Abstract
Recirculating aquaculture and aquaponics are considered sustainable aquaculture models playing important roles in animal-derived protein supply. In these aquaculture systems, microorganisms are crucial for the system stability. The community coalescence by mixing substances and microorganisms from various microhabitats under hydraulic forces is important for shaping the bacterial communities in these small-scale complex systems. However, the influences of community coalescence on bacterial communities remain rarely revealed in these systems. In this study, aquaponics (APS) and recirculating aquaculture (RAS) systems were set up to explore the bacterial community coalescence across different microhabitats, including water, fish feces, biofilter biofilms, and plant rhizosphere environment. Our results showed that diversity and compositions varied across different microhabitats in both systems. However, bacterial transmissions across these microhabitats differed between systems. The core microbiome of the RAS and APS were formed under community coalescence with the highest contribution of bacterial taxa derived from the fish feces. Nevertheless, the plant rhizosphere bacterial community also contributed to the core microbiome of the APS. Furthermore, the core taxa showed a higher average degree than the other nodes in the bacterial community networks in all microhabitats except for the plant rhizosphere environment, implying the important roles of core taxa in maintaining these bacterial community networks. Our results provide new insights into the assembly of bacterial communities under community coalescence in the artificial aquatic ecosystems comprising complex microhabitats, which is vital for developing microbial solutions for regulating the microbial communities to improve system performance in the future.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00248-024-02461-5.
Keywords: Aquaponics, Microhabitats, Community coalescence, Bacterial transmission, Core taxa, Co-occurrence network
Introduction
The demand for animal-derived protein continues to grow, driven by population growth, rising incomes, and urbanization [1]. Aquaculture is expected to play a crucial role in meeting global protein needs [2]. Nevertheless, aquaculture must address sustainability challenges, including water usage, feed efficiency, and environmental impacts [3]. The recirculating aquaculture system (RAS) offers the potential for relatively minimal environmental discharge by removing toxic fish metabolic waste and reusing water [4]. Moreover, aquaponics, the integration of RAS and hydroponics, offers greater benefits in pollution reduction and productivity improvement [5]. Within the aquaponics system (APS), excrement produced by aquatic animals is mineralized by microorganisms into nutrients that can be utilized by plants [6, 7]. In both APS and RAS, microorganisms play important roles in influencing biogeochemical processes, as well as regulating the health of aquatic animals and plants [8–10]. Thus, it is now widely recognized that microorganisms, especially bacterial taxa, play a crucial role in maintaining the stability of both APS and RAS [11, 12], which can be considered biological indicators for system function and stability.
As small-scale complex ecosystems, APS and RAS encompass a variety of microhabitats for microbes to colonize, which are associated with water, biofilms, aquatic animals, and plants [11, 13]. Substances and microorganisms from different microhabitats of these artificial aquatic ecosystems can undergo passive dispersal and mixing under hydraulic forces, leading to community coalescence. Community coalescence refers to merging two or more previously distinct microbial communities into a single environment, resulting in entire communities and their environments interacting [14, 15]. Microbial community coalescence has been reported to show important ecological impacts in other artificial ecosystems [16, 17]. The interchange and interaction of microbial communities and substances across various microhabitats in the APS and RAS could lead to complex ecological implications. For instance, fish feces and their heterotrophic bacteria-dominated microbial communities can coalesce with the nitrifying bacteria-dominated biofilms in the biofilter unit of an RAS under the influence of water flow. This community coalescence may introduce competition between heterotrophic and nitrifying bacteria for oxygen and space, while also protecting the nitrifying bacteria from detachment and grazing [12, 13, 18]. In an APS, with the presence of plants, the ecological effects of community coalescence may become even more complex. It has been suggested that bacterial communities from tilapia recirculating aquaculture systems can serve as a potential species pool of plant growth-promoting microorganisms (PGPMs) [19], which could show positive effects on plant health and production [20–22]. Moreover, the plant-associated microbiome could also influence the compositions of bacterial communities inhabiting the other microhabitats of the APS, although related research remains limited. Hence, understanding the consequences of community coalescence is vital for revealing the role of microbial communities in system function and stability, and in the future, applying microbial techniques to improve system performances.
Various ecological processes, including dispersal, selection, drift, and diversification, jointly drive the assembly of bacterial communities [23]. In the RAS or APS, when community coalescence occurs, it can be hypothesized that the microbial individuals have the chance to arrive in any microhabitat under the passive dispersal driven by hydraulic forces. However, whether the microbial individuals can colonize this microhabitat would be influenced by several ecological processes, for example, the selection of different abiotic forces and biotic interactions [24]. Thus, dispersal and selection processes are recognized as key mechanisms underlying the community assembly during the coalescence [14, 15]. Furthermore, biotic interactions dominated by competition or mutualism can result in varying stability among the new communities. Therefore, characterizing the bacterial transmissions across different microhabitats and the potential biotic interactions within these communities is essential for understanding the influences of community coalescence on the community assembly of these small-scale aquatic ecosystems. Furthermore, the core microbiome can be formed during community coalescence and play a crucial role in regulating the potential biotic interactions among microbes [25, 26]. Core microbiomes are measured as the microbial taxa shared among two or more samples from a particular host or environment [27]. This concept was first proposed in the study of host microbiomes [28]. In recent years, the idea of core microbiome has been widely applied in various studies of environmental microbial ecology [29, 30]. Identifying the core microbiome in a system is crucial for understanding the role of microorganisms in maintaining system stability [27]. For instance, a study reported that core microbes regulated plant health through changing microbial interactions and network complexity during community coalescence [31]. Here, it can be hypothesized that a distinct core microbiome would be formed in the APS from the RAS under community coalescence, as the plant rhizosphere microhabitat of the APS can provide an additional source and sink of bacterial taxa for the bacterial communities inhabiting the other microhabitats compared to the RAS. However, our current understanding of the core microbiome in the RAS and APS is still limited. Knowledge of the biotic interactions among microbes in these systems, as well as the roles of core microbial taxa in these interactions, is even more restricted.
To address these questions, we established small-scale recirculating aquaculture systems (RAS) and aquaponics systems (APS), and bacterial community compositions were assessed in microhabitats associated with fish tank water (FW), fish feces (FF), biofilter biofilms (BT), and plant rhizosphere environment (RZ) during the 78-day aquaculture experiment using the amplicon-based high-throughput sequencing technology. In the present study, thus, we aimed to answer the following questions: (1) How are bacterial community compositions, bacterial transmissions, and co-occurrence patterns characterized in the APS and RAS during community coalescence, and does the presence of plants influence these dynamics? (2) What core bacterial taxa emerge during community coalescence in these systems, and are these core taxa important in maintaining potential biotic interactions among the microbes within these systems?
Materials and Methods
Experiment Design and System Establishment
The experiment was conducted outdoors at the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, in Wuxi of Jiangsu Province, China (31.511126°N, 120.239273°E) from August to October 2023. Three aquaponics systems (APS) and three recirculating aquaculture systems (RAS) were established, each containing a 240-L fish tank, a 48-L filtering tank, a 96-L biofilter tank, and a 360-L hydroponic tank (Fig. S1a). The water flow direction is shown in Fig. S1a.
The details for the establishment of the APS and RAS were presented in the supporting information. The aerated tap water was added to all systems once a week throughout the experiment to compensate for the water loss caused by evaporation. The water in the fish tank, biofilter tank, and hydroponic tank was continuously under aeration in all systems. All systems were powered by solar energy. The timeline of the entire experiment is shown in Fig. S1b.
Sample Collection
Different types of samples were collected on days 16, 30, 50, 64, and 78 from the APS and RAS (Fig. S1b). At each sampling time point, water samples from the fish tanks in all systems were collected and then filtered through the 0.22-μm pore-size polycarbonate membrane filter (47-mm diameter; Millipore) to obtain the bacterial biomass in fish tank water (hereafter referred to as FW). Furthermore, fish feces from the cultured tilapias (Oreochromis niloticus) were collected using a stainless steel net with a 2-mm mesh size 1 h after feeding the fish and then frozen dry to determine the bacterial community compositions of fish feces (hereafter referred to as FF). Ten biofilters were collected randomly from the biofilter tank of each system. The plant roots were also sampled by randomly cutting three water spinach plants (Ipomoea aquatica) in the hydroponic tank of APS. The biofilm bacterial biomass attached to the biofilter and plant root surfaces (hereafter referred to as BT and RZ, respectively) was obtained as follows: (1) the sampled biofilters or plant roots were placed in 35 mL of PBS solution (pH 7.2–7.4; Solarbio) and vortexed at room temperature (~ 20 °C) for 5 min. (2) The biofilters or plant roots were transferred from the initial PBS solution to a fresh 35 mL of PBS solution using sterile tweezers, and the vortexing process was then repeated. (3) The PBS solutions from both steps were filtered through a 0.22-μm pore-size polycarbonate membrane filter (47-mm diameter; Millipore). The bacterial biomass was retained on the filters.
DNA Extraction, PCR, High-Throughput Sequencing, and Sequencing Processing
The DNA extraction for FW, FF, BT, and RZ bacterial biomass was performed using E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to manufacturer’s protocols. The DNA extraction of a total of 105 samples from different microhabitats and time points was conducted. The primers 341F (5’-CCTAYGGGRBGCASCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’) were selected for PCR amplification of the bacterial 16S rRNA gene [32, 33]. PCR amplification was implemented in a total volume of 20-μL mixture containing 4 μL of 5 × FastPfu buffer, 0.4 μL of FastPfu polymerase, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), and 10 ng of template DNA. The amplification program was as follows: an initial denaturation step at 95 °C for 2 min, followed by 25 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. For each sample, PCR was performed three times to minimize technical errors. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA, USA). Purified PCR products were quantified by Qubit®3.0 (Life Invitrogen), and every 24 amplicons whose barcodes were different were mixed equally. The pooled DNA product was used to construct the Illumina Pair-End library following Illumina’s genomic DNA library preparation procedure. Then, the amplicon library was paired-end sequenced (2 × 250) on an Illumina NovaSeq platform (Shanghai BIOZERON Biotech. Co., Ltd). The raw reads were deposited into the National Omics Data Encyclopedia (NODE) database (Accession Number: OEX028684).
The paired-end raw sequences were processed according to our previous study [34]. Generally, sequences that have a low quality (average quality score < 25 and a read length < 200 bp), mismatches with primer matching/comprising blurred characters, and problems with being assembled, were discarded using QIIME (v1.9.1). Chimeric sequences were detected and removed using UCHIME de novo strategy [35]. The operational taxonomic units (OTUs) were then clustered using UCLUST at a 97% similarity [36]. The taxonomic information of each OTU was then assigned based on SILVA database (Release138 http://www.arb-silva.de). Non-bacterial sequences, including chloroplast and mitochondria, were discarded. Rare OTUs with reads < 2 were removed to minimize sequencing errors. Finally, samples were rarefied at 42,618 sequences according to the minimum sequence number.
Statistical Analyses
The alpha diversity of bacterial communities was indicated by the number of observed OTUs (hereafter referred to as richness), phylogenetic diversity, and Pielou’s evenness (hereafter referred to as evenness), which were generated using packages “vegan” and “picante” in R [37, 38]. Three-way ANOVA tests were employed to explore the impacts of systems (i.e., APS vs. RAS), microhabitats (i.e., FW, FF, and BT), and sampling time on alpha diversity of bacterial communities (RZ samples were excluded). The community dissimilarity was calculated based on the Bray–Curtis distance matrix. PERMANOVA tests were performed to detect the differences in bacterial community compositions across different systems, microhabitats, and sampling time points (RZ samples were excluded). Principal coordinates analysis (PCoA) was used to examine the bacterial community dissimilarity across systems and microhabitats, simplified and visualized into a plot of two-dimensional coordinates. PERMANOVA tests and PCoA were all conducted using the package “vegan” in R.
The top ten bacterial classes and genera were selected as the dominant taxa. Biomarker OTUs, which were significantly enriched or depleted in FW, FF, and BT microhabitats of the APS compared to those of the RAS, were identified using negative binomial generalized linear models with the package “DESeq2” of R [39], and then visualized with volcano diagrams. The detection of core OTUs for the APS and RAS was according to the following criteria: (1) The OTU should be presented in all samples of the same system, and (2) the average relative abundance of the OTU across all samples derived from the same system should be > 0.5%. Heatmaps were used to visualize the relative abundances of these biomarker OTUs and core OTUs across different systems and microhabitats with the package “pheatmap” in R. SourceTracker (version 0.9.5) analyses were employed to evaluate the bacterial transmissions across the different microhabitats (i.e., FW, FF, BT, and RZ) in the APS and RAS, respectively. SourceTracker is a Bayesian approach that uses Gibbs sampling to estimate the proportion of each sink sample composed of taxa from a known source environment [40].
Topological networks were constructed for bacterial communities derived from different microhabitats using the package “WGCNA” based on Spearman’s correlation matrices (Spearman r > 0.8 and adjusted p value < 0.05) [41]. Only OTUs with relative abundance > 0.01% and detection rate > 60% were included in the analyses to enhance network reliability. The networks were divided into modules using the fast greedy modularity optimization [42]. Topological properties including modularity, clustering coefficient, average path length, network diameter, and average degree were calculated using the package “igraph” in R [43]. A total of 1000 random networks of equal size were generated for each empirical network using the “igraph” package in R, and all of the indices of the random networks were calculated individually. A statistical Z test was used to verify whether the network indices between the empirical and random networks were significantly different. The robustness and vulnerability of all empirical networks were calculated according to the methods provided by Yuan et al. [44]. The robustness of a network represents the ability of the network to maintain its connectivity after a random failure or an intentional attack, meaning that the nodes (or links) deletions [45]. The network vulnerability depends on the extent to which the removal of a node reduces global efficiency, a measure of the speed and reliability of information, material, or energy flow across the network [44]. The number of core OTUs and their average degree were obtained to estimate the importance of core OTUs in maintaining the bacterial community networks. Gephi (version 0.9.2) was used to depict the network analyses.
Results
The Dynamics of Diversity and Community Structure of Bacterial Communities in the Aquaponics and Recirculating Systems
The alpha diversity of bacterial communities across different microhabitats and systems was represented by indices of richness, phylogenetic diversity, and evenness. In the APS and RAS, the richness, phylogenetic diversity, and evenness were the lowest in the FF bacterial communities, followed by those of the FW bacterial communities (Fig. S2a-c). However, the BT and RZ bacterial communities harbored the highest alpha diversity. Results of three-way ANOVA tests showed that the “microhabitat” factor exhibited overwhelmingly greater influences on richness, phylogenetic diversity, and evenness of bacterial communities than the “system” and “time” factors indicated by much higher F values (Table 1). Furthermore, our analysis revealed significant interaction effects between the “system” and “time” factors on both richness (P < 0.05) and phylogenetic diversity (P < 0.01). Similarly, significant interaction effects were observed between the “microhabitat” and “time” factors on all richness (P < 0.001), phylogenetic diversity (P < 0.01), and evenness (P < 0.001).
Table 1.
The influences of system, microhabitat, time, and their interactions on alpha diversity and community dissimilarity of bacterial communities using three-way ANOVA test and permutational multivariate analysis of variance (PerMANOVA). Plant rhizosphere samples were excluded
| Alpha diversity | Community dissimilarity | ||||
|---|---|---|---|---|---|
| Richness | Phylogenetic diversity | Evenness | Bray–Curtis | ||
| F value | F value | F value | R2 | F value | |
| System | 6.0* | 4.4* | 10.1** | 0.22 | 47.6*** |
| Microhabitat | 1222.5*** | 1227.1*** | 345.1*** | 0.16 | 17.3*** |
| Time | 21.1*** | 21.8*** | 42.6*** | 0.02 | 0.9 |
| System: microhabitat | 0.4 | < 0.1 | 1.8 | 0.27 | 29.5*** |
| System: time | 3.0* | 3.8** | 2.2 | 0.01 | 0.7 |
| Microhabitat: time | 23.9*** | 22.7** | 43.1*** | 0.02 | 0.4 |
| System: microhabitat: time | 1.6 | 1.8 | 0.7 | 0.02 | 0.5 |
*P < 0.05; **P < 0.01; ***P < 0.001
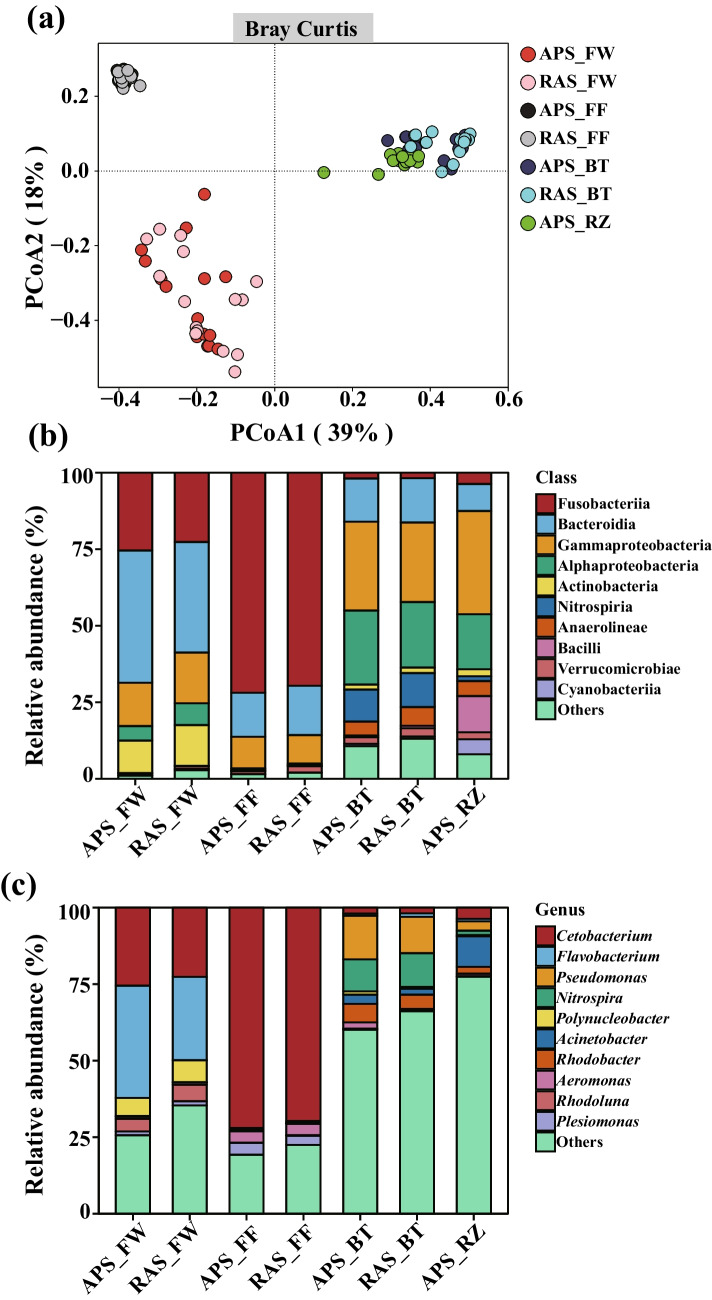
The PCoA plot showed that the first two components (PCoA1 and PCoA2) explained a total of 57% of the variance in the bacterial community compositions across different systems and microhabitats (Fig. 1a). PerMANOVA tests based on Bray–Curtis distance showed significant bacterial community dissimilarity across samples from different systems and microhabitats (Table 1). However, bacterial community dissimilarity across samples from different time points was identified as insignificant. Bacterial community compositions were strongly shaped by microhabitats rather than systems. Furthermore, bacterial communities from RZ samples showed higher similarity with those from BT samples rather than FW and FF samples.
Fig. 1.
Bacterial community structure and compositions of different systems and microhabitats. a Principal coordinate analysis (PCoA) of the bacterial communities in different microhabitats and systems based on Bray–Curtis distance. The bacterial community of each microhabitat of the aquaponics or recirculating aquaculture systems contained samples from all the five time points, with three replicates per time point (n = 15). b–c Taxonomic composition of the bacterial communities of different systems and microhabitats at the class and genus levels, respectively. Only the dominant classes and genera (top 10) were represented. APS, aquaponics system; RAS, recirculating aquaculture system; FW, fish tank water; FF, fish feces; BT, biofilter biofilms; RZ, plant rhizosphere microhabitat
The Dominant Taxa and Differential OTUs of Bacterial Communities Across Different Systems and Microhabitats
The taxa with the top ten highest relative abundance were considered the dominant taxa at both class and genus levels (Fig. 1b and c). The relative abundance of dominant taxa exhibited higher variations between microhabitats than those between systems at both class and genus levels. At the class level, Bacteroidia and Fusobacteriia dominated the FW bacterial communities of both two systems (Fig. 1b). In the FF bacterial communities, the class Fusobacteriia displayed an overwhelmingly dominant abundance of ca. 70%. However, bacterial OTUs belonging to the classes Gammaproteobacteria and Alphaproteobacteria dominated in both the BT and RZ bacterial communities. Furthermore, the class Nitrospiria exhibited a relative abundance of over 10% in the BT bacterial communities of both systems, whereas the relative abundance of the class Bacilli exceeded 10% in the RZ bacterial community. At the genus level, bacterial OTUs affiliated with the genera Flavobacterium and Cetobacterium were the most abundant in the FW bacterial communities of both systems (Fig. 1c). The genus Cetobacterium accounted for approximately 70% of the total reads in the FF bacterial communities of both systems. In the BT bacterial communities, the genera Pseudomonas and Nitrospira exhibited the highest relative abundances, each exceeding 10%. In the RZ bacterial community of the APS, the genus Acinetobacter displayed the highest relative abundance (approximately 10%), followed by the genera Bacillus and Exiguobacterium with the relative abundance of each at about 5%.
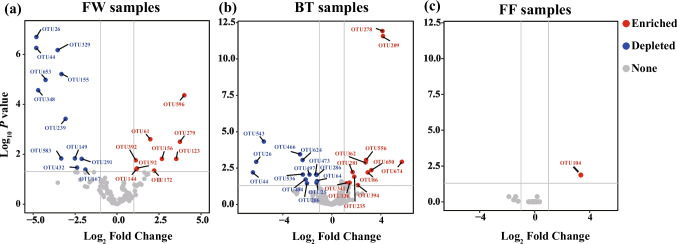
To further identify the significant difference in bacterial community compositions between the two systems, we conducted differential OTU abundance analyses for bacterial OTUs with a relative abundance of > 0.1% in at least one set of samples (Fig. 2). For the FW samples, 9/126 OTUs were significantly enriched in the APS (Fig. 2a), which belonged to the phyla/classes Actinobacteriota, Bacteroidota, and Alphaproteobacteria (Table S1). Moreover, 12/126 OTUs were significantly depleted in the FW samples of aquaponics in comparison with the RAS, which were assigned to the phyla/classes Actinobacteriota, Alphaproteobacteria, Bacteroidota, and Verrucomicrobiota. For the BT samples, 12/228 bacterial OTUs belonging to the phyla/classes Actinobacteriota, Armatimonadota, and Bacteroidota significantly enriched in the APS, whereas 13/228 bacterial OTUs significantly depleted in the APS were affiliated with the phyla/classes Acidobacteriota, Actinobacteriota, Chloroflexi, Bacteroidota, Gammaproteobacteria, and Verrucomicrobiota (Fig. 2b and Table S1). Only one of 42 OTUs (OTU104), which was assigned to the genus Bacteroides, phylum Bacteroidota, was significantly enriched in the APS compared to the RAS found in the FF samples, whereas none was found significantly depleted in the APS (Fig. 2c and Table S1).
Fig. 2.
Volcano plots showing bacterial OTUs significantly enriched or depleted in the aquaponics system considering the recirculating aquaculture system as the reference across different microhabitats. FW, fish tank water; FF, fish feces; BT, biofilter biofilms
Potential Bacterial Sources of Bacterial Communities Colonized in Different Microhabitats and Their Core Microbiome of the Two Systems
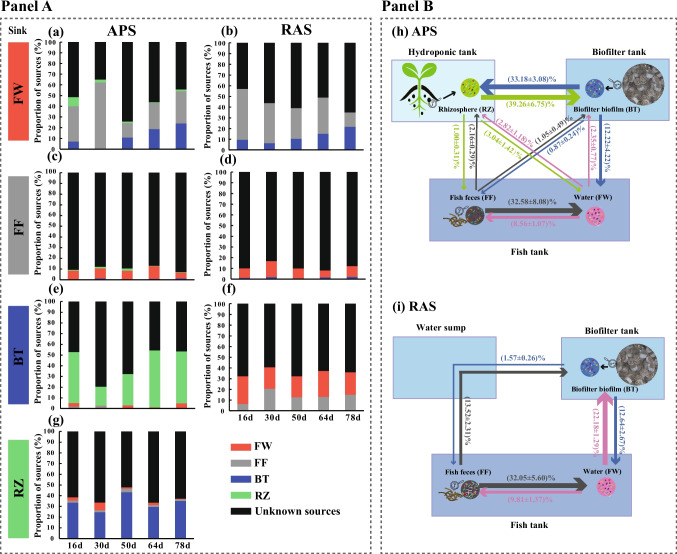
We employed SourceTracker analyses to explore the temporal dynamics of relative contributions of potential bacterial sources to the assembly of bacterial communities colonized in each microhabitat of the APS and RAS (Fig. 3a–g). Furthermore, we quantified the average bacterial transmissions across different microhabitats within these systems over the 78-day period of community coalescence (Fig. 3h and i). Generally, the bacterial sources for structuring community compositions for FW and FF samples were similar between the two systems. In both systems, the FF bacterial source exhibited the highest contribution to the assembly of the FW bacterial community, accounting for an average of over 30%, followed by the BT bacterial source with an average of approximately 12% (Fig. 3h and i). Moreover, we observed an increasing pattern of the relative contribution of BT bacterial source to the FW bacterial communities over time (Fig. 3a and b). In addition, the RZ bacterial source contributed an average of approximately 3% to the total sources of FW bacterial community composition in the APS (Fig. 3h), with a decreasing trend over time (Fig. 3a). For the FF bacterial community compositions in both systems, the contribution of the FW bacterial source to the FF community was around 10% (Fig. 3h and i). Furthermore, the contribution of BT bacterial source to the FF bacterial community was slightly lower in the APS than in the RAS, whereas the RZ bacterial community contributed an average of ca. 1% to the sources of the FF bacterial community in the APS (Fig. 3h and i). The contributions of different bacterial sources to the assembly of BT bacterial communities varied between the two systems (Fig. 3h and i). In the APS, the dominant bacterial source for the BT bacterial community was the RZ bacterial community, contributing an average of approximately 40% (Fig. 3h). Additionally, the contribution of the RZ bacterial sources to the assembly of the BT bacterial communities decreased on day 30, and then increased over time (Fig. 3e). In the RAS, however, the FW bacterial source showed the highest contribution to the BT bacterial community compositions with an average contribution of ca. 20%, followed by the FF (> 10%) (Fig. 3i). For the assembly of the RZ bacterial community in the APS, the BT bacterial community was the most important known source, contributing more than 30%, followed by the FW (2.82%) and the FF (2.16%) (Fig. 3h). Moreover, in the APS, we found that the average contribution of the RZ bacterial source to the BT bacterial community exceeded the average contribution of the BT to RZ (Fig. 3h).
Fig. 3.
Bacterial transmissions across different microhabitats of the aquaponics and recirculating aquaculture systems derived from Source Tracker analyses. Panel A represented the temporal dynamics of the relative contribution of potential sources to the bacterial community compositions of each sink of the aquaponics (a, c, e, and g) and recirculating aquaculture (b, d, and f) systems derived from SourceTracker analyses. (a–b), (c–d), (e–f), and (g) represented the results with bacterial communities derived from fish tank water (FW), fish feces (FF), biofilter biofilms (BT), and plant rhizosphere microhabitat (RZ) as the sink, respectively. Panel B represented the average proportions of bacterial transmissions among different microhabitats of the aquaponics (h) and recirculating aquaculture (i) systems accompanied by the standard errors of the mean (SEM). APS, aquaponics system; RAS, recirculating aquaculture system
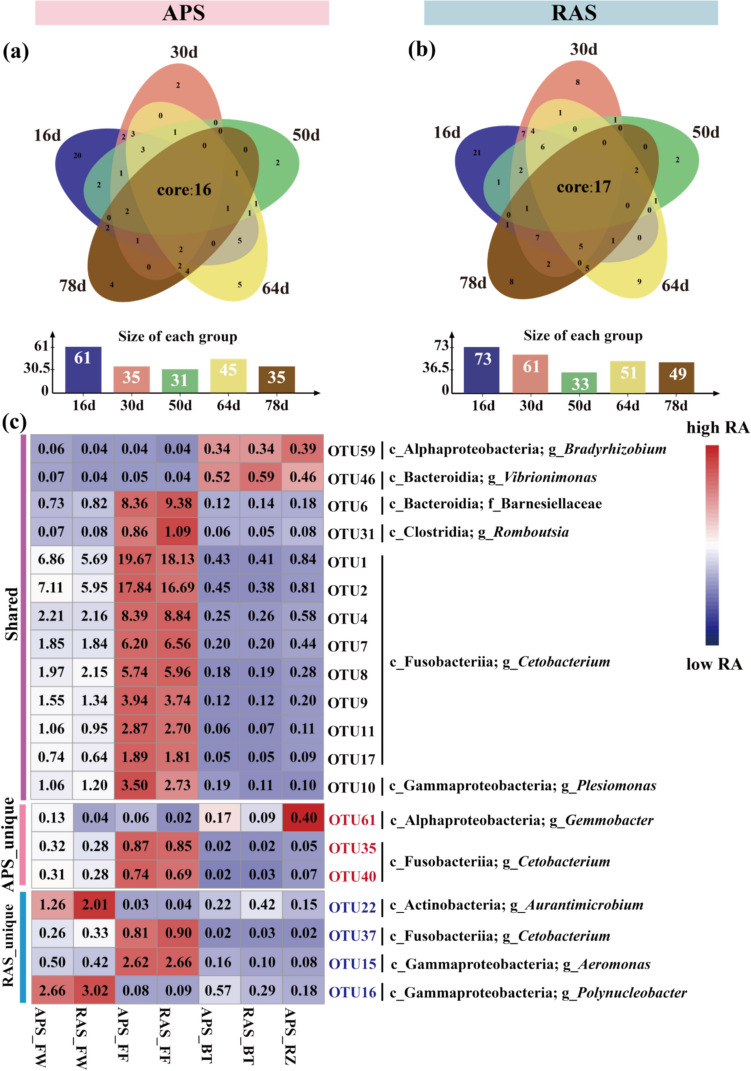
We further detected core OTUs for the APS and RAS, referring to the shared OTUs across samples from different microhabitats and time points. In general, there were 16 and 17 core bacterial OTUs detected in the APS and RAS, respectively, across all microhabitats and time points (Fig. 5a–b). We further divided these core OTUs found in the two systems into three clusters, referring to core OTUs shared in both systems (i.e., “shared” cluster), core OTUs detected only in the APS (i.e., “APS_unique” cluster), and core OTUs detected only in the RAS (i.e., “RAS_unique” cluster) (Fig. 4). Among them, 13 OTUs were assigned to the “shared” cluster, whereas three OTUs and four OTUs belonged to the “APS_unique” and “RAS_unique” clusters, respectively. Furthermore, we found that most of the core OTUs (11/13) from the “shared” cluster showed the highest relative abundance in FF samples in both systems, and the majority of these shared core OTUs were affiliated with the genus Cetobacterium, class Fusobacteriia. The other two core OTUs from the “shared” cluster showed higher relative abundance in BT and RZ samples compared to those in FW and FF samples, which taxonomically belonged to the genera Bradyrhizobium (class Alphaproteobacteria) and Vibrionimonas (class Bacteroidia), respectively. Moreover, two OTUs from the “APS_unique” cluster (OTU35 and OTU40) also showed the highest relative abundance in FF samples of both systems, whereas the other OTU (OTU61) belonging to the genus Gemmobacter (class Alphaproteobacteria) exhibited the highest relative abundance in RZ samples of the APS. For the four core OTUs from the “RAS_unique” cluster, two of them were the most abundant in FF samples, whereas the other two OTUs were the most abundant in FW samples.
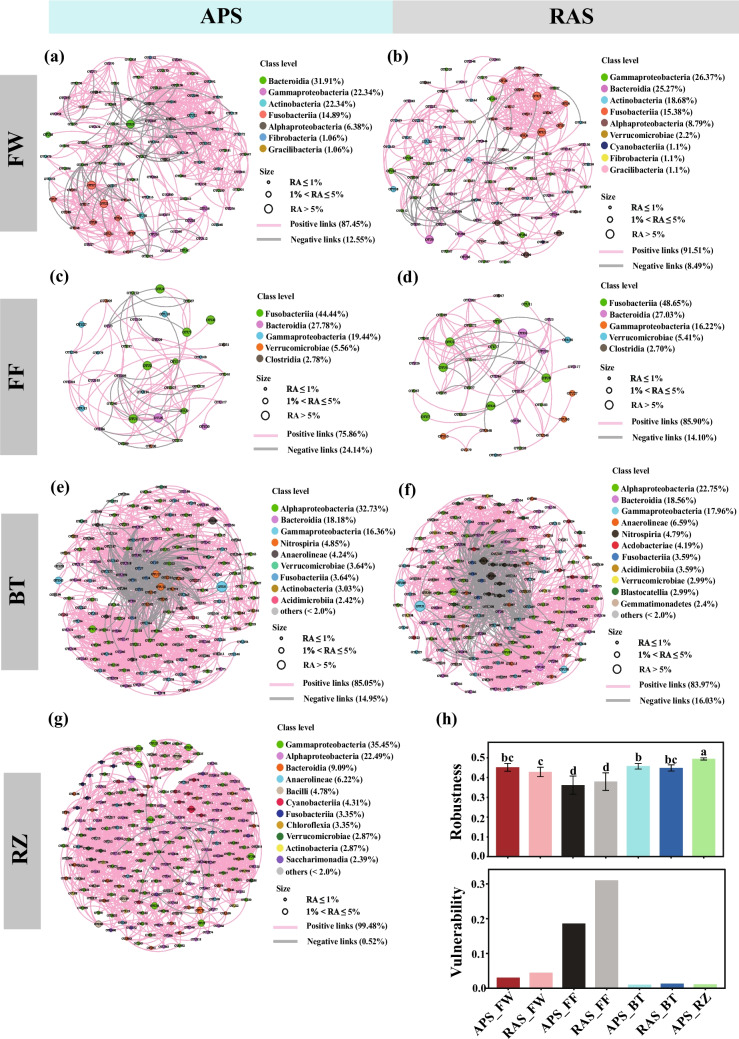
Fig. 5.
Co-occurrence patterns of bacterial communities derived from different microhabitats and systems. a–g Network graphs showing positive and negative relationships (links) between bacterial OTUs (nodes). h Comparisons of robustness and vulnerability of bacterial community networks across different microhabitats and systems. RA, relative abundance; APS, aquaponics system; RAS, recirculating aquaculture system; FW, fish tank water; FF, fish feces; BT, biofilter biofilms; RZ, plant rhizosphere microhabitat
Fig. 4.
The core bacterial taxa formed during the community coalescence of the aquaponics and recirculating aquaculture systems. a–b Venn diagrams showing the number of shared bacterial OTUs across different microhabitats among various time points. Bar charts showing the number of shared bacterial OTUs across different microhabitats at each time point. c Heatmaps showing the relative abundances of core OTUs detected for aquaponics and recirculating aquaculture systems across different microhabitats. Core OTUs were divided into three clusters, referring to core OTUs shared in both systems (i.e., “shared” cluster), core OTUs detected only in the APS (i.e., “APS_unique” cluster), and core OTUs detected only in the RAS (i.e., 'RAS_unique' cluster). RA, relative abundance; APS, aquaponics system; RAS, recirculating aquaculture system; FW, fish tank water; FF, fish feces; BT, biofilter biofilms; RZ, plant rhizosphere microhabitat
Co-occurrence Patterns of Bacterial Communities Derived from Different Microhabitats of the Two Systems
Co-occurrence networks were established at the bacterial community level for the bacterial communities derived from the FT, FF, and BT samples of both the APS and RAS, as well as for the RZ bacterial community of the APS, resulting in a total of seven bacterial community networks (Fig. 5a–g). The topological properties of the seven empirical networks and their associated random networks, including the average degree, modularity, clustering coefficient, network diameter, and average path length, were presented in Table 2. All the bacterial community networks exhibited significant differences from the random networks indicated by Z tests on modularity, clustering coefficient, network diameter, and average path length, implying all the bacterial community networks were non-random (Table 2). In both systems, the BT bacterial community networks had the highest average degree compared to the others, followed by the FW bacterial community networks, while the FF bacterial community networks showed the lowest average degree. In the APS, the average degree of the RZ bacterial community network was in between that of the FW and FF bacterial community networks. Furthermore, the average degree of the FW bacterial community network was higher in the APS than in the RAS. Nevertheless, the average degree indexes of the FF and BT bacterial community networks were comparable between the two systems. Additionally, the higher modularity indexes of the FW, FF, and BT bacterial community networks were observed in the APS than in the RAS. The robustness and vulnerability of all networks were also calculated to compare the stability of these bacterial community networks between different groups. The robustness of the RZ bacterial community network was significantly higher than that of the other, whereas the FF bacterial community network exhibited the lowest robustness (Fig. 5h). Moreover, the vulnerability of the FF bacterial community networks stayed at the highest followed by the FW bacterial community networks. The BT and RZ bacterial community networks’ vulnerability was comparably low. In addition, we observed lower vulnerability of the FW and FF bacterial community networks in the APS than in the RAS.
Table 2.
Topological properties of the empirical network and associated random network for bacterial communities derived from different microhabitats of aquaponics (APS) and recirculating aquaculture (RAS) systems
| Empirical networka | Random networkb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Network size | No. of core OTUs | Average degree of all nodes | Average degree of core OTUs | Modularity | Clustering coefficient | Network diameter | Average path length | Modularity (SD) | Clustering coefficient (SD) | Network diameter (SD) | Average path length (SD) | |
| APS_FW | 94 | 13 | 11.70 | 13.31 | 0.442a | 0.640a | 8a | 3.06a |
0.223 (0.008) |
0.125 (0.007) |
3.29 (0.46) |
2.20 (0.03) |
| RAS_FW | 91 | 16 | 8.29 | 14.00 | 0.593a | 0.689a | 8a | 3.41a |
0.281 (0.010) |
0.092 (0.009) |
4.09 (0.28) |
2.49 (0.04) |
| APS_FF | 36 | 12 | 4.83 | 6.42 | 0.452a | 0.600a | 7a | 3.00a |
0.331 (0.022) |
0.135 (0.027) |
4.63 (0.55) |
2.58 (0.09) |
| RAS_FF | 37 | 14 | 4.22 | 5.57 | 0.539a | 0.688a | 6a | 2.66a |
0.367 (0.023) |
0.114 (0.029) |
5.20 (0.56) |
2.76 (0.12) |
| APS_BT | 165 | 9 | 21.41 | 40.44 | 0.418a | 0.608a | 7a | 2.63a |
0.162 (0.004) |
0.13 (0.003) |
3.00 (0.01) |
2.10 (0.02) |
| RAS_BT | 167 | 12 | 21.52 | 33.92 | 0.425a | 0.578a | 6a | 2.50a |
0.161 (0.005) |
0.129 (0.003) |
3.08 (0.02) |
2.10 (0.02) |
| APS_RZ | 209 | 12 | 14.82 | 8.75 | 0.529a | 0.591a | 8a | 3.34a |
0.212 (0.005) |
0.071 (0.003) |
3.83 (0.37) |
2.32 (0.03) |
FW Fish tank water, FF fish feces, BT biofilter biofilms, RZ plant rhizosphere microhabitat
aSignificant difference between the empirical network and the random network (P < 0.001, Z test)
bRandom networks were generated by rewiring all of the links with the same numbers of nodes and edges to the corresponding empirical network. The numbers in parentheses indicate the standard deviation (SD) of topological properties of 1000 random networks
The dominant taxa of the bacterial communities derived from each group constituted the majority of nodes in the bacterial community networks (Fig. 5a–g). Furthermore, the detected core OTUs were included in the bacterial community networks. The average degree of these core OTUs was much higher than all nodes in the FW, FF, and BT bacterial community networks in aquaponics and control systems (Table 2). On the contrary, a lower average degree of the core OTUs included in the RZ bacterial community network was observed compared to the average degree of all nodes.
Discussions
The plants provide root-associated niches for the aquaponics system, which might contribute to the diversity and compositions of the bacterial community in the system under community coalescence. Based on this assumption, we analyzed differences in bacterial community diversity and compositions across microhabitats between the two systems with and without plants. The results showed that the diversity and compositions of bacterial communities were driven by microhabitat differences, rather than the presence of plants (i.e., system) (Table 1). Furthermore, we also found that the bacterial communities derived from the same microhabitat of the two systems harbored similar dominant bacterial taxa at both class and genus levels (Fig. 1b and c). The effects of microhabitat differences on the diversity and compositions of bacterial communities in aquaponics systems have been reported in several studies [46–49]. Nevertheless, we still observed bacterial OTUs significantly enriched and depleted in the bacterial communities derived from the FW and BT microhabitats under the influence of plant growth (Fig. 2). These contradictory results suggest that growing plants in an aquaculture system could enrich or deplete the bacterial individuals, but show minimal impacts on compositions of the whole bacterial communities.
The relative abundances of dominant bacterial taxa varied across different microhabitats (Fig. 1b and c), highlighting distinct microbial functions in the different units of the APS and RAS. Bacterial OTUs affiliated with the genus Flavobacterium of the class Bacteroidia dominated the FW bacterial communities of both systems. The genus Flavobacterium encompasses a diverse group of Gram-negative, rod-shaped bacteria commonly found in freshwater, marine environments, and soil [50]. These bacteria exhibit versatile metabolic capabilities, including the breakdown of complex organic compounds such as polysaccharides, proteins, and lipids, playing crucial roles in the degradation of organic matter in aquatic systems [51]. In both systems, fish feces accumulated in the fish tank water, where the high levels of organic matter could contribute to the enrichment of the genus Flavobacterium. In the FF bacterial communities, bacterial OTUs belonging to the genus Cetobacterium of the class Fusobacteriia showed an overwhelming dominance. Bacterial taxa belonging to the genus Cetobacterium have been widely reported to dominate the gut bacterial communities of various fish species [52, 53]. The Cetobacterium species play a key role in fish gut health, aiding in nutrient metabolism and contributing essential nutrients like vitamin B12 [54, 55]. Their anaerobic nature and metabolic specialization make them highly suited to the intestinal environment of fish and potentially valuable in aquaculture. Bacterial OTUs belonging to the genera Pseudomonas (class Gammaproteobacteria) and Nitrospira (class Nitrospiria) showed much higher relative abundances in the BT bacterial communities of both systems than the other microhabitats. Bacterial taxa assigned with the genus Pseudomonas can easily form biofilms on various types of surfaces [56], which explains their high relative abundance in the BT bacterial communities of both systems. Moreover, bacterial taxa belonging to the genus Nitrospira are typical nitrifying bacteria [57, 58], which are commonly found in the biofilms of biofilters [11, 46]. Additionally, we found the genus Acinetobacter showed the highest relative abundance in the RZ bacterial communities. Members of the genus Acinetobacter have been isolated from the rhizosphere of different plants. Some of the bacterial strains of the genus Acinetobacter exhibited plant growth–promoting traits such as nitrogen fixation, siderophore production, and mineral solubilization [59].
Bacterial transmissions are important for maintaining system stability and improving system performance in aquaponics systems. In the present study, the contribution of bacterial transmissions from FF microhabitat to FW microhabitat dominated in both systems. Fish feces play important roles in the biogeochemical processes of the aquatic environment as vertical or horizontal transport of feces in the aquatic environment can create fluxes of organic matter that support biological processes [60, 61]. As fish feces are excreted and dispersed through the water column, the microbial community coalescence also occurs by mixing the abiotic and biotic components between the fish feces and the water column [15]. Thus, bacterial transmissions between the fish feces and the water column could be strong under these coalescence events. For instance, bacterial taxa assigned to the genus Cetobacterium, exhibited quite high abundance in both the FW and FF bacterial communities (ca. 70% in the FF samples and 25% in the FW samples; Fig. 1c). As we discussed above, bacterial taxa belonging to the genus Cetobacterium have been widely reported to dominate in the fish gut bacterial communities rather than in the bacterioplankton communities of freshwater ecosystems. Hence, it is inferred that the high abundance of the genus Cetobacterium in the FW bacterial communities can be a result of bacterial transmissions from the FF bacterial communities under community coalescence. Additionally, the genus Cetobacterium was also found colonized in the BT and RZ bacterial communities of the APS, although the bacterial transmission from the FF to the BT and RZ microhabitats was quite low. These results implied that the bacterial taxa affiliated with the genus Cetobacterium might possess a broad ecological niche, enabling them to colonize a variety of environments. We also found that BT bacterial communities contributed more than 10% of the bacterial transmissions to the assembly of FW bacterial communities in both systems. Studies on the potential health risks of biofilms have pointed out that biofilms can be an important source of bacterial communities in the water column [62, 63]. In our study, the high contribution of bacterial transmissions from biofilters to the water column could be attributed to the aging of biofilms on the biofilters. This aging process, coupled with water flushing, dislodges the biofilm, causing bacterial taxa from the biofilter community to migrate into the water column. The bacterial communities inhabiting plant rhizosphere microhabitat primarily influenced the compositions of the BT microbial communities in the APS compared to the RAS. Approximately, 30% of bacterial transmissions between BT and RZ can be observed in the APS. Bacterial communities associated with BT and RZ microhabitats are both derived from biofilms. Both types of biofilms are composed of a complex matrix of extracellular polymeric substances (EPS), which include polysaccharides, proteins, lipids, and nucleic acids [64]. The similar micro-environment results in the recruitment of similar microbial communities throughout the process of homogeneous selection [65]. Therefore, this similarity of microenvironment facilitates the bacterial exchange between the BT and RZ bacterial communities.
Under the bacterial transmissions caused by water flow, 16 and 17 core OTUs were identified in the APS and RAS, respectively (Fig. 4). Among these, 13 core OTUs were shared between the two systems, suggesting a high similarity of core microbiome between the APS and RAS. A core OTU belonging to the genus Gemmobacter (class Alphaproteobacteria), which was more abundant in the RZ samples, was unique to the RAS. Bacterial taxa assigned to the genus Gemmobacter are important for the denitrification processes of aquatic plant rhizosphere environment [66, 67]. Furthermore, 11 core OTUs were affiliated with the class Fusobacteriia, genus Cetobacterium, which are typical bacterial taxa found in the fish gut, as discussed earlier. Other notable OTUs found at higher abundances in the FF samples belonged to the genus Romboutsia (class Clostridia), family Barnesiellaceae (class Bacteroidia), and genus Aeromonas (class Gammaproteobacteria), which are commonly present in gut microbiota [34, 68, 69]. These results further highlight the significant role of aquatic animal metabolic activities in shaping the core bacterial communities in aquaculture systems, particularly in high-density water-recirculating systems with zero water exchange. Nevertheless, the presence of plants still contributes to the core bacterial community compositions in the aquaponics systems.
Exploring the potential interactions between bacterial taxa within the community offers insights into community stability. In the present study, we found that the network complexity of the BT bacterial communities was the highest, no matter in the APS or RAS, indicated by the highest average degree (Table 2). Furthermore, we found that the RZ bacterial community network exhibited the highest robustness and the lowest vulnerability (Fig. 5h). The robustness of a bacterial community network is defined as the proportion of the remaining species in the network after random or targeted node removal [70, 71], where high values indicate high stability of the bacterial community network [44]. On the contrary, network vulnerability is the maximal value of the relative contribution of all nodes to the decline of global efficiency, and the global efficiency can provide information on how fast the consequence of biological/ecological events traverses to parts or the entire network [44]. Thus, low values of network vulnerability suggest high network stability. Hence, our results suggested that the RZ bacterial community network exhibited higher stability than bacterial community networks derived from other microhabitats, implying greater resilience to environmental disturbances. On the contrary, the FF bacterial community networks exhibited the lowest robustness and the highest vulnerability in both the APS and RAS, suggesting the lowest stability of the FF bacterial community networks. It has been hypothesized that lower bacterial diversity might contribute to reducing the community stability for a long time [72]. In general, communities with lower diversity exhibit reduced ecological redundancy [73]. Compared to communities with higher diversity, those with lower diversity exhibit a greater possibility of interaction breakdown due to random species loss, leading to reduced stability within community networks. In the present study, we observed the lowest diversity of the FF bacterial communities compared to those derived from the other microhabitats. The lowest diversity of the FF bacterial communities could be the reason of the lowest stability of community networks. Moreover, we found slightly higher network robustness of FW and BT bacterial communities in the APS than in the RAS, although the differences were tested as insignificant. The network vulnerability of FW, FF, and BT bacterial communities was also lower in the APS than in the RAS systems. These results indicated that the bacterial community networks might exhibit higher stability in the APS than in the RAS. The detected core OTUs were found to be involved in the bacterial community networks. The average degree of these core OTUs was much higher than that of all nodes in all the bacterial community networks in both systems except for the RZ bacterial community network. It meant that these core OTUs showed higher connectivity within the bacterial community networks than the other nodes, suggesting the important roles of these core taxa in maintaining the bacterial community networks. However, it is important to note that these topological networks represent statistically inferred associations among the relative abundances of various bacterial OTUs, indicating only potential positive or negative interactions. The specific relationships between different bacterial species in aquaponics and recirculating aquaculture systems require further investigation at microsite scales.
Conclusions
In the present study, diversity, compositions, transmissions, and co-occurrence patterns of bacterial communities were explored across different microhabitats in the APS and RAS under community coalescence. Generally, we found that the diversity and compositions of bacterial communities was overwhelmingly shaped by microhabitat differences rather than system differences (i.e., the presence of plants), although several bacterial OTUs were found to be significantly different in relative abundance between the two systems. The SourceTracker analyses showed the bacterial transmissions in the APS were different from that in the RAS. The presence of plants led to intense bacterial transmissions between the RZ and BT bacterial communities in the APS. Furthermore, core bacterial taxa were detected for both the APS and RAS under community coalescence. We found that the APS harbored similar core microbial taxa with the RAS, which were mainly derived from the fish feces. Nevertheless, the RZ bacterial community contributed one core bacterial OTU to the core microbiome of the APS. Bacterial communities inhabiting different microhabitats exhibited varied co-occurrence patterns. The RZ bacterial community network exhibited the highest robustness and the lowest vulnerability, suggesting the highest network stability. Furthermore, the presence of plants resulted in a slight increase in the stability of the bacterial community networks of other microhabitats in the APS. The core taxa played important roles in maintaining the bacterial community networks in both systems. These results revealed the distinct influences of fish and plants on bacterial communities under the community coalescence. The various microhabitats within the aquaponics system are structured with diverse bacterial community compositions and distinct bacterial co-occurrence patterns. The metabolic activities of fish profoundly influenced the core microbiome in the aquaponics systems under community coalescence, and the presence of plants also contributed.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
Huimin Xu: Conceptualization, Methodology, Investigation, Data Curation, Formal Analysis, Visualization, Writing-Original Draft, Funding Acquisition. Yi Zhang: Methodology, Investigation, Data Curation, Visualization, Writing-Review and Editing. Dingyue Fan: Investigation, Data Curation, Writing-Review and Editing. Shunlong Meng: Conceptualization, Data Curation, Supervision, Writing-Review and Editing, Funding Acquisition. Limin Fan: Methodology, Investigation, Writing-Review and Editing. Chao Song: Visualization, Writing-Review and Editing. Liping Qiu: Validation, Writing-Review and Editing. Dandan Li: Investigation, Writing-Review and Editing. Longxiang Fang: Investigation, Writing-Review and Editing. Zhuping Liu: Investigation, Writing-Review and Editing. Xuwen Bing: Supervision, Writing-Review and Editing.
Funding
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund, Freshwater Fisheries Research Center, CAFS (No. 2023JBFR01), the earmarked fund for CARS (CARS-46), and Central Public-interest Scientific Institution Basal Research Fund, CAFS(No.2023TD18).
Data Availability
The raw reads were deposited into the National Omics Data Encyclopedia (NODE) database (Accession Number: OEX028684).
Code Availability
Not applicable.
Declarations
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Not applicable.
Consent for Publication
The publisher has the authors’ permission to publish the content presented herein.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shunlong Meng, Email: mengsl@ffrc.cn.
Xuwen Bing, Email: bingxw@ffrc.cn.
References
- 1.Aiking H (2011) Future protein supply. Trends Food Sci Technol 22:112–120. 10.1016/j.tifs.2010.04.005 [Google Scholar]
- 2.Zhang W, Belton B, Edwards P, Henriksson PJG, Little DC, Newton R, Troell M (2022) Aquaculture will continue to depend more on land than sea. Nature 603:E2–E4. 10.1038/s41586-021-04331-3 [DOI] [PubMed] [Google Scholar]
- 3.Boyd CE, D’Abramo LR, Glencross BD, Huyben DC, Juarez LM, Lockwood GS, McNevin AA, Tacon AGJ, Teletchea F, Tomasso JR Jr, Tucker CS, Valenti WC (2020) Achieving sustainable aquaculture: historical and current perspectives and future needs and challenges. J World Aquaculture Soc 51:578–633. 10.1111/jwas.12714 [Google Scholar]
- 4.Edwards P (2015) Aquaculture environment interactions: past, present and likely future trends. Aquaculture 447:2–14. 10.1016/j.aquaculture.2015.02.001 [Google Scholar]
- 5.Somerville C, Cohen M, Pantanella E, Stankus A, Lovatelli A (2014) Small-scale aquaponic food production: integrated fish and plant farming. Food and Agriculture Organization of the United Nations, Rome [Google Scholar]
- 6.Baganz GFM, Junge R, Portella MC, Goddek S, Keesman KJ, Baganz D, Staaks G, Shaw C, Lohrberg F, Kloas W (2022) The aquaponic principle-it is all about coupling. Rev Aquac 14:252–264. 10.1111/raq.12596 [Google Scholar]
- 7.Yep B, Zheng YB (2019) Aquaponic trends and challenges - a review. J Clean Prod 228:1586–1599. 10.1016/j.jclepro.2019.04.290 [Google Scholar]
- 8.Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. 10.1016/j.soilbio.2009.11.024 [Google Scholar]
- 9.Madsen EL (2011) Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol 22:456–464. 10.1016/j.copbio.2011.01.008 [DOI] [PubMed] [Google Scholar]
- 10.Xue S, Xu W, Wei J, Sun J (2017) Impact of environmental bacterial communities on fish health in marine recirculating aquaculture systems. Vet Microbiol 203:34–39. 10.1016/j.vetmic.2017.01.034 [DOI] [PubMed] [Google Scholar]
- 11.Kasozi N, Abraham B, Kaiser H, Wilhelmi B (2021) The complex microbiome in aquaponics: significance of the bacterial ecosystem. Annals of Microbiology 71:1. 10.1186/s13213-020-01613-5 [Google Scholar]
- 12.Blancheton JP, Attramadal KJK, Michaud L, d’Orbcastel ER, Vadstein O (2013) Insight into bacterial population in aquaculture systems and its implication. Aquacult Eng 53:30–39. 10.1016/j.aquaeng.2012.11.009 [Google Scholar]
- 13.Rurangwa E, Verdegem MCJ (2015) Microorganisms in recirculating aquaculture systems and their management. Rev Aquac 7:117–130. 10.1111/raq.12057 [Google Scholar]
- 14.Castledine M, Sierocinski P, Padfield D, Buckling A (2020) Community coalescence: an eco-evolutionary perspective. Philos Trans Royal Soc B: Biol Sci 375:20190252. 10.1098/rstb.2019.0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rillig MC, Antonovics J, Caruso T, Lehmann A, Powell JR, Veresoglou SD, Verbruggen E (2015) Interchange of entire communities: microbial community coalescence. Trends Ecol Evol 30:470–476. 10.1016/j.tree.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 16.Ramoneda J, Le Roux J, Stadelmann S, Frossard E, Frey B, Gamper HA (2021) Soil microbial community coalescence and fertilization interact to drive the functioning of the legume–rhizobium symbiosis. J Appl Ecol 58:2590–2602. 10.1111/1365-2664.13995 [Google Scholar]
- 17.Rillig MC, Tsang A, Roy J (2016) Microbial community coalescence for microbiome engineering. Front Microbiol 7:1967. 10.3389/fmicb.2016.01967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fdz-Polanco F, Méndez E, Urueña MA, Villaverde S, García PA (2000) Spatial distribution of heterotrophs and nitrifiers in a submerged biofilter for nitrification. Water Research 34:4081–4089. 10.1016/S0043-1354(00)00159-7 [Google Scholar]
- 19.Sanchez FA, Vivian-Rogers VR, Urakawa H (2019) Tilapia recirculating aquaculture systems as a source of plant growth promoting bacteria. Aquac Res 50:2054–2065. 10.1111/are.14072 [Google Scholar]
- 20.Bartelme RP, Oyserman B, Blom JE, Sepulveda-Villet OJ, Newton RJ (2018) Stripping away the soil: plant growth promoting microbiology opportunities in aquaponics. Front Microbiol 9:8. 10.3389/fmicb.2018.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil S, Panda P, Ghadamgahi F, Rosberg A, Vetukuri RR (2021) Comparison of two commercial recirculated aquacultural systems and their microbial potential in plant disease suppression. BMC Microbiol 21:205. 10.1186/s12866-021-02273-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stouvenakers G, Massart S, Jijakli MH (2023) First study case of microbial biocontrol agents isolated from aquaponics through the mining of high-throughput sequencing data to control Pythium aphanidermatum on lettuce. Microb Ecol 86:1107–1119. 10.1007/s00248-022-02126-1 [DOI] [PubMed] [Google Scholar]
- 23.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. 10.1128/mmbr.00051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81. 10.1128/mmbr.00002-00017 [DOI] [PMC free article] [PubMed]
- 25.Gao Y, Zhang W, Li Y (2021) Microbial community coalescence: does it matter in the Three Gorges Reservoir? Water Res 205:117638. 10.1016/j.watres.2021.117638 [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Zhang W, Li Y, Gao Y, Niu L, Zhang H, Wang L (2023) Hydrodynamics-driven community coalescence determines ecological assembly processes and shifts bacterial network stability in river bends. Sci Total Environ 858:159772. 10.1016/j.scitotenv.2022.159772 [DOI] [PubMed] [Google Scholar]
- 27.Neu AT, Allen EE, Roy K (2021) Defining and quantifying the core microbiome: challenges and prospects. Proc Natl Acad Sci 118:e2104429118. 10.1073/pnas.2104429118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (2007) The human microbiome project. Nature 449:804–810. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Custer GF, Gans M, Diepen LTAv, Dini-Andreote F, Buerkle CA (2023) Comparative analysis of core microbiome assignments: implications for ecological synthesis. mSystems 8:e01066-01022. 10.1128/msystems.01066-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pershina EV, Ivanova EA, Korvigo IO, Chirak EL, Sergaliev NH, Abakumov EV, Provorov NA, Andronov EE (2018) Investigation of the core microbiome in main soil types from the East European plain. Sci Total Environ 631–632:1421–1430. 10.1016/j.scitotenv.2018.03.136 [DOI] [PubMed] [Google Scholar]
- 31.Qiao Y, Wang T, Huang Q, Guo H, Zhang H, Xu Q, Shen Q, Ling N (2024) Core species impact plant health by enhancing soil microbial cooperation and network complexity during community coalescence. Soil Biol Biochem 188:109231. 10.1016/j.soilbio.2023.109231 [Google Scholar]
- 32.Muyzer G, Waal ECd, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. 10.1128/aem.59.3.695-700.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108:4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng S, Xu H, Qin L, Chen X, Qiu L, Li D, Song C, Fan L, Hu G, Xu P (2023) The gill-associated bacterial bommunity is more affected by exogenous Chlorellapyrenoidosa addition than the bacterial communities of water and fish gut in GIFT Tilapia (Oreochromisniloticus) aquaculture system. Biology 12:1209. 10.3390/biology12091209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 37.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464 [DOI] [PubMed] [Google Scholar]
- 38.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn d, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H (2013) R package version 2.0–10. Community Ecology Package, Vegan, 2013. http://CRAN.R-project.org/package=vegan 18 May 2017, date last accessed
- 39.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. 10.1038/nmeth.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clauset A, Newman MEJ, Moore C (2004) Finding community structure in very large networks. Phys Rev E 70:066111. 10.1103/PhysRevE.70.066111 [DOI] [PubMed] [Google Scholar]
- 43.Csardi G, Nepusz T (2006) The igraph software package for complex network research. InterJ Complex Syst 1695:1–9 [Google Scholar]
- 44.Yuan MM, Guo X, Wu L, Zhang Y, Xiao N, Ning D, Shi Z, Zhou X, Wu L, Yang Y, Tiedje JM, Zhou J (2021) Climate warming enhances microbial network complexity and stability. Nat Clim Chang 11:343–348. 10.1038/s41558-021-00989-9 [Google Scholar]
- 45.Peng G-s, Wu J (2016) Optimal network topology for structural robustness based on natural connectivity. Physica A 443:212–220. 10.1016/j.physa.2015.09.023 [Google Scholar]
- 46.Schmautz Z, Graber A, Jaenicke S, Goesmann A, Junge R, Smits TH (2017) Microbial diversity in different compartments of an aquaponics system. Arch Microbiol 199:613–620. 10.1007/s00203-016-1334-1 [DOI] [PubMed] [Google Scholar]
- 47.Schmautz Z, Walser J-C, Espinal CA, Gartmann F, Scott B, Pothier JF, Frossard E, Junge R, Smits TH (2022) Microbial diversity across compartments in an aquaponic system and its connection to the nitrogen cycle. Sci Total Environ 852:158426. 10.1016/j.scitotenv.2022.158426 [DOI] [PubMed] [Google Scholar]
- 48.Ruiz A, Scicchitano D, Palladino G, Nanetti E, Candela M, Furones D, Sanahuja I, Carbó R, Gisbert E, Andree KB (2023) Microbiome study of a coupled aquaponic system: unveiling the independency of bacterial communities and their beneficial influences among different compartments. Sci Rep 13:19704. 10.1038/s41598-023-47081-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eck M, Szekely I, Massart S, Jijakli MH (2021) Ecological study of aquaponics bacterial microbiota over the course of a lettuce growth cycle. Water 13:2089. 10.3390/w13152089 [Google Scholar]
- 50.Bernardet J-F, Bowman JP (2006) The Genus Flavobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds.) The prokaryotes: volume 7: proteobacteria: Delta, Epsilon Subclass. Springer New York, New York, pp 481-531
- 51.Kim M, Cha I-T, Lee K-E, Li M, Park S-J (2023) Pangenome analysis provides insights into the genetic diversity, metabolic versatility, and evolution of the genus Flavobacterium. Microbiol Spectr 11:e01003-01023. 10.1128/spectrum.01003-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen AM, Mohammed HH, Arias CR (2014) Characterization of the gut microbiota of three commercially valuable warmwater fish species. J Appl Microbiol 116:1396–1404. 10.1111/jam.12475 [DOI] [PubMed] [Google Scholar]
- 53.Mes W, Lücker S, Jetten MSM, Siepel H, Gorissen M, van Kessel M (2023) Comparison of the gill and gut microbiomes of common carp (Cyprinuscarpio) and zebrafish (Daniorerio) and their RAS environment. Sci Total Environ 896:165212. 10.1016/j.scitotenv.2023.165212 [DOI] [PubMed] [Google Scholar]
- 54.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF (2011) Evidence for a core gut microbiota in the zebrafish. ISME J 5:1595–1608. 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuchiya C, Sakata T, Sugita H (2008) Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol 46:43–48. 10.1111/j.1472-765X.2007.02258.x [DOI] [PubMed] [Google Scholar]
- 56.Masák J, Čejková A, Schreiberová O, Řezanka T (2014) Pseudomonas biofilms: possibilities of their control. FEMS Microbiol Ecol 89:1–14. 10.1111/1574-6941.12344 [DOI] [PubMed] [Google Scholar]
- 57.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509. 10.1038/nature16461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, Nielsen PH, Wagner M, Daims H (2015) Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci 112:11371–11376. 10.1073/pnas.1506533112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sachdev D, Nema P, Dhakephalkar P, Zinjarde S, Chopade B (2010) Assessment of 16S rRNA gene-based phylogenetic diversity and promising plant growth-promoting traits of Acinetobacter community from the rhizosphere of wheat. Microbiol Res 165:627–638. 10.1016/j.micres.2009.12.002 [DOI] [PubMed] [Google Scholar]
- 60.Smriga S, Sandin SA, Azam F (2010) Abundance, diversity, and activity of microbial assemblages associated with coral reef fish guts and feces. FEMS Microbiol Ecol 73:31–42. 10.1111/j.1574-6941.2010.00879.x [DOI] [PubMed] [Google Scholar]
- 61.Wotton RS, Malmqvist B (2001) Feces in aquatic ecosystems: feeding animals transform organic matter into fecal pellets, which sink or are transported horizontally by currents; these fluxes relocate organic matter in aquatic ecosystems. Bioscience 51:537–544. 10.1641/0006-3568(2001)051[0537:FIAE]2.0.CO;2 [Google Scholar]
- 62.Huq A, Whitehouse CA, Grim CJ, Alam M, Colwell RR (2008) Biofilms in water, its role and impact in human disease transmission. Curr Opin Biotechnol 19:244–247. 10.1016/j.copbio.2008.04.005 [DOI] [PubMed] [Google Scholar]
- 63.Zhou Z, Zhong D, Zhang Z, Ma W, Chen J, Zhuang M, Li F, Zhang J, Zhu Y, Su P (2023) Biofilm on the pipeline wall is an important transmission route of resistome in drinking water distribution system. Environ Pollut 335:122311. 10.1016/j.envpol.2023.122311 [DOI] [PubMed] [Google Scholar]
- 64.Flemming H-C, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 65.Brandani J, Peter H, Fodelianakis S, Kohler TJ, Bourquin M, Michoud G, Busi SB, Ezzat L, Lane S, Battin TJ (2023) Homogeneous environmental selection structures the bacterial communities of benthic biofilms in proglacial floodplain streams. Appl Environ Microbiol 89:e02010-02022. 10.1128/aem.02010-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J, Li Q, Dzakpasu M, Wang XC, Chang N (2020) Integrating stereo-elastic packing into ecological floating bed for enhanced denitrification in landscape water. Biores Technol 299:122601. 10.1016/j.biortech.2019.122601 [DOI] [PubMed] [Google Scholar]
- 67.Shi M, Li J, Zhou Q, Wang G, Zhang W, Zhang Z, Gao Y, Yan S (2020) Interactions between elevated CO2 levels and floating aquatic plants on the alteration of bacterial function in carbon assimilation and decomposition in eutrophic waters. Water Res 171:115398. 10.1016/j.watres.2019.115398 [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Li X, Li Y, Li J, Zhu S (2022) Gut microbiomes of cyprinid fish exhibit host-species symbiosis along gut trait and diet. Front Microbiol 13:936601. 10.3389/fmicb.2022.936601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Ni J, Li J, Wang C, Li X, Wu S, Zhang T, Yu Y, Yan Q (2014) Comparative study on gastrointestinal microbiota of eight fish species with different feeding habits. J Appl Microbiol 117:1750–1760. 10.1111/jam.12663 [DOI] [PubMed] [Google Scholar]
- 70.Dunne JA, Williams RJ, Martinez ND (2002) Food-web structure and network theory: the role of connectance and size. Proc Natl Acad Sci 99:12917–12922. 10.1073/pnas.192407699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montesinos-Navarro A, Hiraldo F, Tella JL, Blanco G (2017) Network structure embracing mutualism–antagonism continuums increases community robustness. Nat Ecol Evol 1:1661–1669. 10.1038/s41559-017-0320-6 [DOI] [PubMed] [Google Scholar]
- 72.McCann KS (2000) The diversity–stability debate. Nature 405:228–233. 10.1038/35012234 [DOI] [PubMed] [Google Scholar]
- 73.Allison SD, Martiny JBH (2008) Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci 105:11512–11519. 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads were deposited into the National Omics Data Encyclopedia (NODE) database (Accession Number: OEX028684).
Not applicable.