Abstract
Bacillus pumilus plays an essential role in agricultural applications as a beneficial microbe and for sustainable agriculture production. However, the underlying mechanisms of B. pumilus strains remain unclear as to how they are beneficial for plants as stress tolerant and growth promoters. Bacillus pumilus was isolated from the rhizosphere soil of Artemisia vulgaris. NGS (next-generation sequencing) was performed for the strain to gain new insights into the molecular mechanisms underlying plant-microbial interactions. NGS revealed 3,910 genes, 3294 genes with protein-coding, and 11 functional genomic regions related to diverse agronomic traits including stress tolerance. We identified the two possible phytohormone biosynthesis approaches from metabolic regions1(terpense→diterpense→betacarotene→xanthoxin→ABA)2(terpense→diterpense→geranyl diphosphate →C20 →GA). Several gene clusters related to the biosynthesis of phytohormones, stress tolerance, and agricultural diversification were predicted. The genome provides insights into the possible mechanisms of this bacterium for stress tolerance and its future applications. The genomic organization of B. pumilus revealed several hallmarks of its plant growth promotion and pathogen suppression activities. Our results provide detailed genomic information for the strain and reveal its potential stress tolerance mechanisms, laying the foundation for developing effective stress tolerance strategies against abiotic stress.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78227-3.
Keywords: Bacillus pumilus, NGS, Food security, Secondary metabolites
Subject terms: Physiology, Plant sciences
Introduction
Host plants harbor a symbiotic relationship with the microbes. Beneficial microbes are important in enhancing the physiological traits and plant stress tolerance against several stresses1. The agricultural industry has to face dual stress one is to meet the needs of the hungry population, and the other is environmental stress. Both biotic and abiotic stress reduce crop production negatively2. However, several conventional approaches are being practiced enhancing agriculture production. Among them, the most recent approach is the application of beneficial microbes in enhancing agronomic traits and stress reticence. Beneficial microbes, especially bacteria and fungi, are known for their plant growth-promoting potential (PGP) without causing symptoms to the plant cells3. They can colonize plants and exert their beneficial effect on plants to facilitate the growth and development of the plant. The promotion of plant growth by beneficial microbes has been observed to be facilitated in several ways, such as the production of secondary metabolites and enzymes, gene modulation, and defense-related constitution in host cells. Moreover, microbes can affect the host by regulating the antioxidant defense system4,5.
Bacillus pumilus was first isolated from the rhizosphere soil of an arid medicinal plant, Artemisia vulgaris (Pohang Beach, South Korea), and was subsequently found to increase the growth and stress tolerance of rice and soybean. It has been suggested that it can induce the production of phytohormones, organic acids, and biofilms6. It belongs to the genus Bacillus, and members of the Bacillus genus are rod-shaped and sprout in nature. These are gram-positive, chemoheterotrophic, and aerobic microbes. An increasing trend has been observed in the use of microbes such as Bacillus for abiotic stress tolerance7. B. pumilus sp. has been isolated from several different environments; however, only a few reports have described their effect on Plant abiotic stress tolerance8. Bacillus sp. have mostly been reported for their roles in various pollutants and the overexpression of metallothionein in host cells, which alleviates heavy metal toxicity in plants9. This species can also improve salinity10, drought11, and hydrogen peroxide12 stress by regulating the endogenous phytohormones and antioxidant defense system. In addition, this species can produce bioactive secondary metabolites13. Previous studies have suggested that the B. pumilus strain can be used for PGP14. Secondary metabolites produced by beneficial bacteria are not only significant for the producing cells but also affect the host. As bioactive substances, these metabolites have important uses in the pharmaceutics and agriculture industries. Researchers may now prioritize industrially significant secondary metabolites at the genome level and learn more about the genetic groundwork of the strain’s adaptable lifecycle. These specific metabolites are a potential source of stress-tolerant agents and sustainable crop production. For example, previous researchers revealed that B. pumilus contains several genes that promote plant development15–18.
DNA-based investigations of the microbes are highly valuable in answering the aforementioned queries. The agronomic potential and genetic traits of beneficial microbes can be understood through metabolomics investigations of entire microbial colony genome analyses through NGS. This RNA-based research clarified the detailed mechanism of action and regulatory process of the microbes. NGS technologies are significantly possible to answer previously unanswerable issues, mostly about plant-microbial interaction19. Consequently, studying plant-microbe relationships more quickly and thoroughly than ever before is possible20.
This study was performed to determine the complex biological mechanisms by characterizing and identifying the genomic regions that are responsible for stress tolerance. In addition, candidate genes (with agronomic traits that allow plant-microbial interaction) were identified via genome sequencing. It will provide a platform to understand the complex plant-microbial interaction against environmental stress.
Materials and methods
Media cultivation and isolation
The B. pumilus culture was grown from the original stock of SH-9 archived at the Crop Physiology Laboratory at Kyungpook National University, Daegu, South Korea. Previously it was isolated from the rhizospheric soil around the Artemisia plant located Pohang-si, North Gyeongsang, South Korea ( 35.84151 128.99185 36.33363 129.60108 ). Stock was streaked on L.B. agar plates and incubated at 27–30 °C until colonies appeared. After getting the single pure colony was inoculated in 5 mL of L.B. broth cultured overnight at 37 °C, agitated at 100-150 rpm which is the appropriate temperature and agitation according to standard protocol. This procedure was followed according to the method described by21.
DNA extraction and genomic sequencing
Genomic DNA was extracted from 1-mL culture aliquots using the Maxwell 16 DNA/RNA Extraction System (Solgent Company, South Korea). Genomic DNA was extracted for complete genome sequencing using a Qiagen kit (Qiagen, Hilden, Germany). For NGS, the single molecular real-time technique was used by Pacific Bioscience (Pac Bio, Menlo Park, CA, USA). The methodology used was followed according to that described by ADDIN EN.CITE22–24. Sequencing was carried out at the KNU NGS Core Facility utilizing the Illumina MiSeq platform (Illumina, USA) according to the manufacturer’s instructions.
Genome annotation
The NCBI prokaryotic genome annotation pipeline (PGA) was used to perform the complete genome annotation. Rapid annotation using subsystem technology version 3.0, and an integrated microbial genome platform were used for the gene prediction. The assembled and annotated sequences were submitted to GenBank with bio project ID (PRJNA1056679), Locus tag prefix VA028 (SAMN39105494) and the information is available online in the Genome online database https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB13160900/overview.The annotation was performed as described by ADDIN EN.CITE17,24,25.
Genome assembly statistics
NGS was performed at Kyungpook National University, South Korea, and the genome assembly statistics were performed using the build 6494 software. The NGS center at Kyungpook National University, South Korea, provided the annotation. The annotation pipeline used was the NCBI PGA. The adopted annotation methods were the best-placed reference per set and GeneMarkS-2. They provided us with the gene, coding sequence (CDS), rRNA, and tRNA data. The entire protocol was followed as described by26.
Statistical data analysis
The statistical software SPSS (22.0) for Windows (SPSS, Chicago, IL, USA) was utilized to perform a one-way analysis of variance and the general linear technique on the experimental data. To compare mean data and identify treatment differences, Tukey’s test (p ≤ 0.05) was employed. Five replicates, on average, were used for each treatment value.
Results
Isolation and identification of Bacillus pumilus
The rhizosphere soil samples were collected and processed to isolate competent microbes. The objective was to decipher the mechanism of action of stress tolerance and plant-microbial interaction. After subsequent isolation, the rhizobacterial strain was identified via whole genome sequencing (Fig. 1). Moreover, the plant growth-promoting traits of the Bacillus pumilus were also analyzed by using the NGS. The results revealed that it processes multiple plant growth-promoting traits certified by whole genome sequencing technique Table 1.
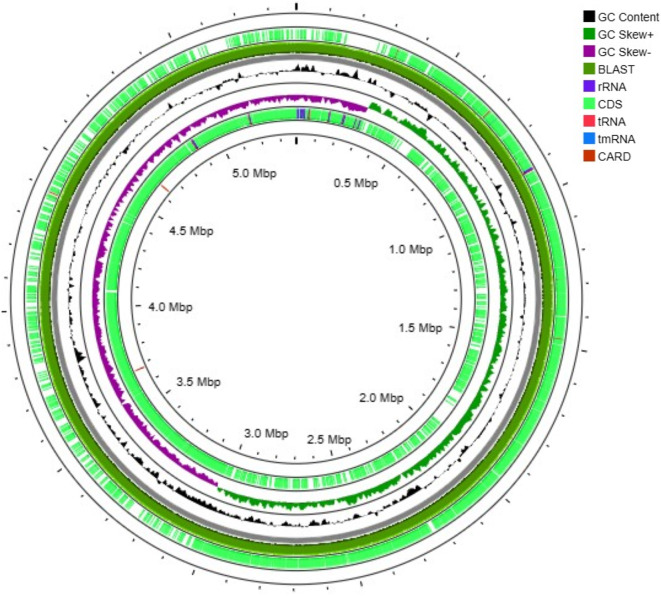
Fig. 1.
Modulation of the gene expression for stress tolerance and agronomic traits of the SH-9 Bacillus pumilus genome. Outer to inner circles show the predicted protein-coding sequence on the plus and minus strands. The third circle shows the distribution of the genes related to the Clusters of Orthologous Genes (COG) categories, and the fourth and fifth circles show the G+C content and G+C skew, respectively.
Table 1.
List of genes attributed to agronomic traits in the Bacillus pumilus genome.
NRP non-ribosomal peptide, TIPKS type 1 polyketides, T3PKS type 3 polyketides, RRE Rev response element, RiPP ribosomal synthesized and post-translationally modified peptide, NA not applicable.
Identification of ABA and GA biosynthesis from precursor of terpenes
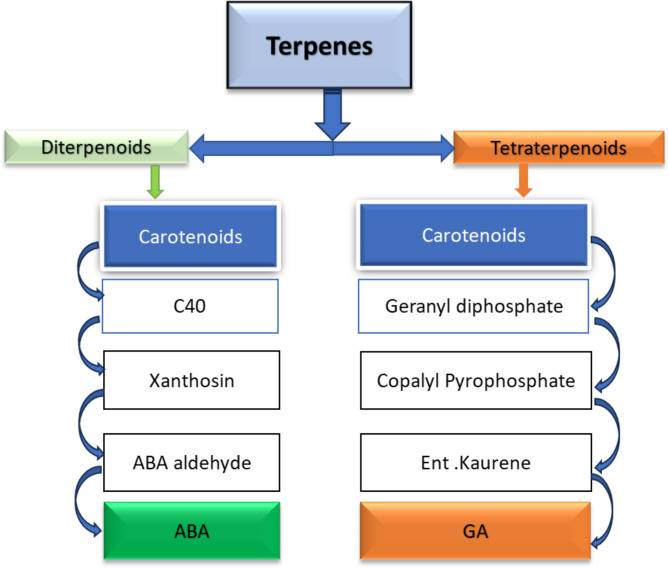
In plants ABA is synthesized in vascular tissue from there, it is transported to the targeted tissue of plants. Therefore, in the present study we detected ABA and GA biosynthesis approach from metabolic precursor terpenes Results revealed that SH-9 has an innate ability to synthesize ABA from precursor of terpenes. Interestingly, Terpenes were present at 2 metabolic region 3 and region 5. Moreover, we also detected that it also can biosynthesize GA. The entire mechanism is explained in (Fig. 2). The related genes to ABA and GA synthesis were certified in WGS of SH-9 (Table 1)0.11 metabolic regions were identified which are shown in Table 2.
Fig. 2.
Possible phytohormones biosynthesis approach from genomic region 5, terpene acts as a bio precursor of the biosynthesis and ultimately gives yield to ABA and GA.
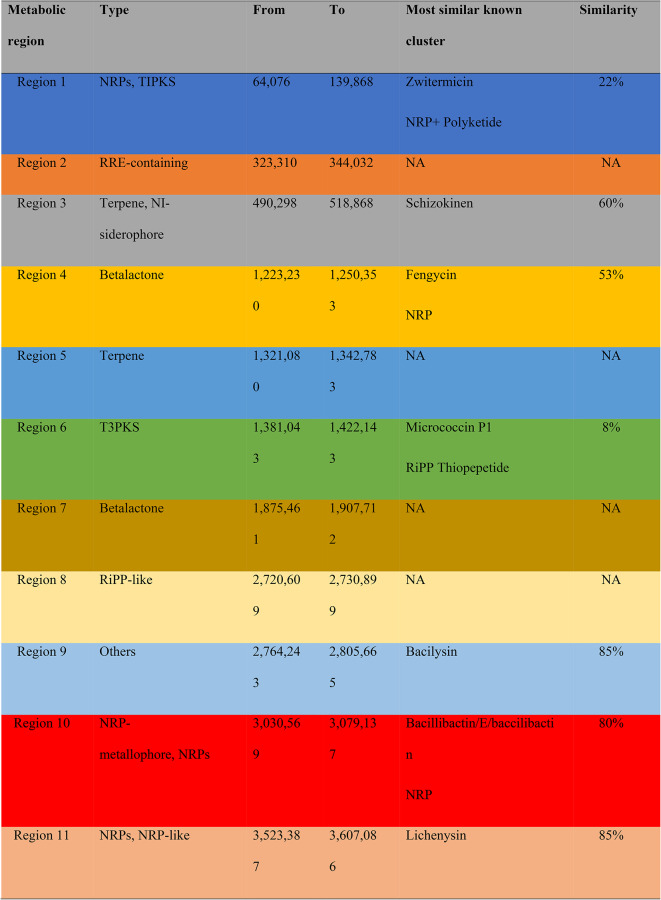
Table 2.
Functional genomic regions of the Bacillus pumilus, genomic region, and production of secondary metabolites.
Gene prediction and annotation summary
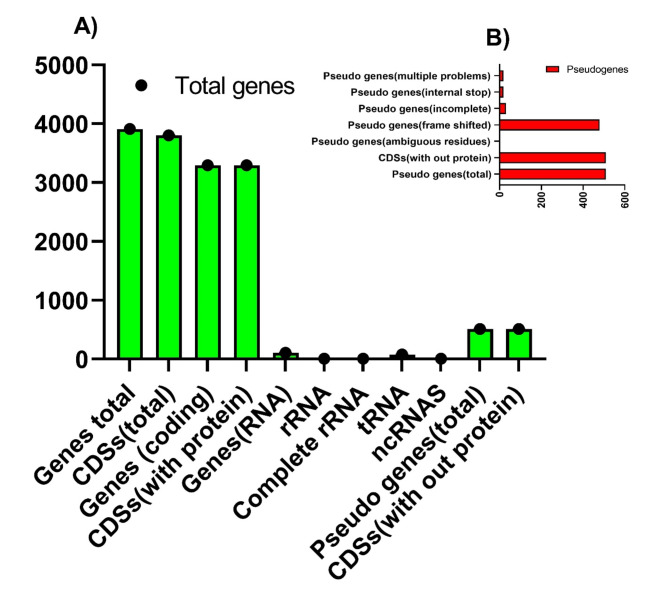
Further analysis of the Bacillus pumilus genome revealed the total number of genes as well as the available protein genes (Fig. 3A and B, respectively). To investigate the stress tolerance mechanisms of B. pumilus, we screened the potential metabolic regions and their corresponding genes. The results revealed that it contains several coding- and pseudogenes, as shown in Fig. 3. Total genes, CDSs, and protein-coding genes were prevalent with close to 3.8 kb of data, as shown in Fig. 3A. However, the total pseudogenes and CDSs without proteins were prolific. The total RNA genes, rRNA, complete rRNA, tRNA, and ncRNAs were present in insignificant amounts in Bacillus pumilus, as shown in Fig. 3. NGS also revealed that it has metabolic regions that produce secondary metabolites. All metabolic regions (n=11) can enhance plant agronomic traits and stress tolerance via several gene modulations.
Fig. 3.
Potential genomic sequences represent corresponding genes of the Bacillus pumilus SH-9. (A) Total number of genes and (B) pseudogenes in the genome sequence.
Genomic regions associated with agronomic traits
Sequence analysis of the rhizosphere Bacillus sp. and its potential for the enhancement of plant growth. A genomic area has a biological function if it has reproducible biochemical activity, such as transcriptional activity or certain chromatin states, which can be observed. NGS has been extensively used in recent years to identify genomic regions and candidate genes for agronomic traits in microbes, and its crosstalk has several diversification qualities27,28. Numerous candidate genes and quantitative regions are associated with agronomic traits29. This study revealed that Bacillus pumilus has 11 functional genomic regions, as shown in Table 2. At genomic region 1, non-ribosomal peptides (NRPs) 64,076–139,868 and type 1 polyketides (TIPKS) were found with 22% similarity to the known cluster of Zwittermicin A. These are a class of secondary peptide metabolites. These are extraordinarily diverse and have various biological and pharmacological features. They are frequently responsible for siderophore and pigment production in plants and also act as a biocontrol agent against plant stress30,31. Similarly, in region 11, they contain NRPs, which have 85% similarity with Lichenysin.
Figure 4 shows the functional genomic regions of the SH-9 strain i.e., SH-9 genomic region and production of secondary metabolites, and the total number of metabolic regions in the SH-9 strain genome. In terms of both structure and biological function, TIPKS represent a diverse class of chemicals. These substances, which include macrolides, polyethers, polyols, and aromatics, are frequently strongly oxygenated. They have various biological effects, including antibacterial, anticancer, antifungal, antiphrastic, and immunosuppressive32,33. Polyketides are present in genomic region 1. The genomics region 2 contains the Rev response element (RRE) nucleotides from 323,310 to 344,032. This is a specialized and cooperative complex of Revs, and the cellular export machinery utilizes RRE as a scaffolding platform for assembly. Creating a high-affinity, export-competent complex depends on the cooperativity that is governed by the RRE structure and sequence34.
Fig. 4.
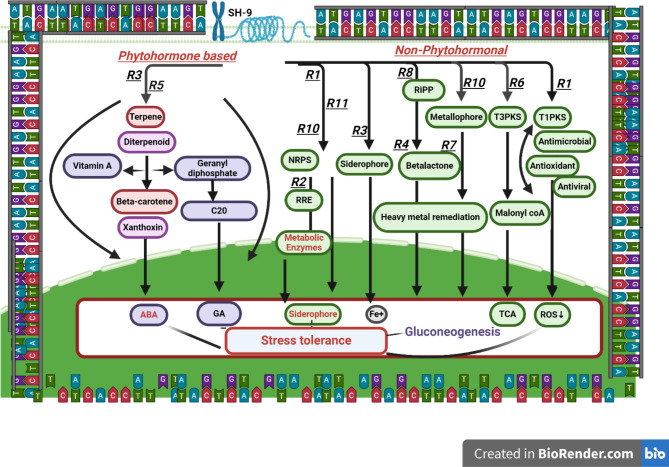
Model elucidating the phytohormonal, transcriptional, and signaling modulation triggered by B. Pumilus that gives resistance to stress.
At genomic region 3, terpene NI siderophores were found from 490,298 to 518,868, which has a 60% similarity with Schizokinenn. Because of their diverse properties, terpenes are essential natural compounds that are important for pollination, fragrance, and biological control. Terpenes are also precursors of gibberellic acid (GA). This genomic region is important because GA is produced here. GA is an important phytohormone that is responsible for the elongation of cells and, ultimately, the growth of plants35,36. Similarly, in the genomic region 5, the production of terpenes was also found. From this region, siderophores are secreted, which can induce iron chelation37. At genomic regions 4 and 7, betalactone is produced with a 53% similarity to fungicide. It acts as a potent, irreversible antagonist of strigolactone receptors. When the Plant is exposed to stressful conditions, betalactone works antagonistically to inhibit branching38. At genomic region 8, ribosomal synthesized and post-translationally modified peptides (RiPPs) are abundant from 2,720,609–2,730,899. These diverse peptide classes are responsible for several biological translation processes and their post-modification in response to stress39,40. In genomic region 10, NRPS metallophores are present from 3,030,569–3,079,137, which shows an 80% similarity with bacillibactin. This diverse class of microbes can chelate metals and be used for heavy metal stress tolerance41,42.
Metabolic regions and their potential to enhance agronomic traits
Reactive oxygen species (ROS) accumulation is enhanced in plants in response to stress conditions, such as oxidative stress. Metabolic reprogramming is performed to reduce stress43,44. However, it is challenging to cope with stress when the duration of stress increases and metabolic reprogramming fails. In this scenario, beneficial microbes produce several secondary metabolites. In this study, SH-9 was screened by NGS to produce secondary metabolites, and the results revealed that in metabolic region one, T1PKS is produced, which can lower oxidative stress and work as an antioxidant. T1PKS also works as an antimicrobial agent. Several studies have suggested that T1PKS has the potential to lower ROS levels, and our results agree that it can lower oxidation stress. In metabolic region 6, type 3 polyketides are present that can activate the TCA cycle. One of the intermediary steps of cellular respiration is the Krebs cycle. Carbohydrates are a group of substrates in which some energy is generated by oxidizing acetyl-CoA and is also obtained by breaking down respiratory substrates such as fats and proteins.
In metabolic region 10, a metallophone is present that remediates heavy metal stress tolerance. In regions 8 and 10, RiPP and NRPS are present, and they are involved in several metabolic and enzymatic processes in plant cells. Metabolic region 5 is important because it contains terpenes, which are important secondary metabolites. Terpenes are converted into diterpenoids and triterpenoids, which are subsequently converted into xanthroin, a precursor of abscisic acid (ABA). Diterpenoid also converts into geranyl diphosphate, which produces gibberellins. ABA and GA are important phytohormones that enhance stress tolerance and improve plant growth even under stressed conditions45.
Discussion
Higher plants are sessile and are exposed to stress throughout their lives. The plant uses several cellular events to protect itself in response to this stress (biotic and abiotic). All plants adopt nonspecific strategies, depending on the stress and duration. However, the general mechanism produces ROS and activates the antioxidant defense system46. The ROS acts as a signaling molecule that initiates several metabolic processes in plants to combat stress, such as the production of phytohormones, secondary metabolites, activation of some genes, and modulation in physio-biochemical processes. However, plants activate their metabolic reprogramming to alleviate stress, but it is up to a certain limit47. After a specific limit, the plant shows some symptoms of stress. Several techniques are being practiced, such as breeding techniques, agrochemicals, intercropping, and application of plant growth regulators and phytohormones. Bio stimulants are getting more attention because of their eco-friendliness and minimal side effects. The application of beneficial microbes is a promising food and energy security approach. Beneficial microbes induce the plant microbial interaction. Plant-microbial interaction facilitates plants’ biological processes and improves plant stress tolerance48.
Several recent studies significantly impacted plant growth and stress tolerance capacities. Recently, our research team conducted experimental work on beneficial microbes (SH-9) and showed excellent results regarding plant growth promotion and stress tolerance capacities49. The results gave us the idea to unravel the hidden mechanism of plant-microbial interaction. In the present study, we did the next-generation sequencing of the microbe SH-9. The results showed that in response to stress, the SH-9 genome encodes several genes that protect against stress. Different genomic regions are responsible for different stress conditions from different genomic regions. The NGS study revealed the presence of 3910 genes with 11 functional genomic regions. A genomic region is the presence of a functional component within the gene responsible for some specific function.
Whenever a plant is exposed to stress, there is a rapid accumulation of ROS that enhances the oxidative stress. Oxidative stress increases the oxidation burst that impedes the damage to the cellular organelles50. In response to oxidative stress, the SH-9 genome encodes ten genes at different regions, namely cdr, rnr, resE, rapA-5, qoxb, ychF, ggt, cyPB, adhA-2, and fhs, to regulate the oxidative stress. The presence of these genes further confirms that SH-9 significantly counteracts the generation of ROS. These findings suggest that the strong antioxidative mechanisms of SH-9 alleviate the oxidative onslaught.
Four genes, namely rnr, Sa1A, SrfAB-1, and g1tA, are found in the genome of SH-9 and are responsible for abiotic stress tolerance. Abiotic stress is the most common phenomenon that limits plant production. Abiotic stress reduces the global yield of plants to 70%. The presence of metabolic regions that contain the genes that enhance abiotic stress tolerance is a vital characteristic of beneficial microbes51. The presence of these genes on the SH-9 plasmid is surprising. This is a promising approach toward stress tolerance and a potentially profitable approach to enhancing plant agronomic traits. Table 2 shows the modulation of the gene expression for stress tolerance and agronomic characteristics of the SH-9 genome.
Iron and magnesium are important micronutrients required for a plant’s physiological growth. Under nutritional or other stressed conditions, the plant faces iron and magnesium deficiency52. Therefore, to cope with this stress, the plasmid of the SH-9 genome encodes for the genes uxaA, dos, and FadF that can bind iron and magnesium. Therefore, it is an essential characteristic of the beneficial microbe to encode the relevant genes that enhance their solubilization. In plant-microbial interactions, biofilm plays a substantial role in facilitating the interaction and alleviating plant stress. Biofilm acts as an interacting medium for plant-microbial interaction. It enhances plant growth, promotion, and production. It also facilitates several biochemical processes53,54. SH-9 encodes for the gene estA to facilitate this mutualistic interaction, which produces glycolipids. Phosphorus is another major molecule that is involved in plant growth and production55. The genome of SH-9 encodes for the six genes Dxs, resE, tagF, SasA-1, CarB-1, and CarB-2 that aid in phosphate solubilization. An important plant growth-promoting trait enhances plant growth and regulation. Microbes can convert insoluble phosphorus to soluble phosphorus and improve plant phosphorus nutrition.
Secondary metabolites are important compounds that are produced in response to stress. That enhances plant growth and improves stress tolerance56. The SH-9 genome also encodes the genes that facilitate metabolite transportation where they are needed. The SH-9 genome encodes for the six genes Csbc, YYCB-1, fruA-1, mdtc, YdfJ, and CYPB, which facilitate the metabolite transportation system. In response to stress, plant-microbial interactions produce several secondary metabolites, such as glycine, betaine, and proline39. These genes are responsible for enhancing the ease of transportation. The SH-9 genome encodes BgIF-1 and mainly acts as a signaling molecule to initiate mutualistic interaction with plant microbes. Signaling molecules act as mainstream to transmit information between microbes and plant cells, activating the cascades of events such as metabolomics expression corresponding to the stress tolerance mechanism57.
Moreover, the nuclease process is also fundamental in bringing the genetic stability that is being incorporated due to the stress, i.e., oxidation stress58 Under oxidation stress, the entire genetic modulation impairs plant cells’ cellular function. The SH-9 genome also encodes for ten genes rnr, leus, va1S, cdr, resE, Sa1A, YhaN, addB, addA2, and ctaD that are directly involved in the nuclease process (DNA and RNA) and improve the genetic stability.
The two most important genes that are encoded by the SH-9 genome are dxs and Acsa-1, which modulate plant phytohormones. Phytohormones are essential for stress tolerance and in several biological processes in plants. Genomic regions 3 and 5 are the most important and responsible for the production of phytohormones. Phytohormones reduce the plant stress and improve the plant production. In genomic regions 3 and 5, terpene has been reported as a major precursor of GA and ABA41. ABA is an important phytohormone that acts as a signaling molecule in response to stress. In addition, it is a versatile hormone that has a role in the stomatal closure in response to stress and reserves the plant activity to combat the level of stress. Moreover, it also has several functions in the physiological process of plants. Gibberellic acid is also an important class of phytohormones that stimulates plant growth, organ development, and cell proliferation59. Our results confirmed that the genome of SH-9 encodes the genes that are liable to respond to stress and improve plant productivity. The results of genomic sequencing and the proposed stress tolerance mechanism are given in Fig. 4.
Conclusion
In the present study, we isolated growth growth-promoting B. pumilus strain SH-9 from a rhizospheric soil field and sequenced its genome. The present study utilized NGS to obtain a comprehensive view of the mechanisms of B. pumilus and the functional diversity that is associated with stress tolerance and improving agronomic traits. The result explains that it can biosynthesis the two important phytohormones ABA and GA. The results revealed that B. pumilus genome encodes the 3910 genes at 11 different genomic regions that are responsible for agronomical traits and stress tolerance via plant-microbial interaction. This study may paw the way to the application of B. pumilus as an alternative sustainable strategy to improve crop yield under unfavorable conditions. However, further transcriptomic and metabolomic analysis is required to confirm the association of the genes highlighted here with the indicated functional potential.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Research Foundation of Korea(NRF) grant funded by the Korea government(MSIT) (No. 2022R1A2C1008993). The authors extend their appreciation to the Deanship of Scientific Research, King Saud University for funding through the Vice Deanship of Scientific Research Chairs; Research Chair of Prince Sultan Bin Abdulaziz International Prize for Water.
Abbreviations
- NGS
Next generation sequencing
- NRP
Non-ribosomal peptide
- TIPKS
Type 1 polyketides
- T3PKS
Type 3 polyketides
- RRE
Rev response element
- RiPP
Ribosomal synthesized and post-translationally modified peptide
Author contributions
Shifa Shaffique conceptualize and write the original draft as the part of her project brain Korea BK21,4TH phase. Dr. Anis Ali Shah, Arjun Adhikari did the critical review editing. Sang-Mo Kang, Abdul Latif Khan, Peter Odongkara, and Hosam O. Elansary did the software manipulation. Prof In-Jung Lee validated the results and supervised the entire project.
Funding
This research was funded by the Deanship of Scientific Research, King Saud University through the Vice Deanship of Scientific Research Chairs; Research Chair of Prince Sultan Bin Abdulaziz International Prize for Water.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Malusá, E., Sas-Paszt, L. & Ciesielska, J. Technologies for beneficial microorganisms inocula used as biofertilizers. The scientific world journal 2012, (2012). [DOI] [PMC free article] [PubMed]
- 2.Hirayama, T. & Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J.61 (6), 1041–1052 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Van Wees, S. C., Van der Ent, S. & Pieterse, C. M. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant. Biol.11 (4), 443–448 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Russo, A. et al. Plant beneficial microbes and their application in plant biotechnology. Innovations Biotechnol.2012:57–72 .
- 5.Porter, S. S. et al. Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Funct. Ecol.34 (10), 2075–2086 (2020). [Google Scholar]
- 6.Shaffique, S. et al. Unraveling the new member Bacillus pumilus SH-9 of Bacillaceae family and its potential role in seed biopriming to mitigate drought stress in Oryza sativa. Plant. Stress. 11, 100318 (2024). [Google Scholar]
- 7.Shafi, Z., Sharma, A. K. & Sahu, P. K. Application of Bacillus species in the alleviation of salinity-stressed agricultural soil: an overview. Appl. Bacillus Bacillus Derived Genera Agric. Biotechnol. Beyond2024:107–131 .
- 8.Govindasamy, V. et al. Bacillus and Paenibacillus spp.: potential PGPR for sustainable agriculture. Plant. Growth Health Promoting bacteria2011:333–364 .
- 9.Rizvi, A., Ahmed, B., Zaidi, A. & Khan, M. S. Heavy metal mediated phytotoxic impact on winter wheat: oxidative stress and microbial management of toxicity by Bacillus subtilis BM2. RSC Adv.9 (11), 6125–6142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vimal, S. R., Gupta, J. & Singh, J. S. Effect of salt tolerant Bacillus sp. and Pseudomonas sp. on wheat (Triticum aestivum L.) growth under soil salinity: a comparative study. Microbiol. Res.9 (1), 7462 (2018). [Google Scholar]
- 11.Vardharajula, S., Zulfikar Ali, S., Grover, M., Reddy, G. & Bandi, V. Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact.6 (1), 1–14 (2011). [Google Scholar]
- 12.Zuber, P. Management of oxidative stress in Bacillus. Annu. Rev. Microbiol.63, 575–597 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Mondol, M. A. M., Shin, H. J. & Islam, M. T. Diversity of secondary metabolites from marine Bacillus species: chemistry and biological activity. Mar. Drugs. 11 (8), 2846–2872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffique, S. et al. Unlocking the potential of newly isolated phytohormone-producing bacterial strains for enhanced plant growth and stress tolerance. Plant. Stress. 10, 100260 (2023). [Google Scholar]
- 15.Stepanov, V. G. et al. Bacillus pumilus SAFR-032 genome revisited: sequence update and re-annotation. PloS One. 11 (6), e0157331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pudova, D. S. et al. Comparative genome analysis of two Bacillus pumilus strains producing high level of extracellular hydrolases. Genes. 13 (3), 409 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iqbal, S., Vollmers, J. & Janjua, H. A. Genome mining and comparative genome analysis revealed niche-specific genome expansion in antibacterial bacillus pumilus strain SF-4. Genes. 12 (7), 1060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Q. et al. Comparative genomic analyses reveal genetic characteristics and pathogenic factors of Bacillus pumilus HM-7. Front. Microbiol.13, 1008648 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park, S. T. & Kim, J. Trends in next-generation sequencing and a new era for whole genome sequencing. Int. Neurourol. J.20 (Suppl 2), S76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci.5, 86904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin, D-S. et al. Plant growth-promoting potential of endophytic bacteria isolated from roots of coastal sand dune plants. J. Microbiol. Biotechnol.17 (8), 1361–1368 (2007). [PubMed] [Google Scholar]
- 22.Asaf, S. et al. Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species. PLoS One. 13 (3), e0192966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iqbal, S., Ullah, N. & Janjua, H. A. In vitro evaluation and genome mining of Bacillus subtilis strain RS10 reveals its biocontrol and plant growth-promoting potential. Agriculture. 11 (12), 1273 (2021). [Google Scholar]
- 24.Iqbal, S. & Begum, F. Identification and characterization of integrated prophages and CRISPR-Cas system in Bacillus subtilis RS10 genome. Brazilian J. Microbiol.55 (1), 537–542 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asaf, S., Khan, A. L., Khan, M. A., Al-Harrasi, A. & Lee, I-J. Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3 Biotech.8, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asaf, S. et al. Chloroplast genomes of Arabidopsis Halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: structures and comparative analysis. Sci. Rep.7 (1), 7556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong, K. H. et al. Analysis of the vaginal microbiome by next-generation sequencing and evaluation of its performance as a clinical diagnostic tool in vaginitis. Annals Lab. Med.36 (5), 441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wensel, C. R., Pluznick, J. L., Salzberg, S. L. & Sears, C. L. Next-generation sequencing: insights to advance clinical investigations of the microbiome. J. Clin. Investig.132(7). (2022). [DOI] [PMC free article] [PubMed]
- 29.Chelliah, R. et al. A review on the application of bioinformatics tools in food microbiome studies. Brief. Bioinform.23 (2), bbac007 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Annaswamy, S. Building a hybrid modular polyketide synthase to investigate ketosynthase selectivity. (2023).
- 31.Hopwood, D. A. & Khosla, C. Genes for polyketide secondary metabolic pathways in microorganisms and plants. In: Ciba Foundation Symposium 171-Secondary Metabolites: their Function and Evolution: Secondary Metabolites: Their Function and Evolution: Ciba Foundation Symposium 171: 2007. Wiley Online Library: 88-112. [DOI] [PubMed]
- 32.Li, S. et al. Polyketide pesticides from actinomycetes. Curr. Opin. Biotechnol.69, 299–307 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Van Heerwaarden, L., Toet, S. & Aerts, R. Current measures of nutrient resorption efficiency lead to a substantial underestimation of real resorption efficiency: facts and solutions. Oikos. 101 (3), 664–669 (2003). [Google Scholar]
- 34.Miller, T. E., Tyre, A. J. & Louda, S. M. Plant reproductive allocation predicts herbivore dynamics across spatial and temporal scales. Am. Nat.168 (5), 608–616 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi, K. Terpene trilactones from Gingko biloba: from ancient times to the 21st century. Bioorg. Med. Chem.13 (17), 4987–5000 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Urbanova, T., Tarkowská, D., Strnad, M. & Hedden, P. Gibberellins–terpenoid plant hormones: biological importance and chemical analysis. Collect. Czechoslov. Chem. Commun.76 (12), 1669–1686 (2012). [Google Scholar]
- 37.Ali, S. S. & Vidhale, N. Bacterial siderophore and their application: a review. Int. J. Curr. Microbiol. App Sci.2 (12), 303–312 (2013). [Google Scholar]
- 38.Wencewicz, T. Beta-lactam and Beta‐lactone antibiotics from Plant Microbiomes. FASEB J.32, 257251–257251 (2018). [Google Scholar]
- 39.Montalbán-López, M. et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep.38 (1), 130–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lima, S. T. et al. A widely distributed Biosynthetic Cassette is responsible for Diverse Plant Side Chain cross-linked Cyclopeptides. Angew. Chem.135 (7), e202218082 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitz, Z. L. & Medema, M. H. Genome mining strategies for metallophore discovery. Curr. Opin. Biotechnol.77, 102757 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Johnston, C. W. et al. Gold biomineralization by a metallophore from a gold-associated microbe. Nat. Chem. Biol.9 (4), 241–243 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Baxter, A., Mittler, R. & Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot.65 (5), 1229–1240 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Huang, H., Ullah, F., Zhou, D-X., Yi, M. & Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci.10, 800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi, S., Kamiya, Y. & Nambara, E. Regulation of ABA and GA levels during seed development and germination in Arabidopsis. Annual Plant Reviews Volume 27: Seed Development, Dormancy and Germination 2007:224-247.
- 46.Ravi, B., Foyer, C. H. & Pandey, G. K. The integration of reactive oxygen species (ROS) and calcium signalling in abiotic stress responses. Plant. Cell. Environ.46 (7), 1985–2006 (2023). [DOI] [PubMed] [Google Scholar]
- 47.Mishra, N. et al. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci.14, 1110622 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhandari, S. et al. Role of beneficial microbes in biotic and abiotic stress. In: Relationship between Microbes and the Environment for Sustainable Ecosystem Services. Elsevier; : 243–259. (2023).
- 49.Shaffique, S. et al. A newly isolated Bacillus pumilus strain SH-9 modulates response to drought stress in soybean via endogenous phytohormones and gene expression (Daegu, South Korea). Plant. Stress. 10, 100279 (2023). [Google Scholar]
- 50.Chaki, M., Begara-Morales, J. C. & Barroso, J. B. Oxidative stress in plants. In., vol. 9: MDPI; : 481. (2020). [DOI] [PMC free article] [PubMed]
- 51.Chu, H. Y., Wegel, E. & Osbourn, A. From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J.66 (1), 66–79 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Cakmak, I. et al. Micronutrients. In: Marschner’s mineral nutrition of plants. Elsevier; : 283–385. (2023).
- 53.Hassani, M. A., Durán, P. & Hacquard, S. Microbial interactions within the plant holobiont. Microbiome. 6, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez, M. B. R. & Gonzalez-Lopez, J. Beneficial plant-microbial Interactions: Ecology and Applications (CRC, 2013).
- 55.Pantigoso, H. A., Newberger, D. & Vivanco, J. M. The rhizosphere microbiome: plant–microbial interactions for resource acquisition. J. Appl. Microbiol.133 (5), 2864–2876 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatla, S. C. & Lal, M. A. Secondary metabolites. In: Plant Physiology, Development and Metabolism. Springer; : 765–808. (2023).
- 57.Khan, M. et al. The key roles of ROS and RNS as a signaling molecule in plant–microbe interactions. Antioxidants. 12 (2), 268 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li, Y. et al. Signal communication during microbial modulation of root system architecture. J. Exp. Bot.75 (2), 526–537 (2024). [DOI] [PubMed] [Google Scholar]
- 59.Hussain, S. et al. Deciphering the role of phytohormones and osmolytes in plant tolerance against salt stress: implications, possible cross-talk, and prospects. J. Plant Growth Regul.43 (1), 38–59 (2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.