroughs of this trend was Mamaprint, a 70-gene panel that was approved by the FDA as a prognostic marker for breast cancer relapse [78, 79]. Following this trend, we trained the 151 cnvd-miRNAs via the 2BDP algorithm to find a putative panel of biomarkers to predict radiation-induced fatality. In the past, we demonstrated the efficacy of 2BDP in grading different stages of Alzheimer’s disease by panels of gene markers [53]. Furthermore, we used this algorithm to determine a panel of gene markers to predict the onset of sepsis (patent pending). In the present study, the deliverables were to find panels of miRNAs to score the risk of radiation-induced fatality or RRiF. As described earlier, 2BDP operated on two parallel routines, which differed in process of determining and computing Training vs. Test sets, but eventually produced the best possible fitting model to explain a clinical event of interest, which was RRiF in the current study. One of the two fitting models of 2BDP, namely the k-fold algorithm, delivered a 3-miRNA panel that could score RRiF with an AUC ~ 0.87. Herein, all three featured miRNAs are linked to cancer; for instance, miR-376c-3p promotes apoptosis in cancer cells [80], miR-342-3p suppresses the invasion and proliferation of cancer cells [81] and miR-363-3p acts as an inhibitor of tumor growth [82]. K-fold methods delivered a few more panels that could be worth further consideration. For instance, there was a 5-miRNA panel with a slightly inferior AUC ~ 0.85, but better McFadden’s R2 ~ 0.19 than that of the above mentioned 3-miRNA panel. This 5-miRNA panel also featured the same three miRNAs, namely miR-376c-3p, miR-342-3p and miR-363-3p; additional candidates included miR-143-3p and miR-301a-3p that function as anti-carcinogenic [83] and pro-carcinogenic [84] factors, respectively. The second fitting model under 2BDP, namely RSBMR, delivered the prognostic panels for RRiF with inferior AUCs than that of the k-fold models. The best performing panel from RSBMR could predict RRiF with AUC ~ 0.82 and McFadden’s R2 ~ 0.28. It was a 7-miRNA panel that included the following: miR-92b-3p [85], miR-363-3p [85] and miR-143-3p [83] function as anti-carcinogenic factors; miR-106b-3p promotes metastasis [86]; miR-23a-3p is an oncogenic marker [70]; miR-28-3p is a marker of pulmonary diseases [87]; and miR-342-3p suppresses tumor cell proliferation [88].
As a limitation of this study, all these panels displayed comparatively low sensitivities, which could be attributed to a small sample size. Nevertheless, we must note that in the space of NHP research, six subjects per time point is not a trivial sample size. In addition, we took several measures to improve the outcome’s translational values. For instance, multi-level screening and in silico comparative routines were applied to curate those miRNAs that would display phylogenetically conserved responses to TBI. The blood samples collected from the pre-irradiated cohort were used as the baseline to minimize the individual sample bias. Furthermore, the 2BDP algorithm applied two unbiased routines to deliver the best fitting models which enabled the ability to score the risk of radiation-induced fatality. Because the present study used pre-irradiated samples as the baseline, the irradiated sample size outnumbered the unirradiated samples. Thus, we decided to refrain from using the 2BDP algorithm to find markers for radiation exposure, since being an ML-based algorithm, 2BDP could overfit the data. In the same token, we have certain confidence that 2BDP would produce viable results in predicting RRiF, since nearly half of the NHP cohort (16 of 36) survived across the radiation doses that were used in the current study.
In conclusion, we presented a pool of 151 serum miRNAs (cnvd-miRNA) that could have high translational potential in the context of radiation. The NHP samples collected from both genders and a wide range of radiation doses coupled with a high-resolution miRNA profiling assay and ML-empowered algorithm upheld the confidence level of the current results. Radiation dose- and time since irradiation-specific miRNA markers were identified; among them, miR-183-5p emerged as a leading candidate. Additional miRNA markers are identified that controls multiple biofunctions, such as suppressed cell proliferation, apoptosis, ion channel modulations, which leads to comorbidities like cancer and fatality. In addition, the 2BDP algorithm delivered candidate panels that can score the risk of radiation-induced fatality. In the clinical space, these panels could be used to triage the radiation exposed patients based on their risk of fatality, henceforth a customized precision care could be ensured. Validation studies of these miRNA panels using independent cohorts are warranted; nevertheless, the translational efficacies of these biomarker panels are expected to be high since these miRNAs affirm a cross-species homology.
Using these miRNA biomarkers developed from the candidate panels, an assay system could be developed to determine the dose of radiation exposure and the time since exposure with a high translational value. These biomarker panels have immense translational ability, and could be used in the clinic to not only triage radiation-exposed victims to determine their risk of death, but could also be used to assist in the development of precise radiation injury treatment strategies. Ultimately, these conservative strategies will optimize scarce resources during any large scale radiological/nuclear scenario and will provide optimal care to radiation exposed victims.
Supporting information
(PDF)
A. Venn diagram showing the sub-clusters that were conserved and unique to the groups as defined in Fig 3A–3D, respectively. The underlined miRNAs were those which emerged most connected in Fig 3B or 3D. B. Venn diagram showing the sub-clusters that were conserved and unique to the groups as defined in Fig 4A–4D, respectively. The underlined miRNAs were those which emerged most connected in Fig 4B or 4D.
(ZIP)
(ii) Human and minipig conserved miRNAs, this list of 92 miRNAs is presented by our earlier work, (Chakraborty, N et al. Scientific Reports 13(1), 2023). (iii) Human-mouse-NHP conserved miRNAs, this list of 7 miRNAs was also published by us (Fendler, W. et al. Sci Transl Med, 2498 (9), 2017).
(PDF)
B. List of 176 miRNAs’ log2 fold change values that emerged significantly expressed in at least one of the 15 analysis models using all dependent and independent variables. C. cnvd-miRNA: A display of the homologue sequences conserved between humans and NHPs.
(ZIP)
(PDF)
B. Correlation matrix of 11 networks that were significantly regulated by RD*TSI. Pearson correlation was calculated across radiation doses at 6-day post-TBI. C. Male vs. female correlation matrix of 7 networks that were significantly regulated by sex*RD*TSI. Pearson correlation was calculated across radiation doses and TSI.
(ZIP)
B. RSBMR results: List of potential panels that can predict RRiF with AUC >0.82.
(ZIP)
(PDF)
Acknowledgments
The authors would like to thank the staff of the Veterinary Science Department, AFRRI; the Comparative Medicine Department of the University of Maryland, Baltimore for animal care; and to the staff of the Radiation Science Department, AFRRI, for dosimetry and radiation exposure to the animals. We also thank Dr. Carmen Rios and Dr. Andrea Dicarlo-Cohen of RNCP, NIAID, NIH for helpful scientific discussions.
Disclaimer
WRAIR
The views, opinions, and/or findings contained in this report are those of the author(s) and should not be construed as official Department of the Army position, policy, or decision, unless so designated by other official documentation. Citations of commercial organizations or trade names in this report do not constitute an official Department of the Army endorsement or approval of the products or services of these organizations. Opinions, interpretations, conclusions, and recommendations are those of the author(s) and are not necessarily endorsed by the US Army.
USUHS
The opinions or assertions contained herein are the private views of the authors and are not necessarily those of the Uniformed Services University of the Health Sciences, or the Department of Defense.
Data Availability
All relevant data are within the manuscript, its Supporting Information files and uploaded on GEO database, GSE247225.
Funding Statement
VKS AAI12044‐001‐07000, Work plan G National Institute of Allergy and Infectious Diseases https://www.niaid.nih.gov/ VKS AFR-B4-10978 and 12080 https://www.usuhs.edu/ Funders played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gale RP. Medical and policy considerations for nuclear and radiation accidents, incidents and terrorism. Curr Opin Hematol. 2017;24(6):496–501. Epub 2017/10/07. doi: 10.1097/MOH.0000000000000380 . [DOI] [PubMed] [Google Scholar]
- 2.Obrador E, Salvador-Palmer R, Villaescusa JI, Gallego E, Pellicer B, Estrela JM, et al. Nuclear and Radiological Emergencies: Biological Effects, Countermeasures and Biodosimetry. Antioxidants (Basel). 2022;11(6). Epub 20220531. doi: 10.3390/antiox11061098 ; PubMed Central PMCID: PMC9219873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ainsbury EA, Bakhanova E, Barquinero JF, Brai M, Chumak V, Correcher V, et al. Review of retrospective dosimetry techniques for external ionising radiation exposures. Radiat Prot Dosimetry. 2011;147(4):573–92. Epub 2010/12/25. doi: 10.1093/rpd/ncq499 . [DOI] [PubMed] [Google Scholar]
- 4.Ainsbury EA, Samaga D, Della Monaca S, Marrale M, Bassinet C, Burbidge CI, et al. Uncertainty on radiation doses estimated by biological and retrospective physical methods. Radiat Prot Dosimetry. 2018;178(4):382–404. Epub 2017/10/06. doi: 10.1093/rpd/ncx125 . [DOI] [PubMed] [Google Scholar]
- 5.Shakyawar SK, Mishra NK, Vellichirammal NN, Cary L, Helikar T, Powers R, et al. A review of radiation-induced alterations of multi-omic profiles, radiation injury biomarkers, and countermeasures. Radiat Res. 2023;199(1). Epub 20221111. doi: 10.1667/RADE-21-00187.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh VK, Newman VL, Romaine PL, Hauer-Jensen M, Pollard HB. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev Mol Diagn. 2016;16:65–81. doi: 10.1586/14737159.2016.1121102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellsworth SG, Yalamanchali A, Lautenschlaeger T, Grossman SA, Grassberger C, Lin SH, et al. Lymphocyte depletion rate as a biomarker of radiation dose to circulating lymphocytes during fractionated partial-body radiation therapy. Adv Radiat Oncol. 2022;7(5):100959. Epub 2022/08/06. doi: 10.1016/j.adro.2022.100959 ; PubMed Central PMCID: PMC9343404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barquinero JF, Fattibene P, Chumak V, Ohba T, Della Monaca S, Nuccetelli C, et al. Lessons from past radiation accidents: Critical review of methods addressed to individual dose assessment of potentially exposed people and integration with medical assessment. Environ Int. 2021;146:106175. Epub 20201016. doi: 10.1016/j.envint.2020.106175 . [DOI] [PubMed] [Google Scholar]
- 9.Raavi V, Perumal V, S FDP. Potential application of gamma-H2AX as a biodosimetry tool for radiation triage. Mutat Res Rev Mutat Res. 2021;787:108350. Epub 20201122. doi: 10.1016/j.mrrev.2020.108350 . [DOI] [PubMed] [Google Scholar]
- 10.Kraus VB. Biomarkers as drug development tools: discovery, validation, qualification and use. Nat Rev Rheumatol. 2018;14(6): Epub 354. doi: 10.1038/s41584-018-0005-9 . [DOI] [PubMed] [Google Scholar]
- 11.Singh VK, Simas M, Pollard H. Biomarkers for acute radiation syndrome: Challenges for developing radiation countermeasures following animal rule. Expert Rev Mol Diagn. 2018;18(11):921–4. Epub 2018/10/06. doi: 10.1080/14737159.2018.1533404 . [DOI] [PubMed] [Google Scholar]
- 12.Singh VK, Seed TM, Cheema AK. Metabolomics-based predictive biomarkers of radiation injury and countermeasure efficacy: Current status and future perspectives. Expert Rev Mol Diagn. 2021;21:641–54. Epub 2021/05/25. doi: 10.1080/14737159.2021.1933448 . [DOI] [PubMed] [Google Scholar]
- 13.Pannkuk EL, Fornace AJ, Jr., Laiakis EC. Metabolomic applications in radiation biodosimetry: exploring radiation effects through small molecules. International journal of radiation biology. 2017;93(10):1151–76. doi: 10.1080/09553002.2016.1269218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicente E, Vujaskovic Z, Jackson IL. A systematic review of metabolomic and lipidomic candidates for biomarkers in radiation injury. Metabolites. 2020;10(6). Epub 2020/06/25. doi: 10.3390/metabo10060259 ; PubMed Central PMCID: PMC7344731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.U.S. Food and Drug Administration. More about biomarkers & qualification 2018 [cited 2018 May 18]. Available from: https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugDevelopmentToolsQualificationProgram/BiomarkerQualificationProgram/ucm535408.htm.
- 16.Cheema AK, Li Y, Moulton J, Girgis M, Wise SY, Carpenter A, et al. Identification of novel biomarkers for acute radiation injury using multiomics approach and nonhuman primate model. Int J Radiat Oncol Biol Phys. 2022;114(2):310–20. Epub 20220605. doi: 10.1016/j.ijrobp.2022.05.046 . [DOI] [PubMed] [Google Scholar]
- 17.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–55. Epub 2012/02/18. doi: 10.1038/nature10888 ; PubMed Central PMCID: PMC3509753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. Epub 2014/09/03. doi: 10.1155/2014/970607 ; PubMed Central PMCID: PMC4142390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11–7. Epub 2019/03/03. doi: 10.1016/j.cbpa.2019.01.024 . [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Lu Q, Chang C. Epigenetics in Health and Disease. Adv Exp Med Biol. 2020;1253:3–55. doi: 10.1007/978-981-15-3449-2_1 . [DOI] [PubMed] [Google Scholar]
- 21.Huang W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol Biol. 2017;1617:57–67. Epub 2017/05/26. doi: 10.1007/978-1-4939-7046-9_4 . [DOI] [PubMed] [Google Scholar]
- 22.Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61(1):56–63. Epub 2014/11/14. doi: 10.1373/clinchem.2014.221341 . [DOI] [PubMed] [Google Scholar]
- 23.Waselenko JK, MacVittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE, et al. Medical management of the acute radiation syndrome: recommendations of the Strategic National Stockpile Radiation Working Group. Ann Intern Med. 2004;140(12):1037–51. Epub 2004/06/16. doi: 10.7326/0003-4819-140-12-200406150-00015 . [DOI] [PubMed] [Google Scholar]
- 24.Tomasik B, Fendler W, Chowdhury D. Serum microRNAs–potent biomarkers for radiation biodosimetry. Oncotarget. 2018;9(18):14038. doi: 10.18632/oncotarget.24381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh VK, Pollard HB. Ionizing radiation-induced altered microRNA expression as biomarkers for assessing acute radiation injury. Expert Rev Mol Diagn. 2017;17(10):871–4. Epub 20170814. doi: 10.1080/14737159.2017.1366316 ; PubMed Central PMCID: PMC6082642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Liu P, Narayanan J, Yang M, Fish BL, Liu Y, et al. Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci Rep. 2017;7:44132. doi: 10.1038/srep44132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acharya SS, Fendler W, Watson J, Hamilton A, Pan Y, Gaudiano E, et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 2015;7(287):287ra69. doi: 10.1126/scitranslmed.aaa6593 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fendler W, Malachowska B, Meghani K, Konstantinopoulos PA, Guha C, Singh VK, et al. Evolutionarily conserved serum microRNAs predict radiation-induced fatality in nonhuman primates. Sci Transl Med. 2017;9(379). Epub 2017/03/03. doi: 10.1126/scitranslmed.aal2408 ; PubMed Central PMCID: PMC5441546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty N, Gautam A, Holmes-Hampton GP, Kumar VP, Biswas S, Kumar R, et al. microRNA and metabolite signatures linked to early consequences of lethal radiation. Sci Rep. 2020;10(1):5424. Epub 2020/03/28. doi: 10.1038/s41598-020-62255-w ; PubMed Central PMCID: PMC7096415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Małachowska B, Tomasik B, Stawiski K, Kulkarni S, Guha C, Chowdhury D, et al. Circulating microRNAs as biomarkers of radiation exposure: a systematic review and meta-analysis. Int J Radiat Oncol Biol Phys. 2020;106(2):390–402. doi: 10.1016/j.ijrobp.2019.10.028 [DOI] [PubMed] [Google Scholar]
- 31.Singh VK, Srivastava M, Seed TM. Protein biomarkers for radiation injury and testing of medical countermeasure efficacy: promises, pitfalls, and future directions. Expert Rev Proteomics. 2023;20(10):221–46. Epub 20231027. doi: 10.1080/14789450.2023.2263652 . [DOI] [PubMed] [Google Scholar]
- 32.Rydzewski NR, Helzer KT, Bootsma M, Shi Y, Bakhtiar H, Sjostrom M, et al. Machine learning & molecular radiation tumor biomarkers. Semin Radiat Oncol. 2023;33(3):243–51. Epub 2023/06/19. doi: 10.1016/j.semradonc.2023.03.002 ; PubMed Central PMCID: PMC10287033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowicka Z, Tomasik B, Kozono D, Stawiski K, Johnson T, Haas-Kogan D, et al. Serum miRNA-based signature indicates radiation exposure and dose in humans: A multicenter diagnostic biomarker study. Radiother Oncol. 2023;185:109731. Epub 2023/06/11. doi: 10.1016/j.radonc.2023.109731 . [DOI] [PubMed] [Google Scholar]
- 34.Tomasik B, Papis-Ubych A, Stawiski K, Fijuth J, Kedzierawski P, Sadowski J, et al. Serum microRNAs as xerostomia biomarkers in patients with oropharyngeal cancer undergoing radiation therapy. Int J Radiat Oncol Biol Phys. 2021;111(5):1237–49. Epub 2021/07/20. doi: 10.1016/j.ijrobp.2021.07.008 . [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, Hawkins PG, Bi N, Dess RT, Tewari M, Hearn JWD, et al. Serum microRNA signature predicts response to high-dose radiation therapy in locally advanced non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2018;100(1):107–14. Epub 2017/10/21. doi: 10.1016/j.ijrobp.2017.08.039 ; PubMed Central PMCID: PMC5736451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei W, He J, Wang J, Ding N, Wang B, Lin S, et al. Serum microRNAs as early indicators for estimation of exposure degree in response to ionizing irradiation. Radiat Res. 2017;188(3):342–54. Epub 2017/06/21. doi: 10.1667/RR14702.1 . [DOI] [PubMed] [Google Scholar]
- 37.Van Norman GA. Limitations of animal studies for predicting toxicity in clinical trials: Is it time to rethink our current approach? JACC Basic Transl Sci. 2019;4(7):845–54. Epub 2020/01/31. doi: 10.1016/j.jacbts.2019.10.008 ; PubMed Central PMCID: PMC6978558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty N H-HG, Gautam A, Kumar R, Hritzo B, Legesse B, Dimitrov G, Ghosh SP, and Hammamieh R. Early to sustained impacts of lethal radiation on circulating miRNAs in a minipig model. Sci Rep. 2023;13:18496. doi: 10.1038/s41598-023-45250-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh VK, Kulkarni S, Fatanmi OO, Wise SY, Newman VL, Romaine PL, et al. Radioprotective efficacy of gamma-tocotrienol in nonhuman primates. Radiat Res. 2016;185(3):285–98. Epub 2016/03/02. doi: 10.1667/RR14127.1 . [DOI] [PubMed] [Google Scholar]
- 40.Singh VK, Fatanmi OO, Wise SY, Carpenter AD, Olsen CH. Determination of lethality curve for cobalt-60 gamma-radiation source in rhesus macaques using subject-based supportive care. Radiat Res. 2022;198(6):599–614. doi: 10.1667/RADE-22-00101.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Singh J, Varghese R, Zhang Y, Fatanmi OO, Cheema AK, et al. Transcriptome of rhesus macaque (Macaca mulatta) exposed to total-body irradiation. Sci Rep. 2021;11(1):6295. Epub 2021/03/20. doi: 10.1038/s41598-021-85669-6 ; PubMed Central PMCID: PMC7973550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Research Council of the National Academy of Sciences. Guide for the care and use of laboratory animals. 8th ed. Washington, DC: National Academies Press; 2011. [Google Scholar]
- 43.Garg S, Garg TK, Wise SY, Fatanmi OO, Miousse IR, Savenka AV, et al. Effects of gamma-tocotrienol on intestinal injury in a GI-specific acute radiation syndrome model in nonhuman primate. Int J Mol Sci. 2022;23(9):4643. Epub 2022/05/15. doi: 10.3390/ijms23094643 ; PubMed Central PMCID: PMC9100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy V. Accuracy considerations in EPR dosimetry. Appl Radiat Isot. 2000;52:1039–50. doi: 10.1016/s0969-8043(00)00052-x [DOI] [PubMed] [Google Scholar]
- 45.International Standardization Organization and ASTM International, editor. Standard Practice for Use of an Alanine-EPR Dosimetry System; 2013; Geneva, Switzerland: ASTM International, ISO and West Conshohocken (US:PA):. [Google Scholar]
- 46.Vellichirammal NN, Sethi S, Pandey S, Singh J, Wise SY, Carpenter AD, et al. Lung transcriptome of nonhuman primates exposed to total- and partial-body irradiation. Mol Ther Nucleic Acids. 2022;29:584–98. Epub 20220804. doi: 10.1016/j.omtn.2022.08.006 ; PubMed Central PMCID: PMC9418744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.American Veterinary Medical Association. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition 2020 [cited 2023 December 29]. Available from: https://www.avma.org/sites/default/files/2020-01/2020-Euthanasia-Final-1-17-20.pdf.
- 48.Garg S, Garg TK, Miousse IR, Wise SY, Fatanmi OO, Savenka AV, et al. Effects of gamma-tocotrienol on partial-body irradiation-induced intestinal injury in a nonhuman primate model. Antioxidants. 2022;11(10):1895. Epub 20220925. doi: 10.3390/antiox11101895 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garg TK, Garg S, Miousse IR, S.Y. W, Carpenter AD, Fatanmi OO, et al. Gamma-tocotrienol modulates total-body irradiation-induced hematopoietic injury in a nonhuman primate model. Int J Mol Sci. 2022;23:16170. doi: 10.3390/ijms232416170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakraborty N, Muhie S, Kumar R, Gautam A, Srinivasan S, Sowe B, et al. Contributions of polyunsaturated fatty acids (PUFA) on cerebral neurobiology: an integrated omics approach with epigenomic focus. J Nutr Biochem. 2017;42:84–94. doi: 10.1016/j.jnutbio.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 51.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 2013;20(12):970–8. Epub 20130820. doi: 10.1089/cmb.2012.0283 ; PubMed Central PMCID: PMC3842884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2003. p. 2.3. 1–2.3. 22. [DOI] [PubMed] [Google Scholar]
- 53.Chakraborty N, Lawrence A, Campbell R, Yang R, Hammamieh R. Biomarker discovery process at binomial decision point (2BDP): Analytical pipeline to construct biomarker panel. Comput Struct Biotechnol J. 2023;21:4729–42. Epub 20230927. doi: 10.1016/j.csbj.2023.09.025 ; PubMed Central PMCID: PMC10562676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. Epub 2007/07/03. doi: 10.1016/j.cell.2007.04.040 ; PubMed Central PMCID: PMC2681231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura T, Fujiwara K, Saitou M, Tsukiyama T. Non-human primates as a model for human development. Stem Cell Reports. 2021;16(5):1093–103. doi: 10.1016/j.stemcr.2021.03.021 ; PubMed Central PMCID: PMC8185448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waseem M, Ahmad MK, Serajuddin M, Bhaskar V, Sankhwar SN, Mahdi AA. MicroRNA-183-5p: A new potential marker for prostate cancer. Ind J Clin Biochem. 2019;34(2):207–12. Epub 2019/05/17. doi: 10.1007/s12291-017-0731-9 ; PubMed Central PMCID: PMC6486929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Pan X, Gui Y, Quan J, Li Z, Zhao L, et al. Upregulation of miR-183-5p predicts worse survival in patients with renal cell cancer after surgery. Cancer Biomark. 2019;24(2):153–8. Epub 2019/01/29. doi: 10.3233/CBM-182047 . [DOI] [PubMed] [Google Scholar]
- 58.Li W, Cui X, Qi A, Yan L, Wang T, Li B. miR-183-5p acts as a potential prognostic biomarker in gastric cancer and regulates cell functions by modulating EEF2. Pathol Res Pract. 2019;215(11):152636. Epub 2019/09/29. doi: 10.1016/j.prp.2019.152636 . [DOI] [PubMed] [Google Scholar]
- 59.Zheng S, Zhong YF, Tan DM, Xu Y, Chen HX, Wang D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J Biosci. 2019;44(4). Epub 2019/09/11. . [PubMed] [Google Scholar]
- 60.Shi DN, Yuan YT, Ye D, Kang LM, Wen J, Chen HP. MiR-183-5p alleviates chronic constriction injury-induced neuropathic pain through inhibition of TREK-1. Neurochem Res. 2018;43(6):1143–9. Epub 2018/05/08. doi: 10.1007/s11064-018-2529-4 . [DOI] [PubMed] [Google Scholar]
- 61.Kalpachidou T, Kummer KK, Kress M. Non-coding RNAs in neuropathic pain. Neuronal Signal. 2020;4(1):NS20190099. Epub 2020/06/27. doi: 10.1042/NS20190099 ; PubMed Central PMCID: PMC7306520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao JM, Li GZ, Han M, Xu HL, Huang KM. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed Pharmacother. 2017;93:554–60. Epub 2017/07/08. doi: 10.1016/j.biopha.2017.06.084 . [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Liu J, Hu W, Zhang Y, Sang J, Li H, et al. miR-424-5p promotes proliferation, migration and invasion of laryngeal squamous cell carcinoma. Onco Targets Ther. 2019;12:10441–53. Epub 2019/12/11. doi: 10.2147/OTT.S224325 ; PubMed Central PMCID: PMC6890199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan X, Liu X.-P., Guo Z.-X., Liu T.-Z. & Li S. Identification of hub genes associated with progression and prognosis in patients with bladder cancer. Front Genet. 2019;10:408. doi: 10.3389/fgene.2019.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Z, Jin Z, Deng Y, Wei L, Yuan X, Zhang M, et al. Co-expression network analysis identifies four hub genes associated with prognosis in soft tissue sarcoma. Front Genet. 2019;10:37. Epub 20190204. doi: 10.3389/fgene.2019.00037 ; PubMed Central PMCID: PMC6369179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsogbadrakh B, Jung JA, Lee M, Lee JA, Seo JH. Identifying serum miRNA biomarkers for radiation exposure in hematopoietic humanized NSG-SGM3 mice. Biochem Biophys Res Commun. 2022;599:51–6. Epub 2022/02/16. doi: 10.1016/j.bbrc.2022.02.010 . [DOI] [PubMed] [Google Scholar]
- 67.Chen C, Liu X, Chen C, Chen Q, Dong Y, Hou B. Clinical significance of let-7a-5p and miR-21-5p in patients with breast cancer. Ann Clin Lab Sci. 2019;49(3):302–8. Epub 2019/07/17. . [PubMed] [Google Scholar]
- 68.Sia J, Szmyd R, Hau E, Gee HE. Molecular Mechanisms of Radiation-Induced Cancer Cell Death: A Primer. Front Cell Dev Biol. 2020;8:41. Epub 20200213. doi: 10.3389/fcell.2020.00041 ; PubMed Central PMCID: PMC7031160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang JS, Wang HJ, Qian HL. Biological effects of radiation on cancer cells. Mil Med Res. 2018;5(1):20. Epub 2018/07/01. doi: 10.1186/s40779-018-0167-4 ; PubMed Central PMCID: PMC6026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quan J, Pan X, Li Y, Hu Y, Tao L, Li Z, et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed Pharmacother. 2019;110:656–66. Epub 2018/12/15. doi: 10.1016/j.biopha.2018.11.065 . [DOI] [PubMed] [Google Scholar]
- 71.Ghafouri-Fard S, Khoshbakht T, Hussen BM, Abdullah ST, Taheri M, Samadian M. A review on the role of mir-16-5p in the carcinogenesis. Cancer Cell Int. 2022;22:342. Epub 2022/11/10. doi: 10.1186/s12935-022-02754-0 ; PubMed Central PMCID: PMC9644515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rausch V, Sala V, Penna F, Porporato PE, Ghigo A. Understanding the common mechanisms of heart and skeletal muscle wasting in cancer cachexia. Oncogenesis. 2021;10(1):1. Epub 2021/01/10. doi: 10.1038/s41389-020-00288-6 ; PubMed Central PMCID: PMC7794402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vanhoutte G, van de Wiel M, Wouters K, Sels M, Bartolomeeussen L, De Keersmaecker S, et al. Cachexia in cancer: what is in the definition? BMJ Open Gastroenterol. 2016;3(1):e000097. Epub 2016/11/16. doi: 10.1136/bmjgast-2016-000097 ; PubMed Central PMCID: PMC5093365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X, Du L, Liu Q, Lu Z. MicroRNAs: Novel players in the diagnosis and treatment of cancer cachexia (Review). Exp Ther Med. 2022;24(1):446. Epub 2022/06/21. doi: 10.3892/etm.2022.11373 ; PubMed Central PMCID: PMC9199081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conway SR, Wong HR. Biomarker Panels in Critical Care. Crit Care Clin. 2020;36(1):89–104. doi: 10.1016/j.ccc.2019.08.007 . [DOI] [PubMed] [Google Scholar]
- 76.Vincent JL, Bogossian E, Menozzi M. The Future of Biomarkers. Crit Care Clin. 2020;36(1):177–87. doi: 10.1016/j.ccc.2019.08.014 . [DOI] [PubMed] [Google Scholar]
- 77.Zaim SR, Li Q, Schissler AG, Lussier YA. Emergence of pathway-level composite biomarkers from converging gene set signals of heterogeneous transcriptomic responses. Pac Symp Biocomput. 2018;23:484–95. ; PubMed Central PMCID: PMC5730363. [PMC free article] [PubMed] [Google Scholar]
- 78.Slodkowska EA, Ross JS. MammaPrint70-gene signature: another milestone in personalized medical care for breast cancer patients. Expert Rev Mol Diagn. 2009;9:417–22. doi: 10.1586/erm.09.32 [DOI] [PubMed] [Google Scholar]
- 79.Audeh W, Blumencranz L, Kling H, Trivedi H, Srkalovic GJAma. Prospective validation of a genomic assay in breast cancer: The 70-gene MammaPrint assay and the MINDACT trial. Acta Med Acad. 2019;48:18–34. [DOI] [PubMed] [Google Scholar]
- 80.Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8(12):5286–97. Epub 2017/01/13. ; PubMed Central PMCID: PMC5209482. [PMC free article] [PubMed] [Google Scholar]
- 81.Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588(17):3298–307. Epub 2014/07/30. doi: 10.1016/j.febslet.2014.07.020 . [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Chen T, Huang H, Jiang Y, Yang L, Lin Z, et al. miR-363-3p inhibits tumor growth by targeting PCNA in lung adenocarcinoma. Oncotarget. 2017;8(12):20133–44. Epub 2017/04/21. doi: 10.18632/oncotarget.15448 ; PubMed Central PMCID: PMC5386750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi H, Shen H, Xu J, Zhao S, Yao S, Jiang N. MiR-143-3p suppresses the progression of ovarian cancer. Am J Transl Res. 2018;10(3):866–74. Epub 2018/04/11. ; PubMed Central PMCID: PMC5883127. [PMC free article] [PubMed] [Google Scholar]
- 84.Xia X, Zhang K, Cen G, Jiang T, Cao J, Huang K, et al. MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4. Oncotarget. 2015;6(25):21046–63. Epub 2015/05/29. doi: 10.18632/oncotarget.4124 ; PubMed Central PMCID: PMC4673249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Long M, Zhan M, Xu S, Yang R, Chen W, Zhang S, et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer. 2017;16(1):167. Epub 2017/10/29. doi: 10.1186/s12943-017-0723-7 ; PubMed Central PMCID: PMC5659029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, et al. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin Sci. 2020;134(4):419–34. Epub 2020/02/18. doi: 10.1042/CS20191087 . [DOI] [PubMed] [Google Scholar]
- 87.Zhou X, Wen W, Shan X, Qian J, Li H, Jiang T, et al. MiR-28-3p as a potential plasma marker in diagnosis of pulmonary embolism. Thromb Res. 2016;138:91–5. Epub 2015/12/26. doi: 10.1016/j.thromres.2015.12.006 . [DOI] [PubMed] [Google Scholar]
- 88.Xie X, Liu H, Wang M, Ding F, Xiao H, Hu F, et al. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36(7):5031–8. Epub 2015/02/11. doi: 10.1007/s13277-015-3154-3 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A. Venn diagram showing the sub-clusters that were conserved and unique to the groups as defined in Fig 3A–3D, respectively. The underlined miRNAs were those which emerged most connected in Fig 3B or 3D. B. Venn diagram showing the sub-clusters that were conserved and unique to the groups as defined in Fig 4A–4D, respectively. The underlined miRNAs were those which emerged most connected in Fig 4B or 4D.
(ZIP)
(ii) Human and minipig conserved miRNAs, this list of 92 miRNAs is presented by our earlier work, (Chakraborty, N et al. Scientific Reports 13(1), 2023). (iii) Human-mouse-NHP conserved miRNAs, this list of 7 miRNAs was also published by us (Fendler, W. et al. Sci Transl Med, 2498 (9), 2017).
(PDF)
B. List of 176 miRNAs’ log2 fold change values that emerged significantly expressed in at least one of the 15 analysis models using all dependent and independent variables. C. cnvd-miRNA: A display of the homologue sequences conserved between humans and NHPs.
(ZIP)
(PDF)
B. Correlation matrix of 11 networks that were significantly regulated by RD*TSI. Pearson correlation was calculated across radiation doses at 6-day post-TBI. C. Male vs. female correlation matrix of 7 networks that were significantly regulated by sex*RD*TSI. Pearson correlation was calculated across radiation doses and TSI.
(ZIP)
B. RSBMR results: List of potential panels that can predict RRiF with AUC >0.82.
(ZIP)
(PDF)
Data Availability Statement
All relevant data are within the manuscript, its Supporting Information files and uploaded on GEO database, GSE247225.