Abstract
Pigs play a central role in human livelihoods in China, but a lack of systematic large-scale whole-genome sequencing of Chinese domestic pigs has hindered genetic studies. Here, we present the 1000 Chinese Indigenous Pig Genomes Project sequencing dataset, comprising 1011 indigenous individuals from 50 pig populations covering approximately two-thirds of China’s administrative divisions. Based on the deep sequencing (~25.95×) of these pigs, we identify 63.62 million genomic variants, and provide a population-specific reference panel to improve the imputation performance of Chinese domestic pig populations. Using a combination of methods, we detect an ancient admixture event related to a human immigration climax in the 13th century, which may have contributed to the formation of southeast-central Chinese pig populations. Analyzing the haplotypes of the Y chromosome shows that the indigenous populations residing around the Taihu Lake Basin exhibit a unique haplotype. Furthermore, we find a 13 kb region in the THSD7A gene that may relate to high-altitude adaptation, and a 0.47 Mb region on chromosome 7 that is significantly associated with body size traits. These results highlight the value of our genomic resource in facilitating genomic architecture and complex traits studies in pigs.
Subject terms: Rare variants, Evolutionary genetics, Haplotypes, Agricultural genetics
Here the authors describe a genomic dataset, the1000 Chinese Indigenous Pig Genomes Project (1KCIGP), based on the whole-genome sequencing of 1,011 Chinese domestic pigs from 50 distinct populations, which provides insights into the genomic architecture of domestic pigs.
Introduction
Pigs are one of the most successful livestock animals and play an important role in worldwide economies and societies1. They provide nutrition and manure, and their bristles can be used to produce brushes. As a member of the family Suidae, Sus scrofa (e.g., domestic pigs and wild boars) originated in Southeast Asia ~ 3–4 million years ago (MYA) and colonized the entire Eurasian mainland over the last one million years2. European and Asian wild boar populations diverged around 1.2 MYA, with European populations experiencing severe bottlenecks during the Last Glacial Maximum3,4. In the Suidae family, domestic pigs are the only domesticated species, and domestication occurred independently in Europe and Asia approximately 10,000 years ago5. At least 235 local pig breeds are recognized across the diverse agroecology of the Asian continent (https://www.fao.org/dad-is). Over one-third of these breeds (83 indigenous breeds) inhabit China and have unique phenotypic and adaptive characteristics (Ministry of Agriculture and Rural Affairs of the People’s Republic of China, http://www.moa.gov.cn/). With the abundance of distinct breeds and diverse phenotypes, studying the genetic basis of Chinese domestic pigs will assist in resolving the role they play in forming the genomic patterns of pigs in East Asia and globally.
Comprehensive catalogs of genetic variations are essential building blocks in population and demographic history research, as well as genotype-phenotype associations. Numerous databases of human genomics and bioinformatics have been developed, including the UKbiobank6, Genome Aggregation Database7, 1000 Genomes project8, UK10K project9, ChinaMAP10, and NyuWa project11. Although many studies have concentrated on pig genomics and created large and diverse population genetic variation resources12–16, the sequencing depth (~ 12 ×) for most samples and un-uniform sequencing platforms in these datasets is insufficient for constructing a high-quality reference panel. Some studies are biased toward European commercial pig breeds or crossbreds like SWIM17. The studies referring to Chinese domestic pig populations have limited breed diversity (20–30 breeds) or sequencing depth (average coverage of sequencing depth < 20 ×) or geographical coverage18–20, and the systematic, high-depth sequencing study of a Chinese domestic pig population cohort is missing, particularly one that provides a population-specific reference panel.
China is the only location outside the Near East with clear archaeological evidence of independent pig domestication21,22. Over the past decade, several studies have investigated the domestication of Chinese pigs, their dispersal, and the overall patterns of admixture13,23–25, focusing on specific local populations, limited samples, or limited genomic data from Chinese domestic pigs, such as genotyping array data. Without access to an integrated, large-scale, and high-quality genomic dataset for domestic Chinese pig populations, the genetic divergence and structure of diverse Chinese indigenous pig breeds cannot be fully elucidated.
Here, we describe a genomic dataset, 1000 Chinese Indigenous Pig Genomes Project (1KCIGP), based on the deep (~25.95 ×) whole-genome sequencing (WGS) of 1011 Chinese domestic pigs from 50 distinct populations across 21 administrative divisions in China. The 1KCIGP dataset includes a total of 50.59 million single nucleotide polymorphisms (SNPs), 12.87 million insertion-deletion polymorphisms (indels), and 168,167 structural variations (SVs). More importantly, we constructed a 1KCIGP panel comprising thousands of samples, which has superior performance for imputation in the Chinese indigenous pig population. Furthermore, we detected an ancient admixture event coinciding with a human immigration climax in the 13th century, potentially contributing to establishing southeast-central Chinese pig populations. Simultaneously, positive selection analyses implicated THSD7A as a candidate gene associated with high-altitude adaptation. Meanwhile, genome-wide association studies (GWAS) revealed another gene with a novel SNP associated with body size traits. In total, our study provides valuable and reliable resources for facilitating genetic studies and the molecular breeding of pigs.
Results
Small variants across the 1011 1KCIGP samples
The 1KCIGP genome resource includes the whole-genome resequencing data of 1011 Chinese domestic pig samples from 50 distinct populations. These samples were collected from 21 administrative divisions in China, including 18 provinces, two autonomous regions, and one municipality directly under the central government (Supplementary Fig. 1). Most of the samples were sequenced at a depth > 20 ×. After genome alignment, the median actual genomic coverage was 25.95 (Supplementary Fig. 2 and Supplementary Data 1). We also used three methods to predict the gender of the samples, including the method offered in the Genome Analysis Toolkit (GATK), calculating the coverage of the sex chromosomes, and assessing the aligned coverage of the porcine SRY gene (Supplementary Methods and Supplementary Data 1). In total, 325 males and 686 females were identified without ambiguous samples (Fig. 1A).
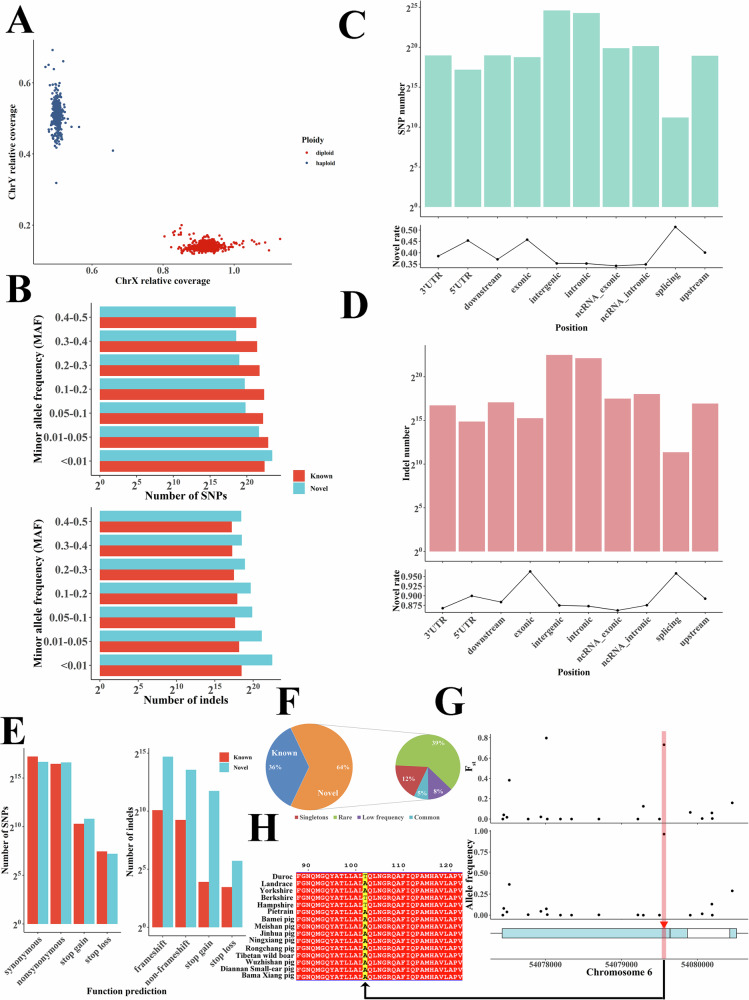
Fig. 1. Overview of the 1KCIGP dataset and small genomic variants.
A The sex of each individual was inferred from sex chromosome coverage and the estimated ploidy of chromosome X. B. The number of variants with different MAF bins, where the upper and lower plots represent SNP and indels, respectively. C Number (top) and novel rates (bottom) of SNPs in different annotation regions. D Number (top) and novel rates (bottom) of indels in the different annotation regions. E Number of functional protein-coding variants for SNPs (left) and indels (right). F The frequency spectrum of deleterious SNPs. G A missense mutation in the FUT1 gene. The top plot shows the Fst between Chinese domestic and European pigs. The middle plot shows the allele frequency of this mutation in Chinese domestic pigs, and the bottom plot shows the gene structure. H The missense mutation induced amino acid change.
The final 1KCIGP database contained a total of 63.46 million small variant loci after strict quality control filtering, including 50.59 million SNPs and 12.87 million indels. We found that 91.29% of these high-quality mutations are biallelic sites, including 1.60 million SNPs and 0.40 million indels from sex chromosomes. A/G (34.76%) transitions made up the majority of the mutation spectrum, followed by C/T (34.72%) transitions (Supplementary Fig. 3). Across all the SNPs and indels in the 1KCIGP dataset, 37.01% of the variants were rare variants (minor allele frequency, MAF < 1%), with singletons, doubletons, and tripletons accounting for 6.76%, 4.79%, and 3.30% of the variants, respectively (Supplementary Fig. 4).
To analyze the novel small variants in Chinese domestic pig populations, we compared the 1KCIGP dataset with the dbSNP (v150)26. Of all the small variants, 29.21 million variants (46.03%), including 17.95 million SNPs and 11.26 million indels, were identified as not being reported in the dbSNP. Among these novel variants, a large number of novel common variants (3.06 million SNPs and 3.04 million indels, MAF > 5%) and low-frequency variants (3.30 million SNPs and 2.29 million indels, MAF = 1%–5%) were identified in 1KCIGP (Fig. 1B). We noticed that singletons accounted for 10.22% of these novel variants, which suggested that the many rare variants were specific to a sample or population27. The Diannan Small-ear pig had the highest number of rare novel variants compared to other populations, followed by the Wujin pig and Tibetan wild boar (Supplementary Fig. 5).
To better characterize the 1KCIGP variant calls, we divided the genome into easy- and difficult-to-sequence regions (Supplementary Methods) according to a previous study28. While making up just 16.70% of the genome, the difficult regions—where sequencing read alignment is particularly difficult, and indel formation is common—included a disproportionately large number of the multiallelic sites (64.76% vs. 16.43% for biallelic sites). In addition, indels were more frequently detected in the difficult regions, with 47.14% of the indels present in these regions compared to just 13.86% of the SNPs. As low-complexity and repetitive components constitute the majority of the difficult regions, the enrichment of multiallelic and indel calls in these regions was consistent with expectations29.
The median numbers of SNPs and indels in the 1KCIGP samples were 12.08 million and 3.50 million, respectively. The numbers of detected SNPs and indels with MAF > 1% per sample showed slight positive correlations with genomic coverage (R2 = 0.017 and 0.071, respectively; P < 0.05 for both) (Supplementary Fig. 6). These results indicate that the detection quality could be improved by increasing the sequencing depth, particularly for indels. This might have been caused by regions that lacked random coverage or were too complex to amplify, and increased sequencing depth improved the variant detection ability in these regions. The mean numbers of SNPs and indels with MAF < 1% per sample were 96,841 (0.80%) and 22,146 (0.63%), respectively. These rare SNPs and indels also had a slightly positive correlation with sequencing depth (Supplementary Fig. 6); however, the correlation level (R2 = 0.025 and 0.025, respectively, P < 0.05 for both) was lower than that of the common variants. This was probably because the number of rare variants varied more widely in different samples than the number of variants with an MAF > 1% (± 62% vs. ± 5%), and the positive correlation was obscured by this large fluctuation.
Predicted function of small variants
To assess the functional consequences of the SNPs and indels in our call set, we annotated these variants based on the Ensembl dataset. The majority (51.16%) of variants were located in intergenic regions, while a total of 27.00 million variants were in protein-coding genes, including 486,756 variants in exon regions, 823,391 variants in untranslated regions (UTRs), 5091 variants within 2-bp of splicing junctions, and 25.69 million mutations in introns (Fig. 1C, D). There was a total of 1.18 million variants in non-coding RNA exon regions. Focusing on variants in the exons of protein-coding genes, 188,948 SNPs were annotated as nonsynonymous SNPs, while 99,336 were novel nonsynonymous SNPs. Other functional protein-coding variants included 255,363 synonymous SNPs, 27,662 frameshift indels, 12,735 non-frameshift indels, 6506 stop gains, and 387 stop losses (Fig. 1E).
The identification and frequency spectrum of deleterious variants contribute to recognizing phenotypic associations. Within the coding region of the genome, 25,237 deleterious SNPs located in 9013 protein-coding genes were identified in the 1KCIGP dataset using SIFT30. Over half of these deleterious SNPs were rare variants (12,602) or singletons (3237). A total of 4659 common and 4739 low frequency (MAF > 1%) deleterious variants were identified, including 3220 novel variants (Fig. 1F). The majority of protein-coding genes (6805 of 9013 genes, 75.50%) had rare deleterious variants (MAF ≤ 1%) in at least one participant. Each sample had an average of 2240 deleterious SNPs, including 658 homozygous and 1582 heterozygous variants. These results demonstrate that the variants in the 1KCIGP dataset could provide useful material to assist in the study of characteristic-related variants in Chinese pigs.
We further examined the frequency distribution in the 1KCIGP individuals of trait-associated and disease-associated variants collected from the Online Mendelian Inheritance in Animals (OMIA)31 database and investigated 71 SNPs and small indels spanning < 20 consecutive bases from the OMIA database. Of these, 12 variants were detected in at least one individual from the 1KCIGP dataset (Supplementary Data 2). As expected, the alternative allele frequencies of the morphology-associated variants ranged from 1.14%–99.85% (eight variants). In addition, four OMIA disease-associated variant positions were detected in KCIGP, especially variant rs335979375, which had a high frequency (96.34%) in 1KCIGP individuals and was associated with resistance to the edema disease (F18 receptor) phenotype. The mutation allele frequency (AF) of this variant in European pigs was lower, only about 43.10%. This variant, located in the FUT1 gene, caused a threonine-to-alanine mutation (Fig. 1H) and might contribute to the ubiquitous disease resistance of Chinese indigenous pigs.
The 1KCIGP imputation reference panel for Chinese domestic pig populations
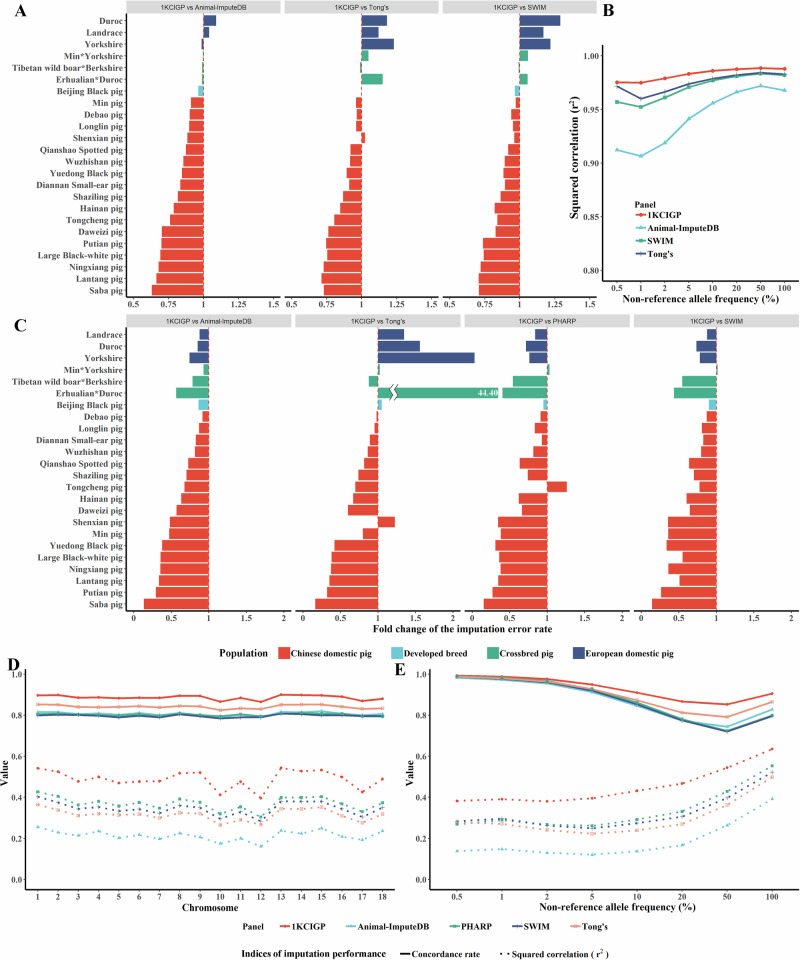
The genome-wide genotype imputation approach is a statistical methodology used to deduce absent genotypic information by inferring missing genotypes based on known haplotype data. This method can assist in genome-wide association studies (GWAS) and genomic prediction using SNP arrays. The 1KCIGP panel used 46.75 million SNPs with a minor allele count of two or more (MAC2) in 1011 independent samples. To evaluate imputation performance, the public sequencing data of 113 Chinese domestic pigs, 30 European domestic pigs, 100 developed pigs, and 19 crossbred pigs were used as test datasets (Supplementary Data 3). We tested two common application scenarios: imputing genotyped variants from low-coverage WGS and SNP chips. In the first scenario, we randomly extracted approximately 1 × reads from each test individual and detected SNPs using these reads (Supplementary Methods). Each SNP set of samples was then imputed, and the imputation performance was compared with the Animal-ImputeDB19, SWIM17, and Tong’s public reference panels20. As expected, for the diverse Chinese domestic pig populations, 1KCIGP outperformed all three comparative reference panels in terms of imputation concordance rates and the squared correlation (r2) between imputed dosages and true genotypes. 1KCIGP indicated a relatively higher concordance rate (average concordance rate: 77.46 %, Fig. 2A and Supplementary Fig. 9) and squared correlation (average r2: 0.987, Fig. 2B and Supplementary Fig. 10) than those of the Animal-ImputeDB (71.34% and 0.962), SWIM (73.24% and 0.979), and Tong’s (73.44% and 0.981) reference panels for the tested Chinese domestic pig breeds.
Fig. 2. Performance of the 1KCIGP haplotype reference panel.
A The fold-change in the imputation error rate in the 262 test WGS data using the 1KCIGP, Animal-ImputeDB, SWIM, and Tong’s reference panels. Imputation error rate = (1 – imputation concordance rate). The different colored bars represent different populations, corresponding to the populations shown in Fig. 2C. B The squared correlation between the imputed allele dosages and the true genotype within the stratified non-reference allele frequency bins was derived from the 113 test WGS data of Chinese domestic pigs using the 1KCIGP, Animal-ImputeDB, SWIM, and Tong’s reference panels. C Fold change in imputation error rate in the simulated commercial 50 K SNP chip using the 1KCIGP, Animal-ImputeDB, SWIM, PHARP, and Tong’s reference panels. Imputation error rate = (1 – imputation concordance rate). D Imputation performance of each chromosome from the simulated commercial 50 K SNP chip of Chinese domestic pigs using the 1KCIGP, Animal-ImputeDB, SWIM, PHARP, and Tong’s reference panels. This and subsequent scenarios only display results for the Chinese domestic pig. The solid line represents the concordance rate, while the dashed line represents the squared correlation. Different colors represent different panels. E The imputation performance within stratified non-reference allele frequency bins derived from the simulated commercial 50 K SNP chip data of Chinese domestic pigs using the 1KCIGP, Animal-ImputeDB, SWIM, PHARP, and Tong’s reference panels.
We also evaluated the performance of the 1KCIGP panel on SNP array data, comparing it to four other reference panels: Animal-ImputeDB, SWIM, Tong’s reference panel, and PHARP18. We simulated three popular commercial SNP chips (50, 60, and 80 K; Supplementary Methods) using sites selected from the aforementioned public sequencing data. The average imputation concordance rates and the average squared correlations (r2) of the three simulated SNP chips using 1KCIGP were significantly higher than those of the other four panels in Chinese domestic pigs (Fig. 2C and Supplementary Fig. 11–13). We also compared these two indices for different chromosomes in the five panels. For example, the imputation concordance rate of the 50 K SNP chip with 1KCIGP ranged from 86.4% to 89.9%, whereas that of the other panels ranged from 78.3% to 85.2% (Fig. 2D). The squared correlation in different chromosomes in the 1KCIGP ranged from 0.396 to 0.543, whereas that of the other panels ranged from 0.160 to 0.426 (Fig. 2D). The 1KCIGP had a decisive advantage over the other panels for Chinese domestic pigs across all AF bins, showing notable improvements for variants with AF from 40% to 90% (Fig. 2E).
Population structure and genetic diversity in Chinese domestic pigs
Precise analysis of the population structure of Chinese domestic pigs with abundant breeds is critical for uncovering their population genetic diversity and characteristics in East Asia. Comparative analysis of Chinese indigenous pig populations and other Suidae populations worldwide might provide insights into the ancestral origins and relationships of different breeds. Therefore, we additionally collected 106 individuals in this study (Supplementary Data 4), including European domestic (EUD) and wild boars (EUW), wild boars from the Near East (NEW) and Island Southeast Asia, the common warthog (Phacochoerus africanus), and six ancient Sus scrofa individuals. We performed principal component analysis (PCA) on the 1011 individuals in the 1KCIGP and these samples to distinguish the genetic and geographic relationships between the Chinese domestic pig and other populations (Fig. 3A).
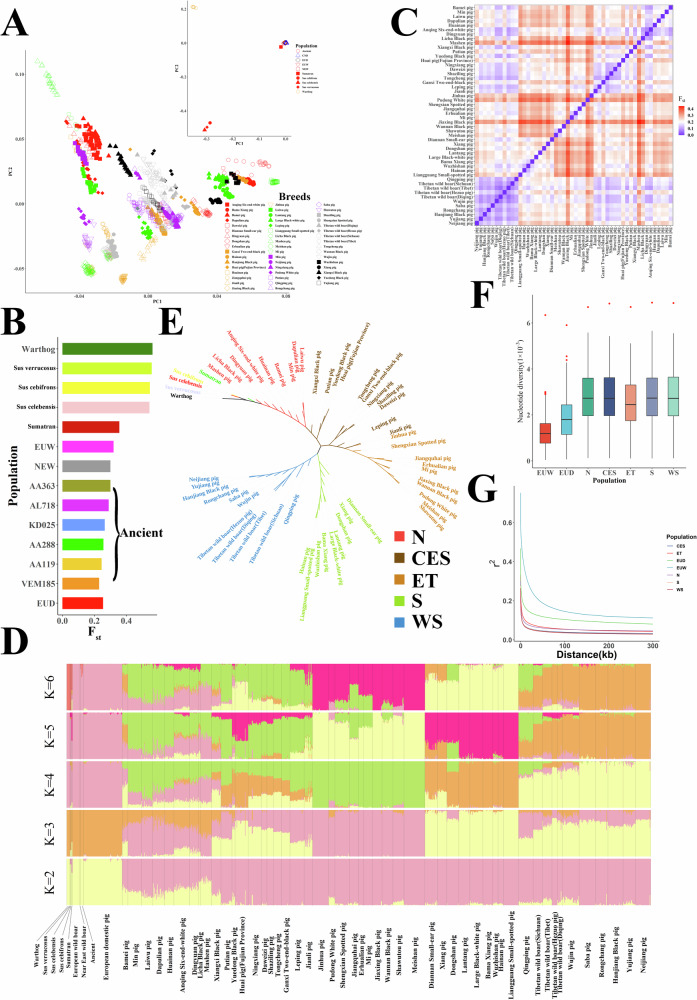
Fig. 3. Population structure analyses of Chinese domestic pig populations.
A PCA of Chinese domestic pig populations and other pig populations across Eurasia. CND: Chinese domestic pigs; EUD: European domestic pigs; EUW: European wild boars; NEW: wild boars from the Near East; Ancient: ancient pigs. B The Fst results between Chinese domestic pigs and other pig populations across Eurasia. C The Fst results among different Chinese domestic pig populations. D The genetic clustering analysis of pigs across Eurasian. E The phylogenetic tree of Chinese domestic pigs. F The nucleotide diversity for Chinese domestic pigs and European pigs. The nucleotide diversity was calculated for each population by a 1 Mb non-overlap window. The individuals in European pigs were divided into EUD (n = 74) and EUW (n = 13), while Chinese indigenous pigs were divided into N (n = 171), CES (n = 193), ET (n = 215), S (n = 179) and WS (n = 253). Boxplots show the median, 25th, and 75th percentile, the whiskers indicate the minima and maxima, and the points laying outside the whiskers of boxplots represent the outliers. G The LD decay for Chinese domestic pigs and European pigs.
The PCA results and pairwise Fst calculations (Fig. 3B) revealed significant differences between the Chinese domestic pig population and the common warthog, Visayan warty pig (Sus cebifrons), Celebes warty pig (Sus celebensis), Javan warty pig from Indonesia (Sus verrucosus). However, European wild and domestic pig populations, as well as the ancient individuals from Europe and the Near East, showed relatively close relationships with the Chinese domestic pig population (Fig. 3B). Within the Chinese domestic pig populations, a clear geographic distribution of the breeds from northern to southern China was identified by the first component. The third component was driven by the geographic distribution of these breeds from western to eastern China. In the genetic clustering analysis (Fig. 3D), when K = 5, four main ancestries were found in the Chinese domestic pig populations. The genetic clustering results showed that Chinese domestic pigs were divided into five subgroups, including breeds mainly distributed in southern (S), southwestern (WS), southeastern-central (CES), eastern China around the Taihu Lake Basin (ET), and northern China (N). The S, WS, and ET groups mainly showed one ancestry each; however, the N and CES groups exhibited at least two distinct ancestries. Interestingly, the N subgroup showed elevated European pig ancestry, whereas the other subgroups did not. A phylogenetic tree constructed using the neighbor-joining (NJ) method identified the same population affinities. Moreover, in the N, WS, and S subgroups, clear divisions were observed in the phylogenetic tree constructed using warthogs as an outgroup (Fig. 3E and Supplementary Fig. 14).
Analysis of the genetic diversity across Eurasian pigs indicated that nucleotide diversity was higher for Chinese domestic pigs (π = 2.43 × 10−3 to 2.73 × 10−3) than for European pigs (π = 1.68 × 10−3 to 1.76 × 10−3, Fig. 3F). Interestingly, unlike other species in which wild breeds have higher nucleotide diversity than domestic breeds, we noticed that the nucleotide diversity of European wild boars was substantially lower than that of European domestic pigs. Among the Chinese domestic pigs, nucleotide diversity was the lowest in the ET group, which reflected their status as a highly selected population when compared to other Chinese domestic pig populations. In addition, the half-life of linkage disequilibrium (LD) decay for Chinese domestic pigs was 332–615 nucleotides (nt), whereas European domestic commercial pigs exhibited an LD decay half-life of 882 nt (Fig. 3G). Consistent with the nucleotide diversity analysis, the ET population had a slower rate of LD decay than other Chinese domestic pig populations, suggesting a substantial decrease in genetic diversity during the domestication of this population.
Evidence of admixture between Chinese domestic pigs and other porcine populations
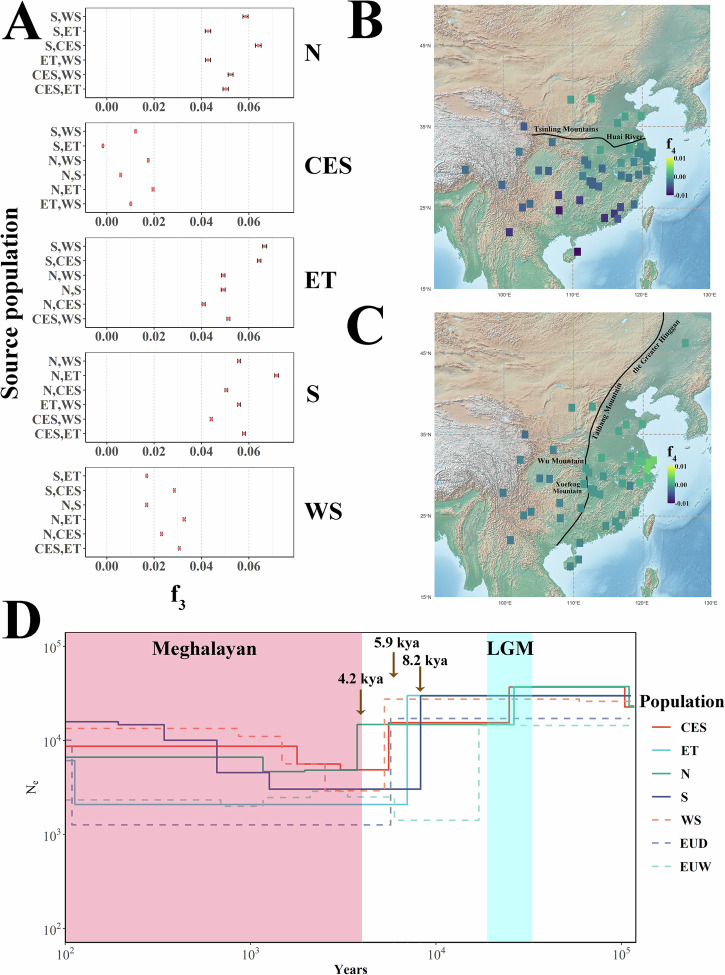
Previous studies have shown that Chinese and European commercial domestic pigs have exchanged genetic material12,15. To further analyze and quantify the admixture levels in Chinese domestic pigs, we examined patterns of allele sharing using a combination of analyses, including f3 statistic, f4 statistic, and Treemix. In the outgroup f3 analyses, European domestic pigs were found to share more genetic similarity with the northern group (f3 = 2.73) of Chinese domestic pigs than with the other four groups (f3 = 2.66, Fig. 4A). Conversely, the non-northern Chinese indigenous porcine population (f3 = 2.36–2.37) showed higher genetic similarity with southeastern Asian warty pigs than northern Chinese pigs (f3 = 2.28, Supplementary Fig. 15).
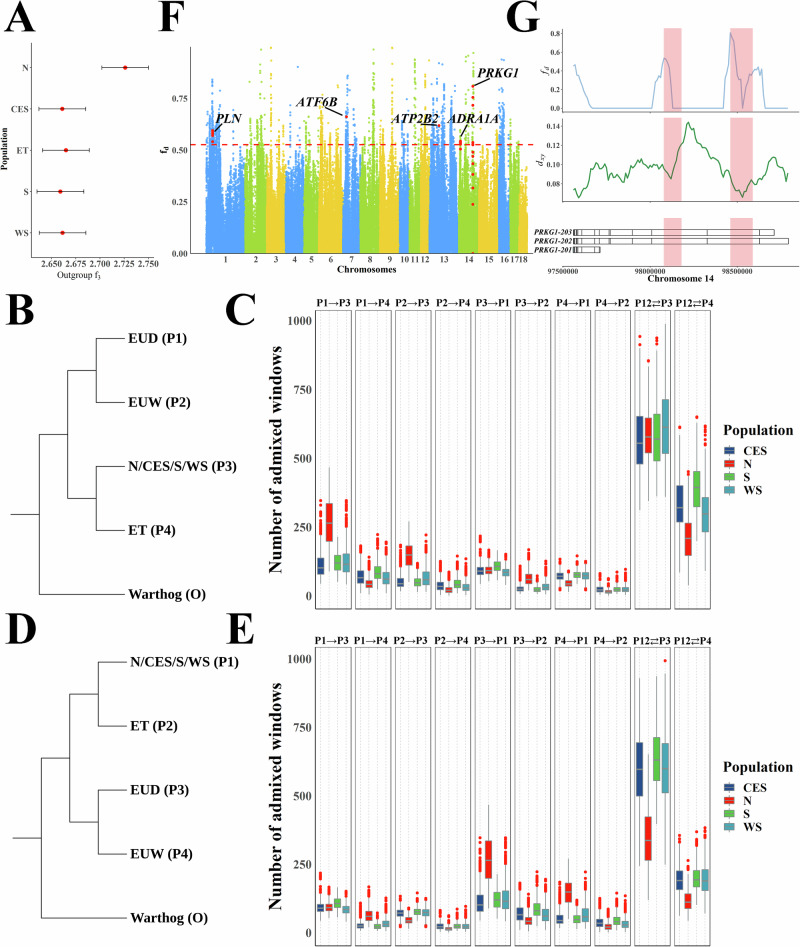
Fig. 4. Admixture between Chinese domestic pig and European pig.
A Outgroup f3 (European domestic pigs, X; Warthog) results for five distinct Chinese domestic pig sub-groups, where X represents the different sub-groups. The individuals of the Chinese domestic pig used in this analysis are the same as in Fig. 3F. The data points are presented as estimated f3 statistics ± s.e. The horizontal bars represent ± 1 s.e. B Schematic diagram of the topology used for the first configuration of the DFOIL analyses applied to population ET. C The DFOIL analyses results of the first configuration for population ET (n = 5 × 1 × 10 × 9 when P3 represents an individual from population N; n = 5 × 1 × 10 × 11 when P3 represents CES; n = 5 × 1 × 10 × 9 when P3 represents S; n = 5 × 1 × 10 × 11 when P3 represents WS). Boxplots show median, 25th and 75th percentile, the whiskers indicate the minima and maxima, and the points laying outside the whiskers of boxplots represent the outliers. D Schematic diagram of the topology used for the second configuration of the DFOIL analyses applied to population ET. E The DFOIL analyses results of the second configuration for population ET (n = 5 × 1 × 10 × 9 when P1 represents an individual from population N; n = 5 × 1 × 10 × 11 when P1 represents CES; n = 5 × 1 × 10 × 9 when P1 represents S; n = 5 × 1 × 10 × 11 when P1 represents WS). Boxplots show the median, 25th, and 75th percentile, the whiskers indicate the minima and maxima, and the points laying outside the whiskers of boxplots represent the outliers. F Degree of introgression from the ET population into European domestic pigs. Five important genes affected by the top introgression signals are shown. The dashed horizontal line indicates the top 1% fd. G Discrete introgression regions in the PRKG1 gene. The top plot shows the fd from ET to European domestic pigs. The middle plot shows the pairwise nucleotide differences per site (dxy) between ET and European domestic pigs. The bottom plot shows the gene structure.
We directly compared distinct Chinese domestic pig populations with European domestic pigs using the following tree topology (Warthog, European domestic pigs; Chinese domestic pig population A, Chinese domestic pig population B). The results indicated that potential introgressions occurred between the European domestic pigs and Chinese domestic pig populations N and ET, respectively (Supplementary Data 5). To further distinguish the direction of the introgression, we ran DFOIL in two configurations for ET and N populations (Fig. 4B–E and Supplementary Fig. 16). First, we ran DFOIL with European domestic and wild boars at positions P1 and P2, the Chinese domestic pig population N or ET at position P4, and other populations at position P3, with the warthog as the outgroup (O) (Fig. 4B, C and Supplementary Fig. 16A, B). Second, we ran DFOIL in a configuration in which we swapped the positions of the European and Chinese pigs in the phylogeny (Fig. 4D, E and Supplementary Fig. 16C, D).
In the first configuration, DFOIL detected admixture in 12.15% and 12.33% of the chromosome windows for populations ET and N, respectively (mean ± s.d. = 1453 ± 173 and 1473 ± 176 windows, respectively). For the N population, most of these windows showed gene flow between population N and European pig ancestors (520 ± 123, Supplementary Fig. 16B). More importantly, the number of windows that showed gene flow from European domestic pigs to the N population was higher than the number of reverse gene flow windows (237 ± 90 vs. 103 ± 24). Intriguingly, the introgression signals between population ET and European pigs were low (310 ± 118, Fig. 4C). We then examined introgression signals in the second configuration for the N and ET populations. In concordance with the first configuration, the number of introgression windows from European domestic pigs to the N population was still higher than that of the reverse gene flow window (237 ± 90 vs. 103 ± 24, Supplementary Fig. 16D). These results indicate that admixtures with European pigs were more common in North Chinese domestic pigs than in other Chinese domestic pig populations. This signal was confirmed by Treemix, with multiple migration events (migration events 1–8, Supplementary Fig. 17). Compared to configuration 1, the results of configuration 2 for the ET population showed that the number of windows supporting introgression between Chinese and European domestic pigs (551 ± 164) exceeded the number of windows that showed gene flow between the ET population and European pigs (Fig. 4E). Moreover, Treemix analyses with seven and eight migration events revealed migration edges from the ET to the European domestic pig population (Supplementary Fig. 17), supporting this admixture.
We also examined the introgression regions between the European domestic pig population and Chinese domestic pig populations N and ET. The proportions of genome introgression (PGI) in populations N and ET were 13.13% and 10.73%, respectively. We then separately detected the strongest introgression signals in the ET and N populations and found that, particularly for population ET, the 807 strongest introgression regions (using the top 1% fd as the cutoff and eliminating the same region in population N) contained 225 genes (Fig. 4F) that were enriched in three Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Supplementary Data 6). Among these three pathways, the cGMP-PKG signaling pathway, which plays a critical role in activating pig oocytes32 and participates in the replication of viruses33,34, included five important genes (Fig. 4F). One of these genes was PRKG1, in which discrete introgression signals were detected (Fig. 4G). These signals were supported by pairwise nucleotide differences per site (dxy), indicating that complex recombination occurred in this gene region.
Gene flow and demographic history of Chinese Indigenous pigs
To further dissect the relationships among Chinese domestic pig populations, we used f3, f4 statistics and Treemix to evaluate the admixture among Chinese indigenous pig breeds. We ran f3 statistics based on all 30 combinations of the five Chinese domestic pig populations. Our test using population CES as the target with populations ET and S as the sources produced a Z-score of − 6.587 (Fig. 5A), which was highly significant, indicating admixture events in population CES. The f4 tests were conducted using warthogs as an outgroup to identify introgression among the five Chinese domestic pig populations. For each population, we retained only those with the highest probability of introgression events with the other populations. The f4 statistic tests also detected strong introgression signals between populations CES and ET, as well as between populations CES and S (Supplementary Data 7). We further inferred the date of this admixture and detected an ancient admixture signal (160.48 ± 22.29 generations) in the CES population. Interestingly, the ET population also showed explicit gene flow between populations N and WS. The Treemix analysis allowed for multiple migration events and identified three migration events among Chinese domestic pigs, including populations ET to N, WS to ET, and S to CES, which were steadily detected when one to eight migration events were allowed (Supplementary Fig. 18). The ET population, in particular, showed gene flow with three of the remaining four Chinese domestic pig populations, indicating its unique position in the long-term domestication of Chinese indigenous pigs. We also examined the genetic relationships among Chinese domestic pigs across space. We used the f4 statistic to set up two symmetry tests to compare the sampled breeds against the northernmost (Min pig) and southernmost (Wuzhishan pig) breeds, as well as against the easternmost (Pudong White pig) and westernmost (Tibetan wild boar) breeds (Fig. 5B, C). The east-west comparison showed a clear discriminative line corresponding to the geographic division line between the third and second steps in China (the Greater Hinggan, Taihang, Wushan, and Xuefeng mountains). For the north-south comparison, most Chinese domestic pigs shared a closer relationship with the Wuzhishan pigs, especially breeds that spread into central China.
Fig. 5. The admixture and demographic history of Chinese domestic pig.
A The f3 results for five distinct Chinese domestic pig sub-groups. The left and right y-axes represent source and target populations, respectively. The individuals of the Chinese domestic pig used in this analysis are the same as in Fig. 3F. The data points are presented as estimated f3 statistics ± s.e. The horizontal bars represent ± 1 s.e. B A heatmap showing f4(Warthog, X; Min pig, Wuzhishan pig), where X represents the different Chinese domestic pig populations. The map in this figure and the subsequent figure is imported from the public 42 domain Natural Earth project (https://www.naturalearthdata.com). C A heatmap showing f4(Warthog, X; Tibetan wild boar, Pudong White pig), where X represents the different Chinese domestic pig populations. D History of the effective population sizes of different sub-groups of Chinese domestic pigs.
The dynamics of effective population size (Ne) offered insight into the repercussions of past environmental factors and human-driven domestication events on the demographic history of different species35. To investigate the geographic history of Chinese domestic pigs, the SMC + + 36 approach with a generation time of g = 5 and a mutation rate per generation of μg = 2.5 × 10−8 was used to infer population size histories (Fig. 5D). We found that only the subgroups N and CES, in which Ne occurred twice, showed a dramatic decrease. The first decline for both groups occurred during the Last Glacial Maximum (LGM) period (33–19 kilo years ago, KYA), which profoundly affected the climate and geography of the Earth37. After this period, the Ne of group S experienced a drastic decline at 8.2 KYA when abrupt climate change occurred38. Interestingly, during the prosperity period of the Majiabang culture (around 7 KYA throughout the Taihu Lake Basin)39, we observed that the Ne of the ET group underwent severe degeneration. Moreover, the first period of Ne degeneration for group WS and the second period for group CES fit well within the 5.9 KYA event, possibly one of the most devastating climate events in the Holocene Epoch. The Ne of all Chinese domestic pig populations recovered to a plateau that was initiated approximately 4000 years ago when the Earth entered the Meghalayan age (4.2 KYA). However, only group N experienced a significant decrease in Ne at approximately 4.2 KYA, during a severe climatic event. During the same period, the famous Longshan culture of northern China faded40.
Maternal and paternal haplogroups found in Chinese domestic pigs
In recent years, studies on mitochondrial DNA (mtDNA) and the Y chromosome have provided useful insight into the migration of the global porcine population, including tracing its origin and its expansion into Europe and Asia41–43. In this study, we inferred maternal lineages using 704 assembled complete mitogenomes (Supplementary Data 1). A previous study divided the mtDNA haplogroups of pigs and wild boars into the A, D, and E haplogroups44. European pigs were haplogroup E, whereas Asian wild boars were haplogroup A. As expected, all the individuals in the 1KCIGP showed haplogroup D, and the D1a1 group, which is the dominant group in the Chinese domestic pigs, accounted for 38.66% of the mtDNA haplogroups. The second most common group was D2 (24.46%). Moreover, mtDNA maps of the Chinese domestic pigs described in our study showed that, except for populations S and WS, the D1a1 haplogroup was the dominant haplotype (> 50%) (Supplementary Fig. 19). Interestingly, within the breeds domesticated in southwestern China, D2 was the most common haplogroup (40.86%; Supplementary Fig. 20A).
We also inferred paternal lineages using the SNPs on the Y chromosome. After quality control, annotation, and filtering, 34,207 high-confidence SNPs were detected in the non-pseudo-autosomal region of the Y chromosome (NPARY) of 392 boars (325 males from the 1KCIGP dataset and 67 males from the aforementioned 106 public individuals used for population structure analyses). We found a large region ranging from 8.94 to 43.55 Mb on Sscrofa11.1, which segregated three distinct haplogroups across Eurasian pigs (Supplementary Fig. 20B). A median-joining haplotype network (Supplementary Fig. 20C) also supported this result, indicating that haplogroup Y1 was specific to Javan warty pigs from Indonesia (Sus verrucosus). European domestic pigs had predominantly Y2 haplogroups, while the Y3 haplogroup was mainly shared among Chinese domestic pigs. Interestingly, when one European sample that belonged to the Y3 haplogroup was excluded, all European pigs belonged to the Y2 haplogroup; many Chinese domestic pigs also showed higher proportions of the Y2 haplogroup, especially in the northern pig populations (Supplementary Fig. 20C). We observed a clade of the Y3 haplogroup, Y3a, which predominantly comprised domesticated pig populations from the Taihu Lake Basin. A previous study also reported that some Chinese pigs contain the European haplotype on the Y chromosome45. Therefore, we calculated the range of male contributions to the European admixture component46 (“Methods”, Supplementary Methods) in 21 Chinese domestic pig populations with the Y2 haplogroup. The results showed that in 15 populations, the observed Y2 haplogroup frequency was higher than expected, even though all European contributions were from males in a binomial test (Supplementary Data 8). This showed that the high Y2 haplogroup frequency in these breeds was not only due to sex-biased hybridization but also to other factors that could induce the frequency of these breeds.
We next examined the SNP distribution in each gene of the NPARY region in detail. Most genes showed haplotypes that were identical to the ~ 34 Mb NPARY region. However, two genes, LOC100625207 and LOC102159347, exhibited distinct haplogroups (Supplementary Fig. 20D, E). Interestingly, the haplogroups of these two genes indicated that at least one haplotype recombination event occurred in each gene. The compound haplotypes appeared to have a high frequency in populations residing around the Taihu Lake Basin, which may correlate with the characteristics of these breeds.
Signatures of positive selection in Chinese Indigenous pigs
Previous studies have reported that some Chinese domestic pigs possess characteristics such as high-altitude adaptation13, distinctive body sizes24, and different ear sizes47. Here, to detect genomic fragments that were significantly differentiated between Chinese indigenous breeds, we selected three typical phenotypes with extreme variance among the different breeds for further analysis. These included adaptation to high altitude, large ear size, and small body size. Using locus-specific branch length (LSBL) and the genomic diversity θπ method, we detected the selection signatures related to these phenotypic characteristics.
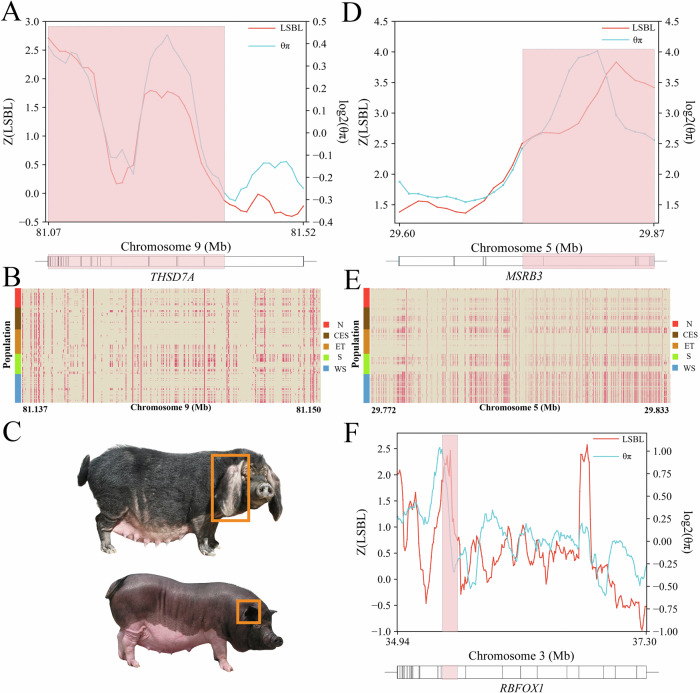
For the high-altitude adaptation trait, we selected breeds from regions with altitudes > 3000 m (Tibetan wild boars) as the high-altitude population, and breeds from regions with altitudes < 100 m (Erhualian, Jiangquhai, Jiaxing Black, Meishan, Mi, Shawutou and Pudong White pigs) as the low-altitude population (Supplementary Data 9). There were 3352 overlapping selection regions for high-altitude adaptation traits (Supplementary Data 10). In these regions, we identified potential functional genes that were under selection, and that may be associated with high-altitude adaptation. Two functionally important genes, THSD7A (Fig. 6A) and HIF1A, participate in the oxidation-reduction process and the mitochondrial respiratory chain pathway. We investigated the genomic patterns in the selection sweeps of these genes across Eurasian pigs and found a 13 kb region of chromosome 9:81.137–81.150 Mb in THSD7A that contained different haplotype patterns across the 1KCIGP individuals (Fig. 6B).
Fig. 6. Selection signatures and haplotype patterns of the high-altitude adaptation, small body size, and large ear size-related genes.
A The top diagram represents the selection signatures detected using the LSBL and θπ methods in the high-altitude adaptation-related gene, THSD7A. The bottom diagram shows the gene structure of the THSD7A gene. The red rectangle indicates the identified region under selection. B Haplotype patterns in regions under selection for THSD7A. Each row represents a phased haplotype, and each column represents a polymorphic SNP variant. The reference and alternative alleles are indicated in cream and red colors, respectively. C Phenotypic variations for ear size, represented by Dapulian (top) and Lantang (bottom) pigs. Images were sourced from the Animal Genetic Resources in China: pigs. D The top diagram represents the selection signatures detected using the LSBL and θπ methods in the large ear size-related gene, MSRB3. The bottom diagram represents the gene structure of MSRB3. The red rectangle represents the identified region under selection. E Haplotype patterns in regions in MSRB3 that are under selection. Each row represents a phased haplotype, and each column represents a polymorphic SNP variant. The reference and alternative alleles are indicated in cream and red colors, respectively. F The top diagram represents the selection signatures detected by LSBL and θπ methods in the small body size-related gene, RBFOX1. The bottom diagram shows the gene structure of RBFOX1. The red rectangle indicates the region under selection.
For the large ear size trait, breeds with large and floppy ears (Dapulian, Erhualian, and Laiwu pigs) were selected as the large ear size population, while breeds with small and erect ears (Lantang, Yuedong Black, and Huai pigs) were selected as the small ear size population (Fig. 6C). There were 4207 overlapping selection regions for different ear size phenotypes (Supplementary Data 11). The MSRB3 (Fig. 6D) and WIF1 genes that are correlated with ear size were identified. We also investigated the genomic patterns of MSRB3 across Eurasian pigs. Specifically, in MSRB3, we identified a 61 kb region of chromosome 5:29.772–29.833 Mb that contained different haplotype patterns across 1KCIGP individuals (Fig. 6E).
Breeds with adult weights < 60 kg (Wuzhishan, Diannan Small-ear, and Bama Xiang pigs) were selected as the small body size population, while breeds with adult pig weights > 160 kg (Min, Neijiang, and Mashen pigs) were selected as the large body size population. A total of 2848 overlapping selection regions were associated with distinct body sizes (Supplementary Data 12), in which the RBFOX1 gene (Fig. 6F) was identified and may have contributed to the growth and development of pigs.
Identification of loci related to biological traits
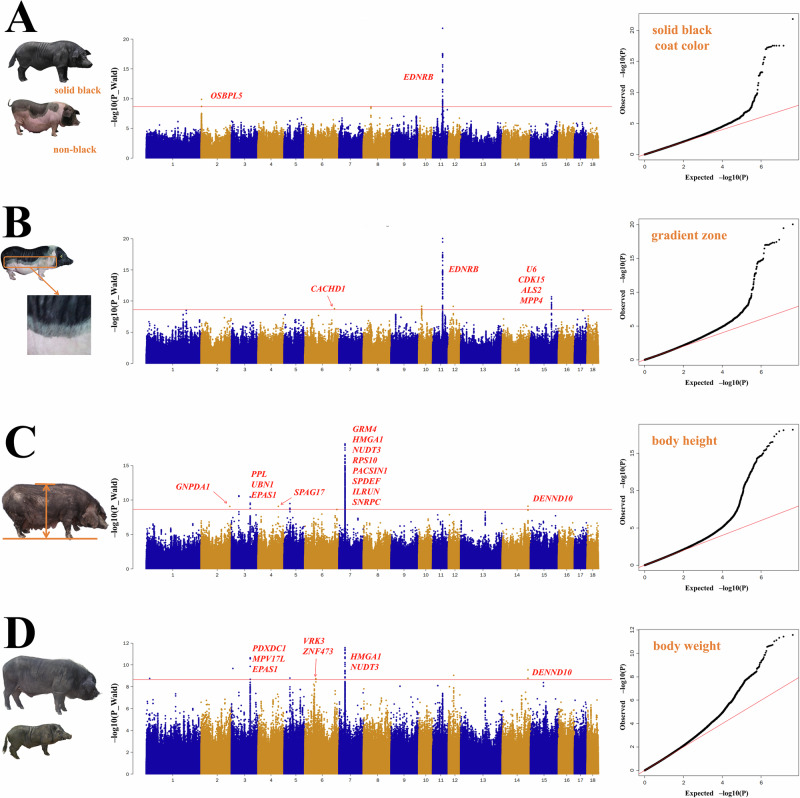
Chinese domestic pigs exhibit diverse characteristics in many biological traits. Traditional breed recognition of distinct Chinese domestic breeds is based on specific biological traits. Using resequencing data, we conducted GWAS across 50 populations and focused on coat color and body size as biological traits (Supplementary Data 13). For the coat color traits, we concentrated on solid black and a gradient zone. The latter describes a coat color phenotype that appears as a strip of light gray separating black and white areas. We identified 96 significant loci associated with the two phenotypes (Supplementary Data 14). Notably, the GWAS results for the solid black and gradient zone phenotypes both highlighted a genomic region on chromosome 11 spanning 50.01 to 50.15 Mb. We found that 42.7% of the total identified significant SNPs were located in this region, and the entire EDNRB gene was contained within this region (Fig. 7A). Previous studies have reported that EDNRB is strongly correlated with coat color, especially the non-black coat color of most Chinese indigenous pigs48,49. Our GWAS also identified candidate genes associated with the gradient zone phenotype, including the CDK15 and ALS2 genes (Fig. 7B).
Fig. 7. GWAS results for the biological traits of Chinese domestic pigs.
A GWAS results for coat color in solid black and non-black pigs. B GWAS results for gradient zone coat color. C GWAS results for body height. D GWAS results for body weight. All left-hand figures represent phenotypes (The images were sourced from the Animal Genetic Resources in China: pigs). All statistical tests were two-sided. In the Manhattan plots, the x-axis represents the chromosomal position, and the y-axis displays the −log10(P-value). The red line indicates the Bonferroni-corrected significance threshold at P = 2.29 × 10−9. In the quantile-quantile (QQ) plots, the x-axis shows the expected −log10(P-value), while the y-axis represents the observed −log10(P-value).
When examining body size traits, we primarily focused on body height, length, weight, and chest girth. We identified 191 significant SNPs associated with body size, most (50.8%) resided in chromosome 7 (Supplementary Data 14). In particular, a 0.47 Mb region on chromosome 7 was significantly associated with both body height (30.25–30.72 Mb) and weight (30.33–30.41 Mb) phenotypes, which contained NUDT3 and HMGA1 genes (Fig. 7C, D). More importantly, after validation by Sanger sequencing, a novel SNP (NC_010449.5:g.30378105 T > C) was identified within NUDT3 and was significantly associated with all body size phenotypes. This locus may be an important candidate marker for body size studies.
Structural variations across the 1011 1KCIGP samples
SVs may play a critical role in environmental adaptation and breed characteristics. Therefore, we generated an SV call set across all 1011 1KCIGP samples using short-read WGS data. To improve the sensitivity and accuracy of SV detection, we used a bioinformatics pipeline that combined four diverse SV-finding algorithms: Delly2, Lumpy, Manta, and Wham50–53. We successfully identified and genotyped 168,167 SVs, including 94,533 deletions, 2758 insertions, 25,031 duplications, 17,140 inversions, and 28,705 translocations. The total number of SVs from 730 randomly selected samples was 166,470, accounting for 99% of the total number of SVs in all 1011 samples. Modeling the SV count by iterative random sampling of individuals revealed that the SV number was relatively finite in Chinese domestic pig populations (Supplementary Fig. 21A), indicating that our SV detection was comprehensive and nearly complete. On average, 30,429 SVs were discovered in each genome, and the distribution of SVs observed per individual showed that the pig populations spread across North China contained a small number of SVs per genome (Supplementary Fig. 21B). Among all the detected SVs, 63,597 (37.82%) SVs had MAF < 0.01 (Supplementary Fig. 22). Most deletions, insertions, and duplications ranged in size from 100 bp to 10 kb, whereas many inversions were > 10 kb in size (Supplementary Fig. 23). To further evaluate the precision of our SV callset, the PacBio reads of ten breeds were downloaded from the public database (Supplementary Data 15). Then, 150 SVs were randomly selected and compared with those detected using the long reads, revealing that 70% of SVs could be validated with these reads (Supplementary Data 16).
Impact of pig SVs on the functional genome
SVs are recognized for their profound impact on genomes and are frequently associated with specific traits. We systematically investigated the association between detected SVs and their genomic features. We first annotated these SVs against features in the Ensembl annotation of Sscrofa11.1, which showed that most SVs were in intergenic and intronic regions (Supplementary Fig. 24), with 15.33%, 7.98%, and 1.78% within 5 kb of a protein-coding gene, long non-coding RNA gene, or pseudogene, respectively. Approximately half (50.08%) of the SVs overlapped one or more Ensembl genes, the majority (82.67%) of which overlapped a single gene, with 29,669 genes overlapping. Approximately 6.66%, 0.22%, 11.40%, and 43.62% of deletions, insertions, duplications, and inversions, respectively, were predicted to have a high impact on genes using SnpEff54 (Supplementary Fig. 25). More importantly, we examined the potential effects of Chinese domestic pig SVs on genome function by aligning seven distinct annotating genome features, including genes, exons, introns, coding sequences (CDSs), promoters, untranslated regions (UTRs), and enhancers. Enrichment analysis of these genome features revealed that most SVs were depleted in genic regions; however, the insertions were enriched in the exonic regions and CDSs (Supplementary Fig. 26). In particular, promoter regions with all SV types showed significant enrichment (t test; P < 0.01) when compared with the random background model.
To discover SVs with candidate functions, 897 tag-SNPs from a previous GWAS atlas55 were obtained from 1011 genomes and used to search for SVs that had strong LD with the tag-SNPs. A total of 95 SVs with high LD (LD > 0.8) overlapped with 36 different genes (Supplementary Data 17). We observed only one deletion located in the 3′UTR region. This deletion had a strong LD (0.94) with a tag-SNP (rs323143577) related to the subcutaneous adipose tissue thickness trait located in the intergenic region. This deletion was ~ 7 kb away from the tag-SNP and overlapped with the 3′UTR of the CASP10 gene (Supplementary Fig. 27A and Supplementary Data 20). The CASP10 gene encodes Caspase-10 and is involved in the apoptotic cell death pathway. This gene is reportedly associated with cholesterol genes and human body mass index phenotype56; therefore, it could be considered a candidate maker in pig development.
Tibetan wild boars native to the Qinghai-Tibet Plateau have adapted to extreme conditions, such as high altitude and strong ultraviolet radiation57. To detect the genomic variants contributing to this environmental adaptation, we identified SVs with large frequency divergences (> 20%) between Tibetan wild boars and other Chinese domestic pig populations, referred to as the Tibetan wild boar-specific SVs (TWBSVs; Supplementary Data 18). In total, 277 TWBSVs that involve 104 genes were discovered, including 264 deletions, five duplications, seven inversions, and one translocation (Supplementary Data 19). We focused on SVs with Tibetan wild boar-specific enrichment and discovered two deletions in the promoters of two genes. A 427 bp deletion in the promoter of the NAV2 gene was enriched in Tibetan wild boars (44.64%) but was relatively rare (11.05%) in other Chinese domestic pig populations (Supplementary Fig. 27B, Supplementary Data 20). This gene was previously reported to play a role in cellular growth and migration, especially in maintaining normal heart function58,59. Another identified 398 bp deletion (Supplementary Fig. 27C and Supplementary Data 20) in the promoter of the POLD3 gene was also enriched in Tibetan wild boars (66.96%) when compared to other indigenous pig populations (30.94%). This gene is associated with DNA repair and is involved in UV irradiation-induced DNA damage response60,61, which may contribute to high-altitude adaptation.
A web-based tool that provides easy access to Chinese Indigenous pig genomic resources
We developed a web-based tool to facilitate ready-access to our Chinese indigenous pig genomic resource (https://1kcigp.com/). This web tool contains a comprehensive catalog of SNPs and indels in Chinese domestic pigs. Summary information for these variants, including the position, reference, and mutated alleles and their frequencies, can be accessed by our 1KCIGP web tool. The 1KCIGP haplotype reference panel and corresponding imputation server are also available on this web tool.
Discussion
Chinese domestic pigs comprise 83 breeds, contributing one-third of Asian local pig breeds, and exhibit highly diverse phenotypes. Constructing a genomic resource for Chinese domestic pig populations will contribute to the genetic diversity of pigs worldwide and facilitate studies on porcine evolution, selection, and breeding. Here, we introduce the 1KCIGP database and provide the largest genomic variation map to date for Chinese domestic pig populations. The 1KCIGP genomic resource offers a valuable reference panel for Chinese domestic pig populations, even the Eurasian pig populations. Using systems genomics analyses, we discovered the divergence, introgression, and specific haplotypes of Chinese domestic pigs and identified genomic regions under significant positive selection in specific Chinese domestic pig breeds.
One significant contribution of this study is that the 1KCIGP resources filled gaps in large-scale WGS-based haplotype reference panels for Chinese domestic pigs. Previous imputation panels17–20, primarily based on European domestic pigs or varied sequencing data collected for Chinese indigenous pigs, exhibited inferior performance in imputing genetic data for Chinese domestic pigs, owing to their divergent genetic backgrounds. Moreover, numerous unique genetic variations were specific to the Chinese domestic pig (Fig. 1B), particularly in rare variants, making imputation problematic for using European domestic pig data. In this study, we systematically evaluate the imputation performance of the 1KCIGP panel across various test scenarios. The results demonstrate that the imputation performance of 1KCIGP exceeded those of Animal-ImputeDB, SWIM, PHARP, and Tong’s public reference panels for most Chinese domestic pig breeds. This superior imputation performance was consistent across almost all AF bins with 1KCIGP. As expected, 1KCIGP did not perform as well as SWIM, PHARP, and Tong’s public reference panels for European pig breeds. However, 1KCIGP has an advantage in certain crossbreds with Chinese domestic pig ancestry. We also noticed that imputation using low-density chips showed a slight decrease in accuracy (60 K vs. 80 K). This may be attributed to fewer SNPs and the erroneous genotyping of heterozygotes. Overall, we provide practically important resources for the imputation of Chinese domestic pigs.
We also noticed that the SNP: indel ratio (1.59) for novel variants in the 1KCIGP dataset is smaller than that for all variants (3.93). This trend is similar to the previous studies, such as SWIM (1.30 vs. 7.39), Tong’s study (3.20 vs. 9.14), IAnimal62 (3.25 vs. 6.30), and Yang’s study63 (2.22 vs. 5.43). This indicated that the public genomic variants dataset of pigs imperatively needs to improve, especially for indels. Simultaneously, the proportion of indels detected in the difficult-to-sequence regions for 1KCIGP (47.14%) is closer to that of the previously reported human study28 (64.3%) than the other studies (SWIM: 24.81%; Tong’s study: 18.28%; IAnimal: 36.31%; Yang’s study: 33.34%). The proportions of SNP detected in difficult-to-sequence regions for IAnimal (13.67%) and Yang’s study (13.96%) are similar to the 1KCIGP (13.86%) and close to the previously reported human study28 (23.1%). These results indicated that the genomics variants in the difficult-to-sequence regions may need higher sequencing depth to disclose, especially for indels.
In this study, we systematically dissected the genetic relationships among Chinese domestic pigs, and between them and European pigs. By combining geographic distribution and genetic relationships among these populations, we divided Chinese domestic pigs into five sub-groups. Introgression analyses demonstrated that the breeds in the Taihu Lake Basin (population ET) showed significant introgression signals in European domestic pigs. This corresponded with historical records, indicating that Asian pigs were introduced to Europe during the 18th and early 19th centuries64,65. This introgression may have also induced higher nucleotide diversity in European domestic pigs than that observed in European wild boars66. We also detected gene flow from the ancestry of European domestic pigs to pigs in northern China (population N), with genetic clustering analysis confirming European ancestry in population N. This finding was concordant with those of previous studies; however, the introgression event times and periods were not confirmed45,67. As an important Chinese domestic pig population, ET showed introgression with three of the four other populations, including the population mainly distributed in southwestern China (WS). The introgression between populations ET and WS may be due to the Yangtze River, which is an important transportation hub that connects eastern and western China. More importantly, in the f3 analysis, the populations in southeastern-central China (population CES) showed significant evidence of admixture, with the population distributed in south China (population S) and population ET as the source populations. This admixture event was estimated to have occurred about 160.48 ± 22.29 generations (802.4 ± 111.45 years) ago and corresponds with large-scale human migration trends in China’s Southern Song Dynasty during the 12th to 13th centuries68. This migration began in the Yangtze River valley and moved southward across the Nanling ranges into the Xi River basin69. It may have stimulated the admixture of ET and S, contributing to the formation of the CES population.
Chinese domestic pigs show extraordinary phenotypic characteristics such as high-altitude adaptation, and their abundant breed diversity provides plentiful phenotypic variance. In this study, to understand the contribution of long-term selection to the distinct phenotypes of Chinese domestic pigs, two positive selective signature detection methods were used to identify genomic regions under selection and the potential candidate genes in these regions. We identified several genes that are potentially under selection and functionally associated with the traits observed in Chinese indigenous pigs. These included THSD7A and HIF1A, which are implicated in high-altitude adaptation; RBFOX1, which is associated with small body size; and MSRB3 and WIF1, which may influence ear sizes. A previous study reported that the HIF1A gene encodes the transcription factor HIF-1, which functions as a master regulator of cellular and systemic homeostatic responses to hypoxia and has an important impact on high-altitude adaptation70. In addition, a high-frequency missense mutation in THSD7A was reported in Tibetan wild boars, indicating the important role of regulatory mutations in their evolution57. Many studies have demonstrated that RBFOX1 is associated with skeletal muscle structure in multiple species, including mice, zebrafish, and humans71–74. This is consistent with our finding that RBFOX1 is related to small body size in pigs. Additionally, two important functional genes correlated with large ear size, MSRB3, and WIF1, were identified in this study, which is consistent with previous reports that these genes represent biological candidates for porcine ear size, with potential applications in breeding programs75–77. Overall, our study detected the footprints of selection in Chinese domestic pigs and mined the important functional genes under selection based on the specific traits of Chinese domestic pigs.
We used GWAS to identify candidate loci that contribute to the biological traits of Chinese domestic pigs. By annotating these loci, we identified the EDNRB gene, which has been reported as a candidate gene for the non-black coat color of Chinese domestic pigs, including the two-end-black, six-white-point, spotted, and white phenotypes48,49,78,79. Although the famous pig coat color-related genes, such as MC1R and KIT, were not identified in our study, previous studies reported that the coding region of MC1R showed low diversity in Chinese domestic pigs80. Only the normal copy number of KIT has currently been identified in Asian pigs, whereas European pigs harbor multiple KIT genotypes that are associated with different coat colors81. Overall, it is plausible that MC1R and KIT are not the primary causative genes for coat color differentiation in Chinese indigenous pigs. In fact, EDNRB might be more likely to contribute to coat color differentiation in Chinese indigenous pigs. As for body size traits, we reported that NUDT3 and HMGA1 on chromosome 7 were associated with all the analyzed body size traits. NUDT3 is an mRNA-decapping enzyme that orchestrates mRNA expression to modulate cell migration82, and its variants have been reported to be associated with the body mass index in humans83. HMGA1 can bind to DNA and modify the chromatin state, affecting the accessibility of regulatory factors to DNA and contributing to overall gene expression tuning84. A previous knock-out study indicated that HMGA1 could decrease body size in mice, leading to a “super pygmy” phenotype85. These findings suggest that NUDT3 and HMGA1 could significantly influence the body size traits of Chinese indigenous pigs.
Collectively, this large and high-quality database of Chinese domestic pig populations will be helpful in examining the effect of known variants on disease and economic traits. The characterized large-scale genomic variations revealed the phylogenetic relationships among Chinese domestic pig populations, signatures of recent positive selection in these populations, and introgression events within or between pigs from other continents. This study offers valuable insights into pig genomic diversity that can inform functional interpretation and facilitate future studies on pig breeding.
Methods
Ethics statement
This study was conducted in strict accordance with the protocol approved by the Institutional Animal Care and Use Committee (IACUC) of China Agricultural University (Beijing, People’s Republic of China; Approval No. AW60604202-1-1). All blood or ear tissue samples were collected from live pigs, without the need for slaughter.
Samples collection and DNA extraction
In this study, 50 Chinese domestic pig populations (Supplementary Data 1), spread approximately two-thirds of China’s administrative divisions, were used to detect genomic variants, construct a population-specific reference panel, and analyze the swine genomic architecture. In total, 1011 pigs from these populations were selected, with all individuals sampled during the wean-to-finish development period. Three methods determined the sexes of these individuals: the method from GATK, calculating the coverage of the sex chromosomes and assessing the aligned coverage of the porcine SRY gene. The blood or ear tissues from the 1011 samples were used for WGS. We extracted DNA from blood or ear tissues using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China).
Sequencing reads mapping, and small variants calling
Sequencing libraries were prepared using the MGISEQ protocol, and sequencing was performed on the MGISEQ 2000 platform (MGI, Shenzhen, China) with 150-bp paired-end reads. Sequence coverage varied from 11.45 × to 33.85 ×, with a mean coverage of approximately 26 ×. Raw reads were filtered and trimmed using TrimGalore (v0.6.1)86. The cleaned reads from all individuals were aligned to Sscrofa11.1, using the Burrows-Wheeler Aligner (BWA v0.7.17-r1188)87. GATK (v4.0.12.0)88 and Samtools (v1.9)89 were used to remove duplicated reads and sort the alignment results. Small variants, including SNPs and indels, were detected and filtered using GATK software. The following criteria were applied for all SNPs: (1) mean sequencing depth for all individuals > 10 × and < 78 ×; (2) variant confidence/unfiltered depth of non-reference samples (QD) > 2.0; (3) RMS mapping quality (MQ) > 40.0; (4) Phred-scaled p-values calculated using Fisher’s exact test were used to detect strand bias in the reads (FS) < 60.0; (5) strand bias was estimated by the symmetric odds ratio test (SOR) < 3.0; (6) the μ-based Z-approximation from the Mann-Whitney rank sum test was used for mapping qualities (MQRankSum) > − 12.5; (7) the μ-based Z-approximation from the Mann-Whitney rank sum test was used to calculate the distance from the end of the read for reads with the alternate allele (ReadPosRankSum) > − 8.0; and (8) no more than three SNPs were clustered in a 10-bp window. For indels, these criteria were used: QD > 2.0, FS < 200.0, SOR < 10.0, MQRankSum > − 12.5, ReadPosRankSum > − 8.0.
Reference panel construction and imputation performance evaluation
SNPs in Chinese domestic pigs were filtered before they were used to construct the reference panel. Sites with missing call rates > 10% and those with a minor allele count of < 2 (MAC2) were removed. The samples with call rates < 10% were excluded from the cohort. Only SNPs on chromosomes 1–18 and X were used to construct the reference panel. Whatshap (v0.18)90 was used to phase the filtered SNPs using the sequencing reads from each sample. The local phase sets were then incorporated into the population-based phasing of 1011 samples using SHAPEIT4 (v4.2.2)91 with the parameter ‘--use-PS 0.0001’. Chromosome X was divided into pseudo-autosomal regions (PARs) and non-PAR regions, and phased.
To evaluate the imputation performance of the 1KCIGP panel, we conducted two assessments under two common scenarios: imputing genotyped variants detected using low-coverage WGS and SNP chips (Supplementary Methods). The 262 test individuals (Supplementary Data 3) that were not included in the five panels (1KCIGP, Animal-ImputeDB, SWIM, Tong’s, and PHARP) in this study were phased using SHAPEIT4, and imputation was conducted using Minimac492. We used two indices to evaluate the imputation performance: (1) the concordance rate, which represented the proportion of correctly imputed genotypes among the total imputed genotypes, and (2) the squared correlation (r2) between the imputed allele dosages and the true genotype. Each test was performed with ten replicates.
Population genetic structure and genetic diversity
Additional 106 pigs from the EBI database (Supplementary Data 4), which included 74 European commercial pigs, 13 European wild boars, two Near East wild boars, five Southeast Asian pigs, six warthog pigs, and six ancient pigs, combined with 1011 1KCIGP samples were used to the population structure and the following admixture analyses. Principal component analysis (PCA) was performed using GCTA (v1.93.2)93 based on the binary SNP data. The neighbor-joining tree was constructed with the same binary SNP set using MEGA (v11)94 based on a pairwise genetic distance matrix calculated with emmax (beta-07Mar2010)95. We also used ADMIXTURE (v1.3.0)96 to estimate individual ancestries, with the number of ancestral component K values ranging from two to six. The Fst statistics and genetic diversity were calculated by VCFtools (v0.1.17)97 using 100 kb sliding windows with 50 kb increments at each step.
Admixture testing and time estimation
We used f3, f4 statistics, and Treemix to analyze admixture levels in Chinese domestic pigs. The f3 and f4 statistics were computed using the ADMIXTTOOLS software (v7.0.1). To dissect the direction of introgression in detail, we used DFOIL98 to measure admixture among the pig populations. DFOIL detects admixture based on a five-taxon phylogeny (((P1, P2),(P3, P4)), O), where O represents an outgroup. We ran DFOIL in 200 kb non-overlapping sliding windows across the whole genome with other default parameters. We selected the sample with the highest sequencing depth to represent each breed. For each computation, one genome was used to represent each P1, P2, P3, and P4. The phylogenetic relationships were determined using Treemix (v1.13)99. We rooted the tree with the warthog, created blocks of 1000 SNPs, and used global rearrangements. The admixture time was estimated using ALDER (v1.03)100.
Demographic analysis
The effective population size (Ne) for each of the seven Eurasian pig populations was inferred using SMC + + (v1.15.3)36 with genome-wide SNPs. The mutation rate and generation time were set at 2.5 × 10−8 and five years, respectively3.
Whole-mitochondrial and paternal analysis
To generate the entire mitochondrial sequence, samples with mitochondrial coverage > 100 were retained. The raw reads aligned to the mitochondrial sequence of Sscrofa11.1 were extracted and assembled using MitoZ (v2.4)101. The mitogenome was eliminated if its topology was not circular. A total of 704 complete mitogenomes were used for the haplogroup analysis, which was conducted using MitoToolPy (v1.0)44.
For paternal analysis, we selected the NPARY regions (0–0.2 Mb and 4.79–43.55 Mb) of the Sscrofa11.1 reference sequence. After removing heterozygous sites and sites with missing genotypes in 5% of the sampled individuals, 34,207 high-confidence SNPs were extracted and applied to construct a median-joining haplotype network with PopART (v1.7)102. The sex-biased hybridization test was performed as previously described45. Briefly, the European-related ancestry (A) in the test populations was calculated using ADMIXTURE, and the range of expected European Y chromosome frequencies in the test population was set as [0, 2 A]. The binomial test was used to assess whether the observed number of European Y haplogroups in the Chinese domestic pig populations was larger than the expected range.
Selective signature detection
Two complementary methods, LSBL103 and θπ, were performed to identify signatures of positive selection in Chinese indigenous pigs. Both methods were performed using 100 kb sliding windows with a shifting increment of 10 kb at each step. The genomic diversity θπ was calculated by VCFtools and log2-transformed. The LSBL in population A was calculated using the following formula for each window:
where A represents the target population, while B represents the control population (Supplementary Data 9), and C represents the outgroup (EUD). The Fst for each pairwise comparison (A and B, A and C, or B and C) was calculated using VCFtools. The LSBL of each window was Z-transformed. For each method, regions with extremely high values in the 5% right-tail were selected as potential candidate regions. Then, the overlapping candidate regions were considered as the final regions under positive selection.
GWAS analysis of biological traits
Before conducting the GWAS, the SNPs were filtered with PLINK (v1.90)104 for MAF > 0.05, missing SNPs and individuals < 0.01. The phenotypic data for the GWAS included ten phenotypes, which were mainly from Animal Genetic Resources in China: pigs, and a previously published study105, including solid black coat color, gradient zone, body height, body length, body weight, chest girth, ear shape, backfat, sexual maturity, and lean meat percentage. The weighted averages of adult pig records for each breed were used for quantitative traits such as body size. Conversely, qualitative traits, such as coat color, were treated as binary characters (e.g., 1 and 0). The detailed phenotypic data used in the GWAS are listed in Supplementary Data 13. GWAS analyses were performed using GEMMA, using the linear mixed model method (Supplementary Methods). Each analysis was corrected using the first five columns of the principal components. The Bonferroni multiple test was used to correct the P-value, and the significance threshold was defined as 0.05/number of variants.
SVs discovery and genotyping
Four diverse tools were selected for SV detection, including Delly2 (v0.8.1)50, Lumpy (v0.3.1)51, Manta (v1.6.0)52, and Wham (v1.7.0)53. For Wham, the SV results were genotyped using SVTyper (v0.7.1)106. For the other three methods, SVs were detected and genotyped using their default parameters because these methods can detect and genotype SVs. The results of all four tools were filtered and merged using SURVIVOR (v1.0.7)107 with the following parameters: “1000 2 1 1 0 50”. To validate the accuracy of SV detection using short-reads, long-reads from ten Chinese domestic pig breeds were used to examine 150 randomly selected SVs. These long-reads were aligned to Sscrofa11.1 using Minimap2 (v2.26)108, and the SVs were detected using Sniffles (v2.2)109. The SVs that showed the same types and overlapping breakpoints were considered validated.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was financially supported by the National Key R&D Program of China (2021YFD1200801), the National Natural Science Foundations of China (31972563 and 32272844), the Earmarked Fund for China Agriculture Research System (No. CARS-pig-35), and the 2115 Talent Development Program of China Agricultural University. We acknowledge the computational support provided by the High-performance Computing Platform of China Agricultural University.
Author contributions
J.F.L. conceived and designed the study, directed the project, provided all data and computational resources, supervised bioinformatic and statistical analyses, and revised the paper. H.D. and L.Z. designed the analytical strategy and performed analysis processes. Z.L. conducted selective sweep analysis and imputation performance evaluation. Y.Z. performed the reference panel analysis. M.Z. conducted the GWAS analysis. Q.H. conducted the population structure analysis. S.L. participated in the PCR and Sanger sequencing experiment. K.X. and L.J. revised this manuscript. All authors read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks Zhihua Jiang, and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw sequencing datasets generated in this study have been deposited in NGDC/GSA under accession code PRJCA030193 (https://ngdc.cncb.ac.cn/gsa). The dbSNP (v150) dataset of pigs was downloaded from the NCBI. The genomic variations, haplotype data, and phasing information can be accessed through Figshare (10.6084/m9.figshare.25847761.v1). Source data are provided in this paper.
Code availability
The list of the software and parameters used in this study is available through GitHub (https://github.com/kimi-du-bio/1KCIGP_V1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Heng Du, Lei Zhou.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54471-z.
References
- 1.Frantz, L. et al. The evolution of Suidae. Annu. Rev. Anim. Biosci.4, 61–85 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Frantz, L. A. F., Madsen, O., Megens, H. J., Groenen, M. A. M. & Lohse, K. Testing models of speciation from genome sequences: divergence and asymmetric admixture in Island South-East Asian Sus species during the Plio-Pleistocene climatic fluctuations. Mol. Ecol.23, 5566–5574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groenen, M. A. M. et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature491, 393–398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frantz, L. A. F. et al. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol.14, R107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groenen, M. A. M. A decade of pig genome sequencing: a window on pig domestication and evolution. Genet. Sel. Evol.48, 10.1186/s12711-016-0204-2 (2016). [DOI] [PMC free article] [PubMed]
- 6.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altshuler, D. M. et al. A global reference for human genetic variation. Nature526, 68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter, K. et al. The UK10K project identifies rare variants in health and disease. Nature526, 82–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, Y. et al. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res.10.1038/s41422-020-0322-9 (2020). [DOI] [PMC free article] [PubMed]
- 11.Zhang, P. et al. NyuWa Genome resource: A deep whole-genome sequencing-based variation profile and reference panel for the Chinese population. Cell Rep.37, 110017 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Bosse, M. et al. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat. Commun.5, 4392 (2014). [DOI] [PMC free article] [PubMed]
- 13.Li, M. Z. et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet.45, 1431–U180 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Ai, H. et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet.47, 217–225 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Frantz, L. A. F. et al. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet.47, 1141–1148 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Nosková, A. et al. Characterization of a haplotype-reference panel for genotyping by low-pass sequencing in Swiss Large White pigs. BMC Genomics22, 290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding, R. et al. The SWine IMputation (SWIM) haplotype reference panel enables nucleotide resolution genetic mapping in pigs. Commun. Biol.6, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Z. et al. PHARP: a pig haplotype reference panel for genotype imputation. Sci. Rep.12, 12645 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, W. et al. Animal-ImputeDB: a comprehensive database with multiple animal reference panels for genotype imputation. Nucleic Acids Res.48, D659–D667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong, X. et al. Accurate haplotype construction and detection of selection signatures enabled by high quality pig genome sequences. Nat. Commun.14, 5126 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flad, R. K., Yuan, J. 袁靖 & Li, S. 李水城 Zooarcheological evidence for animal domestication in northwest China. Dev. Quat. Sci.9, 167–203 (2007).
- 22.Jing, Y. & Flad, R. K. Pig domestication in ancient China. Antiquity76, 724–732 (2002). [Google Scholar]
- 23.Huang, M. et al. The fine-scale genetic structure and selection signals of Chinese indigenous pigs. Evol. Appl.13, 458–475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, Y. L. et al. Signatures of Selection and Interspecies Introgression in the Genome of Chinese Domestic Pigs. Genome Biol. Evol.9, 2592–2603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, Y. et al. Whole-genome analysis reveals the hybrid formation of Chinese indigenous DHB pig following human migration. Evol. Appl.15, 501–514 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res.29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francioli, L. C. et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet.46, 818–825 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Byrska-Bishop, M. et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell185, 3426–3440.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montgomery, S. B. et al. The origin, evolution, and functional impact of short insertion–deletion variants identified in 179 human genomes. Genome Res.23, 749–761 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaser, R., Adusumalli, S., Leng, S. N., Sikic, M. & Ng, P. C. SIFT missense predictions for genomes. Nat. Protoc.11, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Nicholas, F. W. Online Mendelian Inheritance in Animals (OMIA): a comparative knowledgebase of genetic disorders and other familial traits in non-laboratory animals. Nucleic Acids Res.31, 275–277 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petr, J. et al. Nitric-oxide-dependent activation of pig oocytes: the role of the cGMP-signalling pathway. Zygote14, 9–16 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. H., Yedavalli, V. R. & Jeang, K.-T. Activation of HIV-1 expression and replication by cGMP dependent protein kinase type 1-β (PKG1β). Retrovirology4, 91 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, A. et al. Carbon monoxide inhibits porcine reproductive and respiratory syndrome virus replication by the cyclic GMP/protein kinase G and NF-κB signaling pathway. J. Virol.91, 10.1128/jvi.01866-16 (2016). [DOI] [PMC free article] [PubMed]
- 35.Sun, X. et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet.52, 1423–1432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terhorst, J., Kamm, J. A. & Song, Y. S. Robust and scalable inference of population history from hundreds of unphased whole-genomes. Nat. Genet.49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark, P. U. et al. The last glacial maximum. Science325, 710–714 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Kobashi, T., Severinghaus, J. P., Brook, E. J., Barnola, J.-M. & Grachev, A. M. Precise timing and characterization of abrupt climate change 8200 years ago from air trapped in polar ice. Quat. Sci. Rev.26, 1212–1222 (2007). [Google Scholar]
- 39.Long, T. & Taylor, D. A revised chronology for the archaeology of the lower Yangtze, China, based on Bayesian statistical modelling. J. Archaeol. Sci.63, 115–121 (2015). [Google Scholar]
- 40.Gao, H., Zhu, C. & Xu, W. Environmental change and cultural response around 4200 cal. yr BP in the Yishu River Basin, Shandong. J. Geogr. Sci.17, 285–292 (2007). [Google Scholar]
- 41.Larson, G. et al. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc. Natl. Acad. Sci. USA104, 4834–4839 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucchini, V., Meijaard, E., Diong, C. H., Groves, C. P. & Randi, E. New phylogenetic perspectives among species of South-east Asian wild pig (Sus sp.) based on mtDNA sequences and morphometric data. J. Zool.266, 25–35 (2005). [Google Scholar]
- 43.Larson, G. et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science307, 1618–1621 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Peng, M.-S. et al. DomeTree: a canonical toolkit for mitochondrial DNA analyses in domesticated animals. Mol. Ecol. Resour.15, 1238–1242 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Ai, H. et al. Human-mediated admixture and selection shape the diversity on the modern swine (Sus scrofa) Y chromosomes. Mol. Biol. Evol.38, 5051–5065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldberg, A., Verdu, P. & Rosenberg, N. A. Autosomal admixture levels are informative about sex bias in admixed populations. Genetics198, 1209–1229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma, J. et al. A genome scan for quantitative trait loci affecting three ear traits in a White Duroc × Chinese Erhualian resource population. Anim. Genet.40, 463–467 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Zheng, S. et al. Genetic structure and domestication footprints of the tusk, coat color, and ear morphology in East Chinese pigs. J. Genet. Genom.49, 1053–1063 (2022). [DOI] [PubMed] [Google Scholar]
- 49.Lü, M.-D. et al. Genetic variations associated with six-white-point coat pigmentation in Diannan small-ear pigs. Sci. Rep.6, 27534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Layer, R. M., Chiang, C., Quinlan, A. R. & Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol.15, R84 (2014). [DOI] [PMC free article] [PubMed]
- 52.Chen, X. Y. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Prlic, A. et al. Wham: Identifying Structural Variants of Biological Consequence. PLOS Comput. Biol.11, e1004572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, X. et al. GWAS Atlas: an updated knowledgebase integrating more curated associations in plants and animals. Nucleic Acids Res.51, D969–D976 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Relton, C. L. et al. DNA Methylation patterns in cord blood DNA and body size in childhood. PLOS ONE7, e31821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma, Y.-F. et al. Population genomics analysis revealed origin and high-altitude adaptation of tibetan pigs. Sci. Rep.9, 11463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaborit, N. et al. Transcriptional profiling of ion channel genes in Brugada syndrome and other right ventricular arrhythmogenic diseases. Eur. Heart J.30, 487–496 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Harrell, M. D., Harbi, S., Hoffman, J. F., Zavadil, J. & Coetzee, W. A. Large-scale analysis of ion channel gene expression in the mouse heart during perinatal development. Physiol. Genom.28, 273–283 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Cardona, A. et al. Genome-wide analysis of cold adaptation in Indigenous Siberian populations. PLOS ONE9, e98076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Genova, A. et al. Genome sequencing and transcriptomic analysis of the Andean killifish Orestias ascotanensis reveals adaptation to high-altitude aquatic life. Genomics114, 305–315 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Fu, Y. et al. IAnimal: a cross-species omics knowledgebase for animals. Nucleic Acids Res.51, D1312–D1324 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, H. et al. ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs. Nature606, 358–367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giuffra, E. et al. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics154, 1785–1791 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lander, B., Schneider, M. & Brunson, K. A history of pigs in China: From curious omnivores to industrial pork. J. Asian Stud.79, 865–889 (2020). [Google Scholar]
- 66.Ramos-Onsins, S. E., Burgos-Paz, W., Manunza, A. & Amills, M. Mining the pig genome to investigate the domestication process. Heredity113, 471–484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao, P. et al. PRE-1 Revealed previous unknown introgression events in Eurasian Boars during the Middle Pleistocene. Genome Biol. Evol.12, 1751–1764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li, N. The long-term consequences of cultural distance on migration: Historical evidence from China. Aust. Econom. Hist. Rev.58, 2–35 (2018). [Google Scholar]
- 69.Wiens, H. J. Han Chinese Expansion in South China. (Shoe String Press, 1967).
- 70.Prabhakar, N. R. & Semenza, G. L. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol. Rev.92, 967–1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pedrotti, S. et al. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Hum. Mol. Genet.24, 2360–2374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh, R. K., Kolonin, A. M., Fiorotto, M. L. & Cooper, T. A. Rbfox-Splicing factors maintain skeletal muscle mass by regulating calpain3 and proteostasis. Cell Rep.24, 197–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi, D.L. & Grifone, R. RNA-Binding proteins in the post-transcriptional control of skeletal muscle development, regeneration and disease. Front. Cell Dev. Biol. 9, 10.3389/fcell.2021.738978 (2021). [DOI] [PMC free article] [PubMed]
- 74.Gallagher, T. L. et al. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol.359, 251–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen, C. et al. Copy number variation in the MSRB3 gene enlarges porcine ear size through a mechanism involving miR-584-5p. Genet. Select. Evol.50, 72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, L. C. et al. mRNA and protein expression levels of four candidate genes for ear size in Erhualian and Large White pigs. Genet. Mol. Res.16, 10.4238/gmr16029252 (2017). [DOI] [PubMed]
- 77.Zhang, L. et al. Genome-wide scan reveals LEMD3 and WIF1 on SSC5 as the candidates for porcine ear size. PLOS ONE9, e102085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, C. et al. Genome-wide association studies for two exterior traits in Chinese Dongxiang spotted pigs. Anim. Sci. J.89, 868–875 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Zhang, Z. et al. Genomic analysis reveals genes affecting distinct phenotypes among different Chinese and western pig breeds. Sci. Rep.8, 13352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li, J. et al. Artificial selection of the melanocortin receptor 1 gene in Chinese domestic pigs during domestication. Heredity105, 274–281 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Niu, L., Shi, K., Xie, J.-J., Liu, S. & Zhong, T. Divergent evolutional mode and purifying selection of the KIT Gene in European and Asian domestic pig breeds. BioMed. Res. Int.2018, e8932945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grudzien-Nogalska, E., Jiao, X., Song, M.-G., Hart, R. P. & Kiledjian, M. Nudt3 is an mRNA decapping enzyme that modulates cell migration. RNA22, 773–781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kitamoto, A. et al. NUDT3 rs206936 is associated with body mass index in obese Japanese women. Endocr. J.60, 991–1000 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Vignali, R. & Marracci, S. HMGA Genes and proteins in development and evolution. Int. J. Mol. Sci.21, 654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Federico, A. et al. Hmga1/Hmga2 double knock-out mice display a “superpygmy” phenotype. Biol. Open3, 372–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J.17, 10–12 (2011). [Google Scholar]
- 87.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet.43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patterson, M. et al. WhatsHap: Weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol.22, 498–509 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Delaneau, O., Zagury, J.-F., Robinson, M. R., Marchini, J. L. & Dermitzakis, E. T. Accurate, scalable and integrative haplotype estimation. Nat. Commun.10, 5436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet.48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang, J. A., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: A Tool for genome-wide complex trait analysis. Am. J. Hum. Genet88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet.42, 348–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res.19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Danecek, P. et al. The variant call format and VCFtools. Bioinformatics27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pease, J. B. & Hahn, M. W. Detection and polarization of introgression in a five-taxon phylogeny. Syst. Biol.64, 651–662 (2015). [DOI] [PubMed] [Google Scholar]
- 99.Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet.8, 10.1371/journal.pgen.1002967 (2012). [DOI] [PMC free article] [PubMed]
- 100.Loh, P.-R. et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics193, 1233–1254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meng, G., Li, Y., Yang, C. & Liu, S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res.47, e63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leigh, J. W. & Bryant, D. popart: full-feature software for haplotype network construction. Methods Ecol. Evol.6, 1110–1116 (2015). [Google Scholar]
- 103.Wang, M. S. et al. 863 genomes reveal the origin and domestication of chicken. Cell Res.30, 693–701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu, J. et al. Whole genome variants across 57 pig breeds enable comprehensive identification of genetic signatures that underlie breed features. J. Anim. Sci. Biotechnol.11, 115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chiang, C. et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods12, 966–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jeffares, D. C. et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun.8, 14061 (2017). [DOI] [PMC free article] [PubMed]
- 108.Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw sequencing datasets generated in this study have been deposited in NGDC/GSA under accession code PRJCA030193 (https://ngdc.cncb.ac.cn/gsa). The dbSNP (v150) dataset of pigs was downloaded from the NCBI. The genomic variations, haplotype data, and phasing information can be accessed through Figshare (10.6084/m9.figshare.25847761.v1). Source data are provided in this paper.
The list of the software and parameters used in this study is available through GitHub (https://github.com/kimi-du-bio/1KCIGP_V1).