Abstract
Whey protein from bovine milk is highly valued in the food and pharmaceutical industries because of its high protein content and abundance of essential amino acids. The relationship between whey protein and the β-lactoglobulin (BLG) gene has been extensively discussed because BLG is the most abundant whey protein, making up approximately 50 % of the total whey protein in bovine milk. In recent years, researchers have been interested in this gene because of its critical role in healthy milk production, and any genetic polymorphism in this gene may deteriorate the milk quality. In the current study, we identified several deleterious and damaging non-synonymous single nucleotide polymorphisms (nsSNPs) in BLG and analyzed their destabilizing effects using different computational algorithms. Cumulative results from all tools and evolutionary conservation profiles of BLG suggested that four nsSNPs, G17A, W19C, F136S, and C119R, were the most deleterious and could affect the structural integrity of the protein. Detailed molecular dynamics simulation analysis revealed that all variants induced major structural alterations, that affected the ability of the protein to interact with natural and synthetic ligands. Particularly, the G17A, F136S, and C119R variants induced large conformational changes in the EF loop and main α-helix of BLG, which may affect the access of natural and synthetic ligands to the central calyx of BLG. We hope that the suggested nsSNPs will guide future studies and assist researchers in improving the quality of bovine milk.
1. Introduction
The β-lactoglobulin (BLG) is a predominant whey protein found in the milk of most mammals, with notable exception of humans [1,2]. It belongs to the family of lipocalins, a group of transport proteins, and plays a significant role in transporting small hydrophobic molecules, including vitamins and fatty acids [[3], [4], [5]]. Because of its remarkable functional and nutritional value, it is widely used in the food and pharmaceutical industries. It contains essential amino acids required for human health, making it a valuable component of sports nutrition products, infant formula (although care must be taken due to its allergenic potential), and meal replacements [6]. In addition, the ability of BLG to bind and transport various bioactive compounds makes it a suitable candidate for developing drug delivery systems, particularly for hydrophobic drugs [7].
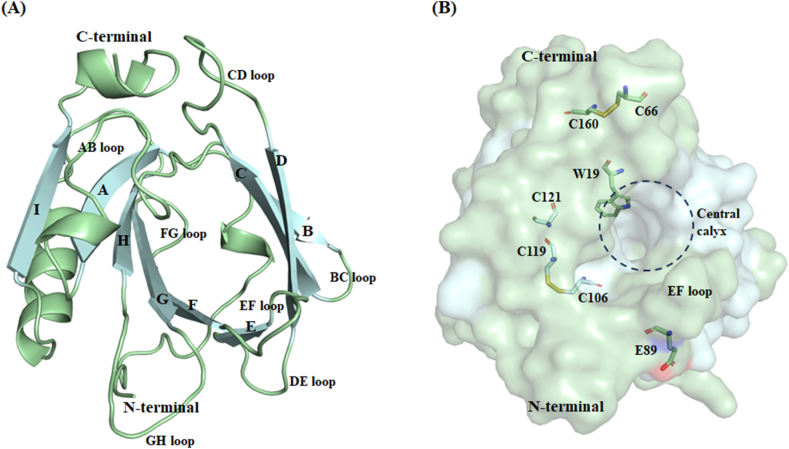
This small globular protein mainly comprises 162 amino acid residues with a molecular weight of approximately 18.3 kDa [4]. The secondary structure of BLG is characterized by nine antiparallel β-strands (A-I) and an external α-helix at the external surface. Eight of these nine flattened β-strands fold up to form a well-suited internal hydrophobic cavity called calyx, which accommodates a range of ligands. This canonical calyx holds two surfaces, i.e., β-strands A to D form one surface, and β-strands E to H form the other one. The ninth β-strand (I) of BLG plays a significant role in dimer interactions. These eight β-strands are connected by seven loops, i.e., AB, BC, CD, DE, EF, FG, and GH loops [7]. Out of these seven loops, three loops (BC, DE, and FG) are short and present at the closed end of the β-barrel. In contrast, the other four loops (AB, CD, EF, and GH) are long, highly flexible, and present at the opened end of the β-barrel (Fig 1A). Although the protein retains its overall compact structure, its sensitivity to pH has been studied in greater detail. At low pH, this protein dissociates into monomers; however, at alkaline pH, the dimeric form is prevalent [[7], [8], [9]]. The “Tanford transition” is of particular significance at a neutral pH because it impacts the protein's capacity to interact with synthetic and natural ligands. Briefly, the protonation state of the carboxyl group in the side chain of E89 at a pH value < 6 causes the EF loop of the β-barrel to close, which prevents the ligand access to the central calyx of BLG. However, at pH > 7, the side chain of E89 is deprotonated, and the EF loop is exposed to the solvent, opening the EF loop (Fig 1B) [10]. Owing to its unique features, BLG has been considered a model protein in numerous studies aimed at explaining the biological significance of protein carrier mechanisms. In their native structures, ligands bind to different protein regions based on their physicochemical properties. Most small molecules are can be hosted in the central calyx, and compete with palmitate (the primary natural ligand binding to the inner cavity of BLG) to stabilize BLG against physical and chemical denaturation [5]. Some molecules bind to another region of the BLG, i.e., between residues near β-strand B and C terminus (involving residues W19, Y20, Y42, Q44, Q59, Q68, L156, E157, E158, H161) or between the main α-helix and β-strand G (involving residues Y102, L104, D129). These compounds do not displace palmitate or stabilize BLG during thermal denaturation [7]. Additionally, BLG contains two tryptophan residues (W19 and W61), with W19 accounting for most of the intrinsic fluorescence intensity of the protein. Hence, it is thought to be the best “reporter” of structural alternations [[11], [12], [13]]. In addition, BLG has five cysteine residues, four of which form disulfide bonds (C66-C160 and C106-C119), whereas the free thiol group of C121, with pH-dependent activity, participates in protein aggregation and denaturation (Fig 1B) [9,14].
Fig. 1.
Features of the mature β-lactoglobulin (BLG) structure. (A) Cartoon representation of BLG and its nine β-strands, i.e., A to I, connected by seven significant loops. (B) Crucial residues of BLG; protonation of E89 triggers the Tanford transition; W19, the “reporter” for structural alterations; C119, C106, C160, and C66 forming disulfide bonds; C121 hidden beneath the main α-helix. The internal hydrophobic cavity, i.e., the central calyx, is represented with a dashed black circle.
The BLG gene, which encodes the prevalent whey protein in ruminant milk, is a significant factor in determining various aspects of milk composition, such as protein, fat, and lactose content. Polymorphisms in BLG may serve as potent molecular makers for evaluating these traits [15]. The distinct calyx structure of BLG, which has hydrophobic walls and several binding sites, affects protein stability, fat-soluble vitamin stability, and other functions. Given the significant impact of milk protein polymorphisms on a range of milk quality factors, it is crucial to investigate genetic variations across this protein. Single nucleotide polymorphisms (SNPs) are the most common single-base pair alterations in DNA sequences, that can be present in both the coding and non-coding regions of a gene. Assessing an individual's SNP genotype may offer a framework for identifying disease susceptibility and the best treatment [16,17]. The non-synonymous SNPs (nsSNPs), also called missense or deleterious SNPs, are of great significance because they cause functional diversity in proteins by substituting an amino acid residue with a single base change in the coding region. These nsSNPs may significantly affect protein function by disturbing the physicochemical characteristics, structural stability, and solubility [18]. Despite playing a crucial regulatory role, non-coding SNPs have received substantially less attention from the functional analysis of genetic variants than coding region SNPs. Thus, in order to gain a better understanding of the clinical significance of these variations, it is necessary to prioritize them based on their functional impact [[19], [20], [21]]. However, demonstrating the functional impact of each variation through experimentation is challenging, expensive, and time-consuming in case-control association studies. Recent advancements in bioinformatics paved the way for analyzing, interpreting and deriving meaningful insights from the vast amounts of genomic and transcriptome data generated through high-throughput sequencing technologies [[22], [23], [24]]. Hence, the current study was designed to explore the most deleterious and damaging nsSNPs in the bovine BLG gene, and how they will affect the structural and functional integrity of the protein using a comprehensive in silico approach.
2. Materials and methods
2.1. Collection of datasets
All SNPs from the bovine BLG gene were assessed and retrieved from the Ensemble (https://ensembl.org/) database (Ensemble gene ID: ENSBTAG00000014678, accessed on Oct 16, 2023), which corresponds to UniProt ID: P02754. With a mass of 19,883 Da, the entire protein is composed of 178 amino acids. Residues 1–16 construct the signaling protein, while residues 17–178 encode the primary BLG protein. A total of 1203 SNPs were found throughout the BLG gene (ensemble transcript ID: ENSBTAT00000019538.6), of which 68 were determined to be non-synonymous SNPs (nsSNPs) or missense variants. All information about these 68 nsSNPs (such as variant ID, position, chromosomal location, alleles, and amino acid changes) was retrieved from the Ensemble database and provided in S1 Table.
BLG was initially synthesized as a precursor protein containing a signal peptide at the N-terminus. This signaling peptide is a short sequence (residue 1–16) that directs the nascent protein to the secretory pathway, where it is subsequently cleaved to produce a mature protein (17–178). Therefore, when previous structural studies refer to residue numbers, they often begin counting from the beginning of the mature protein, i.e., 1–162, which is after the signal peptide [25]. Therefore, we renumbered the selected variants (S2 Table) of the mature protein by ignoring the initial signaling peptide sequence. Of these 68 nsSNPs, seven (rs460853867 (C/W), rs458468150 (A/G), rs477086634 (L/P), rs450652849 (T/P), rs436146677 (G/R), rs448268377 (A/S), and rs448268377 (A/T)) were found in the signaling peptide.
2.2. Identification of most deleterious nsSNPs
Predicting the deleterious nsSNPs using in silico tools is a crucial step in understanding the potential impact of genetic variation on protein function. Hence, the most deleterious or disease-causing nsSNPs in the BLG gene were predicted by using five different bioinformatics tools, such as SIFT (sorting intolerant from tolerant) (https://sift.bii.astar.edu.sg/www/SIFT_dbSNP.html) [26], Phd-SNP (predictor of human deleterious single nucleotide polymorphisms) (https://snps.biofold.org/phd-snp/phd-snp.html) [27], PANTHER (http://pantherdb.org/tools/csnpScoreForm.jsp) [28], SNAP (https://snps.biofold.org/meta-snp) [29], and Meta-SNP [30]. By integrating multiple prediction approaches, researchers can improve the precision and reliability of their predictions, leading to better understanding of the genetic basis of diseases, identifying novel therapeutic targets and advancing personalized medicine strategies. Detailed information on identifying deleterious nsSNPs using these tools is provided in our previous communication [27], and the prediction scores for all variants are presented in (S2 Table).
2.3. Prediction of destabilizing nsSNPs
The stability of a protein structure can be explained by the change in Gibbs free energy upon folding. Proteins are folded into distinct three-dimensional structures that are thermodynamically favorable. Protein stability is determined by the differences in Gibbs free energy (ΔG) between its unfolded and folded states. A negative ΔG suggests that the folded state is more stable, and protein is likely to remain folded. A positive ΔG indicates that the unfolded state is more stable, which may lead to protein misfolding or degradation. In the present study, we accessed six different stability prediction tools, such as I-mutant, DUET, mCSM, SDM2, STRUM, and CUPSAT [[31], [32], [33], [34], [35], [36]]. These tools utilize different algorithms, conservation scores, and integrated structural information to predict the effects of various mutations on protein stability. In addition, we used the MutPred2 webserver (http://mutpred.mutdb.org/) to predict the pathogenicity of selected nsSNPs. MutPred2 has various characteristics, including protein structure, function, and evolution. It uses several structural disorder prediction techniques, such as MARCOIL, TMHMM, and three distinct servers: PSI-BLAST, SIFT, and Pfam [26,[37], [38], [39], [40]]. As a result, MutPred2 aggregates these computational server' scores before delivering the predicted outcomes.
2.4. Conservation profile of BLG
We used the ConSurf database (https://consurfdb.tau.ac.il/) to analyze the evolutionary conservation profile of each amino acid in mature BLG protein. By examining the degree of conservation at each position, ConSurf can help researchers identify functional regions of proteins, such as active sites, ligand-binding sites, or regions important for protein stability, as well as critical regulatory motifs. These regions tend to be more conserved across species than the less functionally critical regions, reflecting the evolutionary pressure to maintain their structure and function [41,42].
2.5. Structural preparation of wild-type (WT) and selected variants of BLG
The WT crystal structure of BLG was downloaded from the RCSB PDB database with PDB ID: 3NQ3 and preprocessed by removing water, ions, and other small molecules [43]. Furthermore, the prepared protein structure was used to model all the selected variants with possible rotamers using the PyMOL mutagenesis plugin. All modeled variants were further subjected to initial energy minimization using the AMBER-ff99SBILDN force field of GROMACS 2022.2. Finally, the stereochemical properties of the modeled complexes were validated using the SAVEs server (S1 Figure).
2.6. Molecular dynamics simulation
To understand the structural impact of these high-risk nsSNPs on the BLG protein, we implemented an all-atom molecular dynamics (MD) simulation to the WT and its selected variants. The MD simulation was performed using the GROMACS 2022.2 package, and the topology for all complexes was generated using the AMBER-ff99SBILDN force field. A dodecahedron periodic boundary with 1.0 nm was filled with TIP3P water molecules, and Na+/Cl− counter ions were added to neutralize the system using the gmx genion module of GROMACS. Next, all WT and variant complexes were minimized using the steepest-descent algorithm with a maximum tolerance of 1000 kJ/mol/nm. Subsequently, all systems were equilibrated at a temperature of 300 K using a Berendsen thermostat (V-rescale) and a pressure of 1.0 bar using the Parrinello-Rahman barostat coupling algorithm. Both the NVT and NPT ensemble processes were performed at 100 ps and 500 ps, respectively. The Particle Mesh Ewald (PME) algorithm was used to sustain long-range interactions, and a cutoff of 1 nm was applied to maintain short and long-range interactions. In addition, the LINCS algorithm was used to restrain all bonds. Finally, all systems were subjected to a 500 ns MD production run with a 2 fs time step, and 2 ps coordinates were saved throughout the MD trajectory. Trajectory information was assessed to calculate the root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent-accessible surface area (SASA), and H-bonds. However, the essential dynamics of all systems were analyzed using the gmx covar and gmx analog modules of GROMACS by considering the last 400 ns trajectories. Furthermore, the Gibb's free energy landscape (FEL) was generated using the first two principal components (PCs) from principal component analysis (PCA), as reported in our previous studies [[44], [45], [46]].
2.7. Protein-protein interaction (PPI) network
We used the search tool for the retrieval of interacting genes/proteins (STRING) database web server (https://string-db.org/) to generate a protein-protein interaction network for BLG. The STRING database is a comprehensive resource used to predict associations between genes and proteins. It covers a vast number of organisms, making it an essential tool for researchers studying various aspects of biology, including the complexities of metabolic pathways, signaling cascades, and mechanisms underlying various diseases. The PPI network for BLG was generated with a high confidence score of 0.700 and no more than 50 indicators in the first shell of the Bos taurus database.
3. Results
3.1. Deleterious and destabilized nsSNPs identified in BLG
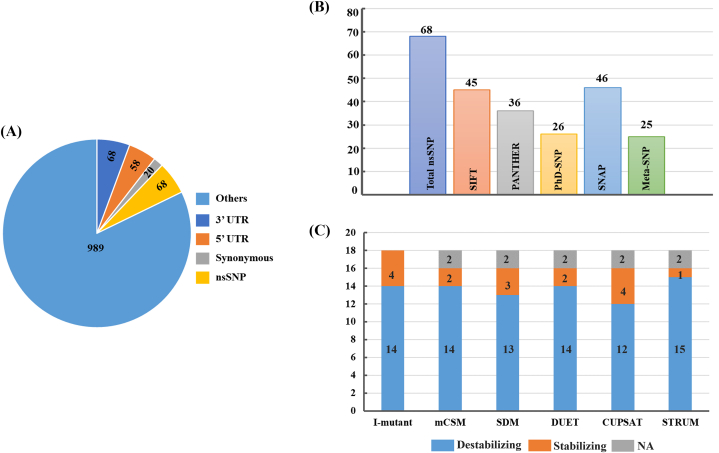
The nsSNPs, also known as missense variants, are recognized as the most significant SNPs that play a vital role in health and livestock industries by altering in vivo protein functions [[47], [48], [49]]. A total of 1203 SNPs were accessed from the Ensemble database, of which 68 SNPs were identified as non-synonymous (Fig 2A). All 68 nsSNPs were further subjected to five nsSNPs prediction algorithms: SIFT, PANTHER, SNAP, PhD-SNP, and Meta-SNP. The SIFT algorithm assigned a score or tolerance index (TI) to each substitution. TI > 0.05 was considered tolerant (neutral), and TI < 0.05 was considered intolerant (deleterious). Similarly, for the four tools, SNAP, PANTHER, PhD-SNP, and Meta-SNP, if the predicted score was >0.5, it was considered disease-causing or deleterious, and if the score was <0.5, it was considered neutral. Of the 68 nsSNPs, the distribution of deleterious nsSNPs in the BLG using SIFT, PANTHER, PhD-SNP, SNAP and Meta-SNP was 66.17 % (45), 52.94 % (36), 38.23 % (26), 67.65 % (46), and 36.76 % (25) respectively (Fig 2B). Out of the 68, common nsSNPs, 18 were predicted to be deleterious by all five tools, hence they were considered for further analysis (S2 Table).
Fig. 2.
(A)Pie chart showing the number of SNPs in BLG gene. Out of 1203 nsSNPs, 68 were identified to be non-synonymous, and 1135 were others (5′ UTR: 58, 3′ UTR: 68, synonymous SNPs: 20, downstream and upstream gene variants: 508, intron variants: 453, splice variants: 23, start lost and stop gained variants: 5). (B) Prediction of deleterious nsSNPs using different computational algorithms. (C) Prediction of destabilizing mutations using different computational methods.
Furthermore, to identify destabilized nsSNPs, we uploaded the 18 identified deleterious variants to six bioinformatics tools. Six different tools, I-mutant, mCSM, SDM, DUET, CUPSAT, and STRUM, predicted 14, 14, 13, 14, 12, and 15 nsSNPs respectively, were destabilized (Fig 2C). Moreover, eight common nsSNPs were found to be destabilized by all six tools (S3 Table). Additionally, we used the MutPred2 tool to predict the pathogenicity of the selected variants. A MutPred2 score >0.5 was considered pathogenic, and a score <0.5, benign. Nevertheless, we discovered that each of the 18 nsSNPs was pathogenic.
3.2. Evolutionary conservation profile of BLG
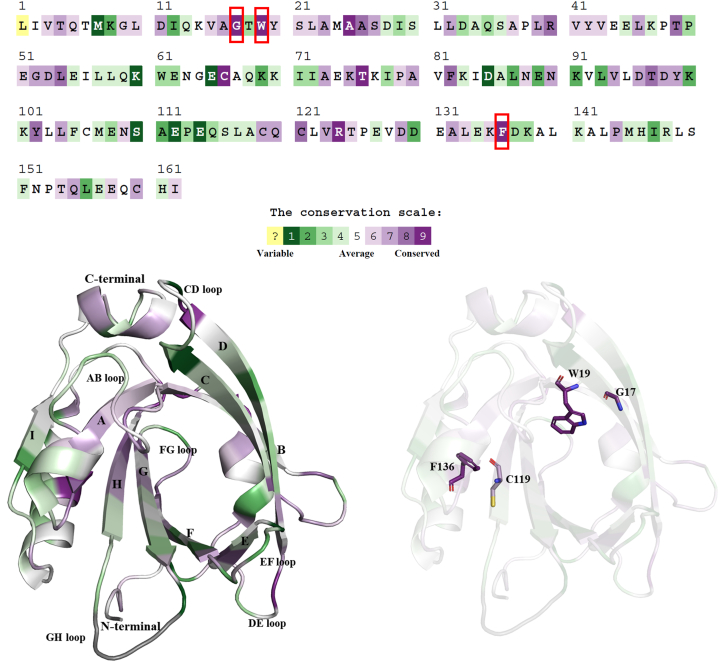
The residual evolutionary conservation profile of a protein is crucial for identifying its structural and functional impacts upon mutations. This provides a better understanding of the significance of particular amino acid residues and their restricted evolution [50]. Hence, the conservation level was assessed using the ConSurf database with the 3D structure of BLG as an input query. The ConSurf results showed that three of the eight variants selected from the protein-destabilizing analysis, G17, W19, and F136, were highly conserved, with a conservation score of 9 (Fig 3). The conservation scores of the other five variants, V15, M24, L39, L54, and C119, were calculated to be 6, 5, 7, 8, and 7, respectively. Since the C119 residue of BLG formed a disulfide bond with residue C106, any mutation at position 119 may interrupt the stability of the protein and its function. Hence, of the eight most deleterious and destabilizing nsSNPs, four variants, (i.e., three highly conserved variants, G17A, W19C, and F136S, along with the C119R variant) were considered for further analysis.
Fig. 3.
Evolutionary conservation profile of BLG gene. The red boxes on the BLG sequence indicate the most deleterious and destabilized conserved variants. The stick model represents the highly conserved variants of these residues (G17A, W19C, F136S, and C119R) that were considered for MD simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Structural consequences through MD simulation
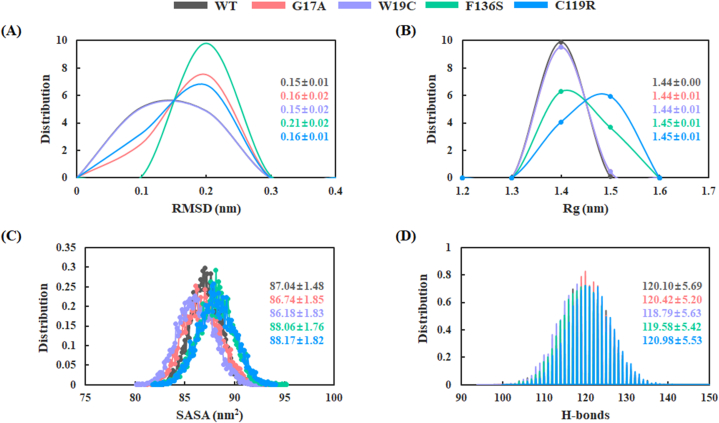
All selected variants and the WT structure of BLG were subjected to MD simulations to study their structural and dynamic behaviors. As described in the methodology, a 500 ns MD simulation was performed, and the structural stability was assessed using the RMSD and Rg values of the protein backbone. The RMSD analysis suggested that after the first 100 ns of MD simulation, all variants maintained their structural stability with an RMSD value between 0.12 and 0.25 nm. In contrast to the other variants, the F136S variant showed a larger RMSD of more than 0.2 nm (S2 Figure). Moreover, we calculated the probability density distribution of RMSD, which showed an average RMSD value of 0.15 ± 0.01, 0.16 ± 0.02, 0.15 ± 0.02, 0.21 ± 0.02, and 0.16 ± 0.01 nm for WT, G17A, W19C, F136S, and C119R respectively. The WT and W19C variants of BLG showed similar unimodal distributions, whereas the F136S variant showed a distinct RMSD distribution (Fig 4A). Furthermore, the C119R variant showed relatively higher Rg values, indicating lower stability than other variants. The probability density distribution of Rg for WT and variants clustered at 0.14 ± 0.05, 0.14 ± 0.07, 0.14 ± 0.07, 0.15 ± 0.07, and 0.15 ± 0.08 nm for WT, G17A, W19C, F136S, and C119R respectively (Fig 4B). Based on the Rg distribution, no significant structural changes were observed in the G17A and W19C variants with respect to the WT BLG structure. In contrast, the F136S and C119R variants exhibited higher Rg values than those of the WT.
Fig. 4.
The Probability density distribution of (A) RMSD, (B) Rg, (C) SASA, and (D) intra H-bonds throughout the trajectories. The average and standard deviation values are represented according to their respective colors. Black, red, purple, green, and blue colors represent the WT, G17A, W19C, F136S, and C119R systems of BLG, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also calculated the SASA to analyze the accessible surface area for biomolecular interactions and represented in S2 Figure. The G17A and W19C variants of BLG showed a similar type of distribution with an average SASA value of 86.74 ± 1.85 nm2 and 86.18 ± 1.83 nm2, respectively. The F136S and C119R variants had relatively higher average SASA value than others (Fig 4D). To predict the effect of BLG variants on intra-molecular interactions, we calculated the number of intra H-bond interactions throughout the trajectory as, represented in Fig 4D. The results revealed that the average number of intra H-bond interactions for WT, G17A, W19C, F136S, and C119R was approximately 120, 120, 119, 120, and 121, respectively (S2 Figure).
3.4. Essential dynamic analysis of BLG variants
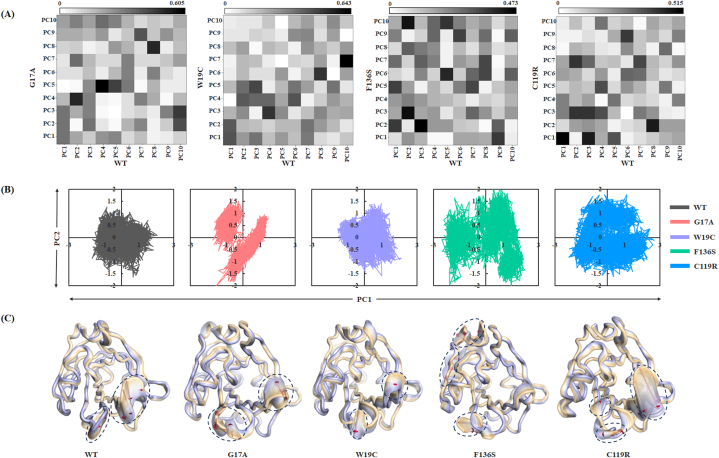
The collective conformational changes, particularly induced by these variants, were studied using PCA. Here, we considered the last 400 ns of trajectories for PCA and identified the principal components (PCs) with larger eigenvalues, which represent the most dominant motions of the protein backbone. The results revealed diagonalized co-variances of 2.81, 3.09, 2.60, 5.13, and 4.26 for the WT, G17A, W19C, F136S, and C119R variants, respectively. Additionally, the first 10 PCs, which accounted for more than 60 % of the total covariance, were used to define the similarities and differences between the essential subspace of the WT and variants through the root mean square inner product (RMSIP). The RMSIP calculation variants showed normalized overlaps of 0.517, 0.573, 0.415, and 0.490 for variants G17A, W19C, F136S, and C119R, respectively, with respect to the WT BLG (Fig 5A). In particular, the RMSIP of the first three PCs differed significantly between the F136S and C119R variants.
Fig. 5.
Conformation changes induced by BLG variants assessed through PCA. (A) Root Mean Square Inner Products (RMSIPs) of the first ten PCs show similarities and differences between the conformational spaces of the WT and variants. (B) Projection of first two PCs for all the systems. Black, red, purple, green, and blue colors represent the WT, G17A, W19C, F136S, and C119R systems of BLG, respectively. (C) Structural changes in all the systems with respect to PC1 are represented using extreme conformations with 30 frames. Black-dashed circles represent large conformational changes, and red arrows represent the size and direction of movements. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The first three PCs represented most of the dominant motions with a cumulative percentage of 31.91 %, 42.46 %, 32.91 %, 48.76 %, and 41.64 % for the WT, G17A, W19C, F136S, and C119R variants, respectively, which indicated that all variants had higher collective flexibility compared to the WT (S3 Figure A, B). Hence, the first two PCs from the PCA were assessed to plot a 2D projection representing the spread in phase space (Fig 5B). Moreover, we assessed the most dominant conformational changes in the collective motion of all systems by considering 30 extreme conformations from the first PC. The results revealed that all systems showed a larger structural alteration in the GH loop region (Fig 5C). The W19C and C119R variants, along with the WT complex of BLG, exhibited similar kinds of conformational changes in the EF loop region by mimicking the Tanford transition. In particular, in the C119R variant, the EF loop was exposed to the solvent by opening access to the central calyx of the BLG. However, the G17A variant causes closure of the EF loop by blocking access to the central calyx. Although the F136S variant did not show any significant changes in the EF loop region, it exhibited large conformational changes in the main α-helix and near C-terminal region (Fig 5C).
3.5. Changes in residual fluctuation and residual cross-correlation
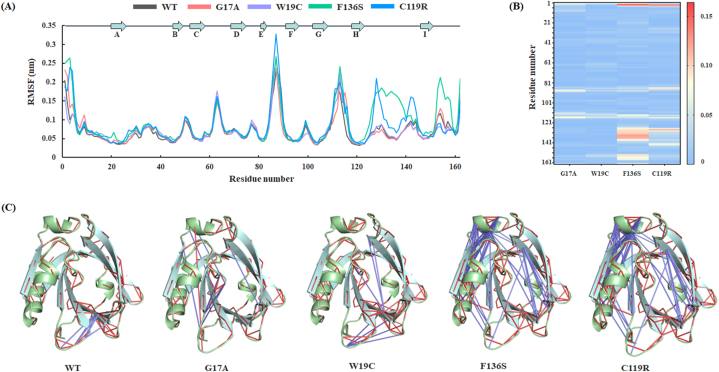
RMSF analysis was performed to assess the residual fluctuation of each amino acid throughout the trajectory and to identify the region that influences structural changes and stability. Fig 6A shows that the F136S and C119R variants increased the residual fluctuation compared to the others, and these fluctuations were pronounced in the EF and GH loop regions. Unlike other variants, the F136S variant showed residual fluctuation in the main α-helix region and near the C terminal region, indicating that this variant may interrupt the intramolecular signaling required for the stability of functional regions like the EF loop region. The average residual fluctuation for WT, G17A, W19C, F136S, and C119R variants was calculated to be 0.07, 0.07, 0.07, 0.09, and 0.08 nm, respectively. Additionally, the △RMSF value for all the variants with respect to the WT were calculated and represented in Fig 6B. The lower △RMSF value represents the rigid region with blue color, and the higher △RMSF value represents the flexible region with dark red color. The heatmap of △RMSF clearly indicates the residual flexibility in the F136S and C119R variants.
Fig. 6.
Residual flexibility throughout the MD simulation. (A) RMSF of backbone atoms throughout the trajectories. (B) Heatmap representing △RMSF of G17A, W19C, F136S, and C119R relative to WT. (C) Structural representation of the strong residual cross-correlation in all the systems. Red and blue lines denote the most correlated and anti-correlated interactions between two residues. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Moreover, the internal dynamics of the WT and other BLG variants were studied using a dynamic cross-correlation map (DCCM). The last 400 ns of MD trajectories of each system were considered to construct a DCCM map using the Cα atoms and represented in (S3 Figure C). The cyan color in the DCCM map represents positively correlated motions (+1.0), and the pink color represents negatively correlated motions (−1.0). Furthermore, the structural residual correlation patterns in all systems were assessed and are presented in Figure: 6C. All systems showed similar types of correlated motions, however, the F136S and C119 variants showed larger anti-correlated motions, than the others. The EF loop region, which exhibited larger RMSF fluctuations, was also observed to have anti-correlated motions with the “C” β-sheet of BLG. In contrast, most of the anti-correlated motions were observed in the main α-helix of F136S and C119R variants.
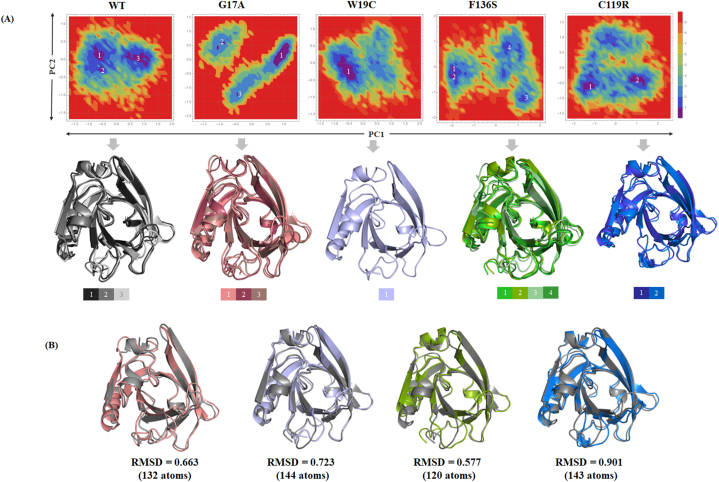
3.6. Free energy landscape (FEL) analysis of all the variants
FEL maps were generated for all complexes by assessing the first two PCs as shown in Fig 7A. FEL analysis revealed more than two global minima clusters in all variants and the WT, except for the W19C variant. The W19C variants showed a single global minima cluster represented by a deep blue color. In contrast to WT, the variants showed evidence of a substantial transition during MD simulations. In particular, the F136S and C119R variants underwent large conformational changes with widely distributed global minima. Furthermore, representative structures from each global minima were retrieved and superimposed to obtain a clear picture the of structural transition. Additionally, the representative structures of each variant were superimposed with respect to the representative WT structure of BLG (Fig 7B). The superimposition results revealed an RMSD of 0.663 Å (132 atoms), 0.723 Å (144 atoms), 0.577 Å (120 atoms), and 0.901 Å (143 atoms) for G17A, W19C, F136S, and C119R respectively, with respect to WT structure. From the superimposition, the molecular interactions of each variant with other amino acids were also analyzed with respect to the WT, as shown in (S4 Figure A). No differences were observed in the interaction pattern upon the substitution of glycine with alanine at position 17, and they formed a common interaction with the L46 residue. Similarly, the W19 residue interacts with E44 and V15; however, when substituted with cysteine, it forms only one interaction with the E44 residue. The C119 residue formed one hydrogen bond and one disulfide bond with the A25 and C106 residues, respectively, but when it mutated to arginine, it formed two hydrogen bonds with the A25 and Q5 residues. Moreover, the F136S variants exhibited large differences in the interaction pattern, suggesting that after mutating to serine, there were two more H-bond interactions with L133 and D137. The increase in the number of hydrogen bonds in the F136S variant may reflect a larger conformational transition in the main helical region of BLG. All the above analyses indicated that the F136S and C119R variants may induce conformational changes, particularly in the main helix and EF loop regions of BLG. Additionally, the G17A variant may induce structural changes in the EF-loop and block access to the central calyx.
Fig. 7.
Free energy landscape of all the systems and their representative structures. (A) Low energy structures were retrieved from their respective global minima and superimposed. (B) Superimposition of WT and representative mutant conformations are shown with RMSD values.
3.7. Analysis of the PPI network
To assess the association between BLG and other proteins, a PPI network was generated using a high confidence score (0.700) in the STRING database (S4 Figure B). The PPI network for BLG (also known as PAEP protein) is comprised 18 nodes, 72 edges, and an average node degree of 8 with an average local clustering coefficient of 0.785. Proteins associated with BLG, and their corresponding scores are listed in S4 Table. The network predicted that the BLG protein was associated with different biological processes, such as response to 11-deoxycorticosterone (GO:1903496), response to dehydroepiandrosterone (GO:1903494), negative regulation of lactation (GO:1903488), response to progesterone (GO:0032570), response to growth hormone (GO:0060416), and response to estradiol (GO:0032355). Hence, any variation in BLG may have a significant impact on several biological processes because they may act as key regulators.
4. Discussion
BLG is the predominant whey protein in bovine milk, constituting about 50 % of the whey protein [9,51]. It plays a major role in the food and pharmaceutical industries due to its ability to bind hydrophobic molecules and vitamins, like retinol (Vitamin A), enhancing nutrient bioavailability [[52], [53], [54]]. Previous studies reported that at a neutral pH, the Tanford transition phase of BLG is of particular interest because it impacts its capacity to interact with both synthetic and natural ligands [10]. In recent years, researchers have become interested in this gene because of its critical role in healthy milk production, and genetic polymorphisms in this gene may affect milk quality. Among the different genetic polymorphisms, nsSNPs are of great significance because a single-base change in the coding region causes functional diversity in the protein by altering the amino acid residue. These nsSNPs may substantially affect the structural stability and solubility of proteins [18]. Preventing the occurrence of deleterious nsSNPs is crucial in efforts to enhance the quality of dairy products and address allergenicity concerns. Several strategies and techniques can be employed to prevent the expression of such SNPs in BLG. One of the most effective methods is to use selective breeding programs by identifying cattle with beneficial genetic variants of the BLG gene. Using Marker-Assisted Selection (MAS), breeders can select animals with favorable gene variants for reproduction, minimizing the passing of deleterious nsSNPs to future generation. Advanced gene editing technologies such as CRISPR-Cas9 have revolutionized genetic engineering. This system can be used to directly edit the harmful variants in the BLG gene, either correcting deleterious nsSNPs or introducing protective mutations to counteract potential damage. Other approaches such as maintaining genetic diversity, environmental and nutritional management can also contribute to reduce the risk and impact of deleterious nsSNPs in BLG. To improve the efficiency of BLG despite of having damaging nsSNPs requires focus on counteracting the negative effects of these mutations. Strategies like directed protein engineering, molecular chaperon-assisted folding, gene editing can enhance BLG function. Furthermore, predictive modelling and MD simulations can identify the unstable regions, enabling targeted interventions like mutagenesis or small molecule design to improve the protein stability and function. However, to understand the clinical significance of these nsSNPs, it is important to prioritize them based on their functional impact. Given the challenges in experimentally assessing the impact of these variations, the current study utilized a series of bioinformatics tools to identify the most deleterious and damaging nsSNPs and assessed their structural and functional impact on BLG.
Using five different tools SIFT, PANTHER, Phd-SNP, Meta-SNP, and SNAP, 18 nsSNPs were identified as the most damaging and deleterious. Furthermore, these 18 nsSNPs were subjected to six different bioinformatics algorithms (I-mutant, mCSM, SDM, DUET, CUPSAT, and STRUM) to identify destabilized nsSNPs in BLG. As a result, eight common nsSNPs were found to be destabilized by all six algorithms. A series of algorithms, including both sequence and structure-based approaches, were used to optimize the prediction accuracy because it is important to note that the prediction of damaging and deleterious nsSNPs using a single tool may lead to false positives. Hence, based on the prediction results of all different tools and residual conservation profiles, four high-risk nsSNPs, G17A, W19C, F136S, and C119R, along with the WT structure of BLG, were selected for MD simulation to study the conformational changes induced by these variants.
The overall MD simulation results suggested that the WT and W19C variants of BLG maintained similar structural integrity throughout the simulation, indicating higher structural stability than the other variants. Moreover, the F136S and C119R variants showed distinct unimodal distributions, indicating lower structural and conformational stability than the other variants. The disulfide bond formed between C119 and C106 preserved the structural integrity and stability of BLG. However, no disulfide bond was formed with C106 when the cysteine residue was substituted with arginine at position 119. This could explain why the C119R variant exhibited lower structural stability than the other variants. The essential dynamics results suggest that the C119R and G17A variants caused severe conformational alterations in the EF loop region, mimicking the deprotonated and protonated states of E89, respectively [10]. Unlike other variants, the F136S variant exhibited larger conformational changes in the main α-helix and near C-terminal region of BLG. Residual flexibility analysis using RMSF and △RMSF revealed that all variants exhibited common fluctuations in the EF loop and GH loop region. However, the F136S and C119 variants exhibited distinct residual fluctuations in the main α-helix residues, which were later observed to exhibit anti-correlated motions.
Although the current study provides detailed structural insights into the impact of high-risk nsSNPs on the BLG, further structural and biochemical comparative analyses of these variants are required to validate our results. Additionally, long-range MD simulations and extensive sampling techniques are required to be performed to come up with definitive conclusions.
5. Conclusion
In the present study, a series of bioinformatics tools were used to identify four high-risk nsSNPs, G17A (rs437398769), W19C (rs474719380), F136S (rs440613020), and C119R (rs3423321025) in the BLG gene from the Ensemble database, which induced major structural changes revealed by molecular dynamic simulation. In particular, the F136S and C119R variants exhibited a larger conformational change in the main α-helix and EF loop of BLG, respectively, indicating lower structural stability. Moreover, the G17A variant induces structural changes in the EF loop region, which causes the EF loop of the β-barrel structure to close, blocking ligand access to the central calyx of the BLG. Although additional experimental validation is needed to confirm these variants, we hope these results will guide future studies and assist in improving milk quality and the management of cattle milk production.
CRediT authorship contribution statement
Sthitaprajna Sahoo: Writing – review & editing, Writing – original draft, Investigation, Formal analysis, Conceptualization. Vijayakumar Gosu: Writing – review & editing. Hak-Kyo Lee: Writing – review & editing, Supervision. Donghyun Shin: Writing – review & editing, Supervision, Conceptualization.
Data availability statement
The raw simulations trajectory data of this study is publicly available on FigShare (https://figshare.com/) with accession number (https://doi.org/10.6084/m9.figshare.26183426.v1).
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Ministry of Education of the Republic of Korea and the National Research Foundation of Korea (NRF-2022R1A2C4002510) and Science and Technology Project Opens the Future of the Region through INNOPOLIS Foundation grant funded by the Ministry of Science and ICT of Korea (2022-DD-UP-0333).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the Jeonbuk National University for providing the facilities necessary for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40040.
Contributor Information
Hak-Kyo Lee, Email: breedlee@jbnu.ac.kr.
Donghyun Shin, Email: sdh1214@gmail.com.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Patel S. Emerging trends in nutraceutical applications of whey protein and its derivatives. J. Food Sci. Technol. 2015;52:6847–6858. doi: 10.1007/s13197-015-1894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majhi P.R., Ganta R.R., Vanam R.P., Seyrek E., Giger K., Dubin P.L. Electrostatically driven protein aggregation: β-lactoglobulin at low ionic strength. Langmuir. 2006;22:9150–9159. doi: 10.1021/la053528w. [DOI] [PubMed] [Google Scholar]
- 3.Sawyer L., Kontopidis G. The core lipocalin, bovine β-lactoglobulin. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000;1482:136–148. doi: 10.1016/S0167-4838(00)00160-6. [DOI] [PubMed] [Google Scholar]
- 4.Brownlow S., Cabral J.H.M., Cooper R., Flower D.R., Yewdall S.J., Polikarpov I., North A.C., Sawyer L. Bovine β-lactoglobulin at 1.8 Å resolution — still an enigmatic lipocalin. Structure. 1997;5:481–495. doi: 10.1016/S0969-2126(97)00205-0. [DOI] [PubMed] [Google Scholar]
- 5.Kontopidis G., Holt C., Sawyer L. Invited review: β-lactoglobulin: binding properties, structure, and function. J. Dairy Sci. 2004;87:785–796. doi: 10.3168/jds.S0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- 6.Varlamova E.G., Zaripov O.G. Beta–lactoglobulin–nutrition allergen and nanotransporter of different nature ligands therapy with therapeutic action. Res. Vet. Sci. 2020;133:17–25. doi: 10.1016/j.rvsc.2020.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Shafaei Z., Ghalandari B., Vaseghi A., Divsalar A., Haertlé T., Saboury A.A., Sawyer L. β-Lactoglobulin: an efficient nanocarrier for advanced delivery systems. Nanomed. Nanotechnol. Biol. Med. 2017;13:1685–1692. doi: 10.1016/j.nano.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Kuwata K., Hoshino M., Forge V., Era S., Batt C.A., Goto Y. Solution structure and dynamics of bovine β-lactoglobulin A. Protein Sci. 1999;8:2541–2545. doi: 10.1110/ps.8.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbiroli A., Iametti S., Bonomi F. Beta-lactoglobulin as a model food protein: how to promote, prevent, and exploit its unfolding processes. Molecules. 2022;27:1131. doi: 10.3390/molecules27031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin B.Y., Bewley M.C., Creamer L.K., Baker H.M., Baker E.N., Jameson G.B. Structural basis of the Tanford transition of bovine β-lactoglobulin. Biochemistry. 1998;37:14014–14023. doi: 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- 11.Geng S., Jiang Z., Ma H., Wang Y., Liu B., Liang G. Interaction mechanism of flavonoids and bovine β-lactoglobulin: experimental and molecular modelling studies. Food Chem. 2020;312 doi: 10.1016/j.foodchem.2019.126066. [DOI] [PubMed] [Google Scholar]
- 12.Yousefi A., Ahrari S., Panahi F., Ghasemi Y., Yousefi R. Binding analysis of the curcumin-based synthetic alpha-glucosidase inhibitors to beta-lactoglobulin as potential vehicle carrier for antidiabetic drugs. J. Iran. Chem. Soc. 2022;19:489–503. doi: 10.1007/s13738-021-02323-8. [DOI] [Google Scholar]
- 13.Liang L., Subirade M. Study of the acid and thermal stability of β-lactoglobulin–ligand complexes using fluorescence quenching. Food Chem. 2012;132:2023–2029. doi: 10.1016/j.foodchem.2011.12.043. [DOI] [Google Scholar]
- 14.Sakai K., Sakurai K., Sakai M., Hoshino M., Goto Y. Conformation and stability of thiol-modified bovine β lactoglobulin. Protein Sci. 2000;9:1719–1729. [PMC free article] [PubMed] [Google Scholar]
- 15.Wodas L., Mackowski M., Borowska A., Puppel K., Kuczynska B., Cieslak J. Genes encoding equine β-lactoglobulin (LGB1 and LGB2): polymorphism, expression, and impact on milk composition. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arshad M., Bhatti A., John P. Identification and in silico analysis of functional SNPs of human TAGAP protein: a comprehensive study. PLoS One. 2018;13 doi: 10.1371/journal.pone.0188143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander E.S. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 18.Tokuriki N., Tawfik D.S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Kaur R., Singh J., Kaur M. Structural and functional impact of SNPs in P-selectin gene: a comprehensive in silico analysis. Open Life Sci. 2017;12:19–33. doi: 10.1515/biol-2017-0003. [DOI] [Google Scholar]
- 20.Krawczak M., Ball E.V., Fenton I., Stenson P.D., Abeysinghe S., Thomas N., Cooper D.N. Human gene mutation database-a biomedical information and research resource. Hum. Mutat. 2000;15:45–51. doi: 10.1002/(SICI)1098-1004(200001)15:1<45::AID-HUMU10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Prokunina L., Alarcón-Riquelme M.E. Regulatory SNPs in complex diseases: their identification and functional validation. Expet Rev. Mol. Med. 2004;6:1–15. doi: 10.1017/S1462399404007690. [DOI] [PubMed] [Google Scholar]
- 22.Okoye C.O., Jiang H., Nazar M., Tan X., Jiang J. Redefining modern food analysis: significance of omics analytical techniques integration, chemometrics and bioinformatics. TrAC, Trends Anal. Chem. 2024;175 doi: 10.1016/j.trac.2024.117706. [DOI] [Google Scholar]
- 23.Manoochehri H., Asadi S., Tanzadehpanah H., Sheykhhasan M., Ghorbani M. CDC25A is strongly associated with colorectal cancer stem cells and poor clinical outcome of patients. Gene Reports. 2021;25 doi: 10.1016/j.genrep.2021.101415. [DOI] [Google Scholar]
- 24.Sheykhhasan M., Ahmadyousefi Y., Seyedebrahimi R., Tanzadehpanah H., Manoochehri H., Dama P., Hosseini N.F., Akbari M., Farsani M.E. DLX6-AS1: a putative lncRNA candidate in multiple human cancers. Expet Rev. Mol. Med. 2021;23:e17. doi: 10.1017/erm.2021.17. [DOI] [PubMed] [Google Scholar]
- 25.Tanzadehpanah H., Bahmani A., Hosseinpour Moghadam N., Gholami H., Mahaki H., Farmany A., Saidijam M. Synthesis, anticancer activity, and β-lactoglobulin binding interactions of multitargeted kinase inhibitor sorafenib tosylate (SORt) using spectroscopic and molecular modelling approaches. Luminescence. 2021;36:117–128. doi: 10.1002/bio.3929. [DOI] [PubMed] [Google Scholar]
- 26.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capriotti E., Fariselli P. PhD-SNPg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017;45:W247–W252. doi: 10.1093/nar/gkx369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H., Ebert D., Muruganujan A., Mills C., Albou L.-P., Mushayamaha T., Thomas P.D. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49:D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bromberg Y., Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capriotti E., Altman R.B., Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genom. 2013;14:S2. doi: 10.1186/1471-2164-14-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pires D.E.V., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pires D.E.V., Ascher D.B., Blundell T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandurangan A.P., Ochoa-Montaño B., Ascher D.B., Blundell T.L. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017;45:W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quan L., Lv Q., Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics. 2016;32:2936–2946. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delorenzi M., Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 38.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes1. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G.A., Tate J., Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben Chorin A., Masrati G., Kessel A., Narunsky A., Sprinzak J., Lahav S., Ashkenazy H., Ben-Tal N. ConSurf-DB: an accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 2020;29:258–267. doi: 10.1002/pro.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loch J., Polit A., Górecki A., Bonarek P., Kurpiewska K., Dziedzicka-Wasylewska M., Lewiński K. Two modes of fatty acid binding to bovine β-lactoglobulin—crystallographic and spectroscopic studies. J. Mol. Recogn. 2011;24:341–349. doi: 10.1002/jmr.1084. [DOI] [PubMed] [Google Scholar]
- 44.Sahoo S., Son S., Lee H.-K., Lee J.-Y., Gosu V., Shin D. Impact of nsSNPs in human AIM2 and IFI16 gene: a comprehensive in silico analysis. J. Biomol. Struct. Dyn. 2023;0:1–13. doi: 10.1080/07391102.2023.2206907. [DOI] [PubMed] [Google Scholar]
- 45.Sahoo S., Lee H.-K., Shin D. Structure-based virtual screening and molecular dynamics studies to explore potential natural inhibitors against 3C protease of foot-and-mouth disease virus. Front. Vet. Sci. 2024;10 doi: 10.3389/fvets.2023.1340126. https://www.frontiersin.org/articles/10.3389/fvets.2023.1340126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahoo S., Samantaray M., Jena M., Gosu V., Bhuyan P.P., Shin D., Pradhan B. In Vitro and in silico studies to explore potent antidiabetic inhibitor against human pancreatic alpha-amylase from the methanolic extract of the green microalga Chlorella vulgaris. J. Biomol. Struct. Dyn. 2024;42:8089–8099. doi: 10.1080/07391102.2023.2244592. [DOI] [PubMed] [Google Scholar]
- 47.Pal L.R., Moult J. Genetic basis of common human disease: insight into the role of missense SNPs from genome-wide association studies. J. Mol. Biol. 2015;427:2271–2289. doi: 10.1016/j.jmb.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han Y., Dorajoo R., Chang X., Wang L., Khor C.-C., Sim X., Cheng C.-Y., Shi Y., Tham Y.C., Zhao W., Chee M.L., Sabanayagam C., Chee M.L., Tan N., Wong T.Y., Tai E.-S., Liu J., Goh D.Y.T., Yuan J.-M., Koh W.-P., van Dam R.M., Low A.F., Chan M.Y.-Y., Friedlander Y., Heng C.-K. Genome-wide association study identifies a missense variant at APOA5 for coronary artery disease in Multi-Ethnic Cohorts from Southeast Asia. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-18214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kęsek-Woźniak M.M., Wojtas E., Zielak-Steciwko A.E. Impact of SNPs in ACACA, SCD1, and DGAT1 genes on fatty acid profile in bovine milk with regard to lactation phases. Animals. 2020;10:997. doi: 10.3390/ani10060997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deeth H., Bansal N. In: Whey Proteins. Deeth H.C., Bansal N., editors. Academic Press; 2019. Chapter 1 - whey proteins: an overview; pp. 1–50. [DOI] [Google Scholar]
- 52.Guo M., Wang G. Whey Protein Production, Chemistry, Functionality, and Applications. John Wiley & Sons, Ltd; 2019. Nutritional applications of whey protein; pp. 141–156. [DOI] [Google Scholar]
- 53.Boscaini S., Skuse P., Nilaweera K.N., Cryan J.F., Cotter P.D. The ‘Whey’ to good health: whey protein and its beneficial effect on metabolism, gut microbiota and mental health. Trends Food Sci. Technol. 2023;133:1–14. doi: 10.1016/j.tifs.2022.12.009. [DOI] [Google Scholar]
- 54.Ha E., Zemel M.B. Functional properties of whey, whey components, and essential amino acids: mechanisms underlying health benefits for active people. J. Nutr. Biochem. 2003;14:251–258. doi: 10.1016/S0955-2863(03)00030-5. submitted for publication. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw simulations trajectory data of this study is publicly available on FigShare (https://figshare.com/) with accession number (https://doi.org/10.6084/m9.figshare.26183426.v1).