Abstract
Analysis of 594 isolates of Escherichia coli sequence type (ST)131 and its single locus variants carrying carbapenemase genes from 17 European Union/European Economic Area countries revealed acquisition of 18 carbapenemase variants, mainly in ST131 clades A and C. Most frequent were bla OXA-244 (n = 230) and bla OXA-48 (n = 224), detected in 14 and 12 countries, respectively. Isolates carrying bla OXA-244 have increased rapidly since 2021. The increasing detection of carbapenemase genes in the E. coli high-risk lineage ST131 is a public health concern.
Keywords: Carbapenem-resistant Enterobacterales, carbapenemase, Escherichia coli, surveillance, whole genome sequencing, cross-border spread, antimicrobial resistance, antibiotics
In March 2024, the European Antimicrobial Resistance Genes Surveillance Network (EURGen-Net) operational contact points from Denmark contacted the European Centre for Disease Prevention and Control (ECDC) with concerns about increasing detection of OXA-244-producing E. coli ST131 in their country. Worldwide, E. coli is the pathogen associated with most deaths attributable to antimicrobial resistance [1]. Sequence type (ST)131 is a high-risk lineage of global distribution, frequently associated with multidrug resistance [2]. To date, there have been only few reports of carbapenemase gene-carrying E. coli ST131 isolates collected from human samples in European Union (EU)/European Economic Area (EEA) countries [3-5].
The aim of this investigation was to determine the epidemiological situation and genomic characteristics of E. coli ST131 and its single locus variants (SLVs) carrying carbapenemase genes in the EU/EEA based on the analysis of epidemiological and whole genome sequencing (WGS) data from national collections.
Data collection and analysis
On 12 April 2024, the ECDC requested, via its EpiPulse platform, national reference laboratories that participate in EURGen-Net to provide WGS and epidemiological data of isolates of E. coli ST131 and its SLVs carrying carbapenemase genes. In response, 17 EU/EEA countries submitted 660 sequence datasets (500 short-read sets, 11 long-read sets, 116 short-read assemblies and 33 hybrid assemblies) from 627 isolates. After quality control and de-duplication, we analysed the sequences of 594 isolates carrying carbapenemase genes covering the period from August 2012 to May 2024 (Table 1).
Table 1. Isolates of Escherichia coli ST131 and its single locus variants carrying carbapenemase genes, by country, EU/EEA, August 2012–May 2024 (n = 594).
| Carbapenemase gene | Number of isolates by country and period covered | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AT | BE | CZ | DE | DK | FI | FR | HU | IE | LT | LU | LV | NL | NO | PT | SE | SI | Total | |
| 2022–2024 | 2023–2024 | 2021–2023 | 2022–2023 | 2014–2024 | 2021–2024 | 2019–2024 | 2023 | 2016–2024 | 2019–2023 | 2019–2024 | 2023 | 2012–2024 | 2012–2023 | 2016–2022 | 2017–2024 | 2022–2023 | 2012–2024 | |
| bla OXA-244 | 8 | 3 | 1 | 32 | 25 | 3 | 85 | 1 | 14 | 0 | 2 | 0 | 28 | 6 | 0 | 21 | 1 | 230 |
| bla OXA-48 | 0 | 2 | 0 | 1 | 9 | 0 | 82 | 0 | 101 | 1 | 1 | 1 | 13 | 3 | 0 | 9 | 1 | 224 |
| bla NDM-1 | 0 | 2 | 2 | 1 | 3 | 1 | 7 | 1 | 4 | 0 | 0 | 0 | 4 | 2 | 3 | 0 | 1 | 31 |
| bla NDM-5 | 0 | 0 | 0 | 1 | 3 | 0 | 8 | 0 | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 2 | 0 | 20 |
| bla KPC-2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 3 | 11 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 19 |
| bla OXA-181 | 0 | 0 | 0 | 1 | 3 | 0 | 8 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 16 |
| bla KPC-31 a | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 15 |
| bla VIM-1 | 0 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 14 |
| bla KPC-3 | 0 | 1 | 0 | 1 | 1 | 2 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 12 |
| bla NDM-7 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| bla OXA-484 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| bla NDM-18 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| bla VIM-4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| bla KPC-53 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| bla KPC-225 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| bla OXA-204 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| bla OXA-232 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| bla OXA-244* b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| bla NDM-5/bla OXA-232 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| bla NDM-5/bla OXA-244 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 8 | 10 | 3 | 40 | 44 | 6 | 202 | 2 | 129 | 12 | 5 | 2 | 60 | 13 | 20 | 35 | 3 | 594 |
AT: Austria; BE: Belgium; CZ: Czechia; DE: Germany; DK: Denmark; EU/EEA: European Union/European Economic Area; FI: Finland; FR: France; HU: Hungary; IE: Ireland; LT: Lithuania; LU: Luxembourg; LV: Latvia; NL: the Netherlands; NO: Norway; PT: Portugal; SE: Sweden; SI: Slovenia; ST: sequence type.
a bla KPC-31 listed in the Beta-Lactamase DataBase (http://bldb.eu, accessed 28 October 2024) as carbapenemase, but reported in literature as extended spectrum beta-lactamase not conferring carbapenem resistance [24].
bbla OXA-244* represents the new bla OXA-48-like variant with C100T substitution leading to H34T amino acid change.
Numbers in bold indicate detection of ≥ 1 isolate.
Short-reads were assembled using SPAdes v3.15.5 [6] and long-reads using Flye v2.9.4 [7]. Alleles were called using ChewBBACA v3.3.4 [8] and the Escherichia/Shigella core genome multilocus sequence typing (cgMLST) scheme from EnteroBase [9]. Serotyping was performed using the E. coli analysis plugin of BioNumerics 7.6.3 (Applied Maths NV/bioMérieux). We assigned ST with the Center for Genomic Epidemiology (CGE) MLST v2.0.9 tool [10], using the 7-gene MLST scheme by Achtman [11]. We used the CGE FimTyper to assign the type 1 fimbriae adhesin fimH allele [12]. We identified antimicrobial resistance genes with ResFinder v4.1.11 with default settings [13]. Clusters were assigned using single-linkage clustering with a cut-off of 10 allelic differences [14].
Distribution of carbapenemase genes
We detected 18 different carbapenemase genes in the E. coli ST131 isolates, including ST131 SLVs. Two carbapenemase genes, bla OXA-244 (n = 230) and bla OXA-48 (n = 224), together accounted for 76% of the isolates (Table 1), followed by bla NDM-1 in 31 (5%) and bla NDM-5 in 20 (3%) isolates. All other carbapenemase genes were detected in fewer than 20 isolates (Table 1). The isolates carrying bla OXA-244 were detected in 14 countries. Isolates carrying bla OXA-48 were detected in 12 countries, although most originated from France and Ireland. Despite the much smaller numbers of isolates carrying bla NDM-1 or bla NDM-5, these isolates were also detected in 12 and seven countries, respectively.
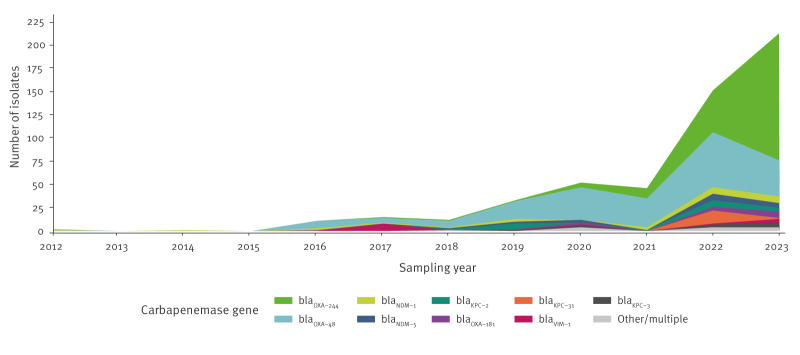
While E. coli ST131 isolates carrying bla OXA-48 appeared earlier than isolates with bla OXA-244 (2012 vs 2017), their frequency of detection increased only moderately over time, with a peak in 2022 followed by a small decrease in 2023 (Figure 1).
Figure 1.
Number of Escherichia coli ST131 isolates, including its single locus variantsa, carrying carbapenemase genes, by year, EU/EEA, 2012–2023b (n = 535)
EU/EEA: European Union/European Economic Area.
a Detected single locus variants included ST8420, ST11358, ST11362, ST11672, ST13133 and ST13730.
b Three isolates without information on year of sampling and 56 isolates from 2024 (as that year was incomplete at time of sampling) were excluded.
In contrast, detection of isolates carrying bla OXA-244 increased sharply between 2021 and 2023. In addition, we observed an increasing diversity of carbapenemase (including metallo-beta-lactamase) genes over time, although without a clear trend for any of the six metallo-beta-lactamase genes detected in this analysis.
Epidemiological and microbiological characteristics
Based on the varying frequency and time trends, we divided the isolates into three groups for further analysis: Group 1: E. coli ST131 isolates, including ST131 SLVs, carrying bla OXA-244; Group 2: isolates carrying bla OXA-48; and Group 3: isolates carrying other carbapenemase genes (Table 2). In the epidemiological analysis, Group 1 stood out with a high proportion of female patients, a relatively low median age, the frequent detection of isolates from urine samples, and slightly more frequent documentation of travel outside the EU/EEA within 12 months before detection (Table 2).
Table 2. Epidemiological and genomic characteristics of carbapenemase genes carrying isolates of Escherichia coli ST131 and its single locus variants, EU/EEA, August 2012–May 2024 (n = 594).
| Characteristic | Group 1: bla
OXA-244
n = 230 |
Group 2: bla
OXA-48
n = 224 |
Group 3: other n = 140 |
|||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Median age (years) | 57 | 77 | 70 | |||
| Sex | ||||||
| Male | 69 | 30 | 81 | 36 | 51 | 36 |
| Female | 135 | 59 | 82 | 37 | 57 | 41 |
| Not available | 26 | 11 | 61 | 27 | 32 | 23 |
| Type of sample | ||||||
| Urine | 115 | 50 | 58 | 26 | 57 | 41 |
| Rectal/faeces | 23 | 10 | 94 | 42 | 17 | 12 |
| Blood | 6 | 3 | 5 | 2 | 8 | 6 |
| Other | 28 | 12 | 11 | 5 | 15 | 11 |
| Not available | 58 | 25 | 56 | 25 | 43 | 31 |
| Travel outside the EU/EEA in the past 12 months | ||||||
| Yes | 35 | 15 | 5 | 2 | 8 | 6 |
| No | 16 | 7 | 3 | 1 | 7 | 5 |
| Not available | 179 | 78 | 216 | 96 | 125 | 89 |
| Destinations (number of travel links to respective destination) | Türkiye (17), Egypt (8), Algeria (3), Morocco (2), Guinea (1), India (1), Jordan (1), Senegal (1), Tunisia (1) | Iran (1), Syria (1), Thailand (1), Venezuela (1), Viet Nam (1) | Ukraine (2), Albania (1), Colombia (1), Morocco (1), India (1), Somalia (1), Senegal (1) | |||
| Serotype | ||||||
| O16:H5 | 148 | 64 | 80 | 36 | 50 | 36 |
| O25:H4 | 73 | 32 | 127 | 57 | 84 | 60 |
| Other | 1 | 0 | 4 | 2 | 1 | 1 |
| Unknown | 8 | 3 | 13 | 6 | 5 | 4 |
| Sequence type | ||||||
| 131 | 182 | 79 | 209 | 93 | 138 | 99 |
| 13730 | 42 | 18 | 0 | 0 | 0 | 0 |
| Other | 6 | 3 | 15 | 7 | 2 | 1 |
| fimH allelea | ||||||
| fimH41 (Clade A marker) | 153 | 67 | 85 | 38 | 51 | 36 |
| fimH30 (Clade C marker) | 75 | 33 | 115 | 51 | 63 | 45 |
| fimH22 (Clade B marker) | 0 | 0 | 9 | 4 | 16 | 11 |
| Other | 2 | 1 | 10 | 4 | 10 | 7 |
| Absent | 0 | 0 | 5 | 2 | 0 | 0 |
| Fluoroquinolone resistance mutation(s) | ||||||
| GyrA S83L; ParC S80I, E84V | 96 | 42 | 1 | 0 | 1 | 1 |
| GyrA S83L, D87N; ParC S80I, E84V | 52 | 23 | 93 | 42 | 69 | 49 |
| GyrA S83L only | 26 | 11 | 42 | 19 | 29 | 21 |
| GyrA S83L, D87N; ParC S80I | 6 | 3 | 4 | 2 | 5 | 4 |
| Other | 13 | 6 | 3 | 1 | 1 | 1 |
| Absent | 37 | 16 | 81 | 36 | 35 | 25 |
| ESBL gene(s) | ||||||
| bla CTX-M-15 | 172 | 75 | 47 | 21 | 44 | 31 |
| bla CTX-M-27 | 7 | 3 | 20 | 9 | 28 | 20 |
| bla SHV-12 | 0 | 0 | 1 | 0 | 16 | 11 |
| Other/multiple | 3 | 1 | 5 | 2 | 12 | 9 |
| Absent | 48 | 21 | 151 | 67 | 40 | 29 |
ESBL: extended-spectrum beta-lactamase; EU/EEA: European Union/European Economic Area; SLV: single locus variant; ST: sequence type.
Genomic characteristics present in less than 10 isolates are grouped into the category ‘other’. Category unknown for serotype signifies that either O antigen or H antigen type, or both, were not assigned by BioNumerics 7.6.3.
a Alleles of fimH correlate with major clades of E. coli ST131. Of 253 isolates carrying fimH30, 251 were assigned to subclades C0 (n = 62), C1 (n = 57) and C2 (n = 132) based on the topology of the cgMLST phylogeny combined with absence (C0) or presence (C1/C2) of mutations in GyrA (S83L, D87N) and ParC (S80I, E84V) as well as detection of specific bla CTX-M genes, i.e. bla CTX-M-27 indicative of C1 or bla CTX-M-15 of C2.
Most Group 1 isolates belonged to serotype O16:H5, while the majority of group 2 and 3 isolates were of serotype O25:H4 (Table 2). Single locus variants of E. coli ST131 were most frequent among Group 1 isolates, all of which belonged to ST13730 (Table 2). Typing of fimH showed that the most frequent fimH allele in Group 1 was the clade A marker fimH41 followed by fimH30 indicative of clade C. In contrast, in Groups 2 and 3, more isolates were carrying fimH30 than fimH41 (Table 2). Of the 251 isolates with fimH30 in clade C, 132 were assigned to subclade C2, followed by C0 (n = 62) and C1 (n = 57). Resistance markers also varied by group, e.g. the extended-spectrum beta-lactamase (ESBL) gene bla CTX-M-15 was markedly more frequent in Group 1 than in Group 2 and 3 isolates. In addition, of 263 isolates co-carrying bla CTX-M-15, more than half (n = 150) had the clade A marker fimH41, followed by isolates with clade C marker fimH30 (n = 110), and clade B marker fimH22 (n = 2) and fimH27 (n = 1). We observed similar variation between groups for fluoroquinolone resistance mutations (Table 2).
Genomic relatedness
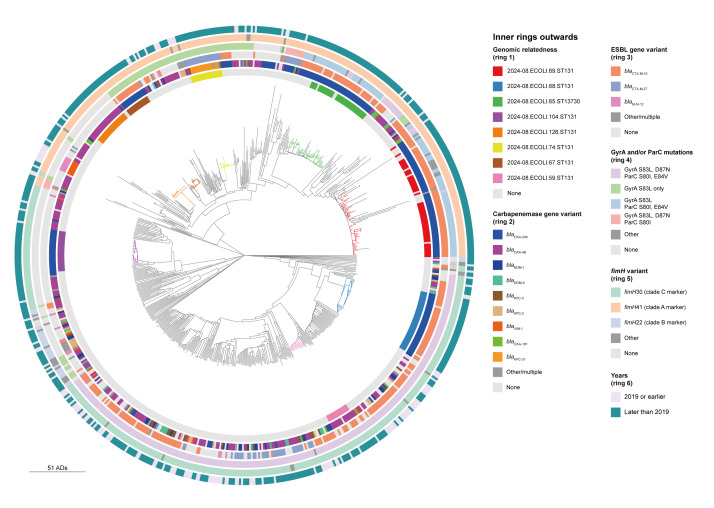
For the investigation of genomic relatedness, we added 93 isolates from ECDC surveys and investigations and four control isolates from the National Center for Biotechnology Information (NCBI) representing clades A, B and C, resulting in a dataset of 691 isolates. Eight larger clusters (n ≥ 10 isolates) were detected, including six clusters with isolates from 2024. Isolates in these eight clusters were carrying bla OXA-244 (five clusters) followed by bla OXA-48 (two clusters) and bla KPC variants (one cluster) (Figure 2).
Figure 2.
Phylogenetic tree of Escherichia coli ST131 isolates, including its single locus variants included in the genomic relatedness analysis, EU/EEA and outside, 2005–2024 (n = 691)
ADs: allelic differences; cgMLST: core genome multilocus sequence typing; ESBL: extended-spectrum beta-lactamase; EU/EEA: European Union/European Economic Area.
The phylogenetic tree was constructed based on cgMLST using neighbour joining algorithm. Of note, a cgMLST-based phylogeny has limitations on resolving E. coli ST131 population structure and division into clades. Categories in each ring are arranged in decreasing order of frequency and represent findings from more than 10 isolates. The eight largest clusters with isolates within 10 ADs in the tree are coloured following the ring 1 legend.
Isolates carrying bla OXA-244 formed multi-country clusters, while clusters of bla OXA-48-carrying isolates were predominantly detected within one country, e.g. France or Ireland. The eight clusters belonged to E. coli ST131 clade A (five clusters) or subclades C0 (one cluster) and C2 (two clusters). From a cladistic point of view, two clusters within clade A contained additional isolates that did not meet the single-linkage cluster definition. These were not counted in the cluster statistics. Four of the five clusters carrying bla OXA-244 included at least one isolate for which available hybrid assemblies confirmed the location of bla OXA-244 on the chromosome. A detailed description of the eight large clusters can be found in the Supplementary Figure.
Discussion
We report the emergence of E. coli ST131 carrying carbapenemase genes based on genomic and epidemiological data from 17 EU/EEA countries. We observed an increasing frequency of detections and diversity of carbapenemase genes from 2012 to 2024. Furthermore, we detected considerable heterogeneity in the geographical distribution and speed of spread of specific carbapenemase genes, in particular for the recent rapid emergence of ST131 isolates carrying chromosomally localised bla OXA-244 associated with large multi-country clusters.
The increasing detection of carbapenemase genes in E. coli ST131 documented in this study is of concern because E. coli can cause a variety of infections in healthcare and community settings, frequently urinary tract infections, but also including bloodstream infection [15]. Worldwide, E. coli ST131 is the predominant extraintestinal pathogenic E. coli (ExPEC) lineage. It has been strongly associated with the global dissemination of the bla CTX-M-15 ESBL gene [2], and there is a high risk that it can play a similar role for the global spread of carbapenemase genes.
While the pooling of data from 17 EU/EEA countries facilitated early detection of this emerging resistance pattern, our study was based on routine national surveillance with differences in sample collection protocols, coverage and data completeness (in particular related to missing data for sample type and travel history), which is a limitation. Nevertheless, the age, sex, sample type and travel history distribution of isolates carrying bla OXA-244 suggest a potential association with community-acquired urinary tract infections (UTI), although this would need to be confirmed by studies with harmonised sampling. Of note, E. coli carrying bla OXA-244 often do not grow on screening media for carbapenemase-producing Enterobacterales (CPE) [16] and are most likely under-detected. The apparent association of E. coli ST131 carrying bla OXA-244 with community-acquired UTIs might therefore only represent the tip of the iceberg in terms of patient colonisation in the community.
Previous global surveys of carbapenemase-producing E. coli covering different geographical areas and time periods have identified only few E. coli ST131 isolates carrying bla OXA-48 and none carrying bla OXA-244 [17,18]. Although E. coli ST131 clade C has been reported as the primary contributor to fluoroquinolone resistance and the spread of ESBL genes globally [19], clade A had a higher rate of increase in estimated effective population size in a longitudinal survey in Norway [20]. Our analysis found large multi-country clusters within clade A and C. Clusters in clade A, where the cluster definition did not capture the full diversity within the cluster-defining branch, may represent clonal expansion over time rather than recent transmission events. We also found frequent co-carriage of bla CTX-M-15 in clade A, although this clade was previously described in some but not all studies as rarely carrying ESBL genes [19-21]. However, co-carriage of bla CTX-M genes may differ by time, geographical region and selection criteria for the studied isolates. It is a limitation of this investigation that we did not analyse a random population of E. coli ST131 but isolates pre-selected for carriage of carbapenemase genes, which probably resulted in an isolate collection with a higher likelihood for co-carriage of other resistance markers.
As ExPEC has been identified in various non-human reservoirs and can be transmitted via the faecal-oral, household, sexual or food-borne routes [15], it is difficult to control its spread within the human population. There are now various examples of increasing dissemination of carbapenemase-producing ExPEC in the EU/EEA, such as E. coli ST131 as shown in this study, but also E. coli ST167, ST405, ST410 and ST648 carrying bla NDM-5 [22] and E. coli ST38 carrying bla OXA-244 [23].
Conclusion
The increasing detection of E. coli ST131 carrying carbapenemase genes with potential community acquisition and dissemination sends another warning about the worsening epidemiological CPE situation in the EU/EEA. Further spread of E. coli carrying carbapenemase genes would mean that carbapenems could no longer be consistently effective for empiric treatment of severe E. coli infections. Urgent public health action is required to improve control of CPE in the EU/EEA and worldwide.
Ethical statement
All data were pseudonymised and collected in accordance with the European Parliament and Council decisions on the epidemiological surveillance and control of communicable disease in the European Community. Ethical approval and informed consent were thus not required.
Funding statement
No specific funding was received for this work.
Use of artificial intelligence tools
None declared.
Data availability
The national whole genome sequencing data collected for this study were deposited in the European Nucleotide Archive under accession numbers PRJEB35685, PRJEB42331, PRJEB56146, PRJEB60743, PRJEB61153, PRJEB75178, PRJEB76821, PRJEB81860, PRJNA295003, and PRJNA1076808. More information can be found in the Supplementary Table.
Supplementary Data
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: Study concept and design: AK, ML. Acquisition, analysis, or interpretation of data: AK, PA, RH, PB, TH, KC, LM, JE, MS, AMH, LR, KR, LD, RAB, AT, KT, CC, MC, AG, KK, MM, BNO, RV, APAH, DWM, ØS, MC, VEM, VIM, BM, UK, MP, DP, DLM, ML. Bioinformatic analysis: OS, EA. Drafting of the manuscript: AK, ML. Critical revision of the manuscript for important intellectual content: all authors.
References
- 1. Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-55. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565-91. 10.1128/CMR.00116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Welker S, Boutin S, Miethke T, Heeg K, Nurjadi D. Emergence of carbapenem-resistant ST131 Escherichia coli carrying bla OXA-244 in Germany, 2019 to 2020. Euro Surveill. 2020;25(46):2001815. 10.2807/1560-7917.ES.2020.25.46.2001815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piazza A, Corbella M, Mattioni Marchetti V, Merla C, Mileto I, Kuka A, et al. Clinical isolates of ST131 blaOXA-244-positive Escherichia coli, Italy, December 2022 to July 2023. Euro Surveill. 2024;29(8):2400073. 10.2807/1560-7917.ES.2024.29.8.2400073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patiño-Navarrete R, Rosinski-Chupin I, Cabanel N, Zongo PD, Héry M, Oueslati S, et al. Specificities and Commonalities of Carbapenemase-Producing Escherichia coli Isolated in France from 2012 to 2015. mSystems. 2022;7(1):e0116921. 10.1128/msystems.01169-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455-77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540-6. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- 8. Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, Santos S, et al. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018;4(3):e000166. 10.1099/mgen.0.000166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M, Agama Study Group . The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30(1):138-52. 10.1101/gr.251678.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355-61. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136-51. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, et al. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol. 2017;55(8):2538-43. 10.1128/JCM.00737-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491-500. 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24(4):350-4. 10.1016/j.cmi.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 15. Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev. 2019;32(3):e00135-18. 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emeraud C, Biez L, Girlich D, Jousset AB, Naas T, Bonnin RA, et al. Screening of OXA-244 producers, a difficult-to-detect and emerging OXA-48 variant? J Antimicrob Chemother. 2020;75(8):2120-3. 10.1093/jac/dkaa155 [DOI] [PubMed] [Google Scholar]
- 17. Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, et al. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis. 2014;20(11):1928-31. 10.3201/eid2011.141388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peirano G, Chen L, Nobrega D, Finn TJ, Kreiswirth BN, DeVinney R, et al. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015-2017. Emerg Infect Dis. 2022;28(5):924-31. 10.3201/eid2805.212535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pitout JDD, Finn TJ. The evolutionary puzzle of Escherichia coli ST131. Infect Genet Evol. 2020;81:104265. 10.1016/j.meegid.2020.104265 [DOI] [PubMed] [Google Scholar]
- 20. Gladstone RA, McNally A, Pöntinen AK, Tonkin-Hill G, Lees JA, Skytén K, et al. Emergence and dissemination of antimicrobial resistance in Escherichia coli causing bloodstream infections in Norway in 2002-17: a nationwide, longitudinal, microbial population genomic study. Lancet Microbe. 2021;2(7):e331-41. 10.1016/S2666-5247(21)00031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio. 2016;7(2):e02162. 10.1128/mBio.02162-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linkevicius M, Bonnin RA, Alm E, Svartström O, Apfalter P, Hartl R, et al. Rapid cross-border emergence of NDM-5-producing Escherichia coli in the European Union/European Economic Area, 2012 to June 2022. Euro Surveill. 2023;28(19):2300209. 10.2807/1560-7917.ES.2023.28.19.2300209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.European Centre for Disease Prevention and Control (ECDC). Rapid risk assessment: Increase in OXA-244-producing Escherichia coli in the European Union/European Economic Area and the UK since 2013, first update. Stockholm: ECDC; 2021. Available from: https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-increase-oxa-244-producing-escherichia-coli-eu-eea
- 24. Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother. 2017;61(5):e02534-16. 10.1128/AAC.02534-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.