Abstract
Background
Immunomagnetic enrichment followed by RT-PCR (immunobead RT-PCR) is an efficient methodology to identify disseminated carcinoma cells in the blood and bone marrow. The RT-PCR assays must be both specific for the tumor cells and sufficiently sensitive to enable detection of single tumor cells. We have developed a method to test RT-PCR assays for any cancer. This has been investigated using a panel of RT-PCR markers suitable for the detection of breast cancer cells.
Methods
In the assay, a single cell line-derived tumor cell is added to 100 peripheral blood mononuclear cells (PBMNCs) after which mRNA is isolated and reverse transcribed for RT-PCR analysis. PBMNCs without added tumor cells are used as specificity controls. The previously studied markers epidermal growth factor receptor (EGFR), mammaglobin 1 (MGB1), epithelial cell adhesion molecule (EpCAM/TACSTD1), mucin 1 (MUC1), carcinoembryonic antigen (CEA) were tested. Two new epithelial-specific markers ELF3 and EphB4 were also tested.
Results
MUC1 was unsuitable as strong amplification was detected in 100 cell PBMNC controls. Expression of ELF3, EphB4, EpCAM, EGFR, CEA and MGB1 was found to be both specific for the tumor cell, as demonstrated by the absence of a signal in most 100 cell PBMNC controls, and sensitive enough to detect a single tumor cell in 100 PBMNCs using a single round of RT-PCR.
Conclusions
ELF3, EphB4, EpCAM, EGFR, CEA and MGB1 are appropriate RT-PCR markers for use in a marker panel to detect disseminated breast cancer cells after immunomagnetic enrichment.
Background
Reverse transcriptase-polymerase chain reaction (RT-PCR) assays have been developed for the detection of disseminated carcinoma cells by targeting epithelial or tumor specific mRNA in bone marrow, peripheral blood, or lymph nodes (reviewed in [1]). However limitations may arise from deficient expression in circulating tumor cells or low level illegitimate expression in haematopoietic cells, particularly if a nested PCR approach is used [2-5].
We developed an immunobead-based technique for the enrichment of epithelial cells to increase the signal to noise ratio for PCR based protocols [6]. This was subsequently adapted to RT-PCR methodology [7]. We have shown immunobead RT-PCR is of sufficient sensitivity to identify 1 tumor epithelial cell in a background of 106 blood cells [7,8]. A recent study compared immunobead RT-PCR with regular RT-PCR using CEA as the marker for colon cancer cells spiked into normal blood samples and confirmed that the immunobead RT-PCR method is more sensitive and specific than the regular RT-PCR method [9].
Up to 100 peripheral blood leukocytes (PBLs) may be concurrently isolated with the tumor cells during the immunobead method [10]. We previously had similar levels of PBL contamination. Now that we carry out incubations at 4°C and take off the cell pellets from the magnets for washing, considerably fewer than 10 cells are isolated (unpublished observations). It is nevertheless important to assess whether this small number of cells can lead to false positives due to illegitimate expression.
In this paper, we introduce a new strategy for the testing of RT-PCR assays using a method in which a single tumor cell is mixed with 100 PBMNCs prior to RT-PCR analysis. 100 PBMNCs without added tumor cells are used as specificity controls. This method has allowed the identification of a panel of RT-PCR markers of sufficient sensitivity and specificity for immunobead RT-PCR. This study includes an assessment of 2 previously untested markers ELF3 (also known as ESX) [11] and EphB4 [12], as well as the previously studied markers mucin 1 (MUC1), carcinoembryonic antigen (CEA), epidermal growth factor receptor (EGFR), epithelial cell adhesion molecule (EpCAM) and mammaglobin 1 (MGB1) markers.
Whether a RT-PCR is useful depends not only on the marker used but also on the assay conditions for that marker. A useful marker will be both sensitive enough to enable detection of a single tumor cell and specific enough to not be amplified from PBLs. However, as immunomagnetic cell isolates have very low contamination by PBLs, it is desirable to check the possibility of illegitimate expression in the very low number of PBLs that are retained after an immunobead harvest. To test both specificity and sensitivity of our chosen markers, we developed an assay in which a single tumor cell is deposited into a microcentrifuge tube containing 100 normal PBMNCs. 100 PBMNCs were chosen as we believe that this number covers the maximum amount of PBMNC contamination that we or others are likely to encounter.
Methods
Patient samples
Peripheral blood was collected from patients with polycythemia vera who presented for treatment at The Queen Elizabeth Hospital, Adelaide, Australia. Informed consent was obtained in all cases and ethics approval was obtained from The Queen Elizabeth Hospital Ethics of Human Research Committee. Mononuclear cells were collected by centrifugation through a Ficoll gradient, washed with phosphate-buffered saline (PBS) and counted using a haemocytometer. In the course of this investigation, PBMNCs from eight different patients were used.
Cell lines
The breast cancer cell lines MDA-MB-453, T47-D, MDA-MB-468, MCF-7, were cultured in DMEM, pH 7.4 (Invitrogen, Carlsbad, CA) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 160 μg/ml L-glutamine and 10% heat-inactivated fetal bovine serum (JRH Biosciences, Lenexa, KS) in 75 cm2 vented tissue culture flasks at 37°C in a 5% CO2 environment. Log phase cells were collected at less than 90% confluency by trypsin digestion and centrifugation for 5 min at 1000 rpm, resuspended in phosphate buffered saline (PBS) and counted using a haemocytometer. The cell lines were purchased from the American Type Culture Collection (ATCC, Rockville, MD).
Single Cell RT-PCR
A dilute suspension of a cell line was prepared in calcium and magnesium free Dulbecco's PBS. Single cells were visualised using an inverted phase microscope and selected by using a micropipette to draw up the cell in a 1 μl volume. The cell was deposited into a 0.5 ml microcentrifuge tube containing 100 peripheral blood mononuclear cells in 1 μl of calcium and magnesium free Dulbecco's PBS. Aliquots that contained 100 mononuclear cells only were included as controls for the specificity of the RT-PCR. The cell mixture was lysed in a 15 μl volume of solution containing 0.3 % v/v Nonidet P-40 detergent (Sigma), 500 ng random hexamers (Pharmacia, Uppsala, Sweden), 20 U of RNasin (Promega, Madison, WI) and 10 mM DTT. Following 70°C denaturation for 3 minutes, reverse transcription was initiated by the addition of 5× First Strand Buffer, 200 U of Superscript II (Invitrogen), 0.5 mM of each deoxynucleotide triphosphate (Roche, Mannheim, Germany), with Ultra-pure water (Fisher Biotech, Perth, Australia) to a final volume of 30 μl. The reaction was incubated at 42°C for 50 minutes, and then the reverse transcriptase was inactivated at 70°C for 10 minutes.
After reverse transcription, 4 μl of cDNA was used as the template in a single round of PCR amplification with 200 nM of each gene specific primer pair (Table 1), 1 U of Hot Star Taq (Qiagen, Valencia, CA), 2.5 mM MgCl2, and 200 μM of each deoxynucleotide triphosphate, in the supplied PCR buffer. Cycling conditions included an initial denaturation step at 95°C for 15 minutes, then 1 minute at each of 94°C, 66–68°C and 72°C for 45–55 cycles and a final extension of 7 minutes at 72°C. Amplification products were visualised by ethidium bromide staining following separation by electrophoresis through agarose gels. In each case an RT negative control, made up of the components of the RT reaction mixture with or without lysis mix, and without the addition of RNA, was used as the template in the PCR reaction (no cDNA made and therefore no amplification expected). The PCR negative control contains the reagents of the PCR reaction but lacks template. Cell line cDNA was included as a positive control for the PCR reaction. Genomic DNA (100 ng) was used to confirm that a product of equal size to the cDNA product would not be amplified from the DNA in the cell lysate.
Table 1.
Primer sequences (sense (s) and anti-sense (as)), melting temperatures (Tm), annealing temperature and expected product size in nucleotides (nt) for each of the seven selected marker genes.
| Name | Sequence 5' – 3' | Tm | Annealing | Size |
|---|---|---|---|---|
| MUC1 s | ACCAAGACTGATGCCAGTAGCACT | 65°C | ||
| MUC1 as | ACCGTTACCTGCAGAAACCTTCT | 63°C | 68°C | 115 nt |
| ESX s | CTCGGAGCTCCCACTCCTCAGA | 68°C | ||
| ESX as | GCTCTTCTTGCCCTCGAGACAGT | 67°C | 68°C | 188 nt |
| CEA s | GGTTGGGGTTGCTCTGATATAGCAGC | 70°C | ||
| CEA as | GCTGTTGCAAATGCTTTAAGGAAGAAGC | 67°C | 66°C | 97 nt |
| EPHB4 s | CCCCAGGGAAGAAGGAGAGCTG | 68°C | ||
| EPHB4 as | GCCCACGAGCTGGATGACTGTG | 68°C | 68°C | 250 nt |
| EGFR s | TGTGAGGTGGTCCTTGGGAATTTGG | 67°C | ||
| EGFR as | TGCTGACTATGTCCCGCCACTGGA | 69°C | 66°C | 339 nt |
| EpCAM s | GGACCTGACAGTAAATGGGGAAC | 65°C | ||
| EpCAM as | CTCTTCTTTCTGGAAATAACCAGCAC | 65°C | 68°C | 186 nt |
| MGB1 s | CGGATGAAACTCTGAGCAATGTTGAG | 66°C | ||
| MGB1 as | CTGCAGTTCTGTGAGCCAAAGGTC | 67°C | 68°C | 110 nt |
Primer sequences (sense (s) and anti-sense (as)), melting temperatures (Tm), annealing temperature and expected product size in nucleotides (nt) for each of the seven selected marker genes. The TM (Salt adjusted) was calculated using the web-based Oligonucleotide Properties calculators http://www.basic.nwu.edu/biotools/oligocalc.html.
Results
Expression of markers in breast cancer cell lines
The expression of each of the RT-PCR markers was assessed for its suitability as a breast cancer marker. RNA was extracted from seven breast cancer cell lines, reverse transcribed into cDNA then amplified using primers specific for the different markers (Table 1). Transcripts corresponding to EpCAM were amplified from all of the cell lines tested (Fig. 1., panel 1). This validates the use of antibodies that recognise epitopes of this protein as the immunobead label for magnetic isolation of the breast carcinoma cells [6] and confirms earlier results which showed uniform bright fluorescence in breast cancer cell lines and biopsies stained with one of these antibodies, Ber-Ep4 [7].
Figure 1.
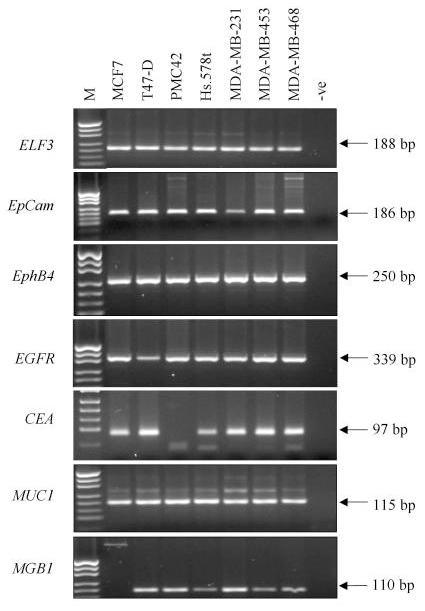
Expression of seven RT-PCR markers in a panel of seven breast cancer cell lines. Each marker is shown in a separate panel and the size of the amplified product is indicated in base pairs (bp) M indicates the pUC19/HpaII marker, -ve indicates the RT-PCR negative control. Weak genomic or primer dimer bands are seen for some RT-PCRs.
ELF3, EphB4 and MUC1 were amplified in all cell lines at consistently high levels (Fig. 1, panels 2, 3 and 4). EGFR expression was also easily detectable (Fig. 1, panel 5) even though several reports have suggested it is expressed at low levels in MCF-7 and T47-D and is not expressed in MDA-MB-453 [13,14]. MGB1 transcripts were amplified from all cell lines except for MCF-7 (Fig. 1, panel 2) and CEA was amplified from all cell lines except PMC42 (Fig. 1 panel 6).
Expression of markers in single tumor cell/100 PBMNC mixes
Tubes that contained 100 PBMNCs and one tumor cell were used to determine the sensitivity of each RT-PCR marker. Most of the sensitivity tests were performed using MDA-MB-453 cells but four different breast cancer cell lines have been included in this part of the study, two that are ER negative (MDA-MB-453, and MDA-MB-468) and two that are ER positive (MCF-7 and T47-D).
Analysing the samples to which a single breast cancer cell had been added to 100 PBMNC showed that we can detect the tumor cell most often with ELF3 (40/43 – 93%) and EpCAM (36/40 – 89%) (Figures 2 and 3, tables 2 and 3). EphB4 specific amplification detected 31/40 (77.5%), MUC1 10/20 (67%), CEA 9/20 (45%), EGFR 19/43 (44%) and MGB1 9/35 (26%).
Figure 2.
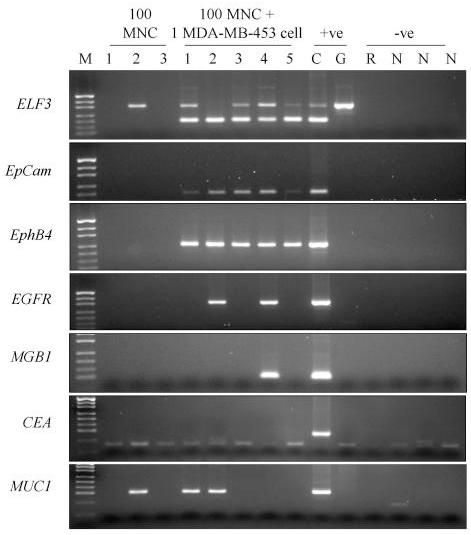
Expression of seven RT-PCR markers in 5 separate single MDA-MB-453 cells lysed with 100 normal PBMNC. Lane M – pUC19/HpaII molecular weight marker. The following 3 lanes are amplifications from 100 PBMNC only. The following 5 lanes are amplifications from one MDA-MB-453 cell in 100 PBMNC, Lane C is the cDNA positive control, Lane G is the genomic DNA control (G), Lane R is the RT – negative control; Lanes labelled N are the PCR negative controls. Genomic bands can be seen for many ELF3 amplifications.
Figure 3.
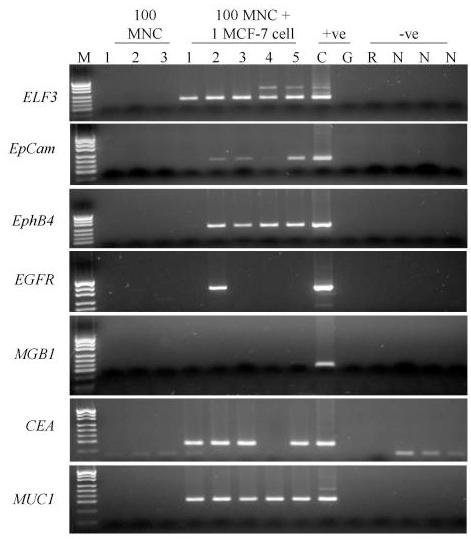
Expression of seven RT-PCR markers in 5 separate single MCF-7 cells lysed with 100 normal MNC. Lane M – pUC19/HpaII molecular weight marker. The following 3 lanes are amplifications from 100 PBMNC only. The following 5 lanes are amplifications from one MCF-7 cell in 100 PBMNC, Lane C is the cDNA positive control, Lane G is the genomic DNA control (G), Lane R is the RT – negative control; Lanes labelled N are the PCR negative controls. Genomic bands can be seen for many ELF3 amplifications,
Table 2.
Comparison of the expression of the seven selected markers in either 100 PBMNCs only, or after the addition of a single tumor cell from one of four different cell lines.
| ELF3 | EpCam | EphB4 | MUC1 | CEA | EGFR | MGB1 | |
|---|---|---|---|---|---|---|---|
| Cycles | 45 | 45 | 45 | 45 | 55 | 45 | 45 |
| 100 PBMNC | 5/30 | 0/21 | 0/21 | 1/6 | 0/18 | 0/27 | 0/24 |
| 453 | 17/18 | 5/5 | 9/10 | 5/10 | 0/10 | 8/18 | 4/10 |
| 468 | 5/5 | 5/5 | 5/5 | ND | ND | 5/5 | ND |
| MCF-7 | 10/10 | 8/10 | 6/10 | 5/5 | 9/10 | 1/5 | 1/10 |
| T-47D | 8/10 | 13/15 | 11/15 | ND | ND | 5/15 | 4/15 |
| TOTAL | 40/43 | 31/35 | 31/40 | 10/15 | 9/20 | 19/43 | 9/35 |
Comparison of the expression of the seven selected markers in either 100 PBMNCs only, or after the addition of a single tumor cell from one of four different cell lines (MDA-MB-453, MDA-MB-468, MCF-7 and T47-D) after 45 or 55 rounds (CEA) of PCR amplification. For example EphB4 was detected in 9 out of 10 single MDA-MB-453 cells added to 100 PBMNC aliquots. ND- test not performed.
Table 3.
Profile of gene expression from 10 different experiments each using 5 single tumour cell/100 PBMNC aliquots from four different cell lines.
| MDA-MB-468 | MDA-MB-453 | MDA-MB-453 | MDA-MB-453 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| ELF3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| EphB4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - |
| EpCam | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| EGFR | + | + | + | + | + | + | - | - | + | + | - | + | - | + | - | + | + | - | - | - |
| MGB1 | - | - | - | - | - | - | - | - | + | - | + | - | - | + | + | |||||
| T-47D | T-47D | T-47D | T-47D | |||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| ELF3 | + | + | + | + | + | + | + | + | + | + | + | + | - | + | - | |||||
| EphB4 | + | + | + | + | + | - | + | + | - | + | + | + | + | + | + | + | + | - | + | - |
| EpCam | + | + | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | - |
| EGFR | - | - | - | - | - | - | + | - | - | - | - | - | - | - | + | + | + | + | - | - |
| MGB1 | - | - | - | - | - | - | - | - | + | + | - | - | - | - | - | + | + | - | - | - |
| MCF-7 | MCF-7 | |||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |||||||||||
| ELF3 | + | + | + | + | + | + | + | + | + | + | ||||||||||
| EphB4 | - | + | + | + | + | - | + | - | + | - | ||||||||||
| EpCam | - | + | + | - | + | + | + | - | + | + | ||||||||||
| EGFR | - | + | - | - | - | + | - | - | - | - | ||||||||||
| MGB1 | - | - | - | - | - | - | - | - | - | - | ||||||||||
Profile of gene expression from 10 different experiments each using 5 single tumour cell/100 PBMNC aliquots from four different cell lines demonstrating heterogeneity of gene expression and utility of several markers for accurate detection of the single tumour cell. A "+" indicates that marker was detected, a "-" indicates that marker was not detected, and a blank indicates that marker was not done.
Expression of markers in a background of 100 PBMNCs
Tubes that contained 100 PBMNCs only were included to determine specificity (Table 2). After combining the data obtained from several different experiments and several different normal individuals in reactions using the appropriate number of amplification cycles, we found no expression of EGFR (0/27), MGB1 (0/24), EphB4(0/21), EpCAM (0/21) or CEA (0/18), (Figures 2 and 3, Table 2). Only ELF3 and MUC1 gave positive bands.
These results were for 45 cycles of amplification except for CEA which was run for 55 cycles. We also examined ELF3, EphB4 and MUC1 at 55 cycles in a small number of experiments. We amplified transcripts that corresponded to ELF3 (1/6), EphB4 (3/12) and MUC1 (3/12) in some of the PBMNC control tubes. The ELF3 and EphB4 bands were visible following ethidium bromide staining but were less intense than the band amplified after the addition of a single tumor cell and in no cases were all three of the PBMNC controls positive in the same experiment.
We assume that the ELF3 band is a result of amplification of illegitimate transcripts. This band was eliminated when only 35 cycles of amplification (0/15) were performed but because only a small amount of product was amplified after 45 or 55 cycles in these controls we could distinguish samples that contained tumor cells and those that did not through comparison of the intensity of bands. Analysing the samples to which a single breast cancer cell had been added to 100 PBMNC showed that we could still detect the tumor cell most often with ELF3 (15/16 – 94%). Tests using single tumour cells in a 100 PBMNC background detected 15/16 (94%) of tumour cells after 35 cycles. Unlike ELF3, the MUC1 PBMNC band was of comparable intensity to those achieved after tumor cell addition making it impossible to distinguish samples that contain tumor cells from those that don't (Figure 2).
Discussion
The development of optimized methods for detecting tumor cells in the peripheral blood and bone marrow of cancer patients may enhance our ability to identify those patients in need of further therapy. Nested RT-PCR of PBMNCs has been the most common method to detect epithelial – specific markers. Due to the high level of sensitivity of nested RT-PCR, even low levels of illegitimate transcription in PBMNCs can cause false-positive results [2-5]. Nested RT-PCR is also more time consuming and stringent procedures need to be observed in order to minimise the risk of false positives due to PCR product cross contamination. It is thus desirable that the RT-PCR is performed in a single round to avoid potential contamination problems.
The use of immunobead enrichment prior to reverse transcription has allowed the use of a single round of PCR amplification to screen a whole blood sample. Our original methodology used blotting and detection with an oligonucleotide to increase sensitivity [7] but this is no longer necessary with the more efficient RT-PCR assays presented here, in particular because of the use of a hot start Taq polymerase. To improve RT-PCR detection further, the identification of epithelial or carcinoma cell specific markers that are expressed at a sufficient level to enable detection of a single cell after only one round of non-nested PCR are needed. For this reason, we have used single tumor cells in a background of 100 PBMNCs to screen seven different RT-PCR assays in order to identify the best ones for inclusion in a panel for the immunobead RT-PCR based detection of circulating tumor cells.
We used breast cancer cell lines as the source of tumor cells. Whereas breast cancer cell lines may have different levels of expression of markers compared to disseminated breast cancer cells, cells from different individuals, and even within an individual, are likely to display considerable heterogeneity. We sought to overcome this difficulty by using a number of cell lines. The use of cell lines has the great advantage of providing reproducible controls to test the sensitivity of the RT-PCR assays during studies of patient material.
The immunobead enrichment step relies on the expression of an epithelial cell adhesion molecule (EpCAM) (current gene symbol, TACTSD1, tumor-associated calcium signal transducer 1), which is recognized by an antibody on immunobeads. We considered this to be an appropriate positive control, as any cell that is detected using the immunobeads should express the EpCAM gene. All of the breast cancer cell lines expressed the EpCAM gene and no message was detected in any of 24 aliquots that contained 100 PBMNC. Tests of the single tumor cell and 100 PBMNC aliquots with EpCAM showed that it was also expressed to a sufficient level to enable detection of the tumor cell in 31/35 (89%) cases after 45 cycles of PCR amplification. We therefore consider EpCAM a useful addition to a marker panel for the detection of breast tumor cells. It is possible that the sensitivity with this marker can be improved if more PCR cycles are performed.
Mucin 1 (MUC1) is another epithelial glycoprotein expressed in breast epithelial cells. It is a commonly used if controversial marker for detection of circulating tumor cells [5]. Recently, it was reported that that immunomagnetic separation followed by MUC1-specific real time RT-PCR allowed the semi-quantitative detection of circulating mammary cells if amplification above a cycle threshold value of 38 was obtained [15]. Using our assay, MUC1 transcripts were detected in 1/6 100 PBMNC only samples and 10/15 tumor cell samples. Although we did not use real-time PCR for our assay, the intensity of the positive bands in the PBMNC only samples was comparable to that obtained with the tumor cell samples and it was concluded that detection of MUC1 transcripts would result in too many false positive results to warrant its inclusion in our panel. Interestingly, during the course of this study it was reported that a majority of CK(+)/MUC1(+) tumor cells circulating in the blood of patients with advanced breast cancer also displayed apoptotic features such as an "inclusion type" cytokeratin staining pattern and nuclear condensation with apoptosis-related DNA strand breaks [16]. It was concluded that the predictive value of such CK(+)/MUC1(+) positive cells should be considered separately when analysing tumor cell dissemination, further supporting our decision not to include MUC1.
Elevated levels of serum carcinoembryonic antigenCEA are commonly used as a clinical tool in the diagnosis and monitoring of metastasis but the use of CEA as an immunobead RT-PCR marker [17] has given varying results. CEA has been considered to show no diagnostic value as a RT-PCR marker for the detection of micrometastases because it is expressed in the blood and lymph nodes of patients without cancer [5]. However, in a comparison of immunobead RT-PCR and regular RT-PCR amplification, it was shown that immunobead isolation increases the specificity of CEA [8]. Using 55 PCR cycles for CEA, we found amplification in none of the PBMNC only samples and gained a positive result for 45% of the single tumor cell samples we tested. Thus, CEA appears to be a very useful marker when immunobead enrichment is performed prior to RT-PCR.
Epidermal growth factor receptor (EGFR, HER1, ErbB1) is a tyrosine kinase whose activation plays an important role in the regulation of cell proliferation, differentiation and survival with a pivotal role in maintenance and repair of epithelial tissues. EGFR is overexpressed in 50–70% of human primary breast, lung, and colon carcinomas [18]. A study using nested RT-PCR and Southern blotting to detect EGFR transcripts in blood samples from patients with metastatic colon cancer found that 10.5% of healthy donors were positive which suggests that their assay detected illegitimate transcription. However, none of the 27 PBMNC only aliquots tested in our assay yielded EGFR amplification products and 19/43 (44%) single tumor cells were detected. A comparison of the results obtained for the two ER positive cell lines (6/20) with those obtained for the two ER negative cell lines (13/23) suggests that ER status will affect the efficacy of this marker but it remains a useful marker.
Mammaglobin 1 (MGB1) expression is limited to mammary epithelium and is often up-regulated in human breast cancer [20]. The gene has been localised to chromosome 11q13 in a region frequently amplified in breast cancer. Using nested RT-PCR, it was shown that MGB1 transcripts are detectable in blood from breast cancer patients [21]. Marchetti et al., (2001) analysed lymph nodes from breast cancer and normal patients and suggest that MGB1 is a more sensitive and more specific RT-PCR marker than a panel of six other markers that included CEA, CK19 and MUC1[22]. MGB1 transcripts were not detected by us in any of the PBMNC only aliquots. In our hands, MGB1 transcripts were only detected in 9/35 (26%) of the single tumor cells tested but because this marker is mammary-specific, its inclusion in a panel of RT-PCR markers is warranted. It is possible that the sensitivity with this marker can be improved if more PCR cycles are performed.
ELF3, also known as Epithelial-Specific with serine bo X (ESX), is a member of the Ets multigene family of transcriptional regulators and is localised to chromosome 1q32 in a region known to be amplified in 50% of early breast cancers [11,23].ELF3 mRNA expression is restricted to tissues of epithelial origin, including normal and malignant human mammary ductal epithelium with no expression reported in lympho-haematopoietic tissues. In our assay, faint bands were visible at 188 bp both after 45 cycles (1/6) and after 55 cycles (5/30) in the PBMNC only aliquots. However the strong intensity of the bands for the 40/43 (93%) samples with a single tumor cell was considered indicative of a positive result in these cases. Decreasing the cycle number to 35 eliminated the weak band in the PBMNC only aliquots (0/15), yet still allowed consistent detection of the tumor cells in our assay (15/16 – 94%) and therefore ELF3 is a useful marker for inclusion in our panel.
EphB4 is a member of the Eph family of receptor protein tyrosine kinases and has recently been reported to be up-regulated in cancers from prostate, breast, endometrium, lung and colon [12,24-27]. It is thought to play an important role in the development and oncogenesis of various tissues through modifications to cell-cell communication and adhesion. Although EphB4 was detected in some PBMNC cell only controls at 55 cycles the level of intensity of the corresponding bands on agarose gels was low and decreasing the cycle number to 45 resulted in no positive samples out of 21 tests. Using EphB4 specific primers with 45 cycles we could detect 77.5% of single tumor cells (31/40) proving this to be an useful marker in this assay.
The various markers used here show different degrees of heterogeneity i.e the numbers of single tumor cells that they are detected in. Interestingly, the more heterogeneous markers also gave the least false positives in the controls. It is unclear whether this is due to the sensitivity of the test or due to stochastic differences in whether a cell expresses a transcript or not. Heterogeneity at the single cell level must be taken into account when designing an RT-PCR assay and this study is the first one to address this. Using a panel of several markers will reduce this source of error. This can be seen in table 3 where nearly every tumour cell is positive for more than one marker. One T-47D cell is negative for all 5 markers tested and it is possible that this cell was not viable at the time of selection.
The need to be both specific and sensitive may entail finding a balance between the specificity and sensitivity. Our findings in the PBMNC suggest that illegitimate transcription occurs in all cells rather than a subset. When large numbers of cells are present or the assay is very sensitive, false positives will be common. ELF3 had a fairly high false positive rate of amplification in PBMNC at 45 cycles presumably due to detecting illegitimate transcription in a very small number of these cells. However, ELF3 was one of the more robust markers giving very strong amplification of single tumour cells at 45 cycles. Amplification from the controls was eliminated when only 35 cycles of amplification were performed. Because only a small amount of product was amplified after 45 cycles in these controls we could distinguish samples that contained tumor cells and those that did not through comparison of the intensity of bands. This suggests that this and similar markers will be useful in a real-time PCR methodology where positive signals can be more readily differentiated from background signals [15].
As mentioned, one question is whether cell lines are truly representative of circulating tumor cells. A large number of circulating tumor cells are believed to be in G0 [1], whereas the cell line controls used are from log phase cells. We compared the expression of ELF3, EphB4 and EGFR in cells grown under conditions of serum starvation, during log phase and at full confluence (data not shown). We saw no appreciable difference in the expression levels of any of these 3 genes under any of the conditions tested. Whereas it is difficult to model the clinical situation, where patterns of gene expression are likely to be heterogeneous, the assay presented in this communication is useful for testing potential RT-PCR markers and as a meaningful control in the determination of the presence of disseminated tumor cells in clinical samples. To fully validate these markers, a clinical study using immunomagnetically enriched cells from patients is needed. Such a study is currently in progress.
Conclusions
Immunobead RT-PCR is a powerful methodology for the detection of disseminated carcinoma cells. We need RT-PCR assays that are sensitive enough to detect very low numbers of captured carcinoma cells As haematopoietic cells may be non-specifically retained during immunomagnetic enrichment of epithelial cells, these RT-PCR markers also need to be tested for their specificity. A set of RT-PCR markers has been tested for their ability to detect a single carcinoma cell using a single round of PCR and ethidium bromide visualization. We have identified six useful markers that include the previously described markers EpCAM, EGFR, CEA and MGB1 as well as two novel markers EphB4, and ELF3. This panel of markers should allow the detection of virtually every tumor cell and this means that a positive result will be obtained each time an immunobead harvest has captured at least one breast cancer cell. Most of these markers will also be useful in the analysis of other carcinomas such as colon, lung and prostate.
Competing interests
None declared.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Michael Raynor, Email: michael.raynor@student.adelaide.edu.au.
Sally-Anne Stephenson, Email: sally.stephenson@eudoramail.com.
David CA Walsh, Email: david.walsh@adelaide.edu.au.
Kenneth B Pittman, Email: Ken.Pittman@nwahs.sa.gov.au.
Alexander Dobrovic, Email: a.dobrovic@pmciunimelb.edu.au.
Acknowledgements
We thank Professor Ed Sage for his support through this project and wish him the best upon his retirement. We thank Ashley Farlow for his proof reading. Funding was obtained from the Queen Elizabeth Hospital Research Foundation and from Dynal, Norway. This work was carried out during the tenure of a grant from the Cancer & Bowel Research Trust.
References
- Pantel K, Cote RJ, Fodstad Ø. Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst. 1999;91:1113–1124. doi: 10.1093/jnci/91.13.1113. [DOI] [PubMed] [Google Scholar]
- Zippelius A, Kufer P, Honold G, Kollermann MW, Oberneder R, Schlimok G, Riethmuller G, Pantel K. Limitations of reverse-transcriptase polymerase chain reaction analyses for detection of micrometastatic epithelial cancer cells in bone marrow. J Clin Oncol. 1997;15:2701–2708. doi: 10.1200/JCO.1997.15.7.2701. [DOI] [PubMed] [Google Scholar]
- de Graaf H, Maelandsmo GM, Ruud P, Forus A, Oyjord T, Fodstad Ø, Hovig E. Ectopic expression of target genes may represent an inherent limitation of RT-PCR assays used for micrometastasis detection: studies on the epithelial glycoprotein gene EGP-2. Int J Cancer. 1997;72:191–196. doi: 10.1002/(SICI)1097-0215(19970703)72:1<191::AID-IJC27>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Lambrechts AC, van't Veer LJ, Rodenhuis S. The detection of minimal numbers of contaminating epithelial tumor cells in blood or bone marrow: use, limitations and future of RNA-based methods. Ann Oncol. 1998;9:1269–1276. doi: 10.1023/A:1008445604263. [DOI] [PubMed] [Google Scholar]
- Bostick PJ, Chatterjee S, Chi DD, Huynh KT, Giuliano AE, Cote R, Hoon DS. Limitations of specific reverse-transcriptase polymerase chain reaction markers in the detection of metastases in the lymph nodes and blood of breast cancer patients. J Clin Oncol. 1998;16:2632–4260. doi: 10.1200/JCO.1998.16.8.2632. [DOI] [PubMed] [Google Scholar]
- Hardingham JE, Kotasek D, Farmer B, Butler RN, Mi JX, Sage RE, Dobrovic A. Immunobead-PCR: a technique for the detection of circulating tumor cells using immunomagnetic beads and the polymerase chain reaction. Cancer Res. 1993;53:3455–3458. [PubMed] [Google Scholar]
- Eaton MC, Hardingham JE, Kotasek D, Dobrovic A. Immunobead RT-PCR: a sensitive method for detection of circulating tumor cells. Biotechniques. 1997;22:100–105. doi: 10.2144/97221st01. [DOI] [PubMed] [Google Scholar]
- Hardingham JE, Hewett PJ, Sage RE, Finch JL, Nuttall JD, Kotasek D, Dobrovic A. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel. Int J Cancer. 2000;89:8–13. doi: 10.1002/(SICI)1097-0215(20000120)89:1<8::AID-IJC2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Park S, Lee B, Kim I, Choi I, Hong K, Ryu Y, Rhim J, Shin J, Park SC, Chung H, Chung J. Immunobead RT-PCR versus regular RT-PCR amplification of CEA mRNA in peripheral blood. J Cancer Res Clin Oncol. 2001;127:489–494. doi: 10.1007/s004320100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cremoux P, Extra JM, Denis MG, Pierga JY, Bourstyn E, Nos C, Clough KB, Boudou E, Martin EC, Muller A, Pouillart P, Magdelenat H. Detection of MUC1-expressing mammary carcinoma cells in the peripheral blood of breast cancer patients by real-time polymerase chain reaction. Clin Cancer Res. 2000;6:3117–3122. [PubMed] [Google Scholar]
- Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, Sayre P, Erny K, Collins C, Gray JW, Benz CC. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene. 1997;14:1617–1622. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- Stephenson S, Slomka S, Douglas E, Hewett PJ, Hardingham JE. Receptor protein tyrosine kinase EphB4 is up-regulated in colon cancer. BMC Mol Biol. 2001;2:15. doi: 10.1186/1471-2199-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden RI, Wilson MA, Chrysogelos SA. Estrogen suppression of EGFR expression in breast cancer cells: A possible mechanism to modulate growth. J Cell Biochem. 2001;81:232–246. doi: 10.1002/jcb.1142. [DOI] [PubMed] [Google Scholar]
- Moulder SL, Yakes FM, Muthuswamy SK, Bianco R, Simpson JF, Arteaga CL. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001;61:8887–8895. [PubMed] [Google Scholar]
- Walsh MD, Luckie SM, Cummings MC, Antalis TM, McGuckin MA. Heterogeneity of MUC1 expression by human breast carcinoma cell lines in vivo and in vitro. Breast Cancer Res Treat. 1999;58:255–266. doi: 10.1023/A:1006345301364. [DOI] [PubMed] [Google Scholar]
- Mehes G, Witt A, Kubista E, Ambros PF. Circulating breast cancer cells are frequently apoptotic. Am J Pathol. 2001;159:17–20. doi: 10.1016/S0002-9440(10)61667-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Mimori K, Ueo H, Karimine N, Barnard GF, Sugimachi K, T Akiyoshi. Molecular detection of circulating solid carcinoma cells in the peripheral blood: the concept of early systemic disease. Int J Cancer. 1996;68:739–743. doi: 10.1002/(SICI)1097-0215(19961211)68:6<739::AID-IJC8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermial growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-I. [DOI] [PubMed] [Google Scholar]
- de Luca A, Pignata S, Casamassimi A, D'Antonio A, Gridelli C, Rossi A, Cremona F, Parisi V, De Matteis A, Normanno N. Detection of circulating tumor cells in carcinoma patients by a novel epidermal growth factor receptor reverse transcription-PCR assay. Clin Cancer Res. 2000;6:1439–1444. [PubMed] [Google Scholar]
- Fleming TP, Watson MA. Mammaglobin, a breast-specific gene, and its utility as a marker for breast cancer. Ann N Y Acad Sci. 2000;923:78–89. doi: 10.1111/j.1749-6632.2000.tb05521.x. [DOI] [PubMed] [Google Scholar]
- Zach O, Kasparu H, Krieger O, Hehenwarter W, Girschikofsky M, Lutz D. Detection of circulating mammary carcinoma cells in the peripheral blood of breast cancer patients via a nested reverse transcriptase polymerase chain reaction assay for mammaglobin mRNA. J Clin Oncol. 1999;17:2015–2019. doi: 10.1200/JCO.1999.17.7.2015. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Bertacca G, Zavaglia K, Bevilacqua G, Angelucci D, Viacava P, Naccarato A, Bonadio A, Barassi F, Felicioni L, Salvatore S, Mucilli F. mRNA markers of breast cancer nodal metastases: comparison between mammaglobin and carcinoembryonic antigen in 248 patients. J Pathol. 2001;195:186–190. doi: 10.1002/path.943. [DOI] [PubMed] [Google Scholar]
- Tymms MJ, Ng AY, Thomas RS, Schutte BC, Zhou J, Eyre HJ, Sutherland GR, Seth A, Rosenberg M, Papas T, Debouck C, Kola I. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene. 1997;15:2449–2462. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- Robinson D, He F, Pretlow T, Kung HJ. A tyrosine kinase profile of prostate carcinoma. Proc Natl Acad Sci USA. 1996;93:5958–5962. doi: 10.1073/pnas.93.12.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova Z, Djonov V, Zuercher G, Andres AC, Ziemiecki A. Cell-type specific and estrogen dependent expression of the receptor tyrosine kinase EphB4 and its ligand ephrin-B2 during mammary gland morphogenesis. J Cell Sci. 1998;111:2741–2751. doi: 10.1242/jcs.111.18.2741. [DOI] [PubMed] [Google Scholar]
- Tang XX, Brodeur GM, Campling BG, Ikegaki N. Coexpression of transcripts encoding EPHB receptor protein tyrosine kinases and their ephrin-B ligands in human small cell lung carcinoma. Clin Cancer Res. 1999;5:455–460. [PubMed] [Google Scholar]
- Takai N, Miyazaki T, Fujisawa K, Nasu K, Miyakawa I. Expression of receptor tyrosine kinase EphB4 and its ligand ephrin-B2 is associated with malignant potential in endometrial cancer. Oncol Rep. 2001;8:567–573. doi: 10.3892/or.8.3.567. [DOI] [PubMed] [Google Scholar]


