Abstract
Background
Cefiderocol, a novel siderophore cephalosporin, is a promising therapeutic option for infections caused by multidrug-resistant Pseudomonas aeruginosa. We evaluated the activity of cefiderocol against carbapenem-resistant P. aeruginosa (Cr-Pa) isolates and investigated the potential mechanisms involved in resistance.
Methods
108 CR-Pa isolates collected from patients without prior exposure to the substance were studied. MICs of cefiderocol were determined by broth microdilution using iron-depleted cation-adjusted Mueller-Hinton broth. Whole genome sequencing was performed to investigate the potential resistance mechanisms by comparing resistant and susceptible P. aeruginosa isolates and identifying unique mutations in the resistant group.
Results
Of the 108 isolates, nine were resistant to cefiderocol with MIC values ranging from 4 to 32 mg/L. The genetic analysis revealed a broad spectrum of mutations in the resistant isolates associated with iron uptake systems, efflux pumps, AmpC β-lactamase and penicillin-binding proteins. The most frequently observed mutations among the resistant isolates were located in fptA, fpvB and chtA. Notably, the presence of carbapenemases did not correlate with cefiderocol resistance.
Conclusions
Our findings show the low prevalence of cefiderocol resistance among CR-Pa isolates, showing its potential as an effective treatment option. However, the complex genetic landscape of resistance mechanisms, particularly mutations affecting iron transport and other TonB-dependent receptors, requires continuous monitoring and functional analyses to identify and manage potential resistance mechanisms. This study provides a foundation for future research to improve antimicrobial resistance prediction and develop targeted therapies against CR-Pa.
Introduction
The rise of multidrug-resistant organisms (MDRO) poses significant clinical challenges worldwide, necessitating novel therapeutic strategies and a deeper understanding of resistance mechanisms. Among these pathogens, Pseudomonas aeruginosa stands out as an opportunistic Gram-negative bacterium, frequently involved in hospital-acquired infections, especially in immunocompromised and multimorbid patients, and known for its capacity to develop resistance to antibiotics, by either acquisition of horizontal resistance determinants and/or through the selection of chromosomal mutations.1,2 Indeed, the WHO classified in 2017 carbapenem-resistant P. aeruginosa (CR-Pa), as one of the priority pathogens for research and development of new antibiotics.3
Infections caused by CR-Pa are associated with poor outcomes and an increased risk of mortality compared to those caused by susceptible strains.4,5 CR-Pa are often multidrug resistant, limiting the treatment options and highlighting the urgent need for effective antimicrobial agents. In this context, cefiderocol, a novel siderophore cephalosporin, has emerged as a promising agent for the treatment of CR-Pa, including carbapenemase-producing P. aeruginosa.6 Its structural similarity to iron scavengers, also called siderophores, provides an alternative mode of entry into the periplasmic space by binding to iron molecules and hijacking the siderophore receptors, bypassing the typical β-lactam resistance mechanisms.7 Indeed, it mostly maintains in vitro activity against P. aeruginosa isolates resistant to the latest β-lactam-β-lactamase inhibitor combinations, including ceftolozane-tazobactam and ceftazidime-avibactam.8,9
Resistance to cefiderocol in P. aeruginosa has been reported to be associated with alterations in iron uptake pathways, structural modifications of AmpC β-lactamase, and changes in efflux systems.10,11,12 As the use of cefiderocol increases due to the increasing prevalence of infections caused by multidrug-resistant P. aeruginosa, we sought to evaluate its activity against CR-Pa isolates and investigate the underlying resistance mechanisms. These isolates were recovered from patients during a period when cefiderocol was not in clinical use (2019–21), thereby providing a unique insight into resistance mechanisms unaffected by clinical use of the drug. This study addresses a critical research gap by providing baseline data on cefiderocol resistance prior to its clinical introduction, enabling a clearer understanding of intrinsic resistance mechanisms. By elucidating the mechanisms and genetic determinants driving resistance among P. aeruginosa, our findings lay a basis for future studies for improving AMR prediction and better knowledge of resistance mechanisms to develop novel therapeutic strategies.
Methods
Sample collection, identification, and antimicrobial susceptibility testing
CR-Pa isolates from patients admitted to the Heidelberg University Hospital between January 2019 and March 2021 were included in this study. All isolates were obtained hospital wide from the routine MDRO admission screening and clinical submissions and were stored at −80°C until further use. These isolates were recovered from rectal screening swabs and clinical samples, including blood cultures, respiratory specimens, urine and wound swabs. Only non-duplicate isolates per patient were included in the analysis.
The hospital's infection control strategy includes admission screening with rectal swabs for all patients considered at ‘risk’ as defined by the Department of hospital hygiene according to the recommendation of the Commission of Hospital Hygiene and Infection Prevention in Germany (KRINKO): (i) admission to intermediate care unit; (ii) past MDRO infection/colonization; (iii) contact with high-prevalence settings/endemic region, including travel and migration; (iv) chronic wounds and catheters; (v) any antibiotic exposure for ≥1 week in the past 6 months; (vi) contact with MDRO patients or MDRO-colonized individuals; and (vii) close contact with animal husbandry. The local screening strategy is designed to detect Gram-negatives with phenotypic resistance towards third-generation cephalosporines and quinolones (the so-called 3MRGN [Multidrug-resistant Gram-negative]) and carbapenem-resistant Gram-negatives (4MRGN).13 The guidelines advise that patients carrying ‘4MRGN’ should be placed in single-room isolation on all wards, while those carrying ‘3MRGN’ should be isolated only on high-risk wards. Therefore, the MDRO screening algorithm was tailored to detect ‘3MRGN and 4MRGN’.14
Briefly, patient screening samples were cultured on Columbia agar with 5% sheep’s blood and ESBL agar (bioMérieux, Germany) for up to 24 h at 37°C. Identification and antimicrobial susceptibility testing (AST) were performed on all representative colonies growing on ESBL agar. Species identification was performed by MALDI-TOF MS (Bruker Daltonics, Germany). In the routine diagnostics, AST for piperacillin/tazobactam, ceftazidime, cefepime, meropenem and imipenem was performed using VITEK 2 (bioMérieux, Germany) and interpreted according to EUCAST clinical breakpoints version 11.0.
Isolates, resistant to all these antibiotics, were included in the study and further evaluated for susceptibility to cefiderocol using iron-depleted cation-adjusted Mueller-Hinton broth (ID-CA-MHB), prepared in-house as previously described.15 Briefly, a bacterial suspension of 0.5 McFarland standard was prepared from fresh colonies grown overnight. This suspension was then diluted 1:10 in ID-CA-MHB and 75 µL of the diluted suspension was distributed into each well, containing 75 µL ID-CA-MHB with cefiderocol at different concentrations (0.03 mg/L to 32 mg/L). To inhibit NDM activity, broth microdilution (BMD) was performed on NDM-positive cefiderocol-resistant isolates, also using ID-CA-MHB supplemented with 100 mg/L dipicolinic acid (DPA), a chemical compound that can chelate metal ions and thus inhibit the activity of metallo-β-lactamase (MBL). The results for cefiderocol were interpreted according to the breakpoints from EUCAST version 11.0 (susceptible ≤2; resistant >2 mg/L). The MIC was defined as the lowest concentration where turbidity was either absent or significantly decreased compared to the growth control due to the ‘trailing effect,’ as described in the EUCAST guidance for cefiderocol MIC testing.16 Tests were performed in duplicate, with the highest MIC recorded in case of discrepancies. Escherichia coli ATCC® 25922 was used as a reference and quality control organism for cefiderocol testing.
DNA extraction and sequencing
Genomic DNA was extracted from the overnight cultures with the DNeasy Blood and Tissue Kit (Qiagen GmbH, Germany) following the manufacturer’s protocol. Sequencing was performed using the Nextera DNA Flex Library Prep Kit (Illumina) and sequenced on the MIseq instrument (2 × 300 bp) at the Department of Infectious Disease, Medical Microbiology and Hygiene in Heidelberg.
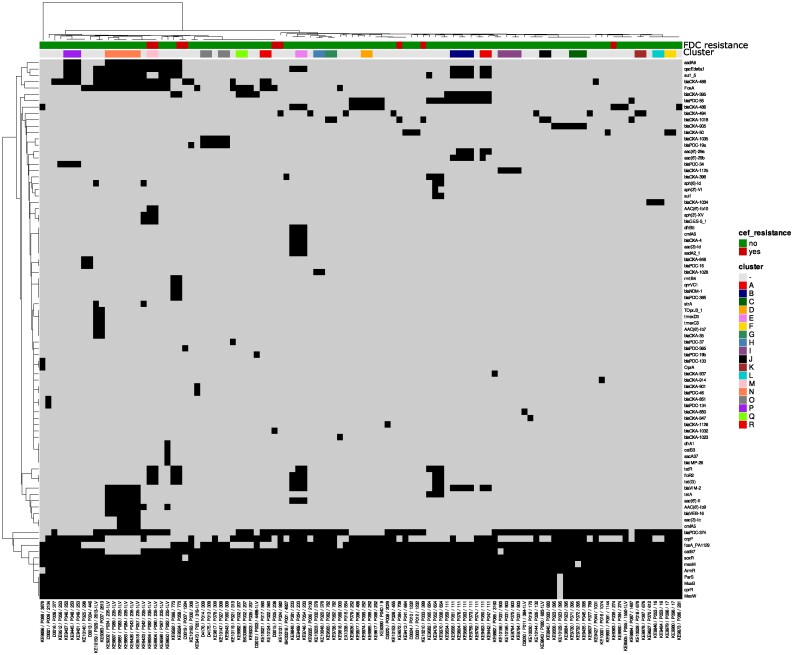
Quality control of the raw sequences was performed using fastp (v0·23·1 with parameters -q = 30 and -l = 45) and assembled with SPAdes 3.15.5 (with the option—careful and—only-assembler).17,18 A curation of the draft genomes was performed by removing contigs with a length <500 bp and/or coverage < 10×. The quality of the final draft was quality-controlled using Quast (v5·0·2).19 The species identification of each draft genome was done using mash (sub-command screen) by screening each draft genome to a database composed of a representative genome of each species present in the Microbial Genomes resource (https://www.ncbi.nlm.nih.gov/genome/microbes/). The complete draft genomes were processed through available databases using Abricate (https://github.com/tseemann/abricate) to identify antimicrobial resistance (NCBI, CARD, ARG-ANNOT, ResFinder and MEGARES databases).20,21 Furthermore, each draft genome was aligned to its representative genome reference from the Microbial Genomes resource using Split Kmer Analysis. The alignment was then analysed with Gubbins 3·1·6 to define hqSNPs distance and phylogenetic relationship.22 Clonal relationship and potential cluster have been evaluated using Average Nucleotide Identity using the threshold of 99.99% identity as previously described (Figure 1).23
Figure 1.
Heatmap representing the AMR genes distribution. The isolates are sorted based on the recombination corrected phylogeny from Gubbins and annotated with the Patient ID and MLST. The AMR genes are sorted hierarchically based on their distribution amongst the population. Gene presence is symbolized by a black square while the absence is symbolized by a grey square.
To identify mutations associated specifically with cefiderocol resistance, a comparative analysis between resistant and susceptible isolates from the cohort was conducted. Initial mapping was performed against the corresponding wild-type reference strains PAO1 and PA14 based on their phylogenetic clustering (Table S1, available as data at JAC-AMR Online) to facilitate the annotation of the SNPs and SNP calling was performed using snippy.24 After identifying these SNPs, we screened for mutations exclusively present in at least one of the resistant isolates by excluding mutations found also in any of the susceptible isolates and/or MBL-producing isolates reverting to a sensitive phenotype under DPA treatment. The reversion of resistance in the presence of DPA indicates that the increased MIC is due to NDM production. This method ensured that only mutations potentially linked to cefiderocol resistance were identified. Mutations uniquely observed in the cefiderocol-resistant isolates have been listed in Table S1. We excluded the mutations in the haeme/haemophore transporters from our analysis due to the absence of evidence supporting their role in mediating cefiderocol uptake.
The draft genome sequence data have been submitted to the NCBI GenBank database under the project number PRJNA1103370. Further details, including sequencing statistics and isolate information, can be found in the supplementary data (Table S2).
Results
During the study period, we identified 108 CR-Pa isolates from 99 patients, with nine of these patients having two different CR-Pa strains. Among these, nine isolates (8.3%, 9/108) from nine different patients were resistant to cefiderocol with a MIC range of 4–32 mg/L, as determined by the BMD method and interpreted following the EUCAST guidelines. No discrepant MICs were observed in resistant isolates in duplicate measurements. Detailed cefiderocol resistance data for all clinical isolates are provided in Table S2.
The isolates were classified into 59 STs. ST235-1LV was the most prevalent MLST (10.2, 11/108). The allelic variant was due to the duplication of the asc gene matching both alleles 38 and 172. ST111 was the second most prevalent clonal lineage (7.4%; 8/108), followed by ST395 (5.6%; 6/108) (Table S2). High-risk clones, such as ST111, ST233, ST253, ST274, ST298, ST308, ST654 and ST773, accounted for 33.3% of our collection (36/108).25,26 Only four resistant isolates belonged to the high-risk clonal lineages (two 235-1LV, one ST274 and one ST773). Among the resistant isolates, only the two 235-1LV isolates formed a distinct cluster (M) as shown in Figure 1.
Impact of carbapenemases on cefiderocol resistance
The collection included 23 carbapenemase-producing isolates (21.3%), with 17 isolates carrying VIM-2 (15.7%), followed by GES-5 (3 isolates, 2.8%), NDM-1 (2 isolates, 1.9%) and IMP-26 (1 isolate, 0.9%). Three of these carbapenemase-producing isolates were resistant to cefiderocol: two with GES-5 and one with NDM-1. Since the overexpression or enhanced NDM activity has previously been attributed to cefiderocol resistance in Enterobacterales,27 we investigated whether the inhibition of NDM by DPA could revert the cefiderocol MIC to susceptible levels. In the presence of DPA, the meropenem MIC of NDM-positive isolate KE9524 was reduced from 32 mg/L to 0.5 mg/L. Notably, all blaVIM-2-carrying isolates remained susceptible to cefiderocol. There was no significant difference in resistance rate and the cefiderocol MIC values between carbapenemase-negative and -positive isolates (Figure 2). Detailed genotypic resistance data for all clinical isolates are provided in Table S3.
Figure 2.
Distribution of cefiderocol MIC (mg/L). Number of isolates by cefiderocol MIC by carbapenemase. All isolates: 91.7% susceptible. Only MBL-positive: 90% susceptible. Only Serine carbapenemase-positive: 85.7% susceptible. Serine carbapenemase- and MBL-positive: 100% susceptible. Carbapenemase negative: 92.2% susceptible.
Mutations potentially associated with cefiderocol resistance
Next, we focused on investigating mutations in potential P. aeruginosa targets associated with other known resistance mechanisms against cefiderocol. Table 1 summarizes the unique mutations found only in cefiderocol-resistant isolates, particularly in genes related to cefiderocol resistance and iron uptake mechanisms. KE9524 was excluded since NDM production was the cause of the resistant phenotype. The full dataset of mutation present only in the resistant isolates is available in the Table S1.
Table 1.
Distinctive genomic mutations in cefiderocol-resistant P. aeruginosa isolates compared to susceptible isolates
| Isolate | D3015 | D3019 | KE9383 | KE9572 | KE9904 | KE9934 | KE10131 | KE10210 |
| ST | 238 | 1284 | 274 | 708 | — | — | 982 | 389 |
| Carbapenemase | — | — | — | — | GES-5 | GES-5 | — | — |
| FDC MIC (mg/L) | 16 | 4 | 8 | 4 | 16 | 16 | 32 | 16 |
| Components of TonB-dependent iron transporters | ||||||||
| FptA | H557N | Y664S | A719V | P558S | ||||
| FpvA | V46I | |||||||
| FpvB | T119I | T119I | I127 V, V271I | D377N, Q660K | ||||
| FemA | T5M | E75D | ALT72VLA | |||||
| PfuA | A164T | P343R | A491 V, V131M | |||||
| CirA | VP339ML | R226L, D236N | T634S | |||||
| PfeA | R267H | D541N, V565I | ||||||
| FecA | A360T | A261T | ||||||
| ChtA | H298Y | D626E | D626E | S173I | ||||
| FiuA | V492fs, E388K | A668S | I578L, P2S | |||||
| PiuA | L15M | |||||||
| PirA | A303V | Q620K, R7H | ||||||
| FepG | V51M | |||||||
| OptE | K607G, S312N | |||||||
| OptJ | D264G | |||||||
| FecR | L107V | Y172H | ||||||
| PA0151 | A39V, P610R | D741E | F750V | F750V | ||||
| MexAB-OprM efflux pump | ||||||||
| MexB | I186V | |||||||
| MexR | R91L | A110T | ||||||
| NalC | S54P | |||||||
| NalD | F51L | |||||||
| MexEF-OprN efflux pump | ||||||||
| MexE | P337A | |||||||
| MexS | F209L | |||||||
| OprN | G364S | |||||||
| MexCD-OprJ efflux pump | ||||||||
| MexC | A330S | E26A | ||||||
| MexD | N775K | A566T, V607A | ||||||
| Other proteins of interest | ||||||||
| AmpD | A29T, H93fs | H36Y | D28G | |||||
| oprD | Initiator codon variant | S338P | A123fs | T105fs | ||||
| PBP1A | H629N | T699I | D309E | E245D | ||||
| PBP2 | A619E | |||||||
| PBP3 | A139S | A467P | L23V | |||||
| CntO | M532L | S333T | ||||||
| BtuB | V11M | AG209SS | F452Y, NV455DI, D458S, GNDP461TYTT, N476F, Q474delinsQGLYV | L490I | ||||
FDC, cefiderocol; ST, sequence type.
In eight of nine resistant isolates, we identified numerous missense mutations in various genes encoding components of the bacterial iron transport systems. The affected genes included pirA, pfeA, cirA, fecA, fpvA, fpvB, fptA, fepA, femA, fiuA, piuA, fecR, exbB, chtA, pfuA, fepG, optE, optJ and PA0151. The most frequently mutated genes were fptA, fpvB, chtA and PA0151 (each in four isolates).
Beyond these tonB-dependent receptors (TBDRs), we identified single amino acid substitutions in three major efflux pump systems: the MexAB-OprM, MexEF-OprN and MexCD-OprJ. Within the MexAB-OprM efflux pump, four resistant isolates exhibited mutations across various components: MexB (the transporter component), nalC (a positive regulator), MexR and nalD (both negative regulators).28 Additionally, mutations in the MexEF-OprN efflux pump were observed in two isolates: a Pro337Ala substitution in MexE, the transporter component, a Gly364Ser substitution in OprN, the porin component and a Phe209Leu substitution in MexS, which regulates the expression of MexEF-OprN.29 In the MexCD-OprJ system, several missense mutations were detected in three resistant isolates. Four isolates lacked mutations in their efflux pumps.
Furthermore, we identified frameshift and missense mutations in the oprD outer membrane protein in five isolates. Other noteworthy mutations include those in ampD, a negative regulator of AmpC β-lactamase expression, in three isolates.30 Lastly, in four resistant isolates, mutations were detected in at least one of the genes encoding penicillin-binding proteins (PBPs) 1A, 2 and 3. These genetic alterations are summarized in Table 1.
Discussion
The increasing prevalence of multidrug-resistant Gram-negative bacteria is leading to more frequent use of novel antimicrobial treatments, such as cefiderocol. This study aimed to evaluate the cefiderocol susceptibility and explore the underlying resistance mechanisms CR-Pa isolates collected in Germany. Through whole genome sequencing and BMD, we assessed 108 CR-Pa isolates, which were recovered from patients prior to the approval of cefiderocol for clinical use by the EMA, ensuring that none of the patients had been exposed to the drug. Our results showed that the majority of the isolates (91.7%, 99/108) were susceptible to cefiderocol, indicating its potential as an effective therapeutic option for CR-Pa infections. However, the necessity of susceptibility testing for cefiderocol remains evident for these multidrug-resistant bacteria. Resistance mechanisms were multifactorial, involving mutations in iron uptake systems, efflux pumps and other bacterial components, highlighting the complexity of cefiderocol resistance. There was no significant association between phenotypic cefiderocol resistance and the genetic background of the isolates. All resistant isolates were of different STs, indicating the broad genetic diversity among P. aeruginosa strains capable of developing resistance. Due to the lack of national molecular surveillance and studies for P. aeruginosa in Germany, we cannot determine to what extent our cohort reflects the overall P. aeruginosa population in the country. However, similar regional studies have shown that ST111, ST395 and ST253 are the most prevalent clones, in line with our findings.31,32
Our findings suggest that the mechanisms underlying cefiderocol resistance are heterogeneous. In this study, resistant isolates had predominantly mutations in the genes related to iron transport. This finding aligns with existing literature showing that alterations in TBDRs such as FpvB, FptA, PfeA, PirA, PiuA, FecA and FepG are associated with elevated cefiderocol MICs.33–38 Furthermore, we identified mutations in additional TBDRs-FemA, ChtA, PfuA, OptE, OptJ PA0151 and FiuA-not previously linked with elevated cefiderocol MICs. However, overexpressing these genes increases the susceptibility of the wild-type PAO1 strain to cefiderocol significantly,36 suggesting that functional deficiencies in these proteins may play a role in the emergence of cefiderocol resistance. Regarding mutations in cirA, it has not been associated with cefiderocol resistance in P. aeruginosa, although changes in this gene are linked to cefiderocol resistance in various other bacterial species.39
These alterations in TBDRs were accompanied by mutations in various efflux pump systems, PBPs, ampD and oprD genes. Previous research has shown that mutations in oprD and ampD are associated with modest increases in cefiderocol MIC, with oprD mutations affecting outer membrane functionality for β-lactams and AmpD mutations potentially leading to overproduction of chromosomal AmpC β-lactamase.34,40,41 Similarly, mutations affecting the regulation of the MexAB-OprM efflux pump system have been associated with slight MIC elevations.33,34,40 Additionally, mutations in MexEF-OprN and MexCD-OprJ efflux pumps were detected in P. aeruginosa isolates that elevated cefiderocol MICs in vivo.33 The specific contribution of these efflux pump mutations to the reduction of cefiderocol efficacy is poorly understood and warrants further exploration. Current evidence suggests a significant impact of the mutations, especially in the case of MexAB-OprM. Notably, we also identified mutations in PBP3 in three resistant isolates, which is significant given cefiderocol’s high affinity for this protein and the association of PBP3 mutations with elevated cefiderocol MICS in P. aeruginosa, highlighting the potential role of PBP3 alterations in contributing to resistance.42,43
A particularly relevant finding was the mutation in the fecR gene of the isolate KE9383. This mutation is interesting as, apart from it, the isolate exhibits mutations only in a regulator of the MexAB-OprM efflux pump and the oprD gene, both of which are known to confer only modest increases in cefiderocol MIC.34 Until now, mutations in fecR have not been observed in cefiderocol-resistant isolates. However, FecR plays a crucial role in the iron uptake process, acting as a necessary regulator for the transcriptional activation of the iron transporter FecA through the modulation of FecI.44,45 Indeed, mutations in FecI, have been identified in an in vitro-derived cefiderocol-resistant strain.10 Given that FecR acts as a regulator of the activator FecI, its alterations could impact the uptake of cefiderocol by affecting FecI-mediated FecA expression.
Cefiderocol resistance in our study appeared largely independent of carbapenemase status. Previous data, including data from the SIDERO-WT, demonstrated that GES-5 does not seem to influence cefiderocol susceptibility in P. aeruginosa.46–48 The two GES-5 positive cefiderocol-resistant isolates also had mutations in their TBDRs, which were likely the primary cause of the elevated cefiderocol MICs. An exception was observed in a single NDM-positive isolate, KE9524, which showed resistance without any notable mutations in iron transport mechanisms. This finding is in line with previous literature, which indicated that NDM production is associated with elevated cefiderocol MICs in both Enterobacterales and P. aeruginosa.42,49
Our study has limitations. While we provide insight into the genotype of cefiderocol resistance in P. aeruginosa, it is limited by not exploring the functional consequences of each specific mutation for cefiderocol susceptibility. Additionally, this study focuses on isolates from a specific geographic region, which may limit the generalizability of our findings to other regions. Furthermore, our findings are based on a small number of cefiderocol-resistant isolates and low-level resistance constrains the power of our conclusions due to the discrepancies between EUCAST and CLSI breakpoints for the resistance threshold (2 and 4 mg/L, respectively). Based on CLSI breakpoints, two of the nine resistant isolates would be considered susceptible and one intermediate. However, even in these cases, the resistance mechanisms appear to be multifactorial, primarily driven by mutations in iron uptake systems.
Nevertheless, our study provides valuable data on cefiderocol resistance and its genomic mechanisms in P. aeruginosa isolates from a specific region in Germany, prior to cefiderocol's clinical approval and use in Europe. These findings offer an important reference for future comparative studies with a larger, geographically diverse cohort of isolates. Further functional research, including targeted mutagenesis,50 serial passage experiments51 and efflux pump activity analysis, using these mutated genes, is needed to fully understand and validate the resistance mechanisms.
In conclusion, we showed here the potential cefiderocol resistance mechanisms in clinical P. aeruginosa isolates from patients with no prior exposure to the antibiotic, indicating that resistance was present before its clinical approval by the EMA. We observed that each resistant isolate presented a distinct combination of genetic alterations. The resistance was likely the result of a complex interplay of factors, including alterations in efflux pumps, porins, ampD and mutations affecting iron uptake systems essential for cefiderocol's entry into P. aeruginosa. The complexity of these interactions highlights the need for further research to analyse the precise impact of each genetic alteration on the development of resistance. Our findings show that cefiderocol is a promising therapeutic option for treating infections caused by multidrug-resistant P. aeruginosa. It may be particularly beneficial in treating severe hospital-acquired infections in high-risk areas, where multidrug-resistant Gram-negative bacteria are prevalent, such as intensive care units and transplant units. As the widespread use of cefiderocol increases, susceptibility testing is important to ensure its appropriate use is and to monitor the emergence of resistance to this novel antibiotic.
Supplementary Material
Contributor Information
Kaan Kocer, Medical Microbiology and Hygiene, Department of Infectious Diseases, Heidelberg University Hospital, Im Neuenheimer Feld 324, 69120 Heidelberg, Germany; Institute of Medical Microbiology, University of Lübeck and University Medical Center of Schleswig-Holstein, Ratzeburger Allee 160, 23562 Lübeck, Germany; German Center for Infection Research (DZIF), Partner Site Hamburg-Lübeck-Borstel-Riems, Lübeck, Germany.
Sébastien Boutin, Institute of Medical Microbiology, University of Lübeck and University Medical Center of Schleswig-Holstein, Ratzeburger Allee 160, 23562 Lübeck, Germany; German Center for Infection Research (DZIF), Partner Site Hamburg-Lübeck-Borstel-Riems, Lübeck, Germany; Airway Research Center North (ARCN), German Center for Lung Research (DZL), Lübeck, Germany.
Maximilian Moll, Medical Microbiology and Hygiene, Department of Infectious Diseases, Heidelberg University Hospital, Im Neuenheimer Feld 324, 69120 Heidelberg, Germany; Medical Clinic III for Oncology, Hematology, Immuno-Oncology and Rheumatology, University Hospital Bonn, University of Bonn, Venusberg-Campus 1, 53127 Bonn, Germany.
Dennis Nurjadi, Institute of Medical Microbiology, University of Lübeck and University Medical Center of Schleswig-Holstein, Ratzeburger Allee 160, 23562 Lübeck, Germany; German Center for Infection Research (DZIF), Partner Site Hamburg-Lübeck-Borstel-Riems, Lübeck, Germany.
Funding
The study was supported by internal funding.
Transparency declarations
DN received speaker’s honorarium from Cepheid and Shionogi outside the scope of this work. All other authors declared no conflicts of interest.
Ethical considerations
The institutional review board of the Medical Faculty of the Ruprecht-Karls University, Heidelberg, Germany, was consulted prior to the start of the study and waived individual informed consent (S-474/2018).
Supplementary data
Tables S1 to S3 are available as Supplementary data at JAC-AMR Online.
References
- 1. Kalashnikov M, Mueller M, McBeth C et al. Rapid phenotypic stress-based microfluidic antibiotic susceptibility testing of Gram-negative clinical isolates. Sci Rep 2017; 7: 8031. 10.1038/s41598-017-07584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 2017; 7: 39. 10.3389/fcimb.2017.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tacconelli E, Carrara E, Savoldi A et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18: 318–27. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 4. Bassetti M, Vena A, Croxatto A et al. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018; 7: 212527. 10.7573/dic.212527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reynolds D, Kollef M. The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: an update. Drugs 2021; 81: 2117–31. 10.1007/s40265-021-01635-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bassetti M, Echols R, Matsunaga Y et al. Efficacy and safety of cefiderocol or best available therapy for the treatment of serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): a randomised, open-label, multicentre, pathogen-focused, descriptive, phase 3 trial. Lancet Infect Dis 2021; 21: 226–40. 10.1016/S1473-3099(20)30796-9 [DOI] [PubMed] [Google Scholar]
- 7. Candel FJ, Santerre Henriksen A, Longshaw C et al. In vitro activity of the novel siderophore cephalosporin, cefiderocol, in Gram-negative pathogens in Europe by site of infection. Clin Microbiol Infect 2022; 28: 447.e1–e6. 10.1016/j.cmi.2021.07.018 [DOI] [PubMed] [Google Scholar]
- 8. Shortridge D, Streit JM, Mendes R et al. In vitro activity of cefiderocol against U.S. and European Gram-negative clinical isolates collected in 2020 as part of the SENTRY antimicrobial surveillance program. Microbiol Spectr 2022; 10: e0271221. 10.1128/spectrum.02712-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakurai A, Dinh AQ, Hanson BM et al. Evolving landscape of carbapenem-resistant Pseudomonas aeruginosa at a single centre in the USA. JAC Antimicrob Resist 2023; 5: dlad070. 10.1093/jacamr/dlad070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ito A, Nishikawa T, Ishii R et al. 696. Mechanism of cefiderocol high MIC mutants obtained in non-clinical FoR studies. Open Forum Infect Dis 2018; 5: S251. 10.1093/ofid/ofy210.703 [DOI] [Google Scholar]
- 11. Nordmann P, Shields RK, Doi Y et al. Mechanisms of reduced susceptibility to cefiderocol among isolates from the CREDIBLE-CR and APEKS-NP clinical trials. Microb Drug Resist 2022; 28: 398–407. 10.1089/mdr.2021.0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kocer K, Boutin S, Heeg K et al. The acquisition of transferable extrachromosomal fec operon is associated with a cefiderocol MIC increase in Enterobacterales. J Antimicrob Chemother 2022; 77: 3487–95. 10.1093/jac/dkac347 [DOI] [PubMed] [Google Scholar]
- 13. Müller J, Voss A, Köck R et al. Cross-border comparison of the Dutch and German guidelines on multidrug-resistant Gram-negative microorganisms. Antimicrob Resist Infect Control 2015; 4: 7. 10.1186/s13756-015-0047-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reinheimer C, Kempf VA, Göttig S et al. Multidrug-resistant organisms detected in refugee patients admitted to a University Hospital, Germany June–December 2015. Euro Surveill 2016; 21. 10.2807/1560-7917.ES.2016.21.2.30110 [DOI] [PubMed] [Google Scholar]
- 15. Hackel MA, Tsuji M, Yamano Y et al. Reproducibility of broth microdilution MICs for the novel siderophore cephalosporin, cefiderocol, determined using iron-depleted cation-adjusted Mueller-Hinton broth. Diagn Microbiol Infect Dis 2019; 94: 321–5. 10.1016/j.diagmicrobio.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 16. Matuschek E, Longshaw C, Takemura M et al. Cefiderocol: EUCAST criteria for disc diffusion and broth microdilution for antimicrobial susceptibility testing. J Antimicrob Chemother. 2022; 77: 1662–9. 10.1093/jac/dkac080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen S, Zhou Y, Chen Y et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018; 34: i884–i90. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gurevich A, Saveliev V, Vyahhi N et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013; 29: 1072–5. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feldgarden M, Brover V, Haft DH et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 2019; 63: e00483-19. 10.1128/aac.00483-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carattoli A, Zankari E, García-Fernández A et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Croucher NJ, Page AJ, Connor TR et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodriguez-R LM, Conrad RE, Viver T et al. An ANI gap within bacterial species that advances the definitions of intra-species units. mBio 2024; 15: e02696-23. 10.1128/mbio.02696-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seemann T. Snippy: rapid haploid variant calling and core genome alignment. GitHub 2020; 10. https://github.com/tseemann/snippy [Google Scholar]
- 25. Chichón G, López M, de Toro M et al. Spread of Pseudomonas aeruginosa ST274 clone in different niches: resistome, virulome, and phylogenetic relationship. Antibiotics 2023; 12: 1561. 10.3390/antibiotics12111561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh S, Pulusu CP, Pathak A et al. Complete genome sequence of an extensively drug-resistant Pseudomonas aeruginosa ST773 clinical isolate from north India. J Glob Antimicrob Resist 2021; 27: 244–6. 10.1016/j.jgar.2021.10.010 [DOI] [PubMed] [Google Scholar]
- 27. Simner PJ, Mostafa HH, Bergman Y et al. Progressive development of cefiderocol resistance in Escherichia coli during therapy is associated with an increase in blaNDM-5 copy number and gene expression. Clin Infect Dis 2022; 75: 47–54. 10.1093/cid/ciab888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morita Y, Cao L, Gould VC et al. Nald encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol 2006; 188: 8649–54. 10.1128/JB.01342-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sobel ML, Neshat S, Poole K. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J Bacteriol 2005; 187: 1246–53. 10.1128/JB.187.4.1246-1253.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidtke AJ, Hanson ND. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2008; 52: 3922–7. 10.1128/AAC.00341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosenboom I, Oguz S, Lüdemann IM et al. Pseudomonas aeruginosa population genomics among adults with bronchiectasis across Germany. ERJ Open Res 2023; 9: 00156-2023. 10.1183/23120541.00156-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wendel AF, Malecki M, Mattner F et al. Genomic-based transmission analysis of carbapenem-resistant Pseudomonas aeruginosa at a tertiary care centre in Cologne (Germany) from 2015 to 2020. JAC Antimicrob Resist 2022; 4: dlac057. 10.1093/jacamr/dlac057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sadek M, Le Guern R, Kipnis E et al. Progressive in vivo development of resistance to cefiderocol in Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 2023; 42: 61–6. 10.1007/s10096-022-04526-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ito A, Sato T, Ota M et al. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob Agents Chemother 2018; 62: e01454-17. 10.1128/AAC.01454-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Q, Neidig N, Chu TY et al. In vitro antibacterial activity of cefiderocol against recent multidrug-resistant carbapenem-nonsusceptible Enterobacterales isolates. Diagn Microbiol Infect Dis 2022; 103: 115651. 10.1016/j.diagmicrobio.2022.115651 [DOI] [PubMed] [Google Scholar]
- 36. Luscher A, Moynié L, Auguste PS et al. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugates. Antimicrob Agents Chemother 2018; 62: e00097-18. 10.1128/AAC.00097-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Padovani M, Bertelli A, Corbellini S et al. In vitro activity of cefiderocol on multiresistant bacterial strains and genomic analysis of two cefiderocol resistant strains. Antibiotics (Basel) 2023; 12: 785. 10.3390/antibiotics12040785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gomis-Font MA, Sastre-Femenia M, Taltavull B et al. In vitro dynamics and mechanisms of cefiderocol resistance development in wild-type, mutator and XDR Pseudomonas aeruginosa. J Antimicrob Chemother 2023; 78: 1785–94. 10.1093/jac/dkad172 [DOI] [PubMed] [Google Scholar]
- 39. Klein S, Boutin S, Kocer K et al. Rapid development of cefiderocol resistance in carbapenem-resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin Infect Dis 2022; 74: 905–8. 10.1093/cid/ciab511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simner PJ, Beisken S, Bergman Y et al. Cefiderocol activity against clinical Pseudomonas aeruginosa isolates exhibiting ceftolozane-tazobactam resistance. Open Forum Infect Dis 2021; 8: ofab311. 10.1093/ofid/ofab311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langaee TY, Gagnon L, Huletsky A. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC beta-lactamase expression. Antimicrob Agents Chemother 2000; 44: 583–9. 10.1128/AAC.44.3.583-589.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang C, Yang D, Wang Y et al. Cefiderocol for the treatment of multidrug-resistant gram-negative bacteria: a systematic review of currently available evidence. Front Pharmacol 2022; 13: 896971. 10.3389/fphar.2022.896971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. López-Causapé C, Maruri-Aransolo A, Gomis-Font MA et al. Cefiderocol resistance genomics in sequential chronic Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin Microbiol Infect 2023; 29: 538.e7–e13. 10.1016/j.cmi.2022.11.014 [DOI] [PubMed] [Google Scholar]
- 44. Ochs M, Veitinger S, Kim I et al. Regulation of citrate-dependent iron transport of Escherichia coli: fecR is required for transcription activation by FecI. Mol Microbiol 1995; 15: 119–32. 10.1111/j.1365-2958.1995.tb02226.x [DOI] [PubMed] [Google Scholar]
- 45. Yokoyama T, Niinae T, Tsumagari K et al. The Escherichia coli S2P intramembrane protease RseP regulates ferric citrate uptake by cleaving the sigma factor regulator FecR. J Biol Chem 2021; 296: 100673. 10.1016/j.jbc.2021.100673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santerre Henriksen A, Jeannot K, Oliver A et al. In vitro activity of cefiderocol against European Pseudomonas aeruginosa and Acinetobacter spp., including isolates resistant to meropenem and recent β-lactam/β-lactamase inhibitor combinations. Microbiol Spectr 2024; 12: e0383623. 10.1128/spectrum.03836-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abouelhassan Y, Gill CM, Nicolau DP. Assessing the in vivo efficacy of rational antibiotics and combinations against difficult-to-treat Pseudomonas aeruginosa producing GES β-lactamases. J Antimicrob Chemother 2023; 78: 1843–7. 10.1093/jac/dkad098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wise MG, Karlowsky JA, Hackel MA et al. In vitro activity of cefiderocol against meropenem-nonsusceptible gram-negative bacilli with defined β-lactamase carriage: SIDERO-WT Surveillance Studies, 2014-2019. Microb Drug Resist 2023; 29: 360–70. 10.1089/mdr.2022.0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kazmierczak KM, Tsuji M, Wise MG et al. In vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptible Gram-negative bacilli, including serine carbapenemase- and metallo-β-lactamase-producing isolates (SIDERO-WT-2014 study). Int J Antimicrob Agents 2019; 53: 177–84. 10.1016/j.ijantimicag.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 50. Csörgő B, Nyerges A, Pál C. Targeted mutagenesis of multiple chromosomal regions in microbes. Curr Opin Microbiol 2020; 57: 22–30. 10.1016/j.mib.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nurjadi D, Kocer K, Chanthalangsy Q et al. New Delhi metallo-beta-lactamase facilitates the emergence of cefiderocol resistance in Enterobacter cloacae. Antimicrob Agents Chemother 2022; 66: e0201121. 10.1128/aac.02011-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.