Summary
Background
Lung cancer and tobacco use pose significant global health challenges, necessitating a comprehensive translational roadmap for improved prevention strategies such as cancer screening and tobacco treatment, which are currently under-utilised. Polygenic risk scores (PRSs) may further motivate health behaviour change in primary care for lung cancer in diverse populations. In this work, we introduce the GREAT care paradigm, which integrates PRSs within comprehensive patient risk profiles to motivate positive health behaviour changes.
Methods
We developed PRSs using large-scale multi-ancestry genome-wide association studies and standardised PRS distributions across all ancestries. We validated our PRSs in 561,776 individuals of diverse ancestry from the GISC Trial, UK Biobank (UKBB), and All of Us Research Program (AoU).
Findings
Significant odds ratios (ORs) for lung cancer and difficulty quitting smoking were observed in both UKBB and AoU. For lung cancer, the ORs for individuals in the highest risk group (top 20% versus bottom 20%) were 1.85 (95% CI: 1.58–2.18) in UKBB and 2.39 (95% CI: 1.93–2.97) in AoU. For difficulty quitting smoking, the ORs (top 33% versus bottom 33%) were 1.36 (95% CI: 1.32–1.41) in UKBB and 1.32 (95% CI: 1.28–1.36) in AoU.
Interpretation
Our PRS-based intervention model leverages large-scale genetic data for robust risk assessment across populations, which will be evaluated in two cluster-randomised clinical trials. This approach integrates genomic insights into primary care, promising improved outcomes in cancer prevention and tobacco treatment.
Funding
National Institutes of Health, NIH Intramural Research Program, National Science Foundation.
Keywords: Lung cancer, Polygenic risk scores, Cancer prevention, Personalised interventions, Translational roadmap, Health behaviour change
Research in context.
Evidence before this study
We systematically searched PubMed for research articles published in English before June 21, 2024, using the search terms “polygenic risk score”, “lung cancer” and “clinical trials”. Several clinical trials, including the WISDOM, GenoVA, and eMERGE studies, have evaluated the utility of polygenic risk scores (PRSs) for diseases such as breast cancer and coronary artery disease. However, these studies do not include lung cancer or tobacco use, both of which present significant global health burdens. While genome-wide association studies (GWASs) have identified several genetic risk factors for both conditions, and PRSs for lung cancer have been developed, they have not yet been implemented in clinical settings.
Added value of this study
Our study introduces a new primary care paradigm that integrates the latest genetic data and research into precision treatment of lung cancer and smoking cessation. We compute ancestry-adjusted PRS using large-scale GWASs and develop standardised risk models appliable across diverse ancestry. We validate our model's accuracy in three ancestrally diverse cohorts, including two large-scale biobanks, finding significant odds ratios for both lung cancer and difficulty quitting smoking between the top and bottom risk groups.
Implications of all the available evidence
Our findings demonstrate that ancestry-adjusted PRS can effectively stratify patient risk for lung cancer and difficulty quitting smoking across diverse backgrounds. This framework will be implemented in two cluster-randomised clinical trials to evaluate the efficacy of PRS-based intervention tools in promoting health behaviour changes related to cancer prevention and tobacco treatment in patients of diverse ancestry.
Introduction
The worldwide burden of lung cancer and tobacco smoking presents major challenges to global health.1 Evidence-based practices to reduce their risk such as cancer screening and tobacco treatment (e.g. smoking cessation medication) have long existed but are infrequently used in most primary care practices. While it is well-established that quitting smoking can dramatically reduce the risk of developing lung cancer,2 patients may face various barriers to participating in smoking cessation programs.3 This presents a critical gap in preventive healthcare, particularly for individuals at high risk of lung cancer.
Polygenic risk scores (PRSs) have emerged as a valuable approach to assess disease susceptibility among populations and pinpoint individuals at higher risk.4, 5, 6 By incorporating PRSs into clinical practice, we can provide patients with personalised information that not only highlights their heightened genetic risk for lung cancer but also expected difficulty quitting smoking. In our previous studies, large majorities of patients who smoke showed high interest in receiving genetically tailored tobacco treatment,7 endorsed the importance of genetic results to guide treatment,8 and found genetics-based tools useful in understanding health risk and taking control of their health.9 With this personalised approach that makes risk more tangible, patients may be more motivated to engage in smoking cessation programs or undergo lung cancer screening.7 However, substantial evidence suggests the complex interplay of fear, fatalism, and risk perception when patients face lung cancer screening10, 11, 12, 13 Therefore, it is important to evaluate the translation potential of personalised genetics in motivating patients for positive behaviour change.
Ongoing studies like eMERGE (electronic MEdical Records and GEnomics),14 GenoVA (Genomic Medicine at Veterans Affairs),15 and WISDOM (Women Informed to Screen Depending on Measures of Risk)16 are leading the implementation of PRS into genetic risk reports (Table 1). These studies aim to personalise medical reports and assess the impact of PRS on screening, diagnostic procedures, and patient behaviour. While the global burden of lung cancer is largely driven by tobacco smoking,17,18 research studies have shown promise for PRS in lung cancer risk independent of traditional clinical risk models.19,20 The unique value proposition of a lung cancer-specific PRS lies in leveraging clear, guideline-based recommendations for smoking cessation and lung cancer screening.20
Table 1.
Research on PRS use in clinical trials.
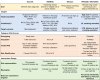 |
We compare the PRECISE and MOTIVATE trials, part of our GREAT framework, with existing PRS-informed trials: GenoVA, eMERGE, and WISDOM. Bolded text in the PRECISE/MOTIVATE column highlights the points where our trials differ from the current trials. Namely, the PRECISE and MOTIVATE trials investigate lung cancer and smoking and will focus on patients at high risk who are smokers or eligible for lung cancer screening. We also look at lung cancer screening, tobacco treatment, and smoking cessation as unique target outcomes. Finally, in addition to genetic and clinical risk messaging, the two trials have a unique emphasis on behaviour mechanisms around lung cancer and smoking.
∗ PCP, primary care provider.
∗∗ BrCa, breast cancer; PrCa, prostate cance; CRCa, colorectal cancer; Afib, atrial fibrillation; CAD, coronary artery/heart disease; T2D, type 2 diabetes; T1D, type 1 diabetes; BMI, body mass index/obesity; CKD, chronic kidney disease; HCL, hypercholesterolemia; LC, lung cancer.
Despite the potential benefits of PRS-based interventions, several practical challenges must be addressed for their effective implementation in the clinic. The include identifying the appropriate PRS to use, determining PRS cutoffs for defining patients at high-risk, understanding the corresponding odds ratios (ORs) for these patients, and standardizing PRS distributions across ancestries to ensure equitable application to diverse populations. Additionally, there remain significant barriers of effectively communicating genetic risk information to patients and ensuring that is understood and acted upon.
To address these challenges and provide practical guidance for the implementation for PRSs in primary care, we introduce the Genomic Informed Care for Motivating High Risk Individuals Eligible for Evidence-based Prevention (GREAT) framework. The GREAT framework integrates PRSs to stratify disease risk and personalise interventions. By delivering personalised risk information, our goal is to empower patients to make informed decisions about their health, thereby increasing the uptake of evidence-based treatments. This personalised information may increase treatment engagement by activating mechanisms based on the Capability-Opportunity-Motivation-Behaviour (COM-B) model and Theoretical Domains Framework (TDF).21, 22, 23
Our proposed approached will be implemented in two cluster randomised clinical trials, which will recruit individuals eligible for lung cancer screening and tobacco treatment. The first, PRECISE (“Precision Approaches to Lung Cancer Screening and Smoking Cessation Treatment in Primary Care”, NCT05627674), will evaluate the effectiveness of a multilevel intervention, RiskProfile, on increasing lung cancer screening and tobacco treatment utilization in primary care. The second trial, MOTIVATE (“Multilevel Intervention to Personalise and Improve Tobacco Treatment in Primary Care”, NCT05846841), will examine the effect of another multilevel intervention, PrecisionTx, on promoting precision tobacco treatment in primary care. These trials aim to demonstrate how personalised risk assessments can significantly improve lung cancer prevention strategies and patient outcomes through PRS-enabled interventions.
Methods
Study design
GREAT framework
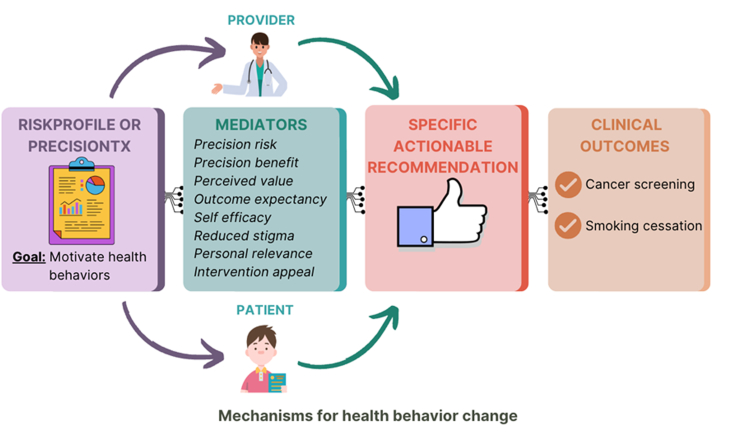
We have developed a GREAT framework to test whether personalised risk assessments can motivate high-risk patients who are eligible for cancer screening and tobacco treatment. The GREAT care paradigm is centred around multi-level intervention tools (RiskProfile and PrecisionTx) that utilise PRS to deliver precision patient risk (Fig. 1). Within the PRECISE and MOTIVATE trials, these PRS-based interventions will be communicated along with traditional clinical risk factors, such as smoking status and living with individuals who smoke.24, 25, 26 The primary goal is to promote positive clinical outcomes, such as increased lung cancer screening rates, enhanced tobacco treatment adherence, and successful smoking cessation within primary care settings. Throughout the trials, we will evaluate shared decision-making processes between patients and clinicians, focusing on actionable recommendations tailored to each patient's risk profile. Additionally, we will assess intermediate health behaviour change mechanisms, including perceived benefit, self-efficacy, and outcome expectancy.
Fig. 1.
Care Paradigm: Genomic Informed Care for Motivating High Risk Individuals Eligible for Evidence-based Prevention (GREAT). The GREAT framework is a primary care paradigm that integrates genetic and clinical risk in precision health. Individuals and their providers in two upcoming trials (PRECISE and MOTIVATE) are enrolled and provided with multilevel interventions (e.g. RiskProfile and PrecisionTx) to promote clinical outcomes of lung cancer screening, tobacco treatment, and successful smoking cessation in primary care settings. Mechanisms of health behaviour changes (e.g. perceived benefit, self-efficacy, and outcome expectancy) will be evaluated. During the specific actionable recommendations phase, personalised shared decision-making will be facilitated by multilevel actions between patients and clinicians for better clinical outcomes.
Translation of genetic risk to clinical practice
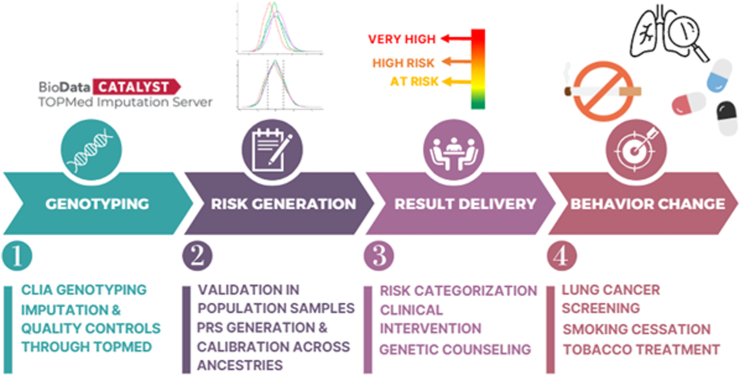
We present the GREAT paradigm within a broader roadmap for translating genetic risk into clinical practice (Fig. 2). This process begins with the Clinical Laboratory Improvement Amendments (CLIA) certified genotyping of enrolled participants' genetic data, conducted by 23andMe. To ensure the integrity and reliability of the genetic data, imputation and quality controls were performed using the Trans-Omics for Precision Medicine (TOPMed) server, which also facilitates the imputation of GWAS variants.
Fig. 2.
Roadmap for translating genetic data to a genetic risk profile as a multilevel intervention in primary care. In step 1, enrolled participants' genetic data are analysed by 23andMe's Clinical Laboratory Improvement Amendments (CLIA) certified genotyping process. Imputation and quality controls are conducted through the Trans-Omics for Precision Medicine (TOPMed) server to ensure the integrity and reliability of the genetic data, as well as to impute the GWAS variants. Step 2 involves identifying available GWAS variants and weights to create the raw Polygenic Risk Scores (PRS). The PRS is adjusted for genetic ancestry using reference data such as the 1000 Genomes Project Phase 3 and applied to validation data such as the UK Biobank to establish risk categories and compute ORs. In step 3, these scores are converted into 3 risk levels based on the established thresholds. In step 4, a report with precision treatment is created and communicated to both the participant and the provider to make informed and educated decisions. Behavioural interventionists (research staff who are trained, certified, and supervised by a team of genetic counsellor, psychologist, and psychiatrist) offer personalised guidance on behaviour change, leveraging the updated genetic insights. The outcome aims to increase lung cancer screening orders, improve participant adherence, promote smoking cessation, and highlight the benefits of tobacco treatment.
Next, available GWAS variants and their corresponding weights were identified to create raw PRSs. These PRSs were then adjusted for genetic ancestry using reference data, such as the 1000 Genomes Project Phase 3 (1000G), and applied to validation datasets like the UK Biobank (UKBB) to establish risk categories and compute ORs. The PRSs were converted into three distinct risk levels based on these established thresholds.
Finally, these risk profiles were integrated into a multi-level intervention strategy, where precision treatment reports were generated and communicated to both participants and their healthcare providers. These reports serve as a foundation for informed, shared decision-making, aimed at motivating health behaviour changes such as increased lung cancer screening, improved participant adherence, and successful smoking cessation. Behavioural interventionists—trained, certified, and supervised by a multidisciplinary team including genetic counsellors, psychologists, and psychiatrists—offer personalised guidance to support behaviour change, leveraging the updated genetic insights.
Ethics
This study was conducted using data from the Genetically Informed Smoking Cessation Trial (GISC),27 UKBB,28 AoU29 and 1000G.30,31 The 1000G was used for genetic ancestry inference. The GISC, UKBB, and AoU cohorts were used for PRS validation. The ethical approval for each dataset was obtained from the respective authorities as follows:
GISC
The study was approved by the Institutional Review Board at Washington University in St Louis (Reference Number: 201305128).
UKBB
Ethical approval was granted by the National Research Ethics Service Committee North West—Haydock (Reference Number: 11/NW/0382).
AoU
The study was approved by the Institutional Review Board of the All of Us Research Program. Detailed information can be found here.
1000G
Ethical approval was granted by the respective ethical review boards of the participating institutions. Detailed ethical considerations for each contributing institution can be found in the project's documentation.
For UKBB, AoU and GISC, all participants provided written informed consent at the time of enrolment, which includes consent for genetic and health data usage in various research purposes. Participants in the 1000G provided written informed consent, allowing their genetic data to be used for research.
Key datasets
1000 Genomes Project Phase 3 reference data and principal components analysis
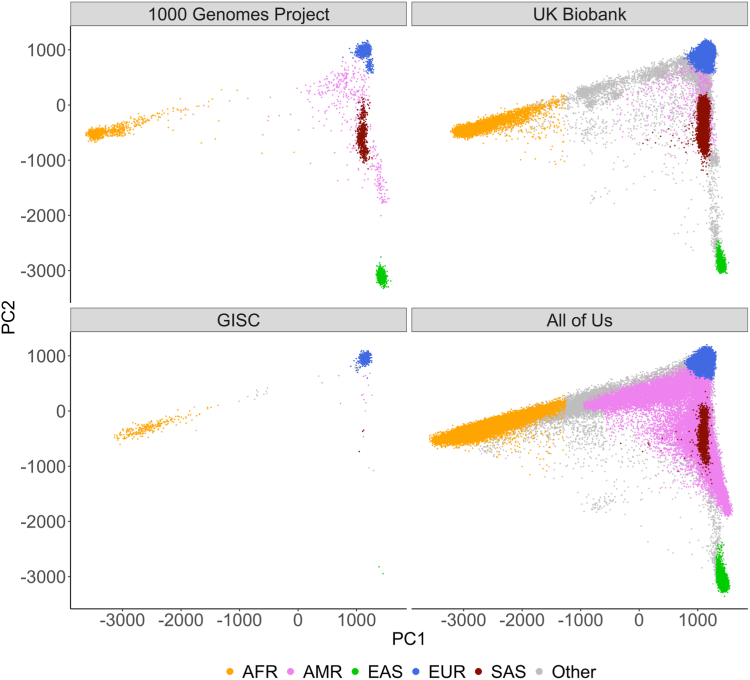
We use the 1000G30,31 as a reference for genetic ancestry inference, as it is publicly accessible and includes genotype data from diverse populations. The 1000G dataset includes 3202 individuals, with 633 Europeans (EUR), 893 Africans (AFR), 585 East Asians (EAS), 601 South Asians (SAS), and 490 Admixed Americans (AMR). We conducted principal components analysis (PCA) in PLINK 2.032 on all 3202 samples, using 55,248 single-nucleotide polymorphisms (SNPs) from the recommended SNPs set by gnomAD33 that were also found in the 1000G reference data, UKBB and GISC validation data, and 23andMe genotyping array used for the trial (Fig. 3, Supplementary Figure S1). From this PCA, we derived SNP weights that are used to compute ancestry PCs, so genotypes from different validation datasets could all be projected onto the same “PC space” (Supplementary Table S1).
Fig. 3.
Cross-dataset distribution of genetic ancestry via PCA Projections in 1000G, GISC, UKBB, and AoU. This figure illustrates the utility of principal components analysis (PCA) loadings obtained from the 1000 Genomes Project Phase 3 (1000G) in discriminating ancestries within external datasets, the Genetically Informed Smoking Cessation (GISC) trial. PCA was initially conducted on the globally diverse genotype data of 1000G. The resultant PCA-space was then used to project genotype data from GISC, UKBB, and AoU. The scatter plot displays the first and second PCs for each individual in these datasets, with points distinctly marked by genetically inferred ancestry.
We ran a random forest classifier to map individuals in 1000G to their respective populations using the first 5 PCs. We then projected our validation data to the 1000G-based “PC space” and applied the previously trained random forest classifier to produce “genetically inferred” ancestry labels. Individuals with a predicted probability less than 90% for any of the five ancestries were labelled as “Other.” While our proposed framework removes the need for labels in the clinic, we use them here for illustrative purposes in validation.
Study dataset and PRS validation
GISC trial
The GISC trial is a prospective, randomized, placebo-controlled trial conducted at Washington University in St. Louis, involving 822 current or previous smokers. Genetic data were available for 796 individuals, comprising 503 of EUR, 257 of AFR, and 36 individuals of “Other” self-reported ethnicity. The average age was 46.5 years (s.d. 11.3 years), with 54.4% female. While this dataset was not used for PRS prediction due to the small sample size and absence of lung cancer outcomes, it was utilized to validate the ancestry-adjustment procedure due to its resemblance to the expected populations in our PRECISE and MOTIVATE trials.
UKBB
The UKBB provides extensive genetic and clinical data from approximately 500,000 British individuals. Our analysis included 340,154 unrelated participants by genetically inferred ancestry: 6844 AFR, 730 AMR, 770 EAS, 313,279 EUR, 7197 SAS, and 11,334 Other. After excluding individuals with missing age or sex information, the average age of the cohort was 56.6 years (s.d. 8.2 years), with 54.1% female. The lung cancer analysis comprised 1830 cases (ICD10 codes C34.0–C34.9) and 338,334 controls, while the smoking cessation analysis involved 152,406 ever-smokers (117,483 former and 34,923 current). Sample characteristics regarding sex, age, and outcomes are provided in Supplementary Table S2a and b.
AoU
The AoU dataset is focused on participants from diverse and historically under-represented backgrounds, with comprehensive genetic and health record data. Our study included 210,826 unrelated individuals with whole-genome sequencing data, comprising 45,108 AFR, 32,563 AMR, 3873 EAS, 110,712 EUR, 1689 SAS, and 16,881 Other by genetically inferred ancestry. After excluding those with missing age or sex information, the average age was 55.3 years (s.d. 14.5 years), with 59.7% female. We identified 1020 lung cancer cases using SNOMED codes and excluded secondary cancer cases. For the smoking analysis, we included 36,507 current and 116,409 former smokers. Additional details regarding sex, age, and outcomes can be found in Supplementary Table 2c and d.
Validation of Ancestry-Adjustment Procedure
To evaluate the robustness of our PRS framework across different populations, we applied our procedure to the GISC, UKBB, and AoU datasets. The GISC trial, although not used for PRS prediction, was essential for validating the ancestry-adjustment procedure. In the UKBB and AoU datasets, we computed adjusted odds ratios (ORs) to assess the effectiveness of our risk stratification categories. Both datasets, with their rich genetic and clinical data, provided strong validation of our PRS model across diverse ancestry groups.
Statistics
Construction of polygenic risk scores
Our PRS models incorporate the latest findings by utilizing recently genome-wide association study (GWAS) summary statistics for lung cancer34 and difficulty quitting smoking.35 The lung cancer GWAS summary statistics were sourced from the International Lung Cancer Consortium (ILCCO), which includes 35,732 cases and 34,424 controls after excluding UKBB samples. The difficulty quitting smoking GWAS summary statistics were derived from the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN), comprising about 1,193,150 individuals overall with 373,510 current smokers. Both sets of summary statistics specifically exclude UKBB samples to avoid overlapping with our validation data. While these GWAS summary statistics are derived from predominantly European ancestry, they also include a substantial proportion of non-European ancestry — about 26% for lung cancer and 21% for difficulty quitting smoking — which enhances the generalizability of the findings.36,37
For lung cancer risk, we started with 128 published SNPs found to be predictive of 5-year and lifetime cumulative risk for lung cancer.20 Out of these, 101 SNPs overlapped with the published summary statistics, reference (1000G), and validation data (UKBB, GISC and AoU), and the 23andMe genotyping array used for the trial (Supplementary Figure S1). These SNPs were assigned effect sizes from the fixed-effect meta-analyses estimates in the latest lung cancer GWAS that includes EUR, AFR, and EAS ancestry.34 For difficulty quitting smoking, we identified 175 SNPs predictive for smoking cessation following the same filtering procedure for lung cancer.35
The PRS construction began with the alignment of genotype data to the summary statistics, ensuring consistent PRS regardless of allele coding. Specifically, for any SNP with reversed alleles, we recoded it as to avoid discrepancies. The raw PRS for an individual with SNPs was computed as
PRS calculations were performed using R, with genotype data input via the genio package.38 PRS SNPs and weights for lung cancer and difficulty quitting smoking are provided in Supplementary Tables S3–S4, respectively. We also used FAVOR (Functional Annotation of Variants Online Resources) to map each variant to a specific gene and functional annotation.39
Standardizing PRS distributions across the continuum of genetic ancestry
We standardised the PRS distributions for lung cancer and difficulty quitting smoking using the 1000G dataset, employing a regression-based method to adjust for distributional differences across ancestries (Supplementary Tables S5–S6).40 This adjustment process involves two key steps:
-
1.
Mean adjustment: we conducted a linear regression of the raw PRS against the top five PCs derived from the PCA:
We then computed residuals of the raw PRS that account for mean differences in PRS distributions across ancestry:
-
2.
Variance adjustment: using the square residuals as a proxy for PRS variance, we ran a secondary linear regression:
The final ancestry-adjusted PRS for each individual was then computed as:
This standardization resulted in a PRS distribution with mean 0 and variance 1 across ancestries, ensuring that genetic risk is accurately reflected independent of ancestry. This method is crucial for individuals with admixed or unknown ancestry, where discrete ancestry-specific models are inappropriate.41,42
Definition of patient risk categories
We converted continuous genetic risks to categorical risk levels to support clinical communication and actionability.43,44 The risk levels were designed for use in 2 ongoing trials in high-risk patients that evaluate the effect of personalised risks on motivating them to engage in cancer screening and tobacco treatment. Therefore, for lung cancer, we defined risk categories as the bottom 20%, middle 60%, and top 20% of the PRS distribution. These thresholds aligned with recent PRS risk stratification analyses for lung cancer.19 It is important to note that all patients who receive this intervention are already at risk, due to factors such as heavy smoking or family history. Therefore, the bottom 20% is categorised as “at risk”, the middle 60% as “at high risk”, and the top 20% as “at very high risk”, which is expected to have approximately two-fold increased risk compared to the “at risk” group.
For difficulty quitting smoking, which is a behavioural phenotype related to substance dependence, we opted for a more agnostic approach by dividing the PRS distribution into equal thirds (top, middle, and bottom 33.3%). Since all patients receiving this intervention are active smokers, the bottom 33.3% are categorised as “at risk”, with the middle and top 33.3% categorised as “at high risk” and “at very high risk”, respectively.
To establish standardised thresholds, we computed PRS and ancestry PCs in the 1000G reference dataset. After running our ancestry-adjustment procedure, we identified percentiles for each ancestry-adjusted PRS distribution, which were applied to our UKBB and AoU validation data. These thresholds can also be used in future clinical applications with new validation datasets.
Adjusted odds ratios of genetic risk
We quantified patient risk as ORs of each outcome (lung cancer and difficulty quitting smoking) for individuals at “high risk” and “very high risk”, relative to individuals in the “at risk” category. Patients enrolled in either of our trials are considered at risk—for instance, meeting eligibility criteria for lung cancer screening or active tobacco smokers.26 Therefore, we used the “at risk” category (i.e. “lowest genetic risk”) as the reference group to further identify individuals at particularly elevated genetic risk, who may benefit most from more comprehensive intervention and treatment strategies.
For each PRS model, we created a categorical variable PRScat taking values according to the three risk categories. Then, we ran logistic regressions with respect to PRScat, using “at risk” as the reference category. For smoking cessation analysis, we adjusted for age, sex, and 20 ancestry PCs. For lung cancer analysis, we additionally adjusted for smoking status (ever-smoker, never-smoker, or no-response). Each regression model yielded coefficients corresponding to “high risk” and “very high risk”, and adjusted ORs for each PRS category (relative to “at risk”) are reported as , along with corresponding 95% confidence intervals (CIs). These adjusted ORs will communicate the genetic risk of lung cancer or difficulty quitting smoking that is independent of the additional covariates. Within UKBB and AoU, we conducted these analyses across all ancestries combined, as well as within specific ancestry groups.
We also compared the risk stratification of our ancestry-adjusted PRS with “ancestry-matched” PRS. That is, we compared an individual's raw PRS with the corresponding ancestry-specific raw PRS distribution in 1000G, i.e. European-only 1000G PRS distribution for individuals of genetically predicted European ancestry. For individuals with “Other” predicted ancestry, we used the overall raw PRS distribution among all 1000G samples.
Role of funders
The funders played no role in the study design, analysis and collection of data, interpretation of results, or writing and submission of the paper.
Results
Harmonization PRS distributions across ancestry
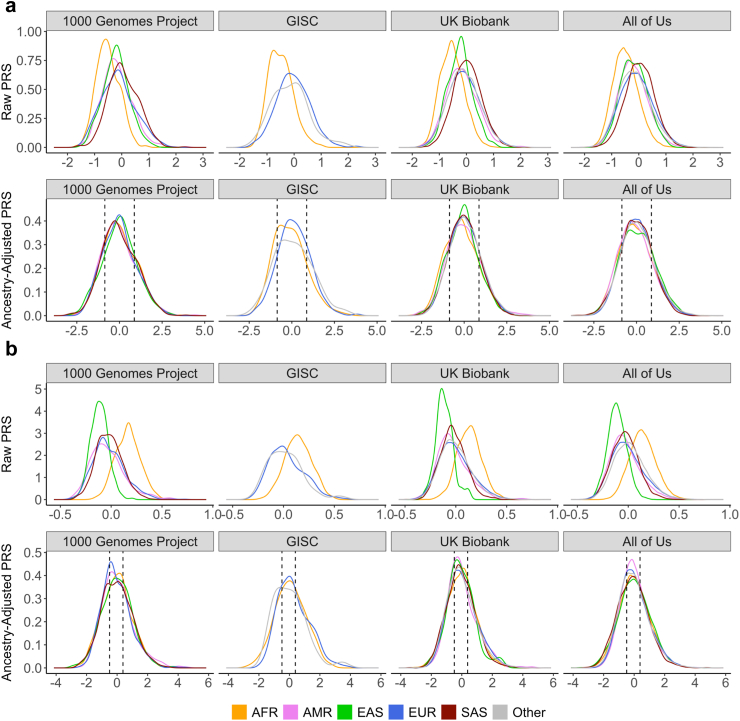
We found notable variation in raw PRS distributions across ancestries, highlighting that applying a universal cutoff for raw PRS without accounting for ancestry can lead to biased risk profiling and inaccurate clinical recommendations (Fig. 4). However, our regression-based ancestry adjustment across all three datasets yields much more standardised distributions across ancestries. Specifically, the adjusted proportions of individuals within each risk category closely align with 20%-60%-20% for lung cancer, and 33.3%-33.3%-33.3% for difficulty quitting smoking. This adjustment ensures that patients of any background can be compared against a unified reference distribution for each outcome (Supplementary Table S6a–f). This standardisation places individuals across all ancestries on the same scale, allowing for a single risk stratification cutoff regardless of ancestral background. Such measures importantly enable fair risk assessment for individuals who may be labelled as “Other”, who may not fit well into a binned ancestry-specific risk model.
Fig. 4.
Ancestry adjustment of PRS for lung cancer and quit difficulty PRS across ancestral populations. We showcase the adjustment process for PRS for (a) lung cancer and (b) difficulty quitting smoking within the 1000 Genomes Project, GISC Trial, UK Biobank, and All of Us datasets. It displays both raw and ancestry-adjusted PRS, with data points color-coded according to genetically inferred ancestries. EAS, AMR, and SAS ancestries were removed for GISC due to their small sample sizes. Ancestry adjustment effectively centres the PRS for different ancestries, mitigating the risk of incorrect stratification due to ancestry-related biases. Dotted vertical lines correspond to the 20th and 80th percentiles for lung cancer PRS distribution and 33rd and 67th percentiles for difficulty quitting smoking PRS among all 3202 samples in the 1000 Genomes Project.
Risk stratification for lung cancer and smoking cessation in UKBB and AoU
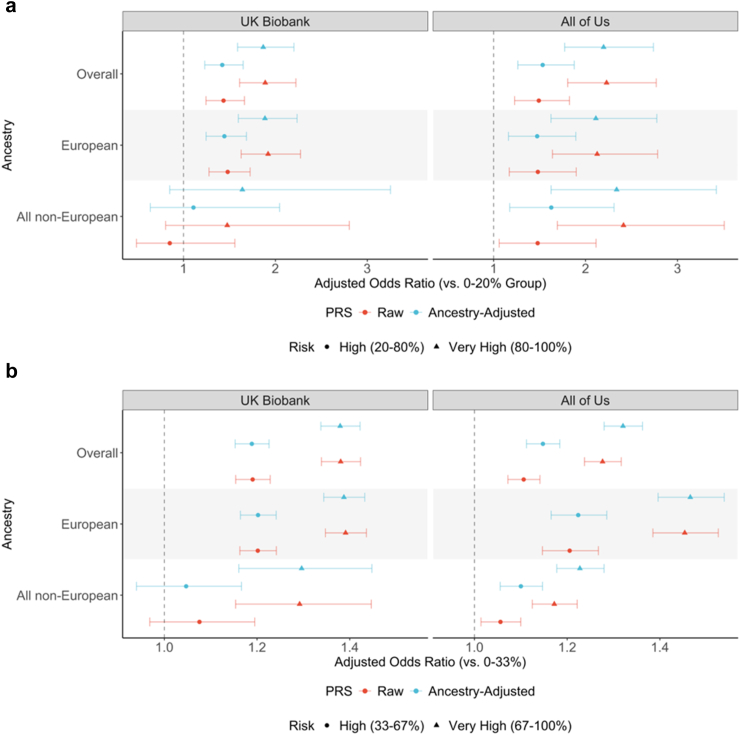
After assigning UKB and AoU participants as “at risk”, “high risk”, or “very high risk” for lung cancer and difficulty quitting smoking, we identified significant ORs for both traits across different ancestry groups (Fig. 5, Supplementary Tables S7–S10). Our ancestry-adjusted PRS yielded similar ORs as using ancestry-matched distributions, demonstrating that a single PRS distribution can be appropriately applied to all individuals. In UKBB, the overall adjusted ORs for lung cancer 1.42 (95% CI: 1.23–1.65) for the “high risk” group and 1.87 (95% CI: 1.59–2.20) for “very high risk” group compared to the “at risk” group (Supplementary Table S7a). In AoU, these adjusted ORs were slightly higher—1.49 (95% CI: 1.23–1.83) for “high risk” group and 2.23 (95% CI: 1.81–2.77) for “very high risk” group. While the adjusted ORs in non-European ancestries within UKBB were not significant, they were significant and even higher than those for European ancestries within AoU (Supplementary Table S8a). Moreover, the ORs for “Other” ancestry group in AoU were much higher and more significant using the ancestry-adjusted PRS compared to raw PRS, where an ancestry-matched risk distribution is least suitable (Supplementary Table S8b).
Fig. 5.
Risk stratification for lung cancer and difficulty quitting smoking using raw and ancestry-adjusted PRS. This figure illustrates adjusted odds ratios with associated 95% confidence intervals of PRSs for (a) lung cancer and (b) difficulty quitting smoking among UK Biobank (N = 340,154 for lung cancer and N = 152,406 for difficulty quitting smoking), and All of Us (N = 210,826 for lung cancer and N = 152,916 for difficulty quitting smoking) participants. For difficulty quitting smoking, we adjusted for age, sex, and 20 ancestry PCs. For lung cancer, we additionally adjusted for smoking status (ever-smoker, never-smoker, or no-response). We compared risk stratification using a raw PRS with ancestry-matched percentiles, and our ancestry-adjusted PRS with the same percentiles for all individuals.
We observed similar patterns for difficulty quitting smoking, where the ancestry-adjusted PRS performed similar or better than using ancestry-matched distributions for raw PRS. In UKBB, the overall adjusted ORs for difficulty quitting smoking were 1.19 (95% CI: 1.15–1.23) for “high risk” group and 1.38 (95% CI: 1.34–1.42) for “very high risk” group (Supplementary Table S9a). In AoU, the adjusted ORs were 1.19 (95% CI: 1.15–1.23) for “high risk” group and 1.38 (95% CI: 1.34–1.42) for “very high risk” (Supplementary Table S10a) group. Notably, we observed slightly higher and more significant ORs in non-European AoU participants when using the ancestry-adjusted PRS compared to the raw PRS. Although the improvement within each specific ancestry group was not as substantial, the aggregate ORs in all non-European groups increased from 1.06 (95% CI: 1.01–1.10) to 1.10 (95% CI: 1.06–1.15) in the “high risk” group, and from 1.17 (95% CI: 1.12–1.22) to 1.23 (95% CI: 1.18–1.28) in the “very high risk” group.
Using ancestry-adjusted PRS ensures accurate risk stratification across all ethnic backgrounds, a critical consideration given the substantial variability in raw PRS distributions across diverse populations. The outcome-based validation in UKBB and AoU further verifies that the ancestry-adjusted PRS provides valid risk stratification, and often yields better risk stratification for non-European ancestries compared to ancestry-matched modelling. These findings collectively facilitate a more robust and standardised application of PRS in clinical reporting.
Translating genetic risk into clinical reports
We have implemented our analytic framework in two recently launched trials—PRECISE and MOTIVATE—which are currently in the preliminary phases of recruitment and aim to engage over 100 physicians and 1600 patients. These trials are designed to promote health behaviour change using genetically informed multi-level interventions, RiskProfile and PrecisionTx, respectively. These interventions incorporate PRS to communicate precision risk of lung cancer and precision benefits of smoking cessation, promoting evidence-based practices such as cancer screening and tobacco treatment in individuals who smoke and/or are eligible for lung cancer screening. Based on functional information from FAVOR, our lung cancer PRS contains putative loss-of-function variants in genes such as CHRNA545 and CHEK2 (Checkpoint kinase 2),46 which are known to be involved in nicotine addiction and DNA damage repair, respectively (Supplementary Table S3). Our PRS for difficulty quitting does not contain similar loss-of-function variants, but still includes variants in smoking-related genes such as DRD2 (Dopamine receptor D2)47 and UBXN2A (UBX domain 2A),48 which are associated with dopaminergic signaling and stress response mechanisms (Supplementary Table S4). These biological insights reinforce the medical relevance of our PRS models by linking genetic variants to key pathways involved in smoking-related diseases.
Most preventive practices such as lung cancer screening and tobacco treatment are still largely underutilised in primary care. Thus, we aim to evaluate whether personalised genetic risk increases provider practices on and patient engagement of these preventive practices. Access to 23andMe genotypes and expanded health information has been a motivating component for the research participants. Once patients provide samples for genotyping, we generate personalised risk profiles for both primary care providers and patients to support behaviour change. We inform patients about their genetic risk category according to the ancestry-adjusted PRS distributions, along with the ORs relative to the “at risk” group. We also provide personalised clinical risks using independent risk models and actionable recommendations to motivate cancer screening and tobacco treatment (Fig. 6, Fig. 7).24, 25, 26 This approach is designed to integrate seamlessly into routine diagnostic workflows, enhancing the utilization of preventive services in primary care.
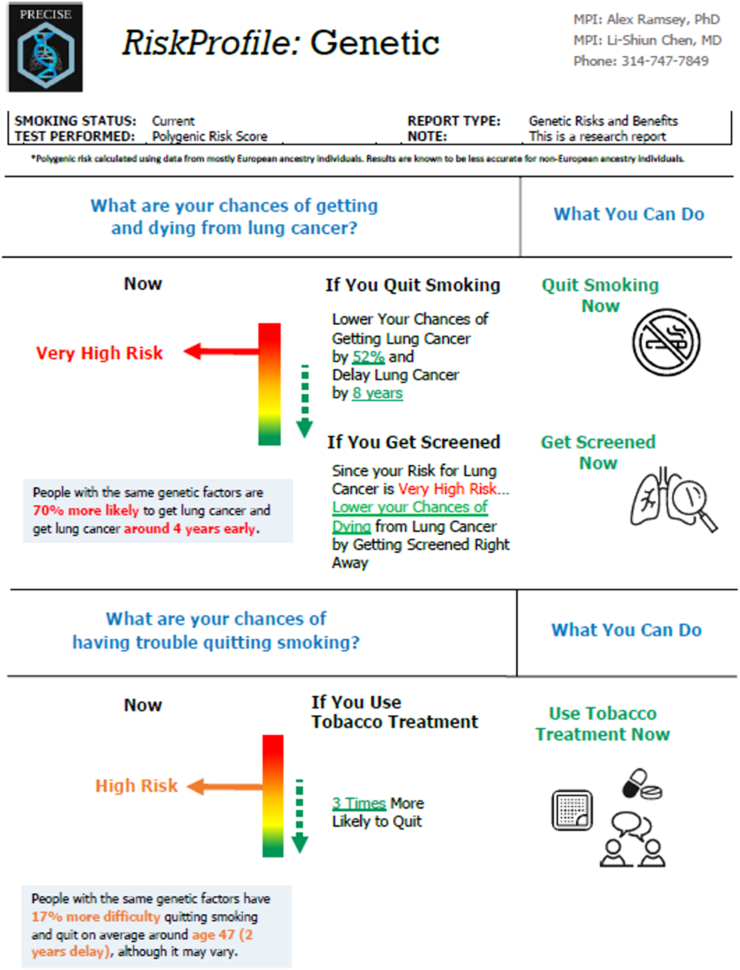
Fig. 6.
Example 1 clinical report: RiskProfile. We present example 1 for genomically informed interventions using the GREAT framework. RiskProfile is designed to motivate lung cancer screening and tobacco treatment among screening-eligible patients. This intervention utilises ancestry-adjusted PRS to stratify patients into “at risk” (yellow), “high risk” (orange), and “very high risk” (red) genetic risk categories. RiskProfile focuses on prevention and expands beyond personalised risk to also provide personalised benefit of cancer screening and use a multilevel intervention design directed to both physicians and patients in clinical settings. In our PRECISE trial (NCT05627674), the effect of RiskProfile on clinician ordering and patient completion of lung cancer screening will be evaluated.
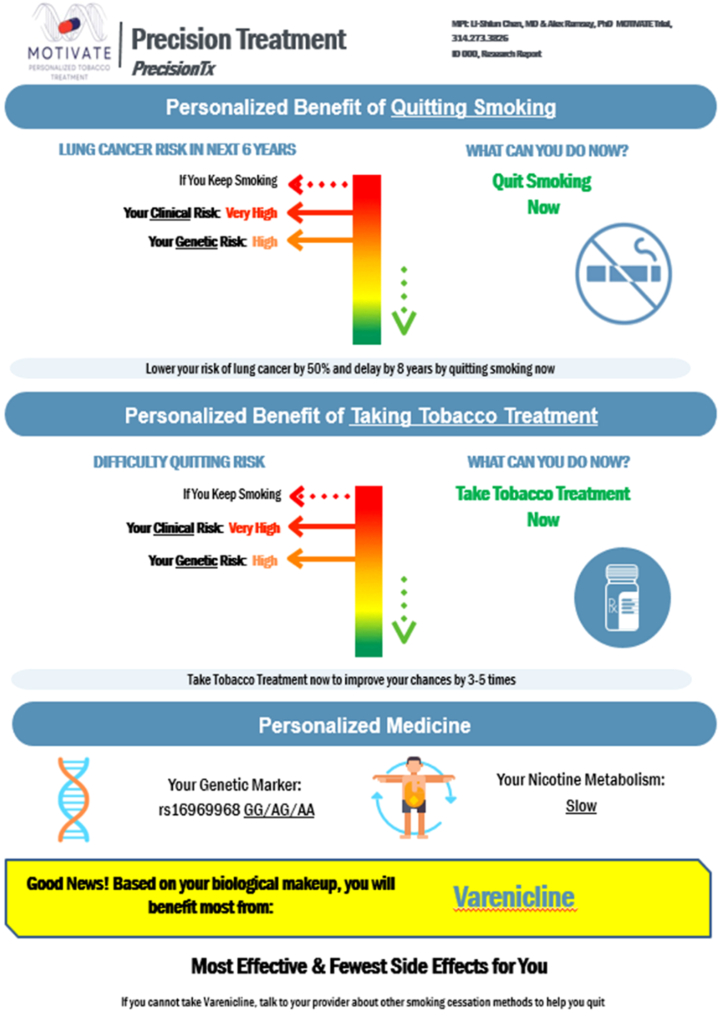
Fig. 7.
Example 2 clinical report: PrecisionTx. We present example 2 for genomically informed interventions using the GREAT framework. PrecisionTx is designed to motivate tobacco treatment among patients who smoke. This intervention utilises ancestry-adjusted PRS to stratify patients into “at risk” (yellow), “high risk” (orange), and “very high risk” (red) genetic risk categories. PrecisionTx focuses on treatment and expands beyond personalised risk to also provide personalised benefit of tobacco treatment and use a multilevel intervention design directed to both physicians and patients in clinical settings. In our MOTIVATE trial (NCT05846841), the effect of PrecisionTx on clinician ordering, patient adherence, and smoking abstinence will be evaluated.
Through our approach, we propose clear and patient-friendly communication strategies, including visual aids and educational materials, to facilitate understanding and meaningful interactions between patients and healthcare providers. Effectively communicating both the risk and precision of the PRS results is challenging but essential to empower patients to make informed decisions about their health. Moreover, it is crucial to consider patient perceived risk, perceived benefit, and personal relevance when discussing PRS results with patients. Patients' understanding and interpretation of PRS may vary, leading to differing levels of engagement in preventive actions. Hence, comprehensive patient education programs can enhance awareness and knowledge about PRS, its implications, and available preventive measures.
Discussion
In this study, we introduce a translational roadmap and analytical framework for implementing PRS within multilevel interventions to communicate precision risk and benefit and ultimately promote health behaviour change. Specifically, we frame our translational message specifically for patients at high risk who have not received guideline-recommended cancer screening or tobacco treatment,49, 50, 51, 52 taking special care to ensure inclusion of and fair risk assessment across diverse ancestries via PC-regression-based PRS adjustment. This framework will be evaluated in two cluster-randomised trials to evaluate the effectiveness of comprehensive risk profiles in motivating positive health decision-making from both patients and primary care providers.
A key feature of our framework is standardisation of PRS distributions across diverse ancestries using widely accessible data from the 1000G dataset, as an alternative to methods in the GenoVA (15) and eMERGE (14) studies that use data from the Mass-General Brigham Biobank and AoU, respectively. We validated the transferability of our 1000G-based standardization in external datasets from the UKBB, GISC and AoU, allowing future trials to adopt a similar methodology irrespective of their specific genetic data. Furthermore, from large-scale biobank analysis, we verified that our ancestry-adjusted PRSs achieved similar or better risk stratification than naïve ancestry-matching, which importantly enables the inclusion of individuals of mixed or uncertain ancestry. By utilizing our provided PC loadings and PRS standardization formula for lung cancer and difficulty quitting smoking, new patients in these trials can receive accurate risk categorization reports, bypassing the inaccuracies of self-reported ethnicity and the need for re-training PCA models.
Unlike most current research that evaluates PRS-enabled interventions in general patient populations, our work uniquely focuses on designing and evaluating these interventions specifically among patients already at high risk due to factors like smoking or family history. While these individuals will benefit tremendously from lung cancer screening and smoking cessation, they may not be fully motivated to quit smoking or make other positive health changes from general medical advice alone. Therefore, we believe that within this context of a high-risk patient population, introducing an additional dimension of genetic risk may further encourage individuals to follow through with cancer screening or tobacco treatment. Importantly, we aim to follow best practices of communicating uncertainties and potential imprecision in risk estimates and thresholds to maintain transparency with patients.
Our work has several limitations, but we hope to contribute to the knowledge pool for the best practices in creating PRS-enabled interventions that may be disease-, population-, or context-specific. Our approach is tailored for unique outcomes, populations, and contexts to optimise health impact. While our framework is broadly applicable within primary care, the specific interplay between genetic and clinical risk, as well as modes of communicating and addressing patient risk, will vary across specific diseases and contexts.53,54 For example, the individuals enrolled in our two trials share similar clinical profiles in line with eligibility for lung cancer screening and tobacco treatment. For other outcomes or populations of interest, domain expertise to identify the key baseline characteristics for high clinical risk is crucial to best communicate comprehensive risk and expected benefits from behaviour change.
Our PRS model also has two key limitations. First is the underrepresentation of non-European populations in the multi-ancestry GWAS used to derive the PRS weights, a challenge that persists across existing GWASs,55, 56, 57, 58 not limited to lung cancer and smoking cessation. This underrepresentation may reduce the predictive power of the PRS in non-European populations. However, with ongoing efforts to recruit individuals of diverse ancestry in genetic studies, we expect this challenge to be resolved in the coming years.
In addition, our current PRS models were constructed from a small set of significant predictive variants. While the PRSs include variants within genes associated with lung cancer and smoking,45, 46, 47, 48 only a small number were coding. Incorporating more biological information59,60 into the model-building process, beyond evaluating genome-wide significance, may further enhance the interpretability and accuracy of the genetic risk models. Furthermore, with the development of new PRS approaches and toolkits to enhance predictive power in diverse populations from multi-ancestry data,42,61, 62, 63, 64 we can iteratively refine PRS implementation in our trial to synchronise with the latest advancements. Maintaining a dynamic PRS framework that aligns with the latest advancements will reduce the implementation gap and maximise impact in preventive healthcare outcomes. Following current genetic counselling recommendations, we have established a process to incorporate new evidence into our intervention for smoking cessation and lung cancer risk. This process will adjudicate new population-specific evidence on genetics and biomarkers, evaluating its impact on personal and population-level risk changes, and effectively communicating the dynamic nature of genetic evidence to patients and providers.
Looking to the future, we must also consider the scalability of precision interventions to real-world clinics and studies. A notable gap in current practice is the absence of genetic information in electronic health records (EHRs) for decision support and the lack of PRS generation in clinical labs. Implementing precision interventions in primary care necessitates a workflow that incorporates EHRs for recruitment, biomarker testing protocols, and standardised processes to generate personalised intervention reports.65 This requires collaborations with primary care stakeholders, community advisory boards, genetic counselling, and health communication to improve intervention clarity, accuracy, and impact.8,9,66, 67, 68 To reduce burden, we need to leverage existing EHR tools (e.g. Best Practice Advisories) and training to efficiently facilitate physician prescribing.69,70 Understanding of mechanistic and implementation outcomes will guide scalable, efficient delivery components for integration into clinic workflows,65 using trained embedded staff, and digital therapeutic tools to enable these PRS-informed behavioural interventions.71
In conclusion, we provide a roadmap that incorporates PRSs using multi-ancestry GWAS-based weights, translates risk into actionable categories, communicates comprehensive risk effectively, considers patient perspectives, and accommodates evolving science is essential for the equitable and pragmatic translation of PRS into clinical care. By addressing the barriers and implementing potential solutions at each stage, we can leverage PRS to improve preventive healthcare and significantly reduce the burden of lung cancer.
Contributors
T.C., H.Z., and L.C. conceived the project, T.C.; G.P., and L.F. carried out all data analyses under the supervision of H.Z. and L.C.; N.A. and X.W. organised data in AoU; G.S. and D.J. provided GWAS summary statistics for difficulty quitting smoking. T.C.; G.P., H.Z., and L.C. drafted the manuscript. All authors reviewed and approved the final version of the manuscript.
Data sharing statement
Genotype data from 1000G are publicly available and can be directly downloaded from the following links:
1. Genotype data: https://www.cog-genomics.org/plink/2.0/resources.
2. Population information: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/.
UKBB genotype and phenotype data are available via application through the UK Biobank website: https://www.ukbiobank.ac.uk/.
AoU genotype and phenotype data are available via application through the All of Us Research Program: https://www.researchallofus.org/.
GISC data is available via application through NIDA Center for Genetic Studies: https://nidagenetics.org/
No additional data were collected for this study. All results are presented in the paper's tables and figures.
Code sharing
R code and plink commands, as well as accompanying data, used for analysis are provided in a walkthrough available on GitHub at https://github.com/chen-tony/GREAT.
Declaration of interests
Laura J. Bierut (LJB) is listed as an inventor on Issued U.S. Patent 8,080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction, LJB receives consulting fees from Research Triangle Institute for grant R01DA048824 “Identifying blood-based DNA methylation biomarkers of cannabis use” is a member of US Food and Drug Administration Tobacco Products Scientific Advisory Committee, and co-chair of National Comprehensive Cancer Network Smoking Cessation Panel. Michael J. Bray (MJB) was an employee at ThinkGenetic, Inc, where he had the option to receive stock options at the time the work was conducted. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
All other authors have no conflict of interests to report.
Acknowledgements
We would like to thank Reeya Joseph for her editorial support with the introduction, Peter Kraft for his advice on our manuscript, and Scott Vrieze for his assistance with summary statistics for difficulty quitting smoking. We would also like to thank and acknowledge the participants enrolled in the UK Biobank (obtained under UK Biobank resource application 52008) and GISC trial for contributing vital data to this work.
This research was supported by NIH Training Grant T32GM135117 and NSF Graduate Research Fellowship DGE-2140743 (T.C.), R01HG011035, R01ES036042, R01HL173869 (D.L.), National Cancer Institute (NCI) R01CA268030, NIDA R01DA056050, National Institute on Drug Abuse (NIDA) R34DA052928, NIDA K12DA041449, Taylor Family Institute for Innovative Psychiatric Research (A.T.R.), NCI R01-CA268030, NIDA R01-DA056050 (L.J.B.), R35-3CA197449, R01-HL163560, U01-HG009088, and U01-HG012064 (X.L.), NIH Intramural Research Program (H.Z.), NIH 5T32-HL007776–25, R01-DA056050, R01-CA268030, P30-CA091842-19S5, P30-CA091842-16S2 and P50-CA244431 (L.C.) and NCI grant U19-CA203654 (Integrative Analysis of Lung Cancer Etiology and Risk Application and Translation).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105441.
Contributor Information
Tony Chen, Email: tonychen@g.harvard.edu.
Haoyu Zhang, Email: haoyu.zhang2@nih.gov.
Li-Shiun Chen, Email: li-shiun@wustl.edu.
Appendix A. Supplementary data
References
- 1.Kocarnik J.M., Compton K., Dean F.E., et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019. JAMA Oncol. 2022;8(3):420–444. doi: 10.1001/jamaoncol.2021.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., Romero-Gutierrez C.W., Kothari J., Shafer A., Li Y., Christiani D.C. Prediagnosis smoking cessation and overall survival among patients with non–small cell lung cancer. JAMA Netw Open. 2023;6(5) doi: 10.1001/jamanetworkopen.2023.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twyman L., Bonevski B., Paul C., Bryant J. Perceived barriers to smoking cessation in selected vulnerable groups: a systematic review of the qualitative and quantitative literature. BMJ Open. 2014;4(12) doi: 10.1136/bmjopen-2014-006414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 5.Lewis C.M., Vassos E. BioMed Central Ltd.; 2020. Polygenic risk scores: from research tools to clinical instruments. Vol. 12, Genome medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adeyemo A., Balaconis M.K., Darnes D.R., et al. Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nat Med. 2021;27:1876–1884. doi: 10.1038/s41591-021-01549-6. [DOI] [PubMed] [Google Scholar]
- 7.Chiu A., Hartz S., Smock N., et al. Most current smokers desire genetic susceptibility testing and genetically-efficacious medication. J Neuroimmune Pharmacol. 2018;13(4):430–437. doi: 10.1007/s11481-018-9818-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsey A.T., Bray M., Acayo Laker P., et al. Participatory design of a personalized genetic risk tool to promote behavioral health. Cancer Prev Res. 2020;13(7):583–592. doi: 10.1158/1940-6207.CAPR-20-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsey A.T., Bourdon J.L., Bray M., et al. Proof of concept of a personalized genetic risk tool to promote smoking cessation: high acceptability and reduced cigarette smoking. Cancer Prev Res. 2021;14(2):253–262. doi: 10.1158/1940-6207.CAPR-20-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quaife S.L., Janes S.M., Brain K.E. The person behind the nodule: a narrative review of the psychological impact of lung cancer screening. Transl Lung Cancer Res. 2021;10(5):2427–2440. doi: 10.21037/tlcr-20-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quaife S.L., Waller J., Dickson J.L., et al. Psychological targets for lung cancer screening uptake: a prospective longitudinal cohort study. J Thorac Oncol. 2021;16(12):2016–2028. doi: 10.1016/j.jtho.2021.07.025. [DOI] [PubMed] [Google Scholar]
- 12.Quaife S.L., Marlow L.A.V., McEwen A., Janes S.M., Wardle J. Attitudes towards lung cancer screening in socioeconomically deprived and heavy smoking communities: informing screening communication. Health Expect. 2017;20(4):563–573. doi: 10.1111/hex.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quaife S.L., McEwen A., Janes S.M., Wardle J. Smoking is associated with pessimistic and avoidant beliefs about cancer: results from the International Cancer Benchmarking Partnership. Br J Cancer. 2015;112(11):1799–1804. doi: 10.1038/bjc.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder J.E., Allworth A., Bland H.T., et al. Returning integrated genomic risk and clinical recommendations: the eMERGE study. Genet Med. 2023;25(4) doi: 10.1016/j.gim.2023.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao L., Kraft P., Berriz G.F., et al. Development of a clinical polygenic risk score assay and reporting workflow. Nat Med. 2022;28(5):1006–1013. doi: 10.1038/s41591-022-01767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shieh Y., Eklund M., Madlensky L., et al. Breast cancer screening in the precision medicine era: risk-based screening in a population-based trial. J Natl Cancer Inst. 2017;109(5):djw290. doi: 10.1093/jnci/djw290. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P., Chen P.L., Li Z.H., et al. Association of smoking and polygenic risk with the incidence of lung cancer: a prospective cohort study. Br J Cancer. 2022;126(11):1637–1646. doi: 10.1038/s41416-022-01736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanwal M., Ding X.J., Cao Y. Familial risk for lung cancer. Oncol Lett. 2017;13(2):535–542. doi: 10.3892/ol.2016.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu M., Lv J., Huang Y., et al. Ethnic differences of genetic risk and smoking in lung cancer: two prospective cohort studies. Int J Epidemiol. 2023;52(6):1815–1825. doi: 10.1093/ije/dyad118. [DOI] [PubMed] [Google Scholar]
- 20.Hung R.J., Warkentin M.T., Brhane Y., et al. Assessing lung cancer absolute risk trajectory based on a polygenic risk model. Cancer Res. 2021;81(6):1607–1615. doi: 10.1158/0008-5472.CAN-20-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michie S. Making psychological theory useful for implementing evidence based practice: a consensus approach. Qual Saf Health Care. 2005;14(1):26–33. doi: 10.1136/qshc.2004.011155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cane J., O'Connor D., Michie S. Validation of the theoretical domains framework for use in behaviour change and implementation research. Implement Sci. 2012;7(1):37. doi: 10.1186/1748-5908-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atkins L., Francis J., Islam R., et al. A guide to using the Theoretical Domains Framework of behaviour change to investigate implementation problems. Implement Sci. 2017;12(1):77. doi: 10.1186/s13012-017-0605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray M., Chang Y., Baker T.B., et al. The promise of polygenic risk prediction in smoking cessation: evidence from two treatment trials. Nicotine Tob Res. 2022;24(10):1573–1580. doi: 10.1093/ntr/ntac043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L.S., Baker T.B., Piper M.E., et al. Interplay of genetic risk (CHRNA5) and environmental risk (partner smoking) on cigarette smoking reduction. Drug Alcohol Depend. 2014;143:36–43. doi: 10.1016/j.drugalcdep.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tammemägi M.C., Katki H.A., Hocking W.G., et al. Selection criteria for lung-cancer screening. N Engl J Med. 2013;368(8):728–736. doi: 10.1056/NEJMoa1211776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L., Baker T.B., Miller J.P., et al. Genetic variant in CHRNA5 and response to varenicline and combination nicotine replacement in a randomized placebo-controlled trial. Clin Pharmacol Ther. 2020;108(6):1315–1325. doi: 10.1002/cpt.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudlow C., Gallacher J., Allen N., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bick A.G., Metcalf G.A., Mayo K.R., et al. Genomic data in the all of us research program. Nature. 2024;627:340–346. doi: 10.1038/s41586-023-06957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auton A., Abecasis G.R., Altshuler D.M., et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrska-Bishop M., Evani U.S., Zhao X., et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022;185(18):3426–3440.e19. doi: 10.1016/j.cell.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S., Francioli L.C., Goodrich J.K., et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625(7993):92–100. doi: 10.1038/s41586-023-06045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byun J., Han Y., Li Y., et al. Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer. Nat Genet. 2022;54(8):1167–1177. doi: 10.1038/s41588-022-01115-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders G.R.B., Wang X., Chen F., et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature. 2022;612(7941):720–724. doi: 10.1038/s41586-022-05477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin A.R., Gignoux C.R., Walters R.K., et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100(4):635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manrai A.K., Funke B.H., Rehm H.L., et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochoa A. Genio: genetics input/output functions. 2023. https://CRAN.R-project.org/package=genio Available from:
- 39.Zhou H., Arapoglou T., Li X., et al. FAVOR: functional annotation of variants online resource and annotator for variation across the human genome. Nucleic Acids Res. 2023;51(D1):D1300–D1311. doi: 10.1093/nar/gkac966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge T., Irvin M.R., Patki A., et al. Development and validation of a trans-ancestry polygenic risk score for type 2 diabetes in diverse populations. Genome Med. 2022;14(1):70. doi: 10.1186/s13073-022-01074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis A.C.F., Molina S.J., Appelbaum P.S., et al. Getting genetic ancestry right for science and society. Science. 2022;376(6590):250–252. doi: 10.1126/science.abm7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kachuri L., Chatterjee N., Hirbo J., et al. Principles and methods for transferring polygenic risk scores across global populations. Nat Rev Genet. 2023;24 doi: 10.1038/s41576-023-00637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belkora J., Moore D.H., Hutton D.W. Assessing risk communication in breast cancer: are continuous measures of patient knowledge better than categorical? Patient Educ Couns. 2009;76(1):106–112. doi: 10.1016/j.pec.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lautenbach D.M., Christensen K.D., Sparks J.A., Green R.C. Communicating genetic risk information for common disorders in the era of genomic medicine. Annu Rev Genomics Hum Genet. 2013;14(1):491–513. doi: 10.1146/annurev-genom-092010-110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krais A.M., Hautefeuille A.H., Cros M.P., et al. CHRNA5 as negative regulator of nicotine signaling in normal and cancer bronchial cells: effects on motility, migration and p63 expression. Carcinogenesis. 2011;32(9):1388–1395. doi: 10.1093/carcin/bgr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cybulski C., Masojc B., Oszutowska D., et al. Constitutional CHEK2 mutations are associated with a decreased risk of lung and laryngeal cancers. Carcinogenesis. 2008;29(4):762–765. doi: 10.1093/carcin/bgn044. [DOI] [PubMed] [Google Scholar]
- 47.Comings D.E., Ferry L., Bradshaw-Robinson S., Burchette R., Chiu C., Muhleman D. The dopamine D2 receptor (DRD2) gene: a genetic risk factor in smoking. Pharmacogenetics. 1996;6(1):73–79. doi: 10.1097/00008571-199602000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Teng Y., Rezvani K., De Biasi M. UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP. Biochem Pharmacol. 2015;97(4):518–530. doi: 10.1016/j.bcp.2015.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.US preventive services task force. 2021. https://www.uspreventiveservicestaskforce.org/uspstf/recommendation/lung-cancer-screening Lung Cancer.
- 50.Agency for healthcare research and quality. 2021. https://www.ahrq.gov/prevention/guidelines/index.html [Clinical Guidelines and Recommendations] [DOI] [PubMed] [Google Scholar]
- 51.Tobacco Use and Dependence Guideline Panel. US Department of Health and Human Services . US Department of Health and Human Services; Rockville, MD: 2008. Tobacco use and dependence guideline Panel. Treating tobacco use and dependence: 2008 update. [Google Scholar]
- 52.Krist A.H., Davidson K.W., Mangione C.M., et al. Screening for lung cancer. JAMA. 2021;325(10):962–970. doi: 10.1001/jama.2021.1117. [DOI] [PubMed] [Google Scholar]
- 53.Arem H., Loftfield E. Cancer epidemiology: a survey of modifiable risk factors for prevention and survivorship. Am J Lifestyle Med. 2018;12(3):200–210. doi: 10.1177/1559827617700600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran K.B., Lang J.J., Compton K., et al. The global burden of cancer attributable to risk factors, 2010–19: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2022;400(10352):563–591. doi: 10.1016/S0140-6736(22)01438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson R.E., Kuchenbaecker K., Walters R.K., et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179(3):589–603. doi: 10.1016/j.cell.2019.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538(7624):161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Need A.C., Goldstein D.B. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25(11):489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 58.Fitipaldi H., Franks P.W. Ethnic, gender and other sociodemographic biases in genome-wide association studies for the most burdensome non-communicable diseases: 2005–2022. Hum Mol Genet. 2023;32(3):520–532. doi: 10.1093/hmg/ddac245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long E., Patel H., Golden A., et al. High-throughput characterization of functional variants highlights heterogeneity and polygenicity underlying lung cancer susceptibility. Am J Hum Genet. 2024;111(7):1405–1419. doi: 10.1016/j.ajhg.2024.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G., Zhou H., Strulovici-Barel Y., et al. Role of OSGIN1 in mediating smoking-induced autophagy in the human airway epithelium. Autophagy. 2017;13(7):1205–1220. doi: 10.1080/15548627.2017.1301327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H., Zhan J., Jin J., et al. A new method for multiancestry polygenic prediction improves performance across diverse populations. Nat Genet. 2023;25 doi: 10.1038/s41588-023-01501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin J., Zhan J., Zhang J., et al. MUSSEL: enhanced Bayesian polygenic risk prediction leveraging information across multiple ancestry groups. Cell Genomics. 2024;4(4) doi: 10.1016/j.xgen.2024.100539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J., Zhan J., Jin J., et al. An ensemble penalized regression method for multi-ancestry polygenic risk prediction. Nat Commun. 2024;15(1):3238. doi: 10.1038/s41467-024-47357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hou K., Gogarten S., Kim J., et al. Admix-kit: an integrated toolkit and pipeline for genetic analyses of admixed populations. Bioinformatics. 2024;40(4) doi: 10.1093/bioinformatics/btae148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayatollahi H., Hosseini S.F., Hemmat M. Integrating genetic data into electronic health records: medical geneticists' perspectives. Healthc Inform Res. 2019;25(4):289. doi: 10.4258/hir.2019.25.4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bourdon J.L., Dorsey A., Zalik M., et al. In-vivo design feedback and perceived utility of a genetically-informed smoking risk tool among current smokers in the community. BMC Med Genomics. 2021;14(1):139. doi: 10.1186/s12920-021-00976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramsey A.T., Chen L.S., Hartz S.M., et al. Toward the implementation of genomic applications for smoking cessation and smoking-related diseases. Transl Behav Med. 2018;8(1):7–17. doi: 10.1093/tbm/ibx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L.S., Baker T.B., Ramsey A., Amos C.I., Bierut L.J. Genomic medicine to reduce tobacco and related disorders: translation to precision prevention and treatment. Addiction Neuroscience. 2023;7 doi: 10.1016/j.addicn.2023.100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L.S., Baker T.B., Korpecki J.M., et al. Low-burden strategies to promote smoking cessation treatment among patients with serious mental illness. Psychiatr Serv. 2018;69(8):849–851. doi: 10.1176/appi.ps.201700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramsey A.T., Chiu A., Baker T., et al. Care-paradigm shift promoting smoking cessation treatment among cancer center patients via a low-burden strategy, Electronic Health Record-Enabled Evidence-Based Smoking Cessation Treatment. Transl Behav Med. 2019;10(6):1504–1514. doi: 10.1093/tbm/ibz107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaphingst K.A., Kohlmann W., Chambers R.L., et al. Comparing models of delivery for cancer genetics services among patients receiving primary care who meet criteria for genetic evaluation in two healthcare systems: BRIDGE randomized controlled trial. BMC Health Serv Res. 2021;21(1):542. doi: 10.1186/s12913-021-06489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.