ABSTRACT
Background
Recently developed rapid real-time reverse transcription PCR (RT-PCR) systems adopting microfluidic thermal cycling technology are ideal for point-of-care (POC) testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Because the RNA extraction step before real-time RT-PCR is rate-limiting, a direct RNA extraction method (direct method) that adopts chemical viral lysis and eliminates RNA purification steps is preferable for rapid real-time RT-PCR. In the direct method, selecting the transport medium is essential because it may be introduced into subsequent real-time RT-PCR steps, but might inhibit PCR. However, the influence of transport medium on the combination of the direct method and rapid real-time RT-PCR has been yet unstudied. In the present study, we examined the influence of various transport mediums when combining the direct method and rapid real-time RT-PCR of GeneSoC® (GeneSoC® RT-PCR), the recently developed compact PCR system that adapts novel microfluidic thermal cycling technology.
Methods
To explore the influence of the transport medium on the GeneSoC® RT-PCR, the concordance of the RNA extraction and direct method was evaluated in the clinical samples collected in viral transport medium (VTM) or eSwab®. The sensitivity of GeneSoC® RT-PCR combined with the direct method was assessed using spiked samples in generic (H2O and PBS) or commercially available transport media (VTM and eSwab®). Analytical sensitivity was examined using clinical specimens collected from the VTM and eSwab®. The inhibitory effect of PCR inhibitors on clinical specimens was assessed using clinical samples diluted 1,000 times.
Results
While only 1 copy/reaction of RNA was detected in H2O and eSwab® of the spiked samples, a minimum of 5 copies/reaction was detected in PBS (-) and VTM. Among the clinical specimens tested using the direct method, the detection of viral RNA was unstable in the samples containing less than 100 copies/reaction viral RNA in VTM, whereas less than 10 copies/reaction viral RNA were detected in eSwab®. The positive, negative, and overall concordance between the RNA extraction and the direct method was 84%, 100%, and 85%, respectively, in eSwab® samples, whereas the values were 35%, 100%, and 38%, respectively, in VTM samples. When the clinical samples were diluted 1,000 times, GeneSoC® RT-PCR could detect as low as 1.15 copies/reaction RNA using direct method, and the sensitivity was comparable to that of RNA extraction.
Conclusion
The combination of the direct method and microfluidic rapid PCR machine GeneSoC® has a high sensitivity for detecting SARS-CoV-2 RNA in clinical samples with eSwab® transport medium.
Keywords: direct method, GeneSoC®, rapid PCR, SARS-CoV-2, transport medium
Numerous testing methods for viral detection have been developed worldwide following the onset of the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Real-time reverse transcription-polymerase chain reaction (RT-PCR) was the first detection method developed for SARS-CoV-21 and is the most used clinical diagnostic method.2 In general, the PCR volumes used for real-time RT-PCR are relatively large, with a turnover time of approximately one hour. Therefore, portable and rapid PCR systems are required for point-of-care (POC) testing. GeneSoC® is a recently developed compact PCR system with a novel microfluidic thermal cycling technology. In this system, the PCR solution shuttles between different heaters through a microflow channel on a small chip at a very high speed. Using this mechanism, GeneSoC® enables rapid real-time RT-PCR for approximately 15 min and can be used for POC testing.
Another vital factor in POC testing for SARS-CoV-2 infection is RNA extraction before real-time RT-PCR. Clinical specimens can be processed using following two methods. The first involves extracting and purifying viral RNA from clinical samples using commercially available kits or automated machines. This methodology involves chemical viral lysis with a detergent and RNA purification using a silica membrane3 or magnetic beads.4 The advantage of this method is that high-purity RNA can be obtained, and the carryover of PCR inhibitors following real-time RT-PCR can be avoided. However, this methodology requires laboratory equipment (kits, centrifuges, or specialized machines) and has a relatively long turnover time (30–60 minutes). The second approach is the ‘direct method’, which involves chemical viral lysis and eliminates RNA purification steps.5 In this methodology, the carryover of PCR inhibitors is reduced by manipulating the formulation of the sample processing solutions. This method is advantageous because it is a simplified procedure that requires only adding a sample processing solution to a clinical specimen. However, the carryover of PCR inhibitors or transport medium in clinical specimens cannot be completely eliminated.
To maximize the leverage of the rapid real-time RT-PCR of GeneSoC® (GeneSoC® RT-PCR) in POC testing, a simplified procedure and short turnover time of the direct method are desirable. A pretreatment kit for GeneSoC®, adopting the direct method, has been developed recently. The pretreatment step for real-time RT-PCR reaction using GeneSoC® comprises adding a single reagent, which is available in the kit, to the clinical specimen, and waiting for 5 min. Despite the convenience and rapid sample processing time, similar to all direct methods, the carryover of PCR inhibitors in clinical samples and the transport medium to the real-time RT-PCR reaction is inevitable when using this kit. Although the carryover of PCR inhibitors cannot be controlled, the adverse impact of transport medium carryover on the subsequent real-time RT-PCR reaction can be adjusted by selecting a suitable transport medium. However, little is known about the influence of transport medium on real-time RT-PCR using the direct method.
In this study, we examined the influence of various transport media when using the direct method and GeneSoC® to formulate a highly sensitive and rapid real-time RT-PCR protocol for POC testing of SARS-CoV-2.
MATERIALS AND METHODS
SARS-CoV-2 viral RNA, clinical specimens, and diluted clinical specimens
To determine the analytical sensitivity of the direct method in the presence of various transport media, we used the EDX SARS-CoV-2 standard (Bio-Rad, Hercules, CA) as the viral RNA for GeneSoC® RT-PCR. Clinical samples were collected at the Tottori University Hospital. A total of 66 nasopharyngeal swabs were obtained from patients with COVID-19. Twenty six swabs were placed in viral transport medium (VTM; SUGIYAMA-GEN Co., Ltd., Tokyo, Japan), and 40 swabs were placed in eSwab® (Copan, Brescia, Italy). Among them, samples for which the viral load were not determined with standard real-time quantitative RT-PCR (real-time qRT-PCR) were excluded, and the remaining 24 swabs in VTM and 34 swabs in eSwab® were used in this study. To prepare artificially low viral copy number samples, we randomly selected 12 samples in eSwab and diluted them 1,000 times with eSwab®. Clinical specimens were tested immediately or stored at –80°C until use.
RNA extraction
As the RNA extraction, we extracted RNA from clinical specimens with the QIAamp Viral RNA Mini Kit (Qiagen, Tokyo, Japan) according to the manufacturer’s instructions.
Direct method
For the direct method without RNA extraction, 3 μL of the clinical specimens were mixed with 15 μL of the Lysis buffer provided by GeneSoC® PCR Pretreatment kit (Kyorin Pharmaceutical Co., Ltd., Tokyo, Japan), and incubated for 5 min at room temperature.
Standard real-time qRT-PCR
A real-time qRT-PCR was performed using a QuantStudio 5 system (Thermo Fisher Scientific, Waltham, MA). Real-time qRT-PCR reactions were performed using 20 μL of reaction mixture, containing 5 μL of TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific), 0.9 µM primers, and 0.25 µM probe. Primers and probes were targeted to the N2 gene RNA of SARS-CoV-26, 7 (Table 1). Amplification was performed under the following conditions: an initial reverse transcription at 50°C for 5 min, followed by an initial denaturation at 95°C for 20 s and 45 cycles each at 95°C for 15 s and 60°C for 1min. N2 gene RNA standard (Nihon Gene Research Laboratories. Inc., Miyagi, Japan) was diluted ten-fold to obtain the standard curve.
Table 1. Primers and probes used in the conventional real-time RT-PCR.
| Name | Sequence |
| 2019 nCoV N F2 primer | AAATTTTGGGGACCAGGAAC |
| 2019 nCoV N R2 primer | TGGCAGCTGTGTAGGTCAAC |
| 2019 nCoV N P2 probe | FAM-ATGTCGCGCATTGGCATGGA-QSY |
GeneSoC® RT-PCR
For the GeneSoC® RT-PCR, we used GeneSoC® SARS-CoV-2 N2 Assay Kit (Kyorin) containing primers and probes targeting N2 gene. After mixing 5 μL of testing samples with 15 μL of the amplification mixture provided by the kit, rapid real-time RT-PCR was performed using GeneSoC® following the manufacturer’s instructions. The reaction conditions were as follows: 42°C for 60 s (reverse transcription) and 96°C for 10 s, followed by 50 cycles of denaturation at 96°C for 10 s, annealing and extension at 58°C for 8s. Amplification results were interpreted as positive when the amplification curve was generated within 50 cycles.
GeneSoC® RT- PCR in spiked samples
Spiked samples were prepared by mixing the EDX SARS-CoV-2 standard (200 copies/µL of SARS-CoV-2 RNA) with various solvents that is generic (H2O and PBS) or commercially available media (VTM and eSwab®). To imitate the direct method, 3 µL of these spiked samples were mixed with 15 µL of H2O, mimicking the Lysis buffer of the GeneSoC® PCR Pretreatment kit (Kyorin). Prepared samples contain SARS-CoV-2 RNA (0.2, 1, 2, and 20 copies/μL) and 17% of the transport medium (H2O, PBS, eSwab®, and VTM) (Fig. 1A, *). We used 5 µL of the prepared samples to make up the 20 µL of the PCR reaction mixture and applied them to the microfluidic chip of GeneSoC®. Therefore, the final inputs of SARS-CoV-2 RNA to GeneSoC® RT-PCR were 1, 5, 10, and 100 copies/reaction, and the final composition of the transport medium was 4.3%. Reactions were performed in quadruplicate.
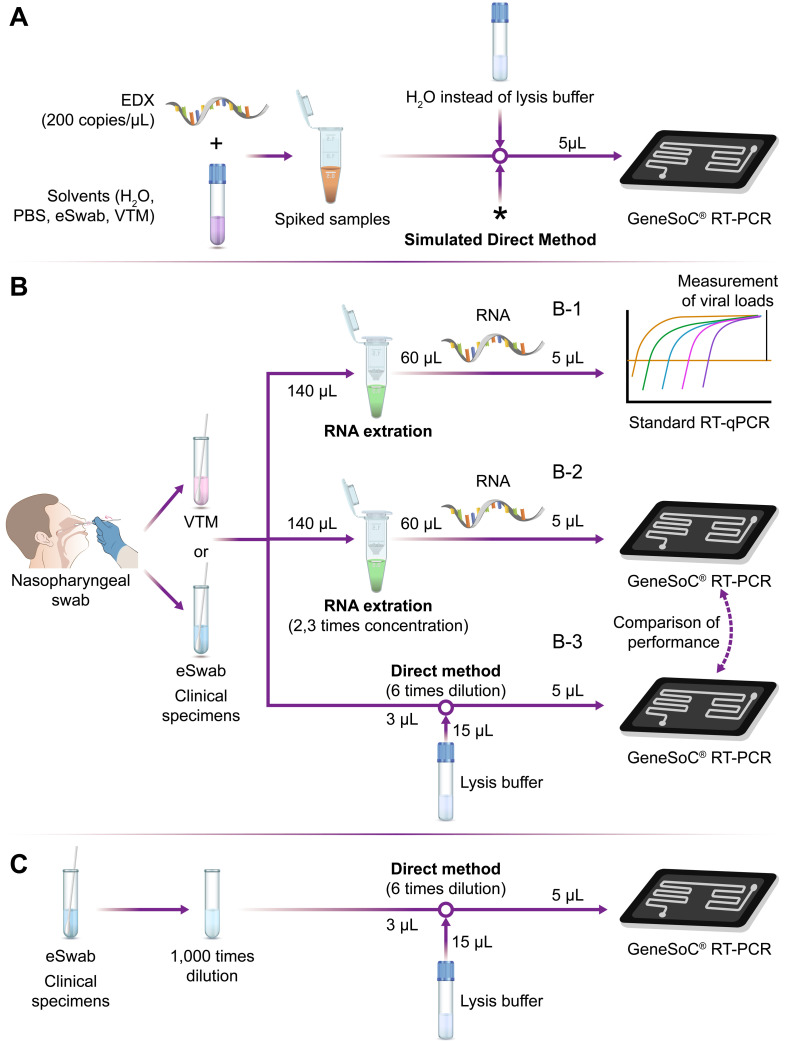
Fig. 1.
Schematic flowchart of the experiments using spiked samples (A), clinical specimens (B), and diluted clinical specimens (C). (A) Spiked samples were prepared by mixing EDX (200 copies/µL of SARS-CoV-2 RNA), various solvents (H2O, PBS, eSwab®, and VTM), and H2O. H2O was used to imitate the Lysis buffer of the GeneSoC® PCR Pretreatment kit. *Prepared samples contain various concentrations of SARS-CoV-2 RNA (0.2, 1, 2, and 20 copies/μL) and 17% of the transport medium (H2O, PBS, eSwab®, and VTM) at this point. As the next step, we used 5 µL of the samples to make up the 20 µL of the PCR reaction mixture and applied them to the microfluidic chip of GeneSoC. Therefore, the final inputs of RNA were 1, 5, 10, and 100 copies/reaction, respectively, and the final composition of the transport medium was 4.3% in the GeneSoC® RT-PCR, which mimicked the direct method. (B) Nasopharyngeal swabs from COVID-19 patients were collected in VTM or eSwab®. The viral loads in each sample were determined by RNA extraction followed by standard real-time qRT-PCR (B-1). Using these samples, we performed GeneSoC® RT-PCR in combination with RNA extraction (B-2) or direct method (B-3) and compared the performance of these two pretreatment methods. Real-time qRT-PCR: Real-time quantitative reverse transcription PCR. (C) Randomly selected clinical specimens in eSwab® were diluted 1,000 times with eSwab®. The diluted samples were treated with direct method and processed to GeneSoC® RT-PCR. The detection limit of this combination was re-evaluated using these diluted samples.
Measurement of viral copy numbers in clinical specimens
The viral copy numbers of clinical specimens were measured by RNA extraction followed by real-time qRT-PCR (Fig. 1B, B-1), according to the protocol of the National Institute of Infectious Diseases in Japan.6, 7
GeneSoC® RT- PCR in clinical specimens
Clinical specimens collected in VTM or eSwab® were tested with GeneSoC® RT-PCR in combination with RNA extraction (Fig. 1B, B-2) or direct method (Fig. 1B, B-3) and compared the performance of these two pretreatment methods in GeneSoC® RT-PCR.
GeneSoC® RT- PCR in 1000 times diluted clinical specimens
Randomly selected 12 clinical specimens collected in eSwab® were 1,000 times diluted with eSwab®. Using these diluted samples that contain a very low copy number of viral RNA and diluted biological materials in the clinical specimens, we performed GeneSoC® RT-PCR with direct method (Fig. 1C).
Statistical analysis
The correlation between the Ct values for standard RT-qPCR and GeneSoC® RT- PCR was calculated using Pearson’s rank correlation. The comparison of the Ct values for RNA extraction and direct method in GeneSoC® RT- PCR was performed with the Wilcoxon Signed-Rank Test. All statistical analysis was conducted with IBM SPSS Statistics 28.0 (IBM Corp., Armonk, NY).
Ethics approval and consent to participate
All study protocols were approved by the Institutional Review Board (IRB) of Tottori University Hospital (approval number: 2673). Written informed consent was obtained from all participants before collecting the clinical samples for this study.
RESULTS
Influence of the transport medium for the sensitivity of GeneSoC® RT-PCR in spiked samples
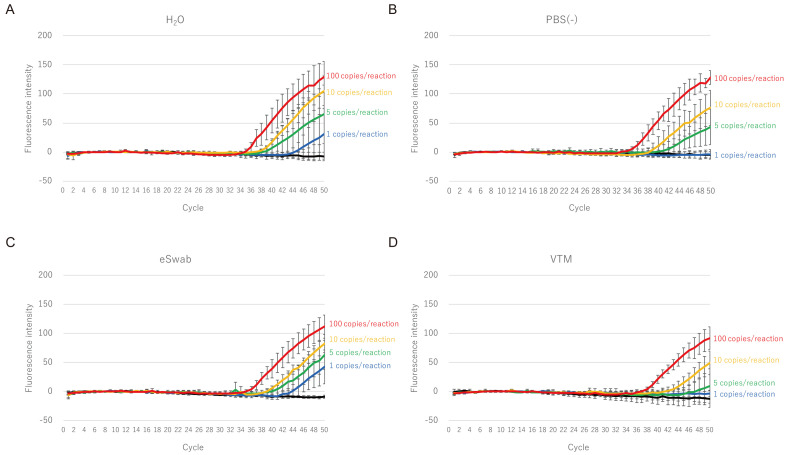
First, we tested the influence of the transport medium for the sensitivity of the GeneSoC® RT-PCR using spiked samples. The samples were prepared by diluting the EDX SARS-CoV-2 standard (1, 5, 10, or 100 copies per reaction). The final concentrations of the different transport media (H2O, PBS, eSwab®, and VTM) in each sample were adjusted to the same concentration (4.3%) as those of the clinical specimens processed using the direct method (Fig. 1A). As shown in Fig. 2, 1 copy/reaction of RNA was detected in H2O and eSwab®, whereas 5 copies/reaction was the minimum detectable amount of RNA in PBS (–) and VTM. In addition, a minimum of 5 copies/reaction of RNA was detected in the eSwab® in a reproducible manner, whereas, in other media (H2O, eSwab®, and VTM), 10 copies/reaction of RNA was required (Table 2).
Fig. 2.
Limit of detection of GeneSoC® RT-PCR using spiked samples; samples containing 4.3% of H2O (A), PBS (–) (B), eSwab® transport medium (C), or VTM transport medium (D) were prepared. Amplification plots were obtained using various concentrations of EDX SARS-CoV-2 standard (1, 5, 10, and 100 copies/reaction). Each experiment was performed in quadruplicate.
Table 2. Cycle threshold (Ct) values to detect various concentrations of template RNA in the spiked samples.
| copies/reaction | H2O | PBS (–) | eSwab | VTM |
| NTC | – (0/4) | – (0/4) | – (0/4) | |
| 1 | 44.3 (2/4) | – (0/4) | 44.0 ± 1.38 (3/4) | – (0/4) |
| 5 | 40.4 ± 2.97 (3/4) | 41.2 ± 3.72 (3/4) | 42.1 ± 2.53 (4/4) | 45.8 (2/4) |
| 10 | 39.0 ± 0.29 (4/4) | 40.0 ± 1.93 (4/4) | 40.5 ± 1.21 (4/4) | 43.7 ± 1.24 (4/4) |
| 100 | 36.3 ± 1.41 (4/4) | 35.6 ± 0.82 (4/4) | 36.7 ± 0.90 (4/4) | 38.6 ± 0.57 (4/4) |
Ct values are indicated as average ± standard deviation (SD). Number of positive samples over tested samples is indicated in parentheses.
Measurement of the viral copy numbers in the clinical specimens
To evaluate the influence of the transport medium in the clinical specimens, we collected the SARS-CoV-2 positive specimens from patients with COVID-19 using commercially available transport media (VTM or eSwab®). The number of viral copies was evaluated using a combination of RNA extraction and real-time qRT-PCR (Fig. 1B, B-1). The range of RNA quantified in each clinical sample was 5.83 × 10−2–1.05 × 106 copies/μL.
Influence of the transport medium to the sensitivity of GeneSoC® RT-PCR in clinical specimens
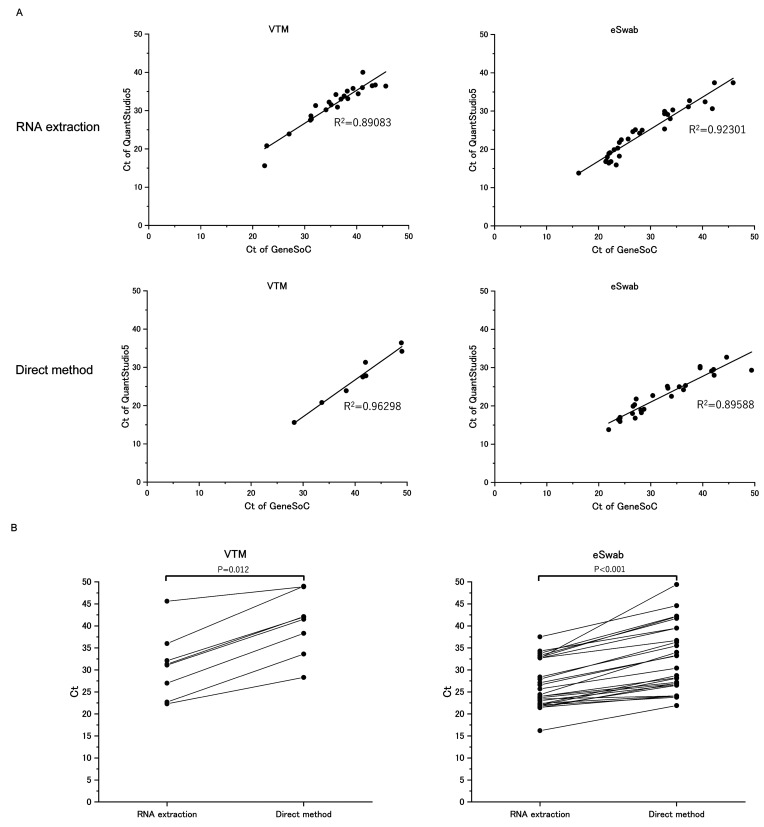
First, we evaluated the correlation between standard real-time qRT-PCR and GeneSoC® RT-PCR. As shown in Fig. 3A, either when using RNA extraction or direct method as a pretreatment method, cycle threshold (Ct) values of GeneSoC® RT-PCR were well correlated with those of real-time qRT-PCR (R2 > 0.89) in both VTM and eSwab samples. These data indicated that the performance of GeneSoC® RT-PCR is similar to that of real-time qRT-PCR through a broad range of Ct.
Fig. 3.
Performance of GeneSoC® RT-PCR with RNA extraction or direct method. (A) Correlation of Ct value between GeneSoC® RT-PCR in combination with RNA extraction or direct method and standard real-time qRT-PCR. Clinical specimens in VTM or eSwab were tested with standard real-time qRT-PCR and GeneSoC® RT-PCR (Fig. 1B). Pretreatment of the GeneSoC® RT-PCR was performed with RNA extraction (upper panel) and direct method (lower panel). Ct values of each method were compared with those of standard real-time qRT-PCR. Samples tested negative in either method were excluded from this analysis. Ct: cycle threshold. (B) Comparison of paired Ct values between RNA extraction and direct method in GeneSoC® RT-PCR. Clinical specimens collected in VTM (left panel) and eSwab® (right panel) were treated with RNA extraction and direct method followed by GeneSoC® RT-PCR. The Ct values of each method in the same samples were compared. Wilcoxon signed-rank sum test found a significant difference between Ct values, as shown by P values.
Moreover, we tested the detection of SARS-CoV-2 RNA using RNA extraction or direct method in GeneSoC® RT-PCR. As shown in Fig. 3B, in the same sample, Ct values are significantly lower in RNA extraction than in the direct method, irrespective of the transport medium, indicating the inferior sensitivity of the direct method compared to the RNA extraction.
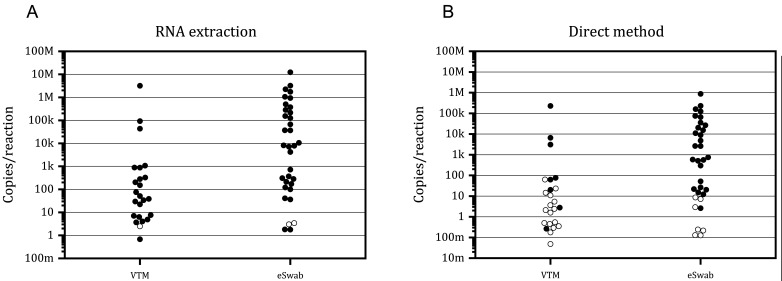
Next, we evaluated the influence of the transport medium on the analytical sensitivity of GeneSoC® RT-PCR. As shown in Fig. 4A, GeneSoC® RT-PCR can detect at least 6.79 × 10−1 copies/reaction in VTM samples and 1.78 copies/reaction in eSwab® samples, whereas three samples with very low copy numbers (one in VTM and two in eSwab®; the copy numbers were 2.47, 3.02, 3.42 copies/reaction) cannot be detected in GeneSoC® RT-PCR. These results suggest that the sensitivity of GeneSoC® RT-PCR is slightly lower than that of real-time qRT-PCR and that the transport medium does not influence the detection ability of GeneSoC® RT-PCR in combination with the RNA extraction method. Next, we tested a combination of the direct method and GeneSoC® RT-PCR (Fig. 4B). In this combination, the detection of viral RNA was unstable in the samples containing less than 100 copies/reaction viral RNA in VTM, whereas less than 10 copies/reaction viral RNA in eSwab®. These data suggest that combination of direct method and GeneSoC® RT-PCR is more sensitive for eSwab® than the VTM.
Fig. 4.
Detection of SARS-CoV-2 RNA in clinical specimens collected using VTM and eSwab®. Clinical samples collected in VTM and eSwab® (X-axis) were tested using GeneSoC® RT-PCR in combination with conventional RNA extraction method (A) and direct method (B). The viral load of each sample was quantified using RNA extraction and real-time qRT-PCR (Y-axis). Black and white circles indicate positive and negative results, respectively, obtained using GeneSoC® RT-PCR. M: 106; k: 103; m: 10−3
Influence of the transport medium to the concordance between RNA extraction and direct method with GeneSoC® RT-PCR
Next, we evaluated the concordance between the RNA extraction and the direct method in clinical specimens using GeneSoC® RT-PCR. The positive concordance between the RNA extraction and the direct method was 84% for eSwab® samples (Table 3A), whereas it was 35% for VTM samples (Table 3B). None of the negative samples showed false-positive results, and the negative concordance was 100% for both the eSwab® and VTM samples. The overall concordance was 85% and 38% for eSwab® and VTM samples, respectively. Therefore, when using eSwab® as a transport medium, the performance of the direct method was comparable to that of the RNA extraction method in GeneSoC® RT-PCR.
Table 3. Correlation between direct method and conventional RNA extraction method using GeneSoC® RT-PCR.
| A. Samples collected in eSwab® | ||||
| eSwab | Direct method | |||
| Positive | Negative | Total | ||
| RNA extraction | Positive | 27 | 5 | 32 |
| Negative | 0 | 2 | 2 | |
| Total | 27 | 7 | 34 | |
| B. Samples collected in VTM | ||||
| VTM | Direct method | |||
| Positive | Negative | Total | ||
| RNA extraction | Positive | 8 | 15 | 23 |
| Negative | 0 | 1 | 1 | |
| Total | 8 | 16 | 24 | |
Influence of the biological materials in the clinical specimens to GeneSoC® RT-PCR
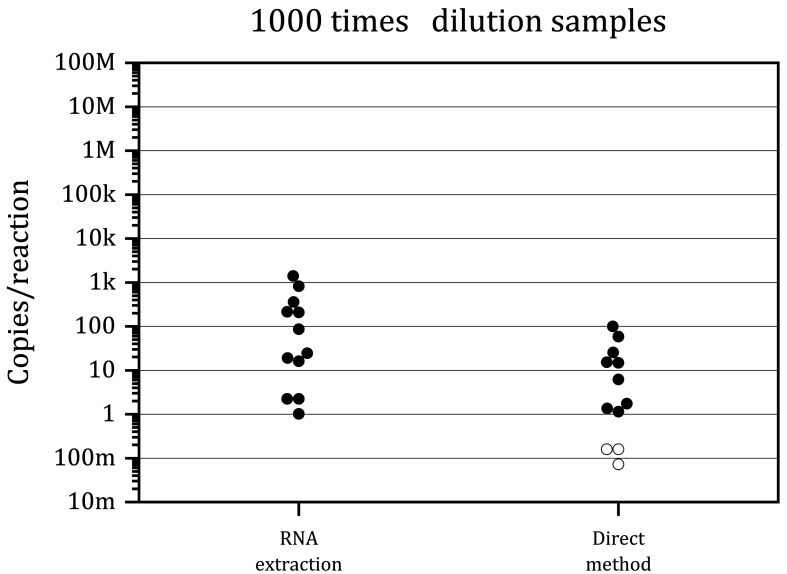
Even when using eSwab®, the sensitivity of the direct method was inferior to that of the RNA extraction method in GeneSoC® RT-PCR. Therefore, we next explored this mechanism. For this purpose, we diluted the clinical samples 1,000 times with eSwab® and prepared artificially low viral copy number samples. As shown in Fig. 5, GeneSoC® RT-PCR could detect at least 1.15 copies/reaction RNA in both the RNA extraction method and direct method. These data indicated that the sample preparation method did not affect the sensitivity of GeneSoC® RT-PCR in diluted samples. Therefore, the reduced sensitivity of the direct method for crude clinical samples may be attributed to the presence of PCR inhibitors in the biological materials of clinical specimens.
Fig. 5.
Detection of SARS-CoV-2 RNA in clinical samples diluted 1,000 times. Clinical samples collected with eSwab® were tested using GeneSoC® RT-PCR in combination with the RNA extraction or direct method. The viral load of each sample was quantified using RNA extraction and quantitative real-time qRT-PCR (Y-axis). Black and white circles indicate positive and negative results obtained using GeneSoC® RT-PCR, respectively. M: 106; k: 103; m: 10−3
DISCUSSION
In this study, we showed that the direct method, combined with the eSwab® transport medium, allows highly sensitive detection of SARS-CoV-2 in the clinical samples by GeneSoC® RT-PCR. GeneSoC® is a newly developed rapid real-time PCR system that adopts the microfluidic principle and enables RT-PCR completion within 15 min. Eliminating the sample preparation time by using the direct method is ideal to ensure high-speed performance. However, the influence of the transport medium on this reaction has not been studied adequately. In the present study, the sensitivity of GeneSoC® RT-PCR was comparable to that of standard real-time qPCR. The transport medium did not influence this detection ability using the RNA extraction method. However, the direct method was less sensitive than the RNA extraction method because PCR inhibitors in biological materials or the transport medium in clinical samples affect the subsequent rapid PCR. We observed that the commercially available transport medium eSwab® minimizes the unfavorable effect of the medium during direct PCR; the sensitivity of the direct method was comparable to that of the RNA extraction method in GeneSoC® RT-PCR.
Similar to GeneSoC®, several PCR systems using microfluidic PCR methodology have recently been developed.8 The microfluidic PCR system, which requires minimal amount of samples and reagents, is advantageous for achieving rapid PCR and footprint downsizing of the PCR machine and is ideal for POC testing for SARS-CoV-2. In the GeneSoC® system, PCR reagents move reciprocally through a very thin flow channel molded on a chip and cycle over multiple preheated zones. Using this mechanism, GeneSoC® enables SARS-CoV-2 detection within 15 min9 and is one of the quickest commercially available PCR machines.10 However, in this mechanism, a very low reaction volume and rapid PCR tend to affect the sensitivity of the test adversely, and the reaction conditions must be strictly adjusted. In line with a former report,11 GeneSoC® could not detect the samples containing very low copy numbers (less than 10 copies/reaction; range 2.47–3.42 copies/reaction). Assuming an RNA extraction efficiency of 100%, we calculated that the viral loads of these undetectable samples before RNA extraction were 2.12–2.93 × 102 copies/mL, which is comparable to the lower limit of the previously reported viral loads of nasopharyngeal samples of the patients with COVID-19 (median viral load was 2.4 log10 copies/mL).12 These data suggest that GeneSoC® potentially has sufficient diagnostic ability for SARS-CoV-2 testing.
This study showed that the direct method was less sensitive than the RNA extraction method in GeneSoC® RT-PCR. Compared with the RNA extraction method, the direct method has several advantages and disadvantages that affect the sensitivity of RT-PCR. The advantage is that all nucleic acids in the samples can be used for PCR. However, considerable amounts of nucleic acids are lost during the absorption or filtration steps during the RNA extraction.13 The disadvantage of the direct method is that the fundamental process involves the dilution of the samples (6 times dilution) using a processing solution (Fig. 1B, B-3). This step may dilute the sample RNA and introduce residual clinical specimens, containing biological PCR inhibitors, and transport medium, into the subsequent RT-PCR. In contrast, the RNA extraction fundamentally involves concentrating (2.3 times concentration) and purifying the nucleic acids in samples (Fig. 1B, B-2). The balance between the advantages and disadvantages affects the sensitivity of the final RT-PCR reaction, and the disadvantages might strongly reduce the sensitivity of the direct method compared to the RNA extraction method. We showed that the sensitivity of the direct method for VTM samples was ten times less than that for eSwab® samples, which is especially important to overcome the reduced sensitivity of the direct method. This result suggests that of transport medium has a strong negative effect on rapid PCR in the direct method. Therefore, the selection of the transport medium is essential for the sensitivity of the direct method combined with rapid PCR.
The composition of the transport medium influences the rapid PCR system. To facilitate viral culture as a downstream application, VTM contains BSA and antibiotics14; however, these components potentially inhibit PCR,15 whereas eSwab® does not contain these materials.16 The residual of these substances in the transport medium might considerably affect the sensitivity of the direct method followed by rapid PCR, because the optimal range of conditions for rapid PCR is narrower than that for conventional PCR. To achieve POC testing for SARS-CoV-2, a combination of the direct method and rapid PCR is ideal. However, we need to evaluate the influence of the transport medium for this combination. The influence of transport media on standard real-time PCR has been extensively investigated.15, 17, 18 To our knowledge, this is the first study to focus on this issue using rapid PCR.
This study has some limitations. We examined only H2O and PBS as the generic media, and VTM and eSwab® as the commercially available transport media. To extend the use of POC testing by combining the direct method with rapid PCR, a broader range of commercially available transport media should be evaluated. Several direct methods, such as heat inactivation19 and the use of lysis amplification buffers of different compositions20 have recently been reported. Therefore, when adopting other direct PCR methodologies, especially for detecting very low amounts of RNA in clinical specimens using rapid PCR, the conditions and transport medium for each method must be optimized.
In conclusion, the combination of the direct method and microfluidic rapid PCR machine GeneSoC® can detect SARS-CoV-2 RNA with high sensitivity in clinical samples using the eSwab® transport medium. This combination enables an ideal POC testing for SARS-CoV-2 with very short hands-on time and eliminates the need for additional procedures in the clinical setting.
Acknowledgments
Acknowledgements: We would like to thank Editage (www.editage.jp) for English language editing.
Footnotes
Conflict of interest: This study was partly funded and provided the study materials (chips and kits) by Kyorin Pharmaceutical Co., Ltd.
REFERENCES
- 1.Corman VM,Landt O,Kaiser M,Molenkamp R,Meijer A,Chu DKW,et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:25. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel R,Babady E,Theel ES,Storch GA,Pinsky BA,St George K,et al. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. MBio. 2020;11:11. 10.1128/mBio.00722-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrosi C,Prezioso C,Checconi P,Scribano D,Sarshar M,Capannari M,et al. SARS-CoV-2: comparative analysis of different RNA extraction methods. J Virol Methods. 2021;287:114008. 10.1016/j.jviromet.2020.114008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein S,Müller TG,Khalid D,Sonntag-Buck V,Heuser AM,Glass B,et al. SARS-CoV-2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses. 2020;12:863. 10.3390/v12080863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin B,Linacre A. Direct PCR: A review of use and limitations. Sci Justice. 2020;60:303-10. 10.1016/j.scijus.2020.04.003 [DOI] [PubMed] [Google Scholar]
- 6.Diseases NIoI. Manual for Detection of Pathogen 2019-nCoV Ver.2.6. NIID; 2020
- 7.Shirato K,Nao N,Katano H,Takayama I,Saito S,Kato F,et al. Development of Genetic Diagnostic Methods for Detection for Novel Coronavirus 2019(nCoV-2019) in Japan. Jpn J Infect Dis. 2020;73:304-7. 10.7883/yoken.JJID.2020.061 [DOI] [PubMed] [Google Scholar]
- 8.Ahrberg CD,Manz A,Chung BG. Polymerase chain reaction in microfluidic devices. Lab Chip. 2016;16:3866-84. 10.1039/C6LC00984K [DOI] [PubMed] [Google Scholar]
- 9.Sakai J,Tarumoto N,Orihara Y,Kawamura R,Kodana M,Matsuzaki N,et al. Evaluation of a high-speed but low-throughput RT-qPCR system for detection of SARS-CoV-2. J Hosp Infect. 2020;105:615-8. 10.1016/j.jhin.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong X,Liu L,Tu Y,Zhang J,Miao G,Zhang L,et al. Rapid PCR powered by microfluidics: A quick review under the background of COVID-19 pandemic. Trends Analyt Chem. 2021;143:116377. 10.1016/j.trac.2021.116377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe R,Asai S,Kakizoe H,Saeki H,Masukawa A,Miyazawa M,et al. Evaluation of the basic assay performance of the GeneSoc® rapid PCR testing system for detection of severe acute respiratory syndrome coronavirus 2. PLoS One. 2021;16:e0248397. 10.1371/journal.pone.0248397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fajnzylber J,Regan J,Coxen K,Corry H,Wong C,Rosenthal A,et al. ; Massachusetts Consortium for Pathogen Readiness. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. 10.1038/s41467-020-19057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Read SJ. Recovery efficiencies of nucleic acid extraction kits as measured by quantitative LightCyclerTM PCR. Mol Pathol. 2001;54:86-90. 10.1136/mp.54.2.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mears MJ,Wallace MJ,Yount JS,Fowler LA,Jones PS,Mohler PJ,et al. Viral transport media for COVID-19 testing. MethodsX. 2021;8:101433. 10.1016/j.mex.2021.101433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkland PD,Frost MJ. The impact of viral transport media on PCR assay results for the detection of nucleic acid from SARS-CoV-2. Pathology. 2020;52:811-4. 10.1016/j.pathol.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumede L,Radebe F,Nhlapo D,Maseko V,Kufa-Chakezha T,Kularatne R. Evaluation of the Copan eSwab®, a liquid-based microbiology transport system, for the preservation of Neisseria gonorrhoeae at different temperatures. S Afr J Infect Dis. 2017;32:96-9. 10.1080/23120053.2017.1313935 [DOI] [Google Scholar]
- 17.Druce J,Garcia K,Tran T,Papadakis G,Birch C. Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J Clin Microbiol. 2012;50:1064-5. 10.1128/JCM.06551-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrera-Avalos C,Luraschi R,Vallejos-Vidal E,Figueroa M,Arenillas E,Barria D,et al. Analysis by real-time PCR of five transport and conservation mediums of nasopharyngeal swab samples to COVID-19 diagnosis in Santiago of Chile. J Med Virol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barza R,Patel P,Sabatini L,Singh K. Use of a simplified sample processing step without RNA extraction for direct SARS-CoV-2 RT-PCR detection. J Clin Virol. 2020;132:104587. 10.1016/j.jcv.2020.104587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castellanos-Gonzalez A,Shelite TR,Lloyd N,Sadiqova A,Ping R,Williams-Bouyer N,et al. Direct RT-PCR amplification of SARS-CoV-2 from clinical samples using a concentrated viral lysis-amplification buffer prepared with IGEPAL-630. Sci Rep. 2021;11:14204. 10.1038/s41598-021-93333-2 [DOI] [PMC free article] [PubMed] [Google Scholar]