Abstract
Background
Currently, there is a lack of biomarkers to identify breast cancer (BC) patients who would benefit from CDK4/6 inhibitors. This study combined machine learning (ML) algorithms based on transcriptomic data with both in vivo and in vitro experiments to identify therapeutic efficacy-related biomarkers of the CDK4/6 inhibitor ribociclib from the perspective of long non-coding RNA (lncRNA).
Methods
We used the Genomics of Drug Sensitivity in Cancer database along with the “oncoPredict” algorithm to calculate the half maximal inhibitory concentration (IC50) values for ribociclib based on transcriptome data. ML algorithms were utilized to select key lncRNAs related to ribociclib and to establish a model which could be used for selection of potential beneficiaries of ribociclib. Cellular experiments were conducted to validate the ML analysis and explore the potential biological mechanisms by which RERE-AS1 influences ribociclib efficacy and malignant phenotype of BC cells. Correlation analysis with clinical pathological factors, RT-qPCR experiments on tissue specimens, and pan-cancer analysis were carried out to explore the expression pattern, and the prognostic and diagnostic potential of RERE-AS1 in cancers.
Results
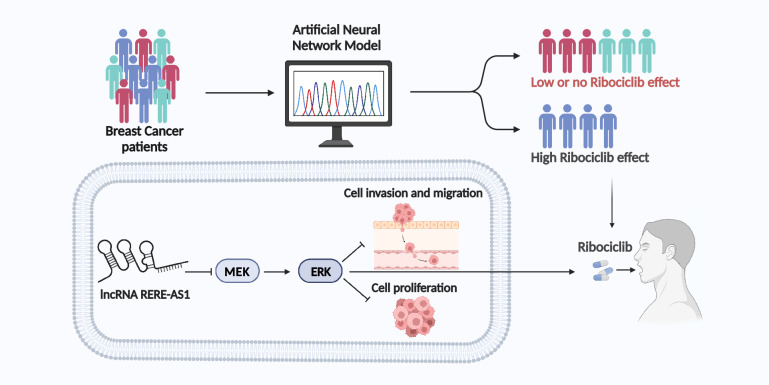
We have identified 11 key ribociclib-related lncRNAs and constructed an artificial neural network model (ANNM) based on lncRNA. Cellular experiments demonstrated that overexpression of RERE-AS1 promoted the anti-tumor activity of ribociclib in BC cells. Furthermore, RERE-AS1 is crucial in suppressing the malignant traits of BC cells through the reduction of MEK and ERK phosphorylation levels. Patients with smaller primary tumors and lower pathological stage exhibited higher levels of RERE-AS1 expression. Lastly, a pan-cancer analysis revealed that RERE-AS1 exhibits distinctly abnormal expression patterns, prognostic significance, and clinical diagnostic value in BC, compared to other cancers.
Conclusions
The ANNM established through ML algorithms can serve as predictive indicators for the efficacy of ribociclib in BC patients. LncRNA RERE-AS1, a newly discovered biomarker, holds significant promise for diagnosis, treatment, and enhancing the therapeutic response to ribociclib in BC.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05828-x.
Keywords: Breast cancer, CDK4/6 inhibitor, Ribociclib, LncRNA, Machine learning algorithm, Malignant phenotype, MEK/ERK pathway
Background
The dysregulated cell cycle, a hallmark feature of cancer cells, are governed by several regulatory proteins [1]. CDK4/6 is particularly crucial in controlling the G1 phase progression during the early stages of the cell cycle. In recent years, novel therapeutics targeting the CDK4/6 protein have garnered significant interest as a promising strategy for cancer treatment [2]. Three inhibitors targeting CDK4/6—ribociclib, palbociclib, and abemaciclib—have undergone development and clinical evaluation, demonstrating significant effectiveness and manageable toxicity profiles in cancer patients, particularly in advanced breast cancer (BC) patients [2]. Phase III studies like MONALEESA-2 have shown that combining ribociclib with letrozole significantly extends progression-free survival (PFS) of BC patients compared to using letrozole alone, with results demonstrating a hazard ratio (HR) of 0.56 and a p-value of 3.29 × 10−6 [3].
Nevertheless, despite the enhanced prognosis from CDK4/6 inhibitor treatments, around 10% of tumors demonstrate initial resistance, resulting in poor outcomes [4]. The biomarkers related to resistance to CDK4/6 inhibitors identified so far include regulators of the cell cycle, genes involved in oncogenic kinase pathways, and genes that influence the tumor microenvironment [5, 6]. At present, the value of these biomarkers in improving drug sensitivity remains limited. Therefore, exploring novel biomarkers to enhance and predict the efficacy of CDK4/6 inhibitors is of ongoing importance in clinical practice [4, 7]. However, research on biomarkers that influence the anti-tumor activity of CDK4/6 inhibitors remains limited. This study aims to explore more potential biomarkers related to ribociclib, providing more preclinical theoretical basis for enhancing patient treatment efficacy.
Non-coding RNAs (ncRNAs) offer a new dimension for exploring cancer mechanisms and devising therapeutic and preventive strategies [8]. Previous studies have shown that targeting miRNA can improve the resistance of CDK4/6 inhibitors [4]. In triple-negative BC (TNBC) cells, miR-29b-3p, negatively regulated by c-myc, further activates CDK6, leading to decreased sensitivity to palbociclib treatment [9]. miR-3613-3p induces cell senescence by directly targeting SMAD2 and EZH2, mediating sensitivity of TNBC cells to Palbociclib [10]. Additionally, in CDK4/6 inhibitor-resistant BC cell lines, miR-432-5p was found to increase CDK6 expression by inhibiting the TGF-β pathway [11]. However, there is scant investigation into the impact of lncRNA on the efficacy of CDK4/6 inhibitors and predictive role of lncRNA in identifying potential beneficiaries of CDK4/6 inhibitors in BC. This study innovatively utilized machine learning (ML) algorithms to identify and screen lncRNAs that can predict the efficacy of ribociclib and to construct a model for screening beneficiary groups. Additionally, through cellular and animal experiments, this research elucidated the mechanisms by which the novel biomarker RERE-AS1 regulates the sensitivity to ribociclib, providing a theoretical basis for clinical research and future medical decision-making.
Materials and methods
Prediction of half-maximal inhibitory concentration (IC50) values for ribociclib
We obtained transcriptome data and clinicopathological factors and survival data of patients with cancers through the Cancer Genome Atlas (TCGA) database. The raw data was then processed by converting the counts into transcripts per million and normalizing these values using the formula log2 (TPM + 1).
Utilizing the normalized transcriptome data of TCGA-BRCA along with the Genomics of Drug Sensitivity in Cancer v2 (GDSC2) database, we determined the IC50 value of ribociclib for each BC patient by employing the “oncoPredict” R package. The “oncoPredict” R package uses in vitro gene expression and drug sensitivity data to train models, which are then used to predict drug sensitivity in new gene expression datasets [12]. Using the R package “survminer”, we identified the optimal cutoff value for classifying patients into groups of drug sensitivity and insensitivity.
Screening of lncRNAs related to ribociclib
We utilized Spearman correlation analysis and the “limma” R package to identify lncRNAs associated with ribociclib. Differential analysis and prognostic analysis were further used to screen lncRNA. The random forest and lasso algorithms were employed utilizing the “randomForest” and “glmnet” R packages, respectively, to identify the lncRNAs that are most critical for classification or prediction. A Venn diagram was created using the “ggplot2” and “VennDiagram” R packages to display the overlap of lncRNAs that were identified using both random forest and lasso algorithms. The receiver operating characteristic (ROC) curve and area under the curve (AUC) value for the lncRNA data was conducted using the “pROC” package.
Construction and validation of artificial neural network model (ANNM)
Patients were randomly divided into training and validation sets at a 7:3 ratio using the “createDataPartition” function from the “caret” package. The “neuralnet” R package was utilized to build ANNM. The “NeuralNetTools” R package facilitated the visualization and evaluation of the constructed models, providing insights into their structure and performance. The accuracy of ANNM was assessed by employing ROC curve analysis, facilitated by the “pROC” package.
Correlation analysis of RERE-AS1 with signatures of cancer hallmarks and clinical pathological characteristics
Cancer hallmark-related gene sets were compiled and analyzed using the “GSVA” package in R, with the “ssgsea” method set as the parameter [13]. Ultimately, the relationship between RERE-AS1 and the scores for cancer hallmarks was evaluated through Spearman correlation analysis.
We used the R package “pheatmap” to generate a heatmap illustrating the differences of clinical pathological characteristics between the high and low expression levels of RERE-AS1. In addition, we analyzed the discrepancies in RERE-AS1 expression between different T and pathological stages with the “stats” and “car” R package. Then, we visualized the results with box plots using the “ggplot2” package. To further explore clinical application of RERE-AS1, we employed the “survival” and “rms” packages to construct nomograms and to plot calibration curves. The “ggDCA” package was used to perform decision curve analysis (DCA).
Construction of RERE-AS1-mediated ceRNA network
We obtained data on the interaction between lncRNAs and miRNAs from miRcode (http://www.mircode.org/index.php), targetscan (http://www.targetscan.org/vert_80/), and miRanda (www.microRNA.org) databases. Additionally, data on the interaction between miRNAs and mRNAs were acquired from miRWalk (http://129.206.7.150/) and the Encyclopedia of RNA Interactomes (http://starbase.sysu.edu.cn/index.php) databases. Finally, based on the results derived from these databases, we visualized the ceRNA interaction network using the software of CytoScape (version 3.9.1).
Functional enrichment analysis
We performed Gene Ontology (GO) functional enrichment analysis and GO classification annotation for mRNA in the ceRNA network using the GO database (https://www.geneontology.org/), and “clusterProfiler” R package. This database was utilized to identify the biological functions represented by enriched GO terms. Additionally, we utilized the “clusterProfiler” R package to conduct Gene Set Enrichment Analysis (GSEA).
Analysis of differentially expressed genes
We used transcriptome sequencing data of RERE-AS1 overexpressing cell lines and control cell lines to obtain differentially expressed genes using the “limma” R package, and created a heatmap to visualize these genes employing the “pheatmap” package. Additionally, we utilized the “ggplot2” package to construct a volcano plot illustrating the upregulation and downregulation of differential genes.
Pan-cancer analysis of RERE-AS1
We used differential analysis to investigate the difference in expression levels of RERE-AS1 between normal and cancerous tissues. The “survival” package was employed to assess the prognostic role, employing both univariate Cox regression analysis and log-rank analysis based on outcomes including overall survival (OS), recurrence-free survival (RFS), disease-specific survival (DSS), and PFS. The diagnostic value across different cancers was evaluated using ROC analysis.
Cell lines, cell culture, and transfection
Human breast cancer cell lines (T47D, MCF-7, BT474, SUM159PT, MDA-MB-231, MDA-MB-453, MDA-MB-468, BT-549, and Hs578T) along with the HEK293T cell line were acquired from Pricella Life Science & Technology Co., Ltd (China). Cell lines such as MCF-7, MDA-MB-231, MDA-MB-453, MDA-MB-468, Hs578T, and HEK293T were propagated in Dulbecco’s Modified Eagle Medium (DMEM, Biological Industries), whereas T47D, BT474, SUM159PT, and BT-549 were grown in Roswell Park Memorial Institute-1640 (RPMI-1640, Biological Industries) medium. Each medium was supplemented with additional fetal bovine serum (Biological Industries) and penicillin/streptomycin (Solarbio, China).
Human OE-RERE-AS1 lentivirus was obtained from GeneChem (China). MCF-7 cells underwent lentiviral infection at a multiplicity of infection (MOI) of 20, and MDA-MB-231 cells were infected at an MOI of 10. Lentivirus containing vector was used as a negative control for all cell infections. Transfection was carried out using HiTransG P transfection reagent (GeneChem, China), followed by treatment with puromycin-containing medium (MedChemExpress, USA) for one week to establish stable RERE-AS1 overexpressing BC cells.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RNA was isolated using TRIzol reagent (TAKARA, Japan) and subsequently converted to cDNA with a reverse transcription kit from TAKARA (Japan). mRNA levels were quantified using the 2−∆∆Ct method, normalized against the housekeeping gene GAPDH. The sequences of the PCR primers are listed in Additional file1: Table S1. Additionally, nuclear and cytoplasmic fractions were separated using the Cytoplasmic and Nuclear RNA Purification Kit from Norgen Biotek Corp (Canada).
Tissue specimens
The frozen tissues from 20 cases of tumor specimens and 13 cases of the non-tumoral surrounding tissue specimens were used for RT-qPCR assay. All samples were stored at − 80 °C from the time of harvest. The study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (bc20240081) and was conducted in accordance with the Declaration of Helsinki.
Cell viability assay
Cells were plated in 96-well plates at a density of 1000 cells per well. Under light-protected conditions, 10 μL of Cell Counting Kit-8 (CCK-8) reagent (Solarbio, China) was added to each well, followed by light-protected incubation in a cell culture incubator for 2 h. cell viability was measured by reading the absorbance at 450 nm using a microplate reader (Bio-Rad, USA). The concentration of ribociclib necessary to achieve a 50% inhibition of cell growth was determined using viability curves obtained from the CCK-8 assay.
Colony formation assay
A total of 1 × 103 cells were placed into each well of 6-well plates and grown for a period of 7 to 21 days. After visible clone formation, the culture medium was discarded, and the cells were fixed with 4% paraformaldehyde for 15 min, stained with 1% crystal violet for 10 min, and then counted. These experiments were conducted in triplicate, with statistical significance determined by the Student’s t-test.
5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay
15 × 104 cells were inoculated into a 12-well plate and cultured overnight. Cells were added with 50 µM EdU reagent (Beyotime, China), incubated in the incubator for 2 h, fixed with 4% paraformaldehyde, and stained with 594 azide. The nucleic acid was stained with Hoechst 33342. Capture images using an inverted fluorescence microscope (Olympus) and analyze staining results using ImageJ software.
Transwell assay
Migration and invasion experiments were conducted using 24-well plates with chambers (Corning, USA) with a pore size of 8 μm. In the invasion assay, the upper chamber was added with 200 μL of MCF-7 at a concentration of 10 × 105 cells/mL and 200 μL of MDA-MB-231 at a concentration of 5 × 105 cells/mL. In the migration assay, the upper chamber was loaded with 200 μL of MCF-7 at a concentration of 5 × 105 cells/mL and 200 μL of MDA-MB-231 at a concentration of 2.5 × 105 cells/mL. The lower chamber was filled with 600 μL of medium containing 20% serum. MCF-7 were cultured for 72 h and MDA-MB-231 cells for 12 h [14, 15]. Cells that migrated through the chambers were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Ten random fields were captured using an inverted optical microscope (Olympus), and cell numbers were quantified using ImageJ software.
Scratch test
Approximately 100 × 104 cells were seeded into each well of a 6-well plate and cultured overnight. Using a 20 µl pipette tip, scratches were made vertically across the confluent cell layer at the bottom of the culture dish. After washing with PBS, images were captured under a microscope. The scratches were photographed again at 24 and 48 h after cultivation. The migration area of the cells was measured using both Adobe Photoshop and ImageJ software.
Western blot
Cell and tissue proteins were extracted using pre-prepared RIPA buffer (Beyotime, Shanghai, China) supplemented with proteinase and phosphatase inhibitors. Following protein denaturation, the cell lysates were subjected to electrophoresis on polyacrylamide gels (Epizyme, Shanghai, China). After electrophoresis, proteins were transferred onto a nitrocellulose membrane (Millipore, Germany). The membrane was blocked with 5% bovine serum albumin, Tris-buffered saline, and 0.2% Tween, and then incubated overnight at 4 °C with primary antibodies. This was followed by incubation with horseradish peroxidase-conjugated secondary antibodies (ZSGB-BIO, #ZB-2305 and ZB-2306) for 45 min at room temperature. The bands were visualized using the Immobilon Western HRP Substrate (Millipore, Germany) and detected with an ImageQuant LAS4000 system (GE Healthcare Life Sciences). Details of the antibodies used are listed in Additional file1: Table S2.
Subcutaneous xenograft models
To establish an ex vivo xenograft tumor model, we selected 16 NOD/ShiLtJGpt mice (NCG mice) from GemPharmatech Co., Ltd. (China) and divided them into two groups (control group and overexpression group). MDA-MB-231 cells were orthotopically inoculated into the abdominal mammary fat pad of 6-week-old female NCG mice, with each mouse receiving an injection of 5 × 106 cells in suspension. Tumor volume was assessed weekly using digital calipers. At the conclusion of the animal experiments, euthanasia was performed on the mice, and tumor tissues were excised for weighing and photography. The mouse experiments were conducted in accordance with the protocol approved by The Animal Ethical and Welfare Committee of Tianjin Medical University Cancer Institute & Hospital.
Statistical analysis
Statistical analyses were conducted using Prism 9.5.1 and R software 4.2.2. The Spearman’s correlation test was employed to assess the relationships between numerical variables. Pairwise comparisons were made using Student’s t-test. One-way analysis of variance (ANOVA) was utilized to identify differences among groups for each assay, with Tukey’s post hoc test applied to determine pairwise differences. p values less than 0.05 were considered significant.
Result
Identification of ribociclib-related lncRNAs and construction of ANNM
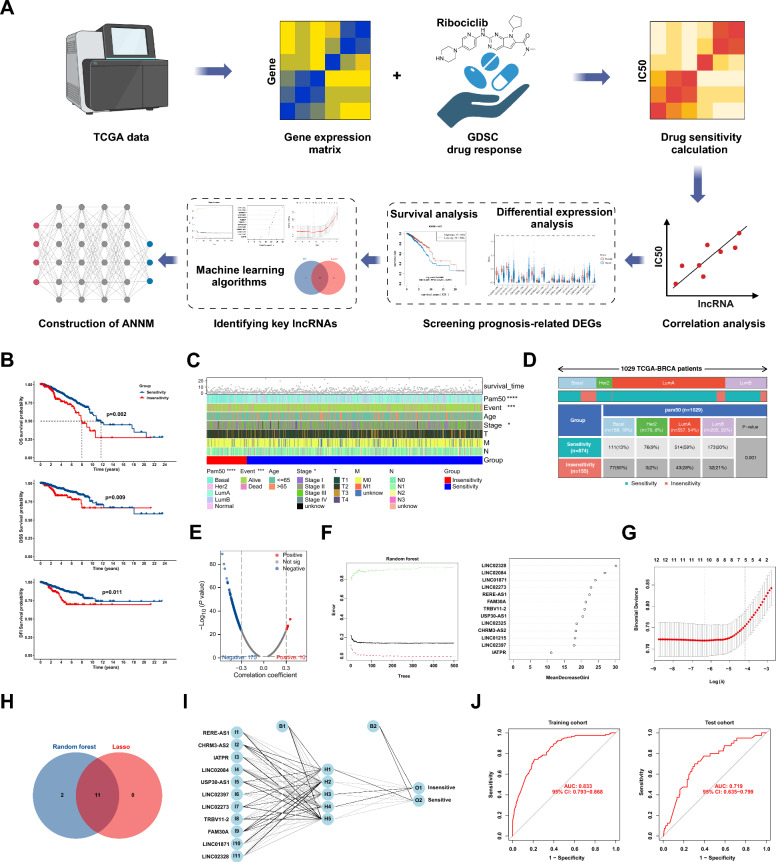
The IC50 value of ribociclib for each BC patient was calculated using transcriptome data from TCGA and drug sensitivity data from GDSC. Subsequently, we divided all patients into ribociclib-sensitive and insensitive groups (Fig. 1A). K–M curve and Log-rank test showed that the OS, DSS, DFI of the sensitive group was better than that of the insensitive group (Fig. 1B). The heatmap revealed that, alongside disparities in survival outcomes, variations in pathological stage and PAM50 subtype were evident between two groups (Fig. 1C). Subgroup analysis of the PAM50 subtype revealed that the luminal A subtype constituted the majority of patients in the sensitive group (54%), contrasting with the Basal subtype, which accounted for 50% of the non-sensitive group (Fig. 1D). Through correlation analysis, we identified 185 ribociclib-related lncRNAs, including 10 positively correlated lncRNAs and 175 negatively correlated lncRNAs (Fig. 1E). Thirteen of these lncRNAs exhibiting differential expression and prognostic value were included in the subsequent analysis (Additional file1: Table S3,4). We employed random forest and lasso algorithms, identifying 11 key lncRNAs (Fig. 1F–H). To predict and screen BC patients potentially benefiting from ribociclib therapy, we established an ANNM based on the training set using the neural network algorithm (Fig. 1I). ROC curves in both the training and test sets indicated the effective prediction of patient response to ribociclib therapy by ANNM, with AUC values of 0.833 (95% CI [0.793–0.868]) and 0.719 (95% CI [0.635–0.799]), respectively (Fig. 1J).
Fig. 1.
Screening of ribociclib-associated lncRNAs and construction of the ANNM. A Bioinformatics analysis flowchart for screening and model construction process; B Differences in prognostic outcomes between ribociclib-sensitive and -insensitive populations; C Differences in clinicopathological factors between ribociclib-sensitive and -insensitive populations; D PAM50 subgroup analysis of ribociclib-sensitive and -insensitive populations; E Correlation analysis between IC50 values of ribociclib and lncRNA expression; F–H Random forest and lasso analysis to screen key lncRNAs; I Construction of ANNM; J ROC analysis to validate the accuracy of the ANNM in the training and test sets. *p < 0.05, ***p < 0.001, ****p < 0.0001
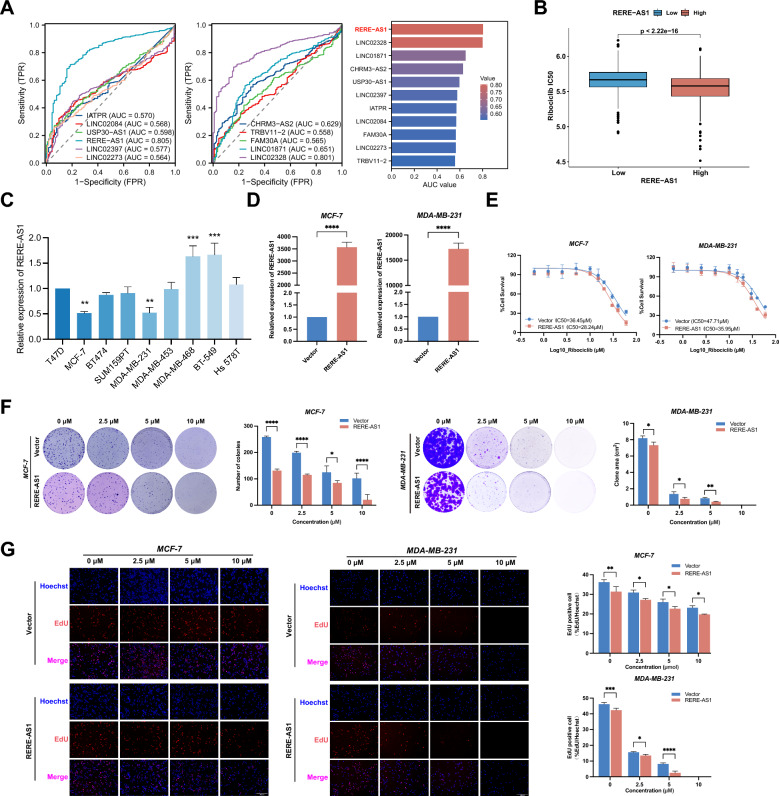
LncRNA RERE-AS1 enhances the drug sensitivity of BC cells to ribociclib
ROC curves showed that RERE-AS1 had the highest AUC value (AUC = 0.805), indicating its highest diagnostic value in BC (Fig. 2A). Hence, we chose RERE-AS1 for subsequent experiments to validate its correlation with the therapeutic response to ribociclib. Patients were stratified into high- and low-expression groups based on the median expression value of RERE-AS1. Significantly lower IC50 values were observed in the high-expression group compared to the low-expression group (p < 0.001, Fig. 2B). Further cellular experiments focused on verifying the relationship between RERE-AS1 and the response to ribociclib treatment. First, Fig. 2C illustrates that MCF-7 and MDA-MB-231 showed lower expression levels of RERE-AS1 compared to other BC cell lines. Previous studies have indicated that these two cell lines harbor functional Rb1 protein and are responsive to ribociclib treatment [16]. Thus, RERE-AS1-overexpressed MCF-7 and MDA-MB-231 cells were developed to validate the correlation between RERE-AS1 expression and the response to ribociclib treatment (Fig. 2D). The IC50 values of ribociclib between the control and overexpression groups were investigated using the cell viability assays. The results indicated that, in MCF-7 cells, the IC50 values for the control and overexpressed groups were 36.45 μM and 28.24 μM, respectively. Similarly, the IC50 values for the control and overexpression groups were 47.71 μM and 35.95 μM, respectively. These findings demonstrated that RERE-AS1 overexpression in BC cells significantly enhanced the anti-tumor efficacy of ribociclib in both MCF-7 and MDA-MB-231 cells (Fig. 2E). Furthermore, we treated BC cell lines in the control group and the RERE-AS1 overexpression group with different concentrations gradients of ribociclib (0, 2.5, 5, 10 μM), and assessed the clonal forming ability and proliferation activity of BC cells through colony formation and EdU experiments. The findings indicated that RERE-AS1 overexpression significantly enhanced the inhibitory effect of ribociclib on the clonal formation and proliferation of BC cells across various drug concentrations (Fig. 2F, G).
Fig. 2.
LncRNA RERE-AS1 enhances anti-tumor effect of ribociclib in BC cells. A Diagnostic value of 11 lncRNAs associated with ribociclib sensitivity, as determined by ROC analysis; B Differences in ribociclib IC50 values between patient groups with low- and high-expression of RERE-AS1; C Levels of RERE-AS1 expression in human BC cell lines; D Construction of MCF-7 and MDA-MB-231 cell lines with stable overexpression of RERE-AS1; E Variations in ribociclib IC50 values between control and overexpressed RERE-AS1 cells; F, G Clone formation and EdU experiments in control and RERE-AS1 overexpressing cells, under various concentrations of ribociclib treatment. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
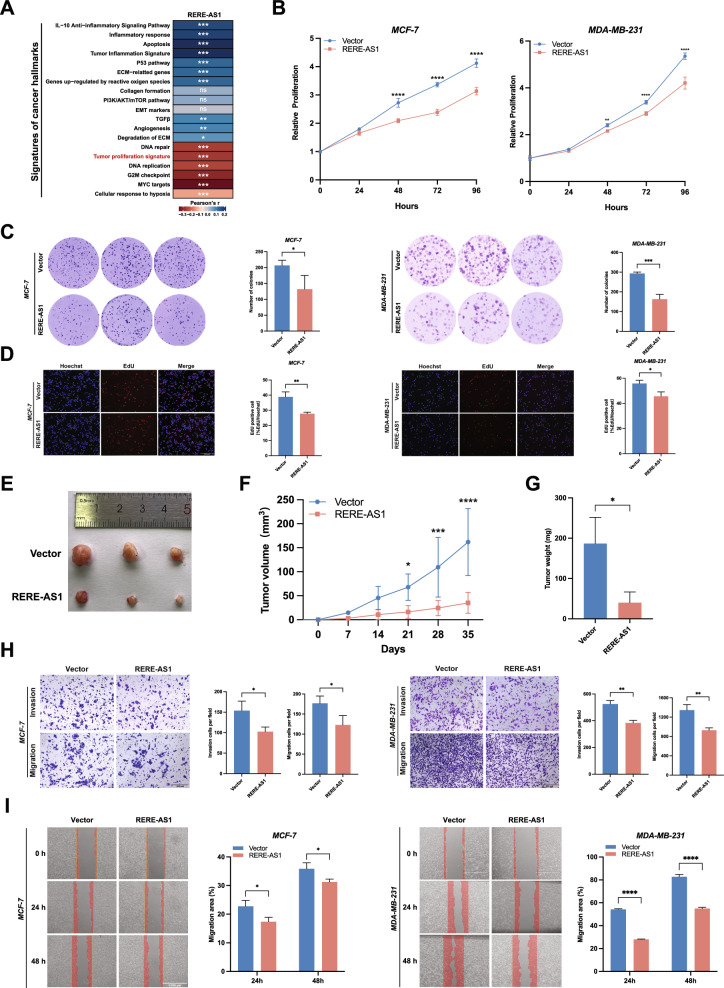
LncRNA RERE-AS1 inhibits proliferation, invasion, and migration of BC cells
The correlation analysis between RERE-AS1 and signatures of cancer hallmark revealed a negative association between RERE-AS1 and tumor proliferation signature as well as cell cycle-related signatures, including DNA replication, G2M checkpoint, and myc targets (Fig. 3A). In cellular experiments, CCK-8, colony formation, and EdU experiments all confirmed that overexpression of RERE-AS1 significantly inhibited the proliferation activity of BC cell (Fig. 3B–D). The tumor xenograft mouse model indicated that the RERE-AS1 overexpressing group exhibited significantly reduced subcutaneous tumorigenicity compared to the control group, aligning with the findings from cellular experiments (Fig. 3E–G). In addition to the proliferative phenotype, we also found that it is involved in the invasion and migration phenotype. Transwell and scratch wound healing experiments also found that high expression of RERE-AS1 inhibited the invasion and migration ability of BC cells (Fig. 3H, I).
Fig. 3.
Overexpression of lncRNA RERE-AS1 suppresses malignant phenotypes of proliferation, invasion, and migration in BC cells. A Heatmap illustrating the correlation between RERE-AS1 expression and signatures of cancer hallmarks; B–D Experiments including CCK8 assays, colony formation, and EdU assays demonstrated the impact of RERE-AS1 overexpression on the proliferative activity of BC cells. E–G MDA-MB-231 cells with stable overexpression RERE-AS1 and control cells were subcutaneously injected into the mammary fat pads of NCG mice. Tumor volumes were measured over time as depicted, and tumors were excised and weighed at the end of the experiment; H, I Transwell and wound healing assays showing the effects of RERE-AS1 overexpression on the invasion and migration capabilities of BC cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
RERE-AS1 regulates the malignant phenotype of BC cells through MER/ERK pathway
The biological mechanisms by which RERE-AS1 influences ribociclib sensitivity are still unknown. Determining cellular localization serves as an informative initial stage in characterizing the potential functions of lncRNAs [17]. Therefore, we initially fractionated nucleoplasm and cytoplasm fractions to extract RNA from the nucleus and cytoplasm of BC cells. The RT-qPCR assay demonstrated that the cellular distribution pattern of RERE-AS1 mirrored that of GAPDH (Additional file 1: Figure S1A), indicating that RERE-AS1 primarily functions in the cytoplasm. CeRNA networks are classical molecular interaction networks in which lncRNAs participate in the cytoplasm, so we employed mircode, targetscan, and miranda databases to detect miRNAs bound by RERE-AS1. Furthermore, utilizing miRWalk and ENCORI databases, we identified mRNAs targeted by miRNA and depicted the ceRNA network using Cytoscape software (Additional file1: Figure S1B). The result of GO showed that these mRNAs were enriched in CDK enzyme activity regulation and dephosphorylation regulation (Additional file1: Figure S1C).
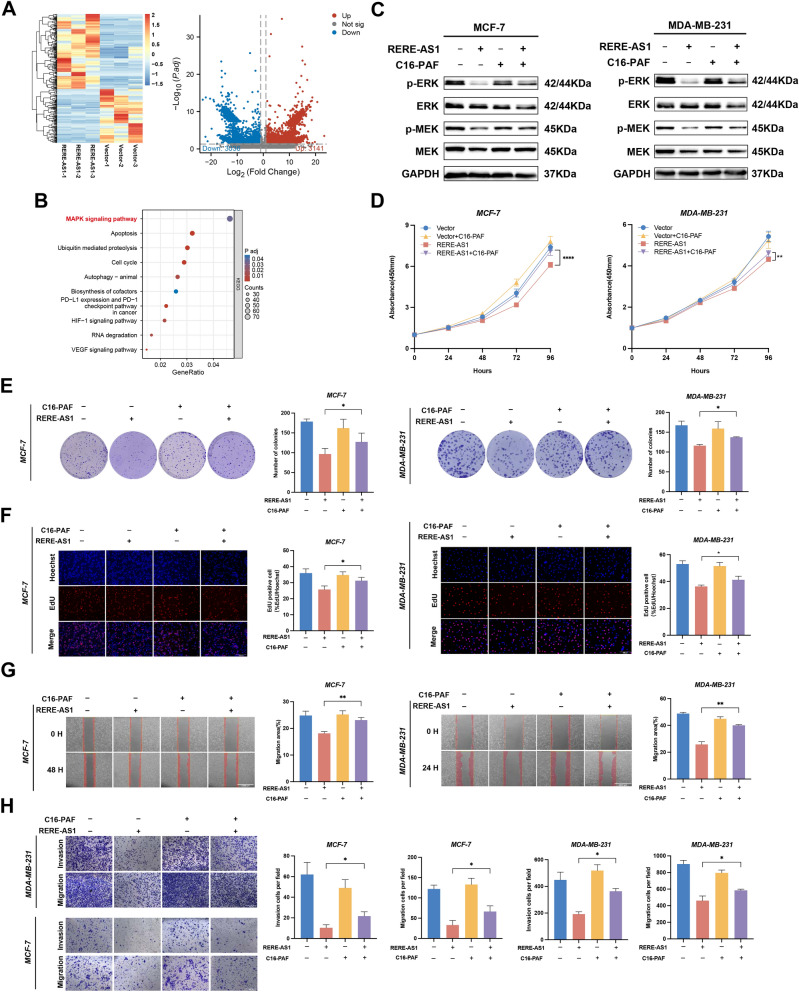
Prior research has indicated the crucial involvement of the MEK/ERK and PI3K/AKT pathways in modulating CDK4/6 drug sensitivity [2]. To further investigate the signaling pathways influenced by RERE-AS1 on ribociclib treatment efficacy, the transcriptome of RERE-AS1 overexpression cells was compared to that of control group. The results revealed 3141 upregulated and 3036 downregulated genes in the RERE-AS1 overexpression group compared to controls (Fig. 4A). The GSEA analysis reveals a significant enrichment of RERE-AS1 in the MAPK signaling pathway (Fig. 4B). Western blot showed that the phosphorylation levels of MEK and ERK were significantly downregulated in the RERE-AS1 overexpression BC cells, and treatment with the MAPK pathway activator C16-PAF partially reversed the downregulation trend (Fig. 4C). Moreover, CCK-8, colony formation, and EdU assays demonstrated that the proliferative activity of RERE-AS1 overexpressing cells was reversed following C16-PAF treatment (Fig. 4D–F). Scratch wound healing and transwell assays indicated that the inhibition of tumor cell invasion and migration by RERE-AS1 overexpression could be reversed by C16-PAF (Fig. 4G, H). Based on these results, RERE-AS1 may affect CDK4/6 drug sensitivity via ceRNA network and MEK/ERK pathway.
Fig. 4.
LncRNA RERE-AS1 mediates malignant phenotype and regulates the anti-tumor activity of Ribociclib in BC via the MEK/ERK signaling pathway. A Transcriptome sequencing was conducted on control and RERE-AS1 stably overexpressed cell lines, with heatmaps and volcano plots showing the differential gene expression between the two groups. B GSEA of differentially expressed genes to investigate potential signaling pathways involving RERE-AS1. C Western blot experiments confirmed the effect of RERE-AS1 overexpression on the phosphorylation levels of MEK/ERK, and the reversal of this effect by the MAPK signaling pathway activator C16-PAF. D–F CCK-8, colony formation, and EdU assays verified that the tumor proliferation suppression mediated by overexpressed RERE-AS1 could be reversed by the MAPK pathway activator C16-PAF. G–H Scratch healing and Transwell assays revealed that C16-PAF could reverse the effects on tumor invasion and migration regulated by overexpressed RERE-AS1. *p < 0.05, ***p < 0.001, ****p < 0.0001
Correlation analysis of RERE-AS1 with clinical pathological factors
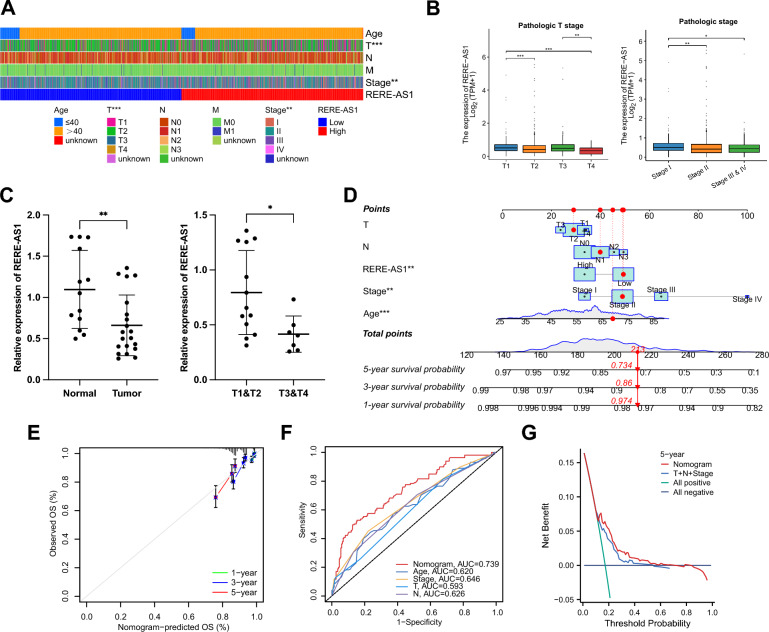
To explore the relationship between RERE-AS1 expression and clinicopathological characteristics in BC patients, we assessed the differences in factors such as age, T, N, M stages, and pathological stages between groups with high and low RERE-AS1 expression. The heatmap showed significant differences in T and pathological stage (p < 0.01, Fig. 5A). Furthermore, subgroup analysis of T and pathological stage showed that larger local tumors and higher pathological stage were associated with lower expression of RERE-AS1 (Fig. 5B). To further validate these results, BC tissues (n = 20) and normal breast tissues (n = 13) were obtained from our hospital for RT-qPCR experiments. As shown in Fig. 5C, compared to normal breast tissues, RERE-AS1 was relatively low-expressed in BC tissues, and its expression was lower in BC patients with higher T staging. We created a nomogram that combines RERE-AS1 with clinical pathological factors, enabling the prediction of 1, 3, and 5-year OS rates for BC patients (Fig. 5D). The calibration curve highlighted the accuracy of the nomogram, with a closer agreement between the predicted and actual survival curves indicating effective calibration (Fig. 5E). The ROC curve demonstrated that the nomogram exhibited the highest diagnostic value compared to other clinicopathological factors, with an AUC of 0.739 (Fig. 5F). DCA indicated that, in comparison to T, N, and stage, the nomogram provided a higher clinical net benefit (Fig. 5G).
Fig. 5.
Correlation analysis of lncRNA RERE-AS1 with clinicopathological factors and construction of nomogram. A Variations in clinicopathological factors between high- and low-expression groups of RERE-AS1. B Expression differences of RERE-AS1 among patients at different pathological T and stages. C Validation of RERE-AS1 expression differences in BC versus normal breast tissue, and among different T stages, using RT-qPCR on tissue samples. D Construction of a nomogram based on RERE-AS1 and clinicopathological characteristics. E Calibration curves assessing the accuracy of the nomogram’s prognosis predictions at 1, 3, and 5 years. F ROC curve analyzes the discriminative ability of the nomogram and clinicopathological factors. G DCA comparing the clinical net benefit of the nomogram versus T, N, and stage. *p < 0.05, **p < 0.01, ***p < 0.001
Pan-cancer analysis of RERE-AS1
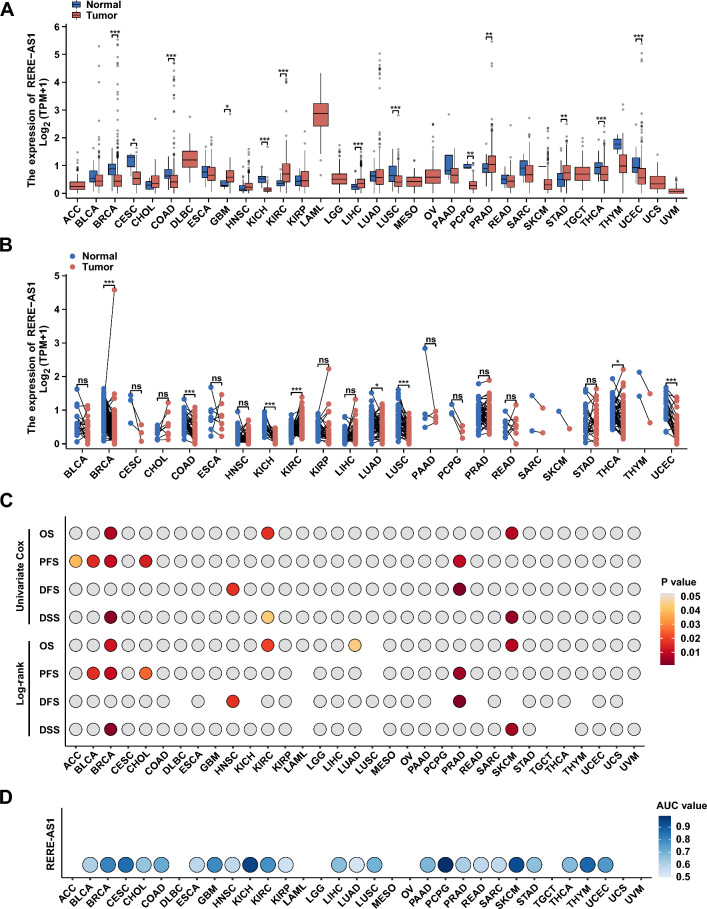
We conducted a comprehensive analysis using pan-cancer transcriptomic data and clinical prognostic information from TCGA to explore the value of RERE-AS1 in cancer pathogenesis, diagnosis, and prognosis. In the differential analysis, the unpaired analysis results showed abnormal expression of RERE-AS1 in most cancers (Fig. 6A). Paired differential analysis indicated that RERE-AS1 was significantly elevated in kidney renal clear cell carcinoma (KIRC) compared to paired normal tissues, but markedly reduced in breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC, Fig. 6B). These results suggest that RERE-AS1 may mainly exert anti-cancer effects in tumors. We further explored the prognostic value of RERE-AS1 in different types of cancer through univariate Cox regression and log-rank test analyses of four prognostic indicators, including OS, PFS, DFS, and DSS. As shown in Fig. 6C, RERE-AS1 has prognostic value in adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), BRCA, cholangiocarcinoma (CHOL), head and neck squamous cell carcinoma (HNSC), KIRC, prostate adenocarcinoma (PRAD), and skin cutaneous melanoma (SKCM), among which BRCA, PRAD, and SKCM show the strongest correlation between RERE-AS1 and prognostic indicators. BRCA is the only type of cancer in which three outcomes (OS, PFS, and DSS) are closely linked with RERE-AS1, highlighting the significant role of RERE-AS1 in impacting the survival outcomes of BC patients. We also performed ROC curve analysis to evaluate the clinical diagnostic value of RERE-AS1. As shown in the bubble plot, RERE-AS1 has significant potential clinical diagnostic value (AUC > 0.7) in patients with BRCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), glioblastoma multiforme (GBM), KICH, pheochromocytoma and paraganglioma (PCPG), SKCM, and thymoma (THYM, Fig. 6D).
Fig. 6.
Pan-cancer analysis of lncRNA RERE-AS1. A, B Unpaired and paired differential analysis of RERE-AS1 across various cancers. C The impact of RRE-AS1 on survival outcomes in pan-cancer was evaluated using univariate Cox regression and Log-rank tests, with all results displayed in a heatmap. D ROC analysis assessing the diagnostic value of RERE-AS1 in pan-cancer. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Discussion
Current therapeutic approaches for BC predominantly encompass endocrine therapy, targeted therapy, immunotherapy, surgical interventions, and various other systemic treatment modalities [18]. CDK4/6 inhibitors, in recent years, have emerged as novel targeted therapy drugs for BC, providing new treatment options and improving prognosis for patients with advanced BC [7]. Although the survival rate of BC is continuously improving, drug resistance issues including CDK4/6 inhibitors are currently significant barriers to further improving patient prognosis [19, 20]. This study innovatively utilizes ML algorithms based on transcriptome data from the perspective of ncRNA to screen for lncRNAs related to the efficacy of ribociclib, and constructs a lncRNA-related ANNM to distinguish potential treatment-beneficial populations. Furthermore, we identified a novel biomarker, lncRNA RERE-AS1, which has demonstrated potential for predicting and promoting the efficacy of ribociclib, as well as serving as a diagnostic and prognostic marker for BC.
LncRNAs are known to serve as biomarkers for predicting and monitoring anti-tumor drug efficacy and as important targets for reversing drug resistance [4, 10, 21, 22]. For instance, the study by Shi et al. demonstrated that lncRNA DILA1 inhibits the phosphorylation of cyclin D1 and preventing its degradation, thereby reversing tamoxifen resistance and serving as a specific therapeutic target for treating BC [23]. However, the investigation of lncRNA biomarkers associated with CDK4/6 inhibitors remains relatively unexplored. In this study, we used ML algorithms, based on RNA sequencing data, to identify 11 lncRNAs associated with ribociclib sensitivity, offering new perspectives for the exploration of biomarkers for CDK4/6 inhibitors. Furthermore, our findings indicate that lncRNA RERE-AS1 enhances the sensitivity of BC cells to ribociclib. This conclusion is substantiated by cell viability and proliferation assays conducted on MCF-7 and MDA-MB-231 cells overexpressing RERE-AS1. LncRNAs perform distinct functions depending on their subcellular localization. LncRNAs situated in the nucleus can modulate gene transcription either in cis or trans, whereas those located in the cytoplasm are primarily engaged in the regulation of ceRNA networks [17]. In this study, nucleoplasmic separation experiments revealed that RERE-AS1 is localized primarily within the cytoplasm, thus, we constructed a ceRNA network and performed functional enrichment analysis, revealing that RERE-AS1 is involved in CDK enzyme activity regulation and dephosphorylation regulation, indicating its important role in regulating CDK4/6 inhibitors sensitivity. The mechanisms underlying resistance to CDK4/6 inhibitors primarily encompass two domains: cell cycle-related processes and upstream signaling pathways, notably the MEK/ERK and PI3K/AKT/mTOR pathways. Previous study has demonstrated that the MEK/ERK pathway influences the sensitivity of CDK4/6 inhibitors by modulating cyclin D1 expression and regulating the cyclin D-CDK4/6-pRb axis [2]. Western blot, gain-of-function experiments, and rescue experiments conducted in our study suggest that RERE-AS1 negatively regulates the MEK/ERK pathway. Therefore, RERE-AS1 might enhance the sensitivity of BC to ribociclib through ceRNA network and the MEK/ERK pathway. Currently, the primary strategies to improve anti-tumor drug resistance encompass optimizing drug delivery systems, developing novel pharmaceutical agents, and employing combination drug therapies [24–26]. Several studies have reported that combining CDK4/6 inhibitors with MEK/ERK pathway inhibitors significantly improves anti-tumor efficacy [27–29]. Thus, RERE-AS1 is a valuable predictor and promotor of ribociclib treatment efficacy. In the future, high-throughput clinical sequencing can help identify potential beneficiaries, enhancing precision medicine in clinical practice [24].
The artificial intelligence models utilized for predicting cancer patient responses to anti-tumor drug therapy predominantly leverage genomics, radiomics, and proteomics data. Nevertheless, the breadth of research in this domain remains constrained. In signet-ring cell carcinoma, Li, Cong, et al. developed a CT-based artificial intelligence model that demonstrated high efficacy in predicting benefits from adjuvant chemotherapy for patients [30]. In lung cancer, Wu, Liangliang, et al. identified key biomarkers, IP-10 and IL-8, through high-throughput sequencing of peripheral blood samples and subsequently constructed a model based on the IP-10/IL-8 ratio to predict responses to combined immunotherapy and chemotherapy [31]. Additionally, a study has reported using the GDSC database combined with ML, to create personalized logic models for BRAF treatment responses in melanoma and colorectal cancer [32]. This study proposes an innovative lncRNA-related ANNM for predicting the treatment response to CDK4/6 inhibitors in BC patients by integrating data from the GDSC and TCGA databases and employing an ANN algorithm. The model demonstrated consistent and robust predictive efficacy in both the training and validation sets, indicating its potential utility in identifying patient groups likely to benefit from CDK4/6 inhibitor therapy in adjuvant clinical settings. In the future, the integration of computational models with high-throughput clinical sequencing data can provide new insights and methodologies for advancing precision medicine.
The widespread and varied functions of lncRNAs are essential in the initiation and progression of cancer [33]. For example, in BC, LINC00115 promotes the stemness and metastasis of chemotherapy-resistant BC stem-like cells through the SETDB1/PLK3/HIF1α pathway [34]. In this study, the transcriptomic data of RERE-AS1 overexpressing cell lines and their parental cells and results of cellular assays suggest that RERE-AS1 mediates the transformation of malignant phenotypes including proliferation, invasion, and migration via downregulating the MEK/ERK pathway. Xenograft tumor experiments verified that overexpression of RERE-AS1 restrained tumorigenesis. The extensive expression, tumor-specific nature, and stability of lncRNAs are crucial for cancer prognosis, predicting treatment efficacy, and therapy [35]. Our study demonstrated that compared to normal breast tissue, RERE-AS1 is downregulated in BC tissues, and its high expression is associated with smaller tumors and lower pathological stages. To apply RERE-AS1 in clinical practice, we constructed a nomogram based on RERE-AS1 and clinical pathological factors to predict patient prognosis. Furthermore, pan-cancer analysis suggests that compared to other tumors, RERE-AS1 exhibits expression specificity, significant prognostic and diagnostic value in BC. In summary, these findings underscore the critical role of RERE-AS1 in BC, suggesting its potential as a biomarker to enhance clinical diagnosis and outcome prediction.
This study has the following limitations. Firstly, the GDSC database presently does not contain sensitivity data for abemaciclib, thereby precluding any further speculation on whether RERE-AS1 similarly enhances the efficacy of all CDK4/6 inhibitors. Secondly, the extremely low expression levels of RERE-AS1 in BC cell lines constrain the investigation into the effects of RERE-AS1 knockdown on ribociclib efficacy. Finally, the potential clinical utility of RERE-AS1 requires further validation through studies involving a large cohort of clinical samples.
Conclusions
This study uses ML algorithms to identify lncRNAs linked to ribociclib efficacy and develops an ANNM to aid in selecting the most beneficial patient population. Research indicates that RERE-AS1 serves as a predictive biomarker for the efficacy of ribociclib and as a diagnostic and prognostic marker for BC. Upregulating RERE-AS1 reduces MEK/ERK phosphorylation, suppressing cancer malignancy and potentially enhancing ribociclib sensitivity.
Supplementary Information
Additional file1: Figure S1.RNA was extracted for RT-qPCR experiments following subcellular fractionation to investigate the cellular distribution of the lncRNA RERE-AS1;Construction of a RERE-AS1 mediated ceRNA interaction network using online databases;GO enrichment analysis of mRNAs within the ceRNA interaction network. Table S1. The sequences of primers for real-time qPCR assays. Table S2. The antibody list used in this study. Table S3. The list of lncRNAs differentially expressed in BC. Table S4. The list of lncRNAs with prognostic value in BC.
Acknowledgements
We thank BioRender (www.biorender.com) for the use of their tools in creating the illustrations presented in this study.
Abbreviations
- ACC
Adrenocortical carcinoma
- ANNM
Artificial neural network model
- AUC
Area under the curve
- BC
Breast cancer
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CCK-8
Cell counting kit-8
- CDKs
Cyclin-dependent kinases
- CESC
Cervical squamous cell carcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DCA
Decision curve analysis
- DFS
Disease-free survival
- DSS
Disease-specific survival
- EdU
5-Ethynyl-2′-deoxyuridine
- ESCC
Esophageal squamous cell carcinoma
- GBM
Glioblastoma multiforme
- GDSC2
Genomics of drug sensitivity in cancer v2
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- HNSC
Head and neck squamous cell carcinoma
- HR
Hazard ratio
- IC50
Half-maximal inhibitory concentration
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- LncRNA
Long non-coding RNA
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- ML
Machine learning
- MOI
Multiplicity of infection
- NcRNAs
Non-coding RNAs
- OS
Overall survival
- PCPG
Pheochromocytoma and paraganglioma
- PFS
Progression-free survival
- PRAD
Prostate adenocarcinoma
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- ROC
Receiver operating characteristic
- SKCM
Skin cutaneous melanoma
- TCGA
The Cancer Genome Atlas
- THCA
Thyroid carcinoma
- THYM
Thymoma
- TNBC
Triple-negative breast cancer
- UCEC
Uterine corpus endometrial carcinoma
Author contributions
HL and DC conceived and designed the study. ZH contributed to experimental design, data analysis, and manuscript writing. ZH, KL and MQ conducted experiments. JW contributed to data analysis and manuscript writing. LL, BS, CW, and XZ revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
This work was supported by the Science and Technology Innovation Joint Fund Project of Fujian Province (Grant number, 2021CXA045), Special Fund for Clinical Research of the Wu Jieping Medical Foundation (Grant number, 320.6750.2021-10-60), and Tianjin Medical University Cancer Hospital Peak Discipline Program—14th Five-Year Plan (Grant number, 7263).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (bc20240081) and was conducted in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Debo Chen, Email: debochensr@fjmu.edu.cn.
Hong Liu, Email: liuhong_submit@tjmuch.com.
References
- 1.Icard P, Fournel L, Wu Z, Alifano M, Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem Sci. 2019;44:490–501. [DOI] [PubMed] [Google Scholar]
- 2.Piezzo M, Cocco S, Caputo R, Cianniello D, Gioia GD, Lauro VD, et al. Targeting cell cycle in breast cancer: CDK4/6 inhibitors. Int J Mol Sci. 2020;21:6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann Oncol. 2018;29:1541–7. [DOI] [PubMed] [Google Scholar]
- 4.Krasniqi E, Goeman F, Pulito C, Palcau AC, Ciuffreda L, Di Lisa FS, et al. Biomarkers of response and resistance to CDK4/6 inhibitors in breast cancer: hints from liquid biopsy and microRNA exploration. Int J Mol Sci. 2022;23:14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, Park K, Kim GM, Jung KH, Kang SY, Park IH, et al. Exploratory analysis of biomarkers associated with clinical outcomes from the study of palbociclib plus endocrine therapy in premenopausal women with hormone receptor-positive, HER2-negative metastatic breast cancer. Breast. 2022;62:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spring LM, Wander SA, Andre F, Moy B, Turner NC, Bardia A. Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: past, present, and future. Lancet. 2020;395:817–27. [DOI] [PubMed] [Google Scholar]
- 7.Morrison L, Loibl S, Turner NC. The CDK4/6 inhibitor revolution—a game-changing era for breast cancer treatment. Nat Rev Clin Oncol. 2024;21:89–105. [DOI] [PubMed] [Google Scholar]
- 8.Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875: 188491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji W, Zhang W, Wang X, Shi Y, Yang F, Xie H, et al. c-myc regulates the sensitivity of breast cancer cells to palbociclib via c-myc/miR-29b-3p/CDK6 axis. Cell Death Dis. 2020;11:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Y, Liao H, Xie R, Zhang Y, Zheng R, Chen J, et al. Overexpression of miRNA-3613-3p enhances the sensitivity of triple negative breast cancer to CDK4/6 inhibitor palbociclib. Front Oncol. 2020;10: 590813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornell L, Wander SA, Visal T, Wagle N, Shapiro GI. MicroRNA-mediated suppression of the TGF-β pathway confers transmissible and reversible CDK4/6 inhibitor resistance. Cell Rep. 2019;26:2667-2680.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22:bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei J, Huang K, Chen Z, Hu M, Bai Y, Lin S, et al. Characterization of glycolysis-associated molecules in the tumor microenvironment revealed by pan-cancer tissues and lung cancer single cell data. Cancers (Basel). 2020;12:1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, He W, Zhang S. Seeking for correlative genes and signaling pathways with bone metastasis from breast cancer by integrated analysis. Front Oncol. 2019;9:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Huang X, Liu J, Chen G, Liu C, Zhang S, et al. New insight into long non-coding RNAs associated with bone metastasis of breast cancer based on an integrated analysis. Cancer Cell Int. 2021;21:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto T, Kanaya N, Somlo G, Chen S. Synergistic anti-cancer activity of CDK4/6 inhibitor palbociclib and dual mTOR kinase inhibitor MLN0128 in pRb-expressing ER-negative breast cancer. Breast Cancer Res Treat. 2019;174:615–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidovic D, Huynh TT, Konda P, Dean C, Cruickshank BM, Sultan M, et al. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2020;27:363–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanker AB, Sudhan DR, Arteaga CL. Overcoming endocrine resistance in breast cancer. Cancer Cell. 2020;37:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh D, Assaraf YG, Gacche RN. Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist Updates. 2022;63: 100851. [DOI] [PubMed] [Google Scholar]
- 20.Antonarelli G, Taurelli Salimbeni B, Marra A, Esposito A, Locatelli MA, Trapani D, et al. The CDK4/6 inhibitors biomarker landscape: the most relevant biomarkers of response or resistance for further research and potential clinical utilityCDK4/6. Crit Rev Oncol Hematol. 2023;192: 104148. [DOI] [PubMed] [Google Scholar]
- 21.Jin X, Ge L-P, Li D-Q, Shao Z-M, Di G-H, Xu X-E, et al. LncRNA TROJAN promotes proliferation and resistance to CDK4/6 inhibitor via CDK2 transcriptional activation in ER+ breast cancer. Mol Cancer. 2020;19:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Z, Shi Q, Li Y, Jin L, Li S, Wong LL, et al. LncRNA EILA promotes CDK4/6 inhibitor resistance in breast cancer by stabilizing cyclin E1 protein. Sci Adv. 2023;9:eadi3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat Commun. 2020;11:5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zang Y-D, Wu H-J, Chen X-Y, Ma Z-L, Li C-J, Ma J, et al. Synthesis and biological evaluation of novel psidium meroterpenoid derivatives against cisplatin-induced acute kidney injury. J Med Chem. 2024;67:14234–55. [DOI] [PubMed] [Google Scholar]
- 26.Zhan X, Liu X, Rang L, Shen M, Zhang J, Tang R, et al. Detection of lenalidomide metabolites in urine to discover drug-resistant compounds. Clin Chim Acta. 2024;553: 117707. [DOI] [PubMed] [Google Scholar]
- 27.Willobee BA, Gaidarski AA, Dosch AR, Castellanos JA, Dai X, Mehra S, et al. Combined blockade of MEK and CDK4/6 pathways induces senescence to improve survival in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2021;20:1246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hart LS, Rader J, Raman P, Batra V, Russell MR, Tsang M, et al. Preclinical therapeutic synergy of MEK1/2 and CDK4/6 inhibition in neuroblastoma. Clin Cancer Res. 2017;23:1785–96. [DOI] [PubMed] [Google Scholar]
- 29.Martin CA, Cullinane C, Kirby L, Abuhammad S, Lelliott EJ, Waldeck K, et al. Palbociclib synergizes with BRAF and MEK inhibitors in treatment naïve melanoma but not after the development of BRAF inhibitor resistance. Int J Cancer. 2018;142:2139–52. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Qin Y, Zhang W-H, Jiang H, Song B, Bashir MR, et al. Deep learning-based AI model for signet-ring cell carcinoma diagnosis and chemotherapy response prediction in gastric cancer. Med Phys. 2022;49:1535–46. [DOI] [PubMed] [Google Scholar]
- 31.Wu L, Xie S, Wang L, Li J, Han L, Qin B, et al. The ratio of IP10 to IL-8 in plasma reflects and predicts the response of patients with lung cancer to anti-PD-1 immunotherapy combined with chemotherapy. Front Immunol. 2021;12: 665147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Béal J, Pantolini L, Noël V, Barillot E, Calzone L. Personalized logical models to investigate cancer response to BRAF treatments in melanomas and colorectal cancers. PLoS Comput Biol. 2021;17: e1007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herman AB, Tsitsipatis D, Gorospe M. Integrated LncRNA function upon genomic and epigenomic regulation. Mol Cell. 2022;82:2252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo F, Zhang M, Sun B, Xu C, Yang Y, Zhang Y, et al. LINC00115 promotes chemoresistant breast cancer stem-like cell stemness and metastasis through SETDB1/PLK3/HIF1α signaling. Mol Cancer. 2024;23:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhan A, Soleimani M, Mandal SS. Long non-coding RNA (LncRNA) and cancer: a new paradigm. Cancer Res. 2017;77:3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file1: Figure S1.RNA was extracted for RT-qPCR experiments following subcellular fractionation to investigate the cellular distribution of the lncRNA RERE-AS1;Construction of a RERE-AS1 mediated ceRNA interaction network using online databases;GO enrichment analysis of mRNAs within the ceRNA interaction network. Table S1. The sequences of primers for real-time qPCR assays. Table S2. The antibody list used in this study. Table S3. The list of lncRNAs differentially expressed in BC. Table S4. The list of lncRNAs with prognostic value in BC.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.