Abstract
Tissue biofabrication mimicking organ-specific architecture and function requires physiologically-relevant cell densities. Bioprinting using spheroids can achieve this, but is limited due to the lack of practical, scalable techniques. This study presents HITS-Bio (High-throughput Integrated Tissue Fabrication System for Bioprinting), a multiarray bioprinting technique for rapidly positioning multiple spheroids simultaneously using a digitally-controlled nozzle array (DCNA). HITS-Bio achieves an unprecedented speed, ten times faster compared to existing techniques while maintaining high cell viability ( > 90%). The utility of HITS-Bio was exemplified in multiple applications, including intraoperative bioprinting with microRNA transfected human adipose-derived stem cell spheroids for calvarial bone regeneration ( ~ 30 mm3) in a rat model achieving a near-complete defect closure (bone coverage area of ~ 91% in 3 weeks and ~96% in 6 weeks). Additionally, the successful fabrication of scalable cartilage constructs (1 cm3) containing ~600 chondrogenic spheroids highlights its high-throughput efficiency (under 40 min per construct) and potential for repairing volumetric defects.
Subject terms: Biomaterials - cells, Tissue engineering, Regenerative medicine, Mesenchymal stem cells
HITS-Bio, a high-throughput bioprinting platform, rapidly assembles spheroids to mimic native tissue architecture. Its intraoperative application in bone shows near-complete calvarial defect repair in rats and fabrication of scalable cartilage.
Introduction
Three-dimensional (3D) bioprinting has been making a progressive impact on medical sciences, which has great potential to facilitate the fabrication of functional tissues not only for transplantation but also for disease modeling and drug screening. It offers great precision for the spatial placement of cells, which is crucial for guiding tissue repair and regeneration1. Despite the significant progress in bioprinting of cells, the standard approach of encapsulating cells within a hydrogel bioink faces major limitations. One key challenge is achieving physiologically-relevant cell densities (100-500 million cells/mL), which is essential for effective tissue repair and regeneration2–4. However, current bioprinting techniques struggle to achieve high cell densities seen in native tissues. For instance, extrusion-based bioprinting (EBB) supports higher densities but often compromises cell viability due to shear stress5. Inkjet bioprinting is constrained by low-viscosity bioinks that limit the cell concentration6. Laser-assisted bioprinting is slow, yields lower cell viability compared to other methods and better suited to small-scale constructs, limiting its use for larger tissues7. Light-based bioprinting usually requires lower cell densities for effective curing of photo-crosslinkable bioinks8, while microvalve bioprinting is limited to bioinks within a narrow range of viscosities and cell concentrations, which increases the risk of clogging9. These limitations hamper the success in development of functional tissues as the resulting bioprinted constructs lack sufficient cell densities. Indeed, most hydrogel bioinks require a compromise between cell density and polymer density to achieve printability, often resulting in suboptimal conditions for the intended application.
In response to these challenges, tissue spheroids are considered a promising candidate. Spheroids are cellular aggregates that have been utilized in tissue fabrication due to their advantages, including native-like cell density and the capability to secrete substantial levels of extracellular matrix (ECM) components with effective communication among cells in a 3D native-like microenvironment10. Due to these advantages, spheroids have been considered as building blocks for bioprinting a variety of tissues10. Spheroid density and spatial arrangement are critical factors in achieving native-like tissue physiology, as they ensure the precise arrangement and high cell concentration necessary for effective tissue formation11. Several bioprinting techniques, each with specific advantages, have been explored for spheroid-based approaches; however, certain challenges limit their applications. For example, EBB offers high throughput by randomly mixing spheroids within a bioink and extruding them into various architectures, but this exposes cells to substantial shear stress and lacks control over the precise number and placement of spheroids5. The Kenzan method, which uses needle arrays for spheroid placement, faces limitations due to its low throughput as it assembles spheroids one at a time, often causing damage, and the fixed needle arrangement restricts its versatility12. Similarly, droplet-based bioprinting (DBB) is constrained by droplet formation, bioink viscosity, and limited precision, making it less suitable for applications requiring high spatial resolution or scalability13. Other spheroid assembly strategies, such as magnetic and acoustic methods, offer innovative approaches but come with challenges14. For example, while the magnetic technique enables spatial patterning of organoids, it requires the use of biocompatible magnetic particles, which may not be consistent across different cell types. Additionally, this method is low-throughput, as it typically manipulates one spheroid at a time, requires specialized molds, and may impact spheroid shape fidelity and viability due to magnetic forces15. On the other hand, the acoustic technique offers non-invasive, high-throughput spheroid patterning. However, it relies on acoustic nodes, which limits the ability to create fully compact cellular structures and restricts interactions between spheroids. It limits the flexibility needed for achieving desired spatial arrangements in complex geometries16. Previously, a high-precision technique, known as aspiration-assisted bioprinting (AAB)17 was introduced, where, an aspiration force is applied to position a biologic (e.g., cells, tissue spheroids or strands) during the bioprinting process. This technique enables picking and placement of biologics ranging from 80 to 800 μm in size into or onto a gel substrate with minimal cellular damage ( > 90% cell viability) and achieving high positional precision ( ~ 11% with respect to spheroid size). AAB has recently inspired applicability in magnetic lifting of neural organoids for the construction of assembloids15, 3D printing of living moving organisms (i.e., beetles18) and bioprinting of high cell-density tissues (i.e., cartilage19 and bone20) and disease models (post-myocardial infarction scarring)3. However, the major limitation of this technique is its reliance on bioprinting one spheroid at a time. While this one-at-a-time approach allows for multiple iterations in the deposition of spheroids to scale up, it significantly prolongs the bioprinting process ( ~ 20 sec/spheroid), similar to other spheroid bioprinting techniques21, which poses major challenges to increasing the scalability of fabricated tissues consistently and efficiently.
In this work, we address a long-standing problem in 3D bioprinting of spheroids and demonstrate a technology named, HITS-Bio (High-throughput Integrated Tissue fabrication System for Bioprinting). HITS-Bio represents a significant advancement in rapid bioprinting of spheroids for scalable tissue fabrication. This technique enabled the bioprinting of scalable tissues via precise positioning of spheroids (and also applicable to organoids) in a high-throughput manner at an unprecedented speed (an order of magnitude faster than the existing techniques) with high cell viability ( > 90%) using a digitally controlled nozzle array (DCNA) for the patterning and spatial distribution of several spheroids simultaneously. The capacity of the DCNA platform to pattern several spheroids at a time enabled the rapid creation of tissues, thus increasing the fabrication rate by a factor of n (the number of nozzles in DCNA) proportionally to the number of nozzles used. Herein, we presented bioinks to support spheroids during bioprinting, analogous to assembling building blocks where the bioink acts as cement and spheroids serve as bricks. To demonstrate the practical application of this technology, we first focused on calvarial bone regeneration, where critical size defects were repaired through intraoperative bioprinting (IOB) of osteogenically-committed bone spheroids. The study introduced IOB with spheroids, enabling on-demand tissue fabrication and reducing surgery time. It used combinatorial micro-RNA (miR) technology to achieve osteogenic differentiation of spheroids, and HITS-Bio enabled simultaneous or sequential aspiration and bioprinting of miR-transfected spheroids on demand. Moreover, the potential of HITS-Bio in the context of scalable tissue fabrication was exemplified through the successful creation of cm3 cartilage tissue constructs, which were precisely assembled using ~600 chondrogenically committed spheroids under 40 min per construct, representing a scale of fabrication with a high efficiency that surpasses the capabilities of current bioprinting technologies.
Results
Working mechanism and characterization of the HITS-Bio process
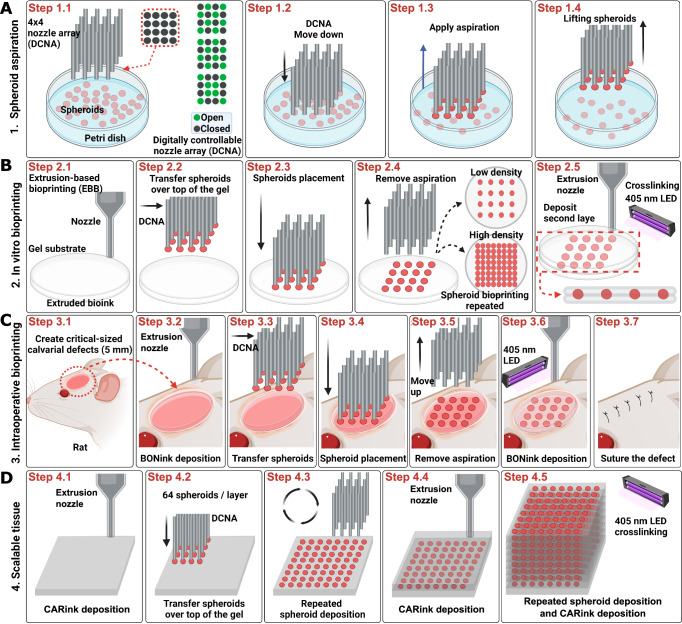
The HITS-Bio platform featured a facile assembly inside a biosafety hood due to its compact footprint, designed as a table-top system equipped with various accessories such as cameras for detailed observation and analysis. The platform had three main components: a multinozzle DCNA (Supplementary Fig. 1), a high-precision XYZ linear stage to move DCNA in 3 axes (X, Y, and Z), and an extrusion head to deposit a gel substrate (Supplementary Fig. 2). It was operated by a custom-made software interface with a control algorithm (Supplementary Movie 1, Supplementary Figs. 3 and 4). To visualize the bioprinting process in real-time and verify the actual position of DNCA in 3D, three microscopic cameras for the isometric, bottom, and side views were integrated (Supplementary Fig. 2). DCNA facilitated picking up multiple spheroids by controlling the aspiration pressure in the selectively opened nozzle depending on the design of applications, as demonstrated in Fig. 1A and Supplementary Fig. 5. To lift spheroids, DCNA was moved to a Petri dish, where spheroids were suspended in a culture medium. The spheroid transfer process was specifically designed to function within the culture medium, eliminating the need for a viscous fluid support bath. This approach simplifies handling and avoids the challenges associated with increased shear and compression forces, ensuring the integrity of spheroids during transfer. After spheroids were successfully aspirated to the end of the selectively opened DCNA, confirmed by the bottom view camera, DCNA with spheroids was gently lifted from the spheroid chamber (Fig. 1A, Step 1.4). As a substrate to place the spheroids, a bioink was extruded (Fig. 1B, Step 2.1). Next, DCNA loaded with spheroids was transferred over the substrate (Fig. 1B, Steps 2.2 and 2.3). Once the spheroids were in contact with the substrate, aspiration pressure was cut off to deposit the spheroids (Fig. 1B, Step 2.4). We exemplified the utility of HITS-Bio in three different configurations. For in-vitro bioprinting of single-layer-spheroids, low density (composed of 16 spheroids) and high density (composed of 64 spheroids) were bioprinted by repeating the process as depicted in Fig. 1A, B (Steps 1.1-1.4 and Steps 2.2-2.4). After the spheroid placement, another layer of the bioink was deposited on top of the bioprinted spheroids to envelop them, followed by photo-crosslinking using a 405 nm light-emitting diode (LED) light source for 1 min (Fig. 1B, Step 2.5). For IOB (Fig. 1C), critical-sized calvarial defects (Step 3.1) were created. Firstly, the bone ink (BONink) was extruded at the defect area (Step 3.2) and DCNA loaded with spheroids was positioned over a defect (Step 3.3). The spheroids were deposited at two different spheroid densities (low − 16 spheroids and high − 64 spheroids) using DCNA (Step 3.4–3.5). Then, another layer of the BONink was extruded over the spheroids (Fig. 1C, Step 3.6), followed by photo-crosslinking and suturing of the skin (Step 3.7). For scalable tissue bioprinting (Fig. 1D), scalable cartilage tissues (SCTs) were created, using a cartilage ink (CARink). Firstly, the CARink was extruded (Step 4.1), followed by the precise placement of 64 chondrogenic spheroids (Steps 4.2–4.3). This iterative process (Steps 4.2–4.4) was repeated nine times to assemble a construct comprising nine stacked layers and a total of 576 spheroids, followed by photo-crosslinking (Step 4.5).
Fig. 1. Schematic of the HITS-Bio process.
A Spheroid loading with DCNA, which selectively enabled or disabled aspiration according to a user-defined design in an iterative manner. B In vitro bioprinting of spheroids including extruded bioink via EBB and spheroid placement using DCNA, having different spheroid loading densities (i.e., low density – 16 spheroids and high density – 64 spheroids) in an iterative manner. C IOB of spheroids into a rat calvarial defect, where spheroids were sandwiched between the extruded BONink layers. D A 1 cm3 of scalable cartilage tissue bioprinted with CARink and ~600 spheroids. Created in BioRender. Ozbolat, I. (2024) BioRender.com/a30b873.
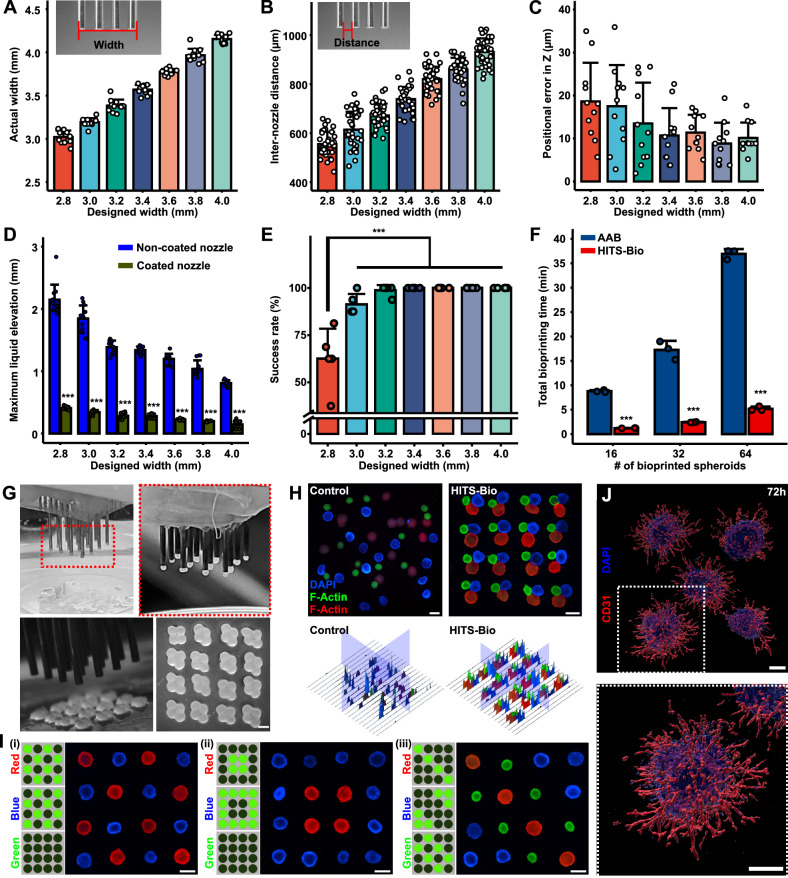
DCNA consisted of stainless-steel needles with predefined spaces between them, which were assembled by precisely stacked multiple acrylic plates (Supplementary Fig. 1). A micro-manufactured 4 × 4 nozzle array with different configurations (various designs with widths from 2.8 to 4.0 mm) was prepared as shown in Supplementary Fig. 6A. To calculate the inaccuracies during micro-manufacturing of DCNA, bottom (Supplementary Fig. 6B) and side (Supplementary Fig. 6C) views of DCNA were captured. These views allowed the measurement of inter-nozzle distance and accuracy of each nozzle against a reference point. The range of widths was determined based on the area, where spheroids could be bioprinted in a circular area with a diameter of 5 mm. As shown in Fig. 2A, B, the actual width and inter-nozzle distance of DCNA were measured and compared with the designed width and inter-nozzle distance, respectively. Considering the error expected during laser cutting, DCNA was micro-manufactured with less than 5% error. The positional errors in XY (Supplementary Fig. 6E) and Z (Fig. 2C) were less than 5% for 300 µm spheroids tested in this study. We observed that lifting spheroids from the culture medium into air resulted in entrapment of liquid (culture medium) between nozzles and its elevation from its surface acted upon by capillary forces due to its surface tension between the liquid and DCNA (Supplementary Fig. 6F, Supplementary Movie 2). The elevated liquid hindered successful spheroid lifting and placement. For example, comparing two different sizes of DCNA (2.8 v.s. 3.4 mm), higher liquid elevation was observed in the closely packed 2.8-mm DCNA, where spheroids experienced resistance in their lifting due to the elevated liquid. In contrast, the 3.4-mm DCNA showed a much lower height of liquid elevation, which resulted in successful spheroid lifting (Supplementary Movie 3). Thus, optimizing parameters, such as the inter-nozzle distance and liquid elevation, was crucial in relation to the likelihood of successful spheroid lifting.
Fig. 2. Analysis of HITS-Bio performance.
Measurement of (A) end-to-end nozzle array width (n = 12 independently prepared DCNA), (B) inter-nozzle distance (n = 36, each data point represents three inter-nozzle distances of 12 independently prepared DCNA), and (C) positional error in the Z-axis (n = 11 independently prepared DCNA). D Maximum liquid elevation in DCNA (n = 12, unpaired two-sided Student’s t-test, p = 1.43 × 10−17, 1.76 × 10−17, 1.83 × 10−20, 1.17 × 10−24, 6.01 × 10−22, 4.96 × 10−16, 3.28 × 10−19 from left to right). E The impact of interstitial distances of the silicon-coated DCNA on the success rate of spheroid lifting (n = 5 independent experiments, one-way ANOVA). F A comparison of elapsed bioprinting time between conventional single-nozzle aspiration-assisted bioprinting (AAB) and HITS-Bio (n = 3 independent experiments, unpaired two-sided Student’s t-test, p = 5.03 × 10−7, 0.00016, 9.30 × 10−7 from left to right). G Lifted spheroids from a spheroid chamber and placed on a GM10 substrate in an iterative manner to bioprint 64 spheroids to generate a rhombus pattern. Scale bar: 300 μm. H Fluorescence images and the corresponding intensity map of three different sizes and colors (stained with DAPI (blue), F-Actin (red), F-Actin (green)) of spheroids (16 spheroids per color) using manually mixed (control) and HITS-Bio. Scale bar: 500 μm. I Selectively patterned spheroids stained with DAPI (blue), F-Actin (red), and F-Actin (green) using the DCNA platform with various configurations. Scale bar: 500 μm. Representative images were obtained from at least five independent repetitions. J Fluorescent staining of iPSC-derived vascular organoids at 72 h after bioprinting into collagen type I-Matrigel mixture for CD31 (red) and DAPI (blue) (scale bar: 200 μm). Representative images were obtained from three independent repetitions. Data are presented as mean ± SD where *p < 0.05, **p < 0.01, and ***p < 0.001. Source data are provided as a Source Data file.
During lifting, a capillary reaction was observed between nozzles, where liquid rose in a narrow gap against gravity (Supplementary Fig. 6F). At equilibrium, the upward force due to surface tension balances the downward force of the liquid weight. The liquid continues to rise until these forces are equal. As detailed in Supplementary Note 1, this relationship can be expressed mathematically, displaying that the height of the elevated liquid is inversely proportional to the inter-nozzle distance and surface tension. To reduce the height of the elevated liquid, one can either increase the inter-nozzle distance or decrease the surface tension. Subsequently, various inter-nozzle distances of DCNA were explored, showing correlation between the quantified elevated liquid length with different sizes of DCNA. Subsequently, silicon coating was applied to DCNA to lower the surface tension by reducing the surface energy. The results indicated that the liquid elevation was significantly decreased after the silicon coating on the DCNA surface (Fig. 2D). In addition, the silicon-coated DCNA was lifted smoothly from the liquid, as opposed to the non-coated DCNA, which tended to drag the liquid (Supplementary Movie 2). Furthermore, as the inter-nozzle distance decreased, the liquid elevation increased. Thus, the success rate of spheroid lifting decreased with reduced designed width due to the liquid elevation (Fig. 2E). In other words, the silicon-coated DCNA with a 3.4-mm designed width showed complete success of spheroid lifting (Fig. 2E and Supplementary Movie 3) and was selected for further consideration in the HITS-Bio process.
To test the performance of HITS-Bio with the optimized DCNA, we benchmarked it against a single nozzle AAB system (Fig. 2F). The results indicated that HITS-Bio was highly efficient when a large number of spheroids were needed to be bioprinted. Using DCNA with 16 nozzles (Fig. 2G), the bioprinting process for 16 spheroids took less than 1 min, which was significantly faster than the conventional AAB system, which took nearly an order of magnitude longer (Fig. 2F) despite the AAB system being upgraded for its bioprinting speed (with full automation) compared to previous publications17,19. As the number of bioprinted spheroids increased, the disparity in total bioprinting time between AAB and HITS-Bio increased exponentially (Fig. 2F and Supplementary Movie 4). To evaluate the performance of HITS-Bio for the patterning of spheroids, various types of spheroids were selectively lifted and patterned on a gelatin methacryloyl (GM) substrate. For example, we compared HITS-Bio with manual loading of spheroids (Fig. 2H). In this regard, a total of 48 spheroids (16 green (~350 μm in diameter), 16 blue (~425 μm in diameter) and 16 red (~500 μm in diameter)) per sample were utilized (Supplementary Fig. 7C). To prepare the control group, spheroids were mixed in 10% GM using a pipette in a 3D-printed mold (Supplementary Fig. 7A, B). For HITS-Bio, spheroids were bioprinted into the GM substrate in a triplet arrangement, where triplets were patterned with precise control on their positioning while the manually-loaded spheroids resulted in random distribution with a lack of control, not just in X- and Y-axis but also Z-axis as most of the spheroids were not in the focal plane (Fig. 2H). HITS-Bio showed 100% spheroid loading efficiency, meaning all spheroids were bioprinted and loaded into GM successfully regardless of their size. In contrast, 80-85% efficiency was attained in the case of manual loading (Supplementary Fig. 7D). The reduced efficiency in manual loading was due to the fact that spheroids were sticking to the wall of the pipette tip during manual deposition (Supplementary Fig. 7B). This was further validated by Hematoxylin and Eosin (H&E) images of the manually loaded group, which revealed uneven spheroid distribution and the presence of air bubbles within the constructs, likely caused by the mixing of spheroids into the viscous bioink (Supplementary Fig. 8). One would expect that the efficiency would further decrease as the number of spheroids increased. Therefore, using HITS-Bio, the number and type of spheroids could be controlled to precisely position them with 100% loading efficiency. To further confirm the spatial placement of spheroids using HITS-Bio, the three types of spheroids were patterned in various configurations (Fig. 2I). Herein, based on the desired location of spheroids, the corresponding valves in DCNA were digitally activated and switched on, and the spheroids were loaded on the nozzles selectively as shown in Supplementary Movie 5. The spheroids were released in the designated positions on demand. Therefore, HITS-Bio facilitated the successful patterning of different types of spheroids at desired positions regardless of their size without loss of spheroids.
To test the broader applicability of HITS-Bio to soft and fragile spheroids and organoids, a wide range of samples made of different cell types were screened. Our findings indicate that spheroids with an elastic modulus greater than ~50 Pa were printable while those below 40 Pa were not (Supplementary Fig. 9). The transfected osteogenic and chondrogenic spheroids had an elastic modulus of 978 ± 96 Pa and 641 ± 110 Pa, respectively. Along with various spheroids, elastic modulus of induced pluripotent stem cell (iPSC)-derived vascular organoids was also tested, which was measured to be 133 ± 20 Pa and fell within the range of printable elastic moduli. The maximum elastic modulus limit for printability was not assessed, as it was beyond the scope of this study. Additionally, it is important to highlight that the method used in this study measured the aspirated length under a fixed aspiration force. While there are specialized instrumentation22,designed for precise stiffness measurements of spheroid based on force-displacement curves, we opted for the aspirated length-based stiffness measurement due to its direct relevance for our specific application involving aspiration forces.
Moreover, the system’s ability to precisely deposit spheroids in tightly packed, fully cellular architectures in order to better replicate tissue-specific structures was demonstrated. Human dermal fibroblast (HDF) spheroids (530 µm) were arranged in rows via bioprinting them onto a pre-crosslinked GelMA (10%) substrate, with gaps filled by smaller spheroids of HDF with human umbilical vein endothelial cells (HUVEC), HDF/HUVEC (300 µm) (Supplementary Fig. 10A). These spheroids fused to form a continuous, densely packed structure, highlighting the potential for creating vascularized tissues, as tdTomato+ HUVECs successfully migrated and formed pre-vascularized-like structures within fused spheroids. This was further supported by Von Willebrand Factor (VWF) and fibronectin (FN) staining (Supplementary Fig. 10B), where VWF staining was used to identify and confirm the presence of endothelial cells and the formation of pre-vascularized-like structures. The integration of HDF and HDF/HUVEC spheroids into a fully cellular architecture was clearly visible at the interface, supporting HITS-Bio’s capability to generate complex tissue constructs. Furthermore, examples were demonstrated for optimization of area-filling models with spheroids of varying sizes to maximize the filled area fraction on the GelMA substrate. As shown in Supplementary Fig. 11, area filling of 37.03% was achieved using large spheroids (~735 µm in diameter), which increased to 63.71% when large and small spheroids (~300 µm) were mixed. This further improved to 86.91% using small spheroids alone and ultimately reached 98.11% by combining two different sizes (~530 µm and ~300µm). These tailored patterns were designed to optimize cellular density while strategically integrating filler hydrogel substrate, utilizing various nozzle configurations to precisely balance cellular content and filler hydrogel distribution, which can be further expanded to 3D volume-filling models.
To demonstrate the further applicability of HITS-Bio, abovementioned iPSC-derived vascular organoids were also bioprinted into a composite hydrogel made of collagen type I (2 mg/mL)-Matrigel (mixed in 4:1 ratio) successfully without any visible structural damage to organoids. These organoids gradually sprouted capillaries after 72 h of culture (Fig. 2J and Supplementary Fig. 12), further validating the suitability of HITS-Bio for bioprinting organoids.
As shown in Supplementary Fig. 13, the capability of HITS-Bio to create complex architectures was also demonstrated by assembling a pyramid construct composed of 171 spheroids, each varying in size and color-tagged for distinction. Specifically, the bottom layer comprised 121 spheroids (300 µm), followed by a 2nd layer of 36 spheroids (540 µm), a 3rd layer containing 9 spheroids (735 µm) and 4 spheroids (540 µm), and the top layer featuring a single 445 µm-spheroid. This intricate structure was meticulously assembled by precisely controlling different nozzles of DCNA. The ability to accurately position and layer this number of spheroids, with such varied sizes and specific arrangements, is not feasible using conventional EBB or casting/molding techniques. This demonstrates HITS-Bio’s potential not only for constructing intricate, multi-layered structures but also for creating complex tissue architectures that require highly controlled spatial organization and varied cellular compositions.
In vitro development and characterization of bioinks as a substrate for spheroid bioprinting
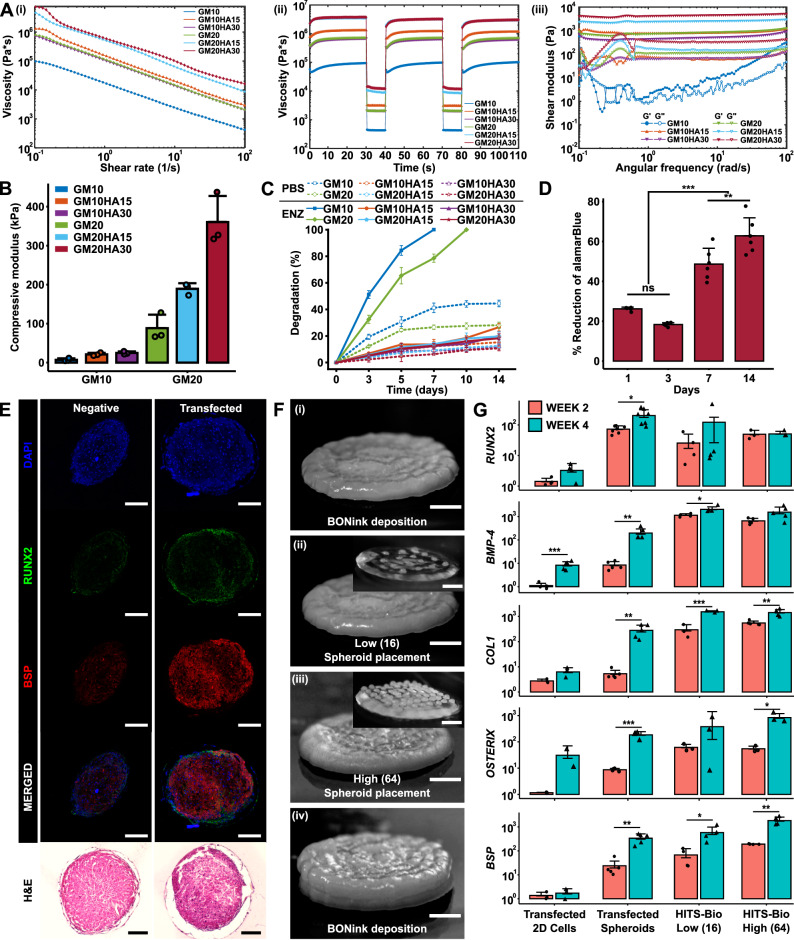
To develop bioinks akin to versatile cement-like substrate capable of assembling spheroids (bricks) into structured building blocks for bioprinting, components were chosen for their compatibility with HITS-Bio in terms of their extrusion, the ability to form a slightly adhesive cement-like substrate to retain spheroids in desired patterns, and to enhance ECM formation. The bioink base was composed of GM for its biocompatibility and tunable mechanical properties, nanohydroxyapatite (HA) to enhance osteoconductivity, and β-glycerophosphate disodium salt hydrate (β-GP) for thermal gelation. Hyaluronic acid sodium salt (HyA) was added to improve cell adhesion and proliferation, fibrinogen (Fib) for its role in promoting cell-matrix interactions, and lithium phenyl (2,4,6-trimethylbenzoyl) phosphinate (LAP) as a photoinitiator to enable light-induced crosslinking, ensuring structural integrity and stability of the bioink. These components were mixed in various ratios to obtain composite bioinks, where the bone ink (Supplementary Fig. 14A) was formulated with the same components as the cartilage ink (Supplementary Fig. 14B), but with the addition of β-GP and HA. GM at a concentration of 10% was denoted as GM10, and at 20% it was labeled GM20. Similarly, HA at 15% concentration was designated as HA15, and at 30%, it was labeled HA30. To assess the bioinks, their rheological properties, mechanical behavior, and degradation profile were evaluated. Rheological analysis was performed to test the flow characteristics of the bioinks. The viscosity profile, determined through a flow sweep across varying shear rates from 0.1 to 100 s−1, revealed a shear thinning behavior in all bioink formulations, making them suitable for extrusion (Figure 3Ai). Bioinks with higher concentrations of GM (GM20) and HA (HA30) exhibited higher viscosities compared to GM10 and HA15 formulations, respectively. The recovery sweep test (Figure 3Aii) showed that the viscosity of all samples recovered rapidly, suggesting that the bioinks exhibited a self-healing property. The viscosity profile of the bioinks during the recovery followed the same trend as that of the flow sweep. Further, the frequency sweep showed that storage modulus (G′) was higher than loss modulus (G″) in the frequency range of 0.1–100 rad/s (Figure 3Aiii). G′ and G″ were independent of frequency for the GM-alone samples, while they were slightly dependent for the composite bioinks. The compression testing of bioinks exhibited a non-linear stress-strain curve. Among the GM20 samples, GM20HA30 exhibited the highest compressive modulus at 360.7 ± 66.6 kPa, followed by GM20HA15 at 189.2 ± 14.4 kPa, and GM20 at 88.3 ± 34.7 kPa. Conversely, GM10 samples displayed lower compressive moduli, with values of 24.7 ± 3.7 kPa, 21.2 ± 4.2 kPa, and 7.2 ± 3.8 kPa for GM10HA30, GM10HA15, and GM10, respectively (Fig. 3B). Although GM20 samples exhibited a higher compressive modulus, they demonstrated a lower fracture strain ranging from 40-45%, whereas the GM10 samples fractured at a higher strain of 65-70% (Supplementary Fig. 14C).
Fig. 3. In vitro characterization of bioinks.
A Rheological characterization of BONink and CARink at different concentrations of HA and GM (n = 3 independent rheological characterizations, data are presented as mean): (i) Flow curves of bioinks from the rotational test at a shear rate ranging from 0.1 to 100 s-1 showing mean plots. (ii) Recovery behavior and viscosity measurements at five intervals at alternating shear rates (shear rate of 0.1 s-1 for 30 s and 100 s-1 for 10 s) showing mean plots. (iii) Measured shear modulus from frequency sweeps at an angular frequency ranging from 0.1 to 100 rad s-1. B Compressive modulus (n = 3 independent samples), (C) % degradation of samples for 14 days in enzyme (ENZ) and PBS (n = 3 independent samples), and (D) % reduction of alamarBlue to assess cell viability with GM20HA30 for 14 days (n = 6 biologically independent samples, one-way ANOVA, Day 1 vs. Day 3 p = 0.140902, Day 7 vs. Day 14 p = 0.003054, all other shown comparisons p < 0.0001). E Histomorphometric characterization of non-transfected and miR-(196a-5p + 21) co-transfected spheroids stained for RUNX2, BSP, and H&E at Week 4 (scale bar: 100 µm). F Steps involved during the bioprinting process for in vitro fabrication of bone constructs at two different spheroid densities: (i) BONink deposition, (ii) bioprinted spheroids at low (16 spheroids) and (iii) high density (64 spheroids), and (iv) overlaying bioprinted spheroids with another layer of BONink. Scale bar: 1 mm. Inset images demonstrate bioprinted spheroids with low and high densities on a transparent gel (CARink) for clear visualization. Scale bar: 1 mm. G Quantification of RUNX2 (n = 4, 7, 4, 3 biologically independent samples (from left to right), unpaired two-sided Student’s t-test, p = 0.06978, 0.01004, 0.42355, 0.92376 from left to right), BMP-4 (n = 5, 5, 4, 5 biologically independent samples (from left to right), unpaired two-sided Student’s t-test, p = 0.00092, 0.00505, 0.03519, 0.06384 from left to right), COL1 (n = 3, 5, 3, 5 biologically independent samples (from left to right), unpaired two-sided Student’s t-test, p = 0.08064, 0.00395, 0.00069, 0.00178 from left to right), OSTERIX (n = 2, 4, 3, 3 biologically independent samples (from left to right), unpaired two-sided Student’s t-test, p = 0.27645, 0.00041, 0.26759, 0.02095 from left to right), and BSP (n = 3, 5, 4, 4 biologically independent samples (from left to right), unpaired two-sided Student’s t-test, p = 0.54175, 0.00147, 0.04690, 0.00367 from left to right) gene expression of bioprinted bone. Unless otherwise noted, data are presented mean ± SD where *p < 0.05, **p < 0.01, ***p < 0.001, and ns not significant. Source data are provided as a Source Data file.
The degradation test results indicated a decrease in enzymatic degradation rate for GM samples upon HA incorporation, with complete degradation occurring in ~7 days for 10% GM and 10 days for 20% GM (Fig. 3C). The HA-based composites demonstrated limited degradation, with only ~18% for GM20HA30 and ~20% for GM20HA15 over 14 days. Similarly, in GM10 samples, degradation amounted to ~20% for GM10HA30 and ~27% for GM10HA15. Conversely, composite bioinks in phosphate-buffered saline (PBS) only underwent 12–15% degradation, while only GM samples in the same medium showed higher degradation levels of 30-40%. After characterizing bioinks for their rheological, mechanical, and degradation properties, the GM20 and GM20HA30 composite were selected and named CARink (cartilage ink) and BONink (bone ink), respectively, for further investigations. GM10 and its composites exhibited poor printability and mechanical properties, whereas GM20HA15 demonstrated inadequate mechanical strength.
After developing the BONink, the metabolic activity of human adipose-derived stem cells (hADSCs) in the BONink was assessed over time, which revealed significantly higher proliferation observed on Day 14 compared to Day 7 (p ≤ 0.01), and lower levels on Days 1 and 3 (p ≤ 0.001) (Fig. 3D). These results demonstrate the biocompatibility of the BONink, affirming its suitability for bone tissue engineering applications. Osteogenically-committed spheroids were then formed using miR-transfected hADSCs as shown in Supplementary Fig. 15A. Specifically, hADSCs were transfected with miR-196a-5p, or miR-21, or in combination (miR-(196a-5p + 21)). The effect of transfection was evaluated via gene expression (Supplementary Fig. 15B) and histomorphometric characterization (IHC and H&E) (Fig. 3E and Supplementary Fig. 16) of osteogenic markers at Weeks 2 and 4. In our preliminary experiments, transfection was assessed in 2D conditions, where hADSCs with and without the miR-transfection were evaluated, and the differentiation in established osteogenic medium was taken as positive control. The results of preliminary tests showed upregulated osteogenic genes (RUNX2, BMP-4, COL-1, OSTERIX, and BSP) when hADSCs were co-transfected with miR-(196a-5p + 21) compared to miR-196a-5p alone and non-transfected hADSCs (negative control) (Supplementary Fig. 15B). The expressions were significantly higher (p ≤ 0.05) at Week 4 as compared to Week 2 and comparable to the positive control. Additionally, IHC assessment of RUNX2 and BSP markers showed intense staining in the miR-(196a-5p + 21) transfected hADSCs, which was comparable to the positive control and higher than the miR-196a-5p alone and non-transfected hADSCs (Fig. 3E and Supplementary Fig. 16). These results were in agreement with real-time polymerase chain reaction (qPCR) results. Further, H&E staining showed a strong dark purple color indicating bone mineralization in the positive group (Supplementary Fig. 16). The transfected group also showed darker color compared to the negative control and miR-196a-5p alone transfected hADSCs. Overall, miR-transfection supported the differentiation of hADSCs into an osteogenic lineage. Based on these results, for the rest of this study, the co-transfection of miR-(196a-5p + 21) was used for osteogenic differentiation of hADSCs for use in spheroids as building blocks for bone tissue fabrication.
Fig. 3F demonstrates the steps for the fabrication of single-spheroid-layer bone constructs with two different densities (16 or 64) of spheroids. Spheroids (350 µm) were placed with a targeted inter-spheroid distance of 670 and 160 µm for the low density (16) and high density (64) spheroids, respectively. The BONink was first extruded in a spiral pattern with 100% infill to lay down a gel substrate with a diameter of ~ 5 mm prior to spheroid placement (Figure 3Fi). On the BONink, spheroids were bioprinted at low (Figure 3Fii) and high (Figure 3Fiii) densities. After the spheroid placement, another layer of BONink was extruded to overlay spheroids (Figure 3Fiv). It is pertinent to note that the BONink deposition did not displace the previously placed spheroids considerably (Supplementary Movies 6 and 7). The final constructs were photo-crosslinked with a 405 nm light source for 1 min and incubated thereafter. qPCR results indicated increased expression of osteogenic markers in the transfected non-bioprinted and bioprinted (low or high density) spheroids (Fig. 3G). There was a significant increase across all groups at Week 4 compared to Week 2. As shown in Supplementary Table 1, the results revealed that RUNX2, an early osteogenic marker, increased up to 4.8-fold, with the highest levels observed in the low-density group. Similarly, BMP-4, another early marker, showed a maximum increase of 23.3-fold in transfected spheroids. COL1, involved in both early and late stages of osteogenic differentiation, increased by up to 52.1-fold in transfected spheroids. OSTERIX, an intermediate to late-stage marker, increased up to 15.4-fold in the high-density spheroids group. Finally, BSP, a late-stage marker crucial for bone strength, increased by 14.1-fold in the transfected spheroids at Week 4 compared to Week 2. These results indicate that the bioprinted constructs, particularly those with lower spheroid density, show increased early osteogenic activity (RUNX2 and BMP-4). In contrast, high-density spheroids and transfected spheroids exhibit significant increases in both intermediate (OSTERIX) and late-stage (BSP) markers, suggesting more mature osteoblast activity and active-matrix mineralization. Furthermore, the gene expression results were corroborated by the IHC staining, where RUNX2 and OSTERIX markers exhibited expression profiles consistent with their roles in osteoblast differentiation (Supplementary Fig. 17). RUNX2 displayed a distinct increase in staining in the low-density group at Week 4 while the high-density group maintained its initial high intensity. On the other hand, OSTERIX showed intense staining in bioprinted constructs particularly in the high-density group, indicating successful active osteoblast differentiation. The IHC staining patterns aligned well with the observed fold-changes in gene expression, reinforcing the dynamic nature of osteogenic differentiation and the sequential involvement of related genes.
Intraoperative bioprinting for bone regeneration in calvarial defects
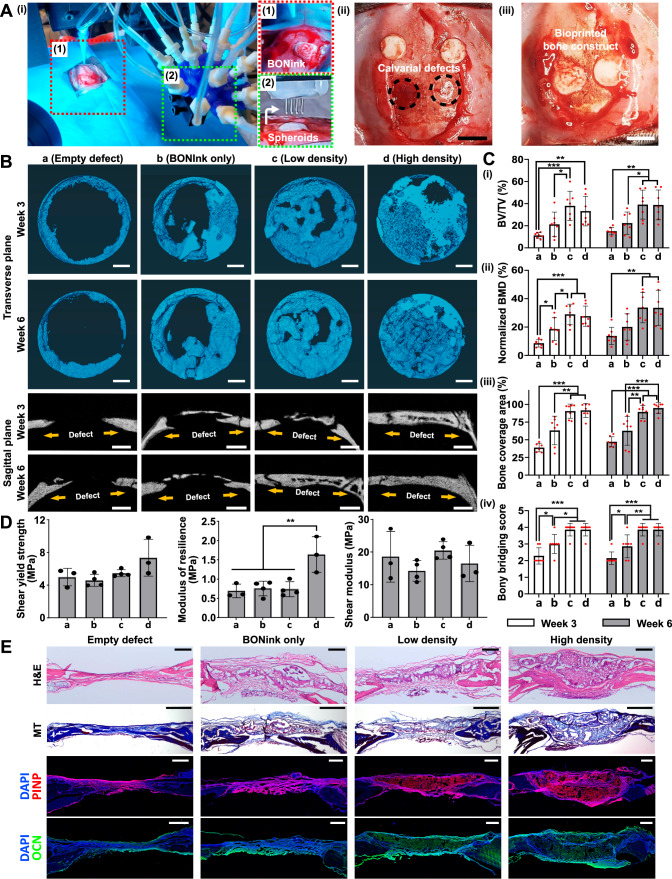
Following the in vitro assessment of physical properties, printability, cytocompatibility, and osteogenic potential of the BONink, we performed animal studies using a rat model. Minimizing the animal numbers, two critical-sized calvaria defects, each 5 mm in diameter, were created on either side of the parietal bone on the rat skull and IOB was performed under aseptic surgical settings (Fig. 4A and Supplementary Fig. 2A). Akin to in vitro settings, the BONink was extruded at the defect site in a spiral pattern with 100% infill (Supplementary Movie 8). Four groups were considered including (i) empty defect (control), (ii) BONInk only, (iii) low density (16 spheroids), and (iv) high density (64 spheroids). Post IOB, bone regeneration was evaluated at 3 and 6 weeks. Micro-computed tomography (µCT) results (Fig. 4B) revealed that the high-density group consisting of 64 spheroids exhibited superior bone regeneration, closing almost the entire defect in Week 6. In contrast, bone regeneration in the empty control group was mainly confined to the periphery of the defects. As a quantitative metric to assess the efficacy and extent of bone tissue formation, bone volume to total volume (BV/TV) was calculated, which revealed significantly higher bone regeneration of ~38 and 33% in Week 3, and ~39 and 39% in Week 6 for low- and high-density group, respectively, compared to the BONink-only (p ≤ 0.05) and empty group (p ≤ 0.01) (Fig. 4Ci). Moreover, the normalized bone mineral density (BMD), reflecting the density of regenerated bone normalized to the native bone density, exhibited ~29 and 28% at Week 3, and ~34 and 34% at Week 6 for low and high-density groups, respectively, compared to the BONink-only (p ≤ 0.05) and empty group (p ≤ 0.01) (Figure 4Cii). Additionally, the bone coverage area (%) for the low-density and high-density groups was ~90 and 91% at Week 3, and ~88 and 96% at Week 6, respectively (Figure 4Ciii). Moreover, the maximum intensity projections generated from µCT data (Fig. 4B) of each group were used for the scoring (1–4) of bony bridging across the defect. The results showed significantly higher scores for spheroid-involved groups compared to empty defect and BONink-only groups at both Weeks 3 and 6 (Figure 4Civ). Furthermore, given the significant role of mechanical properties in bone strength, the regenerated bone samples were subjected to a push-out test following their retrieval after Week 6 (Fig. 4D). The high-density group exhibited a higher shear yield strength, ~1.5 times greater than other groups, indicating enhanced resistance to plastic deformation or failure. Similarly, the modulus of resilience in the high-density group was significantly higher, ~2.4 times greater than the other groups (p ≤ 0.01), signifying higher energy absorption capacity up to the point of yielding. However, there was no significant difference observed among the groups in the shear modulus, representing the material’s resistance to shear deformation.
Fig. 4. IOB of bone constructs into critical-sized rat calvarial defects for bone regeneration.
A HITS-Bio setup under surgical settings. Inset images (1–2) demonstrate deposited BONink and spheroid placement, (ii) created calvarial defects ( ~ 5 mm in a diameter), and (iii) bioprinted bone constructs with BONink and spheroids. Scale bar: 5 mm. B Visualization of newly regenerated bone in calvarial defects with transverse and sagittal planes at Weeks 3 and 6 via µCT, including empty, BONink only, low-density and high-density groups. Scale bar: 1 mm. C Relevant quantification for bone regeneration within the defect examined at Weeks 3 and 6 (n = 7 independent defects, one-way ANOVA, all other shown comparisons p < 0.0001), including (i) BV/TV (new bone volume to total bone volume, %) (Week 3: a vs. b p = 0.35139, a vs. c p = 0.00074, a vs. d p = 0.00545, b vs. c p = 0.04183, b vs. d p = 0.20350, c vs. d p = 0.85065, Week 6: a vs. b p = 0.60072, a vs. c p = 0.00186, a vs. d p = 0.00212, b vs. c p = 0.03614, b vs. d p = 0.04048, c vs. d p = 0.99995), (ii) normalized BMD (bone mineral density, %) (Week 3: a vs. b p = 0.045, b vs. c p = 0.03277, b vs. d p = 0.07177, c vs. d p = 0.98269, Week 6: a vs. b p = 0.63421, a vs. c p = 0.00473, a vs. d p = 0.00528, b vs. c p = 0.07204, b vs. d p = 0.07910, c vs. d p = 0.99996), (iii) bone coverage area (%) (Week 3: a vs. b p = 0.06611, b vs. c p = 0.00656, b vs. d p = 0.00464, c vs. d p = 0.99893, Week 6: a vs. b p = 0.11033, b vs. c p = 0.00259, b vs. d p = 0.00032, c vs. d p = 0.83536) (The outlier was not included in the data plot), and (iv) scores for bony bridging (Week 3: a vs. b p = 0.03791, b vs. c p = 0.01013, b vs. d p = 0.01013, c vs. d p = 1.0, Week 6: a vs. b p = 0.04481, b vs. c p = 0.00326, b vs. d p = 0.00326, c vs. d p = 1.0). D Mechanical properties of the retrieved defect area 6 weeks after the surgery (n = 3, 4, 4, 3 independent defects (from left to right), one-way ANOVA) (Shear yield strength: a vs. b p = 0.97205, a vs. c p = 0.95577, a vs. d p = 0.14142, b vs. c p = 0.74719, b vs. d p = 0.05602, c vs. d p = 0.23294) (Modulus of resilience: a vs. b p = 0.98816, a vs. c p = 0.99588, a vs. d p = 0.00672, b vs. c p = 0.99957, b vs. d p = 0.00692, c vs. d p = 0.00598) (Shear modulus: a vs. b p = 0.65248, a vs. c p = 0.95136, a vs. d p = 0.94709, b vs. c p = 0.31149, b vs. d p = 0.92926, c vs. d p = 0.69672). E Histomorphometric characterization of sectioned defect after decalcification and stained for H&E (scale bar: 500 µm), MT (scale bar: 1 mm), and IHC (P1NP and OCN) (scale bar: 500 µm). Representative images were obtained from at least three independent repetitions. Data are presented as mean ± SD where *p < 0.05, **p < 0.01, and ***p < 0.001. Source data are provided as a Source Data file.
Decalcified sections were then stained with H&E and Masson’s trichrome (MT) to evaluate the morphology of the regenerated bone (Fig. 4E). The H&E images of the high-density group exhibited bridging of the calvarial defect and thicker regenerated bone compared to the soft tissue observed in other groups. MT staining predominantly showed soft tissue formation in the empty defect group, while both spheroid-containing groups showed signs of immature bone formation, which was increased in the high-density group. Thus, histological evaluations of the defects showed that high-density samples illustrated bone formation in 6 weeks, along with active bone healing and mineral deposition. The repair site exhibited characteristics like intramembranous bone formation, aligning with the fact that the development of calvaria is primarily associated with intramembranous ossification23. Procollagen type I N-terminal propeptide (P1NP) and Osteocalcin (OCN), synthesized and secreted by osteoblasts, are well-established markers for bone formation. IHC images revealed that OCN staining intensity was lowest in the empty and BONink-only groups, increased in the low-density group, and was highest in the high-density group (Fig. 4E). Notably, both native and newly formed bone tissues were stained with OCN, suggesting that OCN plays a crucial role not only in the formation of new bone but also in the ongoing maintenance of the existing bone matrix24. This could be attributed to OCN binding to HA, which is more prominent in areas of active bone formation. Similarly, P1NP staining followed a comparable pattern, with increased intensity in spheroid-containing groups, particularly in the high-density group, where the intensity was highest. Importantly, P1NP staining was predominantly observed in the newly formed tissue, suggesting its specific involvement in early collagen synthesis during the initial stages of bone formation25. Furthermore, higher expression levels of RUNX2 were observed in the bioprinted groups compared to the empty defect and BONink-only groups. In contrast, OSTERIX was predominantly localized in the host bone rather than the regenerated bone (Supplementary Fig. 18), suggesting that the newly formed bone was in an early developmental stage, consistent with the MT staining results. Overall, the IHC analysis for P1NP, OCN, RUNX2 and OSTERIX, offer a robust assessment of different stages of osteogenesis, from early osteoblast differentiation to collagen synthesis and mineral deposition, ensuring a thorough evaluation of new bone formation and its integration with the native bone.
Fabrication of scalable cartilage tissues (SCTs)
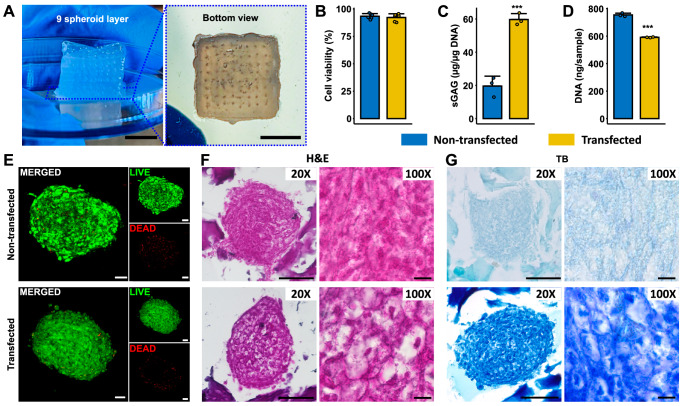
To illustrate the potential of HITS-Bio in generating volumetric tissues, we fabricated a SCTs (as shown in Fig. 1D and Supplementary Movie 9). For this purpose, the CARink (i.e., GM20) was used as a bioink along with miR-(140 + 21) co-transfected chondrogenic spheroids prepared according to Supplementary Fig. 19A, as this combination of miRs has shown promising results for cartilage regeneration in a previous study26. To fabricate the constructs, we extruded a layer of CARink, followed by the precise placement of 64 chondrogenic spheroids. This iterative process was repeated nine times to assemble a construct with a volume of 1 cm3, comprising 9 stacked tissue layers and a total of 576 spheroids taking under 40 min (Fig. 5A). SCTs were then assessed for cell viability and cartilaginous ECM formation. LIVE/DEAD staining revealed that the transfection and tissue fabrication did not impair the cell viability in spheroids, with bioprinted transfected spheroids exhibiting over 90% viability, comparable to non-transfected spheroids (Fig. 5B, E). Further, SCTs with co-transfected spheroids exhibited a significantly higher sulfated glycosaminoglycan (sGAG) content compared to the non-transfected control group (p ≤ 0.001) at Week 2 (Fig. 5C). Interestingly, the DNA content was significantly lower (p ≤ 0.001) in the transfected samples compared to the non-transfected samples (Fig. 5D). At Week 2, the identification and morphology of chondrogenic spheroids within bioprinted constructs were assessed using H&E. The staining showed the presence of spheroids in the CARink bioprinted constructs, with the chondrogenic spheroids exhibiting more intense staining, indicative of a higher density of ECM deposition and displayed a characteristic cobblestone-like morphology in transfected group at Week 2 (Fig. 5F). The compressive modulus of the bioprinted SCTs was found to be 116.8 ± 22.1 kPa (Supplementary Fig. 19C). Additionally, qualitative evaluation of proteoglycans and sGAG was conducted using toluidine blue (TB) staining. In agreement with the quantified sGAG expression (Fig. 5C), images of transfected spheroids stained with TB exhibited intense staining, indicating a higher level of sGAG deposition compared to non-transfected samples (Fig. 5G). From H&E and TB staining, the co-transfected group indicated chondrogenic lacunae-like properties. IHC staining was also performed to identify the expression of chondrogenic markers, ACAN and COLII, in SCTs. The findings revealed stronger fluorescence intensity indicating significantly elevated levels of ACAN (p ≤ 0.01) and COLII (p ≤ 0.001) in SCTs at Week 2, suggesting the formation of new chondrogenic ECM (Supplementary Fig. 19B).
Fig. 5. HITS-Bio for SCT fabrication.
A 1 cm3 of cartilage tissue was bioprinted with 576 spheroids in the CARink. Scale bar: 5 mm. Comparison between non-transfected and co-transfected spheroids cultured in 1 cm3 tissue at Week 2 for (B) cell viability measurements (n = 6 biologically independent samples, unpaired two-sided Student’s t-test, p = 0.51096), C sGAG normalized with DNA content (n = 3 biologically independent samples, unpaired two-sided Student’s t-test, p = 0.00056), and (D) the total DNA content of tissue constructs (n = 3 biologically independent samples, unpaired two-sided Student’s t-test, p = 0.00003). E LIVE/DEAD staining of non-transfected and transfected spheroids at Week 2 in SCT constructs (Scale bar: 50 µm). Histomorphometric characterization of sectioned SCTs stained with (F) H&E with 20X and 100X magnification and (G) TB-stained images captured at 20X and 100X magnification (scale bar: 100 µm (20X) and 10 µm (100X)). Representative images were obtained from at least four independent repetitions. Data are presented as mean ± SD where ***p < 0.001. Source data are provided as a Source Data file.
Discussion
Current bioprinting techniques face critical challenges, such as achieving physiologically-relevant cell densities, enhancing throughput for scalable tissue fabrication, developing bioinks tailored for specific applications, and enabling in situ fabrication capabilities. Towards this, the pursuit of high-throughput bioprinting marks a vital development in achieving rapid and efficient tissue fabrication, driven by automation. These advancements are essential for meeting the increasing demand for complex tissue constructs that replicate native tissue architecture and function. In this study, a High-throughput Integrated Tissue Fabrication System for Bioprinting, termed HITS-Bio, comprising four key components was developed. These include (i) software for customized control, (ii) DCNA facilitating precise and multiple spheroid placement, (iii) compatible bioinks (BONink and CARink) ensuring smooth extrusion and printability, and (iv) miRNA transfected osteogenic and chondrogenic spheroids for de novo tissue fabrication. The platform was assessed for its applicability in in situ osteogenesis of bone tissue and scalability by fabrication of volumetric cartilage tissues. The developed HITS-Bio platform was operated by a custom-made hardware-software interface as shown in Supplementary Fig. 2 and Supplementary Movie 1. The automation control was executed via the LabView Software Control Panel, managing motion stages with high precision ( ~ 1 µm in X, Y, and Z axis). DCNA allowed customization, where selective nozzles could be activated depending on the target design. Initial testing of HITS-Bio was performed using 16 spheroids, which were selectively picked and placed alternately between red and blue dyed spheroids (Fig. 2I). The process was completed under 30 sec, representing a significant speed compared to the existing benchmark in the literature17, which required nearly 30 min. A pressure sensor was used to monitor internal pressure in the platform, but the primary method for spheroid picking relied on a visual confirmation via a camera-based system. This camera-based system provided real-time visual feedback, ensuring precise positioning and adaptability across various spheroid types, regardless of differences in shape, size, or surface texture. While dynamic pressure control could be advantageous in certain situations, such as monitoring pressure changes during spheroid loading, we found the camera-based approach to be more versatile. Additionally, spheroid debris can interfere with pressure-based sensors, leading to false positives during spheroid placement.
The HITS-Bio process was optimized for spheroids within the 300–350 µm range using a 30 G needle, which was proven effective for this size range. It is important to note that the spheroid size must be larger than the inner diameter of the nozzle used. For smaller spheroids, adjustments in nozzle size would be necessary to prevent them from being inadvertently drawn into the nozzles, though this is currently limited by the availability of commercial nozzles with a diameter smaller than 30 G. Larger spheroids may require modifications in nozzle size and spacing to avoid spatial interference and ensure efficient transfer. This can also be managed by selectively controlling (On/Off) adjacent nozzles in DCNA, to accommodate the transfer of larger spheroids. Additionally, while the current 4-mm exposed nozzle length was sufficient for bioprinting onto gel surfaces, extending the exposed nozzle length could optimize the system for embedded bioprinting applications, enabling more complex or deeper spheroid placements. Future iterations of DCNA could benefit from the inclusion of independently height-adjustable nozzles and an increased number of nozzles, enhancing the system’s flexibility and versatility across a broader range of applications.
Spheroids are promising candidates as building blocks for tissue fabrication as they recapitulate the native tissue environment with similar cell density and ECM composition and have the potential to rapidly induce tissue regeneration due to initially-delivered large pre-committed cell numbers17. When spheroids are loaded in gels, they show better cell spreading and proliferation, and tissue-specific differentiation, compared to conventional cell-laden hydrogels27. Until now, spheroid bioprinting techniques enabling the deposition of spheroids onto a gel substrate have been mainly limited to EBB28 and AAB17. EBB with spheroids has been used to overcome the limitation of increasing cell density in an extrudable bioink29,30. However, spheroids are randomly dispersed in the bioink, making their controlled positioning a challenge, resulting in inconsistencies and limitations in loading efficiency in bioprinted constructs, similar to the case shown in Supplementary Figs. 7 and 8. To overcome these limitations, AAB has been utilized for high-precision bioprinting, where various tissue complexes have been fabricated inside functional hydrogels as well as support baths depending on the application19. However, in these strategies, the placement of only one spheroid at a time was feasible, rendering scalable tissue fabrication a challenging task. Alternatively, multiple studies have demonstrated the potential of multi-nozzle bioprinting for rapid tissue fabrication31,32. For example, a study by Hansen et al. used a multi-nozzle array to produce a hierarchically branched, microvascular network and exhibited high-throughput printing of single and multiple extrudable inks over large areas (1 m2). The strategy resulted in a significant reduction in printing time, where a 3D construct that takes a day to print using a single nozzle printhead took only 22 min to print using a system with 64 nozzles33. However, to demonstrate large-scale patterning, the study used wax as an ink to print onto a 1-m2 glass substrate using the 64-nozzle printhead.
In terms of experimental duration, it is pertinent to note that as the complexity increases, particularly with multilayer deposition and non-repeating patterns, the loading process becomes a critical factor influencing the overall efficiency. For physiologically relevant and complex constructs, the experimental duration can vary significantly based on the intricacy of deposition patterns. While the DCNA streamlines simultaneous loading and deposition of multiple spheroids, non-repeating patterns introduce additional challenges. In this context, a total of 50 spheroids were successfully bioprinted in 8 min using two different types and sizes of spheroids (Supplementary Fig. 10). This process involved non-repeating patterns to fill gaps (without a predesigned travel path) for a fully cellular architecture and took ~8 min compared to ~5 min for a repetitive pattern of 64 spheroids using DCNA and ~25 min using a single nozzle AAB. Although the efficiency of non-repeating patterns was not as high as with repeated patterns, it was still significantly faster than the existing benchmark. The use of pre-designed travel path for bioprinting and enhancing the system with more nozzles and incorporating height-adjustable nozzles could significantly improve versatility, particularly for non-repeating patterns.
In the current work, HITS-Bio was used to accelerate the spatial positioning of spheroids via their simultaneous deposition onto a gel substrate for scalable tissue fabrication. Spatial positioning of spheroids is important for mimicking native tissue microenvironments and promoting effective cell-cell communications, ensuring tissue functionality and organization11. The spheroid-based approach offers significant advantages for bone and cartilage tissue engineering by effectively mimicking the hierarchical native structures essential for these tissues20,34. Spheroids enable physiologically relevant cell-cell and cell-matrix interactions, which are crucial for the differentiation and maturation of chondrocytes and osteoblasts10. Moreover, the relative avascularity of cartilage and the minimal vascularization requirements in early-stage bone tissue reduce complexity. Additionally, bone and cartilage are among the most extensively studied tissues, offering a great amount of reference data, making them ideal models for developing and validating new bioprinting technologies. Herein, DCNA enabled the spatial arrangement of miR transfected spheroids. A combination of miR-196a-5p and 21 was used for the co-transfection of hADSCs to create osteogenically-committed spheroids. miR-196a-5p plays a crucial role in bone homeostasis and is highly expressed in osteoclast precursors35. Kim et al. reported that miR-196a-5p regulates the proliferation and osteogenic differentiation of human ADSCs, which may be mediated through HOXC836. Concurrently, miR-21 has been proven to play a role in bone formation by mediating mesenchymal stem cell proliferation and differentiation37,38. It activates the ERK-MAPK (extracellular signal-regulated kinases (ERKs)-mitogen-activated protein kinases (MAPKs)) signaling pathway, promoting osteogenesis by suppressing the expression of its target gene SPRY139. In a study, when combined, miR-196a-5p and -21 exhibit synergistic effects to enhanced osteogenesis, where miR-196a-5p stimulates osteogenic ability, while miR-21 further supports osteoblastic differentiation and amplified proliferation rate, confirming the hypothesis of Abu-laban et al.40 and Celik et al.26 Thus, spheroids with ~8k cells/spheroid were formed in the current work and maintained a week in growth medium in vitro. After a week, spheroids reached ~350 µm in diameter with sufficient structural properties for bioprinting purposes. These spheroids were then used along with the developed BONink for bone bioprinting.
This study introduced bioinks, characterized by their paste-like shear-thinning property enabling both in vitro and in vivo EBB. Bioink comprises GM (10 or 20% w/v), β-GP, HyA, Fib, HA (15 or 30% w/v), and the photocrosslinker LAP. GM and HA served as bulk polymers influencing the bioink’s properties, while HyA and Fib mimic ECM, promoting cell growth and tissue regeneration. β-GP enhances osteogenesis and mineralization, making these polymers ideal for bone regeneration applications41,42. The developed bioinks were characterized rheologically, where flow sweep results indicated that the bioinks had shear-thinning attributes meeting the basic requirement for EBB. The decrease in viscosity upon increasing the shear rate implies a decrease in the extrusion pressure, facilitating smooth extrusion through smaller nozzles43. Additionally, the bioinks possessed self-healing capability, which is essential for maintaining the integrity of bioprinted constructs. Self-healing shear-thinning bioinks stand out as promising materials for EBB44. These bioinks can be extruded as their viscosity decreases under the shear, and subsequently self-heal once the shear is removed. This dual property ensures safe bioprinting of cells and the maintenance of shape fidelity post bioprinting44. Further, the assessment of mechanical properties of bioinks showed that the modulus falls within the range observed in native trabecular or cancellous bone, which typically exhibits elastic moduli of 0.02–2 GPa45. Additionally, construct degradation at physiological conditions is considered advantageous because it allows for the construct to diminish so that the new ECM can slowly replace the degraded portions of the construct. The results indicated that an increase in the concentration of GelMA resulted in slower degradation, which may be attributable to increased methacrylamide crosslinks46. The addition of HA and other components reduced the degradation rate further. This aligns with a prior study by Allen et al., where a GelMA-gelatin-HA bioink was utilized for 3D bioprinting of bone constructs and their degradation with Type IV collagenase was significantly reduced by the addition of HA in a concentration-dependent manner47. Based on these results, GM20HA30 composite was selected as a base cement material and termed ‘BONink,’ which was found to be biocompatible, affirming its suitability for bone tissue engineering applications.
The in vitro assessment results suggested that bioprinted constructs containing spheroids co-transfected with miR-(196a-5p + 21) showed superior upregulation of osteogenic genes. Interestingly, the expression of osteogenic markers may be influenced by the spheroid density. Early-stage markers like RUNX2 and BMP-4 show higher expression in low-density groups, likely due to better nutrient diffusion and efficient paracrine signalling, where factors can diffuse more evenly, promoting early differentiation. Conversely, high-density spheroids exhibit increased expression of intermediate to late-stage markers, such as OSTERIX and BSP, indicating enhanced maturation and mineralization, which may be due to closer cell-cell interactions and higher local concentrations of paracrine factors. These results suggest the role of spheroid density in optimizing osteogenic differentiation through paracrine signalling and mechanical cues. IHC staining confirmed the expression patterns, particularly highlighting intense staining for RUNX2 in the low-density group and OSTERIX in the high-density group, aligning with gene expression profiles. To mimic the native tissue physiology, engineered tissues require optimum cellular density and microstructural complexity48. Studies have shown that spheroids play a crucial role in enhancing osteogenic differentiation. For instance, spheroids facilitate cell-to-cell interactions, leading to increased osteogenic potential10,49. Moreover, the upregulation of osteogenic markers like RUNX2, BSP, and OSTERIX in miR-transfected spheroids has been linked to enhanced osteogenic differentiation26.
Meanwhile, IOB is transforming surgical procedures by offering in situ fabrication of patient-specific tissue constructs directly at the surgical site50. It enables precision customization, minimizing infection risks through the elimination of pre-fabricated implants, and improving healing due to freshly bioprinted tissues, while eliminating storage and transportation concerns. IOB has been previously utilized in repairing calvarial bone defects via laser-based bioprinting (LBB), where nano-HA was combined with collagen and MSCs to serve as the ink and deposited directly onto the calvarial defects in mice, resulting in a significant increase in bone formation observed after 2 months51. However, LBB method is challenged by its slow deposition of biomaterials into a defect site. In another study, IOB was used for the reconstruction of craniomaxillofacial (CMF) tissues, including bone, skin, and composite (hard/soft) tissues. The use of a hybrid IOB approach (EBB and DBB) reconstituted hard/soft composite tissues in a stratified arrangement resulted in ≈80% skin wound closure in 10 days and 50% bone coverage area at Week 652. In the current work, bioprinting was performed intraoperatively using spheroids under surgical settings to repair rat calvarial defects. The results showed that the newly formed bone invaded from one defect edge to another (Fig. 4B) with a bone coverage area of 91 and 96% in 3 and 6 weeks for the high-density group, respectively. This demonstrates substantial bone regeneration in a short timeframe, attributed mainly to the innovative use of IOB with miR-transfected spheroids, which is quite challenging using other similar methods and materials. The maximum intensity projections from µCT data, used to score bony bridging, revealed significantly higher scores (3.85 ± 0.37 out of 4) for spheroid involved groups indicating near-complete bridging and enhanced bone regeneration, likely due to the superior osteogenic properties imparted by the miR transfection and BONink (Fig. 4C). Moreover, the regenerated bone in this group exhibited significantly enhanced shear yield strength and modulus of resilience (Fig. 4D). Histological analysis depicted the connectivity and compactness of the newly formed bone tissue with MT staining revealing dense blue islands indicative of immature bone tissue formation, particularly prominent in the high-density group. The study demonstrated superior regenerated bone in situ, supported by histological analysis showcasing dense, immature bone formation, which can be expected to develop into a mature bone in a longer timeframe in vivo.
Bioprinting using transfected spheroids contributed to earlier defect closure, particularly by Week 3, which is a significant improvement over existing approaches. This early bone formation reduced the difference observed between Weeks 3 and 6, which is consistent with our approach’s ability to expedite bone healing compared to other approaches in the literature. Despite significant bone regeneration, we observed variations in fusion between the implant and host bone, which may be attributed to localized mechanical loading differences and ongoing remodeling. The histological evidence of dense, immature bone tissue suggests that complete maturation and integration could require long-term studies. In this study, a control group with manually loaded spheroids was initially included with randomly distributed spheroids, but this method encountered significant technical challenges, such as spheroid loss during their transfer, spheroid aggregation, and air bubble formation, leading to uneven distribution and reduced reproducibility as also highlighted in in-vitro studies (Supplementary Figs. 7 and 8). These issues compromised the reproducibility and data accuracy. Consequently, we did not pursue this method further in animal studies, in accordance with the principles of the 3Rs, to minimize unnecessary animal experimentation. Instead, the BONink (ink only) was selected as the control, which provided a consistent, cell-free matrix that isolated the effects directly attributable to material properties without interference from the addition of exogenous cells (hADSCs). This approach allowed for a more reliable and clear assessment of the impact of spheroid deposition using HITS-Bio. Additionally, although the spheroids were deposited uniformly, their direct visualization and distribution in vivo was not feasible due to technical limitations associated with embedding spheroids within the tissue matrix. Moreover, the uniformity of the original spheroid distribution was challenging to assess during histological evaluations of retrieved explants, as tissue remodeling and integration processes could alter the initial arrangement of spheroids. Overall, the findings support the feasibility of HITS-Bio as a powerful tool in IOB of spheroids for bone tissue with superior osteogenic potential, and high-throughput and speed, completing each construct in about 4.5 min.
Furthermore, the HITS-Bio platform was explored for the scalability of bioprinted tissues. Thus, SCTs were successfully fabricated by depositing layers of CARink (representing GM20) using EBB followed by the precise placement of chondrogenic spheroids using DCNA, creating constructs with nine stacked tissue layers, comprising around 600 spheroids per SCT, demonstrating the potential for creating intricate, multi-layered tissues. The CARink, being transparent, allowed real-time monitoring of deposited spheroids with favorable extrusion properties (inset images in Figure 3Fii, Fiii, and Supplementary Movie 7). A combination of miR-140 and -21 was used for co-transfection of hADSCs to create chondrogenically-committed spheroids as per a previous report26. Importantly, the results revealed that the process of transfection and tissue fabrication did not compromise spheroid viability ( > 90%) compared to non-transfected spheroids. The dual miR-transfected SCTs exhibited a significantly higher sGAG content compared to the non-transfected control group (Fig. 5C). sGAGs play a critical role in cartilage function, providing mechanical support and maintaining tissue hydration53. The results suggest that HITS-Bio supported enhanced ECM deposition that is essential for cartilage formation. Interestingly, the DNA content was significantly lower in the transfected samples compared to the non-transfected samples (Fig. 5D). This might be due to miR suppressing genes associated with stemness and promoting chondrocyte-specific markers and by downregulating cyclins and cyclin-dependent kinases that limit cell proliferation, favoring differentiation36. However, further investigation is warranted to understand the underlying mechanisms. Histological assessment using H&E and toluidine blue (TB) imaging confirmed the presence of chondrogenic spheroids within bioprinted constructs, where spheroids exhibited intense staining, indicating a higher density of ECM deposition with a developed lacunae-like structure (Fig. 5F, G). Further, TB staining supported the presented quantitative sGAG data, where transfected spheroids displayed intense TB staining, emphasizing enhanced sGAG production (Fig. 5G). Overall, the results demonstrated the efficacy of HITS-Bio and its potential application for the rapid bioprinting of cartilage tissues. Using HITS-Bio, the tissue dimensions were scaled up from a volume of ~30 mm3 (disc with a diameter of 5 mm and a height of 1.5 mm) involving 64 spheroids for bone tissue bioprinted in 4.5 min per construct, to a volume of 1 cm3 (cube of 1 × 1 × 1 cm), incorporating 576 spheroids for cartilage tissue bioprinted in under 40 min (including EBB of the CARink). The ~10-fold increase in the bioprinting speed was achieved through the integration of the aforementioned factors in HITS-Bio. However, because of the thick tissue, hypoxia in SCTs is possible and needs to be investigated in further studies. Nevertheless, in cartilage, it has been reported that the hypoxia condition protects against cartilage loss by regulating Wnt signalling54.
Regarding mechanical properties, the BONink (GM20HA30) formulation without spheroids exhibited a compressive modulus of 360.7 ± 66.6 kPa. When compared to engineered bone reported in the literature, which ranges from 0.1 to 10 MPa55, our in vitro results were within the expected range for early-stage bone constructs. Further, the in vivo retrieved explants demonstrated a significantly higher shear modulus of ~20 MPa, which corresponds to a compressive modulus of ~52 MPa56. For reference, the compressive modulus of cranial or flat bone in rats ranges from 5 to 9 MPa (depending on the age)57–59, while in humans, cortical bone ranges from 10 to 20 GPa and trabecular bone from 0.1 to 2 GPa56,60. It is important to note that mechanical properties of the implanted constructs do not necessarily need to mimic those of native bone, as they can be supplemented with plate osteosynthesis during implantation61. However, the constructs must possess sufficient mechanical strength to provide initial support ensuring stability and facilitating proper integration with the surrounding tissue during the early stages of bone regeneration. For human articular cartilage, the aggregate equilibrium compressive modulus was reported to be in the range of 0.1 to 2 MPa depending on the specific location and health of the tissue62. Reported values for bioprinted cartilage are lower with a compressive modulus typically ranging from 50 to 500 kPa63. In this study, the compressive modulus of CARink (GM20) without spheroids was measured at 88.3 ± 34.7 kPa, which increased in SCTs containing spheroids with a compressive modulus of 116.8 ± 22.1 kPa after 2 weeks of in vitro maturation. This limited increase in compressive modulus was likely due to insufficient localized ECM formation, indicating that longer maturation might be needed for further mechanical improvements. It is also crucial to recognize that standardized protocols for mechanical testing of bioprinted constructs are still being developed, with American Society for Testing and Materials (ASTM) standards yet to be fully established (latest as of April 2024, ASTM F3659-24). Therefore, comparisons with other studies may not be entirely accurate without standardized methodologies.
Still, HITS-Bio has some aspects, where its performance could be enhanced. First, DCNA may have a clogging issue that can delay the bioprinting time. The clogging issue typically arises from cell debris accumulating in the spheroid chamber. Second, all nozzles in DCNA should be positioned on a uniform plane. This alignment is crucial for accurately patterning spheroids, ensuring they are placed at the same level, otherwise, loaded spheroid may penetrate deeper or remain elevated, which could lead to potential damage or imprecise bioprinting due to insufficient contact with the surface. During DCNA manufacturing, the precision along the Z-axis is thus important. As shown in Fig. 2C, although the groups showed no significant differences, a decrease in the inter-nozzle distance led to an increase in variation in Z-axis positional error due to the interference caused by tightly packed nozzles during the micromanufacturing process. The developed DCNA can be conveniently used to bioprint on flat surfaces; however, bioprinting on uneven surfaces like a non-planar defect area during IOB can pose challenges. Thus, it is important to level the defect area on a rat to match the bottom of the DCNA plane before initiating the HITS-Bio process either by angle adjusting the roll pitch yaw of DCNA or by adjusting the rat’s head angle parallel to the DCNA surface. Third, as with most pressure-driven systems, HITS-Bio is susceptible to potential damage to spheroids due to the aspiration forces applied during bioprinting. To mitigate this risk, we optimized several key parameters, including nozzle size and pressure, ensuring that all nozzles in the DCNA are aligned on a uniform plane to minimize stress on spheroids and preserve their structural integrity. Through this optimized approach, we identified a range of elastic moduli suitable for effective bioprinting with HITS-Bio. Specifically, spheroids with an elastic modulus greater than ~50 Pa were successfully bioprinted, while those below 40 Pa were not suitable (Supplementary Fig. 9). Also, we demonstrated the successful bioprinting of iPSC-derived vascular organoids with an elastic modulus of 133 ± 20 Pa (Fig. 2J and Supplementary Fig. 12), showing the system’s applicability to bioprinting of organoids. Fourth, although we limited DCNA to 4 × 4 nozzles in the current study, we can reconfigure it (e.g., 10 × 10 nozzle array) to further expedite the bioprinting process. This would eliminate the periodicity constraints imposed by the current DCNA design while simultaneously leading to improved tissue fabrication efficiency and better capabilities for various applications. Fifth, the inter-nozzle capillary interactions can affect spheroid picking precision, especially for closely spaced nozzles. However, by coating the nozzles with silicon and adjusting their spacing, as demonstrated in our study, and potentially incorporating advanced fluid dynamics management, these interactions can be effectively controlled. These approaches will reduce liquid elevation between nozzles, ensuring accurate and efficient spheroid manipulation. Building on these optimizations, the DCNA setup was further calibrated to correlate with spheroid size, ensuring optimal bioprinting performance. For larger spheroids, increased spacing between nozzles is necessary to prevent physical interference, while smaller spheroids benefit from tighter nozzle spacing and smaller nozzle sizes to maintain precision and avoid unintended suction. In cases where larger spheroids were difficult to lift due to the surface tension, selective controlling of adjacent nozzles was performed, such that only alternate nozzles were active. Using this method, we bioprinting red spheroids (735 µm in diameter) in a pyramidal structure (Supplementary Fig. 13), validating its effectiveness in handling larger spheroids. Lastly, during spheroid aspiration, a minimum pressure of ~3 mmHg was sustained throughout the whole DCNA, preventing media leakage when spheroids were placed with closed channels. It was noted that leaked media on the substrate hindered the placement of spheroids or caused them to move, resulting in inaccurate positioning. Importantly, achieving adequate hemostasis before the IOB process is essential, as bleeding during surgery may reduce accurate spheroid positioning. Thus, based on the potential to improve the aforementioned aspects of DCNA, the next generation of DCNA would ideally feature independently height-adjustable nozzles with an increased number of nozzles as demonstrated for multi-nozzle conformal EBB64. Using the modified DCNA, HITS-Bio can be more accurate on adjustable surfaces and expand their prospective applications and freeform bioprinting of spheroids in support materials for complex shape fabrication.
Using HITS-Bio, a fully cellular architecture composed of HDF/HUVEC spheroids (Supplementary Fig. 10), and iPSC-derived vascular organoids (Fig. 2J and Supplementary Fig. 12) were bioprinted, demonstrating its potential for integration of vascularization within bioprinted constructs. Its precision in co-bioprinting vascular spheroids alongside other tissue-specific cells can support vascularization, which is critical for sustaining the viability of large-scale tissues. Looking ahead, the current work aligns with the urgent need to advance spheroid bioprinting techniques for rapid fabrication of scalable, vascularized tissues. Integrating vascular networks within large-scale bioprinted tissues, particularly for organs with high metabolic demands such as the heart, pancreas, and liver, is a crucial step achieving clinically-relevant tissues for transplantation.
In summary, this study introduces HITS-Bio, a high-throughput bioprinting platform, that enables scalable tissue fabrication by precisely positioning a number of spheroids at an unprecedented speed using the DCNA platform. With a significant increase in the bioprinting performance compared to existing techniques, HITS-Bio enabled the rapid creation of tissue via accelerated spatial positioning of spheroids on a gel substrate. The performance of HITS-Bio was exemplified by bioprinting multiple tissue types including bone and cartilage. The study showed for the intraoperative bioprinting with spheroids, where miR co-transfection enhanced osteogenesis in committed spheroids derived from hADSCs, demonstrating significant potential in repairing rat calvarial defects. Moreover, the potential of HITS-Bio for scalable tissue fabrication was demonstrated by the fabrication of scalable cartilage constructs with an unprecedented speed (under 40 min per construct) that surpasses the capabilities of existing technologies. To further expand its capabilities, future improvements could include incorporating additional nozzles to increase throughput, developing height-adjustable platforms for bioprinting on non-planar surfaces, and increasing automation. These technological enhancements are critical toward fully realizing the potential of HITS-Bio in scalable tissue biofabrication applications.
Methods
Cell culture and spheroid/organoid fabrication
Human adipose-derived stem cells (hADSCs; PT-5006, Lonza) were obtained and cultured in a basal medium consisting of a 1:1 mixture of Dulbecco’s modified Eagle medium (DMEM)/F12 (15-090-CV, Corning) supplemented with 20% fetal bovine serum (FBS; S11150, R&D Systems), 1% penicillin/streptomycin (PS; 30-002-CI, Corning) at 37 °C with 5% CO2 in a humidified incubator. The cell medium was changed every other day. hADSCs from passages 1–3 were used in experiments. For spheroid fabrication, the expanded hADSCs were trypsinized, centrifuged, and transferred to each well of a U-bottom 96-well plate (650970, Greiner Bio One) with 7,000 cells per well to obtain spheroid clusters of 300-350 µm, followed by their incubation in a humidified incubator with 5% CO2 at 37 °C overnight allowing the spheroid formation26. Similarly, other spheroids were prepared by taking 8,000 cells in each well of a cell repellent 96-well plate. In the first group, MDA-MB-231 (MDA; gift from Dr. Danny Welch, University of Kansas) and human lung fibroblasts (HLFs; Lonza) were mixed in ratios of 1:0, 3:1, 1:1, 1:3, and 0:1, respectively. In the second group, MDA and human dermal fibroblasts (HDFs; Lonza) were combined in ratios of 3:1, 1:1, 1:3, and 0:1. In the third group, human umbilical vein endothelial cells (HUVECs; Lonza), transduced to express tdTomato (tdTomato HUVECs) following a previously established protocol65, were mixed with HDFs in a 1:2 ratio. The last group of spheroids consisted of HDFs only. MDAs, HLFs, and HDFs were cultured in the DMEM (10-017-CV, Corning) supplemented with 1% PS, 10% FBS, while HDF/HUVECs were cultured in the MCDB 131 medium (15-100-CV, Corning) supplemented with 10% FBS, 1 mM Glutamine, 1 mM PS, 0.5 mm bovine brain extract (BBE; CC-4092, Lonza), 1200 U mL−1 heparin (H3149-100KU, Sigma-Aldrich) and 0.25 mM endothelial cell growth supplement (ECGS; E2759-5X15MG, Sigma-Aldrich). The cells were then allowed to aggregate and form spheroids over 24-48 h in an incubator set to 37 °C with 5% CO2, ensuring consistent spheroid formation across all groups. For fabrication of pyramidal architectures, spheroids were generated using ADSCs with varying cell numbers. ADSC aliquots containing 7,000, 14,000, 20,000, and 30,000 cells were prepared for spheroid formation by seeding them into low-attachment 96-well plates. The plates were incubated at 37 °C in a humidified atmosphere with 5% CO2 for 24-48 h to allow spheroid formation.
Induced pluripotent stem cell (iPSC)-derived vascular organoids were generated as per an established protocol66. Briefly, iPSCs (IMR90C4 iPSCs, WiCell Research Institute Inc) were aggregated and mesoderm differentiation was induced using CHIR99021 and BMP-4. For vascular differentiation, a combination of growth factors including VEGF-A and forskolin over a period of 3-5 days were used.
Development of the HITS-Bio platform
The HITS-Bio platform integrated X, Y, and Z motion axis for 3D movement, equipped with cameras, a digitally controllable nozzle array (DCNA) consisting of 30 G nozzles and associated controls such as pressure regulators, flow controllers, and positioning systems (Supplementary Fig. 2). In the bioprinting stage, two 35-mm Petri dish were accommodated to be used as spheroid reservoir and tissue fabrication area, respectively. Solenoid valves and a pressure sensor digitally regulated the aspiration force and positive pressure, allowing for selective control and internal pressure monitoring of each nozzle. The hardware component of DCNA consisted of stainless-steel needles (30 G, an inner diameter of 150 µm and an outer diameter of 305 µm) arranged in a 4 × 4 array with dimensions of nozzle array ranging from 2.8 to 4.0 mm in width, and the inter-nozzle distance between 527 and 927 µm (Supplementary Fig. 6D), which was enabled by precisely stacked multiple acrylic plates (10 mm × 10 mm, 2 mm thick), micro-manufactured by laser cutting (Supplementary Fig. 1). These nozzles were carefully inserted through the holes on the plates, and after calibrating the surface and arranging the nozzles on the same plane, were adhered to the acrylic plates for stability. Manufacturing tolerance data (Supplementary Fig. 6E), illustrated by the XY positional error before and after the acrylic plate removal, highlights the importance of plate integrity for maintaining precision. The assembled DCNA was coated with Sigmacote® (SL2, Sigma-Aldrich) to siliconize the surface to reduce the surface tension generated between the cell medium and DCNA. The coated DCNA was attached to the HITS-Bio platform via an adaptor, facilitating exact roll-pitch-yaw adjustments to align the DCNA parallel to the bioprinting workspace. An extrusion head for bioink deposition was integrated into DCNA (Supplementary Fig. 2B), with real-time bioprinting visualization and positional verification of DCNA and extrusion nozzle provided by three microscopic cameras offering isometric, bottom, and side views. Vacuum chambers prevented liquid from damaging the solenoid valves during the HITS-Bio process. The entire platform operated through a custom hardware-software interface (Supplementary Fig. 3 and Supplementary Movie 1), enabling precise operation of motion stages, digital control of solenoid valves, monitoring and recording via a computer vision system, and real-time pressure value monitoring within the pneumatic line through a pressure sensor. The software is available at https://github.com/MHKim-software/HITS-Bio.git. The interface setup, descriptions for each button on the interface, and a detailed algorithm for the entire HITS-Bio were also provided in (Supplementary Note 2 and Supplementary Figs. 3 and 4). Defined positions were saved as a.CSV file for reuse, and G-code-derived motion paths were uploaded for automated bioprinting. The system supported 2D and 3D visualization and analysis of objects for position, quantity, shape, and interspace. It also tracks their movement and incorporated safety features like an emergency stop button and maximum axis limits. The bioprinting process, standardized at 10 mm/s for spheroids 300-350 µm in diameter, was adjusted to 0.2 mm/s for lifting spheroids from the spheroid plates to air. Various DCNA sizes, designed for specific nozzle array widths and spacing, were fabricated, and assessed for positional accuracy and angular deviations, potentially resulting from handling errors during micro-manufacturing. Spheroid lifting percent success rates (SR%) for different DCNAs were calculated using the formula:
| 1 |
where m is the number of successfully lifted spheroids using DCNA and n is the total nozzle count with the platform designed for a 4 × 4 nozzle array, making n = 16. The HITS-Bio platform’s efficacy was benchmarked against a conventional single-nozzle AAB technique20,67,68 using the same bioprinting parameters. Bioprinting time was documented for sets of 16, 32, and 64 spheroids. The platform’s versatility was further demonstrated using three different diameters of spheroids ( ~ 350 μm stained with phalloidin AF488 (green), ~425 μm stained with DAPI (blue) and ~500 μm stained with phalloidin AF647 (red)) deposited on a 10% GelMA (GM10) substrate, as prepared below.
Preparation of Bioinks (BONink and CARink)
Gelatin methacryloyl (GM) was synthesized following a previously established protocol69,70. GM was then reconstituted into 5 and 10% (w/v) solutions using warm PBS and incubated at 37 °C until fully dissolved. For preparation of the cartilage ink (CARink) (Supplementary Fig. 14B), the GM solution was combined with 5 mg/mL fibrinogen (Fib; F8630, Sigma-Aldrich), 3 mg/mL hyaluronic acid sodium salt (HyA; 53747, Sigma-Aldrich), and 0.25% (w/v) lithium phenyl (2,4,6-trimethylbenzoyl) phosphinate (LAP; L0290, TCI Chemicals) as a photoinitiator. Concurrently, the bone ink (BONink) (Supplementary Fig. 14A) was formulated with the same components as CARink, along with the addition of 15 mg/mL β-glycerophosphate disodium salt hydrate (β-GP; G9422, Sigma-Aldrich) and hydroxyapatite (HA; nanoXIM•HAp202, Fluidinova) at 15 and 30% (w/v) concentrations, having a median particle size (d50) of 5.0 ± 1.0 μm. Both bioinks were homogenized using a FlackTek SpeedMixer at 2000 rpm for 3 min, followed by a subsequent 3-min cycle at the same speed to eliminate air bubbles. The resulting materials were carefully transferred into 1 cc syringes, avoiding the incorporation of air, and then wrapped in aluminum foil to shield from light-induced photo-crosslinking. They were stored vertically at room temperature (RT) for 2 h before bioprinting. After bioprinting with the extrusion of CARink and BONink, 25 U/mL of thrombin (T4648, Sigma-Aldrich) was used with the cell culture medium as an enzymatic crosslinker for fibrinogen. The concentrations of the composite materials in BONink (GM and HA) and CARink (GM) were optimized for extrudability, mechanical stability, rheological properties, and biodegradability as described below.
Bioink characterization
Rheological properties
Rheological properties were characterized using a rheometer (MCR 302, Anton Paar). All rheological measurements were performed in triplicates with a 25 mm parallel-plate geometry at RT (22 °C) after having 2 h of resting at RT. The frequency sweep test was carried out in an angular frequency range of 0.1–100 rad/s at a strain in the linear viscoelastic region to examine the storage (G’) and loss (G”) modulus without destroying the sample. To investigate the shear-thinning behavior of prepared bioinks, a flow sweep test was conducted in the shear rate range of 0.1–100 s−1. A recovery sweep test was also performed to evaluate changes in the viscosity after applying low and high shear rates at five intervals: (1) a shear rate of 0.1 s−1 for 30 sec, (2) a shear rate of 100 s−1 for 10 sec, (3) a shear rate of 0.1 s−1 for 30 sec, (4) a shear rate of 100 s−1 for 10 sec, and (5) a shear rate of 0.1 s−1 for 30 sec.
Compression testing of in vitro samples
Unconfined compression tests were conducted using an Instron 5966 series advanced electromechanical testing system with a 10 kN load cell, following American Society for Testing and Materials (ASTM) standard D395–18 guidelines. Briefly, 13 × 6 mm (diameter × height) cylindrical specimens were compressed at a rate of 1.3 mm/min until reaching failure or 70% strain of their original height at RT. The obtained data was converted to stress-strain and the bulk modulus (kPa) was used from the resulting curve.
Biodegradability characteristics
To study the degradation characteristics of the prepared bioinks, we used the traditional gravimetric approach71. Briefly, the initial weight of constructs was recorded, followed by their immersion in PBS with a pH of 7.4 and PBS containing 1 U/mL of collagenase Type II (17-101-015, Thermo Fisher Scientific) at a temperature of 37 °C until they reached a state of equilibrium. The collagenase solution was refreshed every 2-3 days to maintain constant enzyme activity. The weight of constructs was measured at different time points (Days 0, 3, 5, 7, 10, and 14). The degradation study was performed in triplicates per each time point and the percent degradation (D%) was determined using the following equation:
| 2 |
where, and were the initial and measured weight of the construct at each time points, respectively.
Biocompatibility
To evaluate the biocompatibility of BONink and CARink, hADSC spheroids were mixed into the bioinks and cultured at 37 °C in 5% CO2 in a humidified atmosphere. Cell viability in BONink was tested using an alamarBlue HS cell viability reagent (A50100, Thermo Fisher) according to the manufacturer’s instructions on Days 1, 3, 7, and 14. Briefly, samples were treated with 10% (v/v) dye solution and incubated for 3 h. Next, 100 μL of the culture medium was analyzed using a microplate reader (Infinite 200 Pro, Tecan) at 570/600 nm (excitation/emission). The results were expressed as the normalized value of the reduced dye, which correlated with the quantity of viable cells.
Measurement of mechanical properties of spheroids
Elastic moduli of spheroids were assessed using a pulled micropipette as per previous reports65,72, with a final inner diameter of 70–85 µm connected to a vacuum system. The micropipette was mounted on an optical microscope (Motic) and testing was conducted inside the culture medium under a fixed aspiration pressure of 10 mmHg, applied for 5 min. The pressure was precisely controlled using a pressure controller (Ultimus I, Nordson EFD). During the test, videos were recorded to capture the deformation of spheroids, which was measured by tracking the change in the length of aspiration within the micropipette tip from the pre-deformed state of the spheroids. The elastic modulus was calculated using the following equation, under the assumption of the homogeneous half-space model:
| 3 |
where is the elastic modulus, is the inner radius of the micropipette, is the length of aspiration, is the aspiration pressure, and ∅(η) is the geometry of pulled micropipette.
Bioprinting of iPSC-derived vascular organoids
After differentiation, organoids were bioprinted into collagen I (2 mg/mL)-Matrigel (mixed in a 4:1 ratio) and maintained in the StemPro-34 SFM complete medium (10639011, Thermo Scientific) including 15% FBS, 100 ng/mL VEGF-A (100-20, Peprotech) and 100 ng/mL FGF-2 (130-093-841, Miltenyi Biotec) for 3 days.
Fabrication of densely-packed cellular constructs
A 10% GelMA solution containing 0.25% LAP was added to a 35 mm Petri dish and photo-crosslinked using a 405 nm light source to create a gel substrate. For the formation of densely packed cellular constructs, HITS-Bio was used to precisely deposit HDF spheroids (530 µm in diameter) onto the pre-crosslinked GelMA substrate, arranging them in compact rows. To achieve complete cellular architecture and fill any remaining gaps, smaller HDF/HUVEC spheroids (300 µm in diameter) were subsequently deposited. The architecture was then immobilized by overlaying with an additional layer of GelMA solution, followed by a secondary photo-crosslinking for 1 min. The bioprinted constructs were then cultured in a 1:1 DMEM-MCDB mixture medium, allowing the spheroids to fuse for three days.
Fabrication of area-filling models
To fabricate area-filling models, varying spheroid sizes across four groups were used to maximize filled area. The first group used large spheroids ( ~ 735 µm), the second group combined large and smaller spheroids ( ~ 300 µm), the third group used only small spheroids, and the fourth group combined two different sizes ( ~ 530 µm and ~300 µm). The resulting architectures were imaged and processed using thresholding (ImageJ) to distinguish between the spheroid-filled area (white) and the void area (black). The area-filling density was calculated as the percentage of spheroid-covered area relative to the total area, providing a quantitative assessment of filling efficiency for each group.
Fabrication of the pyramidal architecture
The hADSCs-based spheroids were bioprinted using HITS-Bio and a complex pyramid architecture was assembled, consisting of 171 spheroids of varying sizes, each tagged with a different color for distinction. The base layer comprised 121 spheroids (green, stained using Alexa Fluor 488 Phalloidin) with a diameter of 300 µm. The second layer included 36 spheroids (blue, stained using DAPI) with a diameter of 540 µm, followed by a third layer containing 9 spheroids (red, stained using Alexa Fluor 568 Phalloidin) of 735 µm and 4 spheroids (blue) of 540 µm. The top layer featured a single spheroid (green) with a diameter of 445 µm. To ensure accurate positioning and layering, the nozzles in the DCNA were accurately controlled, particularly for the large 735 µm spheroids, where alternate nozzles were activated while adjacent ones were deactivated. The structure was imaged using Zeiss Axio zoom microscope, and the 2.5D rendered image of the assembled construct was reported.
MicroRNA transfection for osteogenic and chondrogenic differentiation
MicroRNA (miR) mimics were used for osteogenic and chondrogenic differentiation according to the protocol reported in a previous work26,73. Custom oligonucleotides (miR-140: 5’-CAGUGGUUUUACCCUAUGGUAG-3’; miR-21: 5’CAACAGCAGUCGAUGGGCUGU 3’; miR-196a-5p: 5’- UAGGUAGUUUCAUGUUGUUGGG- 3’) were ordered from Integrated DNA Technologies. Transfection was performed using lipofectamine RNAiMAX transfection reagent (13778150, Thermo Scientific), where lipofectamine was mixed with miR mimics according to the manufacturer’s protocol26. hADSCs were co-transfected with miR-(196a-5p + 21) mimic for osteogenic differentiation and miR-(140 + 21) mimic for chondrogenic differentiation before being seeded in Opti-MEM medium (31985070, Thermo Scientific) in 175 cm2 cell culture flasks for 24 h. The final concentration of all miR transfections in the opti-MEM medium was determined to be 200 nM for a volume of 10 mL. The 10 mL solution with a final concentration of 200 nM was transferred to 175 cm2 cell culture flasks and incubated at 37 °C and 5% CO2 for 24 h. Transfected cells were collected by trypsinization and formed as spheroids. After transfection, a basal medium, consisting of the 1:1 mixture of DMEM/F12 supplemented with 2% FBS, and 1% PS, was used for miR-transfected spheroids. A commercially available osteogenic differentiation medium (417D-250, Cell Applications) was used as a positive control group for osteogenic differentiation.
In vitro bioprinting process for bone tissue fabrication
Bone constructs were fabricated using osteogenically-committed spheroids and the BONink in a stratified manner utilizing a hybrid approach. First, the BONink was extruded into a disk shape of 5 mm diameter with a 22 G tapered tip (inner diameter of 410 µm) using 80 kPa pneumatic pressure. Subsequently, osteogenic spheroids were precisely bioprinted atop the BONink layer, with densities categorized as 16 spheroids for low density and 64 spheroids for high density. Another layer of BONink was then extruded over the bioprinted spheroids to achieve a total thickness of ~1.5 mm. This assembly was photo-crosslinked using a 405 nm near UV light source for 1 min. The bioprinting process was conducted at RT within a laminar flow biosafety cabinet to maintain sterility. Once bioprinted, the structures were cultured in 12-well plates with the basal or osteogenic differentiation media, incubated at 37 °C in a humidified 5% CO2 atmosphere, and the media refreshed every other day. Samples were collected at Weeks 2 and 4 for further characterization. To ensure high cell viability during bioprinting, the aspiration pressure was optimized, identifying ~10 mmHg as the optimal level. This pressure did not affect the spheroid structural integrity while enabling lifting from the culture medium.
Intraoperative bioprinting of bone
For IOB aimed at bone regeneration in CMF defects, inbred immunodeficient RNU athymic rats (both male and female), acquired at 5 weeks of age from Charles River Laboratories International, Inc., were cared for and aged at our animal facility (Millennium Science Complex, Penn State University) until the age of 12 weeks. This care followed guidelines established by the American Association for Laboratory Animal Science (AALAS) and procedures approved by the Institutional Animal Care and Use Committee (IACUC, protocol #46591). A total of 14 rats (7 males and 7 females) were divided into four groups: empty defect (control), BONink only, low density, and high density. At Week 12, the rats underwent survival surgeries. Anesthesia was induced using isoflurane in a concentration range between 2 and 5% delivered in oxygen at a flow rate of 0.5 to 2 L min−1. In addition, bupivacaine (0.015 mg kg−1) (Centralized Biological Laboratory, PSU) at a concentration of 2.5 mg mL−1 and buprenorphine (0.015 mg kg−1) were injected subcutaneously before the surgery. Once fully anesthetized, artificial tears were applied to the rats’ eyes and their heads were shaved and cleansed with betadine surgical scrub followed by ethanol. A sagittal incision, ~2 cm long, was made on the skin and the periosteum was retracted to expose the calvarium. Two critical size calvaria defects, each 5 mm in diameter, were then created on either side of the parietal bone on the rat skull using a trephine bit, ensuring the Dura mater remained intact. The bone construct was bioprinted using HITS-Bio in a surgical setting (Supplementary Fig. 2A). The BONink was first extruded directly into the defect to create the base layer. Using HITS-Bio, osteogenically-committed spheroids were then bioprinted on this layer in either low (16 spheroids) or high (64 spheroids) densities. A final layer of the BONink was applied over the spheroids and crosslinked with 405 nm near UV light for 1 min. Post bioprinting, the skin was sutured with 4-0 vicryl sutures (Ethicon Inc.). Animals were placed on a warming pad for recovery. They were closely monitored until they regained sternal recumbency. Rats were observed daily and weighed for at least 10 days post-surgery, and weekly thereafter. At 3 weeks, rats were anesthetized and scanned using μCT in the live condition. At 6 weeks post-surgery, rats were euthanized by CO2 inhalation at a flow rate of 2 L min−1 until cessation of breathing, followed by decapitation for tissue collection. The collected tissues underwent histology and immunohistochemistry to evaluate calvarial bone regeneration.
Microcomputed tomography
μCT scanning was conducted to assess new bone formation, utilizing a vivaCT 40 scanner (Scanco Medical) with parameters set at 17.5 µm isometric voxels, 70 kV energy, 114 µA intensity, and 200 ms integration time, as detailed in a previously published work52. Rats at Week 3 were anesthetized and scanned in the live condition as described above, while at Week 6 they were euthanized and scanned. Post-scanning, the data were refined and analyzed using Avizo software (FEI Company) for quantifying bone volume to total volume ratio (BV/TV (%)), normalized bone mineral density (BMD %), and bone coverage area (%). Importantly, the volume fraction of hydroxyapatite in the BONink was pre-calculated before conducting in vivo studies to establish a baseline for mineralization and to mitigate potential artifacts in scanned samples, which was found to be minimal. Further, μCT data was used to assess the extent of bony bridging within the defect, which was scored according to the grading scale74. These scores were determined from maximum intensity projections generated from the μCT datasets in the CT-analyser software (Bruker).
Mechanical testing of calvarial explants
Unconfined compression tests were conducted using an Instron 5966 mechanical testing system with a 10 kN load cell, using a modified fixture as per a previous report75. For the setup, the explants were aligned on a plate with a central hole matching the defects, ensuring an unobstructed path for the probe. Push-out testing was performed to assess the mechanical properties of bone specimens harvested at Week 6. Using a 4.5-mm diameter probe, uniaxial compression was applied to the bone specimens at a crosshead speed of 0.01 mm/s, pushing the probe through the defects and the hole in the plate to measure the force required for bone-implant dislodgement. The test was stopped when the probe passed through the defects. The obtained data was assessed, and the shear yield strength (MPa), modulus of resilience (MPa) and shear modulus (MPa) were calculated from the gradient of the resulting curve.
Histological assessment
In vitro bone tissue samples were fixed in 4% paraformaldehyde (PFA) overnight at RT at Weeks 2 and 4 post bioprinting. Similarly, the manual control group samples (with 64 spheroids randomly mixed into the BONink) were cultured for 28 days and then fixed. The fixed samples were dehydrated in a series of graded alcohol solutions and embedded in paraffin blocks using a Leica TP 1020 automatic tissue processor (Leica). Subsequently, 10 µm sections were cut using a Shandon Finesse Paraffin microtome (Thermo Electron Corporation) and transferred onto positively charged slides. In vivo calvarial tissue samples were harvested at Week 6 following µCT scanning. These tissues were washed with PBS and fixed in 10% PFA for 2 days. Post-fixation, samples were washed again with PBS and subjected to decalcification in 0.5 M ethylenediaminetetraacetic acid (EDTA) disodium salt solution (E14000, Research Products International) for 6 weeks. After decalcification, samples were embedded in O.C.T. cryomatrix (Epredia) and sectioned at 15 µm thickness using a Leica CM1950 cryostat at -20 °C. The resultant sections were used for subsequent histology and immunofluorescence staining. For hematoxylin and eosin (H&E) staining and deparaffinization, a Leica Autostainer ST5010 XL was used. The H&E-stained sections were then mounted and visualized using a Zeiss Axiozoom V16 inverted fluorescence microscope. To evaluate collagen deposition, Masson’s Trichrome (MT) staining was performed following the manufacturer’s protocol (Sigma-Aldrich). After dehydration, the sections were mounted and imaged with a Keyence BZ-9000 microscope.
Immunohistochemistry
For immunohistochemical (IHC) staining, sections from paraffin-embedded samples underwent deparaffinization while cryo-sectioned samples were thawed for 10–20 min at RT from -30 °C. All sections were permeabilized using 0.2% Triton X-100 for 10 min, washed in 1X PBS, and blocked with 2.5% normal goat serum (NGS) for 60 min at RT to prevent non-specific binding. To detect the bone tissue, sections were incubated with a mouse anti-procollagen 1 N-Peptide (P1NP) primary antibody (1:250 in 2.5% NGS, MA5-51183, Invitrogen), mouse anti- osteocalcin (OCN) primary antibody (1:200 in 2.5% NGS, 33-5400, Invitrogen), mouse anti-RUNX2 (runt-related transcription factor 2) primary antibody (1:20 in 2.5% NGS, ab76956; Abcam) and a rabbit anti-Sp7/Osterix (OSTERIX) primary antibody (1:100 in 2.5% NGS; ab209484, Abcam) or a rabbit anti-bone sialoprotein (BSP) primary antibody (1:100 in 2.5% NGS; ab52128, Abcam) overnight. After washing with PBS, sections were incubated with goat anti-mouse IgG (H + L) Alexa Fluor 488 (1:200 in PBS; A11017, Invitrogen) and goat anti-rabbit IgG (H + L) Alexa Fluor 647 secondary antibodies (1:200 in PBS; A21245, Invitrogen) for 3 h. Following additional washes, the sections were mounted with ProlongTM Gold antifade reagent containing DAPI and imaged using an LSM 880 Zeiss confocal microscope. To visualize the cartilage tissue, sections were similarly processed and incubated with mouse anti-Aggrecan (BC-3) primary antibody (1:50 in 2.5% NGS; MA3-16888; Thermo Scientific) and rabbit anti-Col-II primary antibody (1:100 in 2.5% NGS; ab34712, Abcam) overnight, followed by respective secondary antibodies and imaged using the Zeiss LSM 880 confocal microscope. For the fully cellular architecture, samples were fixed in 4% paraformaldehyde, embedded in O.C.T. cryomatrix (Thermo Scientific) and sectioned at 15 µm thickness using a Leica CM1950 cryostat (Leica, Buffalo Grove, IL) at -20 °C. The sections were immunostained with antibodies against Von Willebrand Factor (VWF) (Rabbit, 1:100, 27186-1-AP, Proteintech) and Fibronectin (Sheep, 1:100, AF1918, R&D Systems) to confirm the presence of endothelial cells and pre-vascularized structures. For bioprinted iPSC-derived vascular organoids, the bioprinted constructs were imaged in brightfield using a Zeiss Axio Observer at 36 and 72 h post bioprinting. Further, at 72 h, the samples were fixed and immune-stained for mouse anti-CD31 primary antibody (1:100, ab9498, Abcam), imaged using the Zeiss LSM880 confocal microscope and 3D reconstructed and rendered using Imaris software (Imaris 10.2, Oxford Instruments).
Gene expression using quantitative real-time polymerase chain reaction (qPCR)
To assess osteogenic gene expression profiles through qPCR, RNA was isolated from each sample using TRIzol reagent (Life Technologies) and subsequently purified using the RNA Mini Kit (12183025, Thermo Scientific) as per the manufacturer’s protocol at Weeks 2 and 4. The concentration of RNA from each sample was determined by measuring the absorbance at a ratio of 260/280 nm using a Nanodrop (ND-1000, Thermo Scientific). The isolated RNA was then converted to cDNA using AccuPower® CycleScript RT PreMix (K2047, BIONEER) following the manufacturer’s guidelines. Quantitative analysis of gene expression was performed using PowerUp™ SYBR™ Green Master Mix (A25742, Thermo Scientific) on a Quant Studio 3 PCR system (Thermo Scientific). The set of osteogenic genes analyzed included collagen type-1 (COL-1), runt-related transcription factor-2 (RUNX2), bone sialoprotein (BSP), bone morphogenetic protein-4 (BMP-4), and transcription factor Sp7 (OSTERIX). The primer (Integrated DNA Technologies) details for these genes were provided in Supplementary Table 2. The fold-change in gene expression of the target genes was quantified using the 2−ΔΔCT method and then normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a housekeeping gene. The fold change of negative control (non-transfected hADSCs) on Day 1 was set as 1-fold and values in all groups were normalized to that of the group.
Scalable cartilage tissue fabrication
For the scalable cartilage tissue (SCT) fabrication, CARink and chondrogenic-committed transfected spheroids were alternately bioprinted to create a 1 cm³ construct. A week after the transfection process, spheroids were deposited onto 1 × 1 cm square extruded CARink. The deposition of spheroids and CARink was repeated nine times, layering 64 spheroids into each layer to accumulate 576 spheroids per sample in a stratified manner, resulting in a 1 cm³ cartilage tissue construct. The constructs were then photocrosslinked for 3 min using 405 nm light and cultured in basal medium for two weeks in vitro to assess cartilage development.
Cell viability analysis of SCTs
LIVE/DEAD staining was performed to assess the cell viability of CARink. The fabricated scalable cartilage tissue was collected on Day 14 and washed with PBS, stained with a working solution composed of 2 µM calcein AM (1755-250, BioVision) and 4 µM ethidium homodimer-1 (EthD-1; E1169, Invitrogen) for 45 min in an incubator and then imaged using the Zeiss LSM 880 confocal microscope. Cell viability (%) was determined after the deconvolution process to reduce the background signal based on the number of cells that had green or red fluorescence signal by dividing the number of green-fluorescent cells by the total number of cells and multiplying by 100. Image J software (National Institutes of Health) was used for image analysis.
Physicochemical characterization of SCT
To quantity the sulfated glycosaminoglycan (sGAG) content of SCT, we examined both miR-(140 + 21) co-transfected and non-transfected hADSCs spheroids using a glycosaminoglycans assay kit (6022, Chondrex Inc.). For each group, three samples were collected at Week 2 and then washed with PBS. Proteinase K (P2308, Sigma Aldrich) was added to each sample at a final concentration of 0.5 mg/mL and incubated overnight at 56 °C for enzymatic cell lysis and DNA release. After incubation, 1,9-dimethylmethylene blue (DMMB) solution was added following the manufacturer’s protocol and the absorbance was measured at 525 nm using a spectrophotometer (Infinite 200 Pro, Tecan). The sGAG concentration was then normalized to the dsDNA content of each sample. DNA quantitation was performed using a Quant-iT PicoGreen dsDNA Assay Kit (P7589, Thermo Scientific) according to the manufacturer’s guidelines. Samples were excited at 480 nm and emission was measured at 520 nm using the spectrophotometer. The total DNA concentration was determined by comparison with a standard curve of Lambda DNA standard solution to ascertain DNA amounts. For histomorphometric analysis, SCT samples were fixed at Week 2 post bioprinting, dehydrated, and then sectioned. For H&E staining, resultant sections were stained using the Leica Autostainer ST5010 XL. The H&E-stained sections were then mounted and visualized using the Zeiss Axiozoom V16 inverted fluorescence microscope. For Toluidine blue O (TB) staining, sections were incubated in a 0.1% TB solution in deionized water at RT for 2 min. Following two rinses with deionized water, the sections were dehydrated using 95 and 100% ethanol, cleared with xylene, and mounted using Xylene Substitute Mountant (Epredia™) before imaging with the Keyence BZ-9000 microscope.
Statistics and Reproducibility
All data are presented as mean ± standard deviation from at least three biological replicates or otherwise stated in the manuscript and analyzed by GraphPad Prism 8.4.2. Multiple comparisons were analyzed by one-way analysis of variance (ANOVA) followed by post-hoc Tukey’s multiple-comparison test to determine the individual differences among the groups. For comparisons between two different experimental groups, statistical significance was analyzed using two-sided t-tests. Statistical differences were considered significant at *p < 0.05, **p < 0.01, ***p < 0.001. Details of specific statistical methods and p value results are included within the figure captions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work has been supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) Award R01EB034566 (I.T.O.) and National Institute of Dental and Craniofacial Research (NIDCR) Award R01DE028614 (I.T.O.). We thank Dr. Jian Yang (Penn State) for providing access to material characterization equipment, Ethan M. Gerhard (Penn State) for his assistance in mechanical testing, Vaibhav Pal (Penn State) for his assistance with preparation of DCNA and GelMA, Elisabeth G. Aliftiras (Penn State) for her assistance with illustration for Fig. 1, Dr. Suihong Liu (Penn State) for his help with preparation of spheroids, Ilayda Namli (Penn State) for her help with preparation of vascular organoids, and Jiyang Chen (Penn State) for his assistance in setting up the initial prototype of the system.
Author contributions
M.H.K. conceived ideas, designed and performed the experiments and analyses, assembled the data into figures, and wrote the manuscript. Y.P.S. performed the experiments and analyses and wrote the manuscript. N.C. and M.Y. conducted experiments and analyses. E.R. conducted animal surgeries. D.J.H. provided project guidance and contributed to data interpretation. I.T.O. conceived ideas, supervised the study, and revised the manuscript. All authors approved the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Dominic Rütsche and Dan Xing for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data supporting the findings of this study are included in the manuscript and Supplementary Information. Source data are provided with this paper.
Code availability
The developed platform in this work utilized custom designed software, which can be accessed at GitHub (https://github.com/MHKim-software/HITS-Bio.git). Details of the software interface and control algorithm were included as supplementary information.
Competing interests
I.T.O. has an equity stake in Biolife4D and is a member of the scientific advisory board for Biolife4D and Healshape. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54504-7.
References
- 1.Murphy, S. V. & Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol.32, 773–785 (2014). [DOI] [PubMed] [Google Scholar]
- 2.You, S. et al. High cell density and high-resolution 3D bioprinting for fabricating vascularized tissues. Sci. Adv.9, eade7923 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daly, A. C., Davidson, M. D. & Burdick, J. A. 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun.12, 753 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozbolat, I. T. & Yu, Y. Bioprinting toward organ fabrication: challenges and future trends. IEEE Trans. Biomed. Eng.60, 691–699 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Satpathy, A. et al. Developments with 3D bioprinting for novel drug discovery. Expert Opin. drug Discov.13, 1115–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng, Z. et al. Visible light-induced 3D bioprinting technologies and corresponding bioink materials for tissue engineering: a review. Engineering7, 966–978 (2021). [Google Scholar]
- 7.Bishop, E. S. et al. 3-D bioprinting technologies in tissue engineering and regenerative medicine: current and future trends. Genes Dis.4, 185–195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, W. et al. Stereolithography apparatus and digital light processing-based 3D bioprinting for tissue fabrication. Iscience26, 106039 (2023). [DOI] [PMC free article] [PubMed]
- 9.Ng, W. L., Lee, J. M., Yeong, W. Y. & Naing, M. W. Microvalve-based bioprinting–process, bio-inks and applications. Biomater. Sci.5, 632–647 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Banerjee, D. et al. Strategies for 3D bioprinting of spheroids: a comprehensive review. Biomaterials291, 121881 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Kim, S.-j. et al. Spatially arranged encapsulation of stem cell spheroids within hydrogels for the regulation of spheroid fusion and cell migration. Acta Biomaterialia142, 60–72 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Aguilar, I. N. et al. Scaffold-free bioprinting of mesenchymal stem cells with the regenova printer: optimization of printing parameters. Bioprinting15, e00048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badr, S. et al. Development of a mist-based printhead for droplet-based bioprinting of ionically crosslinking hydrogel bioinks. Bioprinting27, e00207 (2022). [Google Scholar]
- 14.Choi, Y.-J. et al. 3D bioprinting of in vitro models using hydrogel-based bioinks. Polymers13, 366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth, J. G. et al. Spatially controlled construction of assembloids using bioprinting. Nat. Commun.14, 4346 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao, T. et al. High-throughput fabrication of cell spheroids with 3D acoustic assembly devices. Int. J. Bioprinti.9, 106039 (2023). [DOI] [PMC free article] [PubMed]
- 17.Ayan, B. et al. Aspiration-assisted bioprinting for precise positioning of biologics. Sci. Adv.6, eaaw5111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, G. et al. 3D Printed organisms enabled by aspiration-assisted adaptive strategies. Adv. Sci.11, 2404617 (2024). [DOI] [PMC free article] [PubMed]
- 19.Ayan, B. et al. Aspiration-assisted freeform bioprinting of pre-fabricated tissue spheroids in a yield-stress gel. Commun. Phys.3, 183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo, D. N. et al. Aspiration-assisted bioprinting of co-cultured osteogenic spheroids for bone tissue engineering. Biofabrication13, 015013 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Utama, R. H. et al. A 3D bioprinter specifically designed for the high-throughput production of matrix-embedded multicellular spheroids. Iscience23, 101621 (2020). [DOI] [PMC free article] [PubMed]
- 22.Stuart, M. P. et al. Successful low‐cost scaffold‐free cartilage tissue engineering using human cartilage progenitor cell spheroids formed by micromolded nonadhesive hydrogel. Stem cells Int.2017, 7053465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salhotra, A., Shah, H. N., Levi, B. & Longaker, M. T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol.21, 696–711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neve, A., Corrado, A. & Cantatore, F. P. Osteocalcin: skeletal and extra‐skeletal effects. J. Cell. Physiol.228, 1149–1153 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Schini, M., Vilaca, T., Gossiel, F., Salam, S. & Eastell, R. Bone turnover markers: basic biology to clinical applications. Endocr. Rev.44, 417–473 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celik, N. et al. miRNA induced 3D bioprinted-heterotypic osteochondral interface. Biofabrication14, 044104 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heo, D. N., Hospodiuk, M. & Ozbolat, I. T. Synergistic interplay between human MSCs and HUVECs in 3D spheroids laden in collagen/fibrin hydrogels for bone tissue engineering. Acta biomaterialia95, 348–356 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jakab, K., Neagu, A., Mironov, V., Markwald, R. R. & Forgacs, G. Engineering biological structures of prescribed shape using self-assembling multicellular systems. Proc. Natl Acad. Sci.101, 2864–2869 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Y. et al. hESCs‐derived early vascular cell spheroids for cardiac tissue vascular engineering and myocardial infarction treatment. Adv. Sci.9, 2104299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decarli, M. C. et al. Bioprinting of stem cell spheroids followed by post‐printing chondrogenic differentiation for cartilage tissue engineering. Adv. Healthc. Mater.12, 2203021 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, W. et al. Rapid continuous multimaterial extrusion bioprinting. Adv. Mater.29, 1604630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skylar-Scott, M. A., Mueller, J., Visser, C. W. & Lewis, J. A. Voxelated soft matter via multimaterial multinozzle 3D printing. Nature575, 330–335 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Hansen, C. J. et al. High‐throughput printing via microvascular multinozzle arrays. Adv. Mater.25, 96–102 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Kopinski-Grünwald, O., Guillaume, O., Ferner, T., Schädl, B. & Ovsianikov, A. Scaffolded spheroids as building blocks for bottom-up cartilage tissue engineering show enhanced bioassembly dynamics. Acta Biomaterialia174, 163–176 (2024). [DOI] [PubMed] [Google Scholar]
- 35.Takafuji, Y. et al. MicroRNA-196a-5p in extracellular vesicles secreted from myoblasts suppresses osteoclast-like cell formation in mouse cells. Calcif. Tissue Int.108, 364–376 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Kim, Y. J., Bae, S. W., Yu, S. S., Bae, Y. C. & Jung, J. S. miR‐196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone Miner. Res.24, 816–825 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Chen, C., Liu, Y.-M., Fu, B.-L., Xu, L.-L. & Wang, B. MicroRNA-21: an emerging player in bone diseases. Front. Pharmacol.12, 722804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, N. et al. Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR‐21 in estrogen deficiency–induced osteoporosis. J. Bone Miner. Res.28, 559–573 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Mei, Y. et al. miR‐21 modulates the ERK–MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J. Cell. Biochem.114, 1374–1384 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Abu‐Laban, M. et al. Combinatorial delivery of miRNA‐nanoparticle conjugates in human adipose stem cells for amplified osteogenesis. Small15, 1902864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burdick, J. A. & Prestwich, G. D. Hyaluronic acid hydrogels for biomedical applications. Adv. Mater.23, H41–H56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung, C.-H., Golub, E. E., Forbes, E., Tokuoka, T. & Shapiro, I. M. Mechanism of action of β-glycerophosphate on bone cell mineralization. Calcif. Tissue Int.51, 305–311 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Schwab, A. et al. Printability and shape fidelity of bioinks in 3D bioprinting. Chem. Rev.120, 11028–11055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karvinen, J. & Kellomäki, M. Design aspects and characterization of hydrogel-based bioinks for extrusion-based bioprinting. Bioprinting, e00274 (2023).
- 45.Wang, X. et al. Topological design and additive manufacturing of porous metals for bone scaffolds and orthopaedic implants: A review. Biomaterials83, 127–141 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Zhao, X. et al. Photocrosslinkable gelatin hydrogel for epidermal tissue engineering. Adv. Healthc. Mater.5, 108–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allen, N. B. et al. 3D-bioprinted GelMA-gelatin-hydroxyapatite osteoblast-laden composite hydrogels for bone tissue engineering. Bioprinting26, e00196 (2022). [Google Scholar]
- 48.Skylar-Scott, M. A. et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv.5, eaaw2459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffin, K. H., Fok, S. W. & Kent Leach, J. Strategies to capitalize on cell spheroid therapeutic potential for tissue repair and disease modeling. npj Regenerative Med.7, 70 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, Y., Ravnic, D. J. & Ozbolat, I. T. Intraoperative bioprinting: repairing tissues and organs in a surgical setting. Trends Biotechnol.38, 594–605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keriquel, V. et al. In situ printing of mesenchymal stromal cells, by laser-assisted bioprinting, for in vivo bone regeneration applications. Sci. Rep.7, 1778 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moncal, K. K. et al. Intra‐operative bioprinting of hard, soft, and hard/soft composite tissues for craniomaxillofacial reconstruction. Adv. Funct. Mater.31, 2010858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alcaide-Ruggiero, L., Cugat, R. & Domínguez, J. M. Proteoglycans in articular cartilage and their contribution to chondral injury and repair mechanisms. Int. J. Mol. Sci.24, 10824 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouaziz, W. et al. Interaction of HIF1α and β-catenin inhibits matrix metalloproteinase 13 expression and prevents cartilage damage in mice. Proc. Natl Acad. Sci.113, 5453–5458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potyondy, T. et al. Recent advances in 3D bioprinting of musculoskeletal tissues. Biofabrication13, 022001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guimarães, C. F., Gasperini, L., Marques, A. P. & Reis, R. L. The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater.5, 351–370 (2020). [Google Scholar]
- 57.Guan, F. et al. Application of optimization methodology and specimen-specific finite element models for investigating material properties of rat skull. Ann. Biomed. Eng.39, 85–95 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Gefen, A., Gefen, N., Zhu, Q., Raghupathi, R. & Margulies, S. S. Age-dependent changes in material properties of the brain and braincase of the rat. J. neurotrauma20, 1163–1177 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Mao, H., Wagner, C., Guan, F., Yeni, Y. N. & Yang, K. H. Material properties of adult rat skull. J. Mech. Med. Biol.11, 1199–1212 (2011). [Google Scholar]
- 60.Collins, M. N. et al. Scaffold fabrication technologies and structure/function properties in bone tissue engineering. Adv. Funct. Mater.31, 2010609 (2021). [Google Scholar]
- 61.Arlettaz, Y. Augmented osteosynthesis in fragility fracture. Orthop. Traumatology: Surg. Res.109, 103461 (2023). [DOI] [PubMed] [Google Scholar]
- 62.Zhang, L., Hu, J. & Athanasiou, K. A. The role of tissue engineering in articular cartilage repair and regeneration. Crit. Rev. Biomedi. Eng.37, 1–57 (2009). [DOI] [PMC free article] [PubMed]
- 63.Wu, Y. et al. Three-dimensional bioprinting of articular cartilage: a systematic review. Cartilage12, 76–92 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uzel, S. G., Weeks, R. D., Eriksson, M., Kokkinis, D. & Lewis, J. A. Multimaterial multinozzle adaptive 3D printing of soft materials. Adv. Mater. Technol.7, 2101710 (2022). [Google Scholar]
- 65.Dey, M. et al. Chemotherapeutics and CAR‐T cell‐based immunotherapeutics screening on a 3D bioprinted vascularized breast tumor model. Adv. Funct. Mater.32, 2203966 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wimmer, R. A., Leopoldi, A., Aichinger, M., Kerjaschki, D. & Penninger, J. M. Generation of blood vessel organoids from human pluripotent stem cells. Nat. Protoc.14, 3082–3100 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Kim, M. H., Banerjee, D., Celik, N. & Ozbolat, I. T. Aspiration-assisted freeform bioprinting of mesenchymal stem cell spheroids within alginate microgels. Biofabrication14, 024103 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ayan, B., Wu, Y., Karuppagounder, V., Kamal, F. & Ozbolat, I. T. Aspiration-assisted bioprinting of the osteochondral interface. Sci. Rep.10, 13148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, M. et al. Gelatin methacryloyl and its hydrogels with an exceptional degree of controllability and batch-to-batch consistency. Sci. Rep.9, 6863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pal, V. et al. High-throughput microgel biofabrication via air-assisted co-axial jetting for cell encapsulation, 3D bioprinting, and scaffolding applications. Biofabrication15, 035001 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang, Y. et al. Intraoperative bioprinting of human adipose-derived stem cells and extra-cellular matrix induces hair follicle-like downgrowths and adipose tissue formation during full-thickness craniomaxillofacial skin reconstruction. Bioact. Mater.33, 114–128 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Theret, D. P., Levesque, M., Sato, M., Nerem, R. & Wheeler, L. The application of a homogeneous half-space model in the analysis of endothelial cell micropipette measurements.110, 190–9 (1988). [DOI] [PubMed]
- 73.Celik, N., Kim, M. H., Hayes, D. J. & Ozbolat, I. T. miRNA induced co-differentiation and cross-talk of adipose tissue-derived progenitor cells for 3D heterotypic pre-vascularized bone formation. Biofabrication13, 044107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patel, Z. S. et al. Dual delivery of an angiogenic and an osteogenic growth factor for bone regeneration in a critical size defect model. Bone43, 931–940 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawson, Z. T. et al. Methodology for performing biomechanical push-out tests for evaluating the osseointegration of calvarial defect repair in small animal models. MethodsX8, 101541 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data supporting the findings of this study are included in the manuscript and Supplementary Information. Source data are provided with this paper.
The developed platform in this work utilized custom designed software, which can be accessed at GitHub (https://github.com/MHKim-software/HITS-Bio.git). Details of the software interface and control algorithm were included as supplementary information.