Abstract
Many aging clocks have recently been developed to predict health outcomes and deconvolve heterogeneity in aging. However, existing clocks are limited by technical constraints, such as low spatial resolution, long processing time, sample destruction, and a bias towards specific aging phenotypes. Therefore, here we present a non-destructive, label-free and subcellular resolution approach for quantifying aging through optically resolving age-dependent changes to the biophysical properties of NAD(P)H in mitochondria through fluorescence lifetime imaging (FLIM) of endogenous NAD(P)H fluorescence. We uncover age-dependent changes to mitochondrial NAD(P)H across tissues in C. elegans that are associated with a decline in physiological function and construct non-destructive, label-free and cellular resolution models for prediction of age, which we refer to as “mito-NAD(P)H age clocks.” Mito-NAD(P)H age clocks can resolve heterogeneity in the rate of aging across individuals and predict remaining lifespan. Moreover, we spatiotemporally resolve age-dependent changes to mitochondria across and within tissues, revealing multiple modes of asynchrony in aging and show that longevity is associated with a ubiquitous attenuation of these changes. Our data present a high-resolution view of mitochondrial NAD(P)H across aging, providing insights that broaden our understanding of how mitochondria change during aging and approaches which expand the toolkit to quantify aging.
Subject terms: Cell biology, Mitochondria
Endogenous mitochondrial NAD(P)H fluorescence enables label-free and subcellular resolution quantification of aging, prediction of remaining lifespan and tracking of age-dependent changes to mitochondria.
Introduction
As individuals can vary remarkably in their rate of aging, generating effective strategies to quantify aging has become a priority of geroscience research. In response to this need, many groups have generated “aging clocks” by using machine learning to create models that provide a prediction of age based on biomarkers of aging. For example, aging clocks trained on DNA methylation, blood plasma protein, or transcriptomic profiles exhibit the capacity to predict chronologic age (the time an organism has been alive) with relatively high accuracy1–11. While these techniques represent powerful approaches to quantify aging, these approaches also remain limited by a variety of technical constraints that can restrict their use. For example, approaches involving next generation sequencing can take months to complete. Further, many existing techniques also involve destruction of the sample, limiting spatial resolution of the resulting data and the ability to track the same organism longitudinally. Many existing approaches also involve intricate sample processing steps that could confound results. Lastly, although many existing techniques resolve a variety of features of aging, each clock individually captures a narrow subset of aging biology (e.g. epigenetic changes during aging). Thus, it is important to expand the diversity of approaches to gain a comprehensive view of aging.
To address these challenges, we explored the capacity of nicotinamide adenine dinucleotide phosphate ((NAD(P)H), representing spectrally indistinguishable NADPH and NADH)) FLIM to track mitochondria across age throughout an intact organism, using C. elegans as a model of aging. FLIM of metabolic cofactors has been used across many model systems both in vitro and in vivo as a label-free, non-destructive and subcellular resolution technique for monitoring the biophysical properties of NAD(P)H in living samples12–18. NAD(P)H FLIM capitalizes on the observation that as autofluorescent metabolic cofactors, such as NAD(P)H, are used differentially by cells, their optical properties will change. For example, NAD(P)H is autofluorescent when reduced, but not when oxidized as NAD(P)+19. Further, NAD(P)H has a longer lifetime when it is bound to a protein, as opposed to when NAD(P)H is freely diffusing in the cytosol20. Pairing NAD(P)H FLIM with C. elegans is particularly powerful, as C. elegans are translucent and progress through an entire life cycle in 2-3 weeks, allowing for rapid imaging of NAD(P)H autofluorescence across the entire body throughout aging. Despite the wide use of NAD(P)H FLIM as a tool across many model systems, only a small number of studies have briefly explored NAD(P)H FLIM in C. elegans, with focus largely restricted to C. elegans development21–24. Thus, it remains largely unclear how NAD(P)H FLIM could be used to study adult C. elegans and whether this technology could be used to track aging. Here, we expand upon these initial investigations to establish NAD(P)H FLIM in C. elegans as a non-destructive, label-free and subcellular resolution tool for organism-wide tracking of mitochondrial NAD(P)H and apply this technology to present a high-resolution landscape of mitochondria throughout aging, which we use to spatiotemporally deconvolve age-dependent changes to mitochondria and develop approaches to non-invasively quantify aging at the cellular level.
Results
We first set out to determine whether NAD(P)H autofluorescence could be used to visualize mitochondria in C. elegans. As expected, throughout tissues in C. elegans, we found autofluorescent signals reminiscent of mitochondria in our NAD(P)H channel, which we conservatively call the “blue autofluorescence (B.A.)/NAD(P)H channel” to account for NAD(P)H autofluorescence as well as autofluorescence detected in this channel which may not be NAD(P)H; Ex: 750 nm (2 P), Em: 400–480 nm) (Fig. S1A–D). To determine whether these signals were mitochondria, we examined B.A./NAD(P)H autofluorescence in transgenic C. elegans harboring proteins localizing to the outer-membrane of mitochondria fused to green fluorescent protein (GFP) (TOMM-20aa1-49::GFP and tomm-70::GFP). We found that enriched autofluorescent signals in the B.A./NAD(P)H channel exclusively represented mitochondria in all tissues except for the intestine, where mitochondrial B.A./NAD(P)H signals were detected alongside autofluorescent gut granules, which were previously shown to be lysosome related organelles (LROs) (Fig. S1A–D)25–28. Moreover, we found that B.A./NAD(P)H autofluorescence alone was sufficient to visualize mitochondrial lumens in all major tissue classes in C. elegans except for neurons, where mitochondrial matrices are much smaller and lack the space for NAD(P)H autofluorescence to accumulate for visualization consistently (Fig. S1A–D). Verifying that these B.A./NAD(P)H mitochondrial signals were predominantly NAD(P)H, we found that a short treatment with carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), which drives oxidation of NAD(P)H and thus a decline in NAD(P)H (Fig. S1E), resulted in a strong reduction in B.A./NAD(P)H autofluorescence inside the mitochondria of body wall muscle (BWM) cells, and that this effect was largely dependent on mitochondrial respiratory chain complex I (Fig. 1A,B)29. Further, we observed similar emission of B.A./NAD(P)H autofluorescence in C. elegans mitochondria and pure NADH fluorescence in solution (Fig. S1F). Finally, we found that C. elegans with a deletion of T20D3.5/slc-25a51, the predicted C. elegans homolog of the mitochondrial NAD+ transporter, MCART1/SLC25A5130–32, had strongly reduced mitochondrial B.A./NAD(P)H fluorescence (Fig. S1G,H). Thus, NAD(P)H autofluorescence can be used as a label-free marker to visualize mitochondrial lumens across tissues in C. elegans.
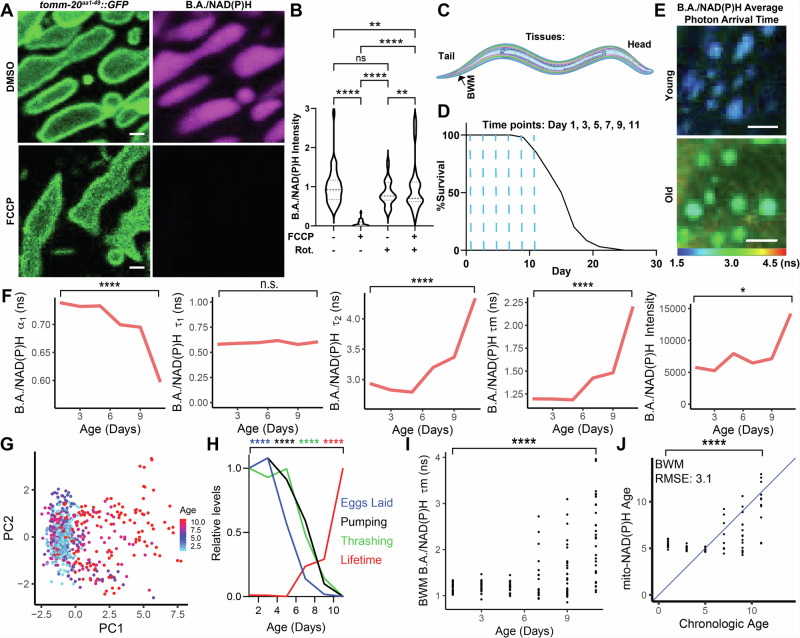
Fig. 1. Non-destructive, label-free and subcellular resolution quantification of aging based on mitochondrial B.A./NAD(P)H FLIM.
A,B. Images and analysis of B.A./NAD(P)H (magenta) and TOMM-20aa1-49::GFP (green) in BWM cells of Day 1 adult C. elegans treated with either 10 mM FCCP and/or 10 mM rotenone for 5–15 min. B shows quantification of mitochondrial B.A./NAD(P)H fluorescence intensity (n = 25–40 C. elegans per condition; Two-way ANOVA with post-hoc Tukey’s test). C,D Schematic of imaging location in the C. elegans body € and time points (D; blue dashed lines) imaged for analyzes presented in Fig. 1E–J. n = 100 C. elegans in the lifespan. E WT B.A./NAD(P)H average photon arrival time images of C. elegans BWM cells in either young (Day 1) or old (Day 11) C. elegans. F C. elegans were imaged using B.A./NAD(P)H FLIM at Day 1, 3, 5, 7, 9 and 11 in BWM cells and then analyzed as described in Fig. S2A. Graphs depict the median value across animals imaged for each endpoint at each age respectively (n = 17–32 C. elegans per condition; moderated t test; median). G PCA of non-redundant BWM B.A./NAD(P)H FLIM endpoints (α1, Ƭ1, Ƭ2 and intensity) of individual mitochondria colored by age. Each dot represents an individual mitochondrion (n = 67-128 mitochondria across 17–32 C. elegans per condition; points represent distinct mitochondria). H Plot of relative BWM B.A./NAD(P)H lifetimes (red), pumping rate (black), thrashing rate (green), and egg laying (blue) across age in C. elegans (n = 17–32 C. elegans per condition; moderated t test (B.A./NAD(P)H FLIM), Student’s t test (all other comparisons); median (B.A./NAD(P)H FLIM), mean (all other comparisons)). I Plot of age versus BWM B.A./NAD(P)H lifetime. Each dot represents an individual C. elegans (n = 17–32 C. elegans per condition; moderated t test). J LASSO regression models generated with B.A./NAD(P)H FLIM endpoints from BWM mitochondria. Plots depict age predicted by the model (Mito-NAD(P)H Age) versus chronological age. The blue line indicates when predicted age hypothetically matches chronologic age. RMSE stands for root mean square error. Each dot represents one C. elegans (n = 157 C. elegans used to train the model; Student’s t test; points represent distinct C. elegans). Scale bars, 1 µm (A) 10 µm (E). ****p < 0.0001, **p < 0.01, *p < 0.05.
To increase the resolution by which we could track NAD(P)H in mitochondria, we next used a FLIM-capable 2-photon microscope to perform FLIM and examined fluorescence signal decay in mitochondria in the germline, the metacorpus of the pharyngeal muscle (PM), BWM and hypodermis of C. elegans (Fig. S2A–G). Mitochondria are dynamic and thus, in many cell types, it is often not possible to track individual mitochondria by FLIM, as mitochondria may move out of an analyzed region of interest in the ~1 minute of time required to acquire an B.A./NAD(P)H FLIM image. However, in C. elegans, we observed that mitochondria were relatively less dynamic, with most mitochondria not moving out of a specified region of interest during data acquisition (Fig. S2C). Thus, we determined that C. elegans mitochondria could be analyzed on an individual level. To quantify the lifetime data, following convention in the field, we used weighted, 2 component, n-exponential reconvolution modeling to account for the freely diffusing and enzyme-bound NAD(P)H lifetime components respectively16 (Fig. S2A). This analysis quantitatively extracts four coefficients that define NAD(P)H lifetimes and hypothetically model the biophysical parameters of NAD(P)H: the α1 coefficient represents the proportion of diffusing NAD(P)H; the α2 coefficient represents the proportion of bound NAD(P)H contributing to the observed lifetimes; τ1 reports the lifetime of diffusing NAD(P)H; and τ2 reports the lifetime of bound NAD(P)H. These values can be compressed into one value referred to as the fluorescence lifetime (τm)16. Thus, this imaging and analysis strategy allows for non-destructive, label-free, quantitative tracking of mitochondrial NAD(P)H across individual mitochondria in live C. elegans.
With a validated system for performing B.A./NAD(P)H FLIM in C. elegans, we next asked whether we could track mitochondria across aging using B.A./NAD(P)H FLIM. We found that C. elegans harbor detectable mitochondrial B.A./NAD(P)H autofluorescence signals which were sensitive to FCCP treatment that can be observed in many tissues up until ~Day 11 of adulthood, a time in the C. elegans life cycle that is well after the onset of many phenotypes of aging, such as a decline in locomotion, fertility, food consumption, and resilience to stress, and just prior to when many C. elegans in a population begin to die (Fig. S3A–E). Therefore, to track mitochondria across age, we imaged wild type (WT) BWM cells in C. elegans at Days 1, 3, 5, 7, 9 and 11 with NAD(P)H FLIM (Fig. 1C, D; Supplementary Data 1 and 2). Excitingly, we identified many changes in mitochondrial B.A./NAD(P)H FLIM endpoints in BWM cells throughout the course of aging (Figs. 1E–G, S3F–H). For example, we found that B.A./NAD(P)H α1 decreased during aging whereas B.A./NAD(P)H τ2 increased during aging. Interestingly, B.A./NAD(P)H FLIM endpoints remained relatively stable throughout young adulthood and began to progressively change starting around Day 5-7 of adulthood, a time when C. elegans are known to progressively sustain a general collapse in many physiologic functions. This finding suggests that B.A./NAD(P)H lifetimes may be linked to organismal fitness and that some components of mitochondria may not begin to change during aging until middle age, similar to the dynamics of other age-dependent changes in C. elegans33. Confirming this association between shifts in B.A./NAD(P)H lifetimes and a decline in physiologic function, we found that B.A./NAD(P)H lifetimes began to change as C. elegans began to sustain a reduction in egg production, motility, and pharyngeal pumping rate (Fig. 1H). We also noted that as C. elegans became older, individuals became more variable, with some individuals displaying more youthful B.A./NAD(P)H lifetimes while other animals displayed more aged B.A./NAD(P)H lifetime profiles (Fig. 1I). Thus, B.A./NAD(P)H FLIM can be used to non-invasively track mitochondria during aging and B.A./NAD(P)H lifetimes may be a proxy for organismal fitness.
As we uncovered an age-dependent shift in B.A./NAD(P)H lifetimes that progressively developed throughout aging, we next used Least Absolute Shrinkage and Selection Operator (LASSO) regression to construct a model for prediction of age based on B.A/NAD(P)H FLIM endpoints. As expected, our model provided increases in predicted age as chronologic age increased (Fig. 1J). Matching our data, we observed that our model was unable to discriminate between C. elegans at young ages, as B.A./NAD(P)H lifetimes did not start to change until later in aging. However, starting at Day 5, our model predicted progressively higher ages on average at each subsequent chronologic age time point. We also found that our models predicted more variance in age at later time points, suggesting that these models may be resolving heterogeneity in the rate of aging.
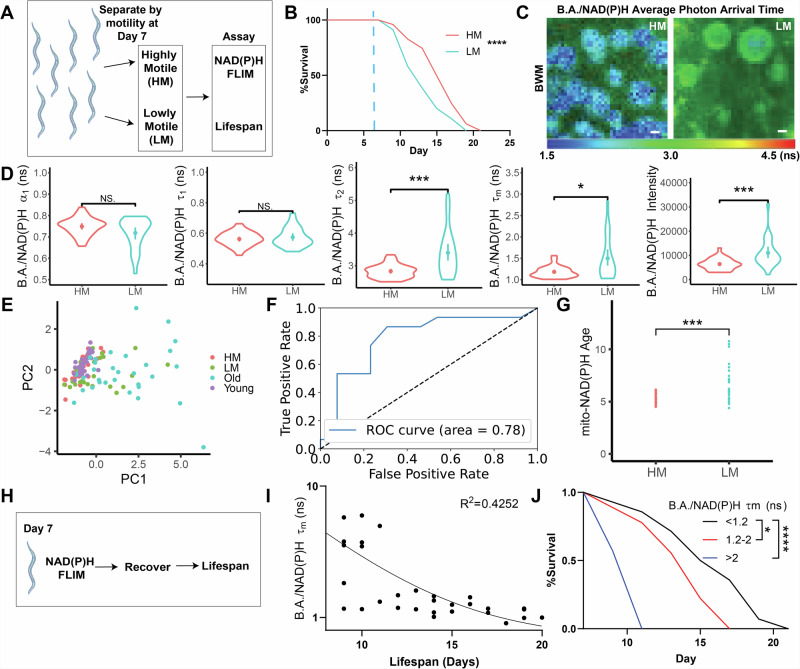
Therefore, we next hypothesized that this spread in B.A./NAD(P)H FLIM endpoints reflected heterogeneity in the rate of aging and that B.A./NAD(P)H FLIM endpoints could be used to predict fitness. Many studies have previously demonstrated the capacity to predict remaining lifespan in transgenic animals based on fluorescent reporter expression patterns in mid-life, such as lipid droplet number34, RNA splicing patterns35, heat shock reporter expression36, microRNA expression37, and “red autofluorescence” of whole C. elegans27. In addition to these systems, similar to a previous report that predicted remaining lifespan based on motility in mid life38, we found that separating C. elegans by mobility at Day 7 was sufficient to predict remaining lifespan, with higher mobility (HM) C. elegans living longer than lower mobility (LM) C. elegans (Fig. 2A, B). To determine if natural variance in motility is associated with changes to B.A./NAD(P)H FLIM endpoints, we performed B.A./NAD(P)H FLIM on the BWM of cohorts of HM and LM C. elegans at Day 7 and found that many B.A./NAD(P)H FLIM endpoints were significantly different between these two groups (Fig. 2C,D), almost completely reflecting changes we saw in B.A./NAD(P)H FLIM endpoints in young and old C. elegans, with the exception of B.A./NAD(P)H α1, which showed a consistent but non-significant trend. Importantly, we observed that HM C. elegans clustered closer to young C. elegans, while LM C. elegans clustered closer to old C. elegans (Fig. 2E). To determine whether B.A./NAD(P)H FLIM could be used to predict whether C. elegans were either in the HM or LM group and thus whether they would be relatively shorter or longer lived, we trained random forest classifier models with Day 7 HM and SL B.A./NAD(P)H endpoints and found that our model provided higher performance in predicting remaining lifespan (area under the receiver-operator characteristic (ROC) curve (AUC): 0.78; Fig. 2F)39. Predicting age with our BWM regression model (Fig. 1J) revealed that LM C. elegans had a higher average predicted age compared to HM C. elegans (Fig. 2G). Finally, to determine whether B.A./NAD(P)H FLIM could be used to directly predict remaining lifespan, we imaged BWM cells of Day 7 adult C. elegans individually using B.A./NAD(P)H FLIM and then recovered animals and tracked lifespan (Figs. 2H–J, S3I–J). We observed that C. elegans with the highest BWM cell B.A./NAD(P)H fluorescence lifetimes were associated with the shortest remaining lifespan, validating the direct predictive power of B.A./NAD(P)H FLIM to determine future lifespan. Thus, B.A./NAD(P)H FLIM represents a label-free and non-destructive approach to track aging at cellular resolution and can determine C. elegans motility and predict future lifespan.
Fig. 2. Mitochondrial B.A./NAD(P)H FLIM can predict remaining lifespan.
A,B Day 7 C. elegans were separated by motility into highly motile (HM; red) and lowly motile (LM; blue) groups and then tracked for the time of death following separation. Blue dotted line indicates the day of separation for imaging at Day 7 (n = 100 C. elegans per condition; Log-rank (Mantel–Cox) test). C,D B.A./NAD(P)H FLIM average photon arrival time images and analysis of Day 7 HM and LM C. elegans BWM (n = 25–31 C. elegans per condition; Student’s t test; mean ± SD). E PCA of WT Day 1 (young; purple), Day 11 (old; blue), HM (red) and LM (green) C. elegans non-redundant B.A./NAD(P)H FLIM endpoints (α1, Ƭ1, Ƭ2 and intensity; n = 17–32 C. elegans per condition). F Receiver operating characteristic curve (ROC) depicting the efficacy of a Random Forest classifier to predict whether C. elegans will be in either the HM or LM group based only on B.A./NAD(P)H FLIM endpoints (n = 56 C. elegans; 39 train, 17 test). G Mito-NAD(P)H Age for HM and LM C. elegans using the BWM LASSO regression model shown in Fig. 1J. Each dot represents an individual C. elegans (n = 25–31 C. elegans per condition; Student’s t test). H Cartoon outlining experimental strategy in which C. elegans were imaged individually using B.A./NAD(P)H FLIM, recovered and then tracked for lifespan. Data are shown in I and J. I,J C. elegans were imaged at Day 7 of adulthood, recovered and then tracked for remaining lifespan. I shows a plot of C. elegans lifespan compared to B.A./NAD(P)H lifetime in BWM cells. Each dot represents one C. elegans. J shows a survival curve for 3 pooled replicates of the data shown reflecting a range of BWM B.A./NAD(P)H FLIM lifetimes (B, n = 30 C. elegans, p < 0.0001, Log-rank (Mantel–Cox) test). Scale bars, 1 µm. ****p < 0.0001, ***p < 0.001, *p < 0.05.
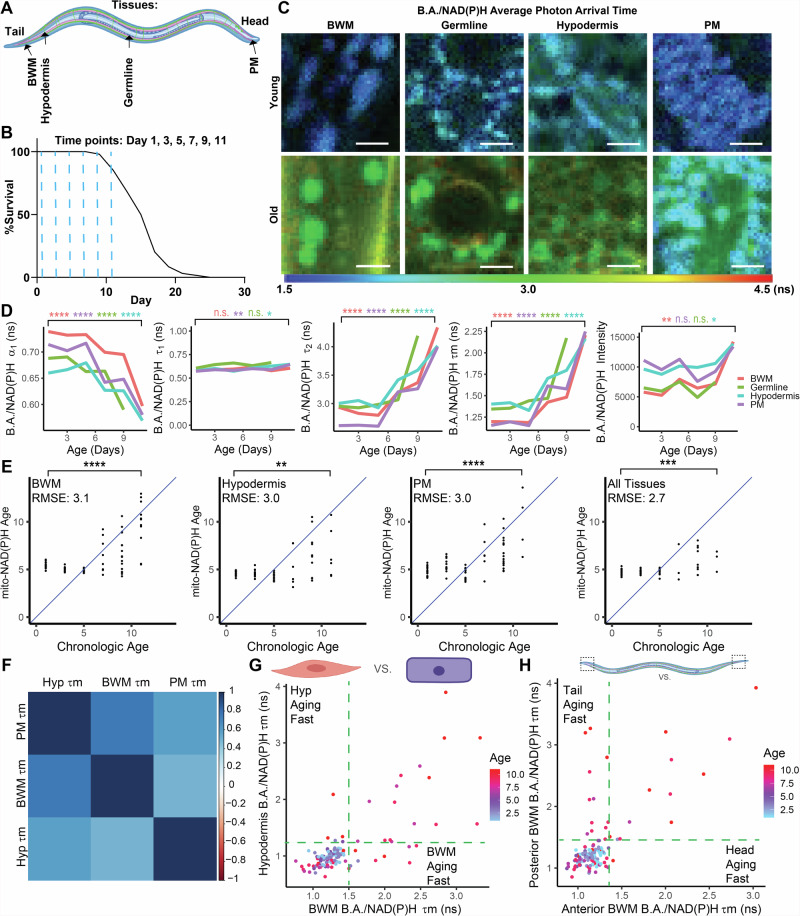
We next asked whether we could analogously use B.A./NAD(P)H FLIM to track aging across other tissues in C. elegans. To this end, we imaged and analyzed B.A./NAD(P)H lifetime endpoints across PM, hypodermal, germline, and BWM cells during aging and used these data to construct LASSO models for prediction of age across cell types in C. elegans (Fig. 3A–E). Interestingly, we found that across age, all tissues analyzed sustained largely similar changes to B.A./NAD(P)H FLIM endpoints with relatively strong correlations in B.A./NAD(P)H lifetimes across tissues within the same animal throughout aging and, similar to our BWM cell data, we found that the changes we resolved did not begin until middle age (Fig. 3C,D). Thus, on a global level, mitochondria may be aging in similar ways across tissues and with similar temporal dynamics. However, by analyzing animals, tissues, cells and mitochondria on an individual level, we observed many instances in which mitochondria changed asynchronously during aging (Fig. 3D–H, Fig. S3K–M). For example, we observed that at Days 7 and 9, some animals exhibited mitochondrial B.A./NAD(P)H lifetimes in the hypodermis that fall within the Day 1 lifetime distribution, while B.A./NAD(P)H lifetimes in the BWM of the same animal fall within the lifetime distribution observed in Day 11 animals (Fig. 3G). Further, analyzing anterior or posterior cells in the same tissue revealed that mitochondria in posterior cells of many C. elegans age faster as assayed by B.A./NAD(P)H FLIM than mitochondria in anterior cells (Fig. 3H). Thus, these results uncover a ubiquitous shift in mitochondria during aging beginning in mid-life, as well as modes by which aging progresses asynchronously across biological levels.
Fig. 3. B.A./NAD(P)H FLIM resolves age-dependent changes to mitochondria across and within tissues.
A,B Schematic of imaging locations throughout the C. elegans body (A) and time points (B; blue dashed lines) imaged for the data presented in Fig. 3. B shows a lifespan of WT (N2) C. elegans (n = 100 C. elegans). C-D) WT B.A./NAD(P)H average photon arrival time images and analysis of C. elegans in germline, PM, BWM, and hypodermal cells in young (Day 1) or old (Day 11) C. elegans (n = 5–32 C. elegans per condition; moderated t test; median). E LASSO regression models generated with B.A./NAD(P)H FLIM endpoints from either PM, BWM, or hypodermal mitochondria individually or altogether. Plots depict age predicted by the models (Mito-NAD(P)H Age) versus chronological age. The blue lines indicate when predicted age hypothetically matches chronologic age. Each dot represents one C. elegans. RMSE stands for root mean square error (n = 13–32 C. elegans per condition; Student’s t test). F Correlation matrix between B.A./NAD(P)H lifetime in PM, BWM, and hypodermal (hyp) cells in individual animals across all ages analyzed in WT C. elegans showing the overall extent to which B.A./NAD(P)H FLIM lifetimes (Ƭm) across cell types in the same animal correlate with each other throughout aging. G Plot of BWM versus hypodermal B.A./NAD(P)H lifetime colored by age. Each dot represents an individual C. elegans with each axis reporting the B.A./NAD(P)H lifetime for each tissue respectively within the same C. elegans. Green lines mark the edge of the young lifetime distribution (n = 124 C. elegans). H Plot of anterior versus posterior BWM B.A./NAD(P)H lifetime (Ƭm) colored by age. Each dot represents an individual C. elegans with each axis reporting the B.A./NAD(P)H lifetime (Ƭm) for each location in the BWM respectively within the same C. elegans. Green lines mark the edge of the young lifetime distribution (n = 131 C. elegans). Scale bars, 10 µm. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
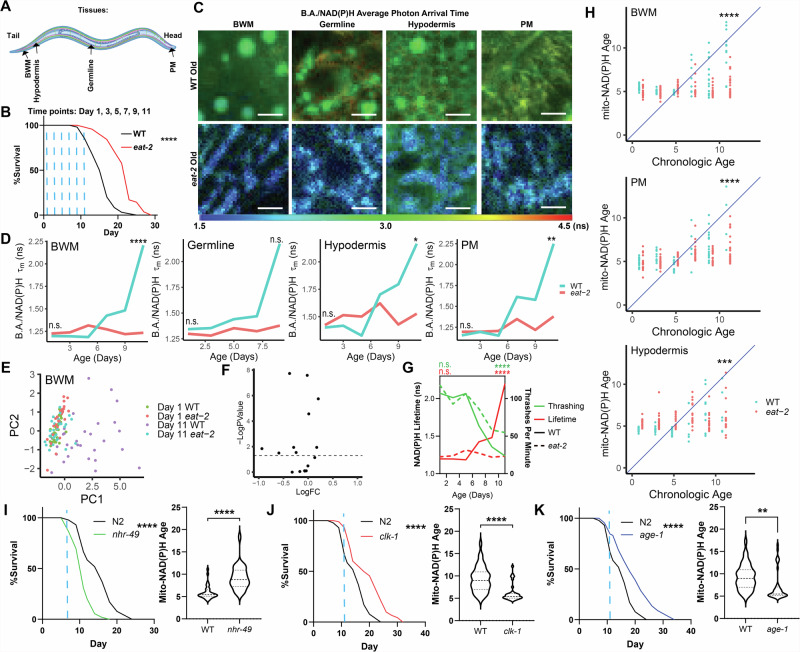
Finally, we asked whether the age-dependent changes we observed in mitochondria would be altered in response to modulating longevity. To this end, we imaged and analyzed WT and long-lived eat-2(ad1116) mutant C. elegans (a genetic model of dietary restriction) throughout aging analogously to our experiments in WT animals (Fig. 4A, B, S4A, B). As we had found earlier that WT animals exhibited a strong shift in mitochondrial NAD(P)H lifetimes during aging, we hypothesized that long-lived eat-2 animals would either display an attenuation of these changes or a different trajectory during aging. We found that up until Day 11 of adulthood, eat-2 animals were largely refractory to age-dependent changes in B.A./NAD(P)H FLIM endpoints across tissues (Fig. 4C–F, S4C–E). For example, in WT animals, B.A./NAD(P)H lifetime increases during aging, starting at Days 5-7. However, in eat-2 animals, we did not observe a similar increase in B.A./NAD(P)H lifetime. Although we observed a small increase in B.A./NAD(P)H lifetime with age in some tissues (e.g. PM mitochondria), these effects were largely diminished when compared to the effect of age observed in WT animals. Further, we found that this attenuation of age-dependent changes to mitochondrial B.A./NAD(P)H FLIM endpoints was matched with an attenuation of age-dependent changes to C. elegans mobility (Fig. 4G). Consistent with our data demonstrating an attenuation of age-dependent changes to B.A./NAD(P)H FLIM endpoints, we also observed an attenuation in the increase in predicted age observed in WT animals in our regression models (Figs. 1, 3), demonstrating that longevity is associated with a reduction in predicted age measured by B.A./NAD(P)H FLIM of mitochondria (Fig. 4H). Finally, to evaluate the extent to which mito-NAD(P)H age clocks could resolve potentiation in longevity across a diverse range of interventions which influence longevity, we examined BWM cells in WT, nhr-49(nr2041)-short-lived, clk-1(qm30)-long-lived and age-1(hx546)-long-lived mutant C. elegans (Fig. 4I–K, S4F). Consistently, we observed that nhr-49 mutants which have shortened lifespan were associated with higher mito-NAD(P)H ages earlier in life relative to WT, whereas clk-1 and age-1 mutants which have extended lifespan were associated with lower mito-NAD(P)H ages. Thus, these data demonstrate the sensitivity of mito-NAD(P)H age clocks to changes in longevity.
Fig. 4. Mitochondrial B.A./NAD(P)H FLIM endpoints are linked to lifespan.
A,B Schematic of imaging locations throughout the C. elegans body (A) and time points (B; blue dashed lines) imaged for data presented in Fig. 4. B shows a lifespan curve for WT (N2) and eat-2 (ad1116) C. elegans (n = 100 C. elegans per condition, p < 0.0001, Log-rank (Mantel–Cox) test). C,D WT (blue) and eat-2 (red) B.A./NAD(P)H average photon arrival time images and analysis of C. elegans in the germline, PM, BWM, and hypodermis throughout age (n = 5–32 C. elegans per condition; moderated t test; median). E PCA plots of non-redundant B.A./NAD(P)H FLIM endpoints (α1, Ƭ1, Ƭ2 and intensity) of Day 1 and Day 11 WT and eat-2 C. elegans BWM mitochondria B.A./NAD(P)H FLIM endpoints. Each dot is an individual C. elegans (n = 24–32 C. elegans per condition). F Volcano plot summarizing statistical analyzes of B.A./NAD(P)H FLIM endpoints comparing Day 11 WT and eat-2 C. elegans to each other. (moderated t test). G Plot of thrashing rate (green) and BWM B.A./NAD(P)H lifetime (red) in WT (solid line) or eat-2 (dashed line) C. elegans versus age (n = 21–32 C. elegans per condition; Two-way ANOVA with post-hoc Tukey’s test; median (B.A./NAD(P)H FLIM), mean (thrashing)). H Mito-NAD(P)H Age for WT (blue) and eat-2 (red) animals using LASSO regression models generated in Figs. 1 and 3 plotted versus age. Each dot represents an individual C. elegans (n = 13–32 C. elegans per condition; Student’s t test). I–K Lifespan and mitochondrial BWM Mito-NAD(P)H Age in WT, nhr-49, clk-1 and age-1 mutants at the indicated time points (blue dashed line – Day 7 for nhr-49 or Day 11 for age-1 and clk-1; n = 100 C. elegans per condition in lifespans and n = 27–40 for B.A./NAD(P)H FLIM analyzes, Log-rank (Mantel–Cox) test (lifespans) Student’s t test (mito-NAD(P)H Age). Scale bars, 10 µm. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
Discussion
Here we developed and applied B.A./NAD(P)H FLIM in adult C. elegans to non-destructively track mitochondrial NAD(P)H in individual mitochondria throughout cells and tissues in C. elegans across aging and used the resulting data sets to develop a next generation of non-destructive, label-free and cellular resolution aging clocks. B.A./NAD(P)H enables both the ability to track the abundance of NAD(P)H by tracking the fluorescence intensity of B.A./NAD(P)H as well as additional properties resolved by tracking the fluorescence lifetime of B.A./NAD(P)H. As such, B.A./NAD(P)H FLIM expands upon previously developed biosensors for imaging NAD(P)(H), which can only image the abundance of NAD(P)(H)40 in living cells that our data suggest would not be predictive of biological aging. This approach to quantify aging is powerful as its strengths complement the weaknesses of many previously existing orthogonal approaches1–11,41–44. For example, B.A./NAD(P)H FLIM is fast, providing the capacity for a prediction of age on the same day as data acquisition. Further, B.A./NAD(P)H FLIM can track individual mitochondria, providing the capacity to trace mitochondria during aging at cellular or subcellular resolution within an intact organism. B.A./NAD(P)H FLIM is also label-free and non-destructive; thus, remedying concerns that reagents used in the assay may be confounding results or that the same individual could be measured at multiple time points throughout life longitudinally. As our approach relies on resolution of biophysical changes to NAD(P)H in mitochondria, mito-NAD(P)H age clocks also provide quantification of aging through a unique lens which diversifies our cohort of approaches to quantify aging. Thus, B.A./NAD(P)H FLIM represents an approach to quantify aging with many powerful advantages over comparable previously existing technologies.
While B.A./NAD(P)H FLIM has many advantages, B.A./NAD(P)H FLIM also has limitations. For example, as other species of autofluorescence eventually overcome mitochondrial signals in the B.A./NAD(P)H channel in animals close to death, mito-NAD(P)H age clocks have an upper limit in ages that they can track. However, future optical aging clocks could integrate these different species of autofluorescence to overcome this limitation. As NAD(P)H lifetimes are a comprehensive representation of the biophysical environment of NAD(P)H and can be sensitive to changes in many physical and chemical parameters, such as enzyme binding, temperature, pH and viscosity, it is also difficult to know exactly what the changes to NAD(P)H we detect during aging in mitochondria specifically represent. Our initial investigation is also limited to analysis of C. elegans. As NAD(P)H FLIM is widely used across many model systems16,45,46 and mammalian aging is known to be associated with changes to autofluorescence across many tissues47–49, application of NAD(P)H FLIM to study mammalian aging is a promising future area of investigation in many model systems. Despite these limitations, the advantages of B.A./NAD(P)H FLIM represent a substantial advance in our capacity to quantify aging and provide a unique cohort of advantages that broaden the toolkit of aging clocks available to the geroscience community. Taken together, here we describe a live-cell, label-free, non-destructive and subcellular resolution approach to track mitochondrial NAD(P)H throughout a multicellular organism on the level of individual mitochondria. Using this technique, we spatiotemporally resolve components within mitochondria which change during aging across cell types and tissues and develop models for the prediction of age, health, and remaining lifespan that expand our technical capacity to quantify aging.
Methods
C. elegans strains and husbandry
N2 Bristol (wild type (WT); from the CGC), DA1116 (eat-2(ad1116) II; from the CGC), PX62750 (fxIs1 I; spe-44(fx110[spe-44::degron]) IV; from the CGC), STE68 (nhr-49(nr2041) I; from the CGC), TJ1052 (age-1(hx546) II; from the CGC), MQ130 (clk-1(qm30) III; from the CGC), WBM1633 (N2, wbmIs153[dpy-7p::3XFLAG::dpy-10::SL2::wrmScarlet::unc-54 3’UTR, *wbmIs88] V), WBM1231 (N2, wbmIs97[eft-3p::tomm20::GFP::unc-54 3’UTR, *wbmIs65]), WBM1444 (N2, tomm-70::GFP), WBM1215 (N2, wbmIs89 [rab-3p::3xFLAG::dpy-10::SL2::wrmScarlet::rab-3 3’UTR, *wbmIs68]), WBM1478 (N2, wbmIs136[nep-17p(mini)::3XFLAG::dpy-10::SL2::wrmScarlet::unc-54 3’UTR, *wbmIs88] V), WBM1153 (N2, wbmIs72 [pie-1p::3XFLAG::GFP::unc-54 3’UTR *wbmIs60]), WBM1133 (N2, wbmIs63 [myo-3p::3XFLAG::wrmScarlet::unc-54 3’UTR *wbmIs61]), RG3152 (T20D3.5(ve652[LoxP + myo-2p::GFP::unc-54 3’ UTR + rps-27p::neoR::unc-54 3’ UTR + LoxP])/tmC5 IV), WBM1214 (N2, eft-3p::SL2::wrmScarlet V) and GRU101 (gnaIs1 [myo-2p::yfp]) were used in this study. To make WBM1231, CRISPR/Cas9 was used to insert tomm-20aa1-49::GFP into WBM114051. To make WBM1444, CRISPR/Cas9 was used to insert GFP amplified from pHW21 on the carboxyl terminus of tomm-70 at its endogenous locus in the C. elegans genome. To make WBM1633, the 868 base pairs of the dpy-7 promoter52,53 was amplified from genomic DNA of WT C. elegans, and then swapped in place of the eft-3 promoter in WBM1214 using CRISPR/Cas9. The strain was first isolated by visual screening and then sequence verified. C. elegans were maintained on standard nematode growth media (NGM) plates seeded with E. coli (OP50-1) at 20 °C. OP50-1 was cultured overnight in LB at 37 °C and then added to NGM plates and allowed to grow for 2 days prior to adding C. elegans to each plate. To generate day 1 adults, a timed egg lay was performed and L4 larvae were picked 24 h prior to imaging.
CRISPR/Cas9 editing
Following established protocols51, homology directed repair templates were generated by PCR with primers that attach at least 35 base pairs of homology on either end of the insert. CRISPR injection mixes were generated with the following composition: 0.375 μl Hepes pH 7.4 (200 mM), 0.25 μl KCl (1 M), 2.5 μl tracrRNA (4 μg/μl), 0.6 μl crRNA (2.6 μg/μl), HDR template 500 ng/μl, water up to 8 μl. Prior to injection, 2 μl purified Cas9 (12 μg/μl) was added before the solution was centrifuged at 13,000 rotations per minute (rpm) and then incubated at 37 °C for 10 min. All newly generated strains were outcrossed at least 6 times prior to using in experiments.
Microscopy
For imaging, C. elegans were placed in 0.1 µm PolyBead Microspheres (PolyScience 00876-15) on a 10% agar pad, covered with a #1.5 coverslip and imaged immediately after mounting. All images were acquired using an upright Leica Stellaris 8 DIVE FALCON 2-photon + confocal microscope outfitted with 4Tune spectral NDD and an InSight X3 dual beam pulsed (80 Hz) infrared laser (Spectra Physics) and a pulsed WLL (80 Hz). To image B.A./NAD(P)H, samples were excited with a 750 nm (2 P) and 400–480 nm emission wavelengths were collected using a HyD NDD 2 detector set to counting mode. GFP was imaged by exciting at 900 nm (2 P) and collecting 490–570 nm wavelengths using a HyD NDD 2 in digital mode at gain 40. Images were acquired with a Plan Apo 63x oil-immersion objective with 1.4 numerical aperture at 5x zoom and 512 × 512 (effective pixel size: 172 nm) pixel images, a pixel dwell of 15.39 µs, and 10 frame integration to collect 500–1000 photons at peak of decay per analyzed pixel for an image collection time of ~50 s. LasX acquisition software was used to control the microscope. Flavin adenine dinucleotide (FAD; 5 mM; Sigma-Aldrich F6625-10MG) or NADH (5 mM; Sigma-Aldrich N8129-100MG) were imaged dissolved in water and placed on an agar pad, analogous to how C. elegans were imaged.
FCCP and rotenone experiment
C. elegans were treated with FCCP (Cayman Chemical Company 15218) and/or rotenone (Sigma-Aldrich R8875) dissolved in DMSO or a vehicle control dose of DMSO as indicated in the figure legends, mounted and imaged as described in the microscopy section, and then analyzed in Fiji. To analyze the data, B.A./NAD(P)H intensity was normalized to region of interest area (ROI) in mitochondrial ROIs.
FLIM analysis
FLIM decays were analyzed to quantify B.A./NAD(P)H lifetime components using Leica Application Suite (LAS) X’s FLIM analysis module, modeling established B.A./NAD(P)H FLIM analyzes16. For each image, a bin of 4×4 pixels was used and a region of interest was drawn around in focus mitochondria to be analyzed. All data was first deconvolved from LAS X’s instrument response function and modeled using a 2-component n-exponential reconvolution model to account for freely diffusing and bound B.A./NAD(P)H components comprising the fluorescence decay: I(t) = α1 × e−t/τ1 + α2 × e−t/τ2 where I(t) is the fluorescence intensity as a function of time (t) following pulsed excitation. Α1 and α2 are the fractional contributions of the short and long lifetime component contributing to the overall fluorescence lifetime such that α1 + α2 = 1. Τ1 and τ2 are the short and long lifetime components. To analyze the data, a region of interest (ROI) was drawn around individual mitochondria and then average lifetime values were obtained per mitochondrial ROI. Tissues were identified morphologically based on the B.A./NAD(P)H channel. Mitochondrial measurements were merged per cell by taking the median of all mitochondrial ROIs analyzed per cell.
TMRE labeling
Mitochondria were visualized with TMRE (ThermoFisher Scientific T669) by placing on C. elegans on OP50-1 seeded NGM plates with 10 µM TMRE in M9 added to the top of the plate for 24 h prior to imaging.
Statistics and reproducibility
“n” denotes the number of C. elegans, cells or mitochondria, as indicated in each figure legend. “N” denotes the number of times an experiment was independently repeated. All experiments were repeated independently at least three times on three separate days. Statistical analyzes were performed in R studio, Spyder, and GraphPad Prism using the statistical tests indicated in figure legends (code and data available at: https://github.com/chrismorrow5/C.-elegans-Mitochondrial-Aging). Data presented represent either individual mitochondria or median values calculated per cell by taking the median of all mitochondria analyzed in a specified cell, as indicated in the figure legends. B.A./NAD(P)H FLIM endpoints were evaluated using hypothesis tests for change across age in WT animals, or across genotype in young or old animals in R version 4.1.2, using package ‘limma’ version 3.50.3 and functions lmFit(), eBayes() and p.adjust() with method = “BH” to adjust for multiple testing. Due to a low number of C. elegans germlines imaged at Day 11, Day 9 was used as the “aged” time point for germline analyzes. Heat maps were made using pheatmap() from R package ‘pheatmap’ version 1.0.12. Correlation matrices were made in R using cor() and corplot() from base R. Principal component analyzes (PCA) were performed using prcomp() with data normalized using scale() prior to analysis in base R. Random forest classifier models were made in Python version 3.7.9 using a training set consisting of 70% of data points (C. elegans or cells), randomly selected, and the models were tested on the remaining 30% using B.A./NAD(P)H α2 and τ1. Data were standardized so that each variable had an average of 0 and a sample standard deviation of 1 prior to model training. LASSO regression models were made in R using glmnet() with package glmnet version 4.1.7. Data were randomly assigned to training (50%) and testing (50%) sets and cv.glmnet() was used to select an optimal regularization parameter λ for LASSO. Data were standardized so that each variable had an average of 0 and a standard deviation of 1 prior to model training. To run the trained models on additional data sets (SL/LL and eat-2), the additional data were standardized using the same standardization parameters that were used to center and scale the original data sets used during training. Model performance was evaluated by calculating the respective root mean square error (RMSE) of each model.
Lifespan and Healthspan Experiments
All experiments were performed on standard NGM with 100 µl OP50-1 seeded 48 h at room temperature prior to adding C. elegans. C. elegans were synchronized by egg lay and then ~48 h later, L4-stage C. elegans were picked to have day 1 adults to start the lifespan the next day. On Day 1 of adulthood, 100 C. elegans per condition per experiment were transferred to NGM plates at 20 C. elegans per plate and then transferred at least once every two days to avoid starvation until ~Day 11. C. elegans were determined to be dead if they did not respond to three taps on the head and tail. C. elegans were censored either due to contamination, eggs hatching inside the adult, leaving the plate, or loss of vulval integrity. Lifespan analyzes were performed in GraphPad Prism using the statistical tests indicated in the figure legends.
Briefly, C. elegans at each genotype and age were generated as described above, and then analyzed for pumping, egg lay, and thrashing rate54. To analyze thrashing rate, C. elegans were suspended in M9 salt solution, allowed to acclimate for 30 s, and then tracked for thrashes per minute under a dissection microscope. To track pumping rate, we scored pumps per minute in C. elegans on bacterial lawns under a dissection microscope. To track egg laying rate, C. elegans were transferred individually to NGM plates and then left to lay eggs for 24 h prior to scoring. For each experiment, at least 10 C. elegans were analyzed per group and each experiment was repeated at least 3 times.
NADH Assays
NADH levels were quantified using a biochemical analysis kit (Millipore Sigma MAK037) following the manufacturer’s instructions. Briefly, 10,000 day 1 adult C. elegans per sample were prepared as described in the figure legends, then mechanically lysed in a Qiagen TissueLyser with 5 mm metal beads (Qiagen 69989) in 100 uL NADH/NAD extraction buffer. Samples were spun at 12,000× g for 10 min at 4 °C. Protein was removed from the samples by centrifugation at 12,000×× g for 20 min at 4 °C (Millipore Sigma UFC501008). All of the sample was then analyzed for NADH level (no fractionation) per the manufacturer’s protocol and the resulting samples were analyzed on a spectrometer at absorbance 450 nm.
Separation into HM and LM groups
HM and LM groups of C. elegans were generated by manually separating C. elegans based on motility at Day 7 of adulthood. C. elegans were considered LM if they were immobile prior to being tapped with a worm pick and only exhibited a head movement in response to being tapped. C. elegans were considered HM if they were already moving without any stimuli. For these experiments, conditionally sterile C. elegans (PX62750 (fxIs1 I; spe-44(fx110[spe-44::degron]) IV) were used to generate animals for selection of LM or HM animals on Day 7 to use for imaging. To induce sterility, PX627 C. elegans were raised starting at hatch on NGM plates supplemented with 0.15 mM auxin (Indole-3-acetic acid, 98+%; Alfa Aesar A10556-06). C. elegans were then transferred at least once every other day to avoid starvation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
We thank the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School, Dr. Amani Gillette (UW-Madison), and Dr. Melissa Skala (UW-Madison) for their support with 2-photon fluorescence lifetime imaging. WBM is funded by NIH/NIA R01AG059595, R01AG044346 and R01AG067106. CSM is funded by NIH/NIA F32AG085703.
Author contributions
C.S.M. conceived the idea for the study, performed the experiments, analyzed the data, and wrote the manuscript with input from all authors. P.V.A. and P.M.L. assisted and consulted on establishing the FLIM. J.W.M. consulted on development of the statistical models for predicting age. P.Y. generated the strains for visualizing mitochondria with GFP. C.A.V.J. made the hypodermal fluorescence strain. B.A.B. consulted on statistical analyzes of the mitochondrial autofluorescence aging data set. W.B.M. oversaw the project and co-wrote the manuscript.
Peer review
Peer review information
This manuscript has been previously reviewed at another Nature Portfolio journal. The manuscript was considered suitable for publication without further review at Communications Biology.
Data availability
Code and data used in this study has been either attached in the supplemental tables (Supplementary Data 1–3) or uploaded to: https://github.com/chrismorrow5/C.-elegans-Mitochondrial-Aging. All additional details related to this manuscript are available upon reasonable request to the corresponding author: Dr. William B. Mair (wmair@hsph.harvard.edu).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07243-w.
References
- 1.Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol.14, 3156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY)10, 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, C. G. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol.20, 249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY)11, 303–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet19, 371–384 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell49, 359–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson, D. J. & Chandra, T. Epigenetic age prediction. Aging Cell20, e13452 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belsky, D. W. et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife9, e54870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka, T. et al. Plasma proteomic signature of age in healthy humans. Aging Cell17, e12799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menni, C. et al. Circulating proteomic signatures of chronological age. J. Gerontol.: Ser. A70, 809–816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun, E. D. et al. Predicting physiological aging rates from a range of quantitative traits using machine learning. Aging (Albany NY)13, 23471–23516 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chance, B., Schoener, B., Oshino, R., Itshak, F. & Nakase, Y. Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J. Biol. Chem.254, 4764–4771 (1979). [PubMed] [Google Scholar]
- 13.Heaster, T. M., Humayun, M., Yu, J., Beebe, D. J. & Skala, M. C. Autofluorescence imaging of 3D tumor-macrophage microscale cultures resolves spatial and temporal dynamics of macrophage metabolism. Cancer Res80, 5408–5423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta, R., Heaster, T. M., Sharick, J. T., Gillette, A. A. & Skala, M. C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt.25, 1–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagar, M. A. K. et al. Microglia activation visualization via fluorescence lifetime imaging microscopy of intrinsically fluorescent metabolic cofactors. Neurophotonics7, 35003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh, A. J. et al. Classification of T-cell activation via autofluorescence lifetime imaging. Nat. Biomed. Eng.5, 77–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirshin, E. A. et al. Label-free sensing of cells with fluorescence lifetime imaging: The quest for metabolic heterogeneity. Proc. Natl Acad. Sci. USA119, e2118241119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrow, C. S. et al. Autofluorescence is a biomarker of neural stem cell activation state. Cell Stem Cell10.1016/j.stem.2024.02.011 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blacker, T. S. & Duchen, M. R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med100, 53–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakowicz, J. R., Szmacinski, H., Nowaczyk, K. & Johnson, M. L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl Acad. Sci. USA89, 1271–1275 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stringari, C., Nourse, J. L., Flanagan, L. A. & Gratton, E. Phasor fluorescence lifetime microscopy of free and protein-bound NADH reveals neural stem cell differentiation potential. PLoS One7, e48014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stringari, C. et al. Multicolor two-photon imaging of endogenous fluorophores in living tissues by wavelength mixing. Sci. Rep.7, 3792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorrells, J. E. et al. Computational photon counting using multithreshold peak detection for fast fluorescence lifetime imaging microscopy. ACS Photonics9, 2748–2755 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yerevanian, A. et al. Riboflavin depletion promotes longevity and metabolic hormesis in Caenorhabditis elegans. Aging Cell21, e13718 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weir, H. J. et al. Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling. Cell Metab.26, 884–896.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coburn, C. & Gems, D. The mysterious case of the C. elegans gut granule: death fluorescence, anthranilic acid and the kynurenine pathway. Front Genet4, 151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pincus, Z., Mazer, T. C. & Slack, F. J. Autofluorescence as a measure of senescence in C. elegans: look to red, not blue or green. Aging (Albany NY)8, 889–898 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajdú, G., Somogyvári, M., Csermely, P. & Sőti, C. Lysosome-related organelles promote stress and immune responses in C. elegans. Commun. Biol.6, 936 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartolome, F. & Abramov, A. Y. Measurement of mitochondrial NADH and FAD autofluorescence in live cells. Methods Mol. Biol.1264, 263–270 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Luongo, T. S. et al. SLC25A51 is a mammalian mitochondrial NAD+ transporter. Nature588, 174–179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kory, N. et al. MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci. Adv.6, eabe5310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girardi, E. et al. Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import. Nat. Commun.11, 6145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labbadia, J. & Morimoto, R. I. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell59, 639–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papsdorf, K. et al. Lipid droplets and peroxisomes are co-regulated to drive lifespan extension in response to mono-unsaturated fatty acids. Nat. Cell Biol.25, 672–684 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintz, C. et al. Corrigendum: splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature547, 476 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Rea, S. L., Wu, D., Cypser, J. R., Vaupel, J. W. & Johnson, T. E. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet37, 894–898 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinser, H. E., Mosley, M. C., Plutzer, I. B. & Pincus, Z. Global, cell non-autonomous gene regulation drives individual lifespan among isogenic C. elegans. eLife10, e65026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herndon, L. A. et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature419, 808–814 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Breiman, L. Random forests. Mach. Learn.45, 5–32 (2001). [Google Scholar]
- 40.Kostyuk, A. I. et al. In vivo imaging with genetically encoded redox biosensors. Int. J. Mol. Sci.21, 8164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee, I. et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science346, 473–477 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Komura, T., Yamanaka, M., Nishimura, K., Hara, K. & Nishikawa, Y. Autofluorescence as a noninvasive biomarker of senescence and advanced glycation end products in Caenorhabditis elegans. npj Aging Mech. Dis.7, 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waqas, K. et al. Skin autofluorescence, a noninvasive biomarker of advanced glycation end-products, is associated with frailty: the Rotterdam study. J. Gerontol.: Ser. A77, glac025 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eigenfeld, M., Kerpes, R., Whitehead, I. & Becker, T. Autofluorescence prediction model for fluorescence unmixing and age determination. Biotechnol. J.17, e2200091 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Miskolci, V. et al. In vivo fluorescence lifetime imaging of macrophage intracellular metabolism during wound responses in zebrafish. eLife11, e66080 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh, A. J. et al. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res74, 5184–5194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Na, R., Stender, I.-M., Henriksen, M. & Wulf, H. C. Autofluorescence of human skin is age-related after correction for skin pigmentation and redness. J. Investig. Dermatol.116, 536–540 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Stillman, J. M. et al. Lipofuscin-like autofluorescence within microglia and its impact on studying microglial engulfment. bioRxiv 2023.02.28.530224 10.1101/2023.02.28.530224 (2023). [DOI] [PMC free article] [PubMed]
- 49.Dong, Y., Digman, M. A. & Brewer, G. J. Age- and AD-related redox state of NADH in subcellular compartments by fluorescence lifetime imaging microscopy. Geroscience41, 51–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dilberger, B., Baumanns, S., Spieth, S. T., Wenzel, U. & Eckert, G. P. Infertility induced by auxin in PX627 Caenorhabditis elegans does not affect mitochondrial functions and aging parameters. Aging Albany NY12, 12268–12284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva-Garcia, C. G. et al. Single-copy knock-in loci for defined gene expression in Caenorhabditis elegans. G3 (Bethesda)9, 2195–2198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilleard, J. S., Barry, J. D. & Johnstone, I. L. Cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol.17, 2301–2311 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paix, A., Folkmann, A., Rasoloson, D. & Seydoux, G. High efficiency, homology-directed genome editing in caenorhabditis elegans using crispr-cas9 ribonucleoprotein complexes. Genetics201, 47–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bansal, A., Zhu, L. J., Yen, K. & Tissenbaum, H. A. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc. Natl Acad. Sci.112, E277–E286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
Code and data used in this study has been either attached in the supplemental tables (Supplementary Data 1–3) or uploaded to: https://github.com/chrismorrow5/C.-elegans-Mitochondrial-Aging. All additional details related to this manuscript are available upon reasonable request to the corresponding author: Dr. William B. Mair (wmair@hsph.harvard.edu).