Abstract
The GRAMD genes are involved in maintaining cholesterol homeostasis, apoptosis, cancer and production traits in livestock. A lipid-binding GRAM domain is implicated in lipid transport and metabolism. The functions of GRAMD proteins remain incompletely understood. The aim of the present study was therefore to investigate the associations between six GRAMD genes in cattle using data from the international genomic evaluation of the Interbull InterGenomics Centre and to evaluate genotype-phenotype associations in human, cattle, pig, mouse and, chicken. Genotyping of 55,013 bulls was performed using DNA microarrays and 11 SNPs were mapped to the five GRAMD genes. A phenome-wide association study (PheWAS) tested associations between the 11 SNPs and 36 traits. The integrated analysis of SNP effects, rankings, and clustering patterns revealed their potential for improving cattle productivity, health, and robustness, and established a baseline for the targeted improvement of cattle traits. This study lays the groundwork for functional experiments aimed at uncovering the mechanism of action of GRAMD genes and to evaluate the potential of using GRAMD sequence variants for selection programs in dairy cattle. The study presents an example of how the combination of GWAS and the PheWAS offers a promising toolbox for the systematic functional annotation of vertebrate genomes.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80117-7.
Keywords: Cattle, GRAM domain, GWAS, PheWAS, Single nucleotide polymorphism (SNP), Mouse
Subject terms: Genetics, Molecular biology
Executive summary of the study.
Understanding GRAMD Genes: The GRAMD group of genes, key players in cholesterol homeostasis, apoptosis, and autophagy, remains partially understood. This study aims to provide further insights into their comprehensive functions.
Methodological Approach: Utilizing GWAS on 55,013 bulls and 55,172 SNPs, followed by PheWAS analysis of 36 phenotypic traits, we conducted a functional annotation of the GRAMD gene family in cattle.
Insights and Implications: Our integrated analysis of SNP effects, rankings, and clustering within the GRAMD family reveals their significance in enhancing cattle productivity, health, and robustness. This provides a foundation for the targeted improvement of cattle traits.
Beyond Cattle - A Broader Perspective: Examination using a mouse obesity model suggests a potential involvement of GRAMD genes in fat deposition, indicating their importance across mammalian species.
Toolbox for Genetic Research: This study reinforces the combined use of GWAS and PheWAS (GWAS-to-PheWAS) as a potent approach for the systematic functional annotation of vertebrate genomes.
Future Directions: The introduction of gene-QTL overlap analysis emerges as a promising method for prioritizing and further functionally characterizing genes, setting the stage for future genetic advancements.
Introduction
GRAM (glucosyltransferases, Rab-like GTPase activators and myotubularin) domain- containing gene group includes genes with various functions, including GRAMD1A, GRAMD1B, and GRAMD1C, which are involved in maintaining cholesterol homeostasis1 and GRAMD4 which is involved in apoptosis2. GRAMD2A and GRAMD2B are less researched but have been shown to be involved in cellular Ca2+ homeostasis3 and serum leukocyte concentration in coronary artery disease (CAD)4 respectively. GRAM domain (GRAMD) genes have been associated with various production and reproduction traits in cattle. For example, GRAMD1B is associated with feed efficiency5. In addition, three intronic polymorphisms have been linked with four reproduction traits in Nordic red cattle: the rs41763261 polymorphism with female fertility index and days from first to last insemination, rs41763326 with the length in days of the interval from calving to first insemination, and rs110670590 with the number of inseminations per conception6. However, intronic polymorphisms of GRAMD1B were not associated with fattening and carcass traits in Slovenian dual-purpose Simmental cattle7.
Genome-wide association study (GWAS) and phenome-wide association study (PheWAS) are approaches to detect associations between genotype and phenotype. Several GWAS have been conducted to identify genetic variants associated with a phenotype (many variants – one phenotype). However, a PheWAS is a reverse approach that aims to identify phenotypes associated with a genotype. The PheWAS technique is, therefore, capable of accessing cross-phenotype associations or pleiotropy8,9. Several PheWAS studies have already been conducted in human, but they are less common in animal breeding. For example, GWAS and PheWAS data present a vital part of the recently developed PigBiobank10.
The function of the GRAMD gene group is not completely elucidated; therefore, the aim of the present study was to perform an association analysis between six members of the GRAMD gene group (GRAMD1A/B/C, GRAM2A/2B, and GRAMD4) and 36 phenotypic and economically important traits in the Brown Swiss cattle population (BSW) using GWAS followed by PheWAS. Additionally, we performed literature and database review regarding the function of GRAMD genes in five vertebrate species: human, cattle, pig, mouse and chicken.
Materials and methods
Databases and bioinformatics tools
In the present study, we focused to function of six genes from the GRAMD gene group (Table 1) in cattle and performed comparative analysis in human, pig, mouse and chicken. Relevant genomic data (genomic location, synteny, sequence variations, Genomic Evolutionary Rate Profiling (GERP), Gene Ontology (GO)) of the GRAMD genes were extracted from the Ensembl genome browser, release 11111 and HGNC database (https://www.genenames.org). Constrained elements, defined as sequences or regions in the genome highly conserved across different species, indicating their importance in biological functions and evolutionary significance, were obtained from the Ensembl database. The alignment between GRAMD genes locations and the positions of SNPs from the commercial BovineSNP50 BeadChip was analyzed using the SNPchiMp v.312. The potential impact of SNPs on GRAMD gene expressions in different bovine tissues was validated using the cattle Genotype-Tissue Expression atlas (cGTEx) database (https://cgtex.roslin.ed.ac.uk/, accessed September 18, 2024). QTLs were retrieved from QTLdb, release 5213.
Table 1.
11 SNPs within a group of five GRAMD genes analyzed in the present study.
| Symbol | SNP rs ID | Commercial SNP ID | variation consequence | SNP in constrained element | GERP |
|---|---|---|---|---|---|
| GRAMD1A | / | / | / | / | / |
| GRAMD1B | rs41761360 | ARS-BFGL-NGS-2713 | intron variant | no | 0.58 |
| GRAMD1B | rs109709275 | ARS-BFGL-NGS-98,724 | splice region variant, splice polypyrimidine tract variant, intron variant | yes | 1.22 |
| GRAMD1B | rs41763278 | ARS-BFGL-NGS-5976 | intron variant | no | -4.73 |
| GRAMD1B | rs41756499 | ARS-BFGL-NGS-111,085 | intron variant | no | -4.73 |
| GRAMD1B | rs41757840 | ARS-BFGL-NGS-116,109 |
downstream gene variant (GRAMD1B), intronic (ENSBTAG00000063982; lncRNA), exonic (ENSBTAG00000061947; lncRNA) |
yes | 0.05 |
| GRAMD1C | rs41616212 | Hapmap44172-BTA-96,950 | intron variant | no | -0.46 |
| GRAMD2A | rs109872173 | ARS-BFGL-NGS-113,726 | intron variant | no | 0.7 |
| GRAMD2B | rs43504641 | ARS-BFGL-NGS-18,240 | intron variant | no | -6.03 |
| GRAMD2B | rs110898337 | ARS-BFGL-NGS-44,139 | intron variant | no | -1.44 |
| GRAMD4 | rs109247065 | ARS-BFGL-NGS-112,849 | intron variant | no | 0.29 |
| GRAMD4 | rs109726105 | ARS-BFGL-NGS-1089 | downstream gene variant | no | 1.07 |
Cattle traits were categorized into trait classes according to the Vertebrate Trait Ontology (VTO)14 and QTLdb. The effects of SNPs and SNP pRank for variants within GRAMD genes associated with various cattle traits were visualized using a heatmap, generated in RStudio (http://www.posit.co/, Version 2024.04.2) with the ComplexHeatmap package (https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html, Version 2.20.0)15,16.
Mammalian phenotype ontology annotations were obtained from the Mouse Genome Informatics (MGI)17. As the genome databases are currently based on different genome assemblies, two genome locations were used for the analysis. For example, for the GRAMD1B gene, we used the current assembly ARS-UCD1.3 (GCA_002263795.3) (15:34039549–34221229) and the former assembly Bos_taurus_UMD_3.1.1 (15:34577459–34764024) for SNP locations on the DNA microarray. SNPs and gene expression of inbred mouse models for polygenic obesity (FLI) and leanness (FHI) were obtained from the studies18,19. Predicted variant consequences were identified using the Ensembl Variant Effect Predictor (VEP). The tool annotates variants located within the gene region, extending to 5000 base pairs (bp) both upstream and downstream. Literature on six GRAMD genes was extracted from the PubMed database.
Samples, genotyping and statistics analysis
The analysis was performed using SNP effect data from the routine international Interbull InterGenomics genomic evaluation for Brown Swiss cattle (https://interbull.org/ib/interbullcentremain)20,21. The study included 55,013 genotyped bulls from Germany, Switzerland, Italy, the United States of America, Austria, France, Slovenia, Canada, the Netherlands, and Luxembourg. Genotyping was performed using various DNA microarrays, including Weatherbys Scientific IDB versa50k and IDBv3 BovineSNP50 BeadChip, and GeneSeek GGP4v2 and GGPv3, with all SNP densities harmonized to the Interbull reference map, which comprises 55,172 SNPs22. Imputation was performed according to the method described by23 using FindHap v3 software (http://aipl.arsusda.gov/software/findhap/). Quality control measures such as call rate threshold, minor allele frequency threshold, the departure from Hardy-Weinberg equilibrium, and removal of animals with missing genotypes were performed as described by22 and 24 to assess the integrity and informativeness of the SNPs.
The InterGenomics evaluation estimates SNP effects for SNPs retained after quality control for each country-trait combination, using a common multi-country reference population25. SNP effect estimates were performed using an iterative, nonlinear GBLUP model with a heavy-tailed prior for marker effects, analogous to Bayes A, as described by22,25,26. In the present study, the estimated SNP effects for the analyzed traits were expressed on the Slovenian scale. A total of 43,322 estimated SNP effects were obtained from the December 2023 routine evaluation and used for further GWAS analyses. For each of the 36 traits included in the genomic evaluation, the proportion of phenotypic variance explained by the selected SNP was calculated as the sum of the absolute values of SNP effects greater than that of the selected SNP. Based on this value, the traits were ranked to determine the highest proportion of phenotypic variance explained by the SNP.
Results
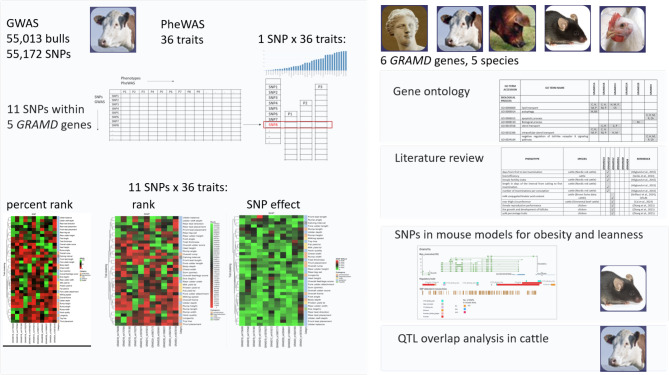
We performed a GWAS followed by a PheWAS approach based on the analysis of 55,172 SNPs and 36 phenotypic traits in 55,013 bulls for the functional annotation of the GRAMD gene family in cattle. Additionally, we summarized published literature and GO databases for genotype-phenotype associations in five vertebrate species and analyzed genotype differences of the GRAMD gene family in inbred mouse models for polygenic obesity and leanness. A graphical summary of the study is shown in Fig. 1.
Fig. 1.
Graphical summary of the study. Legend: cattle (C), human (H), mouse (M), pig (P), chicken (Ch), GO: gene ontology, QTL: quantitative trait locus.
In the present study, we focused on six GRAMD genes in five species (Supplementary Table S1). An example of syntenic maps for the GRAMD4 gene is shown in the Supplementary Figure S1 and visualizes synteny between human chromosome 22 with GRAMD4 gene and chromosomes in cattle, pig, mouse and chicken. We used a two-step approach: GWAS, followed by PheWAS, which we named GWAS-to-PheWAS. The study group for the GWAS and estimation of SNP effects consisted of 55,013 genotyped bulls. Of the polymorphisms present on the cattle DNA-microarrays, 11 SNPs are located at the coordinates of five GRAMD genes: GRAMD1B (five SNPs), GRAMD1C (one SNP), GRAMD2A (one SNP), GRAMD2B (two SNPs) and GRAMD4 (two SNPs); however, SNPs are not present in the GRAMD1A gene (Table 1). Most of the SNPs are intronic, and one is located downstream of the GRAMD4 gene. Sequence variant rs41757840 is located downstream of the GRAMD1B and is also located within two lncRNA genes: the intron of ENSBTAG00000063982 and the exon of ENSBTAG00000061947.
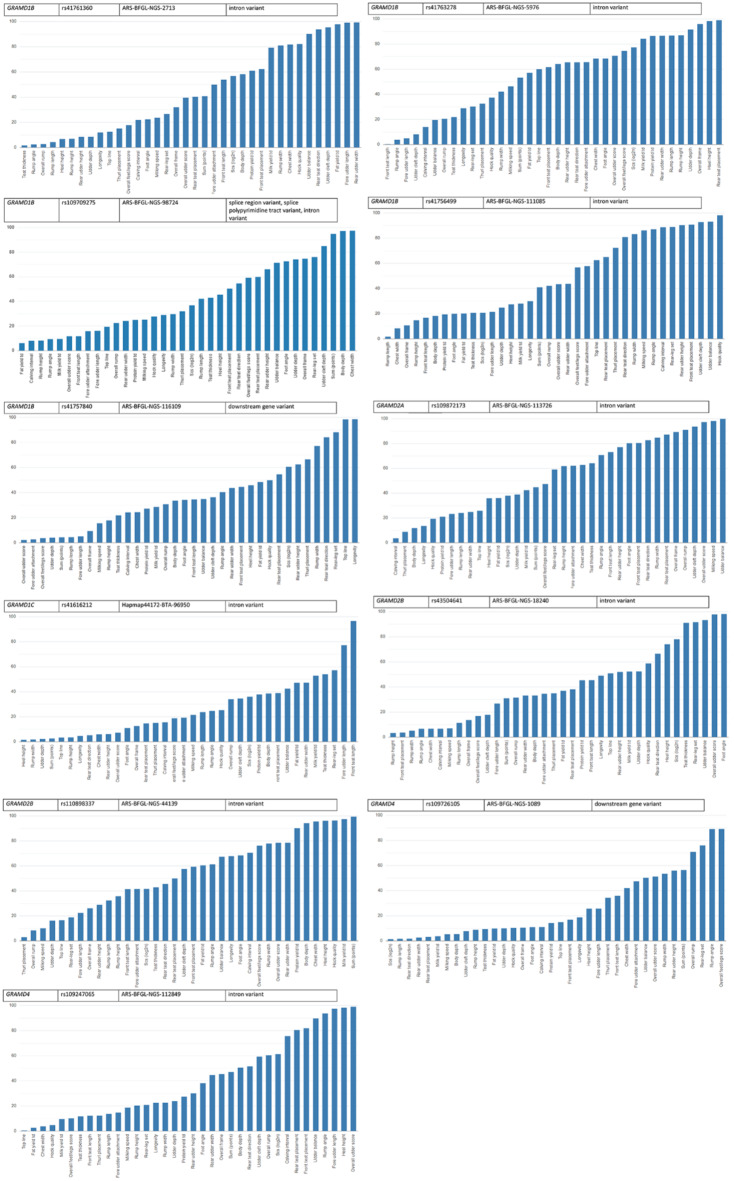
An association analysis was performed between 11 GRAMD group polymorphisms and 36 traits from five trait classes: health (1), milk (4), production (9), reproduction (1), and exterior traits (21). Figure 2 illustrates the percent rank for the effect of the 11 SNPs of the GRAMD gene group on 36 traits (Supplementary Tables S2-S12). For example, of the polymorphisms present on the DNA-microarray, the polymorphism rs109709275 T > C (ARS-BFGL-NGS-98724) maps to the coordinates of the GRAMD1B gene. An association analysis was performed between rs109709275 and 36 traits from five trait classes (Table 2, Supplementary Table S3). The results showed that the polymorphism rs109709275 explained the highest proportion of phenotypic variance in the traits of milk fat yield, calving interval, rump height and rump angle, milk yield, overall udder score, and front teat length. Of the 36 traits analyzed, the seven traits most strongly associated with rs109709275 belong to the milk, reproduction, production, and exterior trait classes.
Fig. 2.
Effects of 11 GRAMD gene SNPs: percentile rankings across 36 traits.
Table 2.
Results of the association analysis between GRAMD1B polymorphism rs109709275 and 36 traits in BSW cattle. *% of phenotypic variability explained with SNP rs109709275 and other SNPs (rank-1) with higher SNP effect for a specific trait.
| Trait | Trait class | SNP effect | Rank | Percent rank | Phenotypic variability* | |
|---|---|---|---|---|---|---|
| 1 | Milk fat yield | milk | 0.003694209 | 2617 | 6.04081 | 20.21531 |
| 2 | Calving interval | reproduction | 0.003240869 | 3430 | 7.91746 | 25.12208 |
| 3 | Rump height | production | 0.003402021 | 3515 | 8.11366 | 25.86083 |
| 4 | Rump angle | exterior | 0.003581134 | 4028 | 9.29782 | 27.62449 |
| 5 | Milk yield | milk | 0.003273226 | 4104 | 9.47325 | 28.24709 |
| 6 | Overall udder score | exterior | 0.002904529 | 5036 | 11.62458 | 32.79526 |
| 7 | Front teat length | exterior | 0.003356773 | 5047 | 11.64997 | 33.40014 |
| 8 | Fore udder attachment | exterior | 0.002325045 | 6808 | 15.71488 | 40.98461 |
| 9 | Fore udder length | exterior | 0.002676451 | 6938 | 16.01496 | 41.25329 |
| 10 | Top line | exterior | 0.002233132 | 8403 | 19.39661 | 46.64559 |
| 11 | Overall rump | production | 0.001993696 | 9703 | 22.3974 | 51.74095 |
| 12 | Rear udder width | exterior | 0.00218361 | 10,419 | 24.05014 | 53.97597 |
| 13 | Milk protein yield | milk | 0.002081498 | 10,780 | 24.88343 | 55.28863 |
| 14 | Milking speed | milk | 0.002126747 | 10,854 | 25.05424 | 55.80904 |
| 15 | Hock quality | exterior | 0.001782385 | 11,983 | 27.66031 | 58.88893 |
| 16 | Longevity | production | 0.001764321 | 12,529 | 28.92064 | 60.73292 |
| 17 | Rump width | production | 0.001706142 | 12,806 | 29.56004 | 61.65832 |
| 18 | Thurl placement | production | 0.001561074 | 13,824 | 31.90988 | 64.43125 |
| 19 | Scs (log2n); somatic cell score | health | 0.001708772 | 15,949 | 36.81501 | 69.75879 |
| 20 | Rump length | production | 0.001325549 | 18,264 | 42.15872 | 75.23024 |
| 21 | Teat thickness | exterior | 0.00123856 | 18,554 | 42.82812 | 76.23165 |
| 22 | Heel height | exterior | 0.001377944 | 19,671 | 45.40649 | 78.19852 |
| 23 | Front teat placement | exterior | 0.001108429 | 21,816 | 50.35779 | 82.25086 |
| 24 | Rear teat direction | exterior | 0.001030407 | 23,646 | 54.58197 | 85.22757 |
| 25 | Overall feet/legs score | exterior | 0.000872135 | 25,604 | 59.10161 | 88.05863 |
| 26 | Rear teat placement | exterior | 0.000798565 | 25,931 | 59.85642 | 88.69103 |
| 27 | Rear udder height | exterior | 0.000675109 | 28,601 | 66.01957 | 91.87504 |
| 28 | Udder balance | exterior | 0.000509724 | 30,888 | 71.29865 | 94.40238 |
| 29 | Foot angle | exterior | 0.000604226 | 31,409 | 72.50127 | 94.83624 |
| 30 | Udder depth | exterior | 0.000597598 | 32,094 | 74.08245 | 95.3957 |
| 31 | Overall frame | production | 0.000478474 | 32,354 | 74.68261 | 95.68689 |
| 32 | Rear-leg set | exterior | 0.000507667 | 32,936 | 76.02604 | 96.02775 |
| 33 | Udder cleft depth | exterior | 0.000310256 | 36,766 | 84.86681 | 98.45448 |
| 34 | Sum (points); overall conformation score | exterior | 0.000090297 | 41,071 | 94.80403 | 99.81904 |
| 35 | Body depth | production | 0.000060817 | 42,046 | 97.05461 | 99.94091 |
| 36 | Chest width | production | 0.000051688 | 42,173 | 97.34777 | 99.95302 |
SNP rs109709275 and 2617 SNPs with higher SNP effects contribute to 20.22% of the phenotypic variability for milk fat yield. Together with 3430 SNPs, it explained 25.12% of phenotypic variance for the calving interval. Together with 3515 SNPs, it explained 25.86% of the phenotypic variance for rump height. Along with 4028 other SNPs, rs109709275 explained 27.62% of phenotypic variance for rump angle. Together with 4104 SNPs, it explained 28.25% of the phenotypic variance for milk yield, while together with 5036 other SNPs, it explained 32.80% of the phenotypic variance for overall udder score. Lastly, together with 5047 other SNPs, rs109709275 explained 33.40% of the phenotypic variance for front teat length. The percent rank of the impact of rs109709275 on 36 traits showed that there are 6.05–11.65% SNPs with greater impact on the seven most significantly associated milk, reproduction, production and exterior traits. There is a total of 14 traits on which rs109709275 has a relatively strong effect (percent rank up to 25.05%), among which exterior traits and milk traits are the most common.
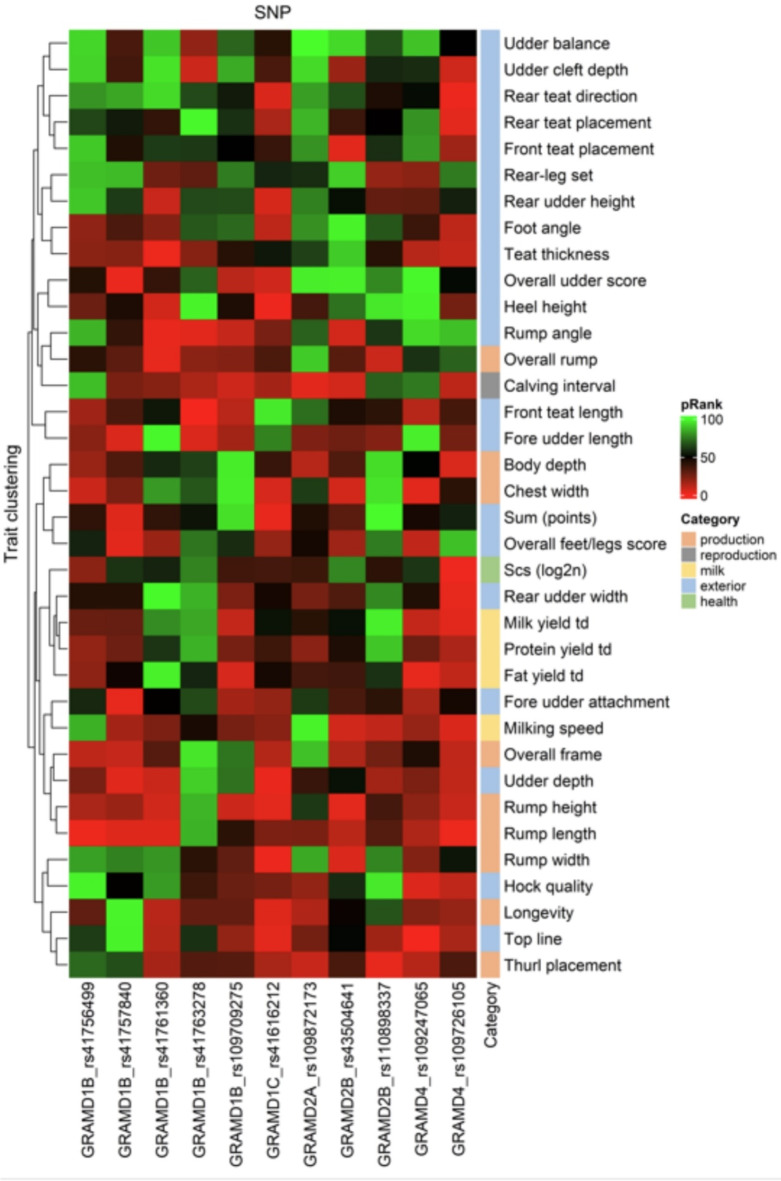
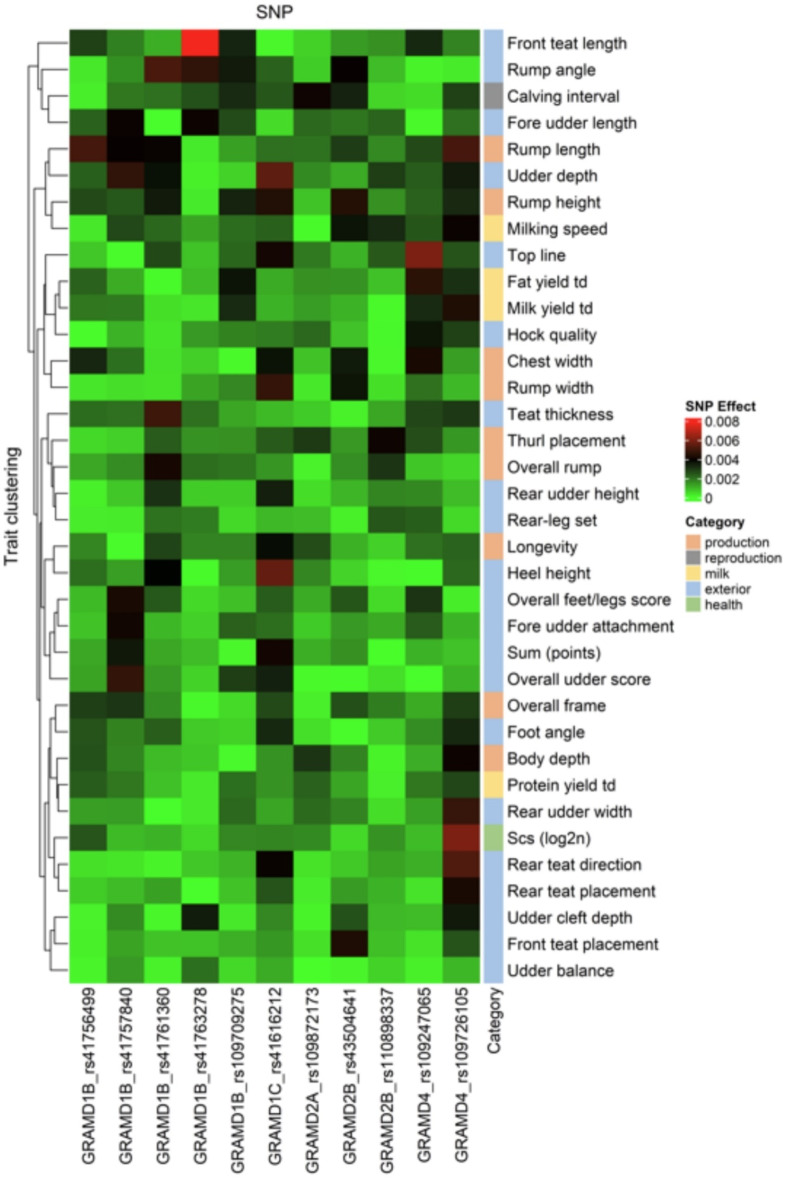
A heatmap of percent rank of 11 GRAMD gene family SNPs with 36 cattle traits is shown in the Fig. 3 and heatmap of SNP effects of GRAMD gene family on 36 cattle traits in the Fig. 4. Integrating the extensive data from SNP effects, their hierarchical clustering and ranking revealed involvement of GRAMD gene family in various cattle traits. Results indicated that three GRAMD genes (GRAMD1B, GRAMD1C and GRAMD4) affect a high number of teat-related and udder-related traits: teat thickness, front teat length, rear teat direction, overall udder score, fore udder attachment, fore udder length, and udder depth. Interestingly, the SNP rs41757840, located within a constrained element in the GRAMD1B gene, is associated with a high number of udder-related traits. Additionally, three genes (GRAMD1B, GRAMD1C, GRAMD2B) were also associated with various rump traits, including rump length, width, height, and angle. On the contrary, milk fat yield is mostly associated with GRAMD1B splice region variant rs109709275 and the SNP within the GRAMD4 gene.
Fig. 3.
Heatmap of percent rank (pRank) of GRAMD gene family within 36 cattle traits. The heatmap was generated in RStudio using ComplexHeatmap package.
Fig. 4.
Heatmap of SNP effects of GRAMD gene family on 36 cattle traits. The heatmap was generated in RStudio using ComplexHeatmap package.
GRAMD1B SNPs might impact front teat length and fore udder attachment, key traits for dairy production due to their influence on milking efficiency and udder health. GRAMD1C may affect both production and exterior traits, such as foot angle and heel height, suggesting a role that could benefit overall animal longevity and welfare. GRAMD2A and GRAMD2B show more specific roles, with GRAMD2A potentially influencing reproductive traits, such as calving interval, while GRAMD2B may affect teat placement. These traits are vital for reproductive efficiency and calf development, respectively, and present potential targets for breeding programs focusing on these specific cattle phenotypic traits. GRAMD4 emerged as a gene that might affect a diverse set of traits across different categories, including milk production traits such as milk yield and somatic cell score, as well as body conformation traits like rump height and width. The broad impact of GRAMD4 across various categories indicates its potential as a target for genomic selection strategies to enhance cattle performance.
Biotype of sequence variants
Most of the SNPs included in the analysis are intronic; one is located downstream of the GRAMD4 gene, and rs41757840 is located downstream of the GRAMD1B and within two lncRNA genes: the intron of ENSBTAG00000063982 and the exon of ENSBTAG00000061947. GRAMD1B sequence variant rs109709275 is classified as a splice region variant, intron variant and splice polypyrimidine tract variant (Table 1). The nucleotide sequence of the transcript ENSBTAT00000074465.2 (GRAMD1B-205) containing rs109709275 is shown in Supplementary Figure S2. The highest population MAF for this SNP is 0.06 and is therefore considered common. The multiple species alignment of 23 genomes revealed that the sequence variant is located in a conserved region; thymine is present in all 23 species. Information on sequence variants is available for the human genome; however, the SNP in human is not located at the orthologous locus as rs109709275 in cattle (Supplementary Figure S3). The GERP values of 11 SNPs range from − 6.03 to 1.22, with rs109709275 being the highest, indicating a conserved position, as the value is positive. Notably, two GRAMD1B SNPs are located within constrained elements. Supplementary Figure S4 shows that the SNP rs109709275 is located within a constrained genomic element with a score of 576.40 and spans 164 bp.
Cis- and trans-acting SNP analysis
We investigated whether SNPs associated with the GRAMD gene family could directly affect their expression as well as expression of other genes, as this could provide valuable insights into their functional roles in trait associations. Although none of the SNPs demonstrated a direct effect on the expression of GRAMD genes, one SNP, rs109726105, located downstream of GRAMD4, exhibited a cis-acting regulatory effect on two other linked genes: ATXN10 (ataxin 10) and ENSBTAG00000009351. Furthermore, several SNPs were found to exert trans-acting effects on various transcriptional regulators, including CCAR2, RPS3, CDK7, CHD3, WAC, FANK1, ZNF484, and TLX2. These findings highlight the complex regulatory landscape associated with the GRAMD gene family and suggest that even SNPs not directly linked to gene expression in the GRAMD gene family can exert broader cis- and trans-regulatory effects, potentially influencing multiple pathways relevant to trait development.
QTL overlap analysis
Gene-QTL overlap analysis represents an important approach for the functional characterization of the genes and we performed this analysis for the GRAMD1B gene. To analyze genomic overlaps between the GRAMD1B gene and QTLs, we downloaded data from the QTLdb database, with genomic location 15:34039549–34,221,229. The results showed 21 overlaps with the GRAMD1B gene, comprising 15 QTLs and 6 associations from SNPs in GWAS studies (Supplementary Table 13). QTLs are associated with different phenotypic traits from six trait classes: reproduction, production, health, meat and carcass, exterior, and milk traits.
GRAMD gene family variability between inbred mouse models for polygenic obesity and leanness
We analyzed whether the SNPs in the GRAMD gene family differ between inbred mouse models for polygenic obesity and leanness. Figure 5 shows the structure of the GRAMD family of genes in a mouse, along with regulatory elements, and the number of SNPs in the Fat and Lean mouse lines. The VEP tool annotates variants located within the gene region, extending to 5000 base pairs (bp) upstream and downstream. In comparison to the reference genome, Gramd1a and Gramd2a have SNPs only in the Fat line (167 and 38 SNPs, respectively), while in Gramd1c, 10 SNPs are present only in the Lean line. In contrast, in Gramd1b, Gramd2b and Gramd4 genes SNPs were present in the Fat, Lean or both lines in comparison to the reference genome. Interestingly, in Gramd1b gene, SNPs in Lean line are located only in the 5’ part and SNPs in the Fat line are located only in the 3’ part of the gene. As shown in Fig. 5, several SNPs are located within regulatory elements. Besides genetic variability, we also reanalyzed our previous expression experiments in mouse model for fatness and leanness. Results revealed that among Gramd genes, the Gramd1b gene had lower expression in muscle tissue in the Fat compared to the Lean line. In the remaining eight tissues, the difference in expression was not significant18.
Fig. 5.
Structure of the GRAMD family of genes in mouse, regulatory elements and number of SNPs in the Fat and Lean mouse lines.
Synthesis of GO analysis in five vertebrate species
We then performed a comparison of GO terms for six GRAMD genes in five vertebrate species: human, cattle, pig, mouse, and chicken (Supplementary Table S14). For example, the molecular functions of GRAMD1B in cattle include phosphatidylserine binding, cholesterol binding, phosphatidic acid binding, and cholesterol transfer activity. In addition, the annotated biological processes include lipid transport, sterol transport, intracellular sterol transport, cholesterol homeostasis, cellular response to cholesterol, and intermembrane lipid transfer. The GO annotations of the GRAMD family highlight a complex array of roles these genes play in vital biological functions. While some annotations are shared across species, indicating conserved functions, others are more sporadically distributed, possibly due to differences in research focus areas. Processes related to lipid and sterol transport are fundamental to cell membrane composition and signaling, with annotations for lipid transport present across all species. Intracellular sterol transport is noted for cattle, humans, and mice, indicating that these species may share mechanisms for managing cholesterol within cells, which could be important for the synthesis of steroids, including reproductive hormones. Only humans and mice have reported roles in autophagy, while apoptotic processes are present across all species. The lack of annotations for autophagy in cattle might reflect an area where more research is needed rather than an absence of this function, which is critical for cellular quality control and health.
Literature review of GRAMD genes and associated phenotypes in five vertebrate species
In the next step, we reviewed the literature on reported associations between six GRAMD genes and phenotypes in five vertebrate species: human, cattle, pig, mouse and chicken. The data were extracted from 37 PubMed publications, QTLdb and the MGI database (Table 3). Literature analysis demonstrated the diverse impact of these genes on health and development across species.
Table 3.
Phenotypes associated with six GRAMD genes in five vertebrate species. Results from the present study include the first three associations for 11 SNPs according to the percent rank.
| Phenotype | Species | GRAMD1A | GRAMD1B | GRAMD1C | GRAMD2A | GRAMD2B | GRAMD4 | Reference |
|---|---|---|---|---|---|---|---|---|
| days from first to last insemination | cattle (Nordic red cattle) | ✓ | [6] | |||||
| feed efficiency | cattle | ✓ | [5] | |||||
| female fertility index | cattle (Nordic red cattle) | ✓ | [6] | |||||
| length in days of the interval from calving to first insemination | cattle (Nordic red cattle) | ✓ | [6] | |||||
| number of inseminations per conception | cattle (Nordic red cattle) | ✓ | [6] | |||||
| milk conjugated linoleic acid content | cattle (Brown Swiss dairy cattle) | ✓ | [27] | |||||
| rear thigh circumference | cattle (Simmental beef cattle) | ✓ | [28] | |||||
| female reproductive performance | chicken | ✓ | [29] | |||||
| the growth and development of follicles | chicken | ✓ | [29] | |||||
| yolk percentage traits | chicken | ✓ | [29] | |||||
| autophagy | human (cell line) | ✓ | [49] | |||||
| mitochondrial bioenergetics | human (cell line) | ✓ | [48] | |||||
| adrenal and testicular steroidogenesis | human | ✓ | [34] | |||||
| autophagy | human | ✓ | [42] | |||||
| autophagy | human | ✓ | [43] | |||||
| breast cancer | human | ✓ | [53] | |||||
| cell migration in breast cancer cells | human | ✓ | [36] | |||||
| hepatocellular carcinoma | human | ✓ | [50] | |||||
| regulation cellular cholesterol distribution | human (cell lines) | ✓ | ✓ | ✓ | [39] | |||
| nonvesicular plasma membrane to endoplasmic reticulum cholesterol transport | human, mouse (cell lines) | ✓ | ✓ | ✓ | [40] | |||
| hepatocellular carcinoma | human | ✓ | [44] | |||||
| apoptosis | human (cell lines) | ✓ | [2] | |||||
| asymmetric cholesterol distribution in plasma membrane | human (cell lines) | ✓ | [41] | |||||
| HPV16 + head and neck squamous cell carcinomas | human | ✓ | [57] | |||||
| inhibiting tumor metastasis in hepatocellular carcinoma | human | ✓ | [58] | |||||
| kidney renal clear cell carcinoma (KIRC) | human | ✓ | [45] | |||||
| kidney renal clear cell carcinoma (KIRC) | human | ✓ | [51] | |||||
| macrocephaly, Phelan–McDermid syndrome (PMS) | human | ✓ | [56] | |||||
| muscle fiber size and strength | human | ✓ | [35] | |||||
| non-cirrhotic hepatocellular carcinoma (NCHCC) | human | ✓ | [52] | |||||
| pediatric sepsis | human | ✓ | [54] | |||||
| prostate cancer | human | ✓ | [46] | |||||
| renal transplantation rejection | human | ✓ | [47] | |||||
| cellular Ca(2+) homeostasis | human (cell lines) | ✓ | [3] | |||||
| serum leucocyte concentration, coronary artery disease (CAD) | human | ✓ | [4] | |||||
| susceptibility to chronic lymphocytic leukemia (CLL) | human | ✓ | [37] | |||||
| total serum IgE levels | human | ✓ | [38] | |||||
| transition between different states in Huntington’s Disease | human | ✓ | [55] | |||||
| TLR9-mediated innate immune responses | mouse | ✓ | [33] | |||||
| nutrient sensing; negative regulator of mTORC1 signaling during nutrient starvation | mouse (cell line) | ✓ | [32] | |||||
| number of animals born alive and dead | pig | ✓ | [30] | |||||
| porcine epidemic diarrhea (PED) | pig | ✓ | [31] | |||||
| abnormal adrenal cortex morphology | mammalian phenotype | ✓ | MGI | |||||
| abnormal adrenal gland morphology | mammalian phenotype | ✓ | MGI | |||||
| abnormal adrenal gland zona fasciculata morphology | mammalian phenotype | ✓ | MGI | |||||
| abnormal cholesterol homeostasis | mammalian phenotype | ✓ | MGI | |||||
| abnormal head shape | mammalian phenotype | ✓ | MGI | |||||
| abnormal lens morphology | mammalian phenotype | ✓ | MGI | |||||
| abnormal liver morphology | mammalian phenotype | ✓ | MGI | |||||
| abnormal pancreas morphology | mammalian phenotype | ✓ | MGI | |||||
| abnormal startle reflex | mammalian phenotype | ✓ | MGI | |||||
| abnormal tail morphology | mammalian phenotype | ✓ | MGI | |||||
| decreased circulating corticosterone level | mammalian phenotype | ✓ | MGI | |||||
| decreased fasting circulating glucose level | mammalian phenotype | ✓ | MGI | |||||
| decreased mean corpuscular hemoglobin concentration | mammalian phenotype | ✓ | MGI | |||||
| decreased startle reflex | mammalian phenotype | ✓ | MGI | |||||
| enlarged lymph nodes | mammalian phenotype | ✓ | MGI | |||||
| enlarged urinary bladder | mammalian phenotype | ✓ | MGI | |||||
| exencephaly | mammalian phenotype | ✓ | MGI | |||||
| hyperactivity | mammalian phenotype | ✓ | MGI | |||||
| impaired glucose tolerance | mammalian phenotype | ✓ | MGI | |||||
| increased circulating aspartate transaminase level | mammalian phenotype | ✓ | MGI | |||||
| increased mean corpuscular hemoglobin | mammalian phenotype | ✓ | ✓ | MGI | ||||
| increased mean corpuscular hemoglobin concentration | mammalian phenotype | ✓ | MGI | |||||
| increased mean corpuscular volume | mammalian phenotype | ✓ | MGI | |||||
| increased urine microalbumin level | mammalian phenotype | ✓ | MGI | |||||
| limb grasping | mammalian phenotype | ✓ | MGI | |||||
| preweaning lethality, incomplete penetrance | mammalian phenotype | ✓ | MGI | |||||
| small uterus | mammalian phenotype | ✓ | MGI | |||||
| unresponsive to tactile stimuli | mammalian phenotype | ✓ | MGI | |||||
| vertebral fusion | mammalian phenotype | ✓ | MGI | |||||
| exterior trait: teat thickness | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| milk trait: milk fat yield | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| exterior trait: front teat length | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| production trait: rump length | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| production trait: chest width | cattle (Brown Swiss cattle | ✓ | ✓ | Present study | ||||
| production trait: overall frame | cattle (Brown Swiss cattle | ✓ | Present study | |||||
| exterior trait: rump angle | cattle (Brown Swiss cattle) | ✓ (2 SNPs) | Present study | |||||
| exterior trait: overall udder score | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: fore udder attachment | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait fore udder length | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: overall feet/legs score | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: heel height | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: udder depth | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| production trait: rump width | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| reproduction trait: calving interval | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| production trait: rump height | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| production trait: body depth | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| production trait: thurl placement | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| production trait: overall rump | cattle (Brown Swiss cattle) | ✓ | ✓ | Present study | ||||
| milk trait: milking speed | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: front teat placement | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: top line | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| exterior trait: rear teat direction | cattle (Brown Swiss cattle) | ✓ | Present study | |||||
| health trait: scs (log2n) | cattle (Brown Swiss cattle) | ✓ | Present study |
Legend: scs: somatic cell score.
The dynamics and focus of GRAMD gene studies differ between species. In cattle, phenotypes related to reproduction, feed efficiency, and milk content are associated with the GRAMD1B5,6 and GRAMD1C gene27,28. In chicken, phenotypes related to reproductive performance and egg traits are associated with the GRAMD1C gene29. Two studies were conducted in pig: GRAMD1B has been studied with the number of animals born alive and dead30 and GRAMD4 with porcine epidemic diarrhea (PED)31. In mouse, GRAMD1C was associated with nutrient sensing32 and GRAMD4 in immune response33. In human GRAMD genes have been studied in association with diverse phenotypes. For example, GRAMD1B has been shown to be involved in adrenal and testicular steroidogenesis34. It has been associated with muscle fiber size and strength, possibly contributing to a greater predisposition to strength and power sports35. It has also been shown to regulate cell migration in breast cancer cells through JAK/STAT and Akt signaling. Gramd1b knockdown led to morphological changes in the cells, suggesting its role in cell migration36. It has also been reported to be associated with susceptibility to chronic lymphocytic leukemia (CLL)37 and total serum IgE levels38. All three GRAM1 genes (GRAM1A, GRAM1B and GRAM1C) have been shown to be involved in the regulation of cholesterol distribution and transport39–41.
Comparative analysis of GRAMD gene functions across species revealed that GRAMD1A has been studied in human and is associated with autophagy42,43 hepatocellular carcinoma44, kidney renal clear cell carcinoma (KIRC)45, prostate cancer46 and renal transplantation rejection47. GRAMD1B shows associations with reproductive phenotypes in cattle, such as influencing the days from first to last insemination6. These associations underscore the gene’s role in cattle fertility and may have implications for breeding programs to enhance reproductive efficiency. In humans, GRAMD1B is associated with breast cancer cell migration36, indicating its role in cancer progression.
In cattle, GRAMD1C is implicated in milk conjugated linoleic acid content27, pointing to its involvement in milk fat composition, a trait of economic importance. In humans, it is associated with mitochondrial bioenergetics48 and autophagy in cell lines49, suggesting a broader role in cellular health and disease. It has also been shown to be involved in hepatocellular carcinoma50, KIRC51, non-cirrhotic hepatocellular carcinoma (NCHNN)52, breast cancer53, pediatric sepsis54 and Huntington’s disease55. GRAMD2A and GRAM2B have only been researched in humans and are shown to be implicated in cellular Ca2+ homeostasis3 and serum leukocyte concentration4. GRAMD4 has been shown to be associated with various human health conditions, including macrocephaly56 and HPV16 + head and neck squamous cell carcinomas57. It has been reported to be involved in inhibiting tumor metastasis in hepatocellular carcinoma58. Its relationship with apoptotic processes in cell lines2 suggests its fundamental role in cell death and survival, which can be crucial for understanding disease mechanisms.
The second section of the Table 3 lists mammalian phenotypes primarily associated with abnormal development and physiological functions, such as abnormal morphology, decreased reflexes, and hyperactivity (MGI database). Table 3 also includes gene-phenotype associations obtained in the present study in cattle. Considering the three phenotypes with the highest association with 11 SNPs according to the percentile rankings, it was revealed that GRAMD genes have the highest association with 24 traits across all five trait groups, including eight traits associated with two genes. Additionally, one trait is associated with two SNPs within one gene; namely, rump angle is linked to two SNPs within the GRAMD1B gene.
Discussion
In the present study, we illustrated using the GRAMD gene group as an example, how the combination of GWAS and the PheWAS offers a promising toolbox for the systematic functional annotation of vertebrate genomes. The analysis was performed using 55,172 SNPs and 36 phenotypic traits in 55,013 bulls. Additionally, we summarized published literature and GO databases for genotype-phenotype associations in five vertebrate species and presented genotype differences in the GRAMD gene family of inbred mouse models for polygenic obesity and leanness.
The data reflect the multifunctional nature of the GRAMD gene family, with members influencing a wide range of biological processes that translate into observable traits across species. The specificity of these associations underscores the potential for using genetic information to guide selective breeding in livestock and provides valuable targets for biomedical research. Additionally, the cross-species prevalence of certain traits suggests evolutionarily conserved functions of these genes that might be exploited to improve animal health and production.
The synthesis of the data from various publications and databases suggests that GRAMD genes play significant roles in cattle physiological processes, manifesting in measurable traits of agricultural importance6,27. The study revealed that SNPs within specific genes correlate with effects on cattle traits, which are supported by their associated biological processes and molecular functions. Comparisons of phenotypic variability across different vertebrate species, including cattle, human, pig, mouse, and chicken, have highlighted potential conserved genetic mechanisms. The integrated analysis of SNP effects, gene ontology, and phenotypic data has unveiled several genes that may significantly influence cattle traits.
The ranking analysis revealed the relative influence of GRAMD gene group SNPs among all variants on the microarray. The study provided an additional dimension to understanding the genetic impact on these traits. For example, high-ranking GRAMD4 SNPs show substantial effects on body depth and potential for genetic improvement targeting stature and robustness. On the other hand, other lower-ranking SNPs highlight the complexity of genetic regulation and the specific contributions of different SNPs to phenotypic traits.
In future studies, several experimental approaches could be employed to functionally test whether the identified SNPs exert the above-proposed cis- or trans-regulatory effects. RNA sequencing (RNA-seq) of tissues relevant to the trait of interest could quantify gene expression in a population of individuals carrying different GRAMD gene family SNP alleles. The resulting expression data could then be mapped to identify significant SNP-expression correlations, allowing for the distinction between cis- (local) and trans- (distant) regulatory effects. Alternatively, a smaller-scale experiment could be conducted to directly test the candidate genes using quantitative PCR (qPCR).
Other in vitro or in vivo experimental methods could also be employed to establish causality rather than mere correlation. For instance, reporter assays could validate cis-regulatory effects of alternative GRAMD SNP alleles in vitro, while CRISPR/Cas9 genome editing could be used to modify SNPs and observe functional outcomes in vivo. Furthermore, perturbing the expression of candidate trans-regulated genes (e.g., CCAR2, RPS3) through knockdown or overexpression techniques could reveal whether these perturbations influence the same pathways affected by the GRAMD-associated SNPs.
Results revealed groups of traits that share a genetic basis and could be improved simultaneously through selective breeding targeting a set of SNPs that have the greatest effect across these traits. For example, clustering of rump-related traits such as height, length, and width indicates a shared genetic background, which could be targeted to improve body conformation. Comparative analysis of the obtained genotype and phenotype data demonstrated that the most potent SNPs for specific traits are not always the highest-ranked across all traits. This finding is crucial for precision breeding as it implies that selective improvement of one trait may affect another.
The present study used GWAS and PheWAS approaches (GWAS-to-PheWAS) to determine the association between sequence variants and phenotypic traits in cattle. The GWAS approach follows a “many variants-one phenotype” design, whereas PheWAS uses a “one variant–many phenotypes” approach, allowing for the analysis of pleiotropy59. Two main advantages of our study are the high number of analyzed animals and the large number of available phenotypic traits that allowed a functional annotation of the GRAMD genes in cattle. The combination of GWAS and PheWAS approaches represents one of the most important research directions in genomics. This combined approach provides valuable insights into the genetic and biological mechanisms underlying various complex traits, marking a significant step towards integrative biology research in animal science10,60.
The gene-QTL overlap analysis presents a preliminary step towards functional annotation. Although most of the overlapped QTLs span large genomic regions, the current data do not yet provide convincing evidence to associate these regions with the GRAMD1B gene. To verify the findings, future studies should be repeated using upcoming new versions of QTLdb, including all members of the GRAMD gene family. Despite the inconclusive nature of these results, this approach offers potential for further functional characterization of these genes.
Comparison between mouse models and livestock is challenging, as some traits, like milk and teat traits, are not measured in mice. However, the mouse model could still be useful for comparisons using anatomical traits or body regions similar to those recorded in livestock, such as exterior traits and expression in the mammary gland and fat tissue. For example, a search of MGI data revealed the following findings: (1) Gramd1b has a strong expression and function in body region determination, also in hind parts (trunk, limbs, tail). (2) Expression of Gramd1a and Gramd1b in mammary gland is found to be medium to strong. (3) Some mutations (i.e., Gramd1bem1(IMPC)J/Gramd1bem1(IMPC)J) show effects on abnormal body shapes, including abnormal tail morphology. (4) Gramd2a is expressed in exocrine organs including mammary gland. (5) Intragenic deletion, Gramd2aem1(IMPC)Tcp (J: 237616) is associated with abnormal body morphology in skeleton17. Therefore, comparison with mice still offers insights for comparative studies focusing on anatomical traits, external features, and expressions in tissues analogous to those found in livestock.
The Gene Ontology offers a species-independent framework for detailing the molecular functions, biological roles, and cellular locations of numerous gene products. Its design facilitates cross-species comparisons, allowing functional annotations from model organisms to be applied to species with less characterized genomes. Consequently, the GO serves as a valuable resource for providing gene annotation in species where complete genomic data are not yet available61. Variations in these annotations between species often reflect the current state of research rather than biological differences. For example, certain functions of the GRAMD family may be well-studied in humans and mice, commonly used model organisms, while equivalent research in livestock species may lag. These annotations offer a guide of potential gene functions that could be associated with important traits. This highlights the need for future research to fill in the gaps and elucidate the roles of these genes in physiology and productivity.
Limitations of the study
Despite several advances, our study also has some limitations. For example, the association study in cattle depended on the available SNPs on the microarray, as the SNPs are not evenly distributed across the genes. Five SNPs are present within the GRAMD1B gene, two SNPs in the GRAMD2B and GRAMD4 genes each, and one SNP is present within the GRAMD1C and GRAMD2A genes. The association was not performed for the GRAMD1A gene as no SNPs map to this gene. Comparison of phenotypes across species is also challenging due to differences in measured traits, especially milk and teat characteristics. For example, in cattle, we performed an association study between five genes and 36 traits. However, in mouse SNP distribution was compared for six genes and two traits between inbred mouse models for polygenic obesity and leanness. The literature review also depended on available studies, mostly performed in human, but only a few in cattle, pig, mouse and chicken.
Conclusions
Our study reinforces the pivotal role of GRAMD genes in animal breeding and genetic selection strategies. The present study revealed an association with several additional traits, suggesting that the identified SNPs may serve as genetic variants for selection programs in Slovenian and possibly global populations of cattle. Biomarkers based on polymorphisms contribute to the study of phenotypic traits of interest in animal husbandry and medicine. Our study presents an example of functional annotation of GRAMD genes using GWAS and PheWAS approaches, which could now be applied to other gene groups.
GRAMD gene family evaluation revealed their potential for improving cattle productivity, health, and robustness. The integrated analysis of SNP effects, rankings, and clustering patterns provides a baseline for the targeted improvement of cattle traits. The study highlighted the importance of understanding relationships between SNPs, genes, traits and their categories, which should lead to advancements in genetic and genomics research and more efficient breeding strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Miran Štepec and Jurij Krsnik for technical assistance, and express gratitude to the Interbull InterGenomics consortium for permitting the use of their results. They extend their thanks to all partners involved in InterGenomics for supplying genomic data that contributed to estimating the SNP effects utilized in this research.
Abbreviations
- BSW
Brown Swiss cattle population
- GERP
genomic evolutionary rate profiling
- GO
gene ontology
- GRAMD1B
GRAM domain containing 1B
- GRAM
glucosyltransferases, Rab-like GTPase activators and myotubularin
- GWAS
genome-wide association studies
- KIRC
kidney renal clear cell carcinoma
- lncRNA
long non-coding RNA
- SNP
Single nucleotide polymorphism
- PheWAS
phenome-wide association study
- VTO
Vertebrate Trait Ontology
Author contributions
Conceptualization: K.P., T.K.; Methodology: T.K., M.Š., B.L., K.P., S.H.; Software: K.P., M.Š., B.L.; Formal Analysis: TK, M.Š., B.L., K.P.; Investigation TK, M.Š., B.L., K.P., S.H.; Resources: KP, Data Curation: TK, M.Š., S.H.; Writing – Original Draft Preparation: T.K., B.L., M.Š., S.H.; Writing – Review & Editing: T.K., S.H., Supervision: K.P., S.H., T.K.
Funding
This work was supported by the Slovenian Research and Innovation Agency (ARIS), research programs P4-0092 and P4-0220.
Data availability
Data is provided within the manuscript and supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tanja Kunej, Martin Šimon.
Contributor Information
Tanja Kunej, Email: tanja.kunej@bf.uni-lj.si.
Klemen Potočnik, Email: klemen.potocnik@bf.uni-lj.si.
References
- 1.Naito, T. & Saheki, Y. GRAMD1-mediated accessible cholesterol sensing and transport., Biochim Biophys Acta Mol Cell Biol Lipids, vol. 1866, no. 8, p. 158957, Aug. doi: (2021). 10.1016/j.bbalip.2021.158957 [DOI] [PubMed]
- 2.John, K., Alla, V., Meier, C. & Pützer, B. M. GRAMD4 mimics p53 and mediates the apoptotic function of p73 at mitochondria. Cell. Death Differ.18 (5), 874–886. 10.1038/cdd.2010.153 (May 2011). [DOI] [PMC free article] [PubMed]
- 3.Besprozvannaya, M. et al. GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife710.7554/eLife.31019 (Feb. 2018). [DOI] [PMC free article] [PubMed]
- 4.Antikainen, A. A. V. et al. Jan., Genome-wide association study on coronary artery disease in type 1 diabetes suggests beta-defensin 127 as a risk locus, Cardiovasc Res, vol. 117, no. 2, pp. 600–612, doi: (2021). 10.1093/cvr/cvaa045 [DOI] [PMC free article] [PubMed]
- 5.Serão, N. V. et al. Single nucleotide polymorphisms and haplotypes associated with feed efficiency in beef cattle., BMC Genet, vol. 14, p. 94, Sep. doi: (2013). 10.1186/1471-2156-14-94 [DOI] [PMC free article] [PubMed]
- 6.Höglund, J. K., Buitenhuis, B., Guldbrandtsen, B., Lund, M. S. & Sahana, G. Genome-wide association study for female fertility in Nordic Red cattle. BMC Genet.16, 110. 10.1186/s12863-015-0269-x (Sep. 2015). [DOI] [PMC free article] [PubMed]
- 7.Malovrh, Š., Kunej, T., Kovač, M. & Dovč, P. The microRNA gene bta-mir-2313 in cattle: an atlas of regulatory elements and an association analysis with growth and carcass traits in the Slovenian Simental cattle breed, Arch Anim Breed, vol. 61, no. 3, pp. 271–278, Jul. doi: (2018). 10.5194/aab-61-271-2018
- 8.Hebbring, S. Genomic and Phenomic Research in the 21st Century. Trends Genet.35 (1), 29–41. 10.1016/j.tig.2018.09.007 (Jan. 2019). [DOI] [PMC free article] [PubMed]
- 9.Robinson, J. R., Denny, J. C., Roden, D. M. & Van Driest, S. L. Genome-wide and Phenome-wide Approaches to Understand Variable Drug Actions in Electronic Health Records., Clin Transl Sci, vol. 11, no. 2, pp. 112–122, Mar. doi: (2018). 10.1111/cts.12522 [DOI] [PMC free article] [PubMed]
- 10.Zeng, H. et al. PigBiobank: a valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res.52, D980–D989. 10.1093/nar/gkad1080 (Jan. 2024). no. D1. [DOI] [PMC free article] [PubMed]
- 11.Martin, F. J. et al. Ensembl., Nucleic Acids Res, vol. 51, no. D1, pp. D933–D941, Jan. 2023, doi: (2023). 10.1093/nar/gkac958 [DOI] [PMC free article] [PubMed]
- 12.Nicolazzi, E. L. et al. SNPchiMp v.3: integrating and standardizing single nucleotide polymorphism data for livestock species.,BMC Genom., 16, 1, 283, doi: 10.1186/s12864-015-1497-1.Apr. (2015). [DOI] [PMC free article] [PubMed]
- 13.Hu, Z. L., Park, C. A. & Reecy, J. M. Bringing the animal QTLdb and CorrDB into the future: meeting new challenges and providing updated services. Nucleic Acids Res.50, D956–D961. 10.1093/nar/gkab1116 (Jan. 2022). no. D1. [DOI] [PMC free article] [PubMed]
- 14.Park, C. A. et al. The Vertebrate Trait Ontology: a controlled vocabulary for the annotation of trait data across species. J. Biomed. Semant.4 (1), 13. 10.1186/2041-1480-4-13 (Aug. 2013). [DOI] [PMC free article] [PubMed]
- 15.Posit team. RStudio: Integrated Development Environment for R, (2024).
- 16.Gu, Z. Complex heatmap visualization, iMeta, vol. 1, no. 3, Sep. doi: (2022). 10.1002/imt2.43 [DOI] [PMC free article] [PubMed]
- 17.Blake, J. A. et al. Mouse Genome Database (MGD): knowledgebase for mouse–human comparative biology. Nucleic Acids Res.49, D981–D987. 10.1093/nar/gkaa1083 (Jan. 2021). no. D1. [DOI] [PMC free article] [PubMed]
- 18.Šimon, M. et al. Integration of genomic and transcriptomic data of inbred mouse models for polygenic obesity and leanness revealed ‘obese’ and ‘lean’ candidate alleles in polyadenylation signals. Gene Rep.35, 101903. 10.1016/j.genrep.2024.101903 (Jun. 2024).
- 19.Šimon, M. et al. Whole genome sequencing of mouse lines divergently selected for fatness (FLI) and leanness (FHI) revealed several genetic variants as candidates for novel obesity genes. Genes Genomics Mar.10.1007/s13258-024-01507-9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorjani, H., Jakobsen, J., Hjerpe, E., Palucci, V. & Dürr, J. Status of genomic evaluation in the Brown Swiss populations. INTERBULL Bull.46, 46–54 (2012). [Google Scholar]
- 21.Jorjani, H. et al. Genomic evaluation of BSW populations. InterGenomics: results and deliverables. Interbull Tech. Workshop Febr. 27–28, (2011).
- 22.Savoia, S. Interbull genomic evaluation of small Holstein populations: InterGenomics-Holstein (IG-HOL). [Online]. Available: https://idea.interbull.org/
- 23.Vanraden, P. M., O’connell, J. R., Wiggans, G. R. & Weigel, K. A. Combining Different Marker Densities in Genomic Evaluation.
- 24.Guo, J., Jorjani, H. & Carlborg, Ö. A genome-wide association study using international breeding-evaluation data identifies major loci affecting production traits and stature in the Brown Swiss cattle breed. BMC Genet.1310.1186/1471-2156-13-82 (Oct. 2012). [DOI] [PMC free article] [PubMed]
- 25.Interbull CoP. - Methods of international evaluation, https://interbull.org/ib/cop_chap5
- 26.VanRaden, P. M. Efficient methods to compute genomic predictions., J Dairy Sci, vol. 91, no. 11, pp. 4414–23, Nov. doi: (2008). 10.3168/jds.2007-0980 [DOI] [PubMed]
- 27.Strillacci, M. G. et al. Quantitative trait loci mapping for conjugated linoleic acid, vaccenic acid and ∆ 9 -desaturase in Italian brown Swiss dairy cattle using selective DNA pooling. Anim. Genet.45 (4), 485–499. 10.1111/age.12174 (Aug. 2014). [DOI] [PubMed]
- 28.Cai, W. et al. The eQTL colocalization and transcriptome-wide association study identify potentially causal genes responsible for economic traits in Simmental beef cattle. J. Anim. Sci. Biotechnol.14 (1), 78. 10.1186/s40104-023-00876-7 (May 2023). [DOI] [PMC free article] [PubMed]
- 29.Zhang, J. et al. Screening and validation of candidate genes involved in the regulation of egg yolk deposition in chicken. Poult. Sci.100 (6), 101077. 10.1016/j.psj.2021.101077 (Jun. 2021). [DOI] [PMC free article] [PubMed]
- 30.Chen, Z. et al. Nov., Genome-wide association studies for the number of animals born alive and dead in duroc pigs., Theriogenology, vol. 139, pp. 36–42, doi: (2019). 10.1016/j.theriogenology.2019.07.013 [DOI] [PubMed]
- 31.Xu, X. et al. GRAMD4 regulates PEDV-induced cell apoptosis inhibiting virus replication via the endoplasmic reticulum stress pathway. Vet. Microbiol.279, 109666. 10.1016/j.vetmic.2023.109666 (Apr. 2023). [DOI] [PubMed]
- 32.Zhang, J. et al. Aster-C coordinates with COP I vesicles to regulate lysosomal trafficking and activation of mTORC1. EMBO Rep.21 (9). 10.15252/embr.201949898 (Sep. 2020). [DOI] [PMC free article] [PubMed]
- 33.Kimura, T. et al. Endoplasmic Protein Nogo-B (RTN4-B) Interacts with GRAMD4 and Regulates TLR9-Mediated Innate Immune Responses, The Journal of Immunology, vol. 194, no. 11, pp. 5426–5436, Jun. doi: (2015). 10.4049/jimmunol.1402006 [DOI] [PubMed]
- 34.Valle, I. D. et al. A genomic atlas of human adrenal and gonad development. Wellcome Open. Res.2, 25. 10.12688/wellcomeopenres.11253.2 (Apr. 2017). [DOI] [PMC free article] [PubMed]
- 35.Guilherme, J. P. L. F. et al. Genomic predictors of testosterone levels are associated with muscle fiber size and strength. Eur. J. Appl. Physiol.122 (2), 415–423. 10.1007/s00421-021-04851-w (Feb. 2022). [DOI] [PMC free article] [PubMed]
- 36.Khanna, P., Lee, J. S., Sereemaspun, A., Lee, H. & Baeg, G. H. GRAMD1B regulates cell migration in breast cancer cells through JAK/STAT and Akt signaling. Sci. Rep.8 (1), 9511. 10.1038/s41598-018-27864-6 (Jun. 2018). [DOI] [PMC free article] [PubMed]
- 37.Slager, S. L. et al. Apr., Genetic susceptibility variants for chronic lymphocytic leukemia., Cancer Epidemiol Biomarkers Prev, vol. 19, no. 4, pp. 1098–102, doi: (2010). 10.1158/1055-9965.EPI-09-1217 [DOI] [PMC free article] [PubMed]
- 38.Yatagai, Y. et al. Genome-wide association study for levels of total serum IgE identifies HLA-C in a Japanese population. PLoS One. 8 (12), e80941. 10.1371/journal.pone.0080941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naito, T. et al. Regulation of cellular cholesterol distribution via non-vesicular lipid transport at ER-Golgi contact sites, Nat Commun, vol. 14, no. 1, p. 5867, Sep. doi: (2023). 10.1038/s41467-023-41213-w [DOI] [PMC free article] [PubMed]
- 40.Sandhu, J. et al. Aster Proteins Facilitate Nonvesicular Plasma Membrane to ER Cholesterol Transport in Mammalian Cells. Cell175 (2), 514–529. 10.1016/j.cell.2018.08.033 (Oct. 2018). .e20. [DOI] [PMC free article] [PubMed]
- 41.Ogasawara, F. & Ueda, K. ABCA1 and cholesterol transfer protein Aster-A promote an asymmetric cholesterol distribution in the plasma membrane. J. Biol. Chem.298, 102702. 10.1016/j.jbc.2022.102702 (Dec. 2022). [DOI] [PMC free article] [PubMed]
- 42.Wu, Y. W. & Waldmann, H. Toward the role of cholesterol and cholesterol transfer protein in autophagosome biogenesis, Autophagy, vol. 15, no. 12, pp. 2167–2168, Dec. doi: (2019). 10.1080/15548627.2019.1666595 [DOI] [PMC free article] [PubMed]
- 43.Laraia, L. et al. Jul., The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis, Nat Chem Biol, vol. 15, no. 7, pp. 710–720, doi: (2019). 10.1038/s41589-019-0307-5 [DOI] [PubMed]
- 44.Luo, Z. et al. Apr., CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway, J Cell Mol Med, vol. 25, no. 8, pp. 3793–3802, doi: (2021). 10.1111/jcmm.16254 [DOI] [PMC free article] [PubMed]
- 45.Liu, Y. et al. GRAMD1A Is a Biomarker of Kidney Renal Clear Cell Carcinoma and Is Associated with Immune Infiltration in the Tumour Microenvironment, Dis Markers, vol. pp. 1–25, Jul. 2022, doi: (2022). 10.1155/2022/5939021 [DOI] [PMC free article] [PubMed]
- 46.Quan, Y., Ping, H., Wang, M. & Zhang, X. RNA-Sequencing Analysis Indicates That N-Cadherin Promotes Prostate Cancer Progression by the Epigenetic Modification of Key Genes, DNA Cell Biol, vol. 42, no. 9, pp. 563–577, Sep. doi: (2023). 10.1089/dna.2023.0089 [DOI] [PubMed]
- 47.Saberi, F., Dehghan, Z., Noori, E. & Zali, H. Identification of Renal Transplantation Rejection Biomarkers in Blood Using the Systems Biology Approach., Iran Biomed J, vol. 27, no. 6, pp. 375–87, Aug. doi: (2023). 10.52547/ibj.3871 [DOI] [PMC free article] [PubMed]
- 48.Ng, M. Y. W. et al. The cholesterol transport protein GRAMD1C regulates autophagy initiation and mitochondrial bioenergetics. Nat. Commun.13 (1), 6283. 10.1038/s41467-022-33933-2 (Oct. 2022). [DOI] [PMC free article] [PubMed]
- 49.Charsou, C., Ng, M. Y. W. & Simonsen, A. Regulation of autophagosome biogenesis and mitochondrial bioenergetics by the cholesterol transport protein GRAMD1C., Autophagy, vol. 19, no. 7, pp. 2159–2161, Jul. doi: (2023). 10.1080/15548627.2022.2155020 [DOI] [PMC free article] [PubMed]
- 50.FAN, X. et al. Potential Common Molecular Mechanisms Between Periodontitis and Hepatocellular Carcinoma: A Bioinformatic Analysis and Validation, Cancer Genomics - Proteomics, vol. 20, no. 6, pp. 602–616, Oct. doi: (2023). 10.21873/cgp.20409 [DOI] [PMC free article] [PubMed]
- 51.Hao, H. et al. Reduced GRAMD1C expression correlates to poor prognosis and immune infiltrates in kidney renal clear cell carcinoma. PeerJ7, e8205. 10.7717/peerj.8205 (Dec. 2019). [DOI] [PMC free article] [PubMed]
- 52.Gong, J. et al. Sep., Development and Validation of a Novel Prognosis Model Based on a Panel of Three Immunogenic Cell Death-Related Genes for Non-Cirrhotic Hepatocellular Carcinoma, J Hepatocell Carcinoma, vol. Volume 10, pp. 1609–1628, doi: (2023). 10.2147/JHC.S424545 [DOI] [PMC free article] [PubMed]
- 53.Li, X. et al. A cholesterogenic gene signature for predicting the prognosis of young breast cancer patients. PeerJ10, e13922. 10.7717/peerj.13922 (Aug. 2022). [DOI] [PMC free article] [PubMed]
- 54.Yao, Y. et al. Identification of a Four-Gene Signature for Diagnosing Paediatric Sepsis, Biomed Res Int, vol. pp. 1–14, Feb. 2022, doi: (2022). 10.1155/2022/5217885 [DOI] [PMC free article] [PubMed] [Retracted]
- 55.Christodoulou, C. C. et al. Oct., Investigating the Transition of Pre-Symptomatic to Symptomatic Huntington’s Disease Status Based on Omics Data, Int J Mol Sci, vol. 21, no. 19, p. 7414, doi: (2020). 10.3390/ijms21197414 [DOI] [PMC free article] [PubMed]
- 56.Sarasua, S. M. et al. Head size in Phelan–McDermid syndrome: a literature review and pooled analysis of 198 patients identifies candidate genes on 22q13. Genes (Basel). 14 (3), 540. 10.3390/genes14030540 (Feb. 2023). [DOI] [PMC free article] [PubMed]
- 57.Berglund, A. et al. Characterization of epigenomic alterations in HPV16 + head and Neck squamous cell carcinomas. Cancer Epidemiol. Biomarkers Prev.31 (4), 858–869. 10.1158/1055-9965.EPI-21-0922 (Apr. 2022). [DOI] [PMC free article] [PubMed]
- 58.Ge, Q. et al. GRAMD4 inhibits tumour metastasis by recruiting the E3 ligase ITCH to target TAK1 for degradation in hepatocellular carcinoma. Clin. Transl Med.11 (11). 10.1002/ctm2.635 (Nov. 2021). [DOI] [PMC free article] [PubMed]
- 59.Denny, J. C. et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics26 (9), 1205–1210. 10.1093/bioinformatics/btq126 (May 2010). [DOI] [PMC free article] [PubMed]
- 60.Kunej, T. Interview: Prof. Tanja Kunej, a Pioneer of Multiomics, on 21st Century Systems Science Prospects and Challenges, Mar. 01, 2021, Mary Ann Liebert Inc. 10.1089/omi.2020.0216 [DOI] [PMC free article] [PubMed]
- 61.Primmer, C. R., Papakostas, S., Leder, E. H., Davis, M. J. & Ragan, M. A. Annotated genes and nonannotated genomes: cross-species use of Gene Ontology in ecology and evolution research., Mol Ecol, vol. 22, no. 12, pp. 3216–41, Jun. doi: (2013). 10.1111/mec.12309 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Nicolazzi, E. L. et al. SNPchiMp v.3: integrating and standardizing single nucleotide polymorphism data for livestock species.,BMC Genom., 16, 1, 283, doi: 10.1186/s12864-015-1497-1.Apr. (2015). [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript and supplementary information files.