Full text
PDF
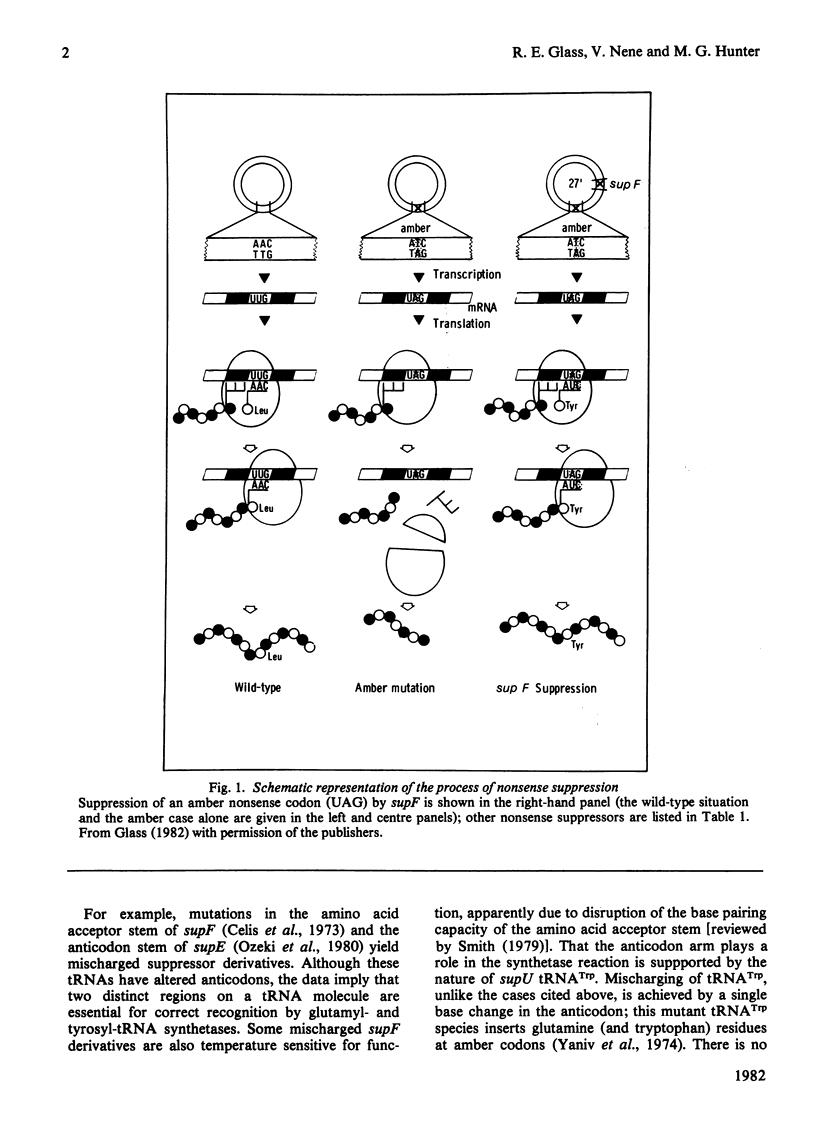
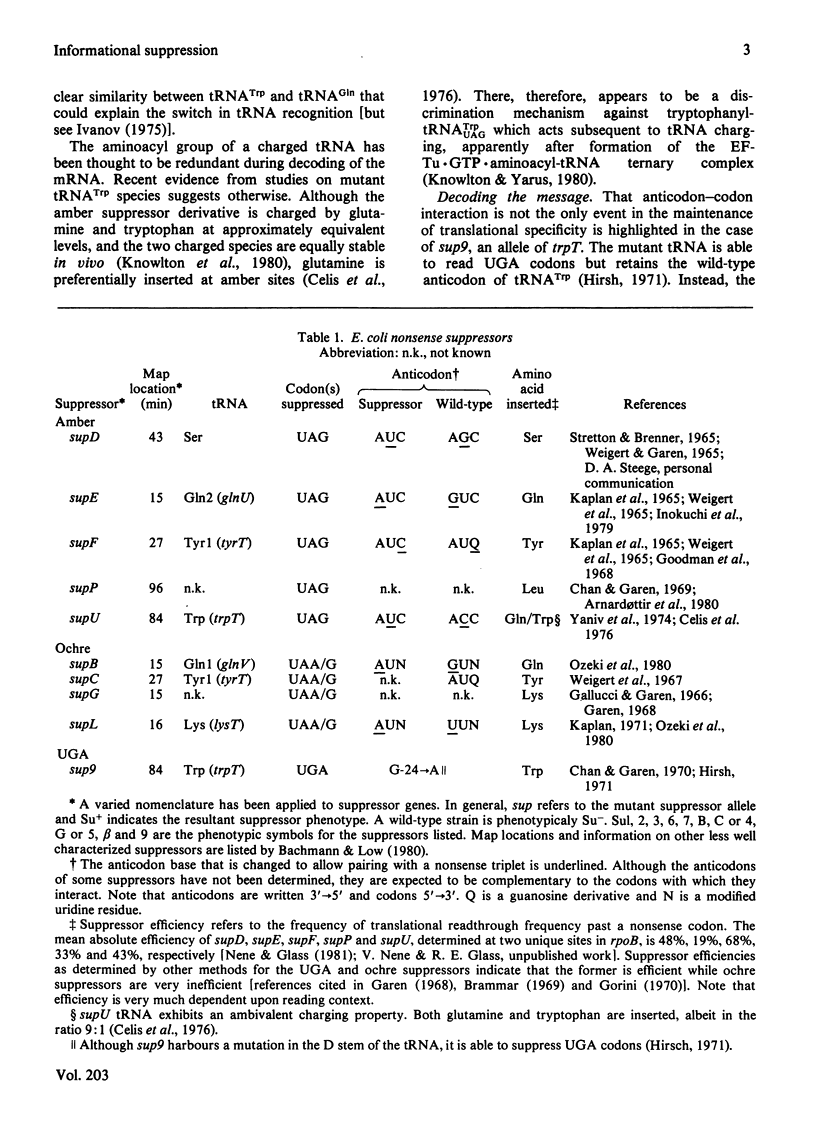
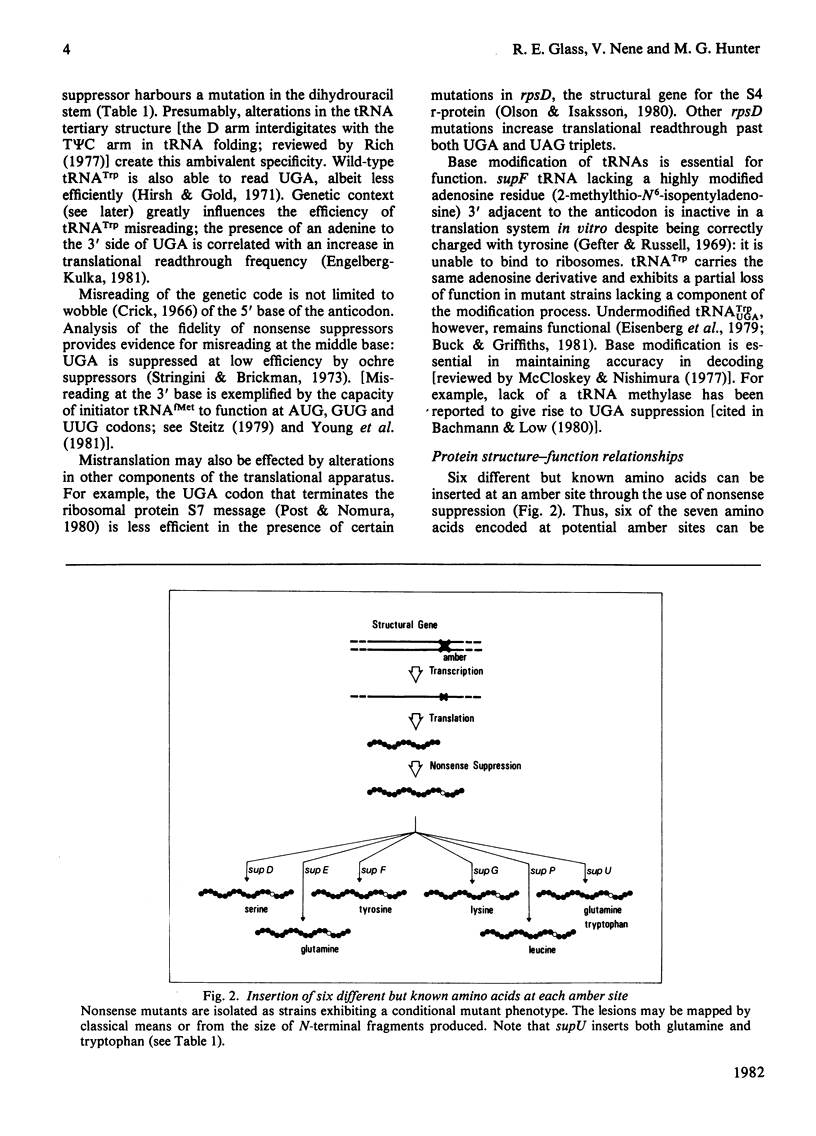
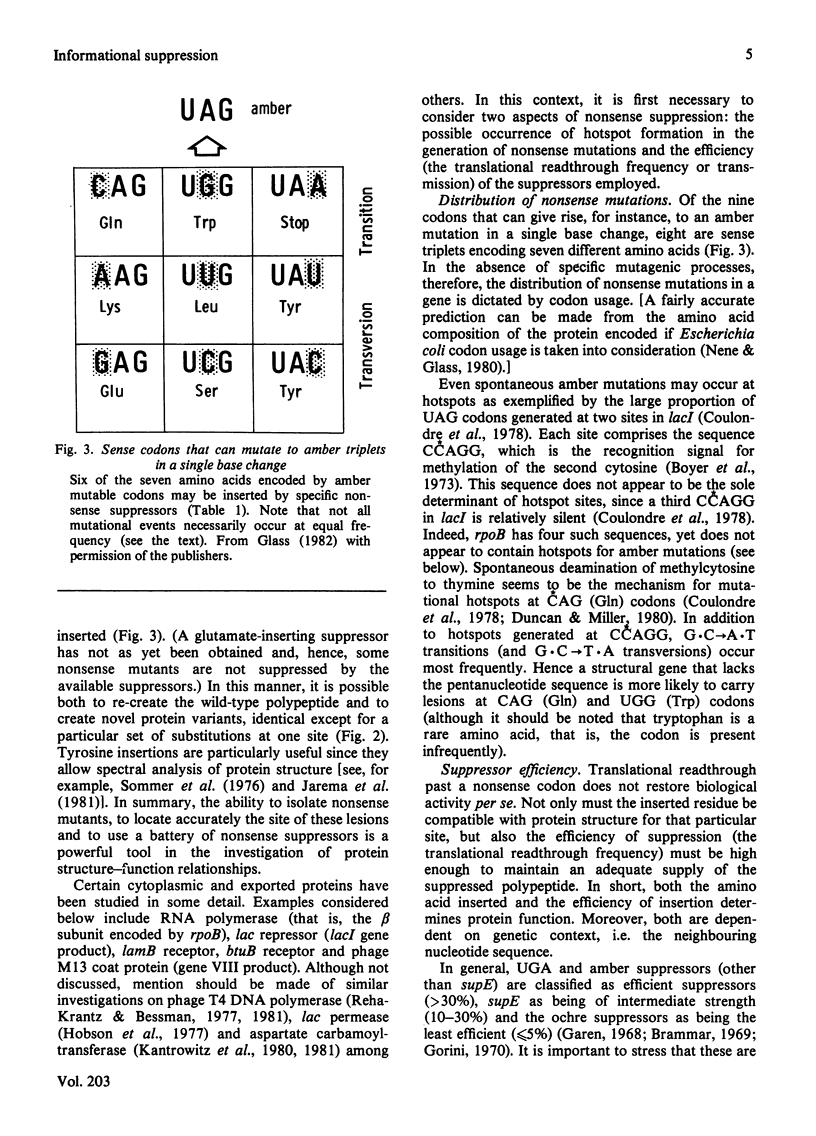








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adetugbo K., Milstein C., Secher D. S. Molecular analysis of spontaneous somatic mutants. Nature. 1977 Jan 27;265(5592):299–304. doi: 10.1038/265299a0. [DOI] [PubMed] [Google Scholar]
- Altman S., Smith J. D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971 Sep 8;233(36):35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- Apirion D., Gegenheimer P. Processing of bacterial RNA. FEBS Lett. 1981 Mar 9;125(1):1–9. doi: 10.1016/0014-5793(81)80984-2. [DOI] [PubMed] [Google Scholar]
- Arnardóttir A., Thorbjarnardóttir S., Eggertsson G. Mapping of the supP (Su6+) amber suppressor gene in Escherichia coli. J Bacteriol. 1980 Feb;141(2):977–978. doi: 10.1128/jb.141.2.977-978.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal J., Kowalska I. E., Maciejko D. M., Wegleński P. Allele specific and locus non-specific suppressors in Aspergillus nidulans. J Gen Microbiol. 1979 Dec;115(2):457–470. doi: 10.1099/00221287-115-2-457. [DOI] [PubMed] [Google Scholar]
- Bassford P. J., Jr, kadner R. J. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J Bacteriol. 1977 Dec;132(3):796–805. doi: 10.1128/jb.132.3.796-805.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Model P. Molecular basis of the am8H1 lesion in bacteriophage M13. Virology. 1979 Jul 15;96(1):299–301. doi: 10.1016/0042-6822(79)90198-3. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Russel M., Model P. Processing of filamentous phage pre-coat protein. Effect of sequence variations near the signal peptidase cleavage site. J Mol Biol. 1980 Dec 5;144(2):103–116. doi: 10.1016/0022-2836(80)90027-3. [DOI] [PubMed] [Google Scholar]
- Boss J. M., Darrow M. D., Zitomer R. S. Characterization of yeast iso-1-cytochrome c mRNA. J Biol Chem. 1980 Sep 25;255(18):8623–8628. [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C., Soll L., Botstein D. Recessive lethal amber suppressors in yeast. Genetics. 1975 Apr;79(4):551–560. doi: 10.1093/genetics/79.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Stewart J. W., Sherman F., Botstein D. Substitution of serine caused by a recessive lethal suppressor in yeast. J Mol Biol. 1976 Apr 15;102(3):467–476. doi: 10.1016/0022-2836(76)90328-4. [DOI] [PubMed] [Google Scholar]
- Buck M., Griffiths E. Regulation of aromatic amino acid transport by tRNA: role of 2-methylthio-N6-(delta2-isopentenyl)-adenosine. Nucleic Acids Res. 1981 Jan 24;9(2):401–414. doi: 10.1093/nar/9.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R., Haar R. A., Capecchi N. E., Sveda M. M. The isolation of a suppressible nonsense mutant in mammalian cells. Cell. 1977 Oct;12(2):371–381. doi: 10.1016/0092-8674(77)90113-1. [DOI] [PubMed] [Google Scholar]
- Capecchi M. R., Klein H. A. Release factors mediating termination of complete proteins. Nature. 1970 Jun 13;226(5250):1029–1033. doi: 10.1038/2261029a0. [DOI] [PubMed] [Google Scholar]
- Celis J. E., Coulondre C., Miller J. H. Suppressor su+7 inserts tryptophan in addition to glutamine. J Mol Biol. 1976 Jul 5;104(3):729–734. doi: 10.1016/0022-2836(76)90132-7. [DOI] [PubMed] [Google Scholar]
- Celis J. E., Hooper M. L., Smith J. D. Amino acid acceptor stem of E. coli suppressor tRNA tyr is a site of synthetase recognition. Nat New Biol. 1973 Aug 29;244(139):261–264. doi: 10.1038/newbio244261a0. [DOI] [PubMed] [Google Scholar]
- Chan T. S., Garen A. Amino acid substitutions resulting from suppression of nonsense mutations. IV. Leucine insertion by the Su6+ suppressor gene. J Mol Biol. 1969 Nov 14;45(3):545–548. doi: 10.1016/0022-2836(69)90311-8. [DOI] [PubMed] [Google Scholar]
- Chan T. S., Garen A. Amino acid substitutions resulting from suppression of nonsense mutations. V. Tryptophan insertion by the Su9 gene, a suppressor of the UGA nonsense triplet. J Mol Biol. 1970 Apr 14;49(1):231–234. doi: 10.1016/0022-2836(70)90388-8. [DOI] [PubMed] [Google Scholar]
- Chang C. N., Blobel G., Model P. Detection of prokaryotic signal peptidase in an Escherichia coli membrane fraction: endoproteolytic cleavage of nascent f1 pre-coat protein. Proc Natl Acad Sci U S A. 1978 Jan;75(1):361–365. doi: 10.1073/pnas.75.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. N., Model P., Blobel G. Membrane biogenesis: cotranslational integration of the bacteriophage f1 coat protein into an Escherichia coli membrane fraction. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1251–1255. doi: 10.1073/pnas.76.3.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Cremer K. J., Bodemer M., Summers W. P., Summers W. C., Gesteland R. F. In vitro suppression of UAG and UGA mutants in the thymidine kinase gene of herpes simplex virus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):430–434. doi: 10.1073/pnas.76.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Downie J. A., Stewart J. W., Brockman N., Schweingruber A. M., Sherman F. Structural gene for yeast iso-2-cytochrome c. J Mol Biol. 1977 Jun 25;113(2):369–384. doi: 10.1016/0022-2836(77)90147-4. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980 Oct 9;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg S. P., Yarus M., Soll L. The effect of an Escherichia coli regulatory mutation on transfer RNA structure. J Mol Biol. 1979 Nov 25;135(1):111–126. doi: 10.1016/0022-2836(79)90343-7. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H. UGA suppression by normal tRNA Trp in Escherichia coli: codon context effects. Nucleic Acids Res. 1981 Feb 25;9(4):983–991. doi: 10.1093/nar/9.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F., Lacroute F. Genetic evidence for the creation of a reinitiation site by mutation inside the yeast ura 2 gene. Mol Gen Genet. 1979 May 23;173(1):109–113. doi: 10.1007/BF00267696. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Gallucci E., Garen A. Suppressor genes for nonsense mutations. II. The su-4 and su-5 suppressor genes of Escherichia coli. J Mol Biol. 1966 Jan;15(1):193–200. doi: 10.1016/s0022-2836(66)80220-6. [DOI] [PubMed] [Google Scholar]
- Garen A. Sense and nonsense in the genetic code. Three exceptional triplets can serve as both chain-terminating signals and amino acid codons. Science. 1968 Apr 12;160(3824):149–159. doi: 10.1126/science.160.3824.149. [DOI] [PubMed] [Google Scholar]
- Gauss D. H., Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1981 Jan 10;9(1):r1–23. [PMC free article] [PubMed] [Google Scholar]
- Gefter M. L., Russell R. L. Role modifications in tyrosine transfer RNA: a modified base affecting ribosome binding. J Mol Biol. 1969 Jan 14;39(1):145–157. doi: 10.1016/0022-2836(69)90339-8. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F., Wills N., Lewis J. B., Grodzicker T. Identification of amber and ochre mutants of the human virus Ad2+ND1. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4567–4571. doi: 10.1073/pnas.74.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F., Wolfner M., Grisafi P., Fink G., Botstein D., Roth J. R. Yeast suppressors of UAA and UAG nonsense codons work efficiently in vitro via tRNA. Cell. 1976 Mar;7(3):381–390. doi: 10.1016/0092-8674(76)90167-7. [DOI] [PubMed] [Google Scholar]
- Gilmore R. A., Stewart J. W., Sherman F. Amino acid replacements resulting from super-suppression of nonsense mutants of iso-1-cytochrome c from yeast. J Mol Biol. 1971 Oct 14;61(1):157–173. doi: 10.1016/0022-2836(71)90213-0. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Goman M., Errington L., Scaife J. Induction of RNA polymerase synthesis in Escherichia coli. Mol Gen Genet. 1975 Dec 30;143(1):79–83. doi: 10.1007/BF00269423. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Abelson J., Landy A., Brenner S., Smith J. D. Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature. 1968 Mar 16;217(5133):1019–1024. doi: 10.1038/2171019a0. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–134. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- Hartman P. E., Roth J. R. Mechanisms of suppression. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C. UGA mutations and UGA suppressors in yeast. Biochimie. 1976;58(1-2):179–182. doi: 10.1016/s0300-9084(76)80368-9. [DOI] [PubMed] [Google Scholar]
- Henderson D., Geisler N., Weber K. A periodic ultrastructure in intermediate filaments. J Mol Biol. 1982 Feb 25;155(2):173–176. doi: 10.1016/0022-2836(82)90444-2. [DOI] [PubMed] [Google Scholar]
- Hobson A. C., Gho D., Müller-Hill B. Isolation, genetic analysis, and characterization of Escherichia coli mutants with defects in the lacY gene. J Bacteriol. 1977 Sep;131(3):830–838. doi: 10.1128/jb.131.3.830-838.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofnung M., Jezierska A., Braun-Breton C. lamB mutations in E. coli K12: growth of lambda host range mutants and effect of nonsense suppressors. Mol Gen Genet. 1976 May 7;145(2):207–213. doi: 10.1007/BF00269595. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper J. E., Rowe L. B. Molecular expression and regulation of the galactose pathway genes in Saccharomyces cerevisiae. Distinct messenger RNAs specified by the Gali and Gal7 genes in the Gal7-Gal10-Gal1 cluster. J Biol Chem. 1978 Oct 25;253(20):7566–7569. [PubMed] [Google Scholar]
- Inokuchi H., Yamao F., Sakano H., Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. I. Amber suppressor su+2, an anticodon mutant of tRNA2Gln. J Mol Biol. 1979 Aug 25;132(4):649–662. doi: 10.1016/0022-2836(79)90380-2. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I. The binary code for protein-nucleic acid recognition with repulsive guanine: application to tRNA case. FEBS Lett. 1975 Nov 15;59(2):282–286. doi: 10.1016/0014-5793(75)80393-0. [DOI] [PubMed] [Google Scholar]
- Jarema M. A., Lu P., Miller J. H. Genetic assignment of resonances in the NMR spectrum of a protein: lac repressor. Proc Natl Acad Sci U S A. 1981 May;78(5):2707–2711. doi: 10.1073/pnas.78.5.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadner R. J., Liggins G. L. Transport of vitamin B12 in Escherichia coli: genetic studies. J Bacteriol. 1973 Aug;115(2):514–521. doi: 10.1128/jb.115.2.514-521.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantrowitz E. R., Foote J., Reed H. W., Vensel L. A. Isolation and preliminary characterization of single amino acid substitution mutants of aspartate carbamoyltransferase. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3249–3253. doi: 10.1073/pnas.77.6.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan S., Stretton A. O., Brenner S. Amber suppressors: efficiency of chain propagation and suppressor specific amino acids. J Mol Biol. 1965 Dec;14(2):528–533. doi: 10.1016/s0022-2836(65)80202-9. [DOI] [PubMed] [Google Scholar]
- Knowlton R. G., Soll L., Yarus M. Dual specificity of su+ 7 tRNA. Evidence for translational discrimination. J Mol Biol. 1980 Jun 5;139(4):705–720. doi: 10.1016/0022-2836(80)90056-x. [DOI] [PubMed] [Google Scholar]
- Knowlton R. G., Yarus M. Discrimination between aminoacyl groups on su+ 7 tRNA by elongation factor Tu. J Mol Biol. 1980 Jun 5;139(4):721–732. doi: 10.1016/0022-2836(80)90057-1. [DOI] [PubMed] [Google Scholar]
- Kohli J., Hottinger H., Munz P., Strauss A., Thuriaux P. Genetic Mapping in SCHIZOSACCHAROMYCES POMBE by Mitotic and Meiotic Analysis and Induced Haploidization. Genetics. 1977 Nov;87(3):471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Kwong T., Altruda F., Söll D., Wahl G. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. J Biol Chem. 1979 Mar 10;254(5):1546–1551. [PubMed] [Google Scholar]
- Konings R. N., Hulsebos T., Van den Hondel C. A. Identification and characterization of the in vitro synthesized gene products of bacteriophage M13. J Virol. 1975 Mar;15(3):570–584. doi: 10.1128/jvi.15.3.570-584.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S. W., Sherman F. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. J Bacteriol. 1979 Sep;139(3):1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S. W., Stewart J. W., Parker J. H., Sherman F. Leucine insertion caused by a yeast amber suppressor. J Mol Biol. 1977 Jan 5;109(1):13–22. doi: 10.1016/s0022-2836(77)80043-0. [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Stewart J. W., Sherman F. Serine substitutions caused by an ochre suppressor in yeast. J Mol Biol. 1975 Jun 5;94(4):595–610. doi: 10.1016/0022-2836(75)90324-1. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masselot M., Robichon-Szulmajster H. Nonsense mutation in the regulatory gene ETH2 involved in methionine biosynthesis in Saccharomyces cervisiae. Genetics. 1972 Aug;71(4):535–550. doi: 10.1093/genetics/71.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Adachi Y., Toh-e A., Oshima Y. Function of positive regulatory gene gal4 in the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae: evidence that the GAL81 region codes for part of the gal4 protein. J Bacteriol. 1980 Feb;141(2):508–527. doi: 10.1128/jb.141.2.508-527.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S. J., McLaughlin C. S. A comparison of small circular DNA molecules in psi+ and psi- strains of Saccharomyces cerevisiae. Biochim Biophys Acta. 1977 Nov 2;479(1):119–121. doi: 10.1016/0005-2787(77)90131-9. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Farabaugh P. J. Correlation of nonsense sites in the lacI gene with specific codons in the nucleotide sequence. Nature. 1978 Aug 24;274(5673):770–775. doi: 10.1038/274770a0. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Miller J. H. Genetic studies of the lac repressor. XI. On aspects of lac repressor structure suggested by genetic experiments. J Mol Biol. 1979 Jun 25;131(2):249–258. doi: 10.1016/0022-2836(79)90075-5. [DOI] [PubMed] [Google Scholar]
- Minet M., Jauniaux J. C., Thuriaux P., Grenson M., Wiame J. M. Organization and expression of a two-gene cluster in the arginine biosynthesis of Saccharomyces cerevisiae. Mol Gen Genet. 1979 Jan 11;168(3):299–308. doi: 10.1007/BF00271500. [DOI] [PubMed] [Google Scholar]
- Murphy E. C., Jr, Wills N., Arlinghaus R. B. Suppression of murine retrovirus polypeptide termination: effect of amber suppressor tRNA on the cell-free translation of Rauscher murine leukemia virus, Moloney murine leukemia virus, and Moloney murine sarcoma virus 124 RNA. J Virol. 1980 May;34(2):464–473. doi: 10.1128/jvi.34.2.464-473.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Properties of a collection of Escherichia coli RNA polymerase mutants. Biochem Soc Trans. 1980 Dec;8(6):732–733. doi: 10.1042/bst0080732. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Loughney K., Hall B. D. Identification of the yeast DNA sequences that correspond to specific tyrosine-inserting nonsense suppressor loci. J Mol Biol. 1979 Aug 15;132(3):387–410. doi: 10.1016/0022-2836(79)90267-5. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Page G. S., Sentenac A., Piper P. W., Worthington M., Weiss R. B., Hall B. D. Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature. 1981 Jun 11;291(5815):464–469. doi: 10.1038/291464a0. [DOI] [PubMed] [Google Scholar]
- Olsson M. O., Isaksson L. A. Analysis of rpsD mutations in Escherichia coli. IV. Accumulation of minor forms of protein S7(K) in ribosomes of rpsD mutant strains due to translational read-through. Mol Gen Genet. 1980 Feb;177(3):485–491. doi: 10.1007/BF00271488. [DOI] [PubMed] [Google Scholar]
- Ono B. I., Stewart J. W., Sherman F. Yeast UAA suppressors effective in psi+ strains serine-inserting suppressors. J Mol Biol. 1979 Feb 15;128(1):81–100. doi: 10.1016/0022-2836(79)90309-7. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Iu A., Monastyrskaia G. S., Gubanov V. V., Gur'ev S. O., Chertov O. Iu. Pervichnaia struktura RNK-polimerazy Escherichia coli. Nukleotidnaia posledovatel'nost' gena rpoB i aminokislotnaia posledovatel'nost' beta-sub"edinitsy. Dokl Akad Nauk SSSR. 1980;253(4):994–998. [PubMed] [Google Scholar]
- Perutz M. F., Lehmann H. Molecular pathology of human haemoglobin. Nature. 1968 Aug 31;219(5157):902–909. doi: 10.1038/219902a0. [DOI] [PubMed] [Google Scholar]
- Philipson L., Andersson P., Olshevsky U., Weinberg R., Baltimore D., Gesteland R. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978 Jan;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- Pieczenik G., Model P., Robertson H. D. Sequence and symmetry in ribosome binding sites of bacteriophage f1 RNA. J Mol Biol. 1974 Dec 5;90(2):191–124. doi: 10.1016/0022-2836(74)90368-4. [DOI] [PubMed] [Google Scholar]
- Piper P. W. A correlation between a recessive lethal amber suppressor mutation in Saccharomyces cerevisiae and an anticodon change in a minor serine transfer RNA. J Mol Biol. 1978 Jun 25;122(2):217–235. doi: 10.1016/0022-2836(78)90037-2. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Wasserstein M., Engbaek F., Kaltoft K., Celis J. E., Zeuthen J., Liebman S., Sherman F. Nonsense suppressors of Saccharomyces cerevisiae can be generated by mutation of the tyrosine tRNA anticodon. Nature. 1976 Aug 26;262(5571):757–761. doi: 10.1038/262757a0. [DOI] [PubMed] [Google Scholar]
- Post L. E., Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980 May 25;255(10):4660–4666. [PubMed] [Google Scholar]
- Pratt D., Tzagoloff H., Beaudoin J. Conditional lethal mutants of the small filamentous coliphage M13. II. Two genes for coat proteins. Virology. 1969 Sep;39(1):42–53. doi: 10.1016/0042-6822(69)90346-8. [DOI] [PubMed] [Google Scholar]
- Rafalski A., Kohli J., Agris P., Söll D. The nucleotide sequence of a UGA suppressor serine tRNA from Schizosaccharomyces pombe. Nucleic Acids Res. 1979 Jun 25;6(8):2683–2695. doi: 10.1093/nar/6.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall-Hazelbauer L., Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973 Dec;116(3):1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Bessman M. J. Studeis on the biochemical basis of mutation. IV. Effect of amino acid substitution on the enzymatic and biological properties of bacteriophage T4 DNA polymerase. J Mol Biol. 1977 Oct 15;116(1):99–113. doi: 10.1016/0022-2836(77)90121-8. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Bessman M. J. Studies on the biochemical basis of mutation VI. Selection and characterization of a new bacteriophage T4 mutator DNA polymerase. J Mol Biol. 1981 Feb 5;145(4):677–695. doi: 10.1016/0022-2836(81)90309-0. [DOI] [PubMed] [Google Scholar]
- Roth J. R. Frameshift mutations. Annu Rev Genet. 1974;8:319–346. doi: 10.1146/annurev.ge.08.120174.001535. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Esposito R. E., Esposito M. S. The effect of ochre suppression on meiosis and ascospore formation in Saccharomyces. Genetics. 1977 Jan;85(1):35–54. doi: 10.1093/genetics/85.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRETTON A. O., BRENNER S. MOLECULAR CONSEQUENCES OF THE AMBER MUTATION AND ITS SUPPRESSION. J Mol Biol. 1965 Jun;12:456–465. doi: 10.1016/s0022-2836(65)80268-6. [DOI] [PubMed] [Google Scholar]
- Seale T. W., Brett M., Baron A. J., Fincham J. R. Amino acid replacements resulting from suppression and missense reversion of a chain-terminator mutation in Neurospora. Genetics. 1977 Jun;86(2 Pt 1):261–274. [PMC free article] [PubMed] [Google Scholar]
- Sommer H., Lu P. Lac Repressor. Fluorescence of the two tryptophans. J Biol Chem. 1976 Jun 25;251(12):3774–3779. [PubMed] [Google Scholar]
- Stewart J. W., Sherman F. Confirmation of UAG as a nonsense codon in bakers' yeast by amino acid replacements of glutamic acid 71 in iso-1-cytochrome c. J Mol Biol. 1973 Jun 25;78(1):169–184. doi: 10.1016/0022-2836(73)90436-1. [DOI] [PubMed] [Google Scholar]
- Strauss A. The genetic fine structure of the complex locus aro3 involved in early aromatic amino acid biosynthesis in Schizosaccharomyces pombe. Mol Gen Genet. 1979;172(3):233–241. doi: 10.1007/BF00271722. [DOI] [PubMed] [Google Scholar]
- Strigini P., Brickman E. Analysis of specific misreading in Escherichia coli. J Mol Biol. 1973 Apr 25;75(4):659–672. doi: 10.1016/0022-2836(73)90299-4. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Sugisaki H., Okamoto T., Takanami M. Studies on bacteriophage fd DNA. IV. The sequence of messenger RNA for the major coat protein gene. J Mol Biol. 1977 Apr 25;111(4):487–507. doi: 10.1016/s0022-2836(77)80065-x. [DOI] [PubMed] [Google Scholar]
- Szmelcman S., Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975 Oct;124(1):112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion J. P., Hofnung M. On some genetic aspects of phage lambda resistance in E. coli K12. Genetics. 1972 Jun;71(2):207–216. doi: 10.1093/genetics/71.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P., Minet M., ten Berge A. M., Zimmermann F. K. Genetic fine structure and function of mutants at the ilv1-gene locus of Saccharomyces cervisiae. Mol Gen Genet. 1971;112(1):60–72. doi: 10.1007/BF00266933. [DOI] [PubMed] [Google Scholar]
- WEIGERT M. G., GAREN A. AMINO ACID SUBSTITUTIONS RESULTING FROM SUPPRESSION OF NONSENSE MUTATIONS. I. SERINE INSERTION BY THE SU-1 SUPPRESSOR GENE. J Mol Biol. 1965 Jun;12:448–455. doi: 10.1016/s0022-2836(65)80267-4. [DOI] [PubMed] [Google Scholar]
- Waldron C., Cox B. S., Wills N., Gesteland R. F., Piper P. W., Colby D., Guthrie C. Yeast ochre suppressor SUQ5-ol is an altered tRNA Ser UCA. Nucleic Acids Res. 1981 Jul 10;9(13):3077–3088. doi: 10.1093/nar/9.13.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner C. K., Jacobson K. B. Mechanisms of suppression in Drosophila. IV. Specificity and properties of tyrosyl-tRNA synthetase. Can J Biochem. 1976 Jul;54(7):650–656. doi: 10.1139/o76-094. [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Brenner S. A suppressor mutation in the nematode acting on specific alleles of many genes. Nature. 1978 Oct 26;275(5682):715–719. doi: 10.1038/275715a0. [DOI] [PubMed] [Google Scholar]
- Weigert M. G., Lanka E., Garen A. Amino acid substitutions resulting from suppression of nonsense mutations. 3. Tyrosine insertion by the Su-4 gene. J Mol Biol. 1967 Feb 14;23(3):401–404. doi: 10.1016/s0022-2836(67)80114-1. [DOI] [PubMed] [Google Scholar]
- Weigert M. G., Lanka E., Garen A. Amino acid substitutions resulting from suppression of nonsense mutations. II. Glutamine insertion by the Su-2 gene; tyrosine insertion by the Su-3 gene. J Mol Biol. 1965 Dec;14(2):522–527. doi: 10.1016/s0022-2836(65)80201-7. [DOI] [PubMed] [Google Scholar]
- Weiss S. R., Hackett P. B., Oppermann H., Ullrich A., Levintow L., Bishop J. M. Cell-free translation of avian sarcoma virus RNA: suppression of the gag termination codon does not augment synthesis of the joint gag/pol product. Cell. 1978 Oct;15(2):607–614. doi: 10.1016/0092-8674(78)90029-6. [DOI] [PubMed] [Google Scholar]
- Wetzel R., Kohli J., Altruda F., Söll D. Identification and nucleotide sequence of the sup8-e UGA-suppressor leucine tRNA from Schizosaccharomyces pombe. Mol Gen Genet. 1979 May 4;172(2):221–228. doi: 10.1007/BF00268286. [DOI] [PubMed] [Google Scholar]
- White B. N., Tener G. M. Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su(s) locus. J Mol Biol. 1973 Mar 15;74(4):635–651. doi: 10.1016/0022-2836(73)90054-5. [DOI] [PubMed] [Google Scholar]
- Wosnick M. A., White B. N. A doubtful relationship between tyrosine tRNA and suppression of the vermilion mutant in Drosophila. Nucleic Acids Res. 1977 Nov;4(11):3919–3930. doi: 10.1093/nar/4.11.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M., Folk W. R., Berg P., Soll L. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J Mol Biol. 1974 Jun 25;86(2):245–260. doi: 10.1016/0022-2836(74)90016-3. [DOI] [PubMed] [Google Scholar]
- Young I. G., Rogers B. L., Campbell H. D., Jaworowski A., Shaw D. C. Nucleotide sequence coding for the respiratory NADH dehydrogenase of Escherichia coli. UUG initiation codon. Eur J Biochem. 1981 May;116(1):165–170. doi: 10.1111/j.1432-1033.1981.tb05314.x. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Hall B. D. Yeast cytochrome c messenger RNA. In vitro translation and specific immunoprecipitation of the CYC1 gene product. J Biol Chem. 1976 Oct 25;251(20):6320–6326. [PubMed] [Google Scholar]


