Abstract
Understanding tumor cell heterogeneity and plasticity is crucial for overcoming drug resistance. Single-cell technologies enable analyzing cell states at a given condition, but catenating static cell snapshots to characterize dynamic drug responses remains challenging. Here, we propose scStateDynamics, an algorithm to infer tumor cell state dynamics and identify common drug effects by modeling single-cell level gene expression changes. Its reliability is validated on both simulated and lineage tracing data. Application to real tumor drug treatment datasets identifies more subtle cell subclusters with different drug responses beyond static transcriptome similarity and disentangles drug action mechanisms from the cell-level expression changes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-024-03436-y.
Keywords: Single-cell, Dynamics, Tumor, Drug response
Background
Drug resistance is one of the major challenges for tumor therapy. High molecular heterogeneity of tumor cells frequently leads to intrinsic drug resistance, while the dynamic plasticity of tumor cells further causes adaptive or acquired resistance over time. Therefore, it is crucial for improving the tumor therapeutic efficacy by investigating the biological mechanisms underlying the tumor cell state dynamics during drug treatment [1–3].
Recent advances in single-cell RNA sequencing (scRNA-seq) technologies provide a great opportunity to characterize and analyze the heterogeneity of cell states at a given condition [4]. However, it is still challenging to understand the tumor cell state dynamics during drug treatment by integrating the cell state “snapshots” (as scRNA-seq data) at different time points [5]. Due to the limitations of current sequencing technologies, cells are destroyed during transcriptome profiling, making it very hard to track the dynamic gene expression of any individual cell at multiple time points. Hence, it is promising if any computational method can virtually align the unpaired single-cell level snapshots between two or more time points as a temporal-like “video”.
Aligning at cluster-level, through direct integrated clustering [6, 7] or nonlinear distribution comparison [8–11], has emerged as a widely utilized strategy and leads to some valuable biological discoveries. But it does not fully consider the variation between cells and is hard to model complex multi-fate characteristics of tumor cell populations in response to drugs. Alternatively, a few cell-level algorithms based on mathematical modeling (such as differential equations) have been proposed to align cells during development and differentiation processes [12–16]. More recently, data-driven strategies have also been developed to cope with the uncertainty of cell fate trajectories when analyzing perturbation responses under diverse stimulation conditions (such as cytokines, drugs, etc.) [17, 18]. However, especially for the tumor cell state dynamics under drug treatment, the high heterogeneity and plasticity of tumor cells lead to more complex drug responses in both cell population abundance and state, which requires special consideration in computational modeling.
Here, we present scStateDynamics, a computational method to thoroughly decipher the tumor cell state dynamics in response to drug treatment by modeling single-cell level expression changes. This method first infers the dynamic characteristics of tumor cell states by minimizing the overall changes in gene expression while also considering distinct proliferation or inhibition rates across cell populations. Then, we identify cluster-shared and cluster-specific components (gene factors) from the cell-level expression changes to dissect the drug action mechanisms. By testing on both the simulated data and the data with lineage tracing information, scStateDynamics shows reliable performance in aligning the cells at different time points. By applying to more real datasets from different tumor types under distinct therapeutic strategies, we highlight the significance of modeling cell-level expression changes in uncovering the intrinsic and acquired intra-cluster heterogeneity of tumor cells in response to drugs and also in dissecting the dynamic mechanisms of drug action.
Results
Overview of scStateDynamics
scStateDynamics is designed to infer tumor cell state dynamics under drug treatment and dissect tumor drug response mechanisms by modeling expression changes. First, we define each tumor cell state with the average expression profiles of several highly similar cells (Additional file 1: Fig. S1a). Then, we align the cells between pre- and post-time points based on the principle of minimizing the overall changes in gene expression, which can be formally modeled using optimal transport (OT) theory. OT was originally developed to determine the most efficient way to move a pile of sands from one location to another, and has been widely applied to compare two probability distributions [19]. Here, we normalize the number of cells in all tumor cell states at each time point into a discrete probability distribution. This allows us to align cells by seeking an optimal transport plan between these two cell state probability distributions. To measure the degree of cell state change (transport cost) and determine which alignment relationship (transport plan) is optimal, we adopt the idea of manifold learning [15, 20, 21], which propagates the local neighbor relationships among cell states to obtain their global distances along the low-dimensional manifold in high-dimensional space. Based on this distance matrix (also called the cost matrix) and the corresponding two cell state probability distributions, we derive an optimal transport plan matrix, which provides an alignment plan between cells. Further, by partitioning the cells into several distinct clusters, we can also derive the dynamic cell flows at the subcluster level (Additional file 1: Fig. S1b). These flows can be evaluated based on their average weighted transport costs, which offer a quantitative measure of the extent of cell state changes (“Methods”).
Notably, ignoring the diverse proliferation or inhibition rates between tumor cell populations may lead to the unreasonable identification of several cell flows. In detail, certain source cells had relatively higher proliferation rates, but their corresponding target cells lacked sufficient probability masses to accommodate them. Consequently, the OT algorithm aligned these increased source cells to other target cells, leading to unreasonable flows with abnormally high transport costs (outliers). Therefore, we categorize the cell flows as either “state-keeping”, “state-changed”, or “unreasonable flows” based on their transport costs and calculate the relative proliferation rates of clusters by correcting the unreasonable flows. In this way, by iteratively performing OT and correcting flows several times until convergence (Additional file 1: Fig. S2 provides an illustrative example of the iterative process for the simulation dataset 1 mentioned later), we obtain the final cell alignment relationships and quantify the changes in cell states and cell abundances (proliferation, inhibition, or death). These results enable the identification of subclusters with distinct drug response fates and facilitate the exploration of intra-cluster intrinsic or acquired heterogeneities (“Methods”).
Furthermore, to dissect the biological mechanisms underlying drug action, we calculated the cell-level changes () in gene expression profiles according to the previously inferred cell alignment results and designed a Bayesian factor analysis (FA) model to decompose the cell-level expression changes into the static components shared within each pre- or post-cluster and the dynamic components (gene factors) reflecting drug action mechanisms across clusters (“Methods”). This provides a new insight of integrating dynamic information to characterize drug action mechanisms and cell cluster heterogeneities (Fig. 1).
Fig. 1.
Overview of the scStateDynamics algorithm. scStateDynamics is designed to decipher the tumor cell state dynamics under drug treatment by modeling cell-level gene expression changes. Given the pre- and post-treatment single-cell gene expression data, scStateDynamics first measures the distances between cell states in low-dimensional manifold space and infers initial alignment relationships between cell states by minimizing the overall changes based on optimal transport theory. Integrating the clustering results allows for the further derivation of the subcluster flows. Then, according to the estimated transport cost of each subcluster flow, we categorize the flow type as either state-keeping, state-changed, or unreasonable flow. By iteratively correcting the unreasonable flows, we finally estimate distinct proliferation or inhibition rates of clusters and determine the types of abundance changes they exhibit. Based on the inferred cell-level dynamics, scStateDynamics implements a Bayesian factor analysis model to decompose the expression changes () into static cluster-specific variations and dynamic cluster-shared gene factors. This provides a novel perspective of integrating dynamic information to dissect drug effects, characterize cell pairs, and compare cluster heterogeneities
Performance validation of scStateDynamics by using simulated data
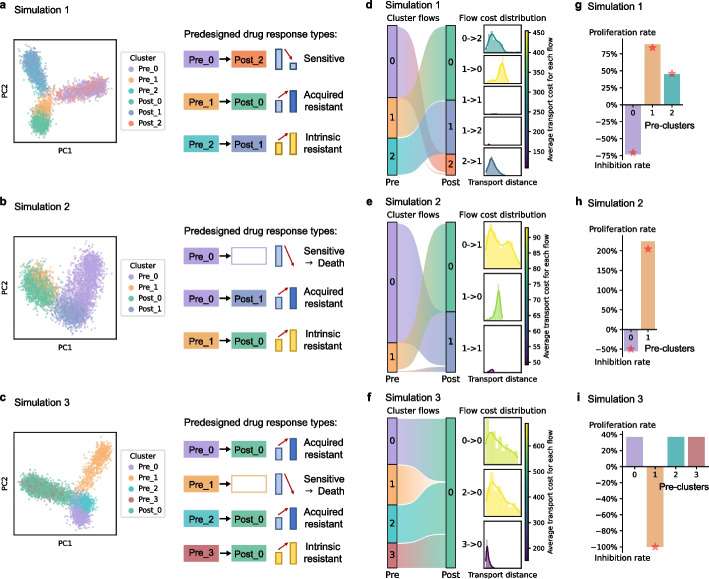
To assess the reliability of scStateDynamics, we simulated the transcriptome profiles of tumor cells in the pre- and post-treatment stages of three distinct drug response scenarios using Splatter [22] (Additional file 2: Table S1). In scenario 1, the Pre_0 cluster was sensitive to the drug and transitioned to Post_2 with a decreased proportion. The Pre_1 cluster adapted to the drug treatment by adjusting its state and transitioned to Post_0. Conversely, the Pre_2 cluster maintained its intrinsic resistance and transitioned to Post_1 (Fig. 2a). In scenario 2, we added a design in which the Pre_0 cluster initially exhibited sensitivity and experienced cell death, but gradually transitioned into a resistant state, resulting in the emergence of the Post_1 cluster (Fig. 2b). In scenario 3, we designed the clusters to either undergo cell death (Pre_1) or be driven towards an intrinsic resistant state (Pre_0, Pre_2, and Pre_3 transitioning to Post_0) (Fig. 2c). By applying scStateDynamics on these data, we successfully inferred the alignment relationships between clusters (Fig. 2d-f), except for three tiny erroneous flows (Pre_1- > Post_1 and Pre_1- > Post_2 in scenario 1, and Pre_1- > Post_1 in scenario 2) due to the inherent biases of unsupervised clustering at cluster boundaries. Furthermore, the inferred proliferation or inhibition rates (Fig. 2g-i), indicative of relative sensitivity or resistance types, were consistent with the simulation settings. The estimated transport distance effectively quantified the extent of cell state changes, thereby facilitating accurate determination of the intrinsic or acquired resistance types (Fig. 2d–f). Further, we benchmarked scStateDynamics against the recently proposed method CINEMA-OT (causal independent effect attribution + optimal transport) and its variant CINEMA-OT-W (adding a reweighting step to overcome differential abundance) [18]. The results showed that scStateDynamics achieved higher accuracy in identifying the simulated alignment relationships between clusters (Additional file 1: Fig. S3).
Fig. 2.
scStateDynamics accurately identifies the drug response characteristics in simulated data. a–c Left side is the low-dimensional PCA representation of cells in three simulation scenarios, with point color indicating the pre- or post-cluster labels. Right side is the corresponding predesigned cluster alignment relationships and drug response types. d–f Left side is a Sankey plot showing the cell subcluster alignments inferred by scStateDynamics. The color of the rectangles corresponds to the pre-clusters and post-clusters, while the height of each rectangle indicates the cluster’s proportion in all cells. Right side is histogram plots showing the distribution of the transport distances (costs) in all flows, with the color representing the average transport cost of each flow. g–i Barplots showing each pre-cluster’s relative proliferation or inhibition rate estimated by scStateDynamics. A positive height indicates an increased proportion (proliferation), while a negative height indicates a decrease (inhibition). Red stars mark the ground truths of the proliferation or inhibition rates predesigned in the simulation
The inferred results of scStateDynamics are supported by lineage tracing information
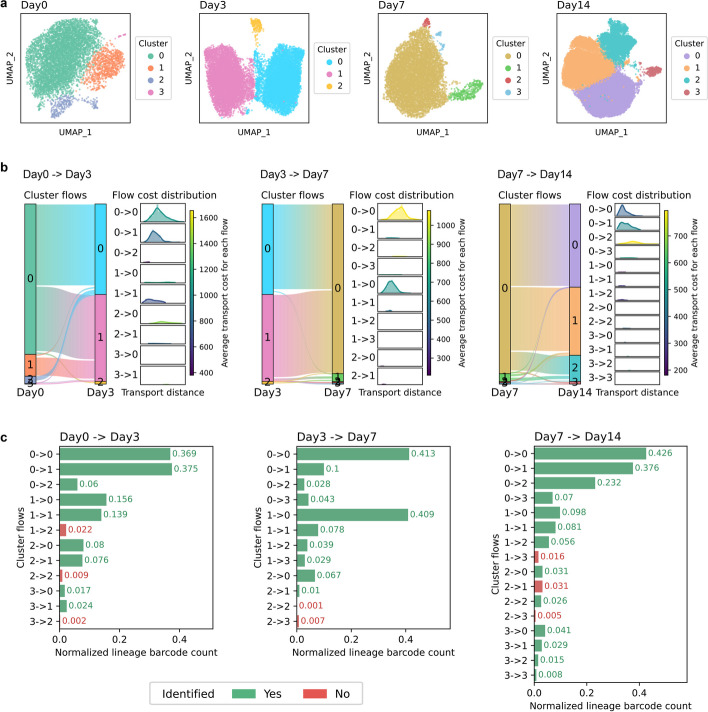
Lineage tracing technologies have emerged as a recent advancement, utilizing inherited DNA sequences (barcodes) to track cell clones [23, 24]. These barcode labels can be seen as strong evidences for linking cells across time [24]. Here, we collected the scRNA-seq data of PC9 lung cancer cell lines at four time points (days 0, 3, 7, and 14) during osimertinib treatment, in which the cell lineages were simultaneously tracked using the Watermelon system [25] (Additional file 2: Table S2). We performed pre-processing and clustering on the data (Fig. 3a) and applied scStateDynamics to infer the dynamic alignment relationships (subcluster flows) between clusters from adjacent time points (Fig. 3b). To evaluate the reliability of the identified subcluster flows, we counted the number of lineage barcodes connecting cells in each pair of clusters. To avoid the influence of cluster sizes, we normalized the lineage barcode counts using the square root of the product of the cell numbers in each cluster pair (“Methods”). Notably, we observed that almost all combinations of pre-clusters and post-clusters were supported by lineage barcodes at varying levels. scStateDynamics successfully identified the majority of these cluster flows. The remaining unidentified flows had only exceedingly few supporting barcodes, which may be due to potential random fate biases with minimal probabilities (Fig. 3c). We compared the cell lineage barcodes with the cell alignments inferred by scStateDynamics, CINEMA-OT, and CINEMA-OT-W and found that scStateDynamics exhibited higher correctness and completeness (Additional file 1: Fig. S4, “Methods”). Besides, we demonstrated that scStateDynamics outperformed the conventional joint-clustering method in aligning cells at cluster level (Additional file 1: Fig. S5, “Methods”).
Fig. 3.
The inferred results of scStateDynamics are supported by lineage tracing information. a UMAP (Uniform Manifold Approximation and Projection) plots of the cells in PC9 lung cancer cell line at days 0, 3, 7, and 14 after osimertinib treatment in Watermelon lineage tracing scRNA-seq data. The color indicates the cluster label of the cells at each time point. b The inferred cell subcluster alignment relationships and the quantified flow costs at each pair of adjacent time points. c Barplots showing the normalized lineage barcode count of each subcluster flow. The height of the bar indicates the strength of confidence supported by lineaging barcodes. The color denotes whether the subcluster flow is identified by scStateDynamics (green for yes, red for no)
Furthermore, we evaluated the performance of scStateDynamics in inferring the proliferation or inhibition rates of cell clusters. We collected a lineage tracing dataset, termed ReSisTrace, which utilizes lentiviral labeling to track the lineage of ovarian cancer cells after treatment of drug olaparib, a PARP enzyme inhibitor [26]. The dataset comprises two replicates (named Olaparib_1 and Olaparib_2), and the cells were sampled before and after olaparib treatment (Additional file 2: Table S2). We firstly clustered the cells (Additional file 1: Fig. S6a and S6e) and analyzed the fraction of lineage barcodes in each pre-cluster that were either inherited or disappeared after drug treatment (Additional file 1: Fig. S6b and S6f). Notably, we found that cluster S1 in Olaparib_1 exhibited a lower inherited fraction compared to other clusters (Additional file 1: Fig. S6b). The analysis of scStateDynamics also inferred that cluster S1 had higher inhibition rates (Additional file 1: Fig. S6c). Furthermore, we compared the expression levels of PARP1 (the target of olaparib) across different clusters and found that cluster S1 exhibited higher expression values (Additional file 1: Fig. S6d), suggesting it may be more sensitive to olaparib. Similar results were also observed for clusters S1 and S2 in Olaparib_2 dataset (Additional file 1: Fig. S6f, S6g and S6h). The consistency of these three aspects of information proves the reliability of the proliferation and inhibition rates estimated by scStateDynamics.
scStateDynamics improves the characterization of tumor subcluster heterogeneity in drug response
To showcase the utility of scStateDynamics in unraveling the underlying heterogeneity and plasticity of tumor drug responses, we collected some real scRNA-seq datasets of tumor cells under drug treatment (Additional file 2: Table S3).
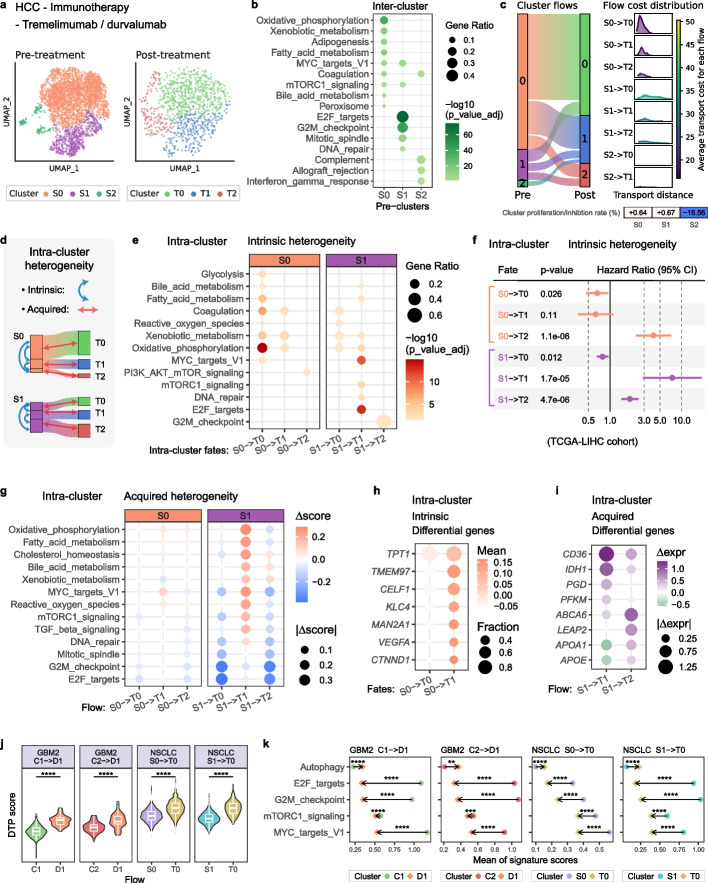
Taking the hepatocellular carcinoma (HCC) dataset as an example, the scRNA-seq data were detected from clinical patient biopsies before and after the immunotherapy with tremelimumab/durvalumab [27]. Through clustering (Fig. 4a) and inter-cluster differential expression analysis, we observed that cluster S0 exhibited higher activity in normal hepatocyte functions, while cluster S1 displayed enhanced malignant proliferation. Cluster S2 expressed immune-related pathways and genes, suggesting potential sensitivity to immunotherapy (Fig. 4b and Additional file 1: Fig. S7a). Cox regression models further confirmed these malignancy patterns (Additional file 1: Fig. S7b, “Methods”).
Fig. 4.
scStateDynamics facilitates comprehensive analysis of both intrinsic and acquired heterogeneities in tumor drug response. a UMAP plots of HCC cells before and after immunotherapy with tremelimumab/durvalumab. The color indicates the clustering results, with labels prefixed by “S” (for pre-treatment) and “T” (for post-treatment). b Bubble heatmap showing the hallmark pathway enrichment analysis results of the differentially expressed genes (DE genes) at inter-cluster level. c The inferred cell subcluster alignment relationships and the quantified flow costs by scStateDynamics. The inferred proliferation or inhibition rates of pre-clusters are presented at the bottom. d Sketch map showing the analysis of intra-cluster heterogeneity. Blue arrowed lines indicate intrinsic heterogeneity analysis, which compares the subclusters with different fates within each pre-cluster. Red arrowed lines indicate acquired heterogeneity analysis, which compares cells before and after treatment in each subcluster flow. e The enrichment analysis results of the DE genes at intra-cluster level. Each panel represents one pre-cluster, and the columns in it indicate its subclusters with different fates. f Forest plot showing the hazard ratio (HR) of the DE gene signature of each subcluster in the TCGA-LIHC cohort. The solid line indicates HR = 1. CI, confidence interval. g Bubble heatmap showing the change values of pathway scores ( score) in each subcluster flow. h Selected DE genes between the subclusters in S0 flowing to T0 and T1. i The expression changes ( expr) of metabolism-related genes in flows S1- > T1 and S1- > T2. j The distribution of the drug tolerant persister (DTP) scores in the source and target cells of four flows. One-sided Wilcoxon rank-sum test. ****p < 0.0001. k The mean of the signature scores in the source (circle) and target (diamond) cells of the identified DTP flows. The directions of the arrows reflect increasing or decreasing trends. One-sided Wilcoxon tests were performed on their distribution. **p < 0.001, ***p < 0.0001, ****p < 0.00001
Beyond routine inter-cluster comparisons, the cell-level alignment via scStateDynamics allowed us to identify subclusters with distinct sources or targets within each cluster (Fig. 4c). This provides novel perspectives for analyzing the intra-cluster intrinsic and acquired heterogeneities of tumor cells in response to drug treatment (Fig. 4d). We performed enrichment analyses on the differentially expressed genes between the subclusters exhibiting different fates within each per-treatment cluster. Within cluster S0, the subcluster transitioning to cluster T2 (S0- > T2) displayed a notable loss of metabolic function compared to the other subclusters (S0- > T0 and S0—> T1). Within cluster S1, the subclusters flowing to clusters T1 and T2 (S1- > T1 and S1- > T2) exhibited higher activity in proliferation-related pathways, in contrast with the subcluster flowing to T0 (S1- > T0) (Fig. 4e). Cox proportional hazards regression analysis confirmed these findings, indicating that the subcluster in S0 flowing to T2 was more malignant potentially due to impaired metabolic function, and the subclusters in S1 flowing to T1 and T2 displayed high malignancy possibly due to enhanced proliferation ability (Fig. 4f). Further, to assess the robustness of our findings, we randomly shuffled the inferred cell fate labels within each cluster 100 times and then performed the same intra-cluster intrinsic heterogeneity analysis. The results showed that none of the genes reached a statistically significantly differential expression (adjusted p-value < 0.05) after any of the 100 permutations, which implied that scStateDynamics effectively assisted in the identification of intra-cluster heterogeneity compared to random shuffling of cell fates (Additional file 1: Fig. S8).
Then, to investigate the acquired heterogeneity arising within each subcluster flow, we calculated the change values of pathway scores ( scores) based on the cell alignment relationships inferred by scStateDynamics (“Methods”). Comparing the average scores of the cells in each subcluster flow revealed distinct drug response characteristics. In cluster S0, the subclusters generally maintained their states, but flow S0- > T1 exhibited slightly increased activity in proliferation-related pathways (Fig. 4g). To investigate the potential molecular causes of this unfavorable drug response in flow S0- > T1, we further checked the intrinsic differences between the subclusters flowing to T0 and T1. The analysis revealed that, although both subclusters resembled normal hepatocytes (as shown in Fig. 4e above), the subcluster flowing to T1 exhibited higher expression of several specific genes (such as TPT1, TMEM97, CELF1, KLC4, MAN2A1, VEGFA, and CTNND1) (Fig. 4h), which have been previously reported to be involved in tumor progression or immunotherapy resistance [28–34]. In cluster S1, the subclusters displayed more pronounced acquired heterogeneity. The flows S1- > T0 and S1- > T2 exhibited significantly reduced activity in proliferation-related pathways, suggesting effective inhibition of tumor cell proliferation. Conversely, the flow S1- > T1 maintained high proliferation ability (intrinsic characteristics of the subcluster in S1 flowing to T1), indicating insensitivity to treatment (Fig. 4g). Although both S1- > T1 and S1- > T2 displayed enhanced activities in metabolism-related pathways, further analysis revealed that S1- > T1 exhibited malignancy-related metabolic reprogramming (increased expression levels of the genes CD36, IDH1, PGD, and PFKM) [35–38], while S1- > T2 exhibited the recovery of normal metabolic function (increased expression levels of the genes ABCA6, LEAP2, APOA1, and APOE) (Fig. 4i). Notably, the original study of this HCC dataset used joint hierarchical clustering analysis and found that the cells overall changed their states after drug treatment [27], while our method of cell-level alignment effectively distinguished the cell subclusters flows with varying levels of acquired heterogeneity, highlighting the values of inferring cell-level dynamics.
We also tested scStateDynamics on two other cancer types under distinct treatment strategies: glioblastoma (GBM) samples undergoing temozolomide chemotherapy [39] and non-small-cell lung carcinoma (NSCLC) samples receiving erlotinib-targeted therapy [40] (Additional file 1: Fig. S9, “Methods”). Notably, acquired heterogeneity analysis identified some intriguing flows, which appeared to enter a state known as drug-tolerant persisters (DTPs) [41–43], including the flows C1- > D1 and C2- > D1 in GBM2 dataset and the flows S0- > T0 and S1- > T0 in NSCLC dataset. DTPs, characterized by tumor cells becoming quiescent or slow cycling to evade drug-induced death while retaining the ability to resume growth upon drug removal, are increasingly recognized as being associated with tumor relapse [43]. To validate this hypothesis, we utilized a DTP signature [42] to score these subclusters and observed indeed higher DTP scores after drug treatment (Fig. 4j). Furthermore, we examined the functional changes in these flows and found that the cells displayed increased autophagy capability [44, 45] but decreased proliferation ability and mTORC1 pathway activity, consistent with previous reports [42] (Fig. 4k and Additional file 1: Fig. S10). This was also in accordance with the findings reported in the original studies of GBM2 and NSCLC datasets [39, 40].
In summary, scStateDynamics improves the resolution of investigating tumor cell heterogeneity from inter-cluster to intra-cluster level. This enables the identification of subclusters with different fates and facilitates the dissection of both intrinsic and acquired heterogeneity of tumor drug response from a dynamic perspective. We observed that despite overall similar expression profiles, some subtle molecular differences may potentially lead to distinct drug responses, which is consistent with the conclusion that diverse resistant clones evolve from homogeneous tumor cells in a recent experimental study [46]. These downstream analyses deepen our understanding about the molecular mechanisms underlying anti-tumor drug responses and provide valuable insights for developing more effective therapeutic strategies by targeting intrinsic drug resistance and drug-induced cell plasticity.
scStateDynamics disentangles the cluster-shared and cluster-specific drug effects
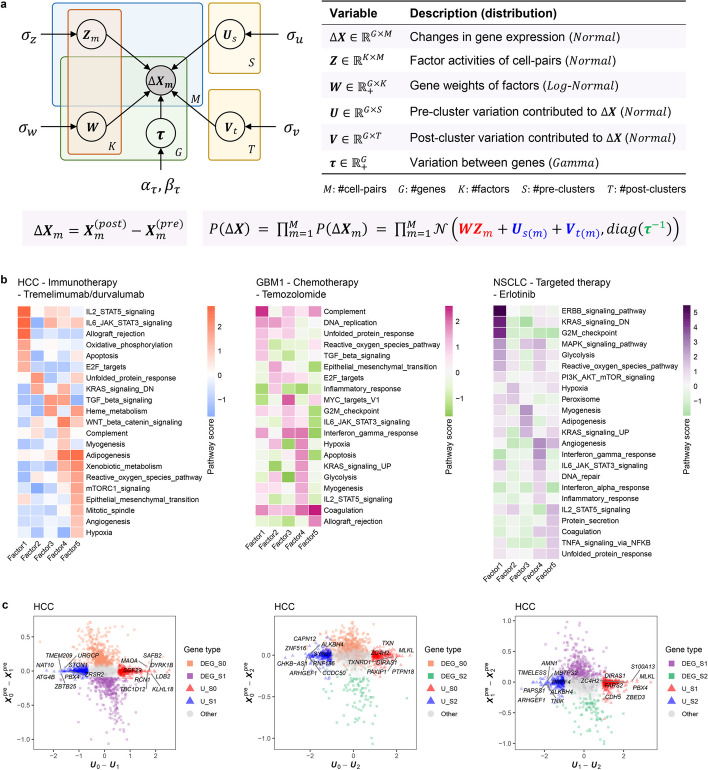
We devised a Bayesian factor analysis (FA) model in scStateDynamics to extract the biological factors (gene signatures) that contribute to the observed cell-level changes in gene expression () before and after treatment. The dynamic changes in expression () are determined by two types of variation: (i) the initial and final static molecular heterogeneity between cells, which can be characterized by cell’s cluster identity ( and ), indicating that the cells belonging to the same cluster shared some common effects and (ii) the dynamic molecular mechanisms of drug action, which are shared by all clusters and can be seen as a combination of multiple expression program factors (). To quantify these static and dynamic variations, we defined probability distributions of the variables and estimated their values by variational inference (Fig. 5a, “Methods”).
Fig. 5.
scStateDynamics disentangles the expression changes under drug action. a The probabilistic graphical model representation of the Bayesian factor analysis (FA) model in scStateDynamics, illustrating hidden random variables as circles and observed variables as shaded circles. Edges denote the statistical dependences between the variables. Boxes (plates) signify independent replications. The table provides a description of the variables and their corresponding probabilistic distributions. b Heatmaps showing the pathway scores of the identified factors in three datasets. c Scatter plots showing the gene comparisons for each pair of pre-clusters in the HCC dataset. The x-axis represents the differences between the vectors of two pre-clusters, while the y-axis represents the differences between the average expression value vectors of two pre-clusters. Square points denote the differential expression genes (DEGs) identified based on pre-treatment expression, with colors corresponding to the cluster colors used in Fig. 4a. Triangular points in red or blue denote the marker genes newly identified by subtracting vectors. Some top genes associated with tumor progression are labeled with their gene symbols. Gray circle points denote other genes
We applied this FA model to previously used real datasets. First, we observed that , as a new embedding of cell pairs, was indeed not relevant to either the pre- or post-cluster labels, proving the successful decomposition of these two types of variations (Additional file 1: Fig. S11). Then, we utilized the gene weights () to calculate the factor scores for some known pathways (“Methods”). Notably, the factors identified in all the datasets were biologically meaningful and associated with the mechanisms of the corresponding drugs (Fig. 5b, Additional file 1: Fig. S12 and S13). For example, in HCC sample undergoing immunotherapy, factor 1 was strongly associated with pathways involved in immune response modulation. In GBM1 sample undergoing chemotherapy, factors 1, 2, and 3 were all related to DNA replication pathway, consistent with the drug’s mechanism of action on DNA. In NSCLC sample undergoing targeted therapy, factor 1 displayed a remarkably strong association with ERBB signaling pathway, which corresponds precisely to the target gene of Erlotinib. In addition, the decomposed static cluster-specific variations ( and ) provide a novel representation for each cluster that considers dynamic information. We observed that some genes with highly positive or negative effects in vectors were associated with tumor progression and prognosis, which may be conventionally ignored due to their relatively low average expression levels (Additional file 1: Fig. S14). By comparing the vectors between each pair of pre-clusters (Fig. 5c), we identified some new cluster-specific and tumor-related genes (red and blue triangular points) that were not captured by conventional differential expression analysis based on static pre-treatment expression (square points). For example, MLKL, a necroptosis regulator, plays a complex but poorly understood role in cancer development and metastasis [47], and it was recently reported that it can promote immune evasion in HCC [48]. High MAOA expression was reported to be associated with an immunosuppressive tumor microenvironment and poor prognosis [49]. And CCDC50 was shown to promote HCC growth via Ras/Foxo4 signaling [50, 51]. These results highlight the profound value of decomposing gene expression changes in characterizing drug mechanisms and cell cluster heterogeneity from a novel dynamic perspective.
Discussion
scStateDynamics is an algorithm to investigate the tumor cell state dynamics in response to drug treatment by modeling single-cell level gene expression changes. To cope with the challenge of unpaired cells, we employed the principle of minimizing the overall weighted changes in gene expression along a low-dimensional manifold to align the cells before and after treatment. Considering the varying drug sensitivities across tumor clusters due to their high heterogeneity and plasticity, we used a data-driven approach to discern the types of cell subcluster flows and estimated the proliferation or inhibition rates of the clusters. To dissect the biological characteristics underlying drug actions by integrating dynamic information, we designed a Bayesian FA model to decompose the gene expression changes into cluster-specific static variations and cluster-shared dynamic gene factors.
For the investigation on the dynamics of complex systems, we think modeling the change values () is very important. Thus, we adopt the strategy of minimizing the overall changes to align cells and decompose the change value matrix to uncover the mechanisms of drug action. The significance of analyzing dynamic changes has been validated on previous studies, such as analyzing differential protein–DNA interactions across different biological conditions by designing dPCA algorithm [52]. Moreover, learning common and specific effects is also an effective strategy to disentangle the characteristics of complex system, which has been applied to compare the patterns across distinct cancer types or multiple differentiation stages via matrix factorization, such as CSMF algorithm [53]. Here, we adopt this strategy and design a Bayesian model to dissect the cluster-common and cluster-specific factors. Bayesian models have numerous advantages. Specifically, they provide increased flexibility and scalability in capturing intricate relationships among variables, enable the quantification of uncertainty associated with variables from a probabilistic view, and enhance the interpretability of the model structure to understand the complex interplay among variables. Besides, the resolution at cell-level is crucial for analyzing the heterogeneity of tumors. Single-cell technologies offer a promising avenue, but its high noise is very annoying. Constructing metacells, as a preprocessing step, effectively reduces the dropout noise, so that a meta-cell can be regarded as a denoised representation of a cell. For the sake of brevity and understandability, we use the term “cell-level” instead of “metacell-level” or “denoised cell-level” in the study. Further, low-noise single-cell sequencing technologies are expected to bring more solid biological insights.
By applying scStateDynamics on real data of tumor cells during drug treatment, we found that some subclusters exhibited generally similar expression profiles, but subtle differences in a few genes may lead to their distinct drug responses. This inspires us to consider not only the conventional static characteristics of cells (what they are like) but also their dynamic information (where they come from and where they go) when clustering cells and analyzing tumor drug responses. Notably, we accurately identified some cell flows entering the drug-tolerant persister states, which has attracted increasing attention for their role in tumor drug resistance. In addition, by disentangling the drug action mechanisms, we uncovered some candidate genes that may affect tumor progression; these genes were easily overlooked when using only conventional differential analysis on pre-treatment expressions.
Currently, we operate under the assumption that short-term drug treatment cannot induce drastic changes in tumor cell states and result in completely different expression profiles. Thus, we infer the cell dynamics by minimizing the overall gene expression changes. However, the dynamics of tumor drug response are inherently complex. If the time interval is very long, tumors cell may undergo substantial alterations in their molecular states. If the drug is more aggressive, a significant proportion of tumor cell clones may be killed, leaving only a small clone to proliferate under drug treatment. In these scenarios, our initial hypothesis may not hold and scStateDynamics may not be very suitable. Therefore, lineage tracing information becomes invaluable for investigating cancer dynamics. By utilizing this information, we can analyze the pattern of change in tumor cell states and population sizes, thereby refining the model to align cells more accurately. Similarly, in cases of temporally unmatched tumor samples (such as from different patients), the high inter-tumor heterogeneity often obscures the gene expression changes induced by drug treatment. Hence, scStateDynamics may also not be suitable in such scenarios. In addition, batch effect noise is very annoying, as it often coincides with real biological signals. Therefore, a reasonable evaluation and correction for batch effect noise are essential to obtain more accurate measurements of cell–cell distances in scStateDynamics. Besides, scStateDynamics may identify an extremely small number of tiny cluster flows. We attribute this phenomenon partly to the inherent biases of unsupervised clustering and also to the potential randomness that exists in cellular fate. How to better model this randomness of cell fates is also a problem worth exploring. Additionally, our primary focus here is on tumor cells, without considering their interaction with the tumor microenvironment, which plays an important role in regulating tumor cell states. Future works can attempt to model the dynamics of tumor cells, immune cells, and stromal cells simultaneously from a systematic perspective, which may be able to offer more insights into tumor therapy.
Overall, deciphering tumor cell state dynamics under drug treatment is highly valuable but challenging. scStateDynamics fully considers the highly heterogeneous and varying molecular characteristics of tumor drug responses and offers a powerful algorithm framework to analyze tumor cell state dynamics by modeling single-cell level gene expression changes. The analysis results pave the way for understanding the dynamic mechanisms of tumor drug resistance, and further explorations hold great potential for developing more effective treatment strategies.
Conclusions
In this study, we present scStateDynamics, a novel algorithm to infer tumor cell state dynamics under drug treatment and dissect tumor drug response mechanisms by modeling gene expression changes. By testing on simulated dataset, we show that scStateDynamics has superior performance on inferring cell cluster dynamics and can accurately identify predesigned drug response types. By testing on the data with lineage tracing labels, we confirm scStateDynamics has higher correctness and completeness in aligning cells and can avoid the influence of random biases to identify more reliable cluster flows. Furthermore, we apply scStateDynamics to real data from different cancer types treated with immunotherapy, chemotherapy, or targeted therapy. The results demonstrate that scStateDynamics facilitates the identification of cell subclusters with distinct drug response fates, enabling the analysis of both intrinsic and acquired intra-cluster heterogeneity. Moreover, scStateDynamics effectively captures known and potential drug action mechanisms in no matter immunotherapy, chemotherapy, or targeted therapy and offers a novel perspective of integrating dynamic information to characterize cell pairs and compare cluster heterogeneities.
Methods
Inferring the cell state dynamic relationships
To characterize the cell states in a high-dimensional gene expression space and identify their alignment relationships, we designed the following computational steps in scStateDynamics.
Identifying cell states and estimating their transcriptomic profiles
Due to the stochastic nature of gene transcription and the limited detection capacity for rare transcripts, scRNA-seq data often exhibit high-frequency dropout events, resulting in a large proportion of zeros in the gene expression matrix (Additional file 1: Fig. S15) [54–56]. To mitigate the influence of dropout event noise, we add a preprocessing step of grouping several similar cells into a metacell to represent a type of cell state [57–59]. In practice, we utilize the “Scanpy” package [60] to perform clustering with a large ‘resolution’ parameter setting (such as 30 or 50). Each resulting cluster is considered as a metacell, generally consisting of approximately three to ten cells, to represent a specific cell state. This adaptive numbers of cells within metacells can better cope with the varying densities of cell distribution in gene expression space. Then, we characterize each metacell by calculating the average expression profiles of its constituent cells. Generally, the cells with very similar expression profiles can be seen as being resampled from a common cell state, and the probability of dropout events occurring simultaneously in these similar cells for a given gene is relatively low. Therefore, this preprocessing step of aggregating similar cells and utilizing their average expression values to represent the corresponding cell states can reduce the dropout noise. The analysis on our testing datasets also validated its necessity and effectiveness (Additional file 1: Fig. S15), thereby a meta-cell can also be regard as a denoised representation of a cell. Besides, it can reduce the computational complexity of subsequent analyses. Notably, to make this preprocessing step more flexible and robust, we provide an interface for users to either supply their own metacell labels or bypass this preprocessing step through setting unique metacell labels.
Measuring the distances between cell states along the low-dimensional manifold
In the high-dimensional gene expression space, cells are usually distributed on a low-dimensional manifold. This observation suggests that direct calculation of global Euclidean distances may not provide an accurate representation of the similarities between cells along the manifold. Hence, we refer to the methodologies of PHATE [21], Diffusion Map [20], and MuTrans [15] algorithms and utilize local distance diffusion to assess the global relationships among different cell states. In detail, we use the Gaussian kernel function to transform the global Euclidean distances between the joint-embedded expression profiles and of metacells and into local affinities. This transformation ensures that the affinity degree between two cell states is greater than 0 only when they are close enough, so that we can obtain the local neighbor relationships based on the affinity degrees (Additional file 1: Fig. S16a). The parameter governs the local bandwidth constrained by the kernel function (Additional file 1: Fig. S16b). Considering that the density of the cell distribution is uneven, the k-nearest-neighbor distance is used as an adaptive bandwidth. Additionally, to control the heavy tail of the Gaussian kernel when is large, the exponent is introduced (Additional file 1: Fig. S16b). Consequently, the resulting kernel function is obtained by taking the average of two kernel values to ensure the symmetry of the affinity matrix: . Then we normalize the affinity matrix by its row sums to generate the following random-walk transition probability matrix among cell states . Furthermore, to propagate the local neighbor relationships and assess the long-range affinity values along the low-dimensional manifold, we perform a -step random walk and calculate the -step diffusion probability . Here, the parameter is a positive integer that denotes how many steps of long-range diffusion are considered acceptable, and it can be determined based to the distribution density of cells. And is an identity matrix. Finally, we measure the distance between two cell states by calculating the -norm distance of their logarithmic -step diffusion probabilities to other cell states: . Notably, at this distance measurement step, we assume that the influence of batch effects on measuring distances between cells along the manifold is slight. In the practice, we advise users to evaluate the extent of batch effect noise using approaches such as low-dimensional co-projection, or quantitative indicators and then determine whether and how to correct the batch effect before utilizing our algorithm. If the batch effect noise is pronounced, it is essential to correct it beforehand.
Aligning the cell states between two time points
Here, we assume that the overall changes in cell states is relatively small during drug treatment and only a portion of cells have significant state changes. Using the distances along manifold calculated above to measure the extent of cell state changes, we adopt the principle of minimizing the overall changes to align cells in a low-dimensional manifold between two time points, which can be achieved by the optimal transport algorithm. At each time point, all cells form a discrete cell state probability distribution in gene expression space. Therefore, aligning the cell states can be regarded as seeking a transport plan between two probability distributions.
In the discrete case, given the source and target probability distributions
and
and a transport cost matrix , where each element indicates the cost when transporting from state to . Then, the objective of the optimal transport problem is to find a transport matrix that minimizes the total cost (overall changes)
subject to and
where indicates the identified probability mass transporting from to . This means that the total cost is the inner product of the cost matrix and the transport matrix , while ensuring that the row sum and column sum of are equal to and , respectively.
In our cell state alignment problem, the number of cells in all metacells (cell states) at the pre- and post- time points can be normalized as probability distributions and , respectively. Taking the distances calculated above (matrix ) as the cost matrix , we can obtain the transport matrix based on the optimal transport algorithm, in which each element indicates the probability mass transforming from the pre-timepoint to the post-timepoint. According to these transport probabilities, we can align the cells in all metacells between the two time points. This step is implemented with the “ot” Python package [61].
Grouping the cell alignment relationships into subcluster flows
To investigate the dynamics at cluster level, we conduct clustering on the cells at each time point individually using “Scanpy” package [60], with a small “resolution” parameter setting (Additional file 1: Fig. S1a). This step of separate clustering, rather than joint clustering, can avoid the influence of batch effect noise. In this way, we connect the clusters between two time points based on cell flows and identify distinct fates among cells. According to the cell fates (which post-cluster the cell transition to), we further divide the pre-clusters into distinct subclusters (Additional file 1: Fig. S1b). This operation of identifying cell subcluster flows can help infer the distinct proliferation or inhibition rates of clusters and support the subsequent differential expression analysis to dissect intra-cluster intrinsic and acquired heterogeneity between distinct cell fates.
Quantifying the dynamic characteristics of cell populations
After drug treatment, distinct clusters of tumor cells may exhibit varying degrees of sensitivity, leading to differences in proliferation or inhibition rates. Besides, some cells can also change their states to adapt to the external environment. Hence, the dynamic characteristics of cell populations include the changes in both cell states and abundances (proportions). The extent of changes in cell states can be quantified by the transport costs. However, for the cell abundances, the classical optimal transport theory we used could not model the increase or decrease in probability mass that reflects the relative proliferation or inhibition of cell populations. To address this limitation, we first identify the unreasonable cell subcluster flows caused by the neglect of cell proliferation and then correct them to obtain more reliable cell dynamics.
Identifying the types of cell subcluster flows
To quantify the extent of changes in cell states and distinguish the pattern of cell subcluster flows, we calculate the average transport cost for each flow. Given that the metacell sets at pre-timepoint and post-timepoint of a cell subcluster flow are and , we define the average weighted transport cost of it as
According to the distribution of average transport costs of all subcluster flows, we think they can be categorized into either state-keeping, state-changed, or unreasonable flows. State-keeping corresponds to the cells that maintain high similarity, resulting in flows with low transport costs. State-changed indicates that cells adaptively adjust their states, leading to flows with relatively high transport costs. Unreasonable flows refer to some incorrect alignments identified by OT algorithm arising from the neglect of distinct proliferation or inhibition rates among cell clusters. As a result, when certain source cell populations are inhibited, their reduced probability masses have to be allocated to other target cell populations exhibiting high proliferation rates. These incorrect alignments lead to some unreasonable flows with abnormally large transport costs. To determine the type of flows, an analysis of the average transport cost distribution for all subcluster flows is conducted through plotting a histogram. If the distribution exhibits a distinct trimodal pattern, a Gaussian mixture model (GMM) can be applied to category the three peaks into state-keeping, state-changed, or unreasonable flows. Instead, if there are several outliers that are clearly distant from the majority of data points, outlier detection approaches (e.g., using 1.5 times the interquartile range above the third quartile as a threshold) can be employed to identify these outliers as unreasonable flow. Besides, manually setting the thresholds to identify unreasonable flows is also optional.
Correcting the unreasonable cell flows
Based on the identified type of cell flows, we aim to correct the unreasonable flows and estimate the proliferation or inhibition rates of clusters.
Given an unreasonable flow with a probability mass of from a subset of cluster Pre_A at pre-timepoint to a subset of cluster Post_B at post-timepoint, we can infer that the cluster Pre_A is inhibited, and the inhibition rate can be calculated by dividing by the total probability masses of cluster Pre_A. Meanwhile, the cluster Post_B should originate from the cells that are more similar to it. Hence, if there are other reasonable sources (without loss of generality, we refer to them as Pre_C and Pre_D) for Post_B, we assign the probability mass to Pre_C and Pre_D based on their relative fractions. If Post_B completely originates from Pre_A, we think the cluster with the highest similarity to Post_B should have a greater proliferation rate. In this way, the probability masses of unreasonable flows can be assigned to more appropriate source clusters. By re-normalizing the probability masses at the pre-timepoint, we obtain the updated source probability distribution, denoted as . Then, we replace with to re-perform optimal transport and re-correct the identified unreasonable flows iteratively, until no outlier flows exist or the results stabilize.
In the end, by comparing the final updated source probability distribution with the initial source distribution , we can estimate the final proliferation or inhibition rate of each cluster at pre-timepoint.
Decomposing the expression changes into static variations and dynamic biological effects
The changes in gene expression profiles between the cells at two time points can provide insights into the dynamic biological mechanisms of drug action. According to the obtained optimal transport matrix , an element greater than 0 indicates a potential dynamic alignment between the th cell state (metacell) at the pre-timepoint and the th cell state at the post-timepoint. Consequently, assuming that there are elements greater than 0 in , we identify alignment relationships among cell states, resulting in the formation of distinct cell pairs. Subsequently, according to the coordinates of these elements in , we can extract the corresponding gene expression vectors at the pre- and post-timepoints. These vectors collectively compose the matrices and , where denotes the number of genes. Then the matrix of gene expression changes can be calculated by . Here, we think these dynamic changes are determined by two types of effects: (i) the initial and final cell expression profiles and (ii) the molecular mechanisms of drug action. To disentangle these two types of variations, we design a Bayesian factor analysis model by characterizing the first type of static variations with the cell cluster identities and decomposing the second type of dynamic effects into a combination of gene factors (signatures).
Model representation
Assuming that there are and clusters at the two time points, then for a cell pair , consisting of a metacell within cluster and a metacell within cluster , we decompose its change vector into a sum of three components and an additive Gaussian noise:
Here, vector denotes the common effects shared by the cells in cluster at pre-timepoint, while vector denotes the effects shared by the cells in cluster at post-timepoint. We model them with normal distributions
The matrix denotes the regulatory weights of factors on genes, and each factor can be seen as a gene signature related to the dynamic biological mechanism of drug action. We constrain the elements in it to be positive and use independent log-normal distribution to model it
The vector denotes the composition coefficients (activities) of the factors for cell pair , and can also be seen as an embedding of this cell pair in the space spanning by these gene factors. We model it with normal distribution
The vector is Gaussian residual noise
We use the precision parameter to capture the gene-specific variation and define an independent conjugate prior for it
In these distributions, , , , , , and are hyperparameters
Hence, we can imply the likelihood as
Inference
Given the observation variable , the latent variables , , , , and are not mutually independent. Exact inference for the posterior distribution requires integrals, which are computationally intractable. Hence, we use the strategy of variational inference to approximate the true posterior distributions. We introduce a parameterized variational distribution , which can be factorized as
where are all the variational parameters in each of the following distributions
Hence, our goal is to optimize the parameters to find the best possible variational distribution that effectively approximates the posterior distribution. When we use Kullback–Leibler divergence to measure the distance between these two probability distributions, the optimization problem can be converted to maximize the evidence lower bound (ELBO)
The optimization and update of variational parameters are performed by using stochastic gradient descent algorithm (Adam optimizer).
Datasets and pre-processing
Simulated data generation
To evaluate the performance of scStateDynamics, we used the “paths” method of the “splatSimulate” function within the Splatter package [22] to generate three scRNA-seq datasets that simulate distinct scenarios of tumor drug responses. To design the characteristics of these dynamic processes, we mainly manipulated the following parameters. We used the “group.prob” to control the sizes of cell clusters and used the “path.from” parameter to determine the dynamic relationships between clusters. Then, to model the extent of changes in cell states, we adjusted the “de.prob” parameter. Besides, we utilized the “path.skew” parameter to fine-tune the distribution of cells towards either the source or target population.
Data pre-processing
The pre-processing for the simulated and real data was performed based on the Scanpy [60], Seurat [62], and scCancer [63, 64] packages. First, data quality control is performed by filtering the potential lysed cells, low-quality cells, and doublets, based on the number of detected transcripts and genes, as well as the percentage of transcripts from mitochondrial genes. Besides, mitochondrial genes, ribosomal genes, and the genes expressed in fewer than three cells are also filtered. For the remaining cells and genes, we calculate the relative expression values by performing data normalization and log-transformation. Next, the highly variable genes of the data at two time points are identified, and their union set is used as the final selected genes. Then, we regress out the unwanted variance sources and perform data centering and scaling. Further, to project the cells of two time points into a shared low-dimensional space, we perform similar pre-processing steps on the combined expression matrix and conduct principal component analysis (PCA). These low-dimensional representations are subsequently used to calculate the distances between the cell states.
Downstream comparison and analyses
In this section, we provide the method details of the downstream analyses based on the results of scStateDynamics.
Comparing the inferred cell alignment relationships with lineage tracing information
We first screen the lineage barcodes that appear at both of the two time points, so that we could leverage the dynamic relationships represented by them to evaluate the confidence level of each cell subcluster flow. For example, if a lineage barcode is observed in cells within cluster Pre_1 at the pre-timepoint and cells within cluster Post_2 at post-timepoint, we interpret this as barcode evidences supporting the Pre_1- > Post_2 flow. To eliminate the influence of clone sizes, we normalize this count by dividing it by the square root of the product of the total number of cells labeled by this barcode at the two time points. Then, we integrate the evidences derived from all lineage barcodes and calculate the sum of their normalized counts for each flow. Further, to avoid the influence of cluster sizes, we also divide these summation results by the square root of the product of the cell numbers in the source cluster and target cluster for each flow. In this way, we obtain the final normalized lineage barcode counts, as shown in Fig. 3c.
Further, we also define two metrics to evaluate the correctness and completeness of the inferred cell alignments by comparing with the cell lineage barcodes, as shown in Fig. S4b of Additional file 1. We first determine the possible target clusters of each cell at the pre-timepoint based on its lineage barcode label. Then, by comparing them with the results inferred by the algorithms (scStateDynamics, CINEMA-OT, or CINEMA-OT-W), we define the correctness metric as the proportion of cells with correct fate inference. Besides, by measuring whether all the fates supported by lineage information in each clone are identified by the algorithms, we define the completeness metric as the average fate recognition rate across all cells at the pre-timepoint.
Besides, we conduct a comparative analysis between the performance of scStateDynamics and joint-clustering, a conventional cluster-level alignment method, based on the Watermelon lineage tracing dataset. In detail, we employ the BBKNN algorithm [65] to integrate the data from adjacent timepoints, and then apply Leiden graph-clustering method [66] to jointly cluster cells based on the Scanpy package [60]. Within each cluster, cells from pre- and post-timepoints are considered temporally aligned. To assess performance, we calculate the mean squared errors (MSEs) between the transition probability matrix (TPM) based on the lineage tracing labels (considered as ground truth) and the TPMs obtained through joint-clustering or scStateDynamics (Fig. S5).
Inter-cluster and intra-cluster heterogeneities analyses
To investigate the intrinsic heterogeneities at inter-cluster and intra-cluster levels, we conduct differential expression analysis between the clusters or the subclusters with distinct fates based on the Wilcoxon rank-sum test method in Scanpy package [60]. Then the significantly differentially expressed genes (DE genes) of each cluster or subcluster are subjected to enrichment analysis on the cancer hallmark pathways in MSigDB [67]. Furthermore, to compare the malignancy degrees among clusters or subclusters, we regard their DE genes as a signature to represent the subcluster and apply them to TCGA bulk samples with the same cancer type. We define the signature scores of the bulk samples by calculating the average expression values of the genes in the signature and utilize Cox proportional hazards regression to analyze the effect of the signatures on survival. In this way, the hazard ratios obtained can be used to quantify the malignancy degrees of the clusters or subclusters (Fig. 4f and Additional file 1: Fig. S7b).
To analyze the acquired heterogeneities, we calculate the change values of gene expression and pathway scores ( in Fig. 4g) by performing subtraction between the post-treatment and pre-treatment cells according to their inferred alignment relationships. Here, the log-normalized gene expression values are adopted. The pathway (signature) scores are defined as the average expression values of the genes within the pathways. In the case of the DTP signature [42], where weights are assigned to genes, we multiply these weights by the gene expression values before calculating the average. In addition, we also perform a Wilcoxon rank sum test on the DTP-related pathway scores to measure the increase or decrease in pathway activity induced by drug treatment (Fig. 4j, 4k and Additional file 1: Fig. S10).
Annotating the factors with signal pathways
We collect the cancer hallmark pathway and drug-related pathway information to provide biological annotations for the identified factors. For each factor in the decomposed gene-factor weight matrix , we define its initial pathway score as the average weight of the genes in the respective pathway. Then to make the scores comparable, we generate a background distribution by randomly shuffling the gene weights 1000 times and calculate the scores as previously described. By utilizing the mean and standard deviation of these 1000 scores, we transform the initial pathway score into its final z-score formation by , as shown in Fig. 5b.
Supplementary Information
Additional file 1: Fig. S1: A schematic diagram illustrating the concepts of cell, metacell, cluster, and subcluster. Fig. S2: Iterative process of correcting unreasonable flows in simulated data 1. Fig. S3: Performance comparison with CINEMA-OT and CINEMA-OT-W on simulated data. Fig. S4: Performance comparison with CINEMA-OT and CINEMA-OT-W on the data with lineage tracing information. Fig. S5: Performance comparison with joint-clustering on the data with lineage tracing information. Fig. S6: Comparing the inferred proliferation or inhibition rates with the lineage barcode information on ReSisTrace dataset. Fig. S7: Tumor heterogeneity and hazard ratio analysis at inter-cluster level in HCC dataset. Fig. S8: Assessing the robustness of the findings about intra-cluster intrinsic heterogeneity by random shuffle experiments. Fig. S9: The analysis results by scStateDynamics on the other three real datasets. Fig. S10: The distribution of the DTP-associated signature scores, related to Fig. 4i. Fig. S11: The Factor Analysis model in scStateDynamics effectively decomposes the dynamic effects from static variations. Fig. S12: The top gene weights of each factor in the four real datasets. Fig. S13: Biological annotation for the factors identified in GBM2 sample. Fig. S14: Comparisons between gene average expression values and vector weights in each pre-cluster. Fig. S15: The dropout rates of all testing datasets before and after constructing metacells. Fig. S16: The shape of Gaussian kernel functions under different parameters
Additional file 2: Table S1: Simulation datasets information. Table S2: Information of datasets with lineage tracing annotation. Table S3: Real tumor drug treatment datasets information
Acknowledgements
We thank Qichen Liao, Baojia Luo, Tianhao Wu, and Dr. Hao Wu from Department of Mathematical Sciences at Tsinghua University for their valuable discussion.
Peer review information
Kin Fai Au and Wenjing She were the primary editors of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3.
Authors' contributions
J.G. conceived this study. W.G. developed the methods, performed the analyses, and drew the figures. J.G, W.G., and X.L. discussed the methods and results. D.W., F.Y., X.Z., and J.Y. provided suggestions for the experiments. W.G., X.L., N.Y., and Q.H. tested the package. W.G. and J.G. wrote the manuscript with the assistance of the other authors. J.G. and J.Y. supervised the project.
Funding
This work was supported by funding from the National Key Research and Development Program of China (nos. 2020YFA0712403 and 2021YFF1200901), the National Natural Science Foundation of China (NSFC) (nos. 62133006 and 92268104), the Tsinghua University Initiative Scientific Research Program (no. 20221080076), and the China Postdoctoral Science Foundation (no. 2022M721839).
Data availability
The datasets supporting the conclusions of this article are available in the Gene Expression Omnibus (GEO) repository. The Watermelon system lineage tracing scRNA-seq data were downloaded from Gene Expression Omnibus (GEO) with accession number GSE150949 [68]. The ReSisTrace data were download from GSE223003 [69]. The HCC data were downloaded from GSE151530 [70]. The GBM data were downloaded from GSE195682 [71]. The NSCLC data were downloaded from GSE134836 [72]. The Python package implementing the scStateDynamics and its tutorial material are freely available at GitHub (http://lifeome.net/software/scStateDynamics/) [73] and Zenodo [74]. The source code is released under the MIT license.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors Fan Yang and Jianhua Yao are from a commercial company (AI Lab, Tencent, Shenzhen, China). All the other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianhua Yao, Email: jianhuayao@tencent.com.
Jin Gu, Email: jgu@tsinghua.edu.cn.
References
- 1.Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell. 2020;37:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19:39–56. [DOI] [PubMed] [Google Scholar]
- 3.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. [DOI] [PubMed] [Google Scholar]
- 4.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, Sharon N, Bar-Joseph Z. Temporal modelling using single-cell transcriptomics. Nat Rev Genet. 2022;23:355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Chen H, Mo H, Hu X, Gao R, Zhao Y, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell. 2021;39:1578-1593.e8. [DOI] [PubMed] [Google Scholar]
- 7.Liu R, Gao Q, Foltz SM, Fowles JS, Yao L, Wang JT, et al. Co-evolution of tumor and immune cells during progression of multiple myeloma. Nat Commun. 2021;12:2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lotfollahi M, Wolf FA, Theis FJ. scGen predicts single-cell perturbation responses. Nat Methods. 2019;16:715–21. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Rivaud P, Park JH, Tsou T, Charles E, Haliburton JR, et al. Dissecting heterogeneous cell populations across drug and disease conditions with PopAlign. Proc Natl Acad Sci USA. 2020;117:28784–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkhardt DB, Stanley JS, Tong A, Perdigoto AL, Gigante SA, Herold KC, et al. Quantifying the effect of experimental perturbations at single-cell resolution. Nat Biotechnol. 2021;39:619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lotfollahi M, Naghipourfar M, Theis FJ, Wolf FA. Conditional out-of-distribution generation for unpaired data using transfer VAE. Bioinformatics. 2020;36:i610–7. [DOI] [PubMed] [Google Scholar]
- 12.Weinreb C, Wolock S, Tusi BK, Socolovsky M, Klein AM. Fundamental limits on dynamic inference from single-cell snapshots. Proc Natl Acad Sci. 2018;115:E2467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell. 2019;176:928-943.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrow A, Schiebinger G. LineageOT is a unified framework for lineage tracing and trajectory inference. Nat Commun. 2021;12:4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Wang S, Li T, Nie Q. Dissecting transition cells from single-cell transcriptome data through multiscale stochastic dynamics. Nat Commun. 2021;12:5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Q, Zhang S, Wan L. Dynamic inference of cell developmental complex energy landscape from time series single-cell transcriptomic data. PLoS Comput Biol. 2022;18: e1009821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunne C, Stark SG, Gut G, Del Castillo JS, Levesque M, Lehmann K-V, et al. Learning single-cell perturbation responses using neural optimal transport. Nat Methods. 2023;20:1759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong M, Wang B, Wei J, De O. Fonseca AH, Perry CJ, Frey A, et al. Causal identification of single-cell experimental perturbation effects with CINEMA-OT. Nat Methods. 2023;20:1769–79. [DOI] [PMC free article] [PubMed]
- 19.Peyré G, Cuturi M. Computational optimal transport. Foundations and Trends® in Machine Learning. 2019;11:355–607.
- 20.Coifman RR, Lafon S. Diffusion maps. Appl Comput Harmon Anal. 2006;21:5–30. [Google Scholar]
- 21.Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, et al. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol. 2019;37:1482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zappia L, Phipson B, Oshlack A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 2017;18:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kester L, van Oudenaarden A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell. 2018;23:166–79. [DOI] [PubMed] [Google Scholar]
- 24.Wagner DE, Klein AM. Lineage tracing meets single-cell omics: opportunities and challenges. Nat Rev Genet. 2020;21:410–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oren Y, Tsabar M, Cuoco MS, Amir-Zilberstein L, Cabanos HF, Hütter J-C, et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature. 2021;596:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai J, Zheng S, Falco MM, Bao J, Eriksson J, Pikkusaari S, et al. Tracing back primed resistance in cancer via sister cells. Nat Commun. 2024;15:1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L, Wang L, Khatib SA, Chang C-W, Heinrich S, Dominguez DA, et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J Hepatol. 2021;75:1397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hangai S, Kawamura T, Kimura Y, Chang C-Y, Hibino S, Yamamoto D, et al. Orchestration of myeloid-derived suppressor cells in the tumor microenvironment by ubiquitous cellular protein TCTP released by tumor cells. Nat Immunol. 2021;22:947–57. [DOI] [PubMed] [Google Scholar]
- 29.Zhu H, Su Z, Ning J, Zhou L, Tan L, Sayed S, et al. Transmembrane protein 97 exhibits oncogenic properties via enhancing LRP6-mediated Wnt signaling in breast cancer. Cell Death Dis. 2021;12:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge W, Chi H, Tang H, Xu J, Wang J, Cai W, et al. Circular RNA CELF1 drives immunosuppression and anti-PD1 therapy resistance in non-small cell lung cancer via the miR-491-5p/EGFR axis. Aging. 2021;13:24560–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek J-H, Yun HS, Kim J-Y, Lee J, Lee Y-J, Lee C-W, et al. Kinesin light chain 4 as a new target for lung cancer chemoresistance via targeted inhibition of checkpoint kinases in the DNA repair network. Cell Death Dis. 2020;11:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi S, Gu S, Han T, Zhang W, Huang L, Li Z, et al. Inhibition of MAN2A1 enhances the immune response to anti–PD-L1 in human tumors. Clin Cancer Res. 2020;26:5990–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bu MT, Chandrasekhar P, Ding L, Hugo W. The roles of TGF-β and VEGF pathways in the suppression of antitumor immunity in melanoma and other solid tumors. Pharmacol Ther. 2022;240: 108211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang B, Tang F, Wang Z, Qi G, Liang X, Li B, et al. Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2016;35:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo X, Zheng E, Wei L, Zeng H, Qin H, Zhang X, et al. The fatty acid receptor CD36 promotes HCC progression through activating Src/PI3K/AKT axis-dependent aerobic glycolysis. Cell Death Dis. 2021;12:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer. 2020;122:1580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu K, He Z, Chen M, Wang N, Zhang D, Yang L, et al. HIF-1α regulates cellular metabolism, and Imatinib resistance by targeting phosphogluconate dehydrogenase in gastrointestinal stromal tumors. Cell Death Dis. 2020;11:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Zou L, Lu G, Grinchuk O, Fang L, Ong DST, et al. PFKP alleviates glucose starvation-induced metabolic stress in lung cancer cells via AMPK-ACC2 dependent fatty acid oxidation. Cell Discov. 2022;8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qazi MA, Salim SK, Brown KR, Mikolajewicz N, Savage N, Han H, et al. Characterization of the minimal residual disease state reveals distinct evolutionary trajectories of human glioblastoma. Cell Rep. 2022;40: 111420. [DOI] [PubMed] [Google Scholar]
- 40.Aissa AF, Islam ABMMK, Ariss MM, Go CC, Rader AE, Conrardy RD, et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat Commun. 2021;12:1628. [DOI] [PMC free article] [PubMed]
- 41.Dhimolea E, de Matos Simoes R, Kansara D, Al’Khafaji A, Bouyssou J, Weng X, et al. An embryonic diapause-like adaptation with suppressed Myc activity enables tumor treatment persistence. Cancer Cell. 2021;39:240–256.e11. [DOI] [PMC free article] [PubMed]
- 42.Rehman SK, Haynes J, Collignon E, Brown KR, Wang Y, Nixon AML, et al. Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell. 2021;184:226-242.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pu Y, Li L, Peng H, Liu L, Heymann D, Robert C, et al. Drug-tolerant persister cells in cancer: the cutting edges and future directions. Nat Rev Clin Oncol. 2023;20:799–813. [DOI] [PubMed] [Google Scholar]
- 44.Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun. 2018;9:1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He B, Zhang H, Wang J, Liu M, Sun Y, Guo C, et al. Blastocyst activation engenders transcriptome reprogram affecting X-chromosome reactivation and inflammatory trigger of implantation. Proc Natl Acad Sci USA. 2019;116:16621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goyal Y, Busch GT, Pillai M, Li J, Boe RH, Grody EI, et al. Diverse clonal fates emerge upon drug treatment of homogeneous cancer cells. Nature. 2023;620:651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens S, Bridelance J, Roelandt R, Vandenabeele P, Takahashi N. MLKL in cancer: more than a necroptosis regulator. Cell Death Differ. 2021;28:1757–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang X, Deng W, Tao S, Tang Z, Chen Y, Tian M, et al. A RIPK3-independent role of MLKL in suppressing parthanatos promotes immune evasion in hepatocellular carcinoma. Cell Discov. 2023;9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y-C, Wang X, Yu J, Ma F, Li Z, Zhou Y, et al. Targeting monoamine oxidase A-regulated tumor-associated macrophage polarization for cancer immunotherapy. Nat Commun. 2021;12:3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Zhang CZ, Lu S-X, Zhang M-F, Liu L-L, Luo R-Z, et al. A coiled-coil domain containing 50 splice variant is modulated by serine/arginine-rich splicing factor 3 and promotes hepatocellular carcinoma in mice by the Ras signaling pathway. Hepatology. 2019;69:179. [DOI] [PubMed] [Google Scholar]
- 51.Hou P, Yang K, Jia P, Liu L, Lin Y, Li Z, et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021;31:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji H, Li X, Wang Q, Ning Y. Differential principal component analysis of ChIP-seq. Proc Natl Acad Sci USA. 2013;110:6789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Zhang S. Learning common and specific patterns from data of multiple interrelated biological scenarios with matrix factorization. Nucleic Acids Res. 2019;47:6606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pierson E, Yau C. ZIFA: dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 2015;16:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrews TS, Kiselev VY, McCarthy D, Hemberg M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat Protoc. 2021;16:1–9. [DOI] [PubMed] [Google Scholar]
- 57.Baran Y, Bercovich A, Sebe-Pedros A, Lubling Y, Giladi A, Chomsky E, et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019;20:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Persad S, Choo Z-N, Dien C, Sohail N, Masilionis I, Chaligné R, et al. SEACells infers transcriptional and epigenomic cellular states from single-cell genomics data. Nat Biotechnol. 2023;41:1746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Badia-i-Mompel P, Wessels L, Müller-Dott S, Trimbour R, Ramirez Flores RO, Argelaguet R, et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat Rev Genet. 2023;24:739–54. [DOI] [PubMed] [Google Scholar]
- 60.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flamary R, Courty N, Gramfort A, Alaya MZ, Boisbunon A, Chambon S, et al. POT: Python optimal transport. J Mach Learn Res. 2021;22:1–8. [Google Scholar]
- 62.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573-3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo W, Wang D, Wang S, Shan Y, Liu C, Gu J. scCancer: a package for automated processing of single-cell RNA-seq data in cancer. Briefings in Bioinformatics. 2021;22:bbaa127. [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, Miao Y, Tan Z, Hu Q, Wu Y, Li X, et al. scCancer2: data-driven in-depth annotations of the tumor microenvironment at single-level resolution. Bioinformatics. 2024;40:btae028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, Park J-E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Traag VA, Waltman L, Van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oren Y, Tsabar M, Cuoco MS, Amir-Zilberstein L, Cabanos HF, Hütter J-C, et al. Cycling cancer persister cells arise from lineages with distinct programs. Datasets. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150949 [DOI] [PMC free article] [PubMed]
- 69.Dai J, Zheng S, Falco MM, Bao J, Eriksson J, Pikkusaari S, et al. Tracing back primed resistance in cancer via sister cells. Datasets. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223003 [DOI] [PMC free article] [PubMed]
- 70.Ma L, Wang L, Khatib SA, Chang C-W, Heinrich S, Dominguez DA, et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Datasets. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151530 [DOI] [PMC free article] [PubMed]
- 71.Qazi MA, Salim SK, Brown KR, Mikolajewicz N, Savage N, Han H, et al. Characterization of the minimal residual disease state reveals distinct evolutionary trajectories of human glioblastoma. Datasets. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE195682 [DOI] [PubMed]
- 72.Aissa AF, Islam ABMMK, Ariss MM, Go CC, Rader AE, Conrardy RD, et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Datasets. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134836 [DOI] [PMC free article] [PubMed]
- 73.Guo W, Li X, Wang D, Yan N, Hu Q, Yang F, et al. scStateDynamics: deciphering the drug-responsive tumor cell state dynamics by modeling single-cell level expression changes. GitHub; 2024. https://github.com/wguo-research/scStateDynamics [DOI] [PMC free article] [PubMed]
- 74.Guo W, Li X, Wang D, Yan N, Hu Q, Yang F, et al. scStateDynamics: deciphering the drug-responsive tumor cell state dynamics by modeling single-cell level expression changes. 2024. Zenodo. 10.5281/zenodo.12697637. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1: A schematic diagram illustrating the concepts of cell, metacell, cluster, and subcluster. Fig. S2: Iterative process of correcting unreasonable flows in simulated data 1. Fig. S3: Performance comparison with CINEMA-OT and CINEMA-OT-W on simulated data. Fig. S4: Performance comparison with CINEMA-OT and CINEMA-OT-W on the data with lineage tracing information. Fig. S5: Performance comparison with joint-clustering on the data with lineage tracing information. Fig. S6: Comparing the inferred proliferation or inhibition rates with the lineage barcode information on ReSisTrace dataset. Fig. S7: Tumor heterogeneity and hazard ratio analysis at inter-cluster level in HCC dataset. Fig. S8: Assessing the robustness of the findings about intra-cluster intrinsic heterogeneity by random shuffle experiments. Fig. S9: The analysis results by scStateDynamics on the other three real datasets. Fig. S10: The distribution of the DTP-associated signature scores, related to Fig. 4i. Fig. S11: The Factor Analysis model in scStateDynamics effectively decomposes the dynamic effects from static variations. Fig. S12: The top gene weights of each factor in the four real datasets. Fig. S13: Biological annotation for the factors identified in GBM2 sample. Fig. S14: Comparisons between gene average expression values and vector weights in each pre-cluster. Fig. S15: The dropout rates of all testing datasets before and after constructing metacells. Fig. S16: The shape of Gaussian kernel functions under different parameters
Additional file 2: Table S1: Simulation datasets information. Table S2: Information of datasets with lineage tracing annotation. Table S3: Real tumor drug treatment datasets information
Data Availability Statement
The datasets supporting the conclusions of this article are available in the Gene Expression Omnibus (GEO) repository. The Watermelon system lineage tracing scRNA-seq data were downloaded from Gene Expression Omnibus (GEO) with accession number GSE150949 [68]. The ReSisTrace data were download from GSE223003 [69]. The HCC data were downloaded from GSE151530 [70]. The GBM data were downloaded from GSE195682 [71]. The NSCLC data were downloaded from GSE134836 [72]. The Python package implementing the scStateDynamics and its tutorial material are freely available at GitHub (http://lifeome.net/software/scStateDynamics/) [73] and Zenodo [74]. The source code is released under the MIT license.