Abstract
Background
The 2020 cancer report states that breast cancer remains a significant cause of death for females, despite the use of various strategies for early detection and treatment. However, there are still gaps in the fight against this disease. Researchers are exploring the hippo pathway, one of eight significant pathways involved in cancer progression, for potential biomarkers to use in personalized therapeutics.
Methods
The current study used bioinformatic tools such as DEGs analysis, Methsurv, Km Plotter to generate data that can predict molecular biomarkers associated with hippo pathway in breast cancer development and treatment. The protein-protein interaction pathway was generated using the STRING database to find associations of hippo pathway genes with other dysregulated genes in breast cancer datasets. A disease enrichment study was also done to explore the potential of the hippo pathway in various aspects.
Results
LATS2 and FAT4 genes of the hippo pathway have shown an interesting association with overall survival, hypermethylation, genetic alterations, and decreased expression levels in the breast cancer cohort. Our findings suggest that both of these genes are associated with breast cancer progression and diagnosis and can be utilized as predictive biomarkers by oncologists for personalized therapy in patients.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79688-2.
Subject terms: Cancer, Computational biology and bioinformatics
Introduction
According to the 2020 study by the World Cancer Research Fund International1, it is not surprising that breast carcinoma (BRCA) continues to be the most prevalent cancer in females and the primary cause of death worldwide2. Although significant advancements have been made in the treatment of breast cancer in the last decade, there is still a need to identify and precisely locate different molecular targets in the remaining gaps. One such recent development in the field of breast cancer treatment is the development of molecular biomarkers, which is strongly impacting the likelihood of the disease, treatment, and decision-making in the oncology sector by utilizing personalized therapeutic strategies3–6. Although various molecular biomarkers have been researched, the treatment of breast cancer is still impeded by the dearth of established molecular biomarkers. The hippo pathway, consisting of hippo pathway genes (MST1/2, LATS1/2, YAP, and TAZ, etc.), has been reported to be potentially dysregulated in various malignancies, including breast cancer7,8. For example, the downregulation of the hippo pathway gene has been considered a predictable biomarker when considering prognosis9. It is a known fact that the Hippo signaling pathway is associated with tumorigenesis and tissue size development. In recent studies it is shown that G protein-couple receptors (GPCRs) activation can lead to possible rearrangements of actin cytoskeleton, regulate the phosphorylation states and activity of YAP and TAZ, two major effectors of the Hippo pathway10.
To further strengthen this, a study found that the YAP/TAZ (hippo pathway gene) down expression reversed the trastzumab resistance in HER2-positive breast cancer, while the overexpression of the YAP/TAZ promoted the resistance11,12. Additionally, analysis based on databases has also showcased the predictive and therapeutic biomarker potential of the hippo pathway genes as biomarkers. An extensive study on The Cancer Genome Atlas (TCGA) data for elucidating the role of hippo pathway genes discovered Large Tumor Suppressor Kinase2 (LATS2) and FAT Atypical Cadherin 4 (FAT4) were linked with the patient’s survival in 5 and 6 datasets out of a total 13 datasets13. An excellent role of hippo pathway genes in breast cancer and their strong link with survival and disease aggressiveness should be explored via biological informatics, and the selected genes can then be made targets for further pre-clinical and clinical studies. The current study exploits the extensive bioinformatics analysis (mention techniques and work) on the hippo pathway genes, aiming to assist in the development of biomarkers for breast cancer management and treatment.
Materials and methods
Expression and stage wise analysis of key hippo pathway genes across BRCA cohort
Gene expression profiling interactive v2 (GEPIA 2)14 (http://gepia2.camcer-pku.cn) was used to access the relative mRNA expression levels of the LATS2, FAT4, YAP1, and SAV1 genes across the TCGA-BRCA cohort and matching TGCA normal and GTEx data. After GEPIA expression data were converted to log2 (TPM + 1) values, differential analysis was conducted. Moreover, pathological plot stages in BC were examined in terms of gene expression using GEPIA 2. For the expression analysis in the GEPIA across the BRCA cohort, the cutoff values were p value < 0.05 and log2 (fold change) > 1.
Prognostic analysis of hippo pathway genes across BRCA cohort
Hippo pathway genes which were found altered in DEGs analysis were queried for prognostic analysis using Kaplan- Meier (KM) plotter15,16 (https://kmplot.com/analysis/). The KM plots were generated for those genes that shows significant OS across BC cohort. Based on median values, the microarray data for the BC patient were split into groups with higher and lower expression. Biased arrays and duplicated data were eliminated in the QC section. The logrank p value, median survival, and the related 95% confidence interval (CI) for the risk ratio were computed. Statistics-wise, the p-value < 0.05 was regarded as significant.
DEGs screening of GEO dataset of breast cancer (BRCA) cohort using ‘R’ as software
We extracted BRCA-associated mRNA expression dataset from NCBI-GEO17 (https://www.ncbi.nlm.nih.gov/geo/). The stated inclusion criteria was used to trim down the results available: (1) Homo sapiens samples were involved. (2) Tissue array data was used. (3) Processed and raw files of data should be available. (4) The dataset should not be older than 10 years. (5) Both healthy and control samples must be included in dataset. (6) The dataset must consist of a minimum of 25 samples. In addition, data obtained from abstracts, case reports, review articles, and cell line-based research were not included. Datasets that lacked healthy controls and studies involving non-human samples were removed from the analysis.
The expression data from the delimited file was divided into tumor and normal group. Batch effects were adjusted and principlal component adjustment analysis (PCA) normalization method was carried out using ARSyNseq method. The “limma” R pakage was used to identify genes, taking (|log FC| > 0.5) and p < 0.05 to be statistically significant. The presence of hippo signaling genes were checked in DEGs analysis. The pairwise correlation of hippo signaling genes were accessed through GEPIA2 across BRCA cohort and normal patients. The cutoff for significant p-value was taken as < 0.05.
PPIN construction and enrichment analysis
The protein-protein interaction (PPI) networks are formed based on functional and structural similarity. This assists in the identification of disease-gene associations and protein function prediction. The well-curated interacting partners deduce gene function by examining system-level characteristics and a high-throughput experimentally validated knowledge database. This knowledge integrates conserved structures and functionally annotated proteins from the same GO functional category. This study utilizes the web-based user-friendly tool, Network Analyst (https://www.networkanalyst.ca), to perform gene set enrichment analysis and evaluate the biological importance of expressed gene sets18. This platform relies on gene expression data of the query genes and experimentally confirmed gene-to-gene interactions to identify up and downregulated genes. Furthermore, disease enrichment of the query genes was performed to identify optimal disease-gene associations.
Tumor infiltration analysis
The mRNA expression levels of modified hippo signaling genes were shown to be associated with the presence of tumor-infiltrating immune cells, specifically B lymphocytes, abundance of CD8 + T cells, macrophages and neutrophils which was assessed in BRCA patients using TIMER 2.019 (http://timer.cistrone.org). Spearman correlation was used to evaluate statistical significance.
Mutational analysis of hippo signaling genes using cBioportal
We utilized cBioportal for cancer genomics to evaluate genetic mutations and alterations. The BRCA dataset from the TCGA Firehose Legacy was selected for analysis.
Analysis of methylation in hippo signaling genes
The MethSurv tool20 (https://biit.cs.ut.ee/methsurv), a web-based tool for multivariate analysis, was employed to conduct prognostic analysis of CpG methylation in patients with BRCA.
Results
Differential expression of hippo pathway genes in BRCA cohort in GEO dataset
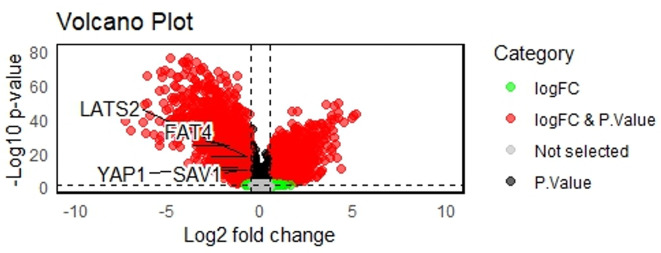
We chose GSE42568 as per specified inclusion exclusion criteria of BRCA-associated mRNA expression profile. The identified DEGs were classified in two upregulated and downregulated subgroups that included 3029 and 2614 genes respectively. The listed Differentially expressed Genes (DEGs) are shown in supplementary data. In the DEGs obtained we identified four hippo signaling genes; LATS2, YAP1, FAT4 and SAV1. All genes were present in the downregulated subgroup (Fig. 1).
Fig. 1.
Volcano plot demonstrating significantly altered Hippo genes.
Hippo signaling genes expression and their correlation with molecular subtype in BRCA cohort
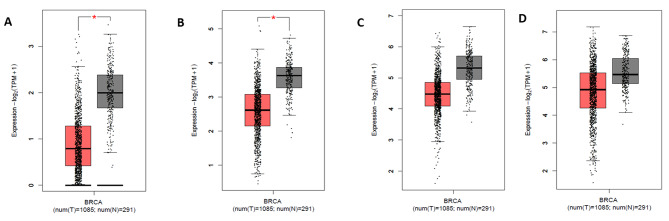
Hippo signaling genes relative expression across TCGA-BRCA cohort was obtained through Gene Expression Profiling Interactive Analysis (GEPIA). The LATS2 and FAT4 expression was found to be significantly downregulated whereas, SAV1 and YAP1 expression levels were also downregulated but are non-significant.
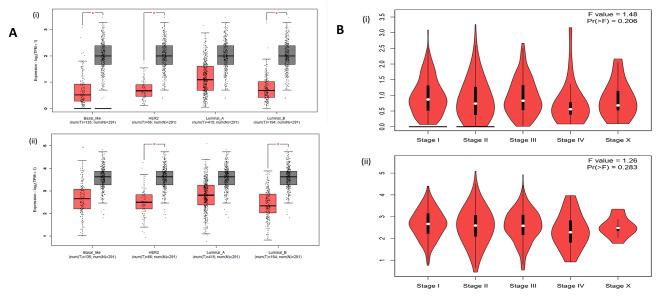
The expression levels were demonstrated by the box-whisker plot in Fig. 2A-D. LATS2 and FAT4 expression levels were further correlated with molecular subtypes and clinical stages as shown in box-whisker plots and volcano plots respectively in Fig. 3(A-B) LATS2 level was significantly correlated with Her2neu and Luminal B subtype whereas, FAT4 levels were significantly correlated with TNBC, Her2neu and Luminal B subtype. The pairwise correlation among LATS2 and FAT4 was demonstrated in Fig. 1 as scatter plot shows high significant value.
Fig. 2.
Box and whisker plots displaying the relative mRNA expression levels of (A) FAT4 (B) LATS2 (C) SAV1 (D) YAP1 across TCGA-BRCA cohort and normal samples. Grey-and red-colored box areas signify normal and tumor patient samples respectively. The top and bottom of the boxes signify 75th and 25th percentile of distribution. Horizontal lines within the boxes represent the median values while minimum and maximum values label the axes endpoints. *p value < 0.05.
Fig. 3.
(A) Association of Molecular subtypes and mRNA expression levels shown in Box and whisker plots (i) FAT4 and (ii) LATS2. (B) The association between clinical stages and mRNA expression levels as shown in volcano plots (i) FAT4 and (ii) LATS2.
Association of FAT4 and LATS2 expression in survival-ship across BRCA cohort
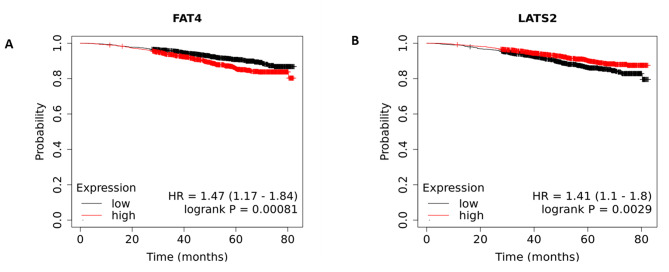
Analyzed using Km Plotter, a prognosis study was conducted on the FAT4 and LATS2 genes to examine the link between their mRNA expression levels and the risk of survival in 2976 samples from BRCA patients. The analysis as demonstrated in Fig. 4 shows significant poor OS of BRCA patients when the mRNA expression levels of FAT4 was high whereas, LATS2 downregulation was found to be associated with poor overall survival. The supplementary Table S1 provides specific information on the median survival time, hazard ratio (HR), 95% confidence interval (95%CI), and logrank p-value for each hippo signaling gene in both the low and high expression cohorts.
Fig. 4.
KM plots showing Overall survival of (A) FAT4 and (B) LATS2 across microarray BRCA cohort. Black and red dots signify lower and higher expression groups.
PPIN construction and enrichment analysis of Hippo signaling genes and top 50 significant DEGs
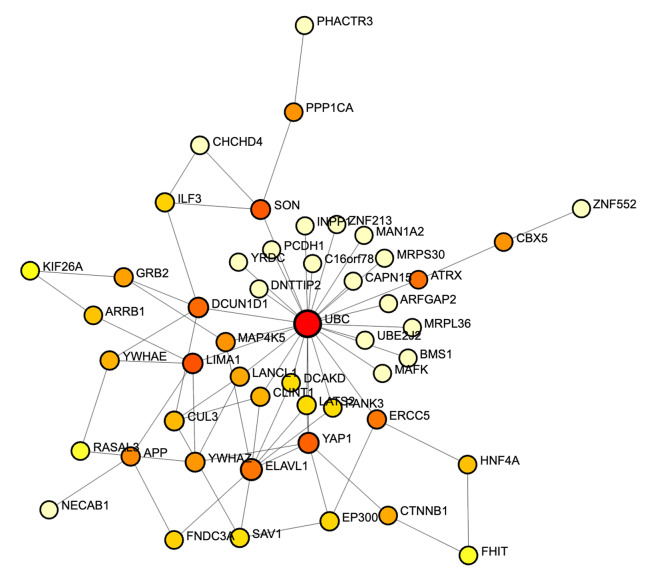
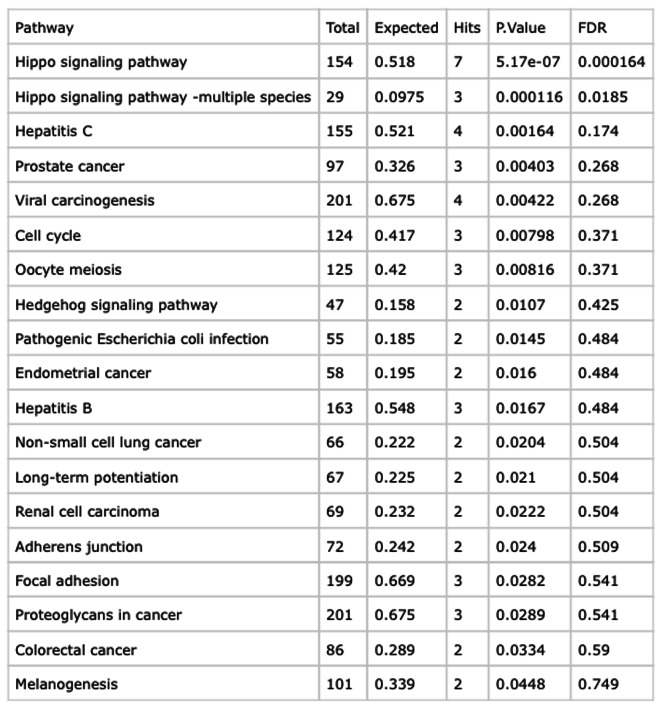
Geneset Enrichment Analysis calculated whether the query genes (top 50 significant DEGs) are statistically overrepresented in any underlying pathway. Therefore, the average FC values are utilized for the generation of interconnected gene-to-gene networks and further gene-disease networks. The query hippo signaling genes were subjected to IMEx database21 as a reference database and two-dimensional network is generated base on the expression of the genes. The colour of the gene expression varies from red to light yellow (where red = upregulated and light yellow as downregulated). Figure 5 highlights the central genes within the network based on computed topological properties and expression data. The majority of genes participating in well-connected modules are differentially expressed with many of them downregulated at experimental level. In support, three-dimensional network visualization is provided in supplementary data. Furthermore, disease enrichment analysis infers the active participation of altered genes in hippo signaling pathway, Hepatitis C, prostate cancer and other types of cancer. Figure 6. In support, the computed enrichment score signifies the level of participating of genes in completing a biological pathway, i.e. Hippo signalling pathway. As per figure, the primary interactors of the BRCA expressed genes are highly enriched towards cancer related pathways.
Fig. 5.
Network analysis of the query gene datasets based on the differential gene expression. The colour of dots indicate level of expression demonstrating High (Red) to low (light Yellow).
Fig. 6.
Listing of the computed p-values and False Discovery Rate (FDR) for query gene dataset participating in network generation.
LATS2 and FAT4 genes correlation with tumor infiltrating immune cells
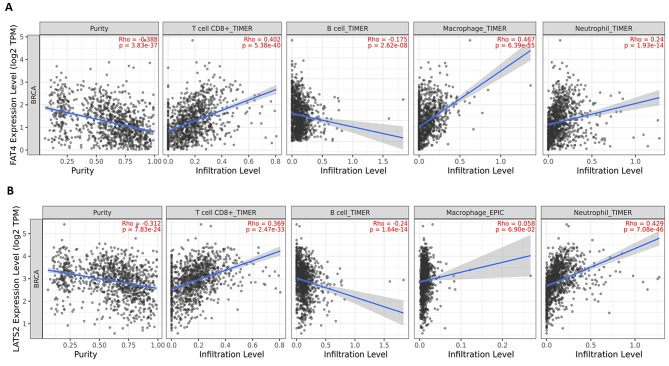
An analysis was conducted to examine the relationship between the expression levels of LATS2 and FAT4 genes and tumor purity, as well as the levels of macrophages, neutrophils, B lymphocytes, and CD8 + T cells infiltrating the TCGA-BRCA cohort. The results of this analysis are presented in scatter plots in Fig. 7.
Fig. 7.
Scatterplots showing significant correlations of (A) FAT4 (B) LATS2 with infiltrating levels of CD8+T cell, B cells, neutrophils, and macrophages across TCGA-BRCA cohort. Spearman’s correlation value and estimated statistical significance were shown as the legends for each scatter plot.
FAT4 exhibits strong positive associations with the numbers of invading CD8 + T cells (r = 0.402, p = 5.38e-40). Macrophages (r = 0.467, p = 6.39e-55) and neutrophils (r = 0.24, p = 1.93e-14). LATS2 also shows significant positive correlation with T cells CD8+ (r = 0.369, p = 2.47e-33), Macrophages (r = 0.058, p = 6.90e-02), neutrophils (r = 0.429, p = 7.08e-46). Both FAT4 and LATS2 shows significant negative correlation with B cell infiltration levels (r=-0.175, p = 2.62e-08) and (r=-0.24, p = 1.64e-14) respectively.
Mutational profile of LATS2 and FAT4 genes using cBioPortal domain
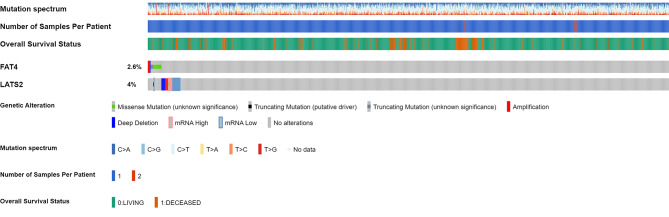
The cBioPortal platform was utilized to verify the exact genetic mutations in both genes within the BRCA cohort, consisting of 1108 samples. The oncoprint findings for the searched genes are presented in Fig. 8. This shows 2.6% of overall genetic alterations in FAT4 and 4% in LATS2 genes. Here, FAT4 has most of genetic alterations as mutation whereas, LATS2 has mRNA dysregulation and deep deletion as major contributors to genetic alterations Fig. 2.
Fig. 8.
OncoPrint summarizing genomic alterations of FAT4 and LATS2 across TCGA-BRCA cohort comprising 1108 patient samples. The bottom row represents frequency of genomic alterations in both genes with red, blue, green, and grey bars signifying amplifications, deep deletions, missense and truncating mutations, respectively.
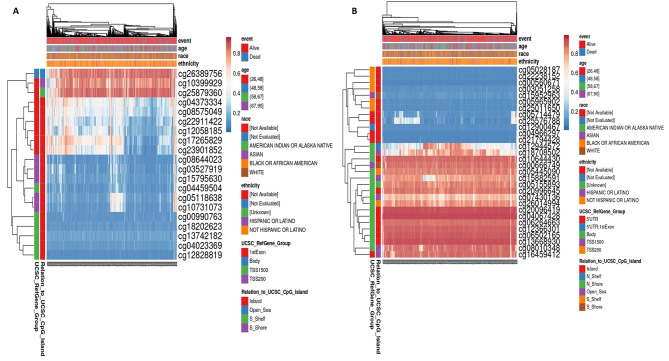
Promoter methylation of LATS2 and FAT4 genes its prognostic significance in BRCA patients
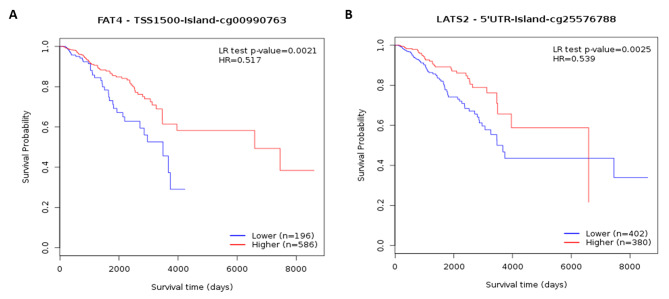
The methylation status of LATS2 and FAT4 genes were analyzed on various CpG island (CGI) using MethSurv. Here, we obtained heatmaps showing methylation expression of both genes across BRCA cohort (Fig. 9). Most of the CpG islands were hypermethylated in case of LATS2 gene. These CpG islands were then analyzed for prognostic significance and most significant CGI is demonstrated as KM plot cg00990763 for FAT4 and cg25576788 for LATS2 (Fig. 10).
Fig. 9.
Heatmaps of CpG methylation levels of (A) FAT4 (B) LATS2 across BRCA patients. Rows indicates the CpGs and columns indicates the patients. Methylation levels (1 = fully methylated; 0 = fully unmethylated) are shown as a continuous variable from red to blue color, high expression to low expression. Various colorful side boxes were used to represent the event, relation to UCSC_CpG_island and UCSC_refGene_Group.
Fig. 10.
KM plot showing single CpG methylation associated with overall survival of (A) FAT4 and (B) LATS2 across BRCA patients. It’s location relative to CpG island, gene sub-region, CpG ID, and gene ID are also shown. Red and blue colors signify higher and lower expression groups.
Discussion
Breast cancer is the most common cancer among females globally22. Despite advancements in the detection and management of the disease, clinicians are now much focused on developing personalized approaches to treat breast cancer. In this sequence, novel biomarkers play a crucial role in accessing the risk factors, stage, subtype, and predicting the treatment strategy23. Biomarkers can be categorized as potent, measurable, and evaluable indicators associated with the advancement of a disease. In recent years, meta-analysis using publicly available datasets has become a popular trend to address complex biological aspects computationally24. Analyzing the big data sets provides bioinformaticians with an opportunity to draw valuable information and identify the key genes associated with breast cancer25.
It is imperative that hippo pathway genes have altered expression levels in many cancers, and LATS2 expression levels were found to be downregulated in breast cancer human samples, establishing it as a major tumor suppressor26,27. As indicated by the cancer genome Atlas, the hippo pathway is one of the eight signaling pathways that are highly dysregulated in cancer28. Also, earlier studies to examine the role of hippo signaling pathway in breast cancer cases suggested the extensive role of hippo pathway genes in progression of ER + breast cancer29. FAT atypical cadherin 4 (FAT4) is known to be associated with hippo signaling by inhibiting the Yes Associated Protein-1 (YAP1) nuclear translocation30. The expression levels of FAT4 in breast cancer are downregulated, making it a promising tumor suppressor in BRCA31.
In our study, we selected a microarray human breast cancer dataset from the GEO database and analyzed them through ‘R’ software for extracting the differentially expressed genes in breast cancer (DEGs). In our findings, four hippo pathway genes were present in DEGs, which include LATS2, FAT4, SAV1, and YAP1. Further, the top 50 most significant DEGs were recognized based on p-values and were subjected to network analysis and pathway enrichment studies.
The network analysis based on the changes in gene expression is termed differential network analysis to unwire biological insights and related complexities. The study includes a network analysis of the obtained gene sets along with related expression values computed by fold change to identify the differentially associated genes based on network-based properties (Fig. 4). The relative fold change values are log transformed and summarized together. The fold change values are computed for all pair-wise comparisons of each data point. The ranks were assigned based on multiple iterations, resulting in a P-value and false discovery rate associated with each gene. Based on the ranks assigned, the enriched gene nodes were highlighted based on the expression ranks. This enables the identification of upregulated and downregulated gene sets among query datasets.
The current study involves the identification of disease-enriched pathways based on the expression of the genes obtained. Such analysis infers the overrepresentation of the specific diseases or conditions provided by the given set of genes or proteins. It includes the support of the reference databases, viz., the Human Gene Mutation Database (HMDB), IMEx, etc., to determine whether certain diseases are significantly enriched in the query gene set, which in turn suggests a potential association or relevance to the disease in question. Such statistical computation assesses the significance of enrichment analysis in disease associations, helping researchers understand the complex genetic underpinnings of cancer. This analysis reveals the enrichment of the majority of cancer-related pathways, hippo-signaling pathways, and viral diseases (Fig. 5).
Moreover, to validate the expression levels of all four hippo signaling genes, the GEPIA2 tool was used. We found that among the query gene sets, LATS2 and FAT4 were significantly downregulated in BRCA, which is in accordance with the previous studies26,27,31. To further establish the role of LATS2 and FAT4 genes in BRCA, the prognostic significance was studied and analysed through a KM plotter. We found that expression of both the genes was significantly associated with patients’ survival, which indicates the relevance of both genes as putative tumor suppressors in the progression of BRCA. This was also in accordance with many studies using patients’ samples and will generate better insights for targeted treatment in cancer therapy27.
Epigenetic regulation plays an important role in dysregulating the expression levels of biomarker genes in cancer. The LATS2 expression level seems to be highly regulated by hypermethylation of CpG islands in the promoter region in different cancer types26,32,33. Whereas, in case of Nasopharyngeal carcinoma the de-methylated LATS2 promotes the growth of tumor cells leading to poor prognosis34. To validate these findings in our study, we used MethSurv and found that both LATS2 and FAT4 gene promoter regions are highly methylated at various CpG islands in Breast Carcinoma. The methylation of different CpG islands in the promoter region of gene can have significant impact on the gene expression. In this order to identify the significantly correlated CpG islands, further analysis was done to explore CpG islands with highest prognostic significance, and the results show a significant association between hypermethylation of the LATS2 promoter region (cg25576788) and poor overall survival in BRCA patients.
Genetic alteration in cancers is widely studied on different platforms; similarly, in the case of breast carcinoma, genetic mutations play an important role in the progression of the disease. The major type of alteration reported in breast cancer is the amplification of proto-oncogenes35 through missense mutations that alter the functionality of proteins. LATS2 and FAT4 genetic mutation status was checked through cBioportal (Firehose Legacy data), and 4% and 2.6% of total genetic alteration were obtained, respectively. This suggests a strong association between genetic mutations in the LATS2 gene and the development of breast cancer, as also reported in earlier studies on lung carcinoma36.
The tumor infiltration analysis was also performed on the LATS2 and FAT4 genes, as tumor-infiltrating immune cells are responsible for maintaining the tumor microenvironment (TME), which influences tumor growth and survival in breast cancer37. Tumor-infiltrating immune cells (TILs) have a significant impact on the immune response against cancer. The major TILs associated with breast cancer include CD8+, CD4+, NK cells, and FOXP3 + cells38. In our analysis, we found a strong positive correlation between both LATS2 and FAT4 and CD8 + T-cells and macrophages, suggesting their role in controlling the TME of breast cancer cells.
Hippo pathway genes play an important role in breast cancer progression through multiple regulations; the LATS2-YAP1 axis is the controlling point in the hippo pathway, whose disruption can lead to cancer aggressiveness. FAT4, being the protein that controls the localization of YAP1 into the nucleus, is another important component. Our study thus suggests that these two can be potential biomarkers of breast cancer for diagnosis as well as prognostic purposes. Breast cancer is hard to treat when patients develop resistance to chemotherapy such as doxorubicin, which is also associated with low expression of LATS239. Therefore, meta-analysis of cancer genomics is an important and useful tool for establishing the role of various genes in cancer progression and treatment through precision medicine.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not Applicable.
Author contributions
Sa, S.M, T.B and M.Z analysed the data and wrote the manuscript. S.A, Malik A.H, A.A, A.A A and K.D performed critical reviews and edits of the manuscript. All authors contributed to the conceptualization of the study and provided final approval to the manuscript submitted for publication consideration.
Funding
This research has been funded by Scientific Research Deanship at University of Ha’il‑Saudi Arabia through project number (RG‑23 131).
Contributions.
Sadaf, Sheersh Massey, Tulika Bhardwaj and Mubashir Zafar analysed the data and wrote the manuscript. Sadaf Anwar, Malik Asif Hussain, Abrar Ali, Abdulaziz A Aloliqi and Kapil Dev performed critical reviews and edits of the manuscript. All authors contributed to the conceptualization of the study and provided final approval to the manuscript submitted for publication consideration.
Data availability
This Manuscript does not report data generation. The data that support the findings of this study are available from corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sadaf, Mubashir Zafar contributed equally to this work.
References
- 1.https://www.wcrf.org/cancer-trends/
- 2.Wu, H. J. & Chu, P. Y. Recent discoveries of Macromolecule- and cell-based biomarkers and therapeutic implications in breast Cancer. Ijms. 22 (2), 636. 10.3390/ijms22020636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Punturi, N. B. et al. Mismatch repair deficiency predicts response to HER2 blockade in HER2- negative breast cancer. Nat. Commun.12 (1), 2940. 10.1038/s41467-021-23271-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venetis, K. et al. Analytical performance of next-generation sequencing and RT-PCR on formalinfixed paraffin-embedded tumor tissues for PIK3CA testing in HR+/HER2 – breast cancer. Cells. 11 (22), 3545. 10.3390/cells11223545 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarantino, P. et al. Prognostic and biologic significance of ERBB2-low expression in early-stage breast cancer. JAMA Oncol.8, 1177–1183. 10.1001/jamaoncol.2022.2286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry, N. L. et al. Biomarkers for systemic therapy in metastatic breast cancer: ASCO guideline update. J. Clin. Oncol.022 (0), 3205–3221. 10.1200/jco.22 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Guimei, M. et al. Inhibition of yes-associated protein-1 (YAP1) enhances the response of invasive breast cancer cells to the standard therapy. Breast Cancer: Targets Therapy, pp.189–199. (2020). [DOI] [PMC free article] [PubMed]
- 8.Skibinski, A. et al. Joshua LaBaer, and Charlotte Kuperwasser. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell reports 6, no. 6 : 1059–1072. (2014). [DOI] [PMC free article] [PubMed]
- 9.Lin, X., Cai, F., Li, X. & Kong, X. Prognostic significance of mammalian sterile 20-like kinase 1 in breast cancer. Tumor Biol.34, 3239–3243 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Luo, J. & Yu, F. X. GPCR-Hippo signaling in cancer. Cells. 8 (5), 426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vici, P. et al. The Hippo transducer TAZ as a biomarker of pathological complete response in HER2-positive breast cancer patients treated with trastuzumab-based neoadjuvant therapy. Oncotarget 5, no. 20 : 9619. (2014). [DOI] [PMC free article] [PubMed]
- 12.Calses, P. C., Crawford, J. J., Lill, J. R. & Dey, A. Hippo pathway in cancer: aberrant regulation and therapeutic opportunities. Trends cancer. 5 (5), 297–307 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Poma, A. M. et al. Hippo pathway affects survival of cancer patients: extensive analysis of TCGA data and review of literature. Sci. Rep.8, 10623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profling and interactive analyses. Nucleic Acids Res.45 (W1), W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyorfy, B. et al. An online survival analysis tool to rapidly assess the efect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat.123 (3), 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Lanczky, A. & Gyorfy, B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res.23(7), e27633 (2021). [DOI] [PMC free article] [PubMed]
- 17.Barrett, T. et al. NCBI GEO: mining millions of expression profles–database and tools. Nucleic Acids Res.33 (Database issue), D562–D563 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, G. et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res.47 (W1), W234–W241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, T. et al. TIMER2.0 for analysis of tumor-infltrating immune cells. Nucleic Acids Res.48 (W1), W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modhukur, V. et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 10 (3), 277–288 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Porras, P., Orchard, S. & Licata, L. IMEx databases: displaying molecular interactions into a single, standards-compliant dataset. Methods Mol. Biol.2449, 27–42 (2022). [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization. (2022).
- 23.Toss, A. & Cristofanilli, M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res.17, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Y. et al. Computer-aided biomarker discovery for precision medicine: data resources, models and applications. Brief. Bioinform.20 (3), 952–975. 10.1093/BIB/BBX158 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Deng, J. L., Xu, Y. H. & Wang, G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front. Genet.10 (JUL), 465362. 10.3389/FGENE.2019.00695/BIBTEX (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadaf, H. M. et al. Hypermethylated LATS2 gene with decreased expression in female breast cancer: a case control study from North India. Gene. 676, 156–163 (2018). Epub 2018 Jul 17. PMID: 30010037. [DOI] [PubMed] [Google Scholar]
- 27.Matsui, S. et al. LATS2 promoter hypermethylation and its effect on gene expression in human breast cancer. Oncol. Lett.15 (2), 2595–2603. 10.3892/ol.2017.7535 (2018). Epub 2017 Dec 6. PMID: 29434979; PMCID: PMC5777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Vega, F. et al. Oncogenic signaling pathways in the Cancer Genome Atlas. Cell. 173, 321–337e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, S. et al. Hippo signalling maintains ER expression and ER + breast cancer growth. Nature. 591 (7848), E1–E0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, W. et al. The novel FAT4 activator jujuboside a suppresses NSCLC tumorigenesis by activating HIPPO signaling and inhibiting YAP nuclear translocation. Pharmacol. Res.170, 105723 (2021). Epub 2021 Jun 9. PMID: 34116210. [DOI] [PubMed] [Google Scholar]
- 31.Qi, C., Zhu, Y. T., Hu, L. & Zhu, Y. J. Identification of FAT4 as a candidate tumor suppressor gene in breast cancers. Int. J. Cancer. 124 (4), 793–798. 10.1002/ijc.23775 (2009). PMID: 19048595; PMCID: PMC2667156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, Y. et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res.11 (4), 1380–1385 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Gu, Y. et al. Hypermethylation of LATS2 promoter and its prognostic value in IDH-mutated low-grade gliomas. Front. Cell. Dev. Biology. 8, 586581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, Y. et al. LATS2 is de-methylated and overexpressed in nasopharyngeal carcinoma and predicts poor prognosis. BMC cancer. 10, 1–3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bièche, I. & Lidereau, R. Genetic alterations in breast cancer. Genes Chromosom. Cancer. 14 (4), 227–251 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Stražišar, M., Mlakar, V., Damjan & Glavač LATS2 tumour specific mutations and down-regulation of the gene in non-small cell carcinoma. Lung cancer. 64 (3), 257–262 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Li, J. J., Julia, Y. & Tsang and Gary M. Tse. Tumor microenvironment in breast cancer—updates on therapeutic implications and pathologic assessment. Cancers 13, no. 16 : 4233. (2021). [DOI] [PMC free article] [PubMed]
- 38.Yang, L. et al. Identifying FL11 subtype by characterizing tumor immune microenvironment in prostate adenocarcinoma via Chou’s 5-steps rule. Genomics 112, no. 2 : 1500–1515. (2020). [DOI] [PubMed]
- 39.Kaur, S., Najm, M. Z., Khan, M. A., Akhter, N. & Shingatgeri, V. M. Mudra Sikenis, Sadaf, and Abdulaziz A. Aloliqi. Drug-resistant breast cancer: Dwelling the hippo pathway to manage the treatment. Breast Cancer: Targets Therapy : 691–700. (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Manuscript does not report data generation. The data that support the findings of this study are available from corresponding author upon reasonable request.