Abstract
Leishmania, a protozoan parasite, is responsible for significant morbidity and mortality worldwide, manifesting as cutaneous, mucocutaneous, and visceral leishmaniasis. These diseases pose a substantial burden, especially in impoverished regions with limited access to effective medical treatments. Current therapies are toxic, have low efficacy, and face growing resistance. Understanding the metabolic pathways of Leishmania, particularly those differing from its host, can unveil potential therapeutic targets. In this study, we investigated the acetyl-CoA synthetase (ACS) enzyme from Leishmania infantum (LiAcs1), which, unlike many organisms, also exhibits acetoacetyl-CoA synthetase (KBC) activity. This dual functionality is unique among acyl-CoA/NRPS/luciferase superfamily enzymes and crucial for the parasite's reliance on leucine catabolism, energy production, and sterol biosynthesis. Our biochemical characterization of LiAcs1 revealed its ability to utilize both acetate and acetoacetate substrates. Additionally, LiAcs1 displayed a distinct CoA substrate inhibition pattern, partially alleviated by acetoacetate. Structural analysis provided insights into the substrate binding flexibility of LiAcs1, highlighting a more promiscuous substrate pocket compared to other ACS- or KBC-specific enzymes. Substrate mimetics elucidated its ability to accommodate both small and large AMP-ester derivatives, contributing to its dual ACS/KBC functionality. These findings not only advance our understanding of Leishmania metabolism but also present LiAcs1 as a promising drug target. The dual functionality of LiAcs1 underscores the potential for developing selective inhibitors that could disrupt critical metabolic pathways across Leishmania spp. as it appears this enzyme is highly conserved across this genus. This paves the way for developing novel effective treatments against this devastating disease.
Keywords: acetyl-CoA synthetase, acetoacetate, Leishmania, metabolism, substrate specificity
Leishmania is a protozoan parasite responsible for causing disease that manifests as skin ulcers called cutaneous leishmaniasis (CL), ulcers of the nose, mouth, and throat called mucocutaneous leishmaniasis, or the more severe organ damage-associated visceral leishmaniasis. World-wide, up to 400,000 cases of CL and 1.2 million cases of visceral leishmaniasis contribute to nearly 40 thousand deaths annually (1). Together, Leishmania spp. are the biggest parasitic killers after the malaria causing parasites. Perhaps as alarming as Leishmaniasis mortality is its morbidity. Survivors of CL frequently face life-long disfigurement, and, in many cultures, this disability serves as a pretext for exclusion from society (2). The global distribution of Leishmania puts 350 million at risk for this devastating disease in primarily impoverished regions with lack of access to medical care (3). As a eukaryotic pathogen and parasite, Leishmania presents unique challenges for treatment. Current therapies include highly toxic and low efficacy options such as pentavalent antimonials, miltefosine, amphotericin B (AmB), and paromomycin. Additionally, each of these therapies are difficult to access and are vulnerable to a growing threat of drug resistance and treatment failure (4). The limited range of treatments, coupled with their significant adverse effects, underscores the urgent need for new therapeutic strategies.
Leishmania is transmitted to humans through the bite of an infected female phlebotomine sandfly. These sandflies are tiny and therefore difficult to see. They are also most active during sleeping hours and particularly resistant to vector control measures, including broad application of insecticides and insecticide treated nets (5). Upon infection, this parasite undergoes a morphological change from the flagellated promastigote found in the sandfly to the nonflagellated amastigote in mainly macrophages but also other mononuclear phagocytic cells. The lifecycle completes upon uptake from a sandfly taking a subsequent blood meal and morphogenesis back to the promastigote form (6). Leishmania has evolved distinct metabolic adaptations given the unique life cycle and transitioning between sandfly and mammalian hosts. Strategies for nutrient acquisition, availability of carbon and nitrogen sources, and the biosynthetic needs for proliferation and morphogenesis each require unique metabolic demands (7, 8, 9, 10). Understanding the metabolic pathways, especially those that differ from its host, can provide insights into potential therapeutic strategies that target processes crucial for the parasite’s survival and pathogenicity.
In general, the trypanosomatids which include Leishmania spp., exhibit an elevated reliance on leucine catabolism for energy production and sterol biosynthesis compared to their mammalian hosts (11). Sterols present a unique metabolic target in the trypanosomatids as this is the metabolic target of one of the few antitrypanosomal therapies, AmB. Like fungi, trypanosomes require ergosterol, the target of AmB, while mammals require cholesterol to maintain proper membrane function (12). Across mammals, fungi, and trypanosomes, the parent sterol generating mevalonate (MVA) metabolic pathway is conserved; however, like the differences in sterol products, the metabolic flux into sterol biosynthesis is distinct. In mammals, the MVA pathway is predominantly supplied carbon equivalents via ATP-citrate lyase generated acetyl-CoA (13). Similar to many parasitic protozoa, Leishmania require acetyl-CoA synthetase (ACS) function for acetyl-CoA production. The lack of alternative viable acetyl-CoA–producing pathways in the cytosol underlies the essentiality of ACS in Leishmania (14).
Leucine catabolism generates two carbon molecules that contribute to sterol biosynthesis: (1) acetyl-CoA and (2) acetoacetate. Acetyl-CoA can directly feed into the MVA pathway. Specifically, the first step of the MVA pathway involves acetoacetyl-CoA production via condensation of two acetyl-CoA molecules to form acetoacetyl-CoA. In addition, some organisms encode a second ACS-like enzyme called acetoacetyl-CoA synthetase or 3-keto-butanoyl-CoA synthetase (KBC) after the systematic IUPAC name of acetoacetate (15, 16, 17, 18, 19). These enzymes allow direct synthesis of acetoacetyl-CoA from acetoacetate and CoA. Curiously, Leishmania spp. lack a separately encoded bona fide KBC despite a high reliance on leucine catabolism and therefore elevated production of acetoacetate (Fig. 1A). We report that the ACS encoded by Leishmania infantum is a novel dual substrate enzyme and serves both ACS and KBC functionality. ACS homologs are highly conserved across Leishmania spp., suggesting that this dual function may also be conserved.
Figure 1.
Route of acetoacetate utilization by Leishmania is not obvious based on encoded CoA ligase homology.A, schematic illustrating leucine catabolism and its end products of acetoacetate and acetyl-CoA. B, CoA ligases encoded by Leishmania spp. and their homology to verified ACS- and KBC-functioning enzymes. The following are identified by their corresponding UniProt ID: Cryptococcus neoformans Acs1 (CnAcs1: J9VFT1), Saccharomyces cerevisiae Acs2 (ScAcs2: P52910), Saccharomyces cerevisiae Acs1 (ScAcs1: Q01574), Streptomyces lividans Acs1 (SlAcs1: A0A7U9HBW6), Homo sapiens Acss2 (HsAcss2: Q9NR19), Leishmania braziliensis Acs1 (LbAcs1: A4HCR9), Leishmania braziliensis Acs2 (LbAcs2: A4HCU1), Leishmania infantum Acs1 (LiAcs1: A4I093), Leishmania infantum Acs2 (LiAcs2: A4I0C2), Leishmania donovani Acs1 (LdAcs1: E9BG78), Leishmania donovani Acs2 (LdAcs2: A0A3S7WXN0), Cryptococcus neoformans Kbc1 (CnKbc,: J9VT24), Homo sapiens Aacs (HsAacs: Q86V21), and Streptomyces lividans Aacs (SlAacs: A0A7U9DRD6). Leishmania amazonensis Acs1 (LaAcs1: LAMA_000440100.1) is identified by its EuPathDB ID and with a modified reading frame as discussed in the text. ACS, acetyl-CoA synthetase; KBC, acetoacetyl-CoA synthetase.
Uncovering a dual function ACS provides new insight into the structural and biochemical features of acyl-CoA/NRPS/luciferase (ANL) family enzymes that will aid the pursuit of ACS as a drug target beyond the fight against Leishmaniasis. Parasites, fungi, and bacteria exhibit similar metabolic vulnerabilities and have been shown to require ACS functionality during infection (17). Additionally, many cancers in humans show a dependence on ACS, making it a focal point in developing anticancer therapies. Expanding our insight into the substrate binding selectivity of ACS could guide the design of disease-specific inhibitors while reducing side effects that may result from off-targeting other ANL family enzymes.
Results
Leishmania spp. lack expression of a KBC via sequence homology detection
A homology detection strategy was implemented using a standard BLAST search of short acyl-CoA–forming enzymes, specifically ACS- or KBC-like enzymes encoded by Leishmania spp. We used previously characterized ACSs and KBCs as reference enzymes (name: UniProt IDs) from humans (HsAcss2: Q9NR19, HsAacs: Q86V21), Streptomyces (SlAcs1: A0A7U9HBW6, SlAacs: A0A7U9DRD6), Cryptococcus (CnAcs1: J9VFT1, CnKbc1: J9VT24), and the model yeast Saccharomyces (ScAcs1: Q01574, ScAcs2: P52910) (15, 17, 18, 20, 21, 22, 23). We identified two copies of short length acyl-CoA–forming enzymes for L. infantum (LiAcs1: A4I093, LiAcs2: A4I0C2), Leishmania donovani (LdAcs1: E9BG78, LdAcs2: A0A3S7WXN0), Leishmania braziliensis (LbAcs1: A4HCR9, LbAcs2: A4HCU1), and only a single copy in Leishmania amazonensis (LaAcs1: LAMA_000440100.1) as identified by its EuPathDB ID. Close inspection of the LaAcs1-annotated reading frame showed an error such that only residues 282 to 615 are currently annotated for a 705 amino acid reading frame. The full-length protein was considered in these studies. We compared sequences using the multiple sequence protein alignment algorithm ClustalOmega along with the previously mentioned reference enzymes (24, 25, 26, 27). The percent identity matrix and sequence clustering indicate that all Leishmania spp. enzymes group with previously characterized ACS enzymes and are more distantly related to KBC-functioning enzymes (Fig. 1B). Among the encoded Leishmania ACS candidates, the sequence homology exceeds 83% identity within species and 78% across species, indicating a higher conservation of function than is seen in yeast, where ScAcs1 and ScAcs2 only share 54% identity despite shared substrate preference. Conversely, these Leishmania ACS candidates exhibit less than 25% identity compared to other KBC functioning enzymes, whereas mammals, yeast, and prokaryotes exhibit >37% identity across their encoded KBCs.
LiAcs1 exhibits dual ACS and KBC functionality
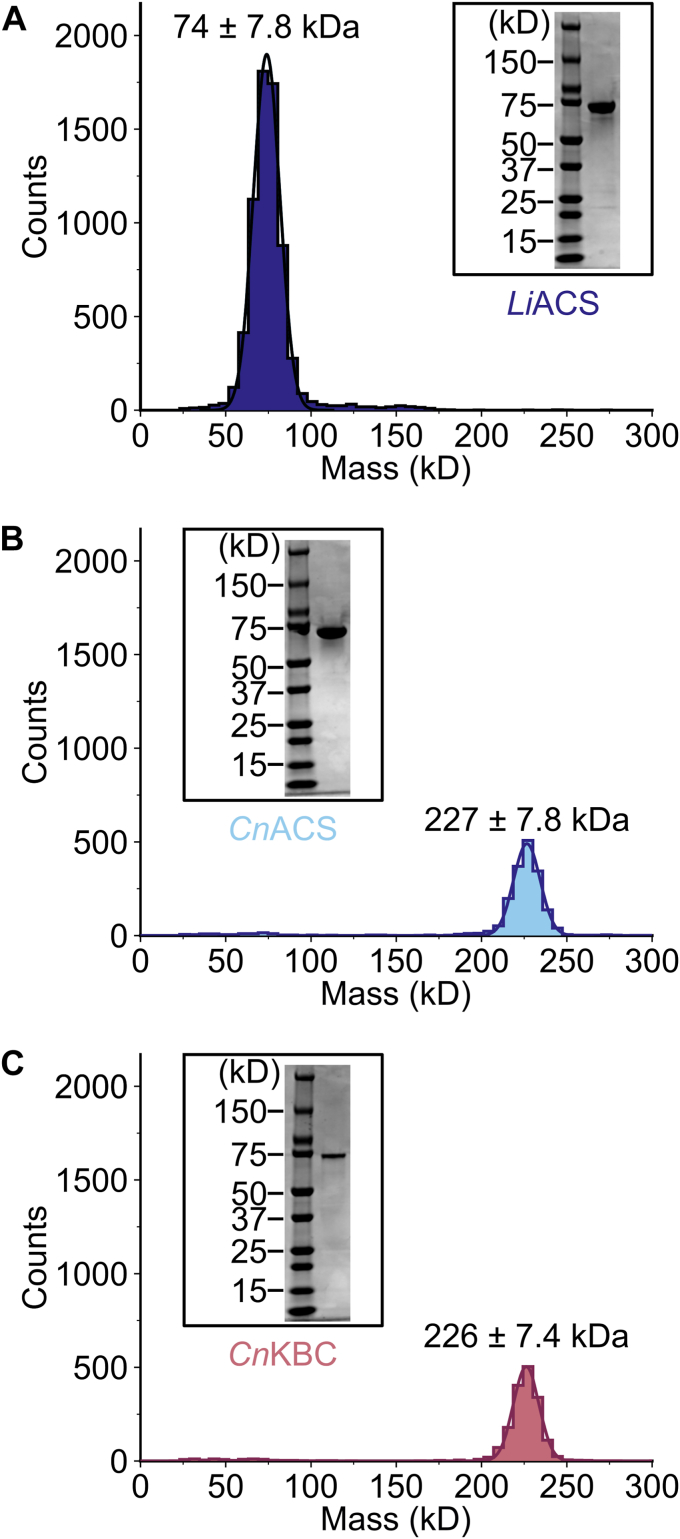
To characterize the biochemical and enzymatic properties of LiACS we expressed an N-terminal His8-tagged fusion protein in Escherichia coli and purified by immobilized metal affinity chromatography and size-exclusion chromatography (Fig. 2A). X-ray crystal structures of ACSs from other species adopt a trimeric form; however, these studies have not assessed oligomer formation using solution based analyses (22). To assess the oligomeric state and particle size of LiACS as well as previously studied CnAcs1 and CnKbc1, we performed dynamic light scattering and single molecule analysis using mass photometry (MP) (17, 22). In agreement with previous crystallography, CnAcs1 forms a particle size consistent with trimer formation at 1 mg/ml (Table 1) and this trimer is stable at low particle density (227 ± 7.8 kDa at 10 nM, Fig. 2B). Similarly, CnKbc1, which has not been structurally characterized, exhibits a particle size consistent with trimer formation at 1 mg/ml (Table 1) that is also stable at low particle density (226 ± 7.4 kDa at 10 nM, Fig. 2C). This is in contrast to LiAcs1 which shows no detectable trimer formation under either high concentrations 1 mg/ml (Table 1) or during single molecule analysis at low concentrations (74 ± 7.8 kDa at 10 nM, Fig. 2A).
Figure 2.
LiACS exists as a monomer in solution. Recombinantly expressed LiAcs1, CnAcs1, and CnKbc1 are pure as indicated by both SDS-PAGE and form a uniform population of monomers in the case of LiAcs1 (A) and trimers in the case of CnAcs1 (B) and CnKbc1 (C) as indicated by single particle analysis via mass photometry. CaAcs, Candida albicans Acs.
Table 1.
Dynamic light scattering data for recombinant proteins
| Enzyme | Radius (nm) | Mw-R (kDa) | Range 1 ange 1 (0.1–20 nm) |
||
|---|---|---|---|---|---|
| Radius (nm) | Mw-R (kDa) | % Mass | |||
| LiACS | 3.69 ± 0.06 | 71.5 ± 1.7 | 4.12 ± 0.10 | 92.3 ± 5.1 | 100.0 |
| CnACS | 9.84 ± 1.21 | 724 ± 208 | 6.43 ± 0.30 | 262 ± 28 | 98.9 ± 0.6 |
| CnKBC | 6.79 ± 0.06 | 296 ± 5.2 | 5.98 ± 0.66 | 225 ± 64 | 99.8 ± 0.1 |
To biochemically characterize LiAcs1, we employed a coupled continuous assay based on pyrophosphate release as previously reported for detecting various acid substrate utilization in the production of their respective acyl-CoA products (17). The specific activity for acetate utilization was 960 nmol/min/mg, which is far higher than our previously reported ACS enzymes ranging from 12 to 836 nmol/min/mg corresponding to a catalytic efficiency of Kcatapp/Kmapp = 20 mM−1s−1 (Fig. 3A) (22). Loss of activity was observed below pH 6.0 as has been previously described for ACS enzymes (Fig. 3B). To characterize (or determine) the substrate selectivity of LiAcs1, we compared the activity of six carboxylic acid substrates: acetate, propionate, butyrate, acetoacetate, 3-hydroxybutyrate, and valerate (smallest to largest). Unlike previously characterized ACSs, LiAcs1 exhibits both acetate and acetoacetate substrate utilization with minimal 3-hydroxybutyrate activity (15% of Vmax) and no appreciable activity for the remaining acid substrates (Fig. 3C). While the pyrophosphate-dependent enzyme activity detection assay only measures the first half of the reaction, intermediate product formation is not released in the absence of CoA and the reaction only progresses when CoA is present (28). To be sure that acyl-CoA products form successfully in the presence of either acetate or acetoacetate, we also performed a hydroxamate detection assay where hydroxylamine reacts with acyl-CoA in the presence of iron to form a hydroxamate product that absorbs at 560 nm (29, 30). We confirm acetyl-CoA and acetoacetyl-CoA formation using this well-established assay (Fig. S1). To be sure that hydroxamate formation was not occurring with the acyl-AMP intermediate, we performed the reaction without CoA to assure hydroxylamine itself was not serving as an electron acceptor and that acyl-AMP release was not occurring in its presence. Together, these data indicate that LiAcs1 is an acyl-CoA synthetase that utilizes both acetate and acetoacetate substrates.
Figure 3.
LiAcs1 is active and exhibits acetoacetate and acetate substrate utilization.A, dilution series indicates enzyme is highly active across a long linear range with respect to detection assay. B, pH sensitivity of LiAcs1 as normalized to pH 6. C, substrate utilization of LiAcs1.
LiAcs1 exhibits strong CoA substrate inhibition that is partially relieved by acetoacetate
ACSs function with an ordered bi-uni-uni-bi ping pong reaction mechanism where ATP binds first and is followed by acetate (31). These substrates are then catalyzed to form the acetyl-AMP reaction intermediate in the adenylate (AD)-forming conformation, while inorganic pyrophosphate is released (22, 32). The enzyme then undergoes a conformational shift to allow CoA binding in the thioester-forming conformation (TE-conf), where the acetyl group from acetyl-AMP is transferred to CoA to form acetyl-CoA (22, 32). Finally, the acetyl-CoA and AMP are released. We were curious if the dual acid substrate utilization of LiAcs1 would result in altered substrate binding affinity compared to other ACS or KBC-functioning enzymes; however, both ATP (Kmapp = 230 μM, confidence interval (95%) = (190, 270)) and acetate (Kmapp = 61 μM, CI (95%) = (45, 83)) exhibited similar kinetics with Kmapp’s comparable to previously characterized ACSs (Fig. 4, A and B) (20, 22, 23). Likewise, acetoacetate exhibited a Kmapp (88 μM, CI (95%) = (71, 110)) comparable to the Km for previously reported KBCs (Fig. 4C) (17). Enzyme kinetics did uncover a mild ATP substrate inhibition yielding higher theoretical Vmax = 1.8 μmol/min/mg, CI (95%) = (1.6, 2.0) not discernible from the acyl or CoA substrate enzyme activity curves. The substrate inhibition for ATP corresponded to a Kiapp = 2.2 μM, CI (95%) = (1.7, 2.7).
Figure 4.
Enzyme kinetics of LiAcs1. Representative Km curves for (A) ATP, (B) acetate, (C) acetoacetate, and (D) CoA. Km curves for CoA were performed with either acetate or acetoacetate provided as cosubstrate. All curves were fit using the GraphPad prism statistical software and Km’s were calculated from a minimum of three experimental replicates as reported in text.
Despite exhibiting dual substrate specificity of LiAcs1, we did not observe an appreciable decrease in its affinity for either acetate or acetoacetate compared to other ACS and KBC enzymes, respectively. Interestingly, LiAcs1 displays stark CoA substrate inhibition that is uncharacteristic of previously described ACS enzymes and more pronounced than previously reported KBCs (Fig. 4D) (17). We fit a nonlinear regression model for substrate inhibition using GraphPad Prism (www.graphpad.com), which indicates a Kiapp = 262 μM, CI (95%) = (110, 460) close to the Kmapp = 265 μM, CI (95%) = (150, 620) for CoA with a Vmax = 2.4 μmol/min/mg, CI (95%) = (1.7, 4.9) (Fig. 4D). The level of CoA substrate inhibition is far more pronounced than was previously described for CnKbc1 based on the observation that Kiapp ≈ Kmapp. Surprisingly, we observed a dramatic reduction in CoA inhibition when the substrate is acetoacetate rather than acetate (Kiapp = 891 μM, CI (95%) = (600, 1275); Kmapp = 171 μM, CI (95%) = (120, 250); Vmax = 2.5 μmol/min/mg, CI (95%) = (2.1, 3.1) where Kiapp > Kmapp. This relationship between Kiapp and Kmapp in the presence of acetoacetate is similar to the CoA substrate inhibition measured for CnKbc1 (17). The differences suggest that small acid substrate binding to LiAcs1 in the first half of the ACS reaction influences CoA binding characteristics in the second half of the reaction.
LiAcs1 substrate pocket accommodates both large and small AMP-ester bisubstrate inhibitors
Acetyl-CoA synthetases are differentiated from KBCs, medium-chain, and long-chain acyl-CoA synthetases by the presence of a stable tryptophan wall, which prevents larger acyl substrates from entering the pocket. Given that LiAcs1 contains this ACS-like tryptophan residue, (W466) we were surprised to observe acetoacetate utilization. The utilization of both acetate and acetoacetate suggests that LiAcs1 contains a more promiscuous substrate binding pocket compared to ACS- and KBC-specific enzymes. We and others have used AMP-ester based, bisubstrate inhibitors of ACS, KBC, and ANL family enzymes to characterize the alkyl group sizes accommodated by the different enzymes (17, 22, 33, 34, 35). We generated a series of AMP-esters with different alkyl chain lengths, alkyl branching, and degrees of unsaturation. The inhibitors mimic the acyl-AMP intermediate following the first half of the reaction. These bisubstrate mimetics allow us to probe the small acid substrate pocket by assessing their ability to bind and inhibit the reaction from progressing.
The profile of inhibition across these various sized and shaped AMP-esters can help us assess the flexibility of the small acid substrate binding pocket for LiAcs1 compared to the acetate-specific CnAcs1 versus acetoacetate-specific CnKbc1 enzymes. By organizing the AMP-esters according to their van der Waals volume from smallest to largest (from left to right), we can directly compare size versus ability to inhibit enzyme activity plotted as the −log10 IC50 (μM) such that a higher value corresponds to an increase in inhibitor potency (Fig. 5 and Table 2). While IC50 is substrate concentration-dependent, it can be used to as a proxy to compare relative Ki’s for inhibitors to a given enzyme when assay conditions hold substrate concentrations constant (36). The inhibition profile of the acetate-specific CnAcs1 shows a peak in inhibition for the ethyl- and cyclopropyl-AMP derivatives with a ∼3-fold drop in inhibition for the propyl- and isopropyl-AMP derivatives and another ∼2-fold drop in inhibition for the allyl- and propargyl-AMP derivatives (Fig. 5 and Table 2). This steady decrease in inhibition as AMP-ester derivatives increase in size indicates CnAcs1 contains a substrate binding pocket that selects for relatively smaller alkyl substituted carboxylic acid substrates. Conversely, the acetoacetate-specific CnKbc1 exhibits a more restrictive substrate binding pocket such that only the larger sized butyl-AMP ester exhibits any inhibition (Fig. 5 and Table 2).
Figure 5.
Visual comparison of AMP-ester inhibition profile according to van der Waals volume. Inverse log of inhibition data from Table 1 plotted according to increasing van der Waals volume from left to right.
Table 2.
Inhibition profile of AMP-ester isosteres
| # | Compound |
CnAcs1 |
CnKbc1 |
LiAcs1 |
|||
|---|---|---|---|---|---|---|---|
| IC50 (μM) | 95% CI [LL, UL] | IC50 (μM) | 95% CI [LL, UL] | IC50 (μM) | 95% CI [LL, UL] | ||
| i | Methyl-AMP | >100 | >100 | 69.9 | [52.8, 98.9] | ||
| ii | Ethyl-AMP | 8.0 | [5.8, 11.1] | >100 | 2.0 | [1.4, 2.8] | |
| iii | Cyclopropyl-AMP | 8.7 | [7.5, 10.2] | >100 | 2.2 | [1.8, 2.7] | |
| iv | Propyl-AMP | 21.1 | [11.3, 48.0] | >100 | 13.5 | [10.7, 17.0] | |
| v | Isopropyl-AMP | 25.3 | [21.6, 29.6] | >100 | 6.7 | [5.5, 8.3] | |
| vi | Allyl-AMP | 41.0 | [30.0, 59.8] | >100 | 12.5 | [10.6, 14.8] | |
| vii | Propargyl-AMP | 55.1 | [48.8, 62.6] | >100 | 10.2 | [7.5, 13.9] | |
| viii | Butyl-AMP | >100 | 7.8 | [5.7, 10.8] | >100 | ||
| ix | Diethyl-AMP | >100 | >100 | >100 | |||
| x | Cyclopentyl-AMP | >100 | >100 | >100 | |||
Consistent with its “hybrid” substrate specificity, the AMP-ester inhibition profile for LiAcs1 is a composite of the CnAcs1 and CnKbc1 profiles such that the peak inhibition occurs with ethyl- and cyclopropyl-AMP derivatives with a ∼3- to 6-fold drop in inhibition for the propyl- and isopropyl-AMP derivatives that does not drop again for the larger allyl- and propargyl-AMP derivatives (Fig. 5 and Table 2). While LiAcs1 can accommodate the larger allyl- and propargyl-AMP derivatives much better than CnAcs1 it does not extend to butyl-AMP as occurs for CnKbc1. Together, the AMP-ester inhibition profile for LiAcs1 does confirm a more flexible substrate binding pocket that, in turn, likely allows for both acetate and acetoacetate utilization and therefore dual ACS/KBC functionality.
LiAcs1 structure is highly homologous to previous ACS structures
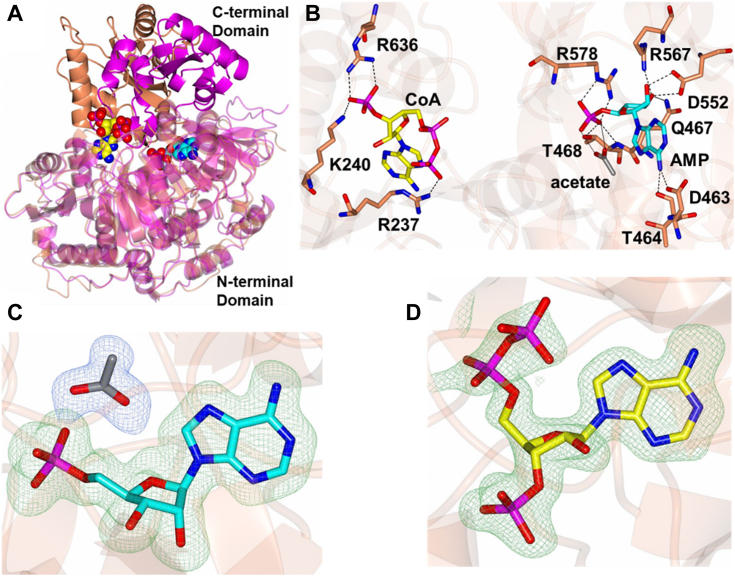
To understand the structural basis for substrate specificity, we performed X-ray crystallography and successfully obtained several structures of LiAcs1 (Table 3). We were able to generate structures in complex with combinations of CoA, AMP, acetate, and/or ethyl-AMP. The AMP-acetate/CoA structure is similar overall to CnAcs1 (PDB: 7L4G) with an RMSD of 1.26 Å between Ca atoms (511 residues). Previously, our group reported the crystal structures for CnAcs1 across each step of the enzyme reaction (22). Consistent with previous structural and biochemical analyses of adenylating enzymes, the structures highlighted a massive conformational shift that occurs in the C-terminal domain (CTD) between the first half of the reaction in the AD-forming conformation forming step to the TE-conf forming second half of the reaction. We captured LiAcs1 in the TE-conf in the presence of CoA, AMP, and acetate (Fig. 6A). The CoA molecule in LiAcs1 is located at an interface between the N-terminal domain and CTD and interacts with the side chains of K240, R237, and R636 and is engaged with the substrate pocket consistent with previous CoA-bound ACS structures (Fig. 6B). However, the pantetheine tail of the CoA molecule was disordered and could not be modeled. We were also able to capture a unique state of the AD-forming pocket with the AMP molecule present with an acetate in the active site. AMP forms hydrogen bond interactions with D463, T464, Q467, T468, D552, R567, and R578, while the acetate molecule appears to remain in contact the phosphate group of AMP. This is a very unusual finding because the presumed mechanism for generation of the acetyl-AMP intermediate of the first reaction is through nucleophilic attack of the acetate on the phosphate of ATP to displace pyrophosphate. ATP and acetate have been shown to bind in an ordered fashion such that the ATP binding event allows for acetate binding. Our structure suggests that the acetate binding pocket is not solely dependent on ATP but rather AMP is sufficient to allow acetate binding, although our crystallization conditions include 200 mM acetate which could preclude any mechanistic explanation for this binding. No previous ACS structure has shown the acid substrate bound alone and the current structures support an ordered binding event. Hydrogen bond interactions and electron density maps for the ligands are shown in Figure 6, B–D.
Table 3.
Crystallographic data for LiAcs1 and CaAcs2 structures
| Sample |
LiAcs1 |
LiAcs1 |
LiAcs1 |
LiAcs1 |
LiAcs1 |
CaAcs2 |
|---|---|---|---|---|---|---|
| Ligand |
AMP-acetate/CoA |
AMP/CoA |
Ethyl-AMP |
AMP-K+/CoA |
Ethyl-AMP-P21 |
Amp-CoA |
| PDB code | (8SF3) | (8U2T) | (8U2R) | (8U2U) | (8U2S) | (8V4R) |
| Data collection | ||||||
| Unit cell parameters (Å,o) | a = 59.02 | a = 59.07 | a = 58.91 | a = 58.75 | a = 91.21 | a = b = 139.41 |
| b = 69.38 | b = 69.92 | b = 74.15 | b = 69.46 | b = 61.81 | c = 542.25 | |
| c = 151.62 | c = 150.72 | c = 151.05 | c = 149.38 | c = 134.81 | ||
| b = 92.98 | ||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P21 | P6122 |
| Resolution (Å)a | 151.62–1.70 (1.74–1.70) | 49.49–1.65 (1.68–1.65) | 46.45–1.55 (1.58–1.55) | 74.69–1.97 (2.02–1.97) | 134.63–2.52 (2.59–2.52) | 49.46–2.70 (2.75–2.70) |
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 | 0.9795 | 0.9795 | 0.9795 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 |
| Observed reflections | 898,647 | 1,022,350 | 1,288,859 | 583,642 | 272,635 | 1,734,018 |
| Unique reflections | 68,961 | 75,992 | 96,781 | 44,111 | 51,168 | 86,857 |
| <I/σ(I)>a | 12.3 (1.8) | 14.5 (1.9) | 16.0 (1.8) | 13.5 (1.6) | 9.3 (1.6) | 14.5 (1.7) |
| Completeness (%)a | 99.5 (98.5) | 100 (100) | 100 (100) | 99.9 (99.6) | 99.9 (100) | 100 (100) |
| Multiplicitya | 13.0 (13.6) | 13.5 (13.9) | 13.3 (13.6) | 7.0 (6.4) | 5.3 (5.6) | 20.0 (19.1) |
| Rmerge (%)ab | 12.8 (175.0) | 11.0 (173.1) | 8.4 (164.2) | 11.2 (161.4) | 13.7 (118.0) | 18.5 (235.2) |
| Rmeas (%)ad | 13.3 (181.7) | 11.5 (179.7) | 8.7 (170.6) | 11.7 (168.7) | 15.2 (129.9) | 18.9 (241.6) |
| Rpim (%)ad | 3.7 (48.8) | 3.1 (47.8) | 2.4 (46.0) | 3.2 (47.5) | 6.4 (53.7) | 4.2 (55.1) |
| CC1/2ae | 0.998 (0.693) | 0.999 (0.762) | 0.999 (0.654) | 0.999 (0.873) | 0.997 (0.596) | 0.999 (0.800) |
| Refinement | ||||||
| Resolution (Å)a | 44.95–1.70 | 45.12–1.65 | 23.84–1.55 | 40.47–1.97 | 91.08–2.52 | 49.46–2.70 |
| Reflections (working/test)a | 65,452/3405 | 72,074/3795 | 91,862/4819 | 64,534/2196 | 48,521/2575 | 82,259/4353 |
| Rfactor/Rfree (%)ac | 15.0/18.7 | 15.4/18.2 | 15.1/17.4 | 16.3/21.6 | 18.6/21.9 | 20.6/23.1 |
| No. of atoms (Protein/ligands/water) | 5248/58/462 | 5196/54/476 | 5210/25/503 | 5192/54/222 | 10,161/50/17 | 14,387/195/41 |
| Model quality | ||||||
| RMSDs | ||||||
| Bond lengths (Å) | 0.009 | 0.009 | 0.008 | 0.012 | 0.002 | 0.003 |
| Bond angles (o) | 1.012 | 0.997 | 0.942 | 1.008 | 0.505 | 0.617 |
| Mean B-factor (Å2) | ||||||
| All atoms | 25.8 | 27.1 | 31.8 | 45.1 | 58.7 | 82.4 |
| Protein | 25.1 | 26.2 | 31.2 | 45.1 | 58.6 | 82.1 |
| Ligand | 27.6 | 32.1 | 20.9 | 44.8 | 41.6 | 106.6 |
| Water | 33.0 | 34.4 | 38.0 | 44.9 | 48.8 | 56.5 |
| Coordinate error (maximum likelihood) (Å) | 0.19 | 0.17 | 0.14 | 0.19 | 0.30 | 0.35 |
| Ramachandran plot | ||||||
| Most favored (%) | 97.6 | 97.6 | 97.1 | 97.9 | 96.1 | 95.6 |
| Additionally allowed (%) | 2.4 | 2.4 | 2.9 | 2.0 | 3.9 | 3.2 |
CaAcs2, Candida albicans Acs2.
Values in parenthesis are for the highest resolution shell.
Rmerge = ∑hkl∑i |Ii(hkl) - <I(hkl)>|/∑hkl∑iIi(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = ∑hkl ||Fobs (hkl) | - |Fcalc (hkl) ||/∑hkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Figure 6.
Structure of LiAcs1.A, CnAcs1 (magenta) superimposed with LiAcs1 (coral). The ligands from Leishmania ACS are colored: AMP (cyan), acetate (gray), and CoA (yellow). B, hydrogen bonds between LiAcs1 and CoA (yellow), AMP (cyan), and acetate (gray). C and D, electron density (Fo-Fc, 3s) for acetate/AMP and CoA, respectively. ACS, acetyl-CoA synthetase.
The structure without acetate bound displays a high degree of similarity compared to the ATP-acetate/CoA-bound structure with an RMSD of 0.27 Å between Ca atoms (661 residues). Despite the absence of the acetate molecule AMP adopts a nearly identical binding mode with only a small shift in the phosphate group as shown in Fig. S2A. We also generated crystals without acetate but in the presence potassium bromide or potassium thiocyanate. These yielded a structure with a potassium ion bound near the AMP molecule and generated very similar structures to the acetate bound and unbound crystals with an RMSD of 0.39 Å between Ca atoms (660 residues). The main difference found in the potassium-bound structure occurs in the loop-containing residues G576 and N573, which is moved away slightly from the AMP molecule. This results in a new contact between AMP and N573 which also coordinates the potassium ion (Fig. S2B).
LiAcs1 acid substrate pocket is wider than the acetate restricted CaAcs2
To better understand any differences that might exist for substrate binding between the dual substrate binding acetate/acetoacetate-utilizing LiAcs1 and an acetate restricted Candida albicans Acs2 (CaAcs2) (PDB: 8V4R), we also crystallized CaAcs2 in the presence of AMP, acetate, and CoA (22). It is clear that the LiAcs1 exhibits an alternative binding orientation for acetate (Fig. S4A) compared to CaAcs2 (Fig. S4B). At first look, the AMP, acetate-containing substrate pockets are not large enough to accommodate acetoacetate. However, the binding pose of AMP relative to the pocket shows a 1.5 Å greater distance between the proximal oriented phosphate of AMP and the classic substrate size restricting tryptophan wall (W466) (Fig. 7, A and B). About half of this distance can be attributed to orientation of the phosphate oxygens as the distance from phosphorous atom to the tryptophan wall is 8.7 Å for LiAcs1 versus 8.1 Å for CaAcs2. Further pocket widening may occur with substrate accommodation as our previously reported CnAcs1 crystal structures highlighted a slight pocket widening that occurs for AMP-ester bisubstrate mimetics. Our and others’ previous studies have illustrated substrate restriction occurs through the presence of a stable tryptophan wall that prevents larger substrates from being accommodated (22, 37). Pocket widening does occur opposite to this tryptophan wall and appears to be determined largely by the overall flexibility of the CTD. This pocket widening can be seen in the previously reported CnAcs1 in complex with ethyl-AMP (Fig. S4C). Therefore, we generated a structure of LiAcs1 in complex with ethyl-AMP to assess whether substrate pocket widening also occurs for LiAcs1 in a manner that may be consistent with acetoacetate utilization.
Figure 7.
LiAcs1 substrate pocket size allows viable pose of docked acetoacetate. Substrate pocket size differences illustrated by distance from conserved tryptophan wall to phosphate of AMP for LiAcs1 8SF3 (coral) (A) and CaAcs2 8V4R (blue) (B). Structures were oriented to illustrate the maximal distance measured without acid substrate present. Molecular docking was performed and acetoacetate was modeled into the acetate substrate pocket for LiAcs1 (8SF3) (C) and CaAcs2 (8V4R) (D). Structures were oriented to illustrate the alternative binding poses of acetoacetate that were adopted when performing a global energy minimization using the Amber10:EHT force field in Molecular Operating Environment (MOE, version 2020.01; Chemical Computing Group). In all panels, only residues within 4 Å of the crystalized acetate ligand that comprised the acid substrate pocket were illustrated. Illustrations rendered by PyMOL (pymol.org). CaAcs2, Candida albicans Acs2.
The structure of LiAcs1 in complex with ethyl-AMP is very similar to the AMP-acetate/CoA-bound structure with an RMSD of 1.00 Å between Ca atoms (651 residues). Since the ethyl-AMP structure was obtained without CoA added during crystallization, there’s a small shift in the CTD relative to the CoA-bound structure as shown in Fig. S3A. As previously observed with ethyl-AMP–bound ACS structures, the ethyl group of the inhibitor occupies the same site as the acetate molecule (Fig. S3B). Not surprisingly, the ethyl-AMP inhibitor forms hydrogen bond interactions with D463, T464, Q467, T468, D552 R567, and R578 similar to AMP, and the ethyl group is positioned with a hydrophobic pocket formed by I362, T363, V438, T468, and W466 (Fig. S3, C and D). The structure of monoclinic P form of the ethyl-AMP complex was also obtained which adopts a nearly identical ligand-binding mode. A direct comparison of the ethyl-AMP–bound LiAcs1 and CnAcs1 illustrates a slightly larger substrate pocket widening for LiAcs1 that results from a shift in the CTD (Fig. S4, D and E).
Molecular docking supports acetoacetate utilization by LiAcs1
To assess acetate versus acetoacetate binding energetics, we performed molecular docking experiments for each substrate in the LiAcs1 (8SF3) and CaAcs2 (8V4R) structures. Both LiAcs1 and CaAcs2 exhibit relatively similar binding energies for acetate, while LiAcs1 exhibits a much greater binding energy for acetoacetate (Table 4). The preferred docking pose for acetoacetate in LiAcs1 allows binding in a reactive pose (Fig. 7C) that is not observed for CaAcs2 (Fig. 7D). Therefore, while acetoacetate may be able to bind in the acetate restricted CaAcs2 enzyme albeit with much reduce binding energy compared to LiAcs1, the preferred binding orientation is not likely to be reactive.
Table 4.
Molecular docking of acetate versus acetoacetate
| Ligand |
CaAcs2 (8V4R) |
LiAcs1 (8SF3) |
|---|---|---|
| (ΔG -kcal/mol) | (ΔG -kcal/mol) | |
| Acetate | −3.95 | −3.86 |
| Acetoacetate | −3.65 | −4.90 |
CaAcs2, Candida albicans Acs2.
Discussion
The lack of a separate KBC enzyme encoded by Leishmania, despite its reliance on leucine catabolism, has been a puzzling aspect of its metabolism (38). Our demonstration that the LiAcs1 has both activities provides an explanation for this metabolic conundrum. Furthermore, the ability of LiAcs1 to serve as both an ACS and KBC in L. infantum is, to our knowledge, unprecedented for this group of adenylating enzymes. This uniquely described metabolic machinery is highly conserved across Leishmania spp. including L. infantum, L. donovani, L. braziliensis, and L. amazonensis, but also highly homologous to other ANL family enzymes spanning a broad range of both prokaryotes and eukaryotes. For Leishmania it appears this dual functionality suggests a unique evolutionary adaptation, allowing efficient utilization of both acetate and acetoacetate as a product of ketogenesis for energy production and biosynthesis.
To understand how the substrate binding pocket of LiAcs1 allows for dual substrate functionality, we tested a series of acyl-AMP intermediate mimicking AMP-esters of varying sizes. We observed a more promiscuous substrate binding pocket than either acetate- or acetoacetate-specific enzymes CnAcs1 or CnKbc1. While CnAcs1 primarily accommodates small AMP-esters and CnKbc1 only accommodates the large butyl-AMP ester, LiAcs1 is able to accommodate both small and large AMP-esters. Looking at either the primary sequence homology and or the structural homology of LiAcs1 to previously characterized ACSs and KBCs, we could not identify an obvious molecular basis that allows for expanded AMP-ester binding or dual ACS/KBC functionality. We attempted to perform a specificity-determining-sites detection strategy utilizing category informed multiple sequence alignment tools: however, we quickly realized that while the annotated pool of ACS and KBC enzymes is quite large the biochemical confirmation of their substrate specificity is sparse. Additionally, given that LiAcs1 operates in a newly described third category with ACS/KBC dual functionality it is currently unclear what fraction of annotated ACSs also contain KBC functionality.
There are multiple pieces of data to suggest that LiAcs1 substrate specificity may be governed by emergent properties that contribute to overall protein flexibility rather than any specific set of substrate-binding residues for this class of enzymes. We already know the CTD flexibility for this class of enzymes is enormous given the dramatic shift between the AD and TE conformations. Overall enzyme and substrate pocket flexibility is likely governed by multiple intramolecular interactions that may include substrate pocket adjacent intramolecular interactions. These interactions can have distal effects to protein stability and substrate accommodation that are not easily discerned. We provide a great example of this in our observation that substrate binding in the AD conformation appears to affect CoA binding in the TE conformation. This became clear as indicated by the difference in CoA substrate inhibition for LiAcs1 when either acetate or acetoacetate is supplied. We suspect these complex interactions contribute to LiAcs1 exhibiting an expanded substrate pocket flexibility and is supported by an expanded AMP-ester–binding profile. This broad substrate accommodation straddles ACS and KBC AMP-ester–binding profiles. The most compelling data to suggest substrate pocket flexibility is our observation that a substrate pocket widening occurs in the ethyl-AMP bound state for LiAcs1 beyond what we had previously described for CnAcs1 and this substrate pocket widening allows for acetoacetate accommodation. Together, this data supports an overall flexibility of LiAcs1 that may contribute to its expanded substrate profile and may also explain why LiAcs1 does not form a trimer like CnAcs1 or CnKbc1. Aside from simply having a larger substrate pocket, acetoacetate accommodation hinges on the acetate binding conformation observed in the LiAcs1 structure but not for CaAcs2.
The regulatory mechanisms governing the switch between ACS and KBC activities in LiAcs1 remain to be elucidated. It is possible substrate availability alone governs its contribution to overall metabolic flux. Alternatively, our observations of CoA substrate inhibition, particularly its modulation by acetoacetate, hint at a complex regulatory interplay between acid substrate availability, utilization, and CoA sensitivity. This substrate-induced modulation could represent a feed-forward mechanism, where acetoacetate accumulation relieves CoA inhibition. Conversely, metabolic demands for CoA may increase during a shift from a ketogenic to glycolytic carbon catabolism such that enhanced ACS inhibition is desired.
The unique dual-function of LiAcs1 represents a potential target for novel small molecule inhibitors that could, in turn, be selective for LiAcs1 over host enzymes given the unique substrate features of LiAcs1. By targeting a single enzyme critical for both acetyl-CoA and acetoacetyl-CoA production, it becomes possible to disrupt two essential metabolic pathways simultaneously. Further studies are required to understand the landscape of ACS enzymes that may also serve a dual ACS/KBC role, which may help elucidate the structural and specificity-determining-sites that dictate ACS versus KBC function.
Experimental procedures
Cloning of expression constructs, expression, and purification
Full-length L. infantum ACS construct (LeinA.00629.b.B1, UniProt A4I093, 1-705) containing a noncleavable His tag at the N terminus (MAHHHHHH) was codon-optimized using ATUM for E. coli expression and cloned by ATUM into ATUM vector pD431-SR via SapI cloning, including a double stop after the ORF. An example of the 5′ adapter just prior to ATG is 5′-TACACGTACTTAGTCGCTGAAGCTCTTCT-3′ and the 3′ adapter just after the double stop is 5′-TAGGTACGAACTCGATTGACGGCTCTTCTACC-3′. Codon optimization excluded restriction sites NcoI, NdeI, XhoI, HindIII, and SapI. The pD431-SR vector is kanamycin-resistant with the p15a origin of replication accepting inserts under the T7 promoter with a lac repressor and strong ribosome binding site. The resulting plasmid was sequence verified and transformed into BL21(DE3) (NEB C2527). An overnight starter culture prepared in LB broth with antibiotic was diluted 1:1000 the following day in LB media also containing antibiotic and grown the A600 between 0.5 and 1.0, where the culture was then cooled to between 18 to 25 °C. Expression was induced with 1 mM IPTG overnight and cell pellets were collected for lysis via sonication as previously described (17, 22). Cleared lysates were subjected to nickel purification and equilibration in overnight dialysis buffer as previously described (17, 22).
Enzyme activity detection
Enzyme activity was measured as previously reported for CnAcs1 and CnKbc1 (17, 22, 39). Briefly, the EnzChek Pyrophosphate assay kit (Thermo Fisher Scientific) was used as indicated by the manufacturer with the addition of 4 mM MgCl2, 10 mM DTT, and concentrations of CoA, ATP, and acid substrates as indicated with a total reaction volume of 50 μl. The final enzyme concentration of LiAcs1 was 3 μg/ml. All reagents were mixed in master mixes as allowed by each assay, aliquoted into 96-well half-well sized plates, incubated for 15 min at 37 °C, with acid substrate provided as the start reagent. The micro plate was then read continuously in a SpectraMax i3X Multi-Mode plate reader (Molecular Devices) at absorbance 360 nm. Michaelis–Menten constants were determined for each substrate such that the other substrates were provided in excess, except where CoA was provided at concentrations up to the maximum reaction velocity but before substrate inhibition. Kinetic constants (Km, Vmax, and Ki) were determined from the nonlinear regression of the slopes generated from the linear reaction curves as calculated using the GraphPad Prism statistical software. Hydroxamate forming reactions were performed under the same conditions as the EnzChek with the addition of 150 mM hydroxylamine pH 7.5. The reactions were carried out such that PPi generation was followed as normal to assure reaction curves were stopped within their linear range using trichloroacetate (200 mM) and FeCl3 (370 mM). Absorbance was then measured at 560 nm with background subtracted from no enzyme controls.
MP and dynamic light scattering
MP experiments were conducted as previously described using a Refeyn TwoMP mass photometer (Refeyn Ltd) (40). Briefly, microscope coverslips (24 mm × 50 mm, Thorlabs Inc.) and Silicon gaskets (Grace Bio-Labs) were cleaned by sequential rinsing with Milli-Q water and isopropanol, followed by drying with a filtered air stream. MP measurements were carried out at room temperature in sterile PBS. Calibration of the instrument was performed using a protein standard mixture: β-amylase (Sigma-Aldrich, 56, 112, and 224 kDa) and thyroglobulin (Sigma-Aldrich, 670 kDa). Movies were recorded for 60 s (3000 frames) using AcquireMP software (acquire.com.au, version 2.3.0; Refeyn Ltd) with standard settings. All recorded movies were processed and analyzed using DiscoverMP software (www.refeyn.com, version 2.3.0; Refeyn Ltd). Dynamic light scattering was performed on the same undiluted sample from mass photometry analysis using a DynaPro NanoStar (Wyatt Technology) equipped with additional static light scattering detector and temperature control.
Crystallization and data collection
Purified LiAcs1 was concentrated to 20 mg/ml in 500 mM NaCl, 20 mM Hepes pH 7.5, 5% glycerol, 2 mM DTT, and 0.025% sodium azide. All crystallization experiments were setup using an NT8 drop-setting robot (Formulatrix Inc) and UVXPO MRC (Molecular Dimensions) sitting drop vapor diffusion plates at 18 °C. One hundred nanoliters of protein and 100 nl crystallization solution were dispensed and equilibrated against 50 ul of the latter. Ligand complexes were prepared by adding 2 mM of the AMP, CoA, and ethyl-AMP compounds to the protein prior to crystallization. Crystals of LiAcs1 were obtained from the following conditions. AMP-acetate/CoA: Crystal Screen HT (Hampton Research) condition A9 (30% (w/v) polyethylene glycol (PEG) 4000, 200 mM ammonium acetate, 100 mM sodium citrate pH 5.6). Samples were vitrified in a fresh drop of crystallant which served as the cryoprotectant. AMP/CoA: JCSG+ E4 (1.26 M ammonium sulfate, 0.2 M lithium sulfate, 100 mM Tris pH 8.5) and cryoprotected in 2.5 M lithium sulfate. Ethyl-AMP: JCSG+ B4 (10% (w/v) PEG 8000, 8% (v/v) ethylene glycol, 100 mM Hepes pH 7.5), and cryoprotected in 20% PEG 200 + 80% crystallant. AMP-K+/CoA: JCSG+ G10 (30% PEG 2000MME, 150 mM KBr) and cryoprotected in a fresh drop of crystallant. Ethyl-AMP (P21 form): JCSG+ B5 (40% (v/v) MPD, 0.1 M Na cacodylate pH 6.5, 5% (w/v) PEG 8000), and cryoprotected in a fresh drop of crystallant. X-ray diffraction data were collected at the National Synchrotron Light Source II (NSLS-II) beamline 19-ID (NYX).
Structure solution and refinement
Intensities were integrated using XDS (www.globalphasing.com) (41) via autoPROC (42) and the Laue class analysis and data scaling were performed with Aimless (43). Structure solution was conducted by molecular replacement with Phaser (44) using previously determined structures of CnACS1 (PDB 7L4G) and Salmonella ACS (PDB 2P2F) for LiACS as the search models. Structure refinement and manual model building were conducted with Phenix (phenix-online.org) (45) and Coot (www.mrc-lmb.cam.ac.uk) (46) respectively. Structure validation was conducted with Molprobity (47) and figures were prepared using the CCP4MG package (48). Structure superpositions were conducted using GESAMT (49). Crystallographic data are provided in Table 3.
Molecular docking
Molecular docking of acetate and acetoacetate into ACS enzymes was conducted using the Molecular Operating Environment (www.chemcomp.com, version 2020.01; Chemical Computing Group). The crystal structures of LiAcs1 (PDB: 8SF3) and CaAcs2 (PDB: 8V4R), which are bound to both acetate and AMP, were used to evaluate substrate preference. Binding energy of acetate was determined via the London dG scoring function, followed by rescoring using the GBVI/WSA dG function within Molecular Operating Environment. Acetoacetate was modeled into the acetate binding site and a global energy minimization was performed using the Amber10:EHT force field. The binding energy of acetoacetate for the ACS structures was evaluated as described above.
Data availability
The coordinates and structure factors for the LiAcs1 and CaAcs2 structures were deposited to the Worldwide Protein Databank with the accession codes 8V4R, 8SF3, 8U2T, 8U2R, 8U2U, and 8U2S as indicated in Table 3. All other data is presented in the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research used resources at the NYX beamline 19-ID, supported by the New York Structural Biology Center, at the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The NYX detector instrumentation was supported by grant S10OD030394 through the Office of the Director of the National Institutes of Health. The HRMS was supported by the Northern Illinois University Molecular Analysis Core Facility and the National Science Foundation under grant No. 1726931. The authors would like to thank Dr Zhen Xu and Dr Nicholas Schnicker of the University of Iowa Protein and Crystallography Facility for the assistance and training in mass photometry.
Author contributions
A. J. J., B. L. S., K. P. B., S. L., T. J. H., and D. J. K. writing–review and editing; A. J. J., K. P. B., and S. L. writing–original draft; A. J. J. visualization; A. J. J., T. E. E., A. J. F., D. D., C. L. III., B. L. S., E. L. W., L. L., K. P. B., and S. L. validation; A. J. J., T. E. E., J. P., A. J. F., D. D., C. L. III., , B. L. S., E. L. W., L. L., K. P. B., and S. L. methodology; A. J. J., T. E. E., A. J. F., B. L. S., E. L. W., L. L., K. P. B., and S. L. investigation; A. J. J., T. E. E., J. P., A. J. F., D. D., C. L. III., , B. L. S., E. L. W., L. L., K. P. B., and S. L. formal analysis; A. J. J., T. E. E., A. J. F., B. L. S., E. L. W., L. L., K. P. B., and S. L. data curation; A. J. J., T. E. E., A. J. F., T. J. H., and D. J. K. conceptualization; J. P. software; B. L. S. and S. L. resources; K. P. B., S. L., T. J. H., and D. J. K. supervision; T. J. H. and D. J. K. project administration.
Funding and additional information
This work was supported by NIH grants 5R01AI161973 (D. J. K.), T32AI007511 (A. J. J.) This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. 75N93022C00036. The purchase of the NMR spectrometer used to obtain results included in this publication was supported by the National Science Foundation under the MRI award CHE-2117776 (T. J. H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
Supporting information
References
- 1.Alvar J., Vélez I.D., Bern C., Herrero M., Desjeux P., Cano J., et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grifferty G., Shirley H., McGloin J., Kahn J., Orriols A., Wamai R. Vulnerabilities to and the socioeconomic and psychosocial impacts of the leishmaniases: a review. Res. Rep. Trop. Med. 2021;12:135–151. doi: 10.2147/RRTM.S278138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres-Guerrero E., Quintanilla-Cedillo M.R., Ruiz-Esmenjaud J., Arenas R. Leishmaniasis: a review. F1000Res. 2017;6:750. doi: 10.12688/f1000research.11120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hefnawy A., Berg M., Dujardin J.-C., De Muylder G. Exploiting knowledge on Leishmania drug resistance to support the quest for new drugs. Trends Parasitol. 2017;33:162–174. doi: 10.1016/j.pt.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Picado A., Dash A.P., Bhattacharya S., Boelaert M. Vector control interventions for visceral leishmaniasis elimination initiative in South Asia, 2005-2010. Indian J. Med. Res. 2012;136:22–31. [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira D.E., Benchimol M., Rodrigues J.C.F., Crepaldi P.H., Pimenta P.F.P., de Souza W. The cell biology of Leishmania: how to teach using animations. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naderer T., McConville M.J. The Leishmania–macrophage interaction: a metabolic perspective. Cell Microbiol. 2008;10:301–308. doi: 10.1111/j.1462-5822.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 8.McConville M.J., Naderer T. Metabolic pathways required for the intracellular survival of Leishmania. Annu. Rev. Microbiol. 2011;6:543–561. doi: 10.1146/annurev-micro-090110-102913. [DOI] [PubMed] [Google Scholar]
- 9.McConville M.J., De Souza D., Saunders E., Likic V.A., Naderer T. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol. 2007;23:368–375. doi: 10.1016/j.pt.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Opperdoes F.R., Coombs G.H. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 2007;23:149–158. doi: 10.1016/j.pt.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Millerioux Y., Mazet M., Bouyssou G., Allmann S., Kiema T.R., Bertiaux E., et al. De novo biosynthesis of sterols and fatty acids in the Trypanosoma brucei procyclic form: carbon source preferences and metabolic flux redistributions. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W., Cross G.A.M., Nes W.D. Cholesterol import fails to prevent catalyst-based inhibition of ergosterol synthesis and cell proliferation of Trypanosoma brucei. J. Lipid Res. 2007;48:665–673. doi: 10.1194/jlr.M600404-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Guerra B., Recio C., Aranda-Tavío H., Guerra-Rodríguez M., García-Castellano J.M., Fernández-Pérez L. The mevalonate pathway, a metabolic target in cancer therapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.626971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soumya N., Panara M.N., Neerupudi K.B., Singh S. Functional analysis of an AMP forming acetyl CoA synthetase from Leishmania donovani by gene overexpression and targeted gene disruption approaches. Parasitol. Int. 2017;66:992–1002. doi: 10.1016/j.parint.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Ohgami M., Takahashi N., Yamasaki M., Fukui T. Expression of acetoacetyl-CoA synthetase, a novel cytosolic ketone body-utilizing enzyme, in human brain. Biochem. Pharmacol. 2003;65:989–994. doi: 10.1016/s0006-2952(02)01656-8. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa S., Noda K., Maeda A., Matsuoka M., Yamasaki M., Fukui T. Acetoacetyl-CoA synthetase, a ketone body-utilizing enzyme, is controlled by SREBP-2 and affects serum cholesterol levels. Mol. Genet. Metab. 2012;107:553–560. doi: 10.1016/j.ymgme.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Alden K.M., Jezewski A.J., Beattie S.R., Fox I.I.I.D., Krysan D.J. Genetic interaction analysis reveals that Cryptococcus neoformans utilizes multiple acetyl-CoA-generating pathways during infection. mBio. 2022;13 doi: 10.1128/mbio.01279-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell C.A., Tucker A.C., Escalante-Semerena J.C., Gulick A.M. The structure of S. lividans acetoacetyl-CoA synthetase shows a novel interaction between the C-terminal extension and the N-terminal domain. Proteins. 2015;83:575–581. doi: 10.1002/prot.24738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vögeli B., Engilberge S., Girard E., Riobé F., Maury O., Erb T.J., et al. Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3380–3385. doi: 10.1073/pnas.1718649115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Den Berg M.A., Steensma H.Y. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur. J. Biochem. 1995;231:704–713. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- 21.De Virgilio C., Bürckert N., Barth G., Neuhaus J.M., Boller T., Wiemken A. Cloning and disruption of a gene required for growth on acetate but not on ethanol: the acetyl-coenzyme a synthetase gene of Saccharmoyces cerevisiae. Yeast. 1992;8:1043–1051. doi: 10.1002/yea.320081207. [DOI] [PubMed] [Google Scholar]
- 22.Jezewski A.J., Alden K.M., Esan T.E., DeBouver N.D., Abendroth J., Bullen J.C., et al. Structural characterization of the reaction and substrate specificity mechanisms of pathogenic fungal acetyl-CoA synthetases. ACS Chem. Biol. 2021;16:1587–1599. doi: 10.1021/acschembio.1c00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luong A., Hannah V.C., Brown M.S., Goldstein J.L. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- 24.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sievers F., Higgins D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sievers F., Barton G.J., Higgins D.G. Multiple sequence alignments. Bioinformatics. 2020;227:227–250. [Google Scholar]
- 27.Madeira F., Madhusoodanan N., Lee J., Eusebi A., Niewielska A., Tivey A.R.N., et al. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024;52:W521–W525. doi: 10.1093/nar/gkae241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manandhar M., Cronan J.E. Proofreading of noncognate acyl adenylates by an acyl-coenzyme a ligase. Chem. Biol. 2013;20:1441–1446. doi: 10.1016/j.chembiol.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg P. Acyl adenylates: an enzymatic mechanism of acetate activation. J. Biol. Chem. 1956;222:991–1013. [PubMed] [Google Scholar]
- 30.Wilson D.J., Aldrich C.C. A continuous kinetic assay for adenylation enzyme activity and inhibition. Anal. Biochem. 2010;404:56–63. doi: 10.1016/j.ab.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrar W.W., Plowman K.M. Kinetics of acetyl-CoA synthetase—I. Mode of addition of substrates. Int. J. Biochem. 1975;6:537–542. doi: 10.1016/0020-711x(79)90019-3. [DOI] [PubMed] [Google Scholar]
- 32.Gulick A.M. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X., Zhang H., Tonge P.J., Tan D.S. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg. Med. Chem. Lett. 2008;18:5963–5966. doi: 10.1016/j.bmcl.2008.07.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupte A., Boshoff H.I., Wilson D.J., Neres J., Labello N.P., Somu R.V., et al. Inhibition of siderophore biosynthesis by 2-triazole substituted analogues of 5′-O-[N-(salicyl) sulfamoyl] adenosine: antibacterial nucleosides effective against Mycobacterium tuberculosis. J. Med. Chem. 2008;51:7495–7507. doi: 10.1021/jm8008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lux M.C., Standke L.C., Tan D.S. Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors. J. Antibiot. (Tokyo) 2019;72:325–349. doi: 10.1038/s41429-019-0171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yung-Chi C., Prusoff W.H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 37.Ingram-Smith C., Woods B.I., Smith K.S. Characterization of the acyl substrate binding pocket of acetyl-CoA synthetase. Biochemistry. 2006;45:11482–11490. doi: 10.1021/bi061023e. [DOI] [PubMed] [Google Scholar]
- 38.Blum J.J. Oxidation of leucine by Leishmania donovani. J. Protozool. 1991;38:527–531. doi: 10.1111/j.1550-7408.1991.tb06075.x. [DOI] [PubMed] [Google Scholar]
- 39.Comerford S.A., Huang Z., Du X., Wang Y., Cai L., Witkiewicz A.K., et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnicker N.J., Xu Z., Amir M., Gakhar L., Huang C.-L. Conformational landscape of soluble α-klotho revealed by cryogenic electron microscopy. bioRxiv. 2024 doi: 10.1101/2024.03.02.583144. [preprint] [DOI] [Google Scholar]
- 41.Kabsch W. Automatic indexing of rotation diffraction patterns. J. Appl. Crystallogr. 1988;21:67–72. [Google Scholar]
- 42.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohkamp B., Scott W.G., Cowtan K., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 49.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 2012;1:76. [PMC free article] [PubMed] [Google Scholar]
- 50.Evans P. Scaling and assessment of data quality. Acta Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 51.Diederichs K., Karplus P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 52.Weiss M.S. Global indicators of X-ray data quality. J. Appl. Crystallogr. 2001;34:130–135. [Google Scholar]
- 53.Karplus P.A., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans P. Biochemistry. Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for the LiAcs1 and CaAcs2 structures were deposited to the Worldwide Protein Databank with the accession codes 8V4R, 8SF3, 8U2T, 8U2R, 8U2U, and 8U2S as indicated in Table 3. All other data is presented in the article.