Abstract
Introduction
We performed RNA sequencing with the primary goal of discovering key placental villous trophoblast (VT) and decidua basalis (DB) transcripts differentially expressed in intra-amniotic infection (IAI)-induced preterm birth (PTB).
Methods
RNA was extracted from 15 paired VT and DB specimens delivered of women with: 1) spontaneous PTB in the setting of amniocentesis-proven IAI and histological chorioamnionitis (n=5); 2) spontaneous idiopathic PTB (iPTB, n=5); and 3) physiologic term pregnancy (n=5). RNA sequencing was performed using the Illumina HiSeq 2500 platform, and a spectrum of computational tools was used for gene prioritization and pathway analyses.
Results
In the VT specimens, 128 unique long transcripts and 7 mature microRNAs differed significantly between pregnancies complicated by IAI relative to iPTB (FDR<0.1). The up-regulated transcripts included many characteristic of myeloblast-derived cells, and bioinformatic analyses revealed enrichment for multiple pathways associated with acute inflammation. In an expanded cohort including additional IAI and iPTB specimens, the expression of three proteins (cathepsin S, lysozyme, and hexokinase 3) and two microRNAs (miR-133a and miR-223) was validated using immunohistochemistry and quantitative PCR, respectively. In the DB specimens, only 11 long transcripts and no microRNAs differed significantly between IAI cases and iPTB controls (FDR<0.1). Comparison of the VT and DB specimens in each clinical scenario revealed signatures distinguishing these placental regions.
Discussion
IAI is associated with a transcriptional signature consistent with acute inflammation in the villous trophoblast. The present findings illuminate novel signaling pathways involved in IAI, and suggest putative therapeutic targets and potential biomarkers associated with this condition.
Keywords: Preterm birth, placenta, infection, RNA-sequencing, transcriptomics, microRNA
1. Introduction
Intra-amniotic infection (IAI) contributes to at least 25–40% of all preterm births (PTBs), with a prevalence that varies inversely with increasing gestational age (GA) [1]. Compelling data suggest that bacteria of the lower genital track invade the choriodecidual maternal-fetal interface and spread to the amniotic fluid (AF) via an ascending pathway [2]. In the setting of maternal bacteremia, hematogenous dissemination of infection is also possible, as demonstrated in humans and various experimental animal models of PTB [3–5]. Invasion of the gestational sac by microorganisms elicits a fetal inflammatory response, as the vast majority of the AF neutrophils are of fetal origin in this setting [6]. This preterm parturition model is complex and, despite significant clinical and research efforts, still requires further evaluation.
Currently, there is agreement that the magnitude of the decidual and placental immune response to microbes can significantly impact pregnancy outcome [7]. Multiple lines of evidence imply that dysregulation of numerous molecular inflammatory networks in the decidua leads to activation of myometrial contractility, ripening of the cervix, and premature delivery [8–10]. This conclusion is supported through identification of differentially expressed genes involved in regulating leukocyte trafficking, cytokine signaling, cell fate, and tissue remodeling (such as IL1B, ICAM1, CXCR4, CD44, TLR4, SOCS3, BCL2A, and IDO). Upstream regulators of these inflammatory pathways include nuclear factor-kappa B (NF-κB), signal transducer and activator of transcription (STAT) proteins, high-mobility group protein B1 (HMGB1), and miRNAs-21, −46, −146, −155, and −200. While significant advances have already been made using hybridization-based microarray technologies, high throughput RNA sequencing (RNA-seq) has advanced transcriptomics research by offering key advantages over hybridization-based methods, including reduced background from hybridization/cross-hybridization artifacts, a significantly expanded dynamic range of resolution, and the ability to detect novel transcripts in an agnostic fashion [11]. In the current study, we leveraged comprehensive RNA-seq profiling and in-depth bioinformatic analyses to test the hypothesis that placental villous trophoblast (VT) and decidual basalis (DB) from pregnancies complicated by IAI harbor characteristic transcriptional signatures that are informative of the pathogenic processes leading to PTB. We also compared iPTB and term specimens to determine the influence of advancing gestational age and spontaneous PTB in the absence of IAI on placental gene expression. Finally, we determined transcriptional signatures that distinguished the VT and DB placental regions.
We report for the first time an inflammation-specific transcriptional signature of the villous tissue (fetal origin) that is distinct from the transcriptional response to inflammation in intrauterine tissues of mostly maternal origin (decidua). This work establishes a foundation on which future studies centered on postnatal complications may build.
2. Materials and methods
2.1. Study population and definitions.
We analyzed the VT and DB transcriptome of 15 women pregnant with singletons grouped as follows: (1) spontaneous PTB in the setting of amniocentesis-proven IAI and histological chorioamnionitis (HCA) (IAI: n=5; GA median [range]: 26 [25–31] weeks); (2) spontaneous idiopathic preterm birth (iPTB: n=5, GA: 32 [30–33] weeks); and (3) term normal pregnancy, that delivered a healthy baby by Cesarean in the absence of labor (Term: n=5; GA: 39 [38–39] weeks). Select targets were validated in an expanded cohort which included 5 additional cases in the IAI and iPTB groups. This study was approved by the Human Investigation Committee of Yale University and the Institutional Review Board of the Ohio State University. All women provided written informed consent.
The definitions of GA, preterm labor, the criteria for establishment of IAI, exclusion criteria and the pathological criteria for evaluation of the placenta are presented in the Supplementary Material.
2.2. IL-6 immunoreactivity.
Amniotic fluid IL-6 was assessed by ELISA (eBioscience, San Diego, CA). The assay was run in duplicate with a minimal detectable concentration of 0.039 pg/ml.
2.3. Total RNA extraction.
Within minutes from the time of delivery of the placenta, DB was dissected from the VT and rinsed in sterile saline. Random placental VT specimens were collected and washed in similar conditions. The technique for RNA extraction is presented in the Supplementary Material.
2.4. RNA sequencing (RNA-seq).
Long total RNA-seq libraries were constructed using the TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold (Illumina, San Diego, CA). Small RNA-seq libraries were generated using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Ipswich, MA). Sequencing was performed using the Illumina HiSeq 2500 platform. Details are presented in the Supplementary Material. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE73714.
2.5. Real-time quantitative PCR (qPCR).
Two miRNAs (miR-133a-3p and miR-223–3p) were selected from the RNA-seq data analysis for validation. Total RNA was reverse transcribed using the Universal cDNA Synthesis Kit II (Exiqon, Woburn, MA). Quantitative PCR was performed using the ExiLENT SYBR Green master mix (Exiqon) in the ABI StepOnePlus real-time PCR system (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. The following Exiqon locked nucleic acid PCR primer sets were used for amplification: hsa-miR-133a-3p (product number 204788) and hsa-miR-223–3p (product number 205986). The miRNA levels were normalized to 5S rRNA (product number 203906, Exiqon) as an internal control, and the relative abundance of each miRNA was calculated by the comparative CT method [12] and statistically evaluated using the two-tailed Mann–Whitney rank-sum test. All reactions were performed in duplicate with melting curve analysis following amplification to confirm the presence of single peaks.
2.6. Immunohistochemistry (IHC).
IHC was performed as previously described [13] on selected formalin-fixed placental specimens using the following antibodies: goat polyclonal anti-cathepsin S (1:50, ab115259, Abcam, Cambridge, MA) , mouse monoclonal anti-hexokinase 3 (1:100, ab115751, Abcam) and rabbit monoclonal anti-lysozyme (1:1,000; NBP1–95509, Novus Biologicals, Littleton, CO). Specificity of staining was confirmed by substituting the primary antibody with species-specific preimmune IgG. Staining intensity of chromogen deposits was evaluated in specific cell types using a semi-quantitative 5-point grading scale, with 1 indicating low or absent staining, and 5 indicating intense staining. Statistical comparisons were conducted using the two-tailed Mann–Whitney rank-sum test. Correlations between variables were examined using Spearman’s rank order correlation.
2.7. RNA-seq Data, microRNA, pathways, placental enrichment, hierarchical clustering and principal component analyses.
A large spectrum of statistical software and bioinformatic tools was used to perform these analyses as detailed in the Supplementary Material.
3. Results
3.1. Study population
Maternal demographic, clinical data and pregnancy outcome characteristics associated with the placental specimens subjected to RNA-seq are presented in Table 1. Characteristics of the subjects from which specimens were obtained for validation studies are presented in Table S1. Women with IAI had AF laboratory analysis consistent with IAI and higher degree of acute histologic chorioamnionitis.
Table 1.
Demographic and clinical characteristics associated with placental specimens subjected to RNA-seq.
| IAI PTB n=5 |
Idiopathic PTB n=5 |
Term delivery n=5 |
p value | |
|---|---|---|---|---|
| Clinical and outcome characteristics | ||||
| Maternal age (years) † | 35 [21–41] | 25 [16–39] | 26 [19–37] | 0. 499 |
| Parity † | 1 [0–4] | 1 [0–2] | 2 [1–3] | 0.316 |
| Gravidity † | 3 [2–5] | 3 [1–5] | 3 [2–5] | 0.544 |
| GA at delivery (weeks) † | 26 [25–31] | 32 [29–33] | 39 [38–39] | 0.003 a |
| PPROM ‡ | 4 (80%) | 2 (40%) | 0 (0%) | 0.010 |
| Clinical chorioamnionitis ‡ | 1 (20%) | 0 (0%) | 0 (0%) | 0.300 |
| Cesarean delivery ‡ | 2 (40%) | 4 (80%) | 5 (100%) | 0.037 |
| Corticosteroid exposure ‡ | 5 (100%) | 5 (100%) | 0 (0%) | < 0.001 |
| Birthweight (grams) † | 830 [680–1,830] | 1965 [1,220–2,210] | N/A | 0.032 |
| Amniotic fluid analysis (microbiological and inflammatory status) | ||||
| Positive AF culture ‡ | 5 (100%) | 0 (0%) | N/A | 0.008 |
| Positive Gram stain ‡ | 3 (60%) | 0 (0%) | N/A | 0.080 |
| Glucose (mg/dL) † | 2 [2–5] | 16 [13–30] | N/A | 0.016 |
| LDH (U/L) † | 846 [422–3,110] | 188 [110–311] | N/A | 0.016 |
| WBC (cells/mm3) † | 294 [0–1,976] | 2 [1–4] | N/A | 0.286 |
| MR score 3 or 4 ‡ | 5 (100%) | 0 (0%) | N/A | 0.008 |
| IL-6 (ng/mL) † | 69 [6–223] | 1 [0–9] | N/A | 0.032 |
| Histological evaluation of the placenta | ||||
| Chorioamnionitis (grade) † | 2 [1–2] | 0 [0–1] | 0 [0–0] | 0.003 b |
| Maternal inflammation (stage) † | 3 [2–3] | 0 [0–1] | 0 [0–0] | 0.003 b |
| Fetal inflammation (stage) † | 2 [1–3] | 0 [0–0] | 0 [0–0] | 0.001 b |
Data presented as median [range] and analyzed by Kruskal-Wallis one-way analysis of variance with Dunn’s post hoc test, or Mann-Whitney rank-sum test, as appropriate.
Data presented as n (%) and analyzed by Fisher’s exact test.
Statistically significant for IAI vs. term comparison.
Statistically significant for IAI vs. iPTB and IAI vs. term comparisons.
Abbreviations: IAI= intra-amniotic infection; PTB=preterm birth; GA=gestational age; PPROM=preterm premature rupture of membranes; AF=amniotic fluid; LDH=lactate dehydrogenase; WBC= white blood cell count; IL-6=interluekin-6; N/A = not available. The MR score ranges from 0 to 4: MR score 0 (“absent” inflammation); MR score 1–2 (“minimal” inflammation); MR score 3–4 (“severe” inflammation). The grade and stage of acute chorioamnionitis are reported as follows: grade 0 (“absent”); grade 1 (“mild-moderate”); grade 2 (“severe”); stage 0 (“absent”); stage 1 (“early”); stage 2 (“intermediate”); and stage 3 (“advanced”).
3.2. Deep sequencing of the VT in the setting of IAI reveals a transcriptional signature consistent with acute inflammation
An overview of the RNA sequencing results is presented in the Supplementary Material. Transcriptional profiling of VT revealed significant differential expression for 128 unique long RNA transcripts and 7 mature miRNAs in pregnancies complicated by IAI compared to iPTB specimens (Table 2, Figure 1, A&B). Of the long transcripts, 88 (69%) were up-regulated, and 40 (31%) down-regulated. Several transcripts characteristic of myeloblast-derived cells, including cathepsin S (CTSS), hexokinase 3 (HK3), lysozyme (LYZ), myeloblastin (PRTN3), and myeloperoxidase (MPO) [14–17], were elevated in IAI VT. Two of the transcripts (S100A8, S100A12) encoding calgranulins A and C were earlier identified as proteomic biomarkers of AF IAI [18]. Based on the differential expression of these long RNAs, the IPA upstream regulator algorithm predicted activation of cytokine signaling networks, such as those responsive to granulocyte and macrophage colony-stimulating factors (CSF2 and CSF3) and interleukins-1A, −1B, and −18 (Table S2).
Table 2.
Transcripts differentially expressed between IAI and iPTB VT specimens by RNA-Seq.
| Long RNA Transcripts | ||||
|---|---|---|---|---|
| Gene Symbol | Linear Fold Change | Description | Raw p value† | FDR† |
| MPO | 3.13 | myeloperoxidase | <0.001 | <0.001 |
| ACTA1 | 3.04 | actin, alpha 1, skeletal muscle | <0.001 | <0.001 |
| S100A12 | 2.99 | S100 calcium binding protein A12 | <0.001 | <0.001 |
| PRTN3 | 2.79 | proteinase 3 (myeloblastin) | <0.001 | 0.002 |
| S100A8 | 2.62 | S100 calcium binding protein A8 | <0.001 | <0.001 |
| SPSB4 | 2.62 | splA/ryanodine receptor domain and SOCS box containing 4 | <0.001 | 0.004 |
| MCEMP1 | 2.37 | chromosome 19 open reading frame 59 (mast cell-expressed membrane protein 1) | <0.001 | 0.023 |
| SCG2 | 2.36 | secretogranin II | <0.001 | 0.003 |
| ST6GALNAC5 | 2.35 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 | <0.001 | 0.022 |
| RRM2 | 2.32 | ribonucleotide reductase M2 | <0.001 | 0.002 |
| ALPL | 2.31 | alkaline phosphatase, liver/bone/kidney | <0.001 | 0.015 |
| FUT9 | 2.31 | fucosyltransferase 9 (alpha (1,3) fucosyltransferase) | <0.001 | 0.005 |
| SDS | 2.28 | serine dehydratase | <0.001 | 0.028 |
| FFAR2 | 2.27 | free fatty acid receptor 2 | <0.001 | 0.028 |
| HK3 | 2.23 | hexokinase 3 (white cell) | <0.001 | 0.017 |
| HIST1H2BM | 2.22 | histone cluster 1, H2bm | <0.001 | 0.010 |
| ANK1 | 2.21 | ankyrin 1, erythrocytic | <0.001 | 0.003 |
| RP1 | 2.19 | retinitis pigmentosa 1 (autosomal dominant) | <0.001 | 0.023 |
| SBSPON | 2.14 | somatomedin B and thrombospondin, type 1 domain containing | <0.001 | 0.005 |
| CNN1 | 2.13 | calponin 1, basic, smooth muscle | <0.001 | 0.014 |
| MMP25 | 2.12 | matrix metallopeptidase 25 | <0.001 | 0.028 |
| S1PR5 | 2.12 | sphingosine-1-phosphate receptor 5 | <0.001 | 0.053 |
| MASP1 | 2.11 | mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) | <0.001 | 0.004 |
| HPGDS | 2.10 | hematopoietic prostaglandin D synthase | <0.001 | 0.005 |
| HIST1H4A | 2.07 | histone cluster 1, H4e | <0.001 | 0.007 |
| C17orf51 | 2.07 | chromosome 17 open reading frame 51 | <0.001 | 0.026 |
| HIST1H2BL | 2.05 | histone cluster 1, H2bl | <0.001 | 0.029 |
| SLC17A9 | 2.04 | solute carrier family 17 (vesicular nucleotide transporter), member 9 | <0.001 | 0.028 |
| MMP9 | 2.04 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | <0.001 | 0.069 |
| SAPCD2 | 2.03 | suppressor APC domain containing 2 | <0.001 | 0.067 |
| HIST1H4L | 2.03 | histone cluster 1, H4e | <0.001 | 0.028 |
| HIST1H2AJ | 2.03 | histone cluster 1, H2aj | <0.001 | 0.017 |
| NETO2 | 2.02 | neuropilin (NRP) and tolloid (TLL)-like 2 | <0.001 | 0.035 |
| HIST1H2BB | 2.00 | histone cluster 1, H2bb | <0.001 | 0.029 |
| DDIAS | 2.00 | DNA damage-induced apoptosis suppressor, nitric oxide-inducible | <0.001 | 0.068 |
| AMICA1 | 1.99 | adhesion molecule, interacts with CXADR antigen 1 | <0.001 | 0.026 |
| CLSPN | 1.99 | claspin | <0.001 | 0.028 |
| HIST1H1B | 1.97 | histone cluster 1, H1b | <0.001 | 0.004 |
| ANKRD65 | 1.97 | ankyrin repeat domain 65 | <0.001 | 0.033 |
| CD180 | 1.96 | CD180 molecule | <0.001 | 0.039 |
| UBE2C | 1.95 | ubiquitin-conjugating enzyme E2C | <0.001 | 0.033 |
| KLHL6 | 1.94 | kelch-like family member 6 | <0.001 | 0.009 |
| HIST1H2AB | 1.93 | histone cluster 1, H2ab | <0.001 | 0.050 |
| HIST1H3H | 1.93 | histone cluster 1, H3a | <0.001 | 0.023 |
| HIST1H3I | 1.90 | histone cluster 1, H3a | <0.001 | 0.036 |
| C3orf58 | 1.89 | chromosome 3 open reading frame 58 | <0.001 | 0.014 |
| PANK1 | 1.89 | pantothenate kinase 1 | <0.001 | 0.018 |
| HIST1H3F | 1.88 | histone cluster 1, H3a | <0.001 | 0.018 |
| IL33 | 1.88 | interleukin 33 | <0.001 | 0.020 |
| PDLIM3 | 1.87 | PDZ and LIM domain 3 | <0.001 | 0.025 |
| KCNJ5 | 1.86 | potassium inwardly-rectifying channel, subfamily J, member 5 | <0.001 | 0.032 |
| NNAT | 1.86 | neuronatin | <0.001 | 0.014 |
| HIST1H3B | 1.85 | histone cluster 1, H3a | <0.001 | 0.026 |
| FAS | 1.85 | Fas cell surface death receptor | <0.001 | 0.036 |
| PHGDH | 1.84 | phosphoglycerate dehydrogenase | <0.001 | 0.039 |
| HIST1H3J | 1.83 | histone cluster 1, H3a | <0.001 | 0.047 |
| LY96 | 1.83 | lymphocyte antigen 96 | <0.001 | 0.071 |
| RAB23 | 1.82 | RAB23, member RAS oncogene family | <0.001 | <0.001 |
| CDC20 | 1.82 | cell division cycle 20 | <0.001 | 0.081 |
| BAG2 | 1.80 | BCL2-associated athanogene 2 | <0.001 | 0.009 |
| RAC2 | 1.80 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 0.001 | 0.090 |
| SERPINA1 | 1.80 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | <0.001 | 0.055 |
| HIST1H2BJ | 1.78 | histone cluster 1, H2bj | <0.001 | 0.049 |
| ROBO1 | 1.77 | roundabout, axon guidance receptor, homolog 1 (Drosophila) | <0.001 | 0.028 |
| SORL1 | 1.77 | sortilin-related receptor, L(DLR class) A repeats containing | <0.001 | 0.028 |
| PRC1 | 1.76 | protein regulator of cytokinesis 1 | <0.001 | 0.026 |
| IGSF6 | 1.76 | immunoglobulin superfamily, member 6 | <0.001 | 0.067 |
| MGP | 1.75 | matrix Gla protein | <0.001 | 0.007 |
| LYZ | 1.74 | lysozyme | <0.001 | 0.039 |
| KCNE3 | 1.74 | potassium voltage-gated channel, Isk-related family, member 3 | <0.001 | 0.033 |
| NUSAP1 | 1.74 | nucleolar and spindle associated protein 1 | <0.001 | 0.042 |
| TRIB3 | 1.73 | tribbles pseudokinase 3 | <0.001 | 0.074 |
| EMILIN1 | 1.73 | elastin microfibril interfacer 1 | <0.001 | 0.029 |
| PRR11 | 1.71 | proline rich 11 | 0.001 | 0.099 |
| HIST1H2AG | 1.71 | histone cluster 1, H2ai | <0.001 | 0.046 |
| DOCK10 | 1.70 | dedicator of cytokinesis 10 | <0.001 | 0.057 |
| BTK | 1.70 | Bruton agammaglobulinemia tyrosine kinase | 0.001 | 0.093 |
| HAVCR2 | 1.70 | hepatitis A virus cellular receptor 2 | <0.001 | 0.059 |
| ARHGAP28 | 1.68 | Rho GTPase activating protein 28 | <0.001 | 0.023 |
| FLNC | 1.67 | filamin C, gamma | <0.001 | 0.026 |
| SPEG | 1.66 | SPEG complex locus | <0.001 | 0.069 |
| PLOD2 | 1.62 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | <0.001 | 0.014 |
| CTSS | 1.62 | cathepsin S | <0.001 | 0.041 |
| LBR | 1.61 | lamin B receptor | <0.001 | 0.025 |
| DOCK2 | 1.60 | dedicator of cytokinesis 2 | <0.001 | 0.064 |
| PDE4B | 1.59 | phosphodiesterase 4B, cAMP-specific | <0.001 | 0.072 |
| CNTLN | 1.54 | centlein, centrosomal protein | 0.001 | 0.098 |
| HIST1H4H | 1.49 | histone cluster 1, H4e | 0.001 | 0.088 |
| GNE | −1.50 | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 0.001 | 0.092 |
| WWC3 | −1.50 | WWC family member 3 | 0.001 | 0.098 |
| CAPN6 | −1.50 | calpain 6 | <0.001 | 0.068 |
| TPTEP1 | −1.51 | transmembrane phosphatase with tensin homology pseudogene 1 | 0.001 | 0.092 |
| EFNB1 | −1.52 | ephrin-B1 | <0.001 | 0.073 |
| MC1R | −1.54 | melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) | <0.001 | 0.072 |
| COASY | −1.55 | CoA synthase | <0.001 | 0.036 |
| ULK4 | −1.57 | unc-51 like kinase 4 | <0.001 | 0.018 |
| PVRL1 | −1.57 | poliovirus receptor-related 1 (herpesvirus entry mediator C) | <0.001 | 0.065 |
| RAPGEF3 | −1.58 | Rap guanine nucleotide exchange factor (GEF) 3 | <0.001 | 0.039 |
| TMEM184A | −1.58 | transmembrane protein 184A | <0.001 | 0.064 |
| EMP2 | −1.60 | epithelial membrane protein 2 | <0.001 | 0.030 |
| GLIS2 | −1.60 | GLIS family zinc finger 2 | <0.001 | 0.015 |
| KALRN | −1.62 | kalirin, RhoGEF kinase | <0.001 | 0.074 |
| EFHD1 | −1.62 | EF-hand domain family, member D1 | <0.001 | 0.053 |
| PACSIN3 | −1.63 | protein kinase C and casein kinase substrate in neurons 3 | <0.001 | 0.028 |
| TMEM108 | −1.67 | transmembrane protein 108 | <0.001 | 0.047 |
| CITED4 | −1.69 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | <0.001 | 0.068 |
| PGAP3 | −1.69 | post-GPI attachment to proteins 3 | <0.001 | 0.016 |
| CCDC113 | −1.71 | coiled-coil domain containing 113 | <0.001 | 0.059 |
| STX1B | −1.75 | syntaxin 1B | 0.001 | 0.094 |
| FAM109A | −1.76 | family with sequence similarity 109, member A | <0.001 | 0.026 |
| CORO6 | −1.85 | coronin 6 | <0.001 | 0.033 |
| ALPP | −1.86 | alkaline phosphatase, placental | <0.001 | 0.016 |
| LOC100129345 | −1.88 | uncharacterized LOC100129345 | <0.001 | 0.019 |
| PNMT | −1.97 | phenylethanolamine N-methyltransferase | <0.001 | 0.028 |
| PRX | −2.04 | periaxin | <0.001 | 0.003 |
| AKNAD1 | −2.05 | AKNA domain containing 1 | <0.001 | 0.001 |
| CDH4 | −2.07 | cadherin 4, type 1, R-cadherin (retinal) | <0.001 | 0.055 |
| CYP1B1 | −2.07 | cytochrome P450, family 1, subfamily B, polypeptide 1 | <0.001 | 0.023 |
| LOC100996634 | −2.12 | transmembrane protein FLJ37396 | <0.001 | <0.001 |
| CCDC162P | −2.20 | coiled-coil domain containing 162, pseudogene | <0.001 | 0.001 |
| L1CAM | −2.24 | L1 cell adhesion molecule | <0.001 | 0.023 |
| ADH1B | −2.24 | alcohol dehydrogenase 1B (class I), beta polypeptide | <0.001 | 0.023 |
| DNASE1L3 | −2.32 | deoxyribonuclease I-like 3 | <0.001 | 0.015 |
| ALPPL2 | −2.40 | alkaline phosphatase, placental-like 2 | <0.001 | <0.001 |
| ATP1A2 | −2.47 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | <0.001 | 0.003 |
| MUC19 | −2.48 | mucin 19, oligomeric | <0.001 | 0.003 |
| LDB3 | −2.54 | LIM domain binding 3 | <0.001 | 0.001 |
| ATP8A2 | −2.66 | ATPase, aminophospholipid transporter, class I, type 8A, member 2 | <0.001 | 0.002 |
| Mature miRNA Transcripts | ||||
| miRNA | Linear Fold Change | miRBase Accession No. | Raw p value† | FDR† |
| miR-887–3p | 2.35 | MIMAT0004951 | <0.001 | 0.036 |
| miR-154–5p | 1.65 | MIMAT0000452 | <0.001 | 0.067 |
| miR-376b-5p | 1.59 | MIMAT0022923 | <0.001 | 0.067 |
| miR-376c-3p | 1.58 | MIMAT0000720 | <0.001 | 0.036 |
| miR-500a-5p | 1.57 | MIMAT0004773 | <0.001 | 0.062 |
| miR-133a-3p | 1.40 | MIMAT0000427 | <0.001 | 0.036 |
| miR-223–3p | 1.20 | MIMAT0000280 | <0.001 | 0.036 |
Only the raw p values and FDRs from the ‘edgeR’ analysis are shown; all transcripts were additionally found to be statistically significant based on ‘DESeq2’ analysis. FDR=false discovery rate.
Figure 1. Differentially expressed placental villous trophoblast (VT) and decidua basalis (DB) transcripts in women with preterm birth (PTB) in the setting of intra-amniotic infection (IAI) compared to idiopathic PTB (iPTB).
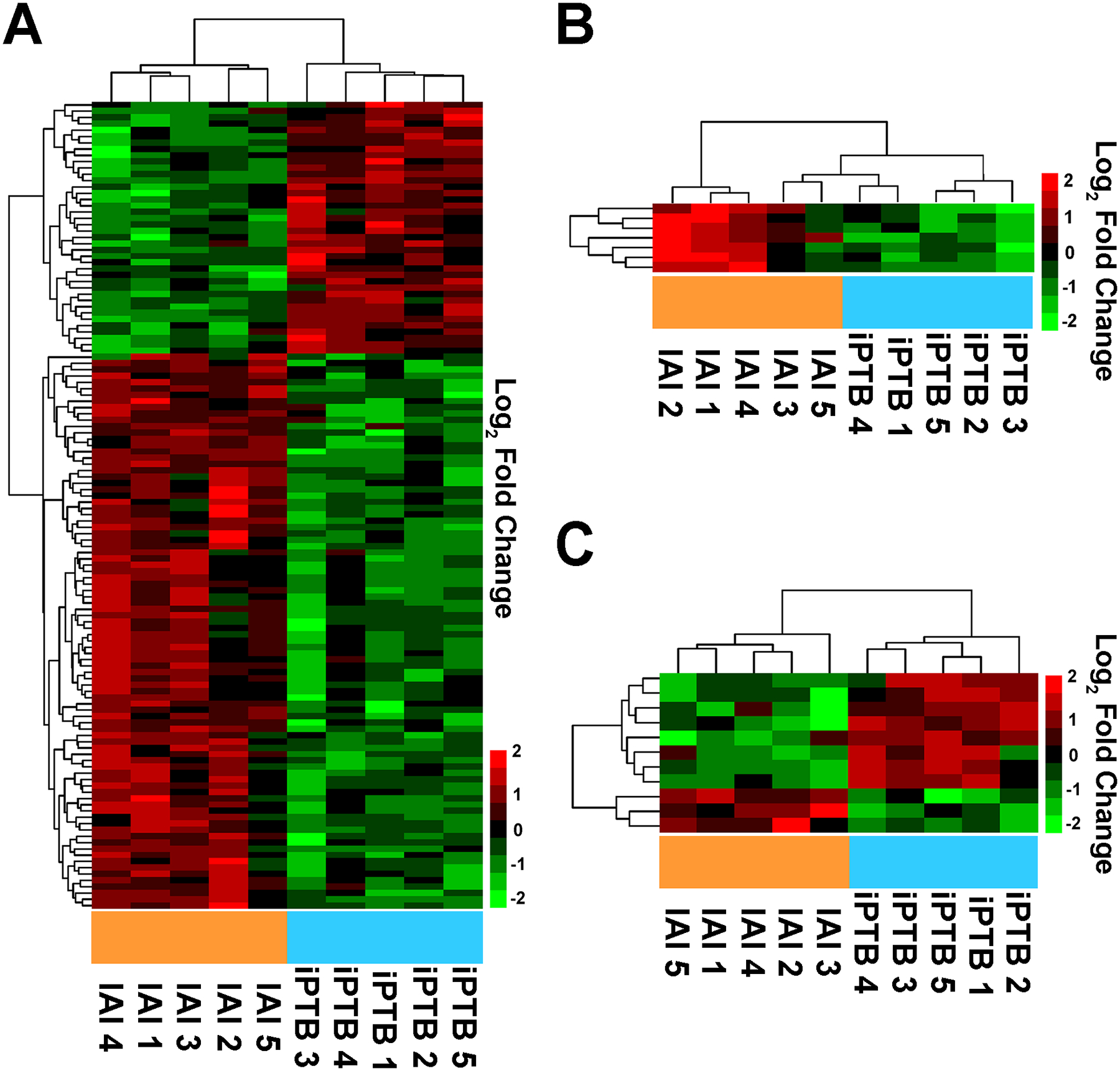
(A) Heat map of differentially expressed long RNA transcripts in the placental VT specimens from pregnancies complicated by IAI compared with iPTB controls. (B) Heat map of differentially expressed miRNA transcripts in VT specimens from pregnancies complicated by IAI compared with iPTB controls. (C) Heat map of differentially expressed long RNA transcripts in the DB specimens from pregnancies complicated by IAI compared with iPTB controls. In each instance, two-dimensional unsupervised hierarchical clustering analysis (using a complete linkage algorithm with Manhattan distance function) was applied to identify RNA expression patterns. Data indicate log2-normalized expression data (relative to the mean) wherein red denotes up-regulation and green denotes down-regulation.
Three targets (cathepsin S, lysozyme, and hexokinase 3) were selected for validation given relevance to host defense and lack of prior data implicating them in VT pathophysiology. Using IHC in an expanded group of specimens, we found that cathepsin S, a lysosomal protease involved in antigen presentation [19], preferentially localized to cells residing in VT stroma likely corresponding to macrophages (Hofbauer cells). Intensity was more conspicuous in IAI placentae relative to iPTB tissue (Figure 2, A–C). Immunoreactivity for lysozyme, an important antimicrobial enzyme that catalyzes the hydrolysis of bacterial cell wall glycosides [20], was also significantly greater in IAI compared to iPTB specimens (Figure 2D). However, the preferential cellular localization of lysozyme was in maternal neutrophils trapped in intervillous vascular spaces (Figures 2E&2F, arrowheads). Lastly, hexokinase 3 also validated as up-regulated in the setting of IAI (Figure 2G), yet localized primarily to villous cytotrophoblasts and syncytiotrophoblast (Figure 2H, open arrows). Inflammatory cells in the maternal intervillous space also stained intensely positive in the setting of IAI (Figure 2I, arrowheads).
Figure 2. Validation of select sequencing results using immunohistochemistry (IHC) and real-time quantitative PCR (qPCR) in an expanded group of specimens.
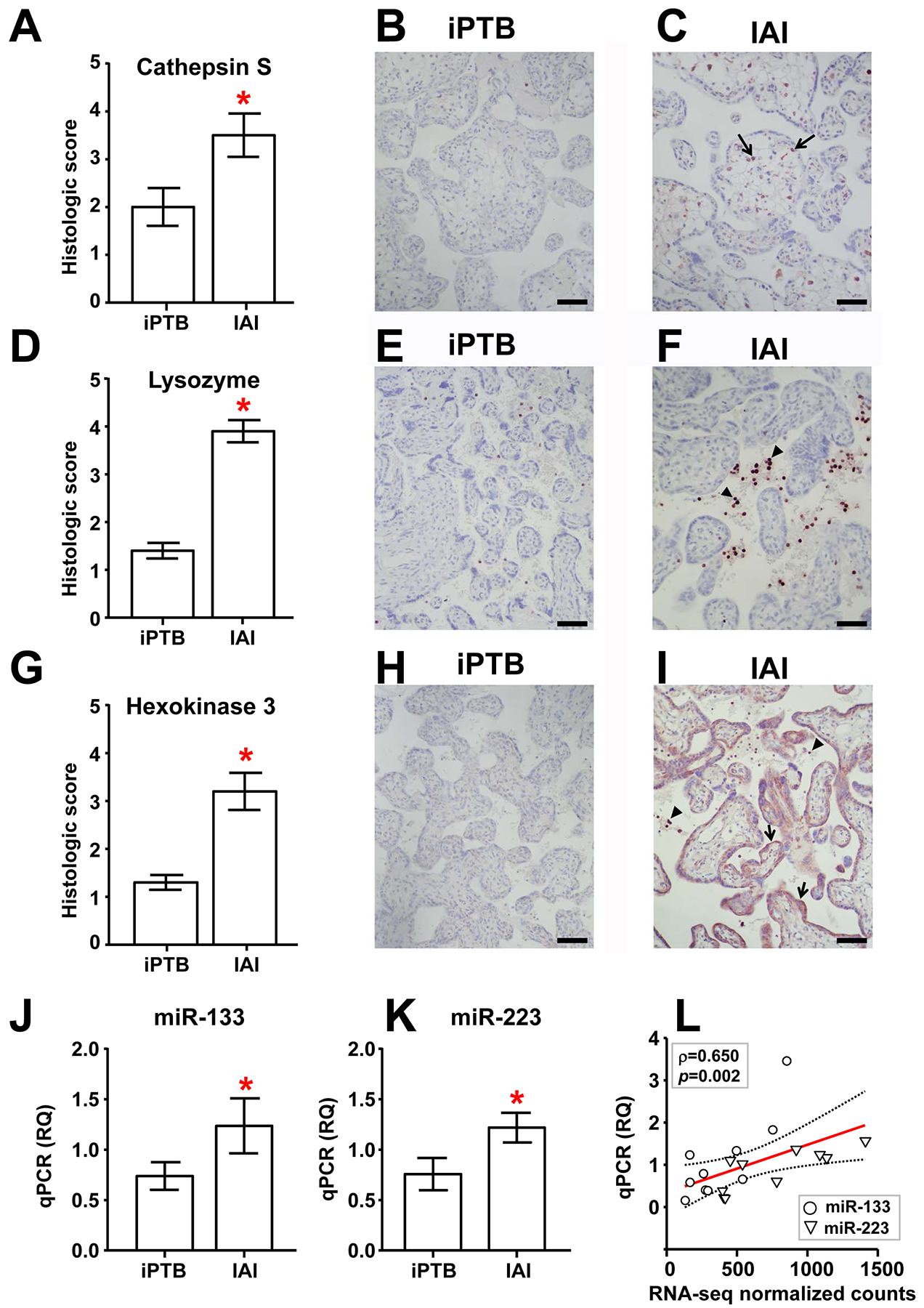
Histological staining intensity (mean ± SEM, n=10/group) and representative photomicrographs are shown for IAI and iPTB villous placental samples labeled with antibodies directed against cathepsin S (A-C), lysozyme (D-F), and hexokinase 3 (G-I). Scale bars = 100 μm. Arrows and arrowheads depict cells exhibiting characteristic immunolabeling in each case (see main text). (J, K) Relative quantification (RQ) of mature miR-133a-3p (J) and miR-223–3p (K) in IAI and iPTB villous trophoblast specimens using qPCR (mean ± SEM, n=10/group). Asterisks indicate p < 0.05 by the Mann-Whitney rank-sum test. (L) Scatterplot and linear regression analysis (line of best fit and 95% confidence interval) comparing qPCR results with normalized RNA-seq feature counts for miR-133a-3p and miR-223–3p in the original IAI and iPTB VT specimens (n=5/group). Note that these were significantly correlated (Spearman ρ = 0.650, p=0.002).
Seven miRNAs were identified as being up-regulated in IAI compared to iPTB VT by RNA-seq. These included miR-223, previously identified as increased in preterm amnio-choriodecidual samples with chorioamnionitis (relative to specimens without chorioamnionitis) [21] and miR-154, previously identified as over-expressed in PTB fetal membrane specimens (compared to term controls) [22]. Database queries revealed a targetome of 102 experimentally verified and 780 predicted unique mRNAs for these miRNAs (Table S3). Two miRNAs (miR-133a-3p and miR-223–3p) were selected for validation by qPCR, and both were found significantly increased in IAI VT samples (Figure 2, J&K). Linear regression analysis revealed that the normalized RNA-seq counts significantly correlated with the corresponding qPCR data (Figure 2L, ρ=0.650, p=0.002).
In comparison to the VT specimens, the DB transcriptome exhibited a modest response to IAI. Only 11 long transcripts differed between IAI cases and iPTB controls (Figure 1C, Table 3). The 3 up-regulated mRNAs in IAI encoded transmembrane 4 superfamily member 5 (TM4SF5), cellular retinoic acid binding protein 1 (CRABP1, a carrier protein involved in vitamin A-directed differentiation [23]), and tetraspanin transmembrane receptor and signal peptide CUB domain epidermal growth factor-like 2 (SCUBE2, a cell surface and secreted glycoprotein implicated in embryonic development, cell adhesions, motility and activation [24, 25]). TM4SF5 and SCUBE2 mRNAs have roles in the control of epithelial-mesenchymal plasticity and angiogenesis [26–29]. Three of the down-regulated DB transcripts (LOC100996634, CCDC162P, ALPPL2) were also down-regulated in the VT specimens comparing IAI to iPTB. Due to the small number of differentially expressed transcripts, IPA upstream regulator analysis failed to yield statistically significant predictions.
Table 3.
Transcripts differentially expressed between IAI and iPTB DB specimens by RNA-Seq.
| Long RNA Transcripts | ||||
|---|---|---|---|---|
| Gene Symbol | Linear Fold Change | Description | Raw p value† | FDR† |
| TM4SF5 | 2.71 | transmembrane 4 L six family member 5 | <0.001 | 0.036 |
| CRABP1 | 2.43 | cellular retinoic acid binding protein 1 | <0.001 | 0.061 |
| SCUBE2 | 2.29 | signal peptide, CUB domain, EGF-like 2 | <0.001 | 0.013 |
| RAB6B | −1.96 | RAB6A, member RAS oncogene family | <0.001 | 0.088 |
| LOC100996634 | −1.96 | transmembrane protein FLJ37396 | <0.001 | 0.067 |
| OPRK1 | −2.06 | opioid receptor, kappa 1 | <0.001 | 0.097 |
| CCDC162P | −2.15 | coiled-coil domain containing 162, pseudogene | <0.001 | 0.061 |
| CD300LG | −2.19 | CD300 molecule-like family member g | <0.001 | 0.043 |
| CSMD1 | −2.24 | CUB and Sushi multiple domains 1 | <0.001 | 0.092 |
| ALPPL2 | −2.50 | alkaline phosphatase, placental-like 2 | <0.001 | 0.001 |
| TPPP3 | −2.56 | tubulin polymerization-promoting protein family member 3 | <0.001 | 0.001 |
Only the raw p values and FDRs from the ‘edgeR’ analysis are shown; all transcripts were additionally found to be statistically significant based on ‘DESeq2’ analysis. FDR=false discovery rate.
To assess the effects of advancing GA on the placental transcriptome, we compared the iPTB specimens with those collected at term. In this analysis the VT transcriptome included 81 differentially expressed long transcripts, of which 41 were up-regulated (Table S4). No mature miRNAs were differentially expressed. When applied to this dataset, the IPA upstream regulator analysis (Table S5) revealed that roughly one-third of the transcripts enriched in iPTB VT were glucocorticoid-responsive, such as BMP2, CD163, FKBP5, FOXO3, and KLF15 [30–34]. In the DB, 14 long RNAs differentiated iPTB specimens from those collected at term (Table S6). Similarly, several glucocorticoid-inducible genes (BMP2, CD163, FKBP5, and KLF15) were up-regulated in the iPTB DB specimens vs. term. No miRNAs were identified as differentially expressed between iPTB and term in either placental region.
3.3. Pathway analysis of the VT and DB transcriptomes reveals novel signaling pathways and putative therapeutic targets that have not been previously interrogated in IAI
To assess patterns of global biological pathway dysregulation in the setting of IAI, we used Gene Set Association Analysis (GSAA) software. In comparing the IAI VT specimens to iPTB, the top-ranking Pathway Interaction Database (PID) gene sets included Toll-like receptor (TLR) and IL-8 receptor signaling pathways, in addition to networks related to integrin binding and cell cycle regulation (polo-like kinase signaling and the Forkhead Box M1 [Foxm1] transcription factor network) (Table S7, Figure 3). The top ranking KEGG and Reactome gene sets included systemic lupus erythematosus, amyloids, transcription and telomere packaging pathways. Of note, these pathways shared consistent core enrichment for numerous histone transcripts, which were abundant in the IAI VT dataset. These IAI VT specimens were also enriched for GO molecular functions such as amine transmembrane transport, cytokine receptor signaling, and steroid binding.
Figure 3. Gene Set Association Analysis (GSAA) of placental villous trophoblast specimens from pregnancies complicated by intra-amniotic infection (IAI) compared to idiopathic preterm birth (iPTB) controls.
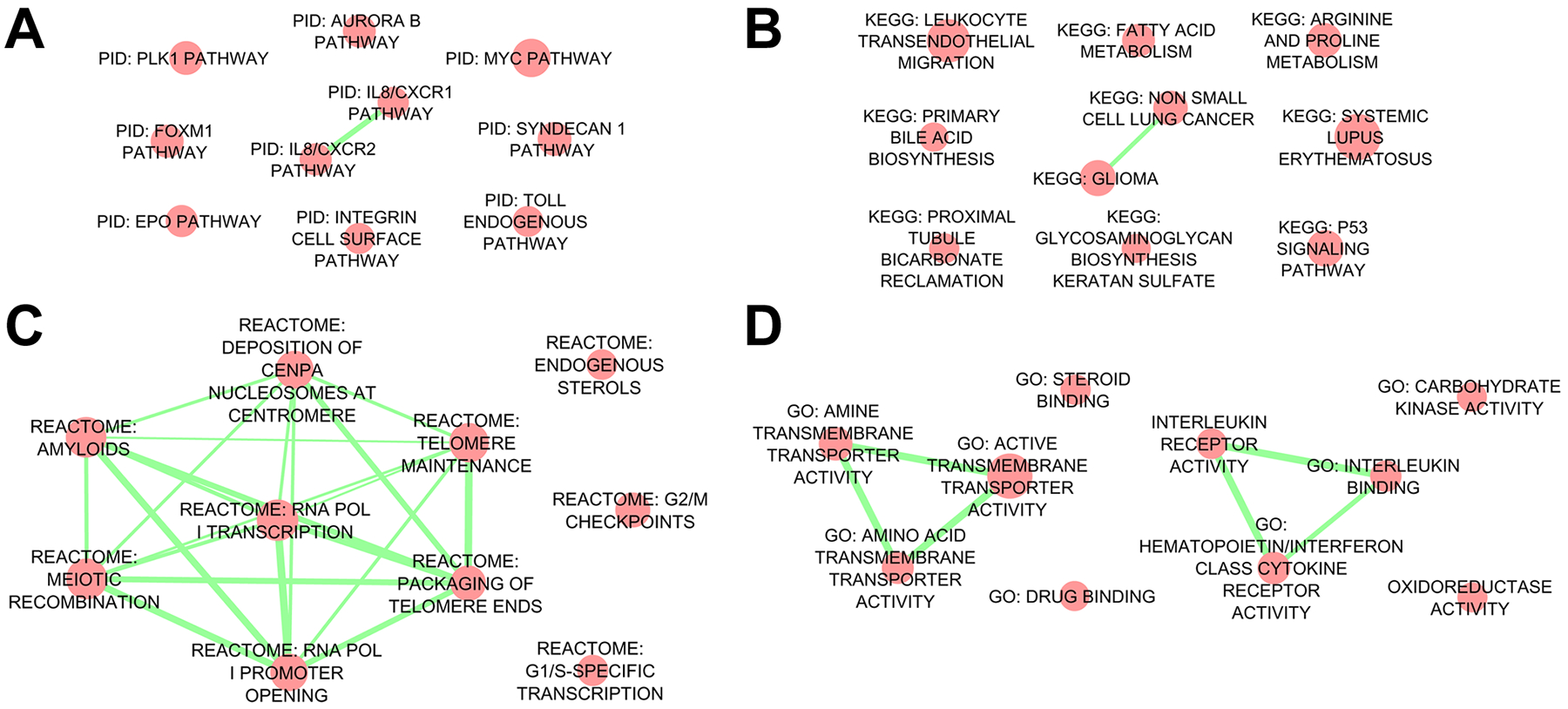
The top ten Pathway Interaction Database (PID) (A), Kyoto Encyclopedia of Genes and Genomes (KEGG) (B), Reactome (C), and Gene Ontology (GO) molecular function (D) gene sets are represented as network diagrams. Red nodes represent enriched gene sets, while edges (green) represent overlap among mapped network objects within individual gene sets. We observed a highly connected Reactome subnetwork (panel C), resulting from numerous common histone transcripts shared among these pathways.
In the IAI DB specimens relative to iPTB controls, the GSAA gene ranking algorithm indicated enrichment for gene sets centering around cell adhesion molecules (particularly integrins) and networks associated with membrane receptors, such as syndecan-1 and Wnt signaling pathways (Table S8, Figure S1). None of these gene sets, however, included mRNAs identified as being differentially expressed by statistical analysis.
Application of GSAA to the transcriptome of iPTB placentas compared with that of term specimens showed just few gene sets having a FDR < 0.1 (Tables S9, S10). Compared to term specimens, the iPTB VT samples were enriched for cell adhesion molecules (including integrins, claudins, selectins, and genes related to hemostasis such as fibrinogens), glycoprotein degradation, lipoprotein binding, and antigen presentation pathways. The iPTB DB specimens, compared to term, showed enrichment for the “SH2 domain binding” GO pathway, inclusive of numerous genes encoding several protein tyrosine kinase receptors.
To complement GSAA, we also applied over-representation analysis to differentially expressed transcripts using MetaCore software. The VT specimens of pregnancies complicated by IAI were enriched for networks related to granulocyte development, leukocyte chemotaxis, and cell cycle control when compared to GA matched controls (Table S11) In the DB samples comparing IAI to iPTB, many of the top ranking MetaCore pathways centered around the transcript encoding opioid receptor kappa 1 (OPRK1) (Table S12). In comparing the iPTB specimens to those delivered at term, MetaCore analysis revealed enrichment for pathways related to cell adhesion (particularly ephrin receptors), axonal guidance, and platelet activation in the VT specimens (Table S13), and over-representation of pathways related to blood vessel morphogenesis and lipoprotein metabolism in the DB specimens (Table S14).
3.4. Identification of regional transcriptional enrichment in placental tissues
We next compared the VT and DB specimens to address whether individual transcripts exhibited preferential expression in either anatomic placental region. We identified 227 transcripts that differentiated the VT and DB specimens in one or more clinical scenario (IAI, iPTB, or term) (Table S15). Unsupervised hierarchical cluster analysis and PCA revealed that all but two of the DB specimens co-clustered, while the remaining DB specimens exhibited a “VT-like” expression signature (Figure 4).
Figure 4. RNA transcripts consistently enriched in placental decidua basalis (DB) specimens compared to villous trophoblast (VT).
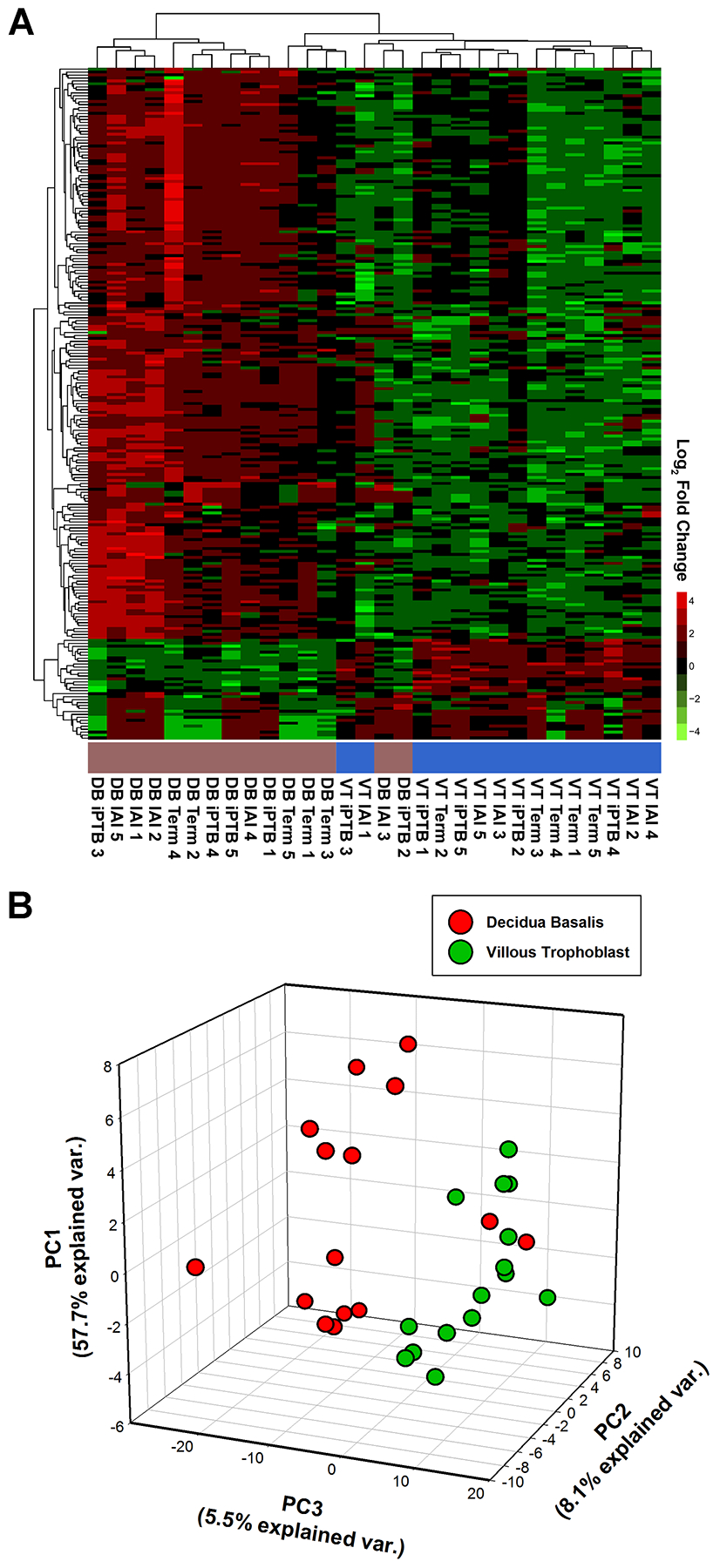
(A) Heat map of RNA transcripts identified as being relatively enriched in the DB or VT in at least one of the three delivery scenarios [intra-amniotic infection (IAI), idiopathic preterm birth (iPTB), and term). (B) Three-dimensional scatterplot of principal component analysis (PCA) results for transcripts enriched in either the DB (red dots) or VT (green dots) specimens. Both methods revealed that 13 of the 15 DB specimens co-clustered, whereas the remainder clustered with the VT specimens.
4. Discussion
Using RNA-seq, we found that VT of pregnancies complicated by IAI-induced PTB, relative to iPTB, exhibited marked differences in long RNA expression. Conversely, RNA expression in DB was minimally affected in the setting of IAI. The IAI VT transcriptome contained elevated levels of numerous mRNAs characteristic of myeloid lineage leukocytes (e.g., monocytes, macrophages, and neutrophils). Bioinformatic analyses of these data revealed enrichment for multiple pathways associated with acute inflammation as anticipated, including cytokine signaling (e.g., interleukin-8 receptor-mediated events), cytokine binding, leukocyte transendothelial migration, chemotaxis, immune response, and neutrophil activation. In an expanded cohort, using IHC, we confirmed at the protein level that cathepsin S, lysozyme, and hexokinase 3 were all elevated in IAI placentas compared to uninfected controls. Whereas lysozyme and hexokinase III were abundant within circulating inflammatory cells (as well as within inflammatory infiltrates in the chorionic plate, not shown), cathepsin S localized predominantly within resident interstitial cells of the villous placenta that resembled tissue macrophages. Interestingly, we also found abundant hexokinase III expression within villous trophoblast cells, which was significantly more abundant in the setting of IAI. Thus, in addition to increased inflammatory cell infiltrate, the RNA-seq results also reflected expression changes within resident placental cells, which was not expected based on the bioinformatic analyses alone. These results thus highlight the continued need for expanded pathway curation within tissues such as the human placenta, since expression patterns within its complex cellular architecture might diverge from those in adult human tissues.
Overall, mature miRNA expression was relatively stable compared to that of the long RNA transcripts. By RNA-seq, differential expression was observed for only 7 miRNAs between the IAI VT specimens and GA-matched controls, with no other comparison yielding statistically significant differences. Interrogation of the verified targets for these miRNAs using the Enrichr application yielded “receptor for advanced glycation end-products (RAGE) signaling” as the top-ranked WikiPathway. This is of relevance insofar as the RAGE system has been implicated in modulating the fetal inflammatory response to intra-amniotic infection [35, 36], and transcripts encoding known RAGE ligands (S100A8 and S100A12) [37] were up-regulated in these specimens. Two miRNAs (miR-133a and miR-223, for which increased expression in VT tissues in the setting of IAI was validated by qPCR) accounted for all of the targets mapping to this RAGE pathway. Fragmentary evidence suggests that miR-133a may have a regulatory role in innate immunity, as it was one of 21 miRNAs strongly induced upon differentiation of THP-1 monoblasts [38], and was recently shown to suppress inflammasome activation in monocytes [39]. MiR-223, a lipopolysaccharide-induced [40], granulocyte/monocyte-specific miRNA [41], serves as an important regulator of granulocyte maturation and activation [42, 43]. Verified miR-133a and miR-223 targets were enriched for the “bacterial invasion of epithelial cells” (ARPC5, CDC42, PIK3R2, and RHOA), “Toll-like receptor signaling” (CHUK and IL6), and “VEGF signaling” KEGG pathways (VEGFA and CASP9), suggesting roles in the regulation of host defense and angiogenesis. Interestingly, prior studies showed that circulating levels of both miR-133 [44] and miR-223 [45] exhibited differential expression in adult patients with sepsis. It is therefore possible that these miRNAs also might have applicability as biomarkers for fetal inflammation and/or neonatal sepsis.
In comparing iPTB and term specimens, we found that in both anatomic placental regions, the iPTB samples were enriched for steroid-responsive genes. This possibly reflects prenatal exposure to corticosteroids to promote lung maturation in the iPTB (but not the term) group. However, we cannot exclude the possible influence of other clinical or subject-specific factors, including the influence of labor, on these results. We also found that the transcriptome of the DB can be distinguished from that of the VT. With additional data, such results may ultimately be useful in the future for computational “deconvolution” [46] of gene expression data from placental samples composed of heterogeneous cell populations. Additional remarks regarding the iPTB vs. term comparisons and regional variation in placental gene expression is presented in the Supplementary Material.
4.1. Conclusions
In summary, in the setting of IAI, the villous placenta was found to harbor a transcriptional signature quite distinct from the cytokine-dominated global transcriptional response characteristic of laboring choriodecidua and myometrium [8, 9]. Overall, the profiling results were consistent with the presence of an acute, tissue-level inflammatory cell infiltrate, as would be expected to accompany hallmark IAI-associated placental lesions such as chorionic vasculitis. We also found that some of these gene expression changes also occurred within resident placental cells. On the other hand, the DB did not exhibit marked transcriptional alterations in this context, which is consistent with the observed infrequency of basal plate pathology in IAI [47]. We recognize that discovery-based transcriptional profiling studies with small sample sizes can be affected by factors such inter-subject variability (even within identical clinical settings), which limits the power of such investigations. Thus, the current results must be interpreted with these limitations in mind, and follow-up studies will be required for verification of many of the current findings. Nevertheless, the present findings illuminate some novel signaling pathways involved in IAI, and suggest putative therapeutic targets and potential biomarkers associated with this condition.
Supplementary Material
Highlights:
The placental response to intra-amniotic infection was profiled using RNA-Seq.
Intra-amniotic infection markedly altered the villous trophoblast transcriptome.
This transcriptional signature suggested an acute inflammatory infiltrate.
Intra-amniotic infection modestly changed the decidua basalis transcriptome.
Regional differences in placental RNA expression were determined.
Acknowledgements
This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University. The authors gratefully acknowledge the technical support of Dr. Pearlly Yan of the Nucleic Acid Shared Resource (The Ohio State University Comprehensive Cancer Center, Columbus, OH) for technical support and helpful discussions. The authors thank Dr. Raymond W. Redline (Case Western Reserve University School of Medicine, Cleveland, OH) and the Placenta Reading Resource of the March of Dimes Prematurity Research Center Ohio Collaborative for histopathological examination of placental specimens. Portions of this work were presented in abstract form at the 35th Annual Pregnancy Meeting of the Society for Maternal-Fetal Medicine, February 2 – 7, 2015, San Diego, CA. The study and participating investigators were supported by funds from the March of Dimes Prematurity Research Center Ohio Collaborative, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD R01 HD084628), as well funds from The Ohio State University College of Medicine (Division of Maternal Fetal Medicine) and The Research Institute at Children’s Hospital (Center for Perinatal Research). The funding sources had no role in study design, data analysis, writing of the report, or decision to submit for publication.
Footnotes
Disclosure statement
The authors declare that they have nothing to disclose.
References
- [1].Goldenberg RL, Culhane JF, Iams JD, Romero R, Epidemiology and causes of preterm birth, Lancet 371(9606) (2008) 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Buhimschi IA, Buhimschi CS, Proteomics/diagnosis of chorioamnionitis and of relationships with the fetal exposome, Semin Fetal Neonatal Med 17(1) (2012) 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shute KM, Kimber RG, Haemophilus influenzae intra-amniotic infection with intact membranes, J Am Board Fam Pract 7(4) (1994) 335–41. [PubMed] [Google Scholar]
- [4].Riggs MA, Maunsell FP, Reyes L, Brown MB, Hematogenous infection of Sprague-Dawley rats with Mycoplasma pulmonis: development of a model for maternal and fetal infection, Am J Obstet Gynecol 198(3) (2008) 318 e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Katz J, Chegini N, Shiverick KT, Lamont RJ, Localization of P. gingivalis in preterm delivery placenta, J Dent Res 88(6) (2009) 575–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sampson JE, Theve RP, Blatman RN, Shipp TD, Bianchi DW, Ward BE, Jack RM, Fetal origin of amniotic fluid polymorphonuclear leukocytes, Am J Obstet Gynecol 176(1 Pt 1) (1997) 77–81. [DOI] [PubMed] [Google Scholar]
- [7].Buhimschi CS, Baumbusch MA, Campbell KH, Dulay AT, Buhimschi IA, Insight into innate immunity of the uterine cervix as a host defense mechanism against infection and preterm birth, Expert Rev Obstet Gynecol 4(1) (2009) 9–15. [Google Scholar]
- [8].Weiner CP, Mason CW, Dong Y, Buhimschi IA, Swaan PW, Buhimschi CS, Human effector/initiator gene sets that regulate myometrial contractility during term and preterm labor, Am J Obstet Gynecol 202(5) (2010) 474 e1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stephen GL, Lui S, Hamilton SA, Tower CL, Harris LK, Stevens A, Jones RL, Transcriptomic profiling of human choriodecidua during term labor: inflammation as a key driver of labor, Am J Reprod Immunol 73(1) (2015) 36–55. [DOI] [PubMed] [Google Scholar]
- [10].Huang SJ, Schatz F, Masch R, Rahman M, Buchwalder L, Niven-Fairchild T, Tang C, Abrahams VM, Krikun G, Lockwood CJ, Regulation of chemokine production in response to pro-inflammatory cytokines in first trimester decidual cells, J Reprod Immunol 72(1–2) (2006) 60–73. [DOI] [PubMed] [Google Scholar]
- [11].Wang Z, Gerstein M, Snyder M, RNA-Seq: a revolutionary tool for transcriptomics, Nat Rev Genet 10(1) (2009) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method, Nat Protoc 3(6) (2008) 1101–8. [DOI] [PubMed] [Google Scholar]
- [13].Dulay AT, Buhimschi CS, Zhao G, Oliver EA, Abdel-Razeq SS, Shook LL, Bahtiyar MO, Buhimschi IA, Amniotic Fluid Soluble Myeloid Differentiation-2 (sMD-2) as Regulator of Intra-amniotic Inflammation in Infection-induced Preterm Birth, Am J Reprod Immunol 73(6) (2015) 507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Itoh K, Okubo K, Utiyama H, Hirano T, Yoshii J, Matsubara K, Expression profile of active genes in granulocytes, Blood 92(4) (1998) 1432–41. [PubMed] [Google Scholar]
- [15].Cowland JB, Borregaard N, The individual regulation of granule protein mRNA levels during neutrophil maturation explains the heterogeneity of neutrophil granules, J Leukoc Biol 66(6) (1999) 989–95. [DOI] [PubMed] [Google Scholar]
- [16].Federzoni EA, Valk PJ, Torbett BE, Haferlach T, Lowenberg B, Fey MF, Tschan MP, PU.1 is linking the glycolytic enzyme HK3 in neutrophil differentiation and survival of APL cells, Blood 119(21) (2012) 4963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].de Kleijn S, Kox M, Sama IE, Pillay J, van Diepen A, Huijnen MA, van der Hoeven JG, Ferwerda G, Hermans PW, Pickkers P, Transcriptome kinetics of circulating neutrophils during human experimental endotoxemia, PLoS One 7(6) (2012) e38255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Buhimschi IA, Christner R, Buhimschi CS, Proteomic biomarker analysis of amniotic fluid for identification of intra-amniotic inflammation, BJOG 112(2) (2005) 173–81. [DOI] [PubMed] [Google Scholar]
- [19].Villadangos JA, Bryant RA, Deussing J, Driessen C, Lennon-Dumenil AM, Riese RJ, Roth W, Saftig P, Shi GP, Chapman HA, Peters C, Ploegh HL, Proteases involved in MHC class II antigen presentation, Immunol Rev 172 (1999) 109–20. [DOI] [PubMed] [Google Scholar]
- [20].King AE, Critchley HO, Kelly RW, Innate immune defences in the human endometrium, Reprod Biol Endocrinol 1 (2003) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Montenegro D, Romero R, Pineles BL, Tarca AL, Kim YM, Draghici S, Kusanovic JP, Kim JS, Erez O, Mazaki-Tovi S, Hassan S, Espinoza J, Kim CJ, Differential expression of microRNAs with progression of gestation and inflammation in the human chorioamniotic membranes, Am J Obstet Gynecol 197(3) (2007) 289 e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Montenegro D, Romero R, Kim SS, Tarca AL, Draghici S, Kusanovic JP, Kim JS, Lee DC, Erez O, Gotsch F, Hassan SS, Kim CJ, Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition, J Pathol 217(1) (2009) 113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chytil F, Ong DE, Intracellular vitamin A--binding proteins, Annu Rev Nutr 7 (1987) 321–35. [DOI] [PubMed] [Google Scholar]
- [24].Xavier GM, Cobourne MT, Scube2 expression extends beyond the central nervous system during mouse development, J Mol Histol 42(5) (2011) 383–91. [DOI] [PubMed] [Google Scholar]
- [25].Lin YC, Chen CC, Cheng CJ, Yang RB, Domain and functional analysis of a novel breast tumor suppressor protein, SCUBE2, J Biol Chem 286(30) (2011) 27039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lin YC, Lee YC, Li LH, Cheng CJ, Yang RB, Tumor suppressor SCUBE2 inhibits breast-cancer cell migration and invasion through the reversal of epithelial-mesenchymal transition, J Cell Sci 127(Pt 1) (2014) 85–100. [DOI] [PubMed] [Google Scholar]
- [27].Lee SA, Lee SY, Cho IH, Oh MA, Kang ES, Kim YB, Seo WD, Choi S, Nam JO, Tamamori-Adachi M, Kitajima S, Ye SK, Kim S, Hwang YJ, Kim IS, Park KH, Lee JW, Tetraspanin TM4SF5 mediates loss of contact inhibition through epithelial-mesenchymal transition in human hepatocarcinoma, J Clin Invest 118(4) (2008) 1354–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang M, Guo M, Hu Y, Jiang Y, Scube regulates synovial angiogenesis-related signaling, Med Hypotheses 81(5) (2013) 948–53. [DOI] [PubMed] [Google Scholar]
- [29].Choi S, Lee SA, Kwak TK, Kim HJ, Lee MJ, Ye SK, Kim SH, Kim S, Lee JW, Cooperation between integrin alpha5 and tetraspan TM4SF5 regulates VEGF-mediated angiogenic activity, Blood 113(8) (2009) 1845–55. [DOI] [PubMed] [Google Scholar]
- [30].Cvoro A, Yuan C, Paruthiyil S, Miller OH, Yamamoto KR, Leitman DC, Cross talk between glucocorticoid and estrogen receptors occurs at a subset of proinflammatory genes, J Immunol 186(7) (2011) 4354–60. [DOI] [PubMed] [Google Scholar]
- [31].Tang Z, Niven-Fairchild T, Tadesse S, Norwitz ER, Buhimschi CS, Buhimschi IA, Guller S, Glucocorticoids enhance CD163 expression in placental Hofbauer cells, Endocrinology 154(1) (2013) 471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lu NZ, Collins JB, Grissom SF, Cidlowski JA, Selective regulation of bone cell apoptosis by translational isoforms of the glucocorticoid receptor, Mol Cell Biol 27(20) (2007) 7143–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR, Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes, Proc Natl Acad Sci U S A 101(44) (2004) 15603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nehme A, Lobenhofer EK, Stamer WD, Edelman JL, Glucocorticoids with different chemical structures but similar glucocorticoid receptor potency regulate subsets of common and unique genes in human trabecular meshwork cells, BMC Med Genomics 2 (2009) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Buhimschi IA, Zhao G, Pettker CM, Bahtiyar MO, Magloire LK, Thung S, Fairchild T, Buhimschi CS, The receptor for advanced glycation end products (RAGE) system in women with intraamniotic infection and inflammation, Am J Obstet Gynecol 196(2) (2007) 181 e1–13. [DOI] [PubMed] [Google Scholar]
- [36].Buhimschi CS, Dulay AT, Abdel-Razeq S, Zhao G, Lee S, Hodgson EJ, Bhandari V, Buhimschi IA, Fetal inflammatory response in women with proteomic biomarkers characteristic of intra-amniotic inflammation and preterm birth, BJOG 116(2) (2009) 257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fritz G, RAGE: a single receptor fits multiple ligands, Trends Biochem Sci 36(12) (2011) 625–32. [DOI] [PubMed] [Google Scholar]
- [38].Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, Yasuda J, Kawai J, Hayashizaki Y, Hume DA, Suzuki H, Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation, Leukemia 24(2) (2010) 460–6. [DOI] [PubMed] [Google Scholar]
- [39].Bandyopadhyay S, Lane T, Venugopal R, Parthasarathy PT, Cho Y, Galam L, Lockey R, Kolliputi N, MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2, Biochem Biophys Res Commun 439(3) (2013) 407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P, MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells, Proc Natl Acad Sci U S A 106(8) (2009) 2735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T, A mammalian microRNA expression atlas based on small RNA library sequencing, Cell 129(7) (2007) 1401–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD, Regulation of progenitor cell proliferation and granulocyte function by microRNA-223, Nature 451(7182) (2008) 1125–9. [DOI] [PubMed] [Google Scholar]
- [43].Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I, A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis, Cell 123(5) (2005) 819–31. [DOI] [PubMed] [Google Scholar]
- [44].Tacke F, Roderburg C, Benz F, Cardenas DV, Luedde M, Hippe HJ, Frey N, Vucur M, Gautheron J, Koch A, Trautwein C, Luedde T, Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients, Crit Care Med 42(5) (2014) 1096–104. [DOI] [PubMed] [Google Scholar]
- [45].Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM, Serum miR-146a and miR-223 as potential new biomarkers for sepsis, Biochem Biophys Res Commun 394(1) (2010) 184–8. [DOI] [PubMed] [Google Scholar]
- [46].Li Y, Xie X, A mixture model for expression deconvolution from RNA-seq in heterogeneous tissues, BMC Bioinformatics 14 Suppl 5 (2013) S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Salafia CM, Popek EJ, Inflammatory and vascular placental pathology, Glob Libr Womens Med doi: 10.3843/GLOWM.10152 (2008). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


