Abstract
Glenea cantor (Fabricius) is an important forest pest that mainly attacks kapok trees, breaking down cellulose and lignin through 3 enzyme activities: endoglucanase, filter paper enzyme, and cellobiase. In this study, we unveiled the cloning and expression of 10 endoglucanase genes, GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, GcEGaseZ9, and Cellulase, all of which exhibit enzymatic activities in G. cantor. These findings indicated that Cellulase shares sequence homology with beetle GHF45, whereas the other 9 endoglucanase genes are homologous to beetle GHF5. GcEGaseZ4 presented the highest expression in the foregut. In contrast, GcEGase5A2 and Cellulase presented peak expression in the midgut. Furthermore, GcEGaseZ7 was identified as the most highly expressed endoglucanase in the hindgut. Functional assays confirmed the ability of GcEGaseZ7 and Cellulase to degrade cellulose, and their cellulase activities were 75.57 ± 1.21 U/mg and 344.79 ± 6.91 U/mg, respectively. These results enhance our understanding of the complex cellulase system in insects and provide insights into the efficient digestion of cellulosic materials by wood-consuming insects. This research also has potential applications in bioenergy production and the development of biomaterials from lignocellulosic biomass.
Keywords: longhorn beetles, glycosyl hydrolase, insect enzymatic activity, prokaryotic expression
Introduction
Lignocellulose is composed of cellulose, hemicellulose, pectin, and lignin, with cellulose being the most abundant polymer in nature (Procópio et al. 2022). Its versatile applications include transparent cellulose-grafted-polylactide nanocomposite films (Amini et al. 2023), cellulose aerosols (Qiu et al. 2023), bioinks (Lin et al. 2023), and enhancing the stability of protein emulsions (Dai et al. 2022). However, owing to the complex structure of cellulose and the limited activity of natural cellulases, efficient degradation and utilization of cellulose pose challenges (Zhu and Pan 2022), which in turn restricts the high-value utilization of cellulose as a resource. Insects particularly their cellulases play a significant role in the breakdown and utilization of cellulose, thus serving as valuable cellulase resource banks (Liu and Fan 2011).
Longhorn beetles (Cerambycidae), comprising an estimated 5,300 genera and 36,300 extant species (Monné et al. 2017), include over 2,000 species reported in China that primarily feed on woody plants (Jin et al. 2019). Moreover, these beetles possess cellulases in their intestines, making them valuable resources for cellulase research (Geib et al. 2010, Tokuda 2019, Shin et al. 2021, 2022). The ability of these beetles to digest cellulose suggests that cellulases are produced by the beetles themselves or/and symbiotic intestinal microorganisms (Willis et al. 2011, Sheng et al. 2012, Mei et al. 2016a). Recently, endogenous cellulases have been isolated from longhorn beetles, and their functions have been confirmed (Busconi et al. 2014, Ko et al. 2015, Li et al. 2020).
The endogenous cellulase system of the longhorn beetle is complex and crucial for its feeding and digestive processes. Three kinds of enzymes, endoglucanase (EC 3.2.1.4), exoglucanase (EC 3.2.1.91 and EC 3.2.1.74), and β-glucosidases (EC 3.2.1.21), play important roles in cellulose degradation (Watanabe and Tokuda 2010). The cellulases of beetles may contain more than one enzyme. For example, 1 GHF (glycosyl hydrolase family) 45 and 7 GHF5 cellulases have been identified in Mesosa myops (Liu et al. 2015). Similarly, in Monochamus alternatus, 14 cellulases from 6 glycosyl hydrolase families were found, including 3 exoglucanase genes from GHF48; 7 endoglucanase genes from GHF5, GHF9, and GHF45; and 4 β-glucosidase genes from GHF1 and GHF3 (Li et al. 2020). The complexity of the cellulase system does not mean that the same cellulases possess only one enzymatic activity; rather, they may have multiple enzymatic activities. Furthermore, cellulases display host specificity and are influenced by different habitats and food sources. For example, the exoglucanase of Anoplophora malasiaca cannot recognize oligosaccharides smaller than cellohexose, which is attributed to the fact that its host has relatively hard long fibers. Moreover, the enzyme not only exhibits exoglucanase activity but also significant endoglucanase activity (Chang et al. 2012). Among the cellulases reported in longhorn beetles, endoglucanases have been identified predominantly, with only a few exoglucanases and β-glucosidases (Chang et al. 2012, Scully et al. 2013). Endoglucanases belonging to 3 glycoside hydrolase families, GHF5, GHF9, and GHF45, have been reported in longhorn beetles. Endoglucanase from GHF5 has been found in Psacothea hilaris, Apriona germari, Monochamus alternatus, and Mesosa myops (Sugimura et al. 2003, Wei et al. 2006, Xu et al. 2011, Ko et al. 2015, Liu et al. 2015). Endoglucanase from GHF9 has been identified in Monochamus alternatus (Li et al. 2020), whereas endoglucanase from GHF45 has been identified in Hylotrupes bajulus, Batocera horsfieldi, and Apriona germari (Lee et al. 2004, 2005, Xia et al. 2013, Busconi et al. 2014, Mei et al. 2016b). Notably, GHF45 is exclusively encoded by the Phytophaga beetle genomes among insects (Busch et al. 2019). In addition, exoglucanase and β-glucosidases have been found in Anoplophora malasiaca and Anoplophora glabripennis, respectively (Chang et al. 2012, Scully et al. 2013).
Glenea cantor (F.) (Coleoptera: Cerambycidae: Lamiinae) can cause severe damage to kapok (Gossampinus malbarica) (Lu et al. 2007). Kapok is extensively cultivated in the Guangdong and Guangxi regions of China, where the mortality rates of newly planted trees can reach as high as 80% (Lu et al. 2011). Beetles possess cellulose-degrading abilities in their intestines, as their intestines contain cellulases that can break down cellulose (Yang et al. 2011). It has also been confirmed that beetle intestinal microorganisms exhibit cellulase activities to aid in the degradation of cellulose (Su et al. 2024). However, it is unclear whether the beetles themselves can encode the proteins necessary for cellulose digestion. This study focused on the gene structure, sequence, and quantitative analysis of the endoglucanase genes of G. cantor belonging to GHF5 and GHF45. The functions of these genes were verified by prokaryotic expression. This research contributes to the understanding of endogenous cellulases in longhorn beetles. The complex system and mechanism of cellulases for efficient digestion of cellulosic materials by wood-feeding insects can promote applications in bioenergy production and the development of biomaterials from lignocellulosic biomass.
Materials and Methods
Insects
In May 2019, G. cantor larvae were originally collected from Qingxiu Mountain in Nanning City, Guangxi Province, China (22°47ʹN, 108°23ʹE). The larvae were obtained by cutting damaged kapok branches and keeping them in cages to facilitate adult eclosion. The beetles were reared according to Dong et al. (2020) and maintained at a temperature of 25 ± 1 °C, and 75 ± 5% relative humidity under a photoperiod of 14:10 [L:D] h.
RNA Extraction and cDNA Synthesis
The fourth-instar larvae and different intestinal segments (foregut (n = 12), midgut (n = 12), and hindgut (n = 12)) were stored at −80 °C, with 3 biological replicates for each segment. The method of intestinal segmentation was performed following the protocol of Su et al. (2024). Total RNA was extracted with RNAiso Plus reagent (9109, Takara Biomedical Technology (Beijing) Co., Ltd.). One microgram of total RNA was reverse-transcribed into cDNA by the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (RR047A, Takara Biomedical Technology (Beijing) Co., Ltd.). PCR primers (Supplementary Table S1) and RT-qPCR primers (Supplementary Table S2) were designed and obtained from NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/; 2021.02), and they were synthesized by Beijing Tsingke Biotech Co., Ltd.
Cloning of the Endoglucanase Genes in G. cantor
The sequences of the endoglucanase genes were identified from the full-length transcriptome of G. cantor (unpublished data). The transcriptome sequencing sample consisted of a mixed collection of RNA extracted from eggs, larvae, female pupae, male pupae, and both male and female adults of G. cantor. The cloned sequences were obtained from fourth-instar larvae.
The complete sequences of the endoglucanase genes were amplified with Premix Taq (Ex Taq Version 2.0 plus dye) (RR902A, Takara Biomedical Technology (Beijing) Co., Ltd.). The PCR program consisted of 30 cycles at the following temperatures: 98 °C for 10 s, 55~60 °C for 30 s and 72 °C for 30 s. The integrity of the nucleic acids was assessed by 1% agarose gel electrophoresis. The PCR products were recovered by the EZNA Gel Extraction Kit (D250002, OMEGA), and the extracted DNA was stored at −20 °C. Ligation and transformation were performed with the pEASY - Blunt Cloning Kit (CB101, Beijing TransGen Biotechnology Co., Ltd.). The transformed bacterial mixture was spread on LB solid media supplemented with ampicillin and cultured at 37 °C overnight. A single white colony was selected from the blue-white colonies and transferred to LB/Amp+ liquid culture medium. The culture was incubated at 37 °C with shaking at 200 rpm for 6 h. PCR of the cloned bacteria was conducted using 1% agarose gel electrophoresis to verify the success of the cloning process. The bacterial mixture was then sent to Beijing Tsingke Biotechnology Co., Ltd. for sequencing, and the sequencing results were compared with the predicted sequences.
Bioinformatics Analysis of Endoglucanase Genes in G. cantor
The ProtScale tool (https://web.expasy.org/protscale/) was employed to predict the hydrophilicity and hydrophobicity of the protein encoded by the cellulase gene. Protein solubility predictions were conducted using Novopro Bio’s online sequence analysis tool (https://www.novopro.cn/tools/prot-sol.html). Protein domain analysis was performed on NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) for prediction analysis. The codon adaptation index (CAI) in Escherichia coli was analyzed via the calculator available at http://www.bioxyz.net/codon-adaptation-index-calculator/index.html. Signal peptide prediction was conducted using SignalP-6.0 (https://services.healthtech.dtu.dk/service.php?SignalP-6.0), whereas transmembrane domain prediction was performed via TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM). NetOGlyc-4.0 (https://services.healthtech.dtu.dk/service.php?NetOGlyc-4.0) and NetNGlyc-1.0 (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0) were used for the analysis and prediction of O-glycosylation and N-glycosylation sites, respectively. Subcellular localization prediction was performed using PSORT II prediction (https://psort.hgc.jp/form2.html).
Phylogenetic Trees of Endoglucanase Genes in G. cantor
Multiple sequence alignment was carried out using DNAMAN 6.0. The amino acid (aa) sequence alignment of endoglucanase genes in G. cantor and the homologous sequences obtained from different beetles, fungi, bacteria, and protists were analyzed by the ClustalW method in MEGA 11.0 software (version 11.0, Mega Limited, Auckland, New Zealand). Phylogenetic trees were constructed by the neighbor-joining method via the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/, accessed on 5 July 2024). The reliability of the tree structure was evaluated by 1,000-fold bootstrap replication. All phylogenetic trees were constructed by FigTree v1.4.3 (Andrew Rambaut Institute of Evolutionary Biology, England) and Adobe Illustrator CC 2022 (Adobe, America).
Quantitative Analysis of Endoglucanase Genes in G. cantor
RT-qPCR analysis was performed using the Green Premix Ex Taq II (Tli RNaseH Plus) kit (RR820A, Takara Biomedical Technology (Beijing) Co., Ltd.) with a real-time fluorescence quantitative PCR instrument (ABI QuantStudio 6 Flex system, Thermo Fisher Scientific). The PCR efficiency and regression coefficient (R2) were calculated for all genes via the gradient dilution method as described. Two reference genes (RPL36 and EF1A1) were used for normalizing the expression levels (Su et al. 2021). The relative expression of genes was calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Construction of Cellulase and GcEGaseZ7 Recombinant Plasmids and Recombinant Enterobacter coli
Signal peptide prediction was performed on the Cellulase and GcEGaseZ7 sequences using SignalP-6.0, and the signal peptides were subsequently removed. The full sequences of Cellulase and GcEGaseZ7 were amplified with specific primers containing the restriction enzyme sites (BamHI and HindIII, Takara Biomedical Technology (Beijing) Co., Ltd.) (Supplementary Table S3). These sequences were then subcloned and inserted into the PET-28a (+) shuttle plasmid (Sangon Biotech (Shanghai) Co., Ltd.). The obtained plasmid constructs, Cellulase-28a and GcEGaseZ7-28a, were subsequently transformed into the Rosetta(DE3) and BL21(DE3) strains. The expression of the proteins was induced with a final concentration of 80 mM IPTG at 37 °C and 220 rpm. Bacterial solutions with and without IPTG induction were collected, and the presence of induced protein bands was confirmed by SDS-PAGE.
Induction and solubility analyses of the recombinant fusion proteins were conducted using a bacterial mixture containing PET-28a(+), Cellulase-28a or GcEGaseZ7-28a. These solutions were inoculated into the fermentation medium 3 and LB medium (with the appropriate antibiotics added), respectively. The mixtures were then inoculated at 37 °C and 220 rpm for 3–4 h until the OD 600 reached 0.4–0.6. Then, IPTG was added at a final concentration of 0.8 mM to induce protein expression. After 4 h, the bacteria were collected and disrupted through sonication. Both the supernatant and pellet were retained, and samples were prepared for SDS-PAGE. The strains that expressed the recombinant proteins Cellulase-28a and GcEGaseZ7-28a in Rosetta(DE3) and BL21(DE3) were named Cellulase-28a-R3, Cellulase-28a-B3, GcEGaseZ7-28a-R3, and GcEGaseZ7-28a-B3, respectively.
The recombinant protein expression strains Cellulase-28a-R3, Cellulase-28a-B3, GcEGaseZ7-28a-R3, and GcEGaseZ7-28a-B3, as well as control 1 (sterile water) and control 2 (the strains of PET-28a (+)-B3) were incubated on carboxymethyl cellulose sodium culture media at 37 °C for 48 h. The strains were subsequently stained with 1% Congo red solution for 30 min and destained with 1 mol/L NaCl solution for 30 min. The presence of transparent circles was observed.
Purification and Cellulase Activity Assays of Cellulase and GcEGaseZ7
Purification of Cellulase-28a and GcEGaseZ7-28a recombinant protein inclusion bodies: 500 ml of the Cellulase-28a and GcEGaseZ7-28a recombinant protein expression strains induced by IPTG were disrupted by sonication, and precipitates were obtained. These precipitates were washed with a washing solution (containing Tris, NaCl, Triton X-100, glycerol, and SDS) to ensure thorough suspension and mixing. The suspension was then incubated on a vibrating shaker for 30 min. This washing process was repeated twice, followed by 3 additional washes of the pellet with 30 ml of 10 mM PBS, as previously described. After washing, the pellet was suspended in 10 mM PBS, and the samples were subjected to SDS-PAGE to determine protein concentration and purity.
Renaturation of Cellulase-28a and GcEGaseZ7-28a recombinant protein inclusion bodies: The 10 mM PBS suspension mentioned above was subjected to centrifugation at 12,000 rpm for 2 min, and the supernatant was discarded to obtain the precipitate. The precipitate was then fully dissolved in 8 M urea and shaken for 30 min. The dissolved liquid was subsequently centrifuged at 12,000 rpm for 5 min. The retained supernatant was subjected to SDS-PAGE to determine the protein concentration and purity. The supernatant was further dissolved in 8 M urea until the protein purity reached more than 80%. The solution was transferred into a dialysis bag and dialyzed with a dialysate consisting 4 M urea for 4 h to replace the liquid. Next, the mixture was dialyzed with a dialysate of 2 M urea and 0.5 M urea for 6–8 h. Finally, the mixture was dialyzed again and changed to 10 mM PBS. The protein obtained as described above was dialyzed with 10 mM PBS 3 times, with each change in the solution lasting 8 h to ensure sufficient dialysis. The solution in the dialysis bag was centrifuged to obtain the supernatant containing the renatured inclusion bodies. This supernatant, the renatured inclusion bodies, was suspended in 10 mM PBS.
The cellulase activity of Cellulase and GcEGaseZ7 was measured by the cellulase (CL) activity detection kit (micromethod) according to the manufacturer’s instructions (Beijing Solarbio Science; Technology Co., Ltd., BC2545). The reaction temperature for this enzyme activity assay was 50 °C, and the pH was 7.2.
Data Statistics
SPSS 26.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis of the data. The endoglucanase genes GcEGaseZ3, GcEGaseZ7, and Cellulase were normally distributed in different intestinal parts of the fourth-instar larvae. These genes were determined by one-way analysis of variance (ANOVA), and multiple comparisons were conducted using Tukey’s HSD test (P < 0.05). The relative expression of GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ4, GcEGaseZ5, GcEGaseZ8, and GcEGaseZ9 exhibited a nonnormal distribution in the foregut, midgut, and hindgut of fourth-instar larvae. The statistical data were transformed via logarithmic transformation to achieve a normal distribution, followed by one-way ANOVA and Tukey’s HSD test for multiple comparisons (P < 0.05). Graphs were generated by GraphPad Prism 8.0.2 software (GraphPad Inc., USA).
Results
Cloning of Endoglucanase Genes in G. cantor
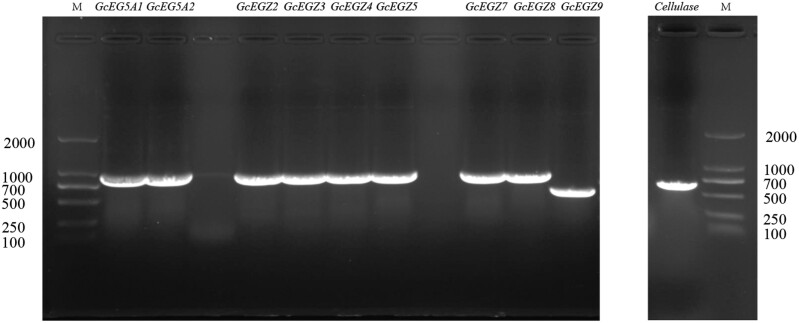
The PCR product bands for GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, GcEGaseZ9, and Cellulase were observed at sizes of 975 bp, 975 bp, 969 bp, 969 bp, 978 bp, 975 bp, 975 bp, 972 bp, 618 bp, and 714 bp, respectively, indicating clear, single bands (Fig. 1). The sequencing results for the cloned bacteria were consistent with the PCR and expected results. Sequence comparison revealed that the 10 endoglucanase genes presented high similarity with the beetles in GenBank (Table 1). All sequences are publicly available, and the corresponding sequence accession numbers are listed in Table 1.
Fig. 1.
Electrophoretic map of endoglucanase genes PCR of Glenea cantor.
Table 1.
Bioinformatics analysis of the endoglucanase gene in Glenea cantor
| Gene family | Gene name | GenBank accession number | Signal peptide | CAI analysis (Escherichia coli) | O-GalNAc (mucin type) glycosylation sites | N-linked glycosylation sites | Subcellular localization | Best similar species, protein name, login number, and similarity (%) with GenBank sequence |
|---|---|---|---|---|---|---|---|---|
| GHF5 | GcEGase5A1 | PQ050618 | 1–20 | 0.6 | 294-294; 300-300; 303-303 | 16 NFST; 299 NTTA | Endoplasmic reticulum | Anoplophora glabripennis; Endoglucanase Z; XP_018562606.1; 80.9 |
| GcEGase5A2 | PQ050618 | 1–20 | 0.6 | 294-294; 300-300 | 16 NFST; 299 NTTA | Endoplasmic reticulum | Anoplophora glabripennis; Endoglucanase Z; XP_018562606.1; 80.9 | |
| GcEGaseZ2 | PQ050618 | 1–21 | 0.6 | – | 17 NLSV | Endoplasmic reticulum | Anoplophora glabripennis; Glycoside hydrolase family 5 subfamily 2 (Gh5-2); XP_018565008.1; 85.8 | |
| GcEGaseZ3 | PQ050618 | 1–19 | 0.6 | 298-298 | 66 NKTT; 211 NQTN; 261 NLTE | Extracellular, including cell wall | Apriona japonica; Gh5-2; AHI15749.1; 89.6 | |
| GcEGaseZ4 | PQ050618 | 1–22 | 0.6 | 295-295; 301-301 | – | Extracellular, including cell wall | Apriona japonica; Gh5-2; AHI15747.1; 84.3 | |
| GcEGaseZ5 | PQ050618 | 1–21 | 0.6 | – | 68 NKTT; 198 NYST; 213 NQTN; 278 NLSY | Extracellular, including cell wall | Anoplophora glabripennis; Endoglucanase Z; XP 018574045.1; 88.5 | |
| GcEGaseZ7 | PQ050618 | 1–20 | 0.6 | 294-294; 300-300 | 136 NYSL; 299 NTTS | Extracellular, including cell wall | Psacothea hilaris; Cellulase; BAB86867.1; 89.6 | |
| GcEGaseZ8 | PQ050618 | 1–22 | 0.6 | 293-293; 299-299 | 258 NGTI | Cytoplasmic | Anoplophora glabripennis; Endoglucanase Z; XP_018565006.1; 85.4 | |
| GcEGaseZ9 | PQ050618 | – | 0.6 | 181-181 | 94 NQTN; 144 NLTE | Cytoplasmic | Apriona japonica; Gh5-2; AHI15749.1; 91.2 | |
| GHF45 | Cellulase | OL757647 | 1–38 | 0.6 | 58-58; 60-60; 64-64; 70-70; 71-71; 74-74; 76-76; 77-77; 80-80 | 98 NETF | Endoplasmic reticulum | Batocera horsfieldi; Cellulase I; AKH90729.1; 76.8 |
Sequence Analysis of Endoglucanase Genes in G. cantor
Multiple comparisons of the amino acid sequences of the proteins encoded by the endoglucanase genes of G. cantor and other beetles revealed distinct conserved domains among the proteins from different glycoside hydrolase families. Each protein, including GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, and GcEGaseZ9 contained two GHF5 conserved catalytic sites at 155-164 (IIYETFNEPT) and 243-253 (GLFLFVTEYGT). In contrast, Cellulase had two GHF45 conserved catalytic sites at 37-48 (TTRYWDCCKPSC) and 101-115 (FALGYVAASFTGGAD) (Fig. 2). The phylogenetic tree revealed that GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, GcEGaseZ9 (Fig. 3A), and other beetle GHF5 proteins clustered into one clade. Similarly, cellulase and other beetle GHF45 proteins clustered into one clade (Fig. 3B), confirming the results of the domain analysis. Importantly, none of the proteins encoded by the endoglucanase genes clustered with microorganisms and protozoa, indicating that these 10 endoglucanase genes originated from insects rather than from intestinal microorganisms.
Fig. 2.
Multiple alignments of amino acid sequences of endoglucanase in Glenea cantor. Note: A) Multiple alignments of amino acid sequences of endoglucanase in GHF5 of G. cantor; B) Multiple alignments of amino acid sequences of endoglucanase in GHF45 of G. cantor.
Fig. 3.
Phylogenetic tree of endoglucanase from other species and Glenea cantor. Note: ‘·’: The endoglucanase of Glenea cantor. The amino acid (aa) sequence alignment of endoglucanase genes in G. cantor and the homologous sequences obtained from different beetles, fungi, bacteria, and protists were analyzed by the Clustal W method. Phylogenetic trees were constructed using the neighbor-joining method and the reliability of the tree structure was evaluated using the 1,000-fold bootstrap replication.
The protein instability coefficients of all the proteins were less than 40, indicating their stability (Supplementary Table S4). The average gross hydrophobic coefficient (GRAVY) of the 10 proteins was less than 0, except for cellulase, which had a positive value of 0.075. Notably, GcEGaseZ5 and cellulase had no obvious hydrophilic and hydrophobic properties. The other 8 proteins had relatively high contents of hydrophilic amino acids, which was consistent with the ProtScale analysis results (Supplementary Table S4; Supplementary Fig. S1). Furthermore, the protein solubility prediction was used to assess the likelihood of soluble proteins in E. coli, and if it is greater than 0.45, it indicates that the protein solubility may be greater than the average value of the data set (Niwa et al. 2009). In this study, all 10 proteins had solubility prediction values greater than 0.45, classifying them as soluble proteins (Supplementary Table S4). Structural domain analysis of the proteins encoded by endoglucanase genes revealed that they belong to endoglucanase domain segments. Specifically, GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, and GcEGaseZ9 were classified as GHF5, whereas Cellulase belonged to GHF45. The classification was consistent with the phylogenetic tree of endoglucanase. Moreover, the CAI was analyzed in E. coli, which revealed that the CAI of all the genes were greater than 0.5. A low CAI would require codon optimization for effective protein expression. In addition, with the exception of GcEGaseZ9, all of the other 9 proteins were found to have signal peptides. Interestingly, the signal peptide of cellulase was the longest, up to 38 amino acids, with a maximum of 9 O-glycosylation sites and a minimum of one N-glycosylation site. Its subcellular location was predicted to be in the endoplasmic reticulum (Table 1).
Quantitative Analysis of Endoglucanase Genes in G. cantor
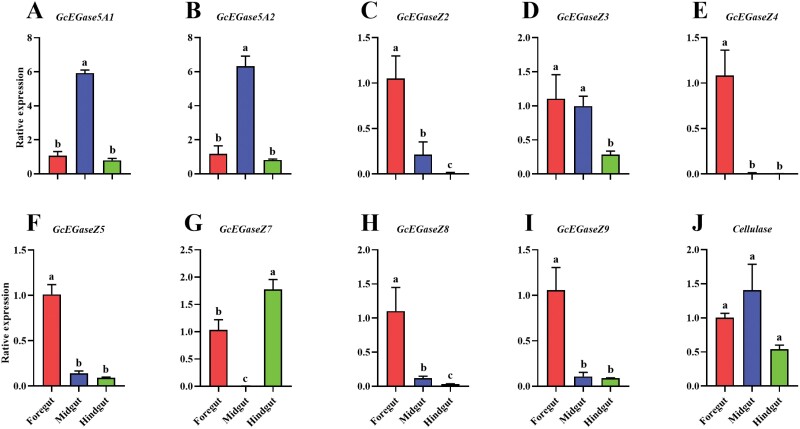
The PCR efficiency and regression coefficient (R2) of all genes were suitable for the requirements of RT-qPCR (Supplementary Table S5). Among the 10 endoglucanase genes, GcEGaseZ2 (F2,6 = 25.7, P = 0.001), GcEGaseZ3 (F2,6 = 10.7, P = 0.01), GcEGaseZ5 (F2,6 = 86.3, P < 0.0001), GcEGaseZ8 (F2,6 = 55.5, P < 0.001), and GcEGaseZ9 (F2,6 = 20.7, P = 0.002) were highly expressed in the foregut, and GcEGaseZ4 (F2,6 = 45.1, P < 0.001) was highly expressed exclusively in the foregut. GcEGase5A1 (F2,6 = 44.0, P = 0.003), GcEGase5A2 (F2,6 = 21.0, P = 0.002), and GcEGaseZ3 (F2,6 = 10.7, P = 0.01) were highly expressed in the midgut, whereas GcEGaseZ7 (F2,6 = 34.4, P < 0.01) was almost not expressed in the midgut. GcEGaseZ7 exhibited high expression levels in the hindgut. The expression of Cellulase did not significantly differ in the intestine (F2,6 = 3.7, P = 0.089) (Fig. 4). In the foregut, GcEGaseZ4 had the highest relative expression level (F9,20 = 19.0, P < 0.0001). In the midgut, GcEGase5A2 and Cellulase had the highest relative expression level (F9,20 = 19.2, P < 0.0001), while GcEGaseZ7 had the lowest (F9,20 = 11.6, P < 0.0001). In the hindgut, GcEGaseZ7 and Cellulase showed the highest relative expression level (Supplementary Fig. S2).
Fig. 4.
Relative expression of endoglucanase genes in different parts of the gut of the fourth-instar larvae of Glenea cantor. Note: The data are average ± standard error in the figure. Different lowercase letters indicate that the relative expression levels of endoglucanase genes in different intestinal positions of fourth-instar larvae are significantly different by Tukey’s HSD test (P < 0.05). A-J: the relative expression levels of GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, GcEGaseZ9 and cellulase in the intestine of fourth instar larvae.
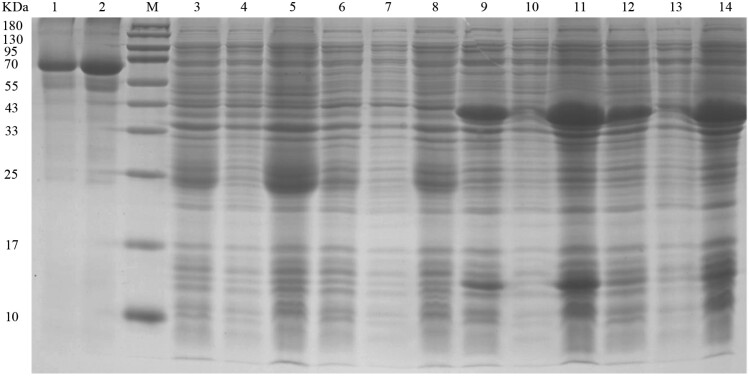
Heterologous Expression of Cellulase and GcEGaseZ7 in Rosetta(DE3) and BL21(DE3)
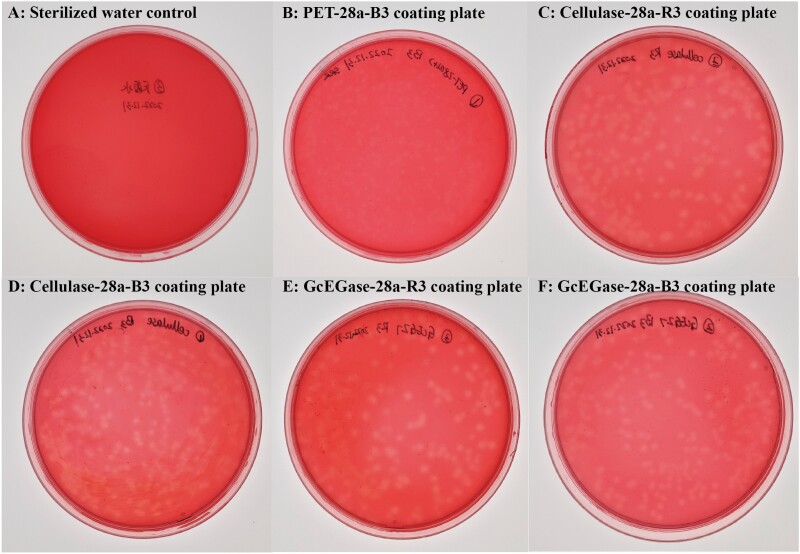
The PCR and sequencing results revealed that the target bands of Cellulase and GcEGaseZ7 were 675 bp and 915 bp, respectively (Supplementary Fig. S3). The expression vectors Cellulase-28a and GcEGaseZ7-28a were successfully constructed, and their plasmids were extracted for protein expression. Functional verification experiments revealed that the recombinant proteins of Cellulase-28a and GcEGaseZ7-28a occurred in inclusion bodies in both Rosetta (DE3) and BL21 (DE3) bacteria, with molecular weights of approximately 24 kDa and 40 kDa, respectively. However, Cellulase-28a had no obvious protein band in Rosetta(DE3) (Supplementary Fig. S4; Fig. 5). Plate verification of the recombinant expression strains showed that Cellulase-28a-R3, Cellulase-28a-B3, GcEGaseZ7-28a-R3, and GcEGaseZ7-28a-B3 had the ability to degrade cellulose (Fig. 6). The enzyme activities of Cellulase-28a-R3, Cellulase-28a-B3, GcEGaseZ7-28a-R3, and GcEGaseZ7-28a-B3 were determined, but no data were obtained.
Fig. 5.
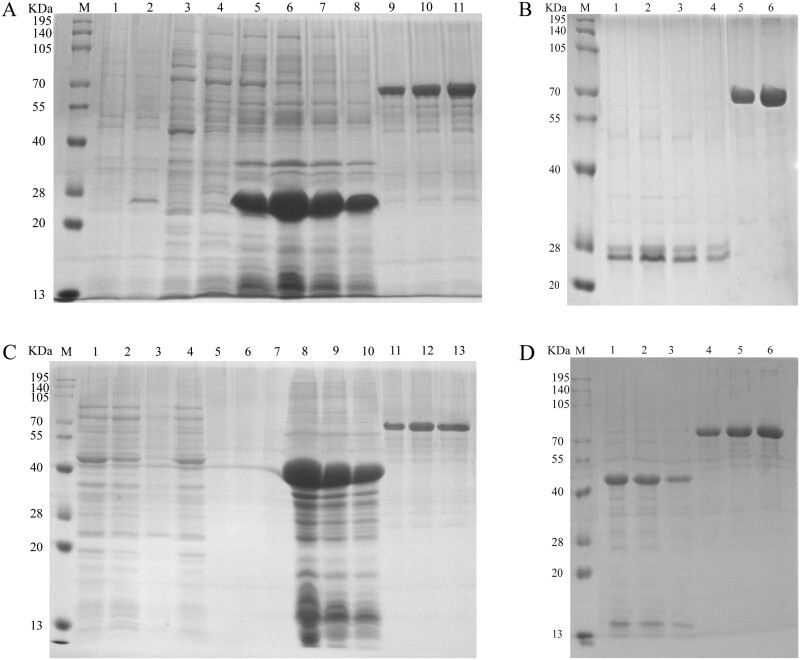
Cellulase-28a and GcEGaseZ7-28a fusion protein of soluble analysis. Note: M: Molecular weight of protein marker, size marked on the left; 1: 0.5 mg/ml BSA; 2: 0.25 mg/ml BSA; 3–5: Cellulase-28a-BL21 bacterial liquid before fragmentation, supernatant after fragmentation, and precipitate after fragmentation; 6–8: Cellulase-28a-Rossetta bacterial liquid before fragmentation, supernatant after fragmentation, and precipitate after fragmentation; 9–11: GcEGaseZ7-28a-Rossetta bacterial liquid before fragmentation, supernatant after fragmentation, and precipitate after fragmentation; 12–14: GcEGaseZ7-28a-BL21 bacterial liquid before fragmentation, supernatant after fragmentation, and precipitate after fragmentation.
Fig. 6.
Plate verification of recombinant protein expression strains of Cellulase-28a and GcEGaseZ7-28a. Note: A-B: the sterilized water control and the control of PET-28a-B3 coating plate; C-F: plate verification of the recombinant expression strains Cellulase-28a-R3, Cellulase-28a-B3, GcEGaseZ7-28a-R3, and GcEGaseZ7-28a-B3.
Purification and Cellulase Activity Assays of Cellulase and GcEGaseZ7
After the disruption of Cellulase-28a, no obvious expression was observed in the supernatant (Fig. 7A). The protein mainly resided in the precipitate (the inclusion bodies) (Fig. 7A). Furthermore, the enzyme activity assay showed that the purified inclusion bodies of Cellulase-28a did not have cellulase activity. However, after renaturation, the cellulase activity of the inclusion bodies of Cellulase-28a was 344.79 ± 6.91 U/mg (Fig. 7B; Table 2).
Fig. 7.
Purification of Cellulase-28a and GcEGaseZ7-28a. Note: M: Molecular weight of protein marker, size marked on the left; A) 1–2: Uninduced and induced expression bacteria of Cellulase-28a; 3–4: The fragmented supernatant of Cellulase-28a; 5–8: Undiluted, 5-fold dilution, 10-fold dilution, and 20-fold dilution of Cellulase-28a precipitates after fragmentation; 9–11: 0.2 mg/ml, 0.3 mg/ml, and 0.4 mg/ml of BSA. B) 1–4: Undiluted, 5-fold dilution, 10-fold dilution, and 20-fold dilution of Cellulase-28a dissolved in 8M urea and precipitated after fragmentation; 5–6: 0.2 mg/ml and 0.4 mg/ml of BSA. C) 1–3: Untreated, the supernatant of GcEGaseZ7-28a purified by His tag and the eluate of the supernatant of GcEGaseZ7-28a fragmented and purified by His tag; 4–7: Untreated, 5-fold dilution, 10-fold dilution, and 20-fold dilution after dialysis and concentration of the eluted protein obtained from the supernatant of GcEGaseZ7-28a purified by His tag; 8–10: 5-fold dilution, 10-fold dilution, and 20-fold dilution of GcEGaseZ7-28a precipitates after fragmentation; 11–13: 0.2 mg/ml, 0.3 mg/ml, and 0.4 mg/ml BSA. D) 1–3: 5-fold dilution, 10-fold dilution, and 20-fold dilution of GcEGaseZ7-28a dissolved in 8M urea and precipitated after fragmentation; 4–6: 0.2 mg/ml 0.3 mg/ml and 0.4 mg/ml BSA.
Table 2.
The cellulase activity assays of the purified Cellulase-28a and GcEGaseZ7-28 in Glenea cantor
| Protein | Cellulase activity (U/mg) |
|---|---|
| Cellulase | 344.79 ± 6.91 |
| GcEGaseZ7 | 75.57 ± 1.21 |
A small quantity of GcEGaseZ7-28a was expressed in the supernatant and was present mainly in inclusion bodies after fragmentation (Fig. 7C). After purification, the protein concentration in the supernatant decreased, indicating that the inclusion bodies were renatured (Fig. 7D). The renatured GcEGaseZ7-28a exhibited cellulase activity of 75.57 ± 1.21 U/mg. Moreover, there was no cellulase activity of the inclusion bodies of GcEGaseZ7-28a (Table 2).
Discussion
The complex structure of cellulose and the limited activity of natural cellulases pose challenges in efficiently degrading and utilizing cellulose (Zhu and Pan 2022). Therefore, further research and the development of more effective cellulases are crucial to overcome these challenges. Cellulose-degrading bacteria that can produce cellulase have been discovered in the intestine of G. cantor, but whether G. cantor can degrade cellulose remains uncertain (Su et al. 2024). Our study addresses this mystery by investigating whether G. cantor itself possesses endoglucanase proteins involved in cellulose degradation. Through cloning and expression experiments, we identified 10 endoglucanase proteins. The functional verification of GcEGaseZ7 and Cellulase clearly demonstrated their cellulose degradation capabilities.
GcEGaseZ4 showed the highest expression level in the foregut, while GcEGase5A2 and Cellulase exhibited the highest expression level in the midgut. GcEGaseZ7 presented the highest expression level in the hindgut. Additionally, the measurement of intestinal enzyme activity in the larvae of G. cantor indicated that cellulases (endo-β-1,4-glucanase, cellobiase, and filter paper enzymes) were predominantly concentrated in the midgut, with no enzyme activity detected in the foregut or hindgut (Yang et al. 2011). This finding suggests that highly expressed endoglucanase genes in the foregut may be involved in initial food decomposition without requiring a significant amount of cellulases; no cellulase enzymes were detected. Conversely, the highly expressed Cellulase in the midgut may play a crucial role in cellulose degradation, while its expression in the foregut and hindgut ranked second compared with that of other endoglucanase genes. The foregut and midgut are the major sites for high levels of endogenous cellulase expression in coleopteran species such as Apriona germari (Wei et al. 2006), and cellulases are expressed mainly in the midgut of P. hilaris (Sugimura et al. 2003). The gut region that corresponds to the midgut is likely a site where large quantities of endoglucanase are expressed for the digestion of cellulose (Ko et al. 2015). This phenomenon is attributed to the structural integrity of the midgut, which is characterized by numerous types of infolding to increase the surface area for cellulose digestion (Shelomi et al. 2014). The hindgut contributes to further food decomposition. In some wood-feeding panesthiine cockroaches, the hindgut contributes around one-fifth of cellulase and xylanase activity, which may originate from symbiotic microorganisms (Schwarz et al. 2023). Moreover, many endogenous cellulases have been identified in the hindguts of wood-feeding beetles, suggesting the coordination of symbiotic bacteria and endogenous cellulase (Lee et al. 2004, 2005, Delalibera et al. 2005, Li et al. 2008, Ko et al. 2015). Notably, GcEGaseZ7 exhibited significantly higher expression in the hindgut compared to the other 9 endoglucanases, suggesting its potential key role in the further decomposition of food in this intestinal region.
Solubility analysis revealed that the target proteins of Cellulase-28a-R3, Cellulase-28a-B3, GcEGaseZ7-28a-R3, and GcEGaseZ7-28a-B3 were found in inclusion bodies. The results of plate verification showed transparent circles, but owing to the relatively low enzyme activity, no enzyme activity data were measured in the fermentation broth. Notably, many cellulases are expressed as inclusion bodies in the E. coli expression system. For example, in the termite Nasutitermes takasagoensis, the xylanase gene was expressed in JM109 as inclusion bodies (Chang et al. 2021), and the expression of the endo-β-1,4-glucanase gene of Apriona germari in E. coli was also as inclusion bodies (Li et al. 2020). Genes expressed in inclusion bodies cannot exhibit cellulase activity unless they are renatured. The endoglucanase gene EG146 was expressed in Rosetta (DE3), and the expression product was present in inclusion bodies but showed no enzyme activity (Chen 2020). Similarly, the endo-β-1,4-glucanase gene of Clostridium thermosporum was expressed in BL21(DE3), but extracellular secretion was not achieved (Chang et al. 2006). Although some cellulase genes can be successfully expressed in the E. coli prokaryotic expression system, their enzyme activity is often weak. For example, Yang et al. (2010) expressed the endoglucanase gene in BL21(DE3), and the resulting enzyme activity was as low as 6.78 U/ml. The endoglucanase I of Trichoderma koningii is expressed in BL21(DE3), but its enzyme activity was not high (Huang et al. 2008).
Although the inclusion bodies of GcEGaseZ7 and Cellulase exhibited no enzymatic activity and displayed minimal activity in plate verification tests, we successfully renatured the proteins from these inclusion bodies into the supernatant, confirming that GcEGaseZ7 and Cellulase can effectively degrade cellulose. The enzyme activities were measured at 75.57 ± 1.21 U/mg and 344.79 ± 6.91 U/mg, respectively, further demonstrating their ability to degrade cellulose. These results validate the functions of these 2 proteins. Notably, renaturation is necessary for their activation, which consequently limits their potential applications. Prokaryotic expression often leads to the formation of insoluble inclusion bodies, complicating the processes of extraction and purification. In contrast, the Pasteurian yeast expression system is an efficient method for the secretion of recombinant proteins (Cregg et al. 2000). The system can be expressed at high yields outside or inside cells under good culture conditions and has the natural ability to express recombinant proteins, including those in the intestine. For example, the gh5-2 enzyme from Rhamnusium bicolor, the endo-β-1,4-mannanase derived from phytophagous beetles, the cellulase from the beetle Exocentrus adspersus, Apriona japonica, and Phytophaga beetles were expressed in insect Sf9 cells (Pauchet et al. 2014, Busch et al. 2017, 2019, Shin et al. 2022, Shin and Pauchet 2023). Additionally, the endoglucanase from Diabrotica virgifera virgifera was expressed in the GS115 methylotrophic strain of Pichia pastoris (Valencia et al. 2014). Further purification of GcEGaseZ7 and cellulase on large scale via yeast or cell line expression systems should be conducted to increase their application value.
In conclusion, we cloned 1 GHF45 and 9 GHF5 endoglucanase genes from G. cantor and analyzed their expression characteristics in different parts of the intestine. We have also investigated their possible functions in the process of cellulose degradation. Furthermore, we conducted prokaryotic expression studies and plate verification on 2 specific genes, GcEGaseZ7 and Cellulase. Our findings suggest that G. cantor is capable of producing cellulase involved in the digestion of cellulose. Furthermore, the presence of endogenous cellulase and intestinal cellulose-degrading bacteria (Su et al. 2024) indicates that both the beetle’s intestinal microorganisms and its own cellulases are involved in the cellulose degradation process in G. cantor. This research enhances our understanding of the complex cellulase system in insects and contributes to the understanding of how wood-eating insects efficiently digest cellulosic materials. Moreover, these findings have implications for the development of bioenergy production and biomaterials from lignocellulosic biomass.
Supplementary Material
Acknowledgments
We would like to acknowledge Guanxin Wu and Zhongyan Huang for their help in insect rearing.
Contributor Information
Ran-Ran Su, Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, China.
Tai-Hui Lan, Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, China.
Bi-Qiong Pan, Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, China.
Xia-Lin Zheng, Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, China.
Wen Lu, Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, China.
Xiao-Yun Wang, Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, China.
Author contributions
Ran-Ran Su (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Investigation [equal], Methodology [equal], Software [equal], Validation [equal], Visualization [equal], Writing—original draft [equal]), Tai-Hui Lan (Methodology [equal], Validation [equal], Writing—review & editing [equal]), Bi-Qiong Pan (Validation [equal], Writing—review & editing [equal]), Xia-Lin Zheng (Conceptualization [equal], Supervision [equal], Writing—review & editing [equal]), Wen Lu (Conceptualization [equal], Project administration [lead], Resources [lead], Supervision [equal], Writing—review & editing [equal]), and Xiaoyun Wang (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Funding acquisition [lead], Investigation [equal], Methodology [equal], Software [equal], Supervision [equal], Validation [equal], Visualization [equal], Writing—original draft [equal])
Funding
This research was funded by High-level Talents Introduction Project of Guangxi University (China), grant number A3310051008; Scientific Research and Technology Development Program Project of Guangxi Forestry Administration (China), 2023GXLK39.
References
- Amini E, Valls C, Yousefi H, et al. 2023. Ionic liquid/ZnO assisted preparation of high barrier cellulose nanocomposite films by in Situ Ringring-Opening polymerization of lactide monomers. J. Polym. Environ. 31(6):2576–2594. https://doi.org/ 10.1007/s10924-022-02740-7 [DOI] [Google Scholar]
- Busch A, Kunert G, Heckel DG, et al. 2017. Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles. PLoS One 12(8):e0184305. https://doi.org/ 10.1371/journal.pone.0184305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Danchin EGJ, Pauchet Y.. 2019. Functional diversification of horizontally acquired glycoside hydrolase family 45 (GH45) proteins in Phytophaga beetles. BMC Evol. Biol. 19(1):1–14. https://doi.org/ 10.1186/s12862-019-1429-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busconi M, Berzolla A, Chiappini E.. 2014. Preliminary data on cellulase encoding genes in the xylophagous beetle, Hylotrupes bajulus (Linnaeus). Int. Biodeterior. Biodegrad. 86(1):92–95. https://doi.org/ 10.1016/j.ibiod.2013.09.009 [DOI] [Google Scholar]
- Chang QL, Sun JY, Xu YL, et al. 2006. Expression of a thermostable endoglucanase gene celD from Clostridium thermocellum in Escherichia coli and characterization of its recombinant enzyme. J. Agric. Biotechnol. 14(6):1000–1001. (In Chinese with English abstract). [Google Scholar]
- Chang CJ, Wu CP, Lu SC, et al. 2012. A novel exo-cellulase from white spotted longhorn beetle (Anoplophora malasiaca). Insect Biochem. Mol. Biol. 42(9):629–636. https://doi.org/ 10.1016/j.ibmb.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Chang HZ, Chang J, Zhan JB, et al. 2021. Comparison of various methods for purification of recombinant inclusion body protein. Chin. J. Biol. 34(07):862–867. https://doi.org/ 10.13200/j.cnki.cjb.003385. (In Chinese with English abstract). [DOI] [Google Scholar]
- Chen YW. 2020. Cloning and expression of endoglucanase gene EG146 from Rhizoctonia solani, the pathogen of rice sheat blight. Yangzhou Univ. https://doi.org/ 10.27441/d.cnki.gyzdu.2020.001459. (In Chinese with English abstract). [DOI] [Google Scholar]
- Cregg JM, Cereghino JL, Shi J, et al. 2000. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 16(1):23–52. https://doi.org/ 10.1385/MB:16:1:23 [DOI] [PubMed] [Google Scholar]
- Dai HJ, Luo YY, Huang Y, et al. 2022. Recent advances in protein-based emulsions: the key role of cellulose. Food Hydrocolloid. 136(1):108260. https://doi.org/ 10.1016/j.foodhyd.2022.108260 [DOI] [Google Scholar]
- Delalibera IJ, Handelsman J, Raffa KF.. 2005. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetle, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ. Entomol. 34(3):541–547. https://doi.org/ 10.1603/0046-225x-34.3.541 [DOI] [Google Scholar]
- Dong ZS, Yang YB, Dou FB, et al. 2020. Observations on the ultrastructure of antennal sensilla of adult Glenea cantor (Cerambycidae: Lamiinae). J. Insect Sci. 20(2):7. https://doi.org/ 10.1093/jisesa/ieaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib SM, Tien M, Hoover K.. 2010. Identification of proteins involved in lignocellulose degradation using in gel zymogram analysis combined with mass spectroscopy-based peptide analysis of gut proteins from larval Asian longhorned beetles, Anoplophora glabripennis. Insect Sci. 17(3):253–264. https://doi.org/ 10.1111/j.1744-7917.2010.01323.x [DOI] [Google Scholar]
- Huang Y, Ling M, Qin YL, et al. 2008. Cloning and expression of Endoglucanase I of T. Knoningii. Biotechnol. (Harbin, China) (2):10–13. https://doi.org/ 10.16519/j.cnki.1004-311x.2008.02.003. (In Chinese with English abstract). [DOI] [Google Scholar]
- Jin MX, Liu XH, Yan YY, et al. 2019. Advances in research on gut content of longhorned beetles. Plant Prot. (Beijing, China) 39(12):23–27 + 36. (In Chinese with English abstract). [Google Scholar]
- Ko H, Kwon HM, Ko HJ, et al. 2015. Heterologous expression and characterization of endocellulase from the Japanese pine sawyer Monochamus saltuarius (MsGHF5). Entomol. Res. 45(1):26–30. https://doi.org/ 10.1111/1748-5967.12089 [DOI] [Google Scholar]
- Lee SJ, Kim SR, Yoon HJ, et al. 2004. cDNA cloning, expression, and enzymatic activity of a cellulase from the mulberry longicorn beetle, Apriona germari. Comp. Biochem. Physiol. Part B, Biochem. Mol. Biol. 139(1):107–116. https://doi.org/ 10.1016/j.cbpc.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee KS, Kim SR, et al. 2005. A novel cellulase gene from the mulberry longicorn beetle, Apriona germari: gene structure, expression, and enzymatic activity. Comp. Biochem. Physiol. Part B, Biochem. Mol. Biol. 140(4):551–560. https://doi.org/ 10.1016/j.cbpc.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Li XJ, Yan XF, Luo YQ, et al. 2008. Cellulase in Anoplophora glabripennis adults emerging from different host tree species. For. Ecosyst. 10(1):27–31. https://doi.org/ 10.1007/s11632-008-0004-z [DOI] [Google Scholar]
- Li YC, Chen H, Chu X, et al. 2020. Molecular cloning and expression analysis of the endogenous cellulase gene MaCel1 in Monochamus alternatus. Forests 11(12):1372. https://doi.org/ 10.3390/f11121372 [DOI] [Google Scholar]
- Lin L, Jiang SL, Yang J, et al. 2023. Application of 3D-bioprinted nanocellulose and cellulose derivative-based bio-inks in bone and cartilage tissue engineering. Int. J. Bioprint 9(1):637. https://doi.org/ 10.18063/ijb.v9i1.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WA, Fan HQ.. 2011. Determination of cellulase activity in vivo of Chalia larminati Heylaerts after feeding on different mangrove species. J. Forest Eng. 25(04):114–116. https://doi.org/ 10.3969/j.issn.1000-8101.2011.04.031. (In Chinese with English abstract). [DOI] [Google Scholar]
- Liu J, Song KQ, Teng HJ, et al. 2015. Endogenous cellulolytic enzyme systems in the longhorn beetle Mesosa myops (Insecta: Coleoptera) studied by transcriptomic analysis. Acta Biochim. Biophys. Sin. 47(9):741–748. https://doi.org/ 10.1093/abbs/gmv070 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. https://doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu W, Wang Q, Tian MY, et al. 2007. Mate location and recognition in Glenea cantor (Fabr.) (Coleoptera: Cerambycidae: Lamiinae): roles of host plant health, female sex pheromone, and vision. Environ. Entomol. 36(4):864–870. https://doi.org/ 10.1603/0046-225x(2007)36[864:mlarig]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- Lu W, Wang Q, Tian MY, et al. 2011. Phenology and laboratory rearing procedures of an asian longicorn beetle, Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). J. Econ. Entomol. 104(2):509–516. https://doi.org/ 10.1603/ec10345 [DOI] [PubMed] [Google Scholar]
- Mei HZ, Chen AL, Xia DG, et al. 2016a. Identification and determination of enzyme activity of endoglucanase in the midgut of Psacothea hilaris. Jiangsu Agrci. Sci. 44(11):165–168. https://doi.org/ 10.15889/j.issn.1002-1320.2016.11.049. (In Chinese). [DOI] [Google Scholar]
- Mei HZ, Xia DG, Zhao QL, et al. 2016b. Molecular cloning, expression, purification and characterization of a novel cellulase gene (Bh-EGaseI) in the beetle Batocera horsfieldi. Gene 576(1 Pt 1):45–51. https://doi.org/ 10.1016/j.gene.2015.09.057 [DOI] [PubMed] [Google Scholar]
- Monné ML, Monné MA, Wang Q.. 2017. General morphology, classification, and biology of Cerambycidae. In: Wang Q, editor. Cerambycidae of the world: biology and pest management. Boca Raton (FL): CRC Press; p. 1–70. [Google Scholar]
- Niwa T, Ying BW, Saito K, et al. 2009. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. U.S.A. 106(11):4201–4206. https://doi.org/ 10.1073/pnas.0811922106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauchet Y, Kirsch R, Giraud S, et al. 2014. Identification and characterization of plant cell wall degrading enzymes from three glycoside hydrolase families in the cerambycid beetle Apriona japonica. Insect Biochem. Mol. Biol. 49(1):1–13. https://doi.org/ 10.1016/j.ibmb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- Procópio DP, Kendrick E, Goldbeck R, et al. 2022. Xylo-oligosaccharide utilization by engineered Saccharomyces cerevisiae to produce ethanol. Front. Bioeng. Biotechnol. 10(1):105. https://doi.org/ 10.3389/fbioe.2022.825981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JH, Guo XZ, Lei W, et al. 2023. Facile preparation of cellulose aerogels with controllable pore structure. Nanomaterials 13(3):613. https://doi.org/ 10.3390/nano13030613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Tokuda G, Osaki H, et al. 2023. Reevaluating symbiotic digestion in cockroaches: unveiling the Hindgut’s contribution to digestion in wood-feeding Panesthiinae (Blaberidae). Insects 14(9):768. https://doi.org/ 10.3390/insects14090768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully ED, Hoover K, Carlson JE, et al. 2013. Midgut transcriptome profiling of Anoplophora glabripennis, a lignocellulose degrading cerambycid beetle. BMC Genomics 14(1):850. https://doi.org/ 10.1186/1471-2164-14-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelomi M, Watanabe H, Arakawa G.. 2014. Endogenous cellulase enzymes in the stick insect (Phasmatodea) gut. J. Insect Physiol. 60(1):25–30. https://doi.org/ 10.1016/j.jinsphys.2013.10.007 [DOI] [PubMed] [Google Scholar]
- Sheng P, Huang SW, Wang Q, et al. 2012. Isolation, screening, and optimization of the fermentation conditions of highly cellulolytic bacteria from the hindgut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). Appl. Biochem. Biotechnol. 167(2):270–284. https://doi.org/ 10.1007/s12010-012-9670-3 [DOI] [PubMed] [Google Scholar]
- Shin NR, Pauchet Y.. 2023. First evidence of a horizontally-acquired GH-7 cellobiohydrolase from a longhorned beetle genome. Arch. Insect. Biochem. Physiol. 114(2):1–14. https://doi.org/ 10.1002/arch.22039 [DOI] [PubMed] [Google Scholar]
- Shin NR, Shin S, Okamura Y, et al. 2021. Larvae of longhorned beetles (Coleoptera; Cerambycidae) have evolved a diverse and phylogenetically conserved array of plant cell wall degrading enzymes. Syst. Entomol. 46(4):784–797. https://doi.org/ 10.1111/syen.12488 [DOI] [Google Scholar]
- Shin NR, Doucet D, Pauchet Y.. 2022. Duplication of horizontally acquired GH5_2 enzymes played a central role in the evolution of longhorned beetles. Mol. Biol. Evol. 39(6):msac128. https://doi.org/ 10.1093/molbev/msac128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su RR, Huang ZY, Qin CW, et al. 2021. Evaluation of reference genes in Glenea cantor (Fabricius) by using qRT-PCR. Genes 12(12):1984. https://doi.org/ 10.3390/genes12121984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su RR, Pan BQ, Lou YX, et al. 2024. Characterization of bacterial diversity and screening of cellulose-degrading bacteria in the gut system of Glenea cantor (Fabricius) larvae. Front. Bioeng. Biotechnol. 12(1):1340168. https://doi.org/ 10.3389/fbioe.2024.1340168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura M, Watanabe H, Lo N, et al. 2003. Purification, characterization, cDNA cloning and nucleotide sequencing of a cellulase from the yellow‐spotted longicorn beetle, Psacothea hilaris. Eur. J. Biochem. 270(16):3455–3460. https://doi.org/ 10.1046/j.1432-1033.2003.03735.x [DOI] [PubMed] [Google Scholar]
- Tokuda G. 2019. Plant cell wall degradation in insects: recent progress on endogenous enzymes revealed by multi-omics technologies. Adv. Insect Physiol. 57(1):97–136. https://doi.org/ 10.1016/bs.aiip.2019.08.001 [DOI] [Google Scholar]
- Valencia JA, Wang H, Siegfried BD.. 2014. Expression and characterization of a recombinant endoglucanase from western corn rootworm, in Pichia pastoris. J. Insect Sci. 14(1):242. https://doi.org/ 10.1093/jisesa/ieu104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Tokuda G.. 2010. Cellulolytic systems in insects. Annu. Rev. Entomol. 55(1):609–632. https://doi.org/ 10.1146/annurev-ento-112408-085319 [DOI] [PubMed] [Google Scholar]
- Wei YD, Lee KS, Gui ZZ, et al. 2006. Molecular cloning, expression, and enzymatic activity of a novel endogenous cellulase from the mulberry longicorn beetle, Apriona germari. Comp. Biochem. Phys. Part B, Biochem. Mol. Biol. 145(2):220–229. https://doi.org/ 10.1016/j.cbpb.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Willis JD, Oppert B, Oppert C, et al. 2011. Identification, cloning, and expression of a GHF9 cellulase from Tribolium castaneum (Coleoptera: Tenebrionidae). J. Insect Physiol. 57(2):300–306. https://doi.org/ 10.1016/j.jinsphys.2010.11.019 [DOI] [PubMed] [Google Scholar]
- Xia DG, Wei YD, Zhang GZ, et al. 2013. cDNA cloning, expression, and enzymatic activity of a novel endogenous cellulase from the beetle Batocera horsfieldi. Gene 514(1):62–68. https://doi.org/ 10.1016/j.gene.2012.08.044 [DOI] [PubMed] [Google Scholar]
- Xu WJ, Han WJ, Li W, et al. 2011. Enzyme analysis, gene CDs cloning and expression in E. coli of Mulberry Borer (Apriona germari hope) endogen β-1, 4-endoglucanase. J. Northwest. A & F Univ. (Nat. Sci. Ed.) 39(1):29–35. https://doi.org/ 10.13207/j.cnki.jnwafu.2011.01.003. (In Chinese with English abstract). [DOI] [Google Scholar]
- Yang DL, Weng HB, Wang ME, et al. 2010. Cloning and expression of a novel thermostable cellulase from newly isolated Bacillus subtilis strain I15. Mol. Biol. Rep. 37(4):1923–1929. https://doi.org/ 10.1007/s11033-009-9635-y [DOI] [PubMed] [Google Scholar]
- Yang DF, Guan N, Mi HZ, et al. 2011. Research on Glenea cantor cellulase characteristics. Guangxi Sci. 18(03):261–263 + 268. https://doi.org/ 10.13656/j.cnki.gxkx.2011.03.032. (In Chinese with English abstract). [DOI] [Google Scholar]
- Zhu JY, Pan X.. 2022. Efficient sugar production from plant biomass: current status, challenges, and future directions. Renew. Sustain. Energy Rev. 164(1):112583. https://doi.org/ 10.1016/j.rser.2022.112583 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.