Abstract
Glioblastoma (GBM) cells are highly heterogeneous and invasive, leading to treatment resistance and relapse. However, the molecular regulation in and distal to tumors remains elusive. Here, we collected paired tissues from the tumor core (TC) and peritumoral brain (PTB) for integrated snRNA-seq and snATAC-seq analyses. Tumor cells infiltrating PTB from TC behave more like oligodendrocyte progenitor cells than astrocytes at the transcriptome level. Dual-omics analyses further suggest that the distal regulatory regions in the tumor genome and specific transcription factors are potential determinants of regional heterogeneity. Notably, while activator protein 1 (AP-1) is active in all GBM states, its activity declines from TC to PTB, with another transcription factor, BACH1, showing the opposite trend. Combined inhibition of AP-1 and BACH1 more efficiently attenuates the tumor progression in mice and prolongs survival than either single-target treatment. Together, our work reveals marked molecular alterations of infiltrated GBM cells and a synergy of combination therapy targeting intratumor heterogeneity in and distal to GBM.
The heterogeneity of tumor cells in and distal to GBM was identified for combination therapy targeting region-specific regulators.
INTRODUCTION
Glioblastoma (GBM) is the most frequent and lethal primary brain tumor (1). Now, the standard of care consists of surgical resection, followed by radio- and chemotherapy, yet tumors recur inevitably, and the median survival is only 15 months (1–3). Residual malignant cells that escape surgical resection in neighboring regions would serve as seeds for tumor recurrence (4–8), and a greater extent of resection may improve survival (9). Therefore, the design of more effective therapies for glioma shall take malignant cells both in and away from the primary tumor into consideration (10–12). However, the comprehensive characterization of the peritumor regions remains to be established, partially due to the scarcity of tumor cells there, making it challenging to separate them from the abundant normal cells. In addition, collecting normal tissues away from the tumor core (TC) is restricted by its specific location and the need to preserve essential functions.
Regarding the source of malignant cells disseminated into unresected regions, previous studies have identified malignant clones in the subventricular zone (SVZ) that often contain tumor precursor cells and therefore pointed out that astrocyte-like neural stem cells in the SVZ are the origin of glioma cells (13). On the other hand, malignant cells from the primary tumor always invade distal regions during the early stage (5, 6, 14), and the infiltrated cells remain quiescent for a period of time before reentering the aggressive growth stage (15, 16). This phenotypic distinction suggests that the underlying molecular regulation of malignant cells alters from TC to peritumoral brain (PTB). Mutations detected in the primary tumor were found to remain at recurrence, and little evidence of recurrence-specific gene alterations was found, suggesting that the contribution of the genetic lesion to invasion and relapse remains to be determined (17). In contrast, infiltrated GBM cells appear to express distinct genes compared to cells at TC, including ones in cell migration/metabolic pathways (10, 12, 18, 19). It is also reported that the GBM infiltration resembles the neuronal migration mechanisms based on molecular and functional characterizations (20). Considering the essentiality of infiltrating cells in treatment resistance and relapse, a full understanding of their molecular features at different regions will help determine key targets to eradicate these “seed” cells.
GBM is highly inter- and intratumor heterogeneous (21–27). On the basis of the transcriptome variation of tumor tissues uncovered by bulk RNA sequencing (RNA-seq) data, The Cancer Genome Atlas (TCGA) categorizes patients with GBM into four divergent subtypes (proneural, neural, classical, and mesenchymal) and suggests specific treatments for each subtype (21). Moreover, our understanding of the intratumor heterogeneity was markedly advanced with single-cell technologies. Using single-cell RNA-seq (scRNA-seq), Patel et al. (28) have revealed that GBM cells with different TCGA subtypes coexist within individual tumors. A following study hasconstructed a cellular state model and suggested that each GBM tumor contains four cellular states [neural progenitor–like (NPC-like), oligodendrocyte progenitor–like (OPC-like), astrocyte-like (AC-like), and mesenchymal-like (MES-like)], which are interconvertible from one to another (22). Meanwhile, the dual states of glioma stem-like cells (GSC), including the mesenchymal and proneural states, have been proposed (26). These new concepts of cellular states were subsequently validated and adopted by other studies (23–25, 29). Tumors with various genetic backgrounds are found to converge on limited numbers of transcriptional states, suggesting genetic makeup influences, but do not determine cellular states (17, 30, 31), warranting further elucidation of potential regulators from the perspectives of the epigenetic, transcriptional, and microenvironmental control.
The transcription factor (TF) occupancy and activation at specific cis-regulatory elements (CREs) constitute the major regulatory force for lineage commitment and cellular state transition (32, 33). The recent development of single nucleus assay for transposase-accessible chromatin using sequencing (snATAC-seq) enables robust profiling of chromatin accessibility landscape for individual cell types or cellular states. In this study, we collected samples from paired TC and PTB regions in patients with GBM for simultaneous snRNA-seq and snATAC-seq to define the regulatory programs, especially for malignant cells in both regions. By correlating the chromatin accessibility with gene expression, we identified cell type–specific candidate CREs and revealed that distal regulatory genomic regions were most closely related to cellular identity. Next, we resolved the regulatory programs associated with four cellular states in GBM. We observed a notable increase of OPC-like state, yet a decrease of AC-like state as tumor cells infiltrated into PTB. Specifically, we identified TFs associated with each of these cellular states, noting that their activity levels often varied between TC and PTB. Last, we evaluated the therapeutic potential of simultaneous targeting of two regulators, AP-1 and BACH1, which showed a preferential activity at TC and PTB in our dataset, respectively.
RESULTS
Single-cell multi-omics profiles of paired GBM patient tissues
To comprehensively characterize and compare malignant cells in the primary tumor and distal brain regions, we collected TC and PTB tissues separately from five patients with GBM (i.e., RM01 to RM05) by the extended resection according to the established surgery protocol (Fig. 1A). The pathological characterization of each sample was done by combining the results of HE staining, magnetic resonance imaging (MRI), and molecular diagnosis (fig. S1, A and B, and table S1). Notably, RM05 showed peritumoral edema and infiltrative tumor growth, indicating the PTB tissue of this patient is likely from the tumor edge or edema region. The cell nuclei of each sample were collected and split into two halves for snRNA-seq and snATAC-seq by the DNBelab system, respectively (34).
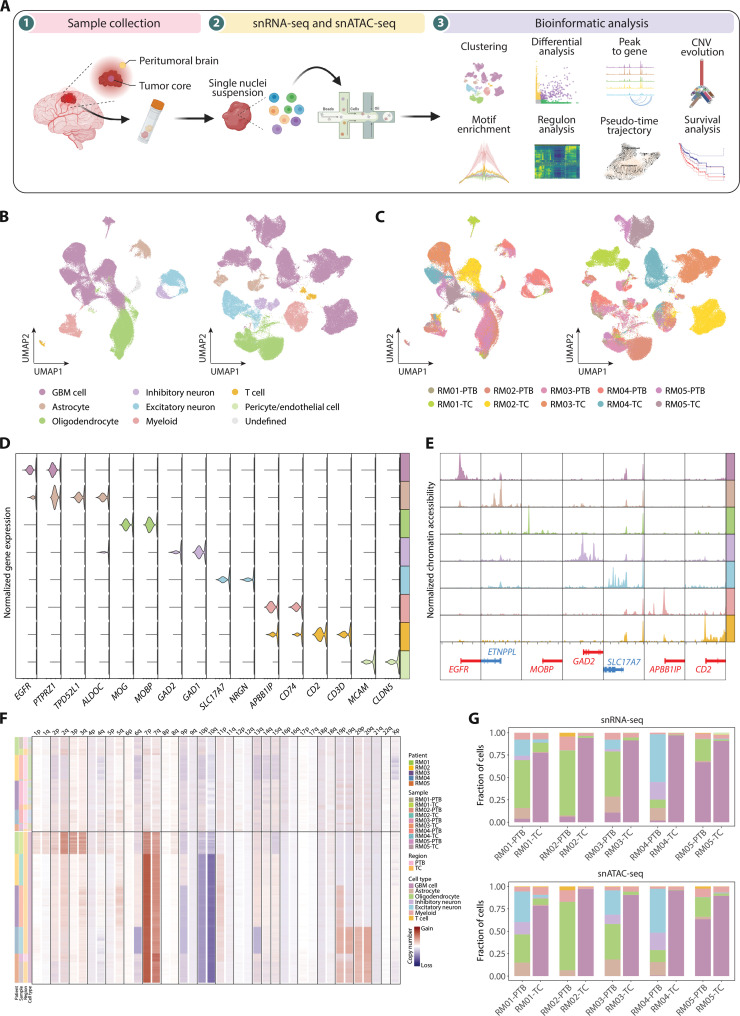
Fig. 1. Single-cell atlas of gene expression and chromatin accessibility in glioblastoma.
(A) Schematic representation of the experimental workflow. Primary surgical samples were obtained and then dissociated for single-cell sequencing. Clustering and other downstream bioinformatic analyses were performed. (B) Uniform manifold approximation and projection (UMAP) visualization colored by cell types for snRNA-seq (left) and snATAC-seq (right). (C) UMAP plot colored by samples for snRNA-seq (left) and snATAC-seq (right). (D) Violin plot for expression of canonical cell-type markers in snRNA-seq. (E) Pseudo-bulk chromatin accessibility profiles for canonical marker genes. Chromosome positions are shown below, with genes on the plus strand colored in red and minus strand in blue. Chromosome coordinates are: EGFR (chr7: 55009020–55029021), ETNPPL (chr4: 108753053–108773054), MOBP (chr3: 39457197–39477198), GAD2 (chr10: 26206306–26226307), SLC17A7 (chr19: 49432359–49452360), APBB1IP (chr10: 26428202–26448203), and CD2 (chr1: 116744384–116764385). (F) CNV profile inferred from snATAC-seq. Rows correspond to cells and columns represent the chromosomal locations. (G) Stacked bar charts showing the proportional composition of cell types across samples in snRNA-seq (top) and snATAC-seq (bottom).
After filtering low-quality nuclei, a total of 90,504 nuclei for snRNA-seq and 100,565 nuclei for snATAC-seq were retained (fig. S1, C to F, and tables S2 and S3). For snRNA-seq, we grouped the cells according to their transcriptome similarity and then determined cell types based on the expression of canonical marker genes (Fig. 1, B to D; fig. S2A; and table S4). The copy number variations (CNVs) deduced by inferCNV were further considered to distinguish malignant from nonmalignant cells according to the presence of GBM hallmarks (chromosome 7 gains and chromosome 10 losses) (fig. S2, B and C) (28). In total, eight cell groups were identified, including the GBM cell, astrocyte, oligodendrocyte, inhibitory neuron, excitatory neuron, myeloid, T cell, and pericyte/endothelial cell (Fig. 1B). A small population (1.81% of total cells) with no associated marker genes were considered as “undefined” and excluded from the following analyses. For snATAC-seq, the cell-type annotation was performed based on the level of chromatin accessibility around canonical marker genes, and malignant cells were assigned by Copy-scAT (Fig. 1, E and F, and fig. S3, A and B) (35). All cell types, except the pericyte/endothelial cell, were identified in snATAC-seq and validated by the expression scores of marker genes (Fig. 1B and fig. S3C).
In line with the previous observations (22, 36), both snRNA-seq and snATAC-seq data showed that GBM cells from different patients were separated, while the same type of nonmalignant cells was clustered together, indicative of the interpatient heterogeneity of tumor cells (Fig. 1, B and C). We next calculated the cell-type composition in two regions (Fig. 1G and table S5). TC tissues were mainly composed of GBM cells, whereas nonmalignant cells, such as the oligodendrocyte and neuron, constituted most of cells in PTB. A small number of tumor cells were detected in PTB, allowing us to investigate the molecular features of tumor cells distal to the defined tumor territory. Notably, the cell-type compositions revealed by the snRNA-seq and snATAC-seq were similar to each other (Fig. 1G and fig. S3D), implicating a high concordance of single-nucleus transcriptomic and epigenomic profiles for paired TC and PTB samples.
Cell type–specific regulation of the chromatin accessibility in GBM
The above results prompted us to further characterize the cell type–specific gene regulation with snRNA-seq and snATAC-seq. By correlating the chromatin accessibility with the gene expression, we identified 52,485 gene-linked CREs (gl-CREs) with a high degree of cell-type specificity (Fig. 2A). There were 9042 CRE-linked genes, and each was linked to a median of 5 CREs, suggesting a complex interaction between genes and distal regulatory sites (Fig. 2B). We observed that most of the CRE-linked genes were expressed in a cell type–specific manner or shared by closely related cell types, such as inhibitory and excitatory neurons (Fig. 2C). Encouraged by this result, we assessed the correlation of CRE-linked genes with differentially expressed genes (DEGs), which showed a notable overlap (fig. S4A), indicating a potential role of CREs in directing the expression of cell type–specific genes.
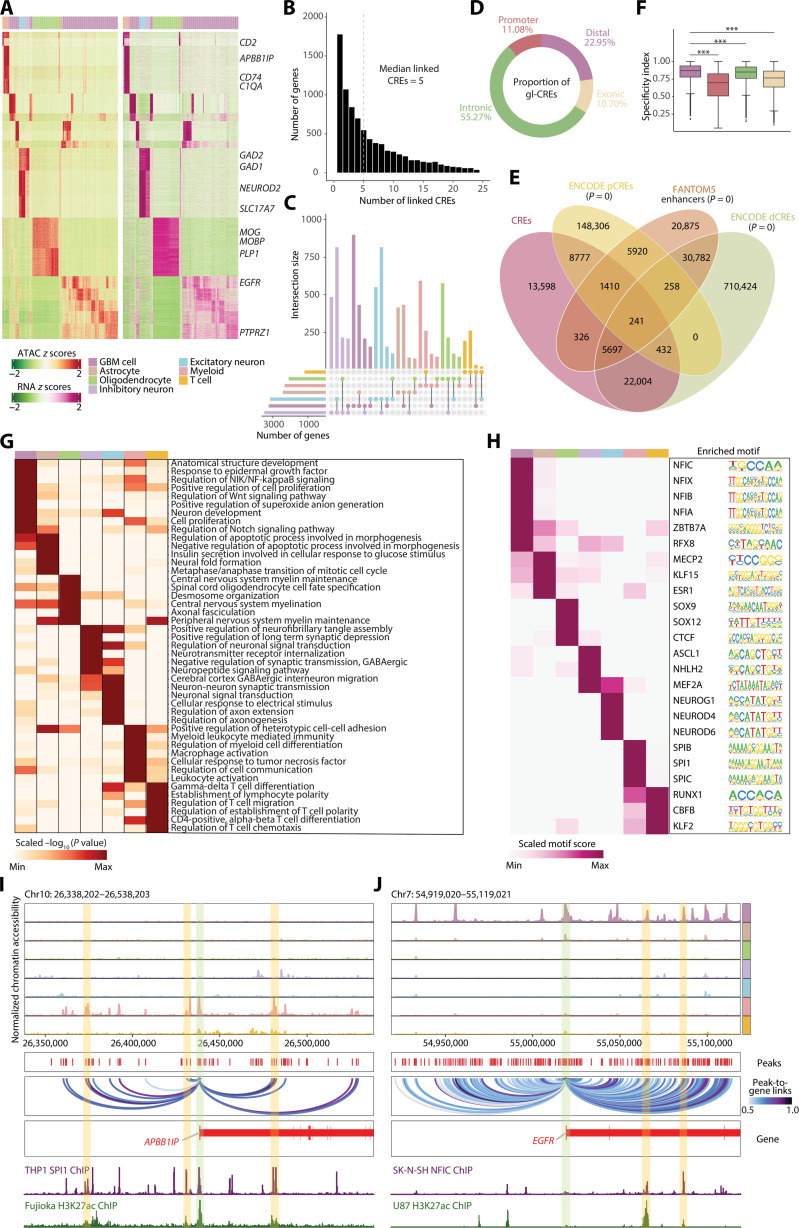
Fig. 2. Distal peaks confer cell type specificality and reveal the underlying cell type–specific TF regulation.
(A) Heatmaps showing chromatin accessibility (left) and gene expression (right) of significantly linked CRE-gene pairs. (B) Histogram showing the number of genes that with 1 to 25 linked CREs. (C) Upset visualization of the overlaps between CRE-linked genes identified in each cell type. The size of CRE-linked genes of each cell type is shown on the left bar plot, and the number of overlapping genes between two cell types or unique genes in one cell type is shown on the top bar plot. (D) Donut diagram showing the genomic distribution of gl-CREs. (E) Venn diagram showing the overlaps of CREs with proximal and distal enhancer-like elements (pCREs and dCREs) revealed by ENCODE (37) or enhancers by FANTOM5 (38). (F) Boxplot showing cell-type specificity of differentially accessible peaks across distal, promoter, intronic, and exonic regions. The cell-type specificity index was measured on scaled, log2 transformed averaged peak accessibility. Wilcoxon statistical test was used. For boxplot, the line and box boundaries correspond to the median and the interquartile range (IDR), respectively. Whiskers extend to 1.5 times the IDR. (G) Heatmap showing the GREAT terms enriched in differential distal peaks. The value corresponds to row normalized −log10 corrected P values (Benjamini-Hochberg) from binomial test on the enriched list of GREAT terms. (H) Heatmap of regulators per cell type determined by the motif enrichment analysis for differential distal peaks. The binding motif logos are shown right. (I and J) Genome track visualization of loci for the selected markers. Inferred peak-to-gene links across the GBM dataset are shown in the middle, and chromatin immunoprecipitation sequencing (ChIP-seq) tracks for identified TFs (SPI1 for myeloid and NFIC for GBM cell) and H3K27ac are shown bottom. ***P < 0.001.
Across the genome, gl-CREs were largely mapped to intronic (55.27%), followed by distal (22.95%), promoter (11.08%), and exonic regions (10.70%) (Fig. 2D), 74.09% (38,887 of 52,485) of which were overlapped with putative enhancers identified by The Encyclopedia of DNA Elements (ENCODE) consortium and Functional Annotation of the Mammalian Genome Project 5 (FANTOM5) projects (Fig. 2E) (37, 38). When calculating the specificity index for each group of gl-CREs (39), distal peaks appeared to contribute the most to cell-type specificity (Fig. 2F). Next, to characterize the importance of distal regulatory elements, we parsed differentially accessible peaks (131,904) across cell types (fig. S4, B and C), among which 46,046 (34.91%) distal peaks were subjected to GREAT analysis (40), leading to the identification of biological processes unique to corresponding cell types (Fig. 2G). For example, cancer-related terms, including response to epidermal growth factor and regulation of nuclear factor κB (NF-κB)–inducing kinase/NF-κB signaling, were enriched in GBM cells. Myeloid cell differentiation and myeloid leukocyte mediated immunity were enriched in myeloid cells.
To determine whether the regulatory distal regions contribute to tumorigenesis, we deduced TF binding motifs within differentially accessible distal peaks for each cell type (Fig. 2, H to J; fig. S5, A and B; and table S6). CTCF, which is critical for myelination and repair (41), was enriched in oligodendrocytes. ASCL1, a regulator in neuron generation (42), was enriched in inhibitory neurons. SPI1, which is implicated in myeloid differentiation (43), was enriched in myeloid cells (Fig. 2, H and I). We captured members of the NFI TFs in GBM cells, including NFIA, NFIB, NFIC, and NFIX involved in glioma tumorigenesis (Fig. 2, H and J) (44–46). It therefore suggests that this protein complex may function in GBM cells through a mechanism at the distal regulatory regions. Together, our data demonstrate the power of chromatin accessibility profiling by snATAC-seq in identifying potential regulatory regions linked to tumor-related genes and trans-acting factors that mediate GBM progression.
Infiltrated malignant cells at PTB with distinct transcriptome profiles
To explore the potential origin of GBM cells at PTB (PTB_G) and its relationship with GBM cells at TC (TC_G), we retrieved GBM cells from both regions to dissect the genome architecture of each cell by inferCNV and Uphyloplot2 analyses (Fig. 3A and fig. S6A) (28, 47–50). The aggregated CNV profiles enabled us to construct the GBM cell evolutionary tree for each patient (Fig. 3B, fig. S6B, and table S7) (47). The results showed that while canonical CNVs on chromosome 7 or 10 dominated the evolutionary tree from the root to branches, individual noncanonical CNVs appeared from specific branches. Taking RM01 as an example, 12q/14q gain was shown on branch B, and 3q/7p gain emerged on branch G. Notably, the root of the evolutionary tree only contained cells from TC, and PTB_G appeared from middle points of the branches, thus suggesting that malignant cells at PTB were originated from TC (Fig 3B, inserts at bottom right). On the other hand, because no SVZ sample was included in this study, the possibility that certain infiltrated tumor cells were from the SVZ could not be ruled out.
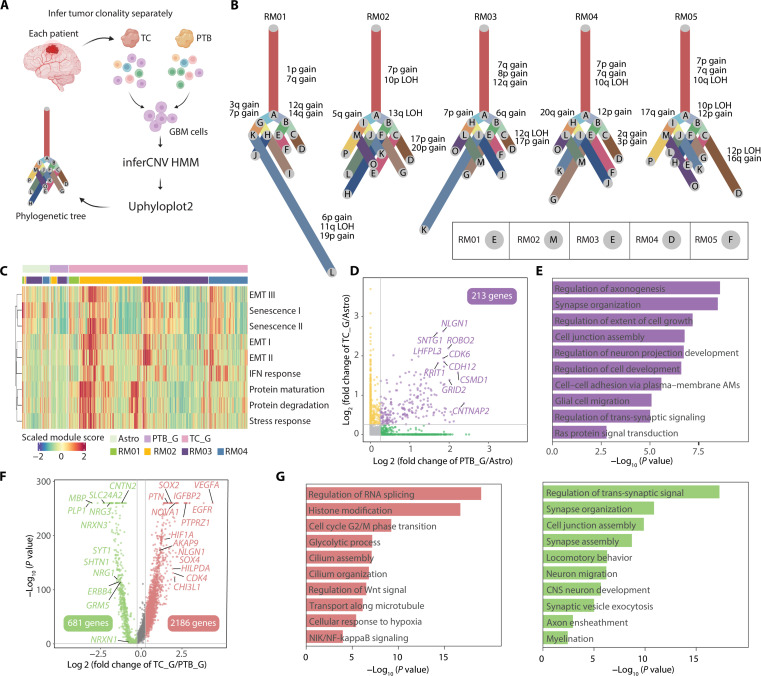
Fig. 3. Infiltrated GBM cells harbor different characteristics compared with GBM cells in the TC.
(A) Schematic diagram depicting the process of tumor evolution analysis. (B) Phylogenetic trees of GBM cells for each patient. Branch lengths are proportional to the number of cells in the subclone harboring the corresponding CNVs. The canonical CNV events of each branch were labeled on the side. The lower right panel shows the branch, which GBM cells in PTB first emerged. (C) Heatmap showing the expression scores for pan-cancer signatures. Cells are categorized into three broad groups: astrocyte in PTB (Astro), PTB_G, and TC_G. (D) Scatter diagram of the differential genes up-regulated in PTB_G and TC_G compared with Astro. (E) Bar plot of the GO terms (biological processes) enriched by genes that were both up-regulated in PTB_G and TC_G. Color of the bar plot corresponds to (D). (F) Scatter plot of differential genes between PTB_G and TC_G. (G) Bar plot of the GO terms (biological processes) separately enriched in TC_G (left) and PTB_G (right). Color of the bar plot denotes the cluster assignment, corresponding to (F).
We next compared the overall malignancy of PTB_G with TC_G by scoring the expression of pan-cancer signatures for each cell (Fig. 3C and table S8) (51). Astrocytes were used as the reference. RM05 was excluded from the analysis, as its PTB was spatially inseparable from TC (Fig. 1G and fig. S1, A and B). The result showed that PTB_G exhibited weaker pan-cancer signatures in general despite widespread inter- and intratumor heterogeneity, consistent with the general view that infiltrated tumor cells in normal tissues become less active or enter quiescence at early tumorigenesis stage (15).
The observation of distinct malignancy levels for PTB_G with TC_G encouraged us to further characterize the molecular features of GBM cells in general and alterations of infiltrated cells away from the tumor. By comparing the transcriptome of TC_G and PTB_G to astrocytes, respectively, 213 genes were specifically up-regulated in GBM cells for both regions (Fig. 3D and table S9). They were enriched in pathways such as synapse organization (NLGN1 and ROBO2), cell adhesion (CDH12 and GRID2), and cell growth (DISC1 and SEMA5A) (Fig. 3, D and E). We next directly compared the gene expression of GBM cells in two regions. The result revealed 681 up- and 2186 down-regulated genes for PTB_G (Fig. 3F and table S10). Genes repressed after invasion were enriched in the regulation of glycolysis (VEGFA and HIF1A), cell cycle (CDK4 and IGFBP2), and Wnt signal (SOX2 and SOX4) (Fig. 3, F and G), indicating that PTB_G adopted a less active state in metabolism and proliferation, as reported previously (10). Nevertheless, genes in synapse organization– (SLC24A2 and NRG3) and neuron development– (MBP and SHTN1) related pathways were induced in PTB_G. As no marker genes have been characterized to represent infiltrated GBM cells, we extracted the gene signature of PTB_G by filtering the DEGs of PTB_G with intrinsically expressed genes in GBM (52), followed by the STRING analysis to generate a 32-gene module (fig. S6, C and D) (53). Gene Ontology (GO) analysis highlighted the signature enriched in neuronal development–related pathways, such as synapse organization and ERBB signaling, implicating the microenvironment of the healthy brain region has an impact in shaping the transcriptome of PTB_G (fig. S6E) (54–56). We next assessed the microenvironment difference between TC and PTB. Besides GBM cells, the results revealed the altered expression for a large number of genes in all nonmalignant cell types, and the changes in the inhibitory/excitatory neuron and myeloid cells were most evident (fig. S7). In summary, our data suggest that infiltrated malignant cells exhibited distinct profiles, and the surrounding normal cells play important roles in shaping the microenvironment.
Diverse TFs underlying the GBM cellular states
It has been increasingly acknowledged that GBM cells are highly plastic (54). Multiple cellular states coexist in every tumor irrespective of the genetic characteristics (22, 26) and are reversible as modulated by tumor microenvironment (57). When we assessed the composition of four cellular states defined previously (22), not only high interpatient but also intrapatient heterogeneity between TC and PTB regions was observed (Fig. 4A). Specifically, the percentage of OPC-like cells at PTB was significantly higher than that at TC, at the expense of decreased proportion of AC-like cells (Fig. 4, B and C, and table S11), consistent with the above DEG analysis and recent reports (20, 24). In line with the deduced lineage relationship between PTB_G and TC_G (Fig. 3B), our results suggest that malignant cells acquire the proneural feature as they infiltrate into the neighboring region and build connections with local nonmalignant cells (Fig. 3, F and G), or cells at higher proneural state are more prone to disseminate.
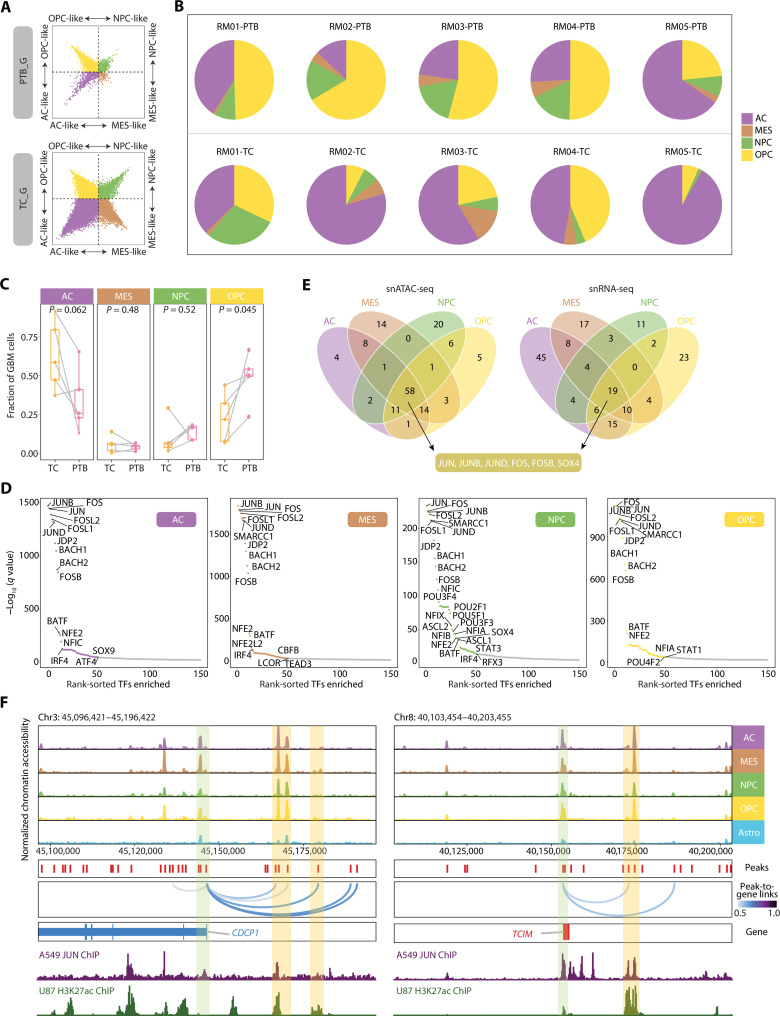
Fig. 4. GBM cells in four cellular states share the regulator AP-1.
(A) Two-dimensional representation of cellular states for PTB_G (top) and TC_G (bottom). The exact position of dots (GBM cells) reflects their relative scores for each cellular state signatures, and dots are colored by corresponding states. (B) Pie charts showing the proportions of GBM cells in four cellular states in each sample. The color corresponds to that in (A). (C) Percentages of AC, MES, NPC, and OPC GBM cells in patient-matched TC and PTB. P value from paired t test. (D) Scatter plots of overrepresented TF motifs in four cellular states inferred from snATAC-seq. The top 50 TFs are colored by corresponding states, same as (A). (E) Venn plots showing the intersection of motifs enriched in each cellular state based on snATAC-seq (left) and the intersection of regulons in each cellular state based on snRNA-seq (right). (F) Genome track visualization of loci for CDCP1 and TCIM. GBM cells with four cellular states and astrocytes from PTB are included. Inferred peak-to-gene links across the GBM dataset are shown in the middle, and ChIP-seq tracks for JUN and H3K27ac are shown bottom.
The heterogeneous cellular states are attributed to diverse regulatory programs in individual cells. We next analyzed differentially accessible peaks and identified tens of state-specific TFs (Fig. 4, D and E, and table S12). For example, GFI1, identified in the OPC-like state, is involved in oncogenesis by inhibiting neuronal differentiation in medulloblastoma (58). FOXK1, a pivotal TF involved in epithelial-to-mesenchymal transition and invasion (59), is enriched in MES-like cells. We noticed that several TFs previously reported to promote glioma progression might be activated only in certain states, such as OPC-like–specific HOXC9 that is implicated in autophagy suppression (60), and AC-like–specific SOX3 for glioma proliferation and invasion (61). Thus, these results indicate that tumor-promoting TFs might be active only in a proportion of malignant cells, leading to cautions to target specific factors for GBM eradication.
On the other hand, multiple AP-1 family regulators—including JUN, JUNB, JUND, FOS, FOSL1/2, JDP2, and FOSB—were revealed among the top enriched factors in all four states (Fig. 4D and table S12). We also performed pySCENIC analysis to characterize the regulons for each state and found that JUN, JUNB, JUND, FOS, FOSB, and SOX4 were shared by both snRNA-seq and snATAC-seq data (Fig. 4E) (62). As exemplified at the CDCP1 and TCIM gene loci, the high chromatin accessibility peaks were highlighted at both promoters and distal enhancers, where AP-1 may bind and in turn activate gene expression in cells from all four states (Fig. 4F and table S6) (63). AP-1 is a key factor for glioma progression and therefore suggested as a promising target for disease treatment (64–67).
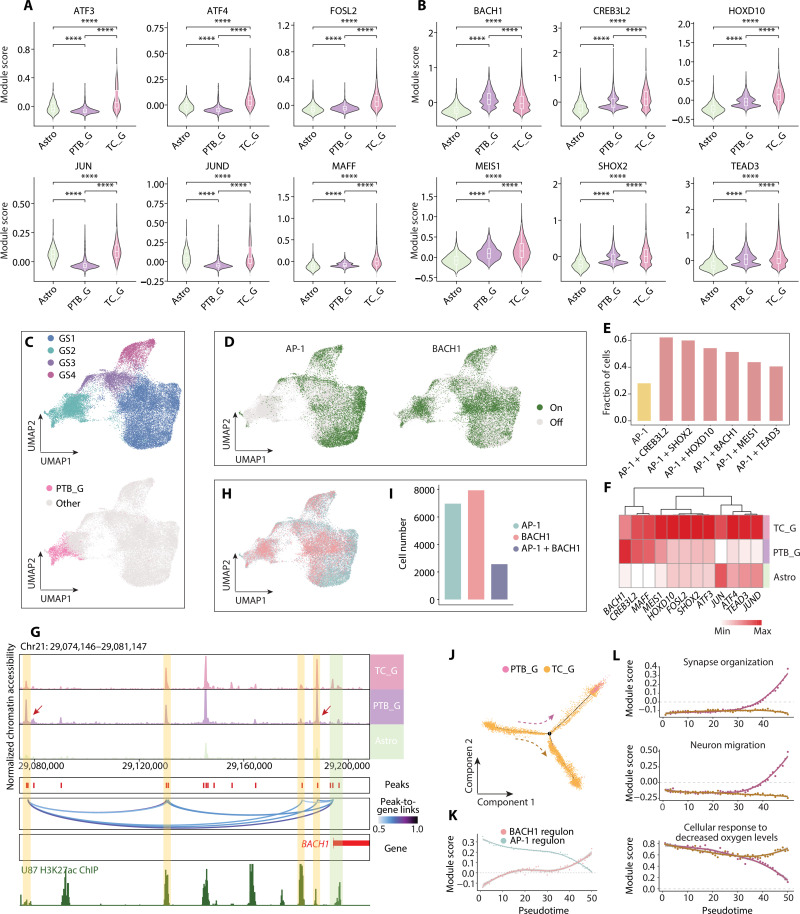
Declined AP-1 yet increased BACH1 activity in the PTB region
To assess whether AP-1 is the all-cell regulator for optimal GBM targeting, we evaluated the activity of AP-1 in PTB_G and TC_G at the single-cell level. While the activity of AP-1 regulon was higher in TC_G compared to astrocytes, it significantly declined in PTB_G, an indication of regional heterogeneity (Fig. 5A). This raised an intriguing question as to whether inhibition of the AP-1 could efficiently block the growth of residual tumor cells following surgery.
Fig. 5. The altered regulation of GBM cells in the PTB.
(A) Violin plots of AP-1 regulons scoring in three groups, including Astro, PTB_G, and TC_G. (B) Violin diagrams showing the regulons with higher activity in PTB_G compared with Astro. (C) UMAP plots showing GBM cell populations (i.e., GS1, GS2, GS3, and GS4). The top panel shows the subclusters of GBM cells, and the bottom panel highlights the GBM cells from PTB in pink. (D) UMAP diagrams of the activation of AP-1 (left) and BACH1 (right) regulons in GBM cells. (E) Bar plot showing the fraction of cells with activated regulon for AP-1 or combination with other candidate regulons. (F) Heatmap showing the expression of candidate TFs in three groups. (G) Genome track visualization of locus for BACH1 in three groups. Inferred peak-to-gene links across the three groups are shown in the middle, and ChIP-seq track for H3K27ac is shown bottom. (H) UMAP showing the activation status for AP-1 and BACH1 regulons in GBM cells. (I) Bar plot showing the number of GBM cells with different activation status. (J) Monocle trajectory for GBM cells from the two regions in RM04, colored by the origin tissues. (K) Scatter plot for selected regulon scores along the PTB_G branch. The scores were averaged for cells in each of the 50 trajectory bins. Solid lines represent loess regressions for the regulon. (L) Scatter plots showing module scores of selected GO terms along the two branches. ****P < 0.0001.
To search for potential regulators for infiltrated malignant cells, we screened the regulons in PTB_G and selected ones with significantly higher activity than those in astrocytes (table S13). The top six TFs identified were BACH1, CREB3L2, HOXD10, MEIS1, SHOX2, and TEAD3 (Fig. 5B), among which only BACH1 showed higher regulon activity in PTB_G compared to TC_G. Notably, the expression levels of these TFs were significantly higher in tumor tissue than the paired normal tissue according to TCGA RNA-seq data analyzed by GEPIA2 (fig. S8A) (68–71), suggesting that these TFs are more tumor specific. Among them, CREB3L2 is implicated in the survival of malignant glioma (72); HOXD10 is a pivotal regulator for GBM invasion (73); MEIS1 maintains stemness in leukemia and drives leukemogenesis (74, 75); SHOX2 is a survival marker for World Health Organization grades II to III diffuse gliomas (76). Notably, BACH1 is reported to contribute to cell pluripotency reprograming and the maintenance of stem cell identity (77) and drive glioma progression and therapy resistance (78–80).
To comprehensively evaluate the potential of targeting these TFs for tumor prevention compared to AP-1, we extracted all malignant cells from both regions and clustered them on the two-dimensional uniform manifold approximation and projection (UMAP). PTB_G was grouped into one cluster that also contained a portion of TC_G (GS2) (Fig. 5C and fig. S8B). The analysis of the public data from a previous study confirmed this clustering results (fig. S8C) (10). GO analysis showed that the PTB_G-containing cluster was enriched in synapse assembly associated pathways (fig. S8D and table S14), corroborative with the above finding that infiltrated malignant cells are from TC.
We then profiled the activity of each putative TF on the UMAP in addition to AP-1 (Fig. 5D and fig. S9A). Specifically, AP-1–positive cells were mainly distributed in GS1, GS3, and GS4, predominantly composed of TC_G, but not in GS2. In contrast, cells with positive regulon activity of the six candidate TFs were distributed not only in GS2 to various extents but also in TC_G-enriched clusters. Combining cells positive for either AP-1 or one of these candidate TFs accounted for around 40 to 60% of total GBM cells, much higher than AP-1 alone (Fig. 5E and table S15).
Notably, the expression level of BACH1 was higher in PTB_G than TC_G in our dataset (Fig. 5F and tables S9 and S10). We then examined the chromatin accessibility level at the BACH1 gene locus (Fig. 5G and table S6). It revealed that two known BACH1 enhancers were more active in PTB_G compared to TC_G and astrocytes (81), suggesting that the up-regulated BACH1 expression may be attributed to transcriptional control by distal enhancers. Moreover, the binding motif of BACH1, but not other candidate TFs, was enriched in all four cellular states (Fig. 4D). We also noticed, relative to AP-1/BACH1 double-positive cells, much more malignant cells are positive for either AP-1 or BACH1 regulon activity (Fig. 5, H and I, and table S16). BACH1 activity was the most inversely correlated with AP-1 compared to other TFs in all tumor cells from our data (fig. S9B), and their opposite trend was also confirmed by public datasets (fig. S9, C to E). Thus, our results raised a possibility that BACH1 is complementary to AP-1 for more comprehensive targeting of GBM cells.
Dynamic transcriptome regulation during GBM cell invasion
We next deduced how cells change the transcriptome as they infiltrate into neighboring regions. To do so, we constructed a pseudo-time trajectory for all malignant cells based on their expression profile similarity (Fig. 5J and fig. S10, A to C) (82). Malignant cells from TC occupied the trajectory starting point and evolved into two branches. In contrast, malignant cells from PTB were enriched at the end point of one branch. Similar results were obtained with the trajectory deduced by RNA velocity (fig. S10, D to F) (83). The cell distribution by pseudo-time trajectory was considered a predictor for lineage transition (49, 84). Our results here corroborate with the lineage deduction by inferCNV (Fig. 3B).
AP-1 activity was high at the starting point of the trajectory yet underwent a sharp decline toward the PTB_G containing branch, where a progressively increased regulon activity of BACH1 was seen (Fig. 5K and fig. S10G). The regulon of NFE2L2, which is negatively regulated by BACH1 (77), became less active as the BACH1 activity increased along the PTB_G branch (fig. S10G). Moreover, we checked the expression of AP-1 members and BACH1 and their downstream target genes, all of which displayed corroborative changes (fig. S10H). Thus, these data reinforced the anticorrelation of AP-1 and BACH1 activities in our study.
To characterize what other molecular features are changed during the transition from high AP-1 to high BACH1 state, we analyzed genes displaying a high correlation with the trajectory and clustered them according to the expression pattern (fig. S11A). Genes altered along with the PTB_G branch in three patients were further intersected to obtain consensus gene sets, resulting in 1801 up-regulated and 298 down-regulated genes (table S17). Consistently, GO analysis for up-regulated genes revealed regulatory functions in microtubule-based movement, neuron migration, synaptic transmission, cell junction assembly, and extracellular matrix organization (Fig. 5L and fig. S11, B to D) In contrast, the enriched biological processes for down-regulated genes included hypoxic response, double-strand break repair, and neuron death (Fig. 5L and fig. S11, E to G). In summary, these results indicate that accompanied with decreased AP-1 and increased BACH1 activity, infiltrated malignant cells may acquire more neuronal-like migration capability and less hypoxia stress and cell death.
The combination therapy targeting both AP-1 and BACH1
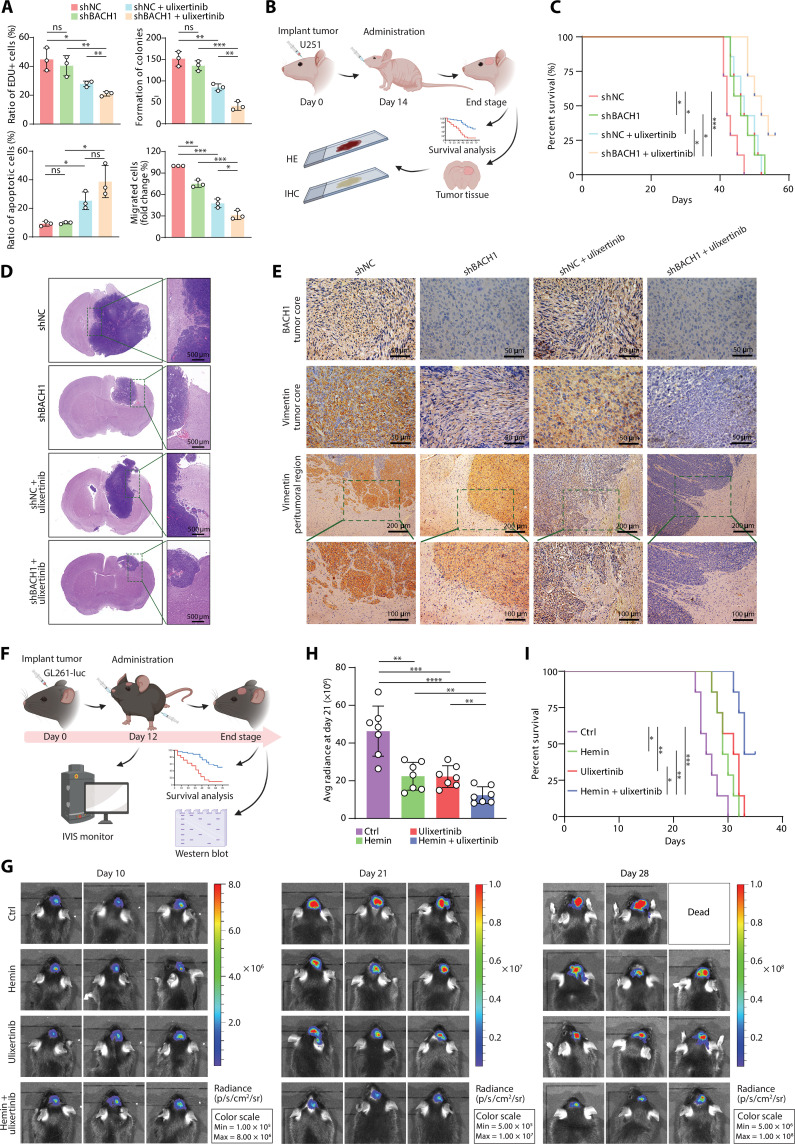
Following the above results, we were intrigued to assess if the finding of anticorrelated AP-1 and BACH1 activity from our data would predict a synergistic tumor repression effect by dual-target inhibition. To test this possibility first in vitro, the GBM cell line U251 was subjected to short hairpin RNA (shRNA)–mediated BACH1 knockdown and AP-1 inhibition by the potent first-in-class extracellular signal-regulated kinase 1/2 (ERK1/2) inhibitor ulixertinib (BVD-523) currently in clinical trials (85–87). While ulixertinib treatment alone resulted in significant suppression of cell proliferation and migration, as well as an increase of apoptosis, consistent with previous reports (85–87), the extent of which was further promoted by the combination treatment with BACH1 depletion (Fig. 6A and fig. S12, A to D). These in vitro observations were further consolidated by examining protein markers for apoptosis and migration by Western blot (fig. S12, E to H).
Fig. 6. Combination therapy suppresses tumor growth and improves survival in GBM mouse models.
(A) Quantification of BrdU incorporation, colony-formation, flow cytometry, and transwell migration assays of the indicated groups of U251 cells. Cells with or without BACH1 depletion were treated with or without ulixertinib (1 μM). Data represent the means ± SD from three independent assays. (B) Illustration depicting the therapy schedule. U251 cells (with or without BACH1 depletion) were intracranially injected into nude mice on day 0. Treatment started on day 14, with or without ulixertinib (80 mg/kg) orally twice daily (every 12 hours) for 5 days (n = 7 for each group). (C) Kaplan-Meier survival curves of the indicated groups of mice in (B). [(D) and (E)] Representative hematoxylin and eosin staining (D) and immunohistochemistry (E) images of U251-derived tumors of mice in (B). Scale bars were indicated in the plots. (F) Schematic representation of the treatment strategy. Luciferase-expressing GL261 cells were intracranially implanted in C57BL/6 mice on day 0. Treatment started on day 12 with hemin (50 mg/kg, every 12 hours) or (80 mg/kg, every 24 hours) or in combination for 5 days (n = 7 for each group). (G) Luminescence intensity showing tumors in representative mice over time. (H) Quantitative radiance in the experimental group on day 21. (I) The survival time of the indicated groups of mice in (F). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next for in vivo examination, we intracranially injected the immunodeficient mice with U251 cells with or without BACH1 depletion (Fig. 6B and fig. S12I). After tumor cell implantation, mice were treated with ulixertinib or the control vehicle. Similar to the in vitro results, combination treatment resulted in substantially improved tumor control and prolonged survival compared to either treatment alone (Fig. 6, C to E). Notably, scattered tumor cells around the tumor edge were observed in shNC- and ulixertinib-treated mice, indicating an invasive potential of tumor cells, which were eradicated when BACH1 was depleted (Fig. 6, D and E), thus suggesting a possible role of BACH1 in regulating tumor cell infiltration in vivo (79).
To exclude the possibility that our observation is due to the U251 model–specific effect, we established another preclinical mouse model by intracranially injecting the luciferase-expressing GL261 cells into immunocompetent C57BL/6 mice (fig. S13A). BACH1 depletion in the GL261 model was achieved by expressing shBach1 (fig. S13, B and C). Consistent with the results from the U251 model, BACH1 depletion inhibited tumor growth and improved survival, and the combination with ulixertinib exhibited a significantly improved tumor suppression effect than either treatment alone (fig. 13, D to F). To leverage the clinical potential of BACH1 inhibition for tumor control, we accomplished BACH1 suppression by the treatment of hemin, a Food and Drug Administration–approved small molecule that causes BACH1 degradation (Fig. 6F) (88, 89). After tumor formation, we treated mice with hemin or ulixertinib, or the combination, resulting in consistent observations with shBACH1 knockdown model (Fig. 6, G to I, and fig. S13G). These data demonstrate the effectiveness of dual-target therapy (AP-1 and BACH1) for GBM both in vitro and in vivo, warranting further evaluation in more clinically relevant tests.
DISCUSSION
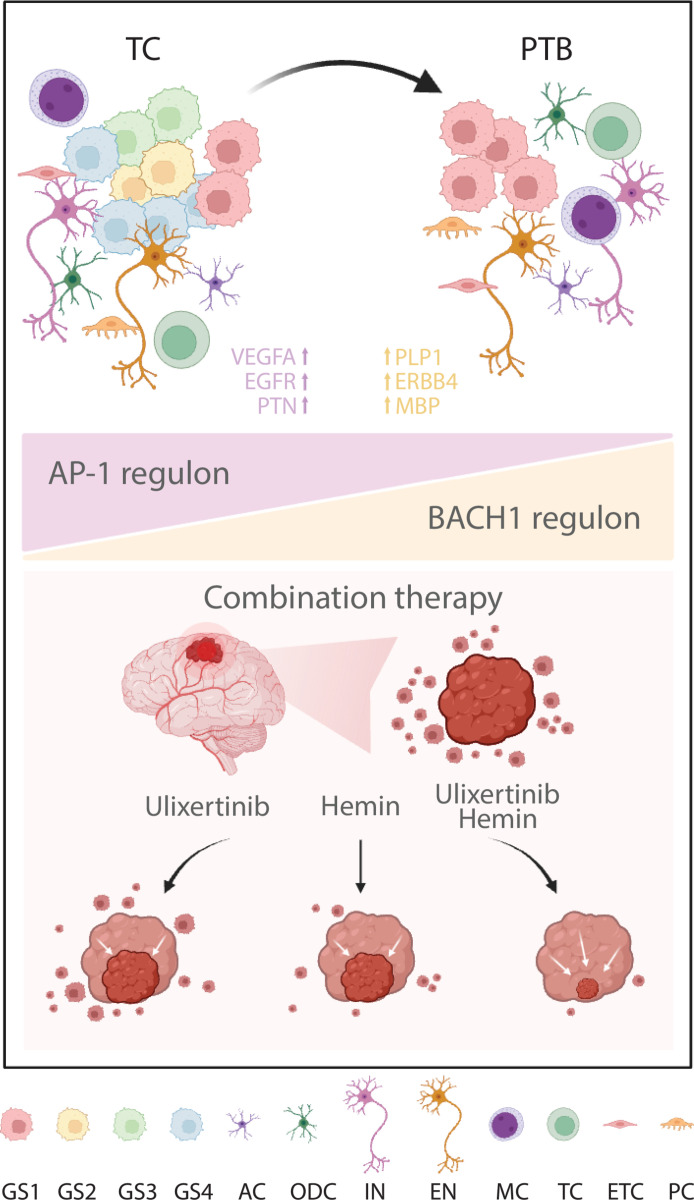
Intratumor heterogeneity is a notorious characteristic of glioma and a well-recognized contributor to treatment resistance, which has been attributed to genetic, epigenetic, and environmental factors (21–26, 54). However, recent efforts in elucidating the genetic contribution to GBM have not established a clear association of hypermutation with overall survival, cellular state classification, or guided satisfactory progress in treatment development (17, 30, 90, 91). On the other hand, insights regarding cellular state plasticity, determined by internal epigenetic and transcriptional programs with microenvironment modulation, have been proposed to account for treatment resistance and relapse (22, 92–94). While our understanding of cell plasticity within tumor regions is increasing (54), how infiltrated cells in the normal region behave remains elusive. In the current study, we have performed integrative analyses with snRNA-seq and snATAC data from both TC and PTB tissues, through which we have identified a cellular state shift toward proneural state as tumor cells infiltrate into distal regions and dissected potential transcriptional regulators for diverse cellular states (Fig. 7). We further demonstrate that targeting tumor cells of all states and regions give rise to synergistic therapeutic effects, thus providing insights into researches in regional heterogeneity and treatment development.
Fig. 7. Scheme explaining the altered characteristics of infiltrated GBM cells lurked in PTB.
AC, astrocyte; ODC, oligodendrocyte; IN, inhibitory neuron; EN, excitatory neuron; MC, myeloid; TC, T cell; ETC, endothelial cell; PC, pericyte.
Regional sampling and bulk tissue analyses have shown that the leading edge of glioma tissues exhibit proneural subtype and stem-like malignant cell characteristics, with up-regulated genes related to neuronal signaling (18, 19, 92). Moreover, malignant cells isolated from the tumor edge exhibit a higher rate of infiltrative growth, while cells at the TC show greater therapy resistance in animal models (95). To specifically characterize the infiltrative malignant cells, early studies performed scRNA-seq with peritumor tissues. The early study identified DEGs in metabolic and cell migration, yet the number of captured malignant cells is limited, restricting the dissection of cellular state dynamics in space (10, 84, 96). Our work here isolated thousands of tumor cells at PTB, thus allowing us to observe a marked transition toward the OPC state and induced the expression of genes particularly active in neuron development and synapse organization. This is reminiscent of the previous study, in which invasive tumor cells were mainly enriched for OPC/NPC cellular states and neurodevelopment-related transcriptional program (20).
These findings consolidate that the microenvironment is indispensable in shaping the transcriptional plasticity in and around GBM. Tumor-host interdependence has been shown to cause changeable transcriptional programs to fit the local environment, such as an inflammatory or hypoxia region, leading to GBM heterogeneity (97–101). Furthermore, tumor cells may interact with surrounding cells to promote the intricacy of transcriptome (20, 102–105), for example, the MES transition of GBM cells driven by immune cells (54, 93, 97). Neuronal activity, through paracrine signaling and glutamatergic synapses integrated into tumor cells, has been reported to reshape tumor cells with increased proliferation and invasion activities (20, 104, 106–108). Our finding of transcriptome dynamics in the context of the infiltration zone adds one more layer of complexity to the recent debate of brain tumor cellular state transitions, leading to cautions against therapies only targeting genetic lesions or GSC (20, 22, 26, 30, 57, 109, 110).
Despite distinct genotypes and cellular states, our work has revealed that GBM cells in different regions share a dependency on core transcriptional programs, yielding an attractive possibility to address the intratumor heterogeneity challenge by identifying more effective reagents or combination treatments that simultaneously and effectively target all cellular states and regions (30, 46, 57, 111). Specifically, we have explored the regulatory factors underlying each GBM state and shown that the AP-1 family is shared across all four states. The importance of AP-1 has been emphasized previously, especially regarding its role in inducing mesenchymal transition during tumor recurrence (64–67). Notably, the ERK inhibitor ulixertinib is currently being tested on pediatric patients with solid tumors, including glioma, in phase 2 clinical trials (NCT03698994 and NCT03155620) (87). However, variable responses to mitogen-activated protein kinase inhibitor (MAPKi) and rebound growth after treatment remain a clinical challenge, necessitating searching for new compounds or synergy with combination treatments (87). Our finding that AP-1 activity at PTB is decreased compared to that at TC provides a perspective to comprehend the partial effectiveness of MAPKi in glioma treatment. Meanwhile, it further endorses the exploration of specific tumor regulators at PTB, leading us to find that BACH1 and AP-1 are complementary targets for synergistic therapy in vivo, particularly in reducing tumor growth and invasion.
Regarding the limitation of this study and future perspectives, note that tumor invasion appears as a dynamic process, with varying molecular regulations adapting to internal needs and the microenvironment (55). Our data suggest that the cellular state changes as tumor cells disseminate or that cells in a specific state are more prone to invade neighboring areas. Therefore, to fully understand the cellular states of tumor cells continuously in space and deduce the invasion mechanism, collection of transcriptome and regulatory information for GBM cells at regions between TC and PTB, accompanied by characterizing the microenvironment from more patients, is needed. The recently developed spatial transcriptomics technologies shall be included in future studies. They will help researchers reveal the spatiotemporal features of GBM cells with distinct neighborhoods more clearly (112, 113) and address intriguing questions, including where and how tumor cells are colonized and regulated (55, 114). Second, while our data have shown that the microenvironment and altered TF activity are implicated in the cellular state transition, the identity of neuronal subtypes and tumor subpopulations driving this phenomenon as well as the regulatory mechanism and functions of shifted TF activity related to OPC state transition are incompletely understood. Future studies to elucidate key cell players and molecular events for tumor infiltration and cellular state shift may help identify effective targets for therapy. Third, the hemin and ulixertinib used in our study are both undergoing clinical trials (85–89), thus warranting this combination strategy to be further evaluated in preclinical and clinical trials for GBM. Our work consolidates the concept that therapies targeting cellular state has the potential to enhance the efficacy of surgical and cytotoxic treatments, as suggested previously (54). Last, we have provided access to a comprehensive single-cell multi-omics dataset through an online interface for researchers (https://db.cngb.org/cdcp/visualization?project=CNP0003766), which will be continuously updated to include data from more patients and regions.
MATERIALS AND METHODS
Study design
The first aim of this study was to identify the cell origin and molecular mechanism of the invasive cells located in the PTB. The second aim was to investigate most efficient combinatorial treatment considering both invasiveness and heterogeneity for GBM. To achieve the aims, GBM patients with extended resection were enrolled, and two regions (PTB and TC) from each patient were collected. The snRNA-seq and snATAC-seq were performed for each sample to dissect the regulatory mechanisms. To evaluate the effectiveness of the combination therapy, GBM cell line U251 was applied for in vitro studies. Two xenograft GBM models were generated for in vivo studies, including immunodeficient mice injected with U251 cells and immunocompetent mice injected with luciferase-expressing GL261 cells. All data in this study were reproduced in biological replicates for cell culture and in independent cohorts of mice. This study was approved by the Institutional Ethics Committee of the Faculty of Medicine at Renmin Hospital, Wuhan University (approval number: WDRY2021-K046). All patient tissues in this study were performed with patient-informed consents.
Statistical analysis
Statistical analysis between groups was performed using Student’s t test. In Kaplan-Meier survival curves, statistical significance was calculated by the log-rank (Mantel-Cox) test. Significance values are indicated in figures or marked as ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
Acknowledgments
We thank the other doctors in Renmin Hospital of Wuhan University who assisted in the operation and testing. We thank BGI for its single-cell technology and sequencing platform. We also thank people in BGI who provided analytical support, such as Q. Shi, S. Liu, S. Hao, and Q Yu, and who provided assistance with single-cell experiments, such as Y. Yuan, Q. Deng, Z. Wang, and G. Dong. We also thank Biorender for the schema drawing tool (www.Biorender.com) and all researchers who developed experimental methods and relevant single-cell analysis software.
Funding: This work was supported by the National Key Research and Development Program of China (2021YFA1100500 to L.C. and 2021YFF1200500 to Y.G.), the Shenzhen Science and Technology Program (RCJC20221008092804002 to Y.G.), the National Natural Science Foundation of China (32171289 to L.C., 81502175 to B.L., and 82072764 to Q.C.), the Fundamental Research Funds for the Central Universities (2042022rc0008 to L.C.), and the Interdisciplinary Innovative Talents Foundation from Renmin Hospital of Wuhan University.
Author contributions: Project design: L.C., B.L., and Y.G. Analysis design: X.W., L.C., Y.G., and T.L. Project supervision: L.C., Y.G., and B.L. Surgery resection: B.L., Q.C., J.W., D.T., and F.Y. Sample collection: Q.S. and H.L. Hematoxylin and eosin/immunohistochemistry/Western blot experiment: Q.S. and H.L. In vivo/vitro experiments: Q.S. and H.L. Single-cell library construction and sequencing: T.L., X.L., X.W., and Y.H. Single-cell data analysis: X.W. Interpretation of analysis results: X.W., L.C., Y.G., B.L., T.L., and Q.S. Analysis technology and platform support: Y.L., G.H., and H.S. Single-cell platform support: Y.G. and L.L. Result arrangement and visualization: X.W. and Q.S. Online webpage construction: W.W. and B.W. Writing—original draft: X.W. Writing—review and editing: L.C., X.W., B.L., Y.G., Q.S., and T.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All snRNA-seq and snATAC-seq data are available at China National GeneBank DataBase (https://db.cngb.org/cnsa/; CNP0003766) and National Genomics Data Center (https://ngdc.cncb.ac.cn/; PRJCA025943). The web interface for exploration of these datasets was also available (https://db.cngb.org/cdcp/visualization?project=CNP0003766). The publicly available data have been filed in The National Genomics Data Center (BF2024050815145).
Supplementary Materials
The PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S13
Legends for tables S1 to S17
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S17
REFERENCES AND NOTES
- 1.Price M., Neff C., Nagarajan N., Kruchko C., Waite K. A., Cioffi G., Cordeiro B. B., Willmarth N., Penas-Prado M., Gilbert M. R., Armstrong T. S., Barnholtz-Sloan J. S., Ostrom Q. T., CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 26, iii1–iii53 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R., Mason W. P., Van Den Bent M. J., Weller M., Fisher B., Taphoorn M. J. B., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O.; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 3.van Solinge T. S., Nieland L., Chiocca E. A., Broekman M. L. D., Advances in local therapy for glioblastoma - taking the fight to the tumour. Nat. Rev. Neurol. 18, 221–236 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milano M. T., Okunieff P., Donatello R. S., Mohile N. A., Sul J., Walter K. A., Korones D. N., Patterns and timing of recurrence after temozolomide-based chemoradiation for glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 78, 1147–1155 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Kallenberg K., Goldmann T., Menke J., Strik H., Bock H. C., Stockhammer F., Buhk J. H., Frahm J., Dechent P., Knauth M., Glioma infiltration of the corpus callosum: early signs detected by DTI. J. Neurooncol. 112, 217–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prados M. D., Byron S. A., Tran N. L., Phillips J. J., Molinaro A. M., Ligon K. L., Wen P. Y., Kuhn J. G., Mellinghoff I. K., de Groot J. F., Colman H., Cloughesy T. F., Chang S. M., Ryken T. C., Tembe W. D., Kiefer J. A., Berens M. E., Craig D. W., Carpten J. D., Trent J. M., Toward precision medicine in glioblastoma: The promise and the challenges. Neuro Oncol. 17, 1051–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrecca K., Guiot M. C., Panet-Raymond V., Souhami L., Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J Neurooncol. 111, 19–23 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Odde D., Glioblastoma cell invasion: Go? Grow? Yes. Neuro Oncol 25, 2163–2164 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown T. J., Brennan M. C., Li M., Church E. W., Brandmeir N. J., Rakszawski K. L., Patel A. S., Rizk E. B., Suki D., Sawaya R., Glantz M., Association of the extent of resection with survival in glioblastoma: A systematic review and meta-analysis. JAMA Oncol. 2, 1460–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darmanis S., Sloan S. A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., Li G., Chang S. D., Connolly I. D., Li Y., Barres B. A., Gephart M. H., Quake S. R., Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep 21, 1399–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shekarian T., Zinner C. P., Bartoszek E. M., Duchemin W., Wachnowicz A. T., Hogan S., Etter M. M., Flammer J., Paganetti C., Martins T. A., Schmassmann P., Zanganeh S., Le Goff F., Muraro M. G., Ritz M. F., Phillips D., Bhate S. S., Barlow G. L., Nolan G. P., Schürch C. M., Hutter G., Immunotherapy of glioblastoma explants induces interferon-γ responses and spatial immune cell rearrangements in tumor center, but not periphery. Sci. Adv. 8, eabn9440 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrieux G., Das T., Griffin M., Straehle J., Paine S. M. L., Beck J., Boerries M., Heiland D. H., Smith S. J., Rahman R., Chakraborty S., Spatially resolved transcriptomic profiles reveal unique defining molecular features of infiltrative 5ALA-metabolizing cells associated with glioblastoma recurrence. Genome Med. 15, 48 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J. H., Lee J. E., Kahng J. Y., Kim S. H., Park J. S., Yoon S. J., Um J. Y., Kim W. K., Lee J. K., Park J., Kim E. H., Lee J. H., Lee J. H., Chung W. S., Ju Y. S., Park S. H., Chang J. H., Kang S. G., Lee J. H., Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560, 243–247 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Spiteri I., Caravagna G., Cresswell G. D., Vatsiou A., Nichol D., Acar A., Ermini L., Chkhaidze K., Werner B., Mair R., Brognaro E., Verhaak R. G. W., Sanguinetti G., Piccirillo S. G. M., Watts C., Sottoriva A., Evolutionary dynamics of residual disease in human glioblastoma. Ann. Oncol. 30, 456–463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soureas K., Papadimitriou M. A., Panoutsopoulou K., Pilala K. M., Scorilas A., Avgeris M., Cancer quiescence: Non-coding RNAs in the spotlight. Trends Mol. Med. 29, 843–858 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Antonica F., Santomaso L., Pernici D., Petrucci L., Aiello G., Cutarelli A., Conti L., Romanel A., Miele E., Tebaldi T., Tiberi L., A slow-cycling/quiescent cells subpopulation is involved in glioma invasiveness. Nat Commun. 13, 4767 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barthel F. P., Johnson K. C., Varn F. S., Moskalik A. D., Tanner G., Kocakavuk E., Anderson K. J., Abiola O., Aldape K., Alfaro K. D., Alpar D., Amin S. B., Ashley D. M., Bandopadhayay P., Barnholtz-Sloan J. S., Beroukhim R., Bock C., Brastianos P. K., Brat D. J., Brodbelt A. R., Bruns A. F., Bulsara K. R., Chakrabarty A., Chakravarti A., Chuang J. H., Claus E. B., Cochran E. J., Connelly J., Costello J. F., Finocchiaro G., Fletcher M. N., French P. J., Gan H. K., Gilbert M. R., Gould P. V., Grimmer M. R., Iavarone A., Ismail A., Jenkinson M. D., Khasraw M., Kim H., Kouwenhoven M. C. M., Laviolette P. S., Li M., Lichter P., Ligon K. L., Lowman A. K., Malta T. M., Mazor T., McDonald K. L., Molinaro A. M., Nam D. H., Nayyar N., Ng H. K., Ngan C. Y., Niclou S. P., Niers J. M., Noushmehr H., Noorbakhsh J., Ormond D. R., Park C. K., Poisson L. M., Rabadan R., Radlwimmer B., Rao G., Reifenberger G., Sa J. K., Schuster M., Shaw B. L., Short S. C., Smitt P. A. S., Sloan A. E., Smits M., Suzuki H., Tabatabai G., Van Meir E. G., Watts C., Weller M., Wesseling P., Westerman B. A., Widhalm G., Woehrer A., Yung W. K. A., Zadeh G., Huse J. T., de Groot J. F., Stead L. F., Verhaak R. G. W.; The GLASS Consortium , Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576, 112–120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puchalski R. B., Shah N., Miller J., Dalley R., Nomura S. R., Yoon J. G., Smith K. A., Lankerovich M., Bertagnolli D., Bickley K., Boe A. F., Brouner K., Butler S., Caldejon S., Chapin M., Datta S., Dee N., Desta T., Dolbeare T., Dotson N., Ebbert A., Feng D., Feng X., Fisher M., Gee G., Goldy J., Gourley L., Gregor B. W., Gu G., Hejazinia N., Hohmann J., Hothi P., Howard R., Joines K., Kriedberg A., Kuan L., Lau C., Lee F., Lee H., Lemon T., Long F., Mastan N., Mott E., Murthy C., Ngo K., Olson E., Reding M., Riley Z., Rosen D., Sandman D., Shapovalova N., Slaughterbeck C. R., Sodt A., Stockdale G., Szafer A., Wakeman W., Wohnoutka P. E., White S. J., Marsh D., Rostomily R. C., Ng L., Dang C., Jones A., Keogh B., Gittleman H. R., Barnholtz-Sloan J. S., Cimino P. J., Uppin M. S., Keene C. D., Farrokhi F. R., Lathia J. D., Berens M. E., Iavarone A., Bernard A., Lein E., Phillips J. W., Rostad S. W., Cobbs C., Hawrylycz M. J., Foltz G. D., An anatomic transcriptional atlas of human glioblastoma. Science 360, 660–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill B. J., Pisapia D. J., Malone H. R., Goldstein H., Lei L., Sonabend A., Yun J., Samanamud J., Sims J. S., Banu M., Dovas A., Teich A. F., Sheth S. A., McKhann G. M., Sisti M. B., Bruce J. N., Sims P. A., Canoll P., MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 111, 12550–12555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkataramani V., Yang Y., Schubert M. C., Reyhan E., Tetzlaff S. K., Wißmann N., Botz M., Soyka S. J., Beretta C. A., Pramatarov R. L., Fankhauser L., Garofano L., Freudenberg A., Wagner J., Tanev D. I., Ratliff M., Xie R., Kessler T., Hoffmann D. C., Hai L., Dörflinger Y., Hoppe S., Yabo Y. A., Golebiewska A., Niclou S. P., Sahm F., Lasorella A., Slowik M., Döring L., Iavarone A., Wick W., Kuner T., Winkler F., Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell 185, 2899–2917.e31 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Verhaak R. G. W., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P., Alexe G., Lawrence M., O’Kelly M., Tamayo P., Weir B. A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H. S., Hodgson J. G., James C. D., Sarkaria J. N., Brennan C., Kahn A., Spellman P. T., Wilson R. K., Speed T. P., Gray J. W., Meyerson M., Getz G., Perou C. M., Hayes D. N.; Cancer Genome Atlas Research Network , Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neftel C., Laffy J., Filbin M. G., Hara T., Shore M. E., Rahme G. J., Richman A. R., Silverbush D., Shaw M. L., Hebert C. M., Dewitt J., Gritsch S., Perez E. M., Gonzalez Castro L. N., Lan X., Druck N., Rodman C., Dionne D., Kaplan A., Bertalan M. S., Small J., Pelton K., Becker S., Bonal D., Nguyen Q. D., Servis R. L., Fung J. M., Mylvaganam R., Mayr L., Gojo J., Haberler C., Geyeregger R., Czech T., Slavc I., Nahed B. V., Curry W. T., Carter B. S., Wakimoto H., Brastianos P. K., Batchelor T. T., Stemmer-Rachamimov A., Martinez-Lage M., Frosch M. P., Stamenkovic I., Riggi N., Rheinbay E., Monje M., Rozenblatt-Rosen O., Cahill D. P., Patel A. P., Hunter T., Verma I. M., Ligon K. L., Louis D. N., Regev A., Bernstein B. E., Tirosh I., Suvà M. L., An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couturier C. P., Ayyadhury S., Le P. U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., Lee C., Wang Y. C. D., Wee Yong V., Guiot M. C., Najafabadi H., Misic B., Antel J., Bourque G., Ragoussis J., Petrecca K., Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 11, 3406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garofano L., Migliozzi S., Oh Y. T., D’Angelo F., Najac R. D., Ko A., Frangaj B., Caruso F. P., Yu K., Yuan J. Z., Zhao W. T., Di Stefano A. L., Bielle F., Jiang T., Sims P., Suva M. L., Tang F. C., Su X. D., Ceccarelli M., Sanson M., Lasorella A., Iavarone A., Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Cancer 2, 141–156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards L. M., Whitley O. K. N., MacLeod G., Cavalli F. M. G., Coutinho F. J., Jaramillo J. E., Svergun N., Riverin M., Croucher D. C., Kushida M., Yu K., Guilhamon P., Rastegar N., Ahmadi M., Bhatti J. K., Bozek D. A., Li N., Lee L., Che C., Luis E., Park N. I., Xu Z., Ketela T., Moore R. A., Marra M. A., Spears J., Cusimano M. D., Das S., Bernstein M., Haibe-Kains B., Lupien M., Luchman H. A., Weiss S., Angers S., Dirks P. B., Bader G. D., Pugh T. J., Gradient of developmental and injury response transcriptional states defines functional vulnerabilities underpinning glioblastoma heterogeneity. Nat. Cancer 2, 157–173 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Wang L., Babikir H., Müller S., Yagnik G., Shamardani K., Catalan F., Kohanbash G., Alvarado B., Di Lullo E., Kriegstein A., Shah S., Wadhwa H., Chang S. M., Phillips J. J., Aghi M. K., Diaz A. A., The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 9, 1708–1719 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Sun Q., Wang W., Liu B., Gu Y., Chen L., Decoding key cell sub-populations and molecular alterations in glioblastoma at recurrence by single-cell analysis. Acta Neuropathol. Commun. 11, 125 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Louis D. N., Rozenblatt-Rosen O., Suvà M. L., Regev A., Bernstein B. E., Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guilhamon P., Chesnelong C., Kushida M. M., Nikolic A., Singhal D., MacLeod G., Madani Tonekaboni S. A., Cavalli F. M., Arlidge C., Rajakulendran N., Rastegar N., Hao X., Hassam R., Smith L. J., Whetstone H., Coutinho F. J., Nadorp B., Ellestad K. I., Luchman H. A., Chan J. A., Shoichet M. S., Taylor M. D., Haibe-Kains B., Weiss S., Angers S., Gallo M., Dirks P. B., Lupien M., Single-cell chromatin accessibility profiling of glioblastoma identifies an invasive cancer stem cell population associated with lower survival. eLife 10, e64090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson K. C., Anderson K. J., Courtois E. T., Gujar A. D., Barthel F. P., Varn F. S., Luo D., Seignon M., Yi E., Kim H., Estecio M. R. H., Zhao D., Tang M., Navin N. E., Maurya R., Ngan C. Y., Verburg N., de Witt Hamer P. C., Bulsara K., Samuels M. L., Das S., Robson P., Verhaak R. G. W., Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response. Nat. Genet. 53, 1456–1468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson J. G., Fine H. A., Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov. 11, 575–590 (2021). [DOI] [PubMed] [Google Scholar]
- 32.C. H. Waddington, The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology (Allen & Unwin, 1957).
- 33.Flavahan W. A., Gaskell E., Bernstein B. E., Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu L., Yang P., Zhao Y., Zhuang Z., Wang Z., Song R., Zhang J., Liu C., Gao Q., Xu Q., Wei X., Sun H., Ye B., Wu Y., Zhang N., Lei G., Yu L., Yan J., Diao G., Meng F., Bai C., Mao P., Yu Y., Wang M., Yuan Y., Deng Q., Li Z., Huang Y., Hu G., Liu Y., Wang X., Xu Z., Liu P., Bi Y., Shi Y., Zhang S., Chen Z., Wang J., Xu X., Wu G., Wang F., Gao G., Liu L., Liu W., Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity 53, 685–696.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolic A., Singhal D., Ellestad K., Johnston M., Shen Y., Gillmor A., Morrissy S., Cairncross J. G., Jones S., Lupien M., Chan J. A., Neri P., Bahlis N., Gallo M., Copy-scAT: Deconvoluting single-cell chromatin accessibility of genetic subclones in cancer. Sci. Adv. 7, eabg6045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couturier C. P., Nadaf J., Li Z., Baig S., Riva G., Le P., Kloosterman D. J., Monlong J., Nkili Meyong A., Allache R., Degenhard T., Al-Rashid M., Guiot M. C., Bourque G., Ragoussis J., Akkari L., Quintana F. J., Petrecca K., Glioblastoma scRNA-seq shows treatment-induced, immune-dependent increase in mesenchymal cancer cells and structural variants in distal neural stem cells. Neuro Oncol. 24, 1494–1508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J. E., Purcaro M. J., Pratt H. E., Epstein C. B., Shoresh N., Adrian J., Kawli T., Davis C. A., Dobin A., Kaul R., Halow J., Van Nostrand E. L., Freese P., Gorkin D. U., Shen Y., He Y., Mackiewicz M., Pauli-Behn F., Williams B. A., Mortazavi A., Keller C. A., Zhang X. O., Elhajjajy S. I., Huey J., Dickel D. E., Snetkova V., Wei X., Wang X., Rivera-Mulia J. C., Rozowsky J., Zhang J., Chhetri S. B., Zhang J., Victorsen A., White K. P., Visel A., Yeo G. W., Burge C. B., Lécuyer E., Gilbert D. M., Dekker J., Rinn J., Mendenhall E. M., Ecker J. R., Kellis M., Klein R. J., Noble W. S., Kundaje A., Guigó R., Farnham P. J., Cherry J. M., Myers R. M., Ren B., Graveley B. R., Gerstein M. B., Pennacchio L. A., Snyder M. P., Bernstein B. E., Wold B., Hardison R. C., Gingeras T. R., Stamatoyannopoulos J. A., Weng Z., Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., Ntini E., Arner E., Valen E., Li K., Schwarzfischer L., Glatz D., Raithel J., Lilje B., Rapin N., Bagger F. O., Jørgensen M., Andersen P. R., Bertin N., Rackham O., Burroughs A. M., Baillie J. K., Ishizu Y., Shimizu Y., Furuhata E., Maeda S., Negishi Y., Mungall C. J., Meehan T. F., Lassmann T., Itoh M., Kawaji H., Kondo N., Kawai J., Lennartsson A., Daub C. O., Heutink P., Hume D. A., Jensen T. H., Suzuki H., Hayashizaki Y., Müller F., Forrest A. R. R., Carninci P., Rehli M., Sandelin A., An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Lancet D., Shmueli O., Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005). [DOI] [PubMed] [Google Scholar]
- 40.McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Wang J., Yang L., Zhao C., Wu L. N., Xu L., Zhang F., Weng Q., Wegner M., Lu Q. R., CTCF-mediated chromatin looping in EGR2 regulation and SUZ12 recruitment critical for peripheral myelination and repair. Nat. Commun. 11, 4133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuguchi R., Kriks S., Cordes R., Gossler A., Ma Q., Goulding M., Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat. Neurosci. 9, 770–778 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Maiques-Diaz A., Spencer G. J., Lynch J. T., Ciceri F., Williams E. L., Amaral F. M. R., Wiseman D. H., Harris W. J., Li Y., Sahoo S., Hitchin J. R., Mould D. P., Fairweather E. E., Waszkowycz B., Jordan A. M., Smith D. L., Somervaille T. C. P., Enhancer activation by pharmacologic displacement of LSD1 from GFI1 induces differentiation in acute myeloid leukemia. Cell Rep. 22, 3641–3659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glasgow S. M., Carlson J. C., Zhu W., Chaboub L. S., Kang P., Lee H. K., Clovis Y. M., Lozzi B. E., McEvilly R. J., Rosenfeld M. G., Creighton C. J., Lee S. K., Mohila C. A., Deneen B., Glia-specific enhancers and chromatin structure regulate NFIA expression and glioma tumorigenesis. Nat Neurosci. 20, 1520–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J., Hoxha E., Song H. R., A novel NFIA-NFκB feed-forward loop contributes to glioblastoma cell survival. Neuro Oncol. 19, 524–534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raviram R., Raman A., Preissl S., Ning J., Wu S., Koga T., Zhang K., Brennan C. W., Zhu C., Luebeck J., Van Deynze K., Han J. Y., Hou X., Ye Z., Mischel A. K., Li Y. E., Fang R., Baback T., Mugford J., Han C. Z., Glass C. K., Barr C. L., Mischel P. S., Bafna V., Escoubet L., Ren B., Chen C. C., Integrated analysis of single-cell chromatin state and transcriptome identified common vulnerability despite glioblastoma heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 120, e2210991120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtenbach S., Cruz A. M., Rodriguez D. A., Durante M. A., Harbour J. W., Uphyloplot2: Visualizing phylogenetic trees from single-cell RNA-seq data. BMC Genomics 22, 419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhaduri A., Di Lullo E., Jung D., Muller S., Crouch E. E., Espinosa C. S., Ozawa T., Alvarado B., Spatazza J., Cadwell C. R., Wilkins G., Velmeshev D., Liu S. J., Malatesta M., Andrews M. G., Mostajo-Radji M. A., Huang E. J., Nowakowski T. J., Lim D. A., Diaz A., Raleigh D. R., Kriegstein A. R., Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 26, 48–63.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu L., Wu W., Zhang J., Zhao Z., Li L., Zhu M., Wu M., Wu F., Zhou F., Du Y., Chai R.-C., Zhang W., Qiu X., Liu Q., Wang Z., Li J., Li K., Chen A., Jiang Y., Xiao X., Zou H., Srivastava R., Zhang T., Cai Y., Liang Y., Huang B., Zhang R., Lin F., Hu L., Wang X., Qian X., Lv S., Hu B., Zheng S., Hu Z., Shen H., You Y., Verhaak R. G. W., Jiang T., Wang Q., Natural coevolution of tumor and immunoenvironment in glioblastoma. Cancer Discov. 12, 2820–2837 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lv S. Q., Fu Z., Yang L., Li Q. R., Zhu J., Gai Q. J., Mao M., He J., Qin Y., Yao X. X., Lan X., Wang Y. X., Lu H. M., Xiang Y., Zhang Z. X., Huang G. H., Yang W., Kang P., Sun Z., Shi Y., Yao X. H., Bian X. W., Wang Y., Comprehensive omics analyses profile genesets related with tumor heterogeneity of multifocal glioblastomas and reveal LIF/CCL2 as biomarkers for mesenchymal subtype. Theranostics 12, 459–473 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinker G. S., Greenwald A. C., Tal R., Orlova Z., Cuoco M. S., McFarland J. M., Warren A., Rodman C., Roth J. A., Bender S. A., Kumar B., Rocco J. W., Fernandes P., Mader C. C., Keren-Shaul H., Plotnikov A., Barr H., Tsherniak A., Rozenblatt-Rosen O., Krizhanovsky V., Puram S. V., Regev A., Tirosh I., Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., Carvalho A. C., Lyu S., Li P., Li Y., Barthel F., Cho H. J., Lin Y. H., Satani N., Martinez-Ledesma E., Zheng S., Chang E., Sauvé C. E. G., Olar A., Lan Z. D., Finocchiaro G., Phillips J. J., Berger M. S., Gabrusiewicz K. R., Wang G., Eskilsson E., Hu J., Mikkelsen T., DePinho R. A., Muller F., Heimberger A. B., Sulman E. P., Nam D. H., Verh R. G. W.aak, Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., Mering C., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yabo Y. A., Niclou S. P., Golebiewska A., Cancer cell heterogeneity and plasticity: A paradigm shift in glioblastoma. Neuro Oncol. 24, 669–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heuer S., Winkler F., Glioblastoma revisited: From neuronal-like invasion to pacemaking. Trends Cancer 9, 887–896 (2023). [DOI] [PubMed] [Google Scholar]
- 56.Brooks L. J., Ragdale H. S., Hill C. S., Clements M., Parrinello S., Injury programs shape glioblastoma. Trends Neurosci. 45, 865–876 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Dirkse A., Golebiewska A., Buder T., Nazarov P. V., Muller A., Poovathingal S., Brons N. H., Leite S., Sauvageot N., Sarkisjan D., Seyfrid M., Fritah S., Stieber D., Michelucci A., Hertel F., Herold-Mende C., Azuaje F., Skupin A., Bjerkvig R., Deutsch A., Voss-Bohme A., Niclou S. P., Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 10, 1787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee C., Rudneva V. A., Erkek S., Zapatka M., Chau L. Q., Tacheva-Grigorova S. K., Garancher A., Rusert J. M., Aksoy O., Lea R., Mohammad H. P., Wang J., Weiss W. A., Grimes H. L., Pfister S. M., Northcott P. A., Wechsler-Reya R. J., Lsd1 as a therapeutic target in Gfi1-activated medulloblastoma. Nat. Commun. 10, 332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Y., Zhang P., Huang X., Yan Q., Wu M., Xie R., Wu Y., Zhang M., Nan Q., Zhao J., Li A., Xiong J., Ren Y., Bai Y., Chen Y., Liu S., Wang J., Direct regulation of FOXK1 by C-jun promotes proliferation, invasion and metastasis in gastric cancer cells. Cell Death Dis. 7, e2480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xuan F., Huang M., Liu W., Ding H., Yang L., Cui H., Homeobox C9 suppresses Beclin1-mediated autophagy in glioblastoma by directly inhibiting the transcription of death-associated protein kinase 1. Neuro Oncol. 18, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su R., Cao S., Ma J., Liu Y., Liu X., Zheng J., Chen J., Liu L., Cai H., Li Z., Zhao L., He Q., Xue Y., Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer 16, 171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van de Sande B., Flerin C., Davie K., De Waegeneer M., Hulselmans G., Aibar S., Seurinck R., Saelens W., Cannoodt R., Rouchon Q., Verbeiren T., De Maeyer D., Reumers J., Saeys Y., Aerts S., A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 63.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X., Zhou R., Xiong Y., Zhou L., Yan X., Wang M., Li F., Xie C., Zhang Y., Huang Z., Ding C., Shi K., Li W., Liu Y., Cao Z., Zhang Z. N., Zhou S., Chen C., Zhang Y., Chen L., Wang Y., Sequential fate-switches in stem-like cells drive the tumorigenic trajectory from human neural stem cells to malignant glioma. Cell Res. 31, 684–702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Jung J., Babikir H., Shamardani K., Jain S., Feng X., Gupta N., Rosi S., Chang S., Raleigh D., Solomon D., Phillips J. J., Diaz A. A., A single-cell atlas of glioblastoma evolution under therapy reveals cell-intrinsic and cell-extrinsic therapeutic targets. Nat. Cancer 3, 1534–1552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z., Wang S., Li H. L., Luo H., Wu X., Lu J., Wang H. W., Chen Y., Chen D., Wu W. T., Zhang S., He Q., Lu D., Liu N., You Y., Wu W., Wang H., FOSL1 promotes proneural-to-mesenchymal transition of glioblastoma stem cells via UBC9/CYLD/NF-κB axis. Mol. Ther. 30, 2568–2583 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marques C., Unterkircher T., Kroon P., Oldrini B., Izzo A., Dramaretska Y., Ferrarese R., Kling E., Schnell O., Nelander S., Wagner E. F., Bakiri L., Gargiulo G., Carro M. S., Squatrito M., NF1 regulates mesenchymal glioblastoma plasticity and aggressiveness through the AP-1 transcription factor FOSL1. eLife 10, e64846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang Z., Kang B., Li C., Chen T., Zhang Z., GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R. M., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., Stuart J. M., The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., Foster B., Moser M., Karasik E., Gillard B., Ramsey K., Sullivan S., Bridge J., Magazine H., Syron J., Fleming J., Siminoff L., Traino H., Mosavel M., Barker L., Jewell S., Rohrer D., Maxim D., Filkins D., Harbach P., Cortadillo E., Berghuis B., Turner L., Hudson E., Feenstra K., Sobin L., Robb J., Branton P., Korzeniewski G., Shive C., Tabor D., Qi L., Groch K., Nampally S., Buia S., Zimmerman A., Smith A., Burges R., Robinson K., Valentino K., Bradbury D., Cosentino M., Diaz-Mayoral N., Kennedy M., Engel T., Williams P., Erickson K., Ardlie K., Winckler W., Getz G., De Luca D., Arthur D. M., Kellis M., Thomson A., Young T., Gelfand E., Donovan M., Meng Y., Grant G., Mash D., Marcus Y., Basile M., Liu J., Zhu J., Tu Z., Cox N. J., Nicolae D. L., Gamazon E. R., Im H. K., Konkashbaev A., Pritchard J., Stevens M., Flutre T., Wen X., Dermitzakis E. T., Lappalainen T., Guigo R., Monlong J., Sammeth M., Koller D., Battle A., Mostafavi S., Carthy M. M., Rivas M., Maller J., Rusyn I., Nobel A., Wright F., Shabalin A., Feolo M., Sharopova N., Sturcke A., Paschal J., Anderson J. M., Wilder E. L., Derr L. K., Green E. D., Struewing J. P., Temple G., Volpi S., Boyer J. T., Thomson E. J., Guyer M. S., Ng C., Abdallah A., Colantuoni D., Insel T. R., Koester S. E., Little A. R., Bender P. K., Lehner T., Yao Y., Compton C. C., Vaught J. B., Sawyer S., Lockhart N. C., Demchok J., Moore H. F., The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.GTEx Consortium , The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheng Z., Li L., Zhu L. J., Smith T. W., Demers A., Ross A. H., Moser R. P., Green M. R., A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat. Med. 16, 671–677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yachi K., Tsuda M., Kohsaka S., Wang L., Oda Y., Tanikawa S., Ohba Y., Tanaka S., miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition. Signal Transduct. Target. Ther. 3, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar A. R., Sarver A. L., Wu B., Kersey J. H., Meis1 maintains stemness signature in MLL-AF9 leukemia. Blood 115, 3642–3643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yokoyama T., Nakatake M., Kuwata T., Couzinet A., Goitsuka R., Tsutsumi S., Aburatani H., Valk P. J. M., Delwel R., Nakamura T., MEIS1-mediated transactivation of synaptotagmin-like 1 promotes CXCL12/CXCR4 signaling and leukemogenesis. J. Clin. Invest. 126, 1664–1678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y.-A., Zhou Y., Luo X., Song K., Ma X., Sathe A., Girard L., Xiao G., Gazdar A. F., SHOX2 is a potent independent biomarker to predict survival of WHO grade II-III diffuse gliomas. EBioMedicine 13, 80–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ahuja M., Ammal Kaidery N., Attucks O. C., McDade E., Hushpulian D. M., Gaisin A., Gaisina I., Ahn Y. H., Nikulin S., Poloznikov A., Gazaryan I., Yamamoto M., Matsumoto M., Igarashi K., Sharma S. M., Thomas B., Bach1 derepression is neuroprotective in a mouse model of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 118, e2111643118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu T., Wang Y., Wang Y., Cheung S. K., Or P. M., Wong C. W., Guan J., Li Z., Yang W., Tu Y., Wang J., Ho W. L., Gu H., Cheng A. S., Tsui S. K., Chan A. M., The mitotic regulator RCC2 promotes glucose metabolism through BACH1-dependent transcriptional upregulation of hexokinase II in glioma. Cancer Lett. 549, 215914 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Cong Z., Yuan F., Wang H., Cai X., Zhu J., Tang T., Zhang L., Han Y., Ma C., BTB domain and CNC homolog 1 promotes glioma invasion mainly through regulating extracellular matrix and increases ferroptosis sensitivity. Biochim. Biophys. Acta. Mol. Basis Dis. 1868, 166554 (2022). [DOI] [PubMed] [Google Scholar]
- 80.Nie E., Jin X., Wu W., Yu T., Zhou X., Zhi T., Shi Z., Zhang J., Liu N., You Y., BACH1 promotes temozolomide resistance in glioblastoma through antagonizing the function of p53. Sci. Rep. 6, 39743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M., Lancet D., Cohen D., GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, bax028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bergen V., Lange M., Peidli S., Wolf F. A., Theis F. J., Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Yu K., Hu Y. Q., Wu F., Guo Q. F., Qian Z. H., Hu W. E., Chen J., Wang K. Y., Fan X. Y., Wu X. L., Rasko J. E. J., Fan X. L., Iavarone A., Jiang T., Tang F. C., Su X.-D., Surveying brain tumor heterogeneity by single-cell RNA-sequencing of multi-sector biopsies. Natl. Sci. Rev. 7, 1306–1318 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smalley I., Smalley K. S. M., ERK inhibition: A new front in the war against MAPK pathway-driven cancers? Cancer Discov. 8, 140–142 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Sullivan R. J., Infante J. R., Janku F., Wong D. J. L., Sosman J. A., Keedy V., Patel M. R., Shapiro G. I., Mier J. W., Tolcher A. W., Wang-Gillam A., Sznol M., Flaherty K., Buchbinder E., Carvajal R. D., Varghese A. M., Lacouture M. E., Ribas A., Patel S. P., DeCrescenzo G. A., Emery C. M., Groover A. L., Saha S., Varterasian M., Welsch D. J., Hyman D. M., Li B. T., First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: Results of a phase I dose-escalation and expansion study. Cancer Discov. 8, 184–195 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Sigaud R., Rösch L., Gatzweiler C., Benzel J., von Soosten L., Peterziel H., Selt F., Najafi S., Ayhan S., Gerloff X. F., Hofmann N., Büdenbender I., Schmitt L., Foerster K. I., Burhenne J., Haefeli W. E., Korshunov A., Sahm F., van Tilburg C. M., Jones D. T. W., Pfister S. M., Knoerzer D., Kreider B. L., Sauter M., Pajtler K. W., Zuckermann M., Oehme I., Witt O., Milde T., The first-in-class ERK inhibitor ulixertinib shows promising activity in mitogen-activated protein kinase (MAPK)-driven pediatric low-grade glioma models. Neuro Oncol. 25, 566–579 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zenke-Kawasaki Y., Dohi Y., Katoh Y., Ikura T., Ikura M., Asahara T., Tokunaga F., Iwai K., Igarashi K., Heme induces ubiquitination and degradation of the transcription factor bach1. Mol. Cell. Biol. 27, 6962–6971 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dhar G. J., Bossenmaier I., Petryka Z. J., Cardinal R., Watson C. J., Effects of hematin in hepatic porphyria. Further studies. Ann. Intern. Med. 83, 20–30 (1975). [DOI] [PubMed] [Google Scholar]
- 90.Brennan C. W., Verhaak R. G., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., Van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O’Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G., Chin L.; TCGA Research. Network , The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakthikumar S., Sakthikumar S., Roy A., Haseeb L., Pettersson M. E., Sundström E., Marinescu V. D., Lindblad-Toh K., Lindblad-Toh K., Forsberg-Nilsson K., Whole-genome sequencing of glioblastoma reveals enrichment of non-coding constraint mutations in known and novel genes. Genome Biol. 21, 127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Varn F. S., Johnson K. C., Martinek J., Huse J. T., Nasrallah M. P., Wesseling P., Cooper L. A. D., Malta T. M., Wade T. E., Sabedot T. S., Brat D., Gould P. V., Wöehrer A., Aldape K., Ismail A., Sivajothi S. K., Barthel F. P., Kim H., Kocakavuk E., Ahmed N., White K., Datta I., Moon H. E., Pollock S., Goldfarb C., Lee G. H., Garofano L., Anderson K. J., Nehar-Belaid D., Barnholtz-Sloan J. S., Bakas S., Byrne A. T., D’Angelo F., Gan H. K., Khasraw M., Migliozzi S., Ormond D. R., Paek S. H., Van Meir E. G., Walenkamp A. M. E., Watts C., Weiss T., Weller M., Palucka K., Stead L. F., Poisson L. M., Noushmehr H., Iavarone A., Verhaak R. G. W., Ormond D. R., Paek S. H., Alfaro K. D., Amin S. B., Ashley D. M., Bock C., Brodbelt A., Bulsara K. R., Castro A. V., Connelly J. M., Costello J. F., de Groot J. F., Finocchiaro G., French P. J., Golebiewska A., Hau A. C., Hong C., Horbinski C., Kannan K. S., Kouwenhoven M. C., Lasorella A., LaViolette P. S., Ligon K. L., Lowman A. K., Mehta S., Miletic H., Molinaro A. M., Ng H. K., Niclou S. P., Niers J. M., Phillips J. J., Rabadan R., Rao G., Reifenberger G., Sanai N., Short S. C., Smitt P. S., Sloan A. E., Smits M., Snyder J. M., Suzuki H., Tabatabai G., Tanner G., Tomaszewski W. H., Wells M., Westerman B. A., Wheeler H., Xie J., Yung W. K. A., Zadeh G., Zhao J.; GLASS. Consortium , Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell 185, 2184–2199.e16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Castro L. N. G., Liu I., Filbin M., Characterizing the biology of primary brain tumors and their microenvironment via single-cell profiling methods. Neuro Oncol. 25, 234–247 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoogstrate Y., Draaisma K., Ghisai S. A., van Hijfte L., Barin N., de Heer I., Coppieters W., van den Bosch T. P. P., Bolleboom A., Gao Z., Vincent A. J. P. E., Karim L., Deckers M., Taphoorn M. J. B., Kerkhof M., Weyerbrock A., Sanson M., Hoeben A., Lukacova S., Lombardi G., Leenstra S., Hanse M., Fleischeuer R. E. M., Watts C., Angelopoulos N., Gorlia T., Golfinopoulos V., Bours V., van den Bent M. J., Robe P. A., French P. J., Transcriptome analysis reveals tumor microenvironment changes in glioblastoma. Cancer Cell 41, 678–692.e7 (2023). [DOI] [PubMed] [Google Scholar]
- 95.Bastola S., Pavlyukov M. S., Yamashita D., Ghosh S., Cho H., Kagaya N., Zhang Z., Minata M., Lee Y., Sadahiro H., Yamaguchi S., Komarova S., Yang E., Markert J., Nabors L. B., Bhat K., Lee J., Chen Q., Crossman D. K., Shin-Ya K., Nam D. H., Nakano I., Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun. 11, 4660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmassmann P., Roux J., Dettling S., Hogan S., Shekarian T., Martins T. A., Ritz M. F., Herter S., Bacac M., Hutter G., Single-cell characterization of human GBM reveals regional differences in tumor-infiltrating leukocyte activation. eLife 12, RP92678 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ravi V. M., Will P., Kueckelhaus J., Sun N., Joseph K., Salie H., Vollmer L., Kuliesiute U., von Ehr J., Benotmane J. K., Neidert N., Follo M., Scherer F., Goeldner J. M., Behringer S. P., Franco P., Khiat M., Zhang J., Hofmann U. G., Fung C., Ricklefs F. L., Lamszus K., Boerries M., Ku M., Beck J., Sankowski R., Schwabenland M., Prinz M., Schuller U., Killmer S., Bengsch B., Walch A. K., Delev D., Schnell O., Heiland D. H., Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639–655.e13 (2022). [DOI] [PubMed] [Google Scholar]
- 98.Hara T., Chanoch-Myers R., Mathewson N. D., Myskiw C., Atta L., Bussema L., Eichhorn S. W., Greenwald A. C., Kinker G. S., Rodman C., Gonzalez Castro L. N., Wakimoto H., Rozenblatt-Rosen O., Zhuang X., Fan J., Hunter T., Verma I. M., Wucherpfennig K. W., Regev A., Suvà M. L., Tirosh I., Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 39, 779–792.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mathewson N. D., Ashenberg O., Tirosh I., Gritsch S., Perez E. M., Marx S., Jerby-Arnon L., Chanoch-Myers R., Hara T., Richman A. R., Ito Y., Pyrdol J., Friedrich M., Schumann K., Poitras M. J., Gokhale P. C., Gonzalez Castro L. N., Shore M. E., Hebert C. M., Shaw B., Cahill H. L., Drummond M., Zhang W., Olawoyin O., Wakimoto H., Rozenblatt-Rosen O., Brastianos P. K., Liu X. S., Jones P. S., Cahill D. P., Frosch M. P., Louis D. N., Freeman G. J., Ligon K. L., Marson A., Chiocca E. A., Reardon D. A., Regev A., Suvà M. L., Wucherpfennig K. W., Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 184, 1281–1298.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia-Diaz C., Poysti A., Mereu E., Clements M. P., Brooks L. J., Galvez-Cancino F., Castillo S. P., Tang W. H., Beattie G., Courtot L., Ruiz S., Roncaroli F., Yuan Y. Y., Marguerat S., Quezada S. A., Heyn H., Parrinello S., Glioblastoma cell fate is differentially regulated by the microenvironments of the tumor bulk and infiltrative margin. Cell Rep. 42, 112472 (2023). [DOI] [PubMed] [Google Scholar]
- 101.Gangoso E., Southgate B., Bradley L., Rus S., Galvez-Cancino F., McGivern N., Guc E., Kapourani C. A., Byron A., Ferguson K. M., Alfazema N., Morrison G., Grant V., Blin C., Sou I., Marques-Torrejon M. A., Conde L., Parrinello S., Herrero J., Beck S., Brandner S., Brennan P. M., Bertone P., Pollard J. W., Quezada S. A., Sproul D., Frame M. C., Serrels A., Pollard S. M., Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 184, 2454–2470.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Osswald M., Jung E., Sahm F., Solecki G., Venkataramani V., Blaes J., Weil S., Horstmann H., Wiestler B., Syed M., Huang L. L., Ratliff M., Jazi K. K., Kurz F. T., Schmenger T., Lemke D., Gommel M., Pauli M., Liao Y. X., Haring P., Pusch S., Herl V., Steinhauser C., Krunic D., Jarahian M., Miletic H., Berghoff A. S., Griesbeck O., Kalamakis G., Garaschuk O., Preusser M., Weiss S., Liu H. K., Heiland S., Platten M., Huber P. E., Kuner T., von Deimling A., Wick W., Winkler F., Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Venteicher A. S., Tirosh I., Hebert C., Yizhak K., Neftel C., Filbin M. G., Hovestadt V., Escalante L. E., Shaw M. L., Rodman C., Gillespie S. M., Dionne D., Luo C. C., Ravichandran H., Mylvaganam R., Mount C., Onozato M. L., Nahed B. V., Wakimoto H., Curry W. T., Iafrate A. J., Rivera M. N., Frosch M. P., Golub T. R., Brastianos P. K., Getz G., Patel A. P., Monje M., Cahill D. P., Rozenblatt-Rosen O., Louis D. N., Bernstein B. E., Regev A., Suvà M. L., Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venkatesh H. S., Morishita W., Geraghty A. C., Silverbush D., Gillespie S. M., Arzt M., Tam L. T., Espenel C., Ponnuswami A., Ni L., Woo P. J., Taylor K. R., Agarwal A., Regev A., Brang D., Vogel H., Hervey-Jumper S., Bergles D. E., Suvà M. L., Malenka R. C., Monje M., Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dai X., Ye L., Li H., Dong X., Tian H., Gao P., Dong J., Cheng H., Crosstalk between microglia and neural stem cells influences the relapse of glioblastoma in GBM immunological microenvironment. Clin. Immunol. 251, 109333 (2023). [DOI] [PubMed] [Google Scholar]
- 106.Krishna S., Choudhury A., Keough M. B., Seo K., Ni L., Kakaizada S., Lee A., Aabedi A., Popova G., Lipkin B., Cao C., Nava Gonzales C., Sudharshan R., Egladyous A., Almeida N., Zhang Y., Molinaro A. M., Venkatesh H. S., Daniel A. G. S., Shamardani K., Hyer J., Chang E. F., Findlay A., Phillips J. J., Nagarajan S., Raleigh D. R., Brang D., Monje M., Hervey-Jumper S. L., Glioblastoma remodelling of human neural circuits decreases survival. Nature 617, 599–607 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Venkatesh H. S., Johung T. B., Caretti V., Noll A., Tang Y., Nagaraja S., Gibson E. M., Mount C. W., Polepalli J., Mitra S. S., Woo P. J., Malenka R. C., Vogel H., Bredel M., Mallick P., Monje M., Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell 161, 803–816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venkatesh H. S., Tam L. T., Woo P. J., Lennon J., Nagaraja S., Gillespie S. M., Ni J., Duveau D. Y., Morris P. J., Zhao J. J., Thomas C. J., Monje M., Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 549, 533–537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fine H. A., Malignant Gliomas: Simplifying the Complexity. Cancer Discov. 9, 1650–1652 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Weller M., van den Bent M., Preusser M., Le Rhun E., Tonn J. C., Minniti G., Bendszus M., Balana C., Chinot O., Dirven L., French P., Hegi M. E., Jakola A. S., Platten M., Roth P., Rudà R., Short S., Smits M., Taphoorn M. J. B., von Deimling A., Westphal M., Soffietti R., Reifenberger G., Wick W., EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 18, 170–186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chaligne R., Gaiti F., Silverbush D., Schiffman J. S., Weisman H. R., Kluegel L., Gritsch S., Deochand S. D., Castro L. N. G., Richman A. R., Klughammer J., Biancalani T., Muus C., Sheridan C., Alonso A., Izzo F., Park J., Rozenblatt-Rosen O., Regev A., Suva M. L., Landau D. A., Epigenetic encoding, heritability and plasticity of glioma transcriptional cell states. Nat. Genet. 53, 1469–1479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heumos L., Schaar A. C., Lance C., Litinetskaya A., Drost F., Zappia L., Lücken M. D., Strobl D. C., Henao J., Curion F.; Single-cell Best Practices Consortium, Schiller H. B., Theis F. J., Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vandereyken K., Sifrim A., Thienpont B., Voet T., Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ratliff M., Karimian-Jazi K., Hoffmann D. C., Rauschenbach L., Simon M., Hai L., Mandelbaum H., Schubert M. C., Kessler T., Uhlig S., Azorin D. D., Jung E., Osswald M., Solecki G., Maros M. E., Venkataramani V., Glas M., Etminan N., Scheffler B., Wick W., Winkler F., Individual glioblastoma cells harbor both proliferative and invasive capabilities during tumor progression. Neuro Oncol. 25, 2150–2162 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bakken T., Hodge R., Miller J., Yao Z., Nguyen T., Aevermann B., Barkan E., Bertagnolli D., Casper T., Dee N., Garren E., Goldy J., Graybuck L., Kroll M., Lasken R., Lathia K., Parry S., Rimorin C., Scheuermann R., Schork N., Shehata S., Tieu M., Phillips J., Bernard A., Smith K., Zeng H., Lein E., Tasic B., Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLOS ONE 13, e0209648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu Y., Wei X., Deng Q., Lan Q., Guo Y., Han L., Yuan Y., Fan P., Wu P., Shangguan S., Liu Y., Lai Y., Volpe G., Esteban M., Liu C., Hou Y., Liu L., Single-nucleus chromatin accessibility landscape reveals diversity in regulatory regions across distinct adult rat cortex. Front. Mol. Neurosci. 14, 651355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi Q., Liu S., Kristiansen K., Liu L., The FASTQ+ format and PISA. Bioinformatics 38, 4639–4642 (2022). [DOI] [PubMed] [Google Scholar]
- 118.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lun A. T. L., Riesenfeld S., Andrews T., Dao T. P., Gomes T.; participants in the 1st Human Cell Atlas Jamboree, Marioni J. C., EmptyDrops: Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Sys. 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.L. M. Innes, J. Healy, J. Melville, UMAP: uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 [stat.ML] (2018).
- 123.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 124.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G., clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A., Fan J., Borm L. E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P. V., RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN] (2013).
- 127.Granja J. M., Corces M. R., Pierce S. E., Bagdatli S. T., Choudhry H., Chang H. Y., Greenleaf W. J., ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wolf F. A., Angerer P., Theis F. J., SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S13
Legends for tables S1 to S17
References
Tables S1 to S17