Abstract
Developing a generalist radiology diagnosis system can greatly enhance clinical diagnostics. In this paper, we introduce RadDiag, a foundational model supporting 2D and 3D inputs across various modalities and anatomies, using a transformer-based fusion module for comprehensive disease diagnosis. Due to patient privacy concerns and the lack of large-scale radiology diagnosis datasets, we utilize high-quality, clinician-reviewed radiological images available online with diagnosis labels. Our dataset, RP3D-DiagDS, contains 40,936 cases with 195,010 scans covering 5568 disorders (930 unique ICD-10-CM codes). Experimentally, our RadDiag achieves 95.14% AUC on internal evaluation with the knowledge-enhancement strategy. Additionally, RadDiag can be zero-shot applied or fine-tuned to external diagnosis datasets sourced from various medical centers, demonstrating state-of-the-art results. In conclusion, we show that publicly shared medical data on the Internet is a tremendous and valuable resource that can potentially support building strong models for image understanding in healthcare.
Subject terms: Diseases, Experimental models of disease, Diagnosis, Medical imaging
Medical imaging has transformed clinical diagnostics. Here, authors present RadDiag, a foundational model for comprehensive disease diagnosis using multi-modal inputs, demonstrating superior zero-shot performance on external datasets compared to other foundation models and showing broad applicability across various medical conditions.
Introduction
In the landscape of clinical medicine, the emergence of radiology techniques, for example, X-ray, CT, and MRI, has truly revolutionized the medical field, which enables diagnosis by reading radiological images. Recent literature has demonstrated significant potential for developing AI models for disease diagnosis in the medical field1,2. However, a notable limitation of the existing models is their specialization, as these specialist models often focus on a narrow range of disease categories, and thus usually fail to accommodate the breadth and complexity of clinical scenarios. Conversely, an ideal radiology diagnosis system is expected to comprehensively analyze the combination of arbitrary historical scans across any anatomical regions and imaging modalities for a wide range of diseases.
In this paper, we aim to build a foundational radiology diagnostic model, that supports information fusion from an arbitrary number of 2D and 3D inputs across different modalities and anatomic sites. Generally speaking, there are three major challenges. First, lack of datasets for training. Despite the large volume of radiology data generated per day across the world, publicly available ones are extremely limited due to privacy issues. Existing open-source datasets are normally collected from a certain modality or anatomy with very limited diseases, thus significantly hindering the development of powerful disease diagnosis models. Second, lack of unified architecture formulation. Models for diagnosing radiological diseases are often tailored to particular datasets, which are optimized for a narrow spectrum of diseases linked with particular imaging modalities3,4. In contrast, developing a foundation model for disease diagnosis requires the architecture to be capable of fusing information from an arbitrary number of images of various modalities, for example, X-ray, CT, and MRI. Third, lack of benchmark to monitor progress. Benchmarking the models’ performance on disease diagnosis relies on datasets that well simulate real-world scenarios, for instance, containing many diseases, long-tail distribution, etc. Existing benchmarks typically focus on a very limited number of diseases, with single modality inputs5,6. To comprehensively measure the progress in disease diagnosis, a high-quality benchmark has yet to be established.
To overcome the above-mentioned challenges, we consider to exploit the widely available data on Internet, especially those focusing on high-quality radiology samples from professional institutions and educational websites, that have undergone meticulous de-identification, and reviewed by a panel of experienced clinicians. Specifically, we develop a flexible data collection pipeline, with each sample associated with an International Classification of Diseases code, i.e., ICD-10-CM7, indicating the diagnostic category. The dataset presents a long-tailed distribution across disease categories. The case numbers range from 1 to 964 with some rare diseases. It includes multi-modal scans per case, with modalities varying from 1 to 5 and image counts between 1 and 30. As a result, we collect a multi-scan medical disease classification dataset, with 40,936 cases (195,010 scans) across 7 anatomy regions and 9 modalities, covering 930 ICD-10-CM codes and 5568 disorders, including a broad spectrum of medical conditions.
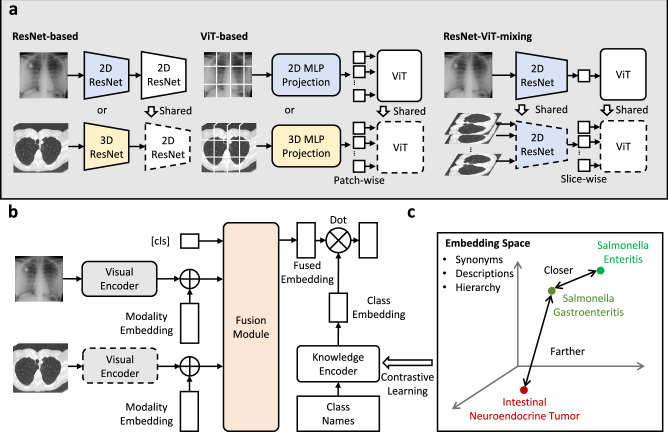
Architecturally, we conduct preliminary explorations on the design that supports both 2D and 3D input from various modalities, together with a transformer-based fusion module for comprehensive diagnosis. Specifically, for visual encoding, we explore three variants of backbones, namely, ResNet-based, ViT-based and ResNet-ViT-mixing to perform 2D or 3D unified encoding. Then, we fuse the multi-scan information with a transformer-based fusion module, treating each image embedding as an input token. At training time, we adopt knowledge-enhanced training strategy1,8,9, where we first leverage the rich domain knowledge to pre-train a knowledge encoder with natural language, and use it to guide the visual representation learning for disease diagnosis.
In experiments, we evaluate our model on a benchmark, constructed by images from online resources, featuring large-scale, multi-modal, multi-scan, long-tailed disease diagnosis. Additionally, we demonstrate that our proposed model can function as a general-purpose image encoder to capture a better image representation for radiology, making it possible to train and classify new sets via data-efficient fine-tuning or zero-shot transferring, leading to improved performance across diverse disease diagnosis tasks on numerous datasets, regardless of their image dimensions, modalities and anatomies. This paper demonstrates that publicly shared medical information is a tremendous resource that can be harnessed to advance the development of general AI for healthcare.
Results
In this section, we start by briefly introducing our considered problem formulation, followed with showing the characteristics of our dataset for case-level multi-modal, multi-scan, long-tailed disorder/ICD-10-CM diagnosis. Then, we leverage a test split of the dataset to carry out a series of studies on diagnostic performance and establish a case-level diagnosis benchmark. Lastly, we train and assess the developed model accordingly, along with its transferring ability to various external datasets.
Problem formulation
In this paper, our objective is to develop a radiology diagnostic model capable of integrating information from various images within a single case. We define a case as encompassing a collection of both 2D and 3D radiological images from different modalities and anatomical sites of a patient, who may be diagnosed with multiple conditions.
More specifically, we represent a case and its associated disorder labels as , where each xi signifies a specific scan (either 2D or 3D) from a radiological examination such as MRI, CT, or X-ray. Here, S is the number of scans for the case, and c is the total number of diagnostic labels. It is noteworthy that a patient’s case might include multiple scans across the same or different modalities.
To illustrate this concept, consider a case where a patient undergoes a physical examination, including a chest CT and a chest X-ray. In this instance, S = 2 reflects the two scans involved in this case. If the patient is diagnosed with nodule and pneumothorax, the labels for these disorders are set to 1, reflecting their presence in the patient’s diagnosis.
Our goal is to train a model that can solve the above multi-class, multi-scan diagnosis problem:
| 1 |
where the model is composed of a visual encoder Φvisual, a fusion module Φfuse and a classifier Φcls.
RP3D-DiagDS Statistics
Here, we provide a detailed statistical analysis of our collected dataset, namely RP3D-DiagDS, from three aspects: modality coverage, anatomy coverage, and disorder coverage. As shown in Supplementary Section E, compared with existing datasets, our dataset has great advantages in terms of anatomy richness, modal richness, and the number of images and categories. Furthermore, we split the dataset based on the number of cases on different levels.
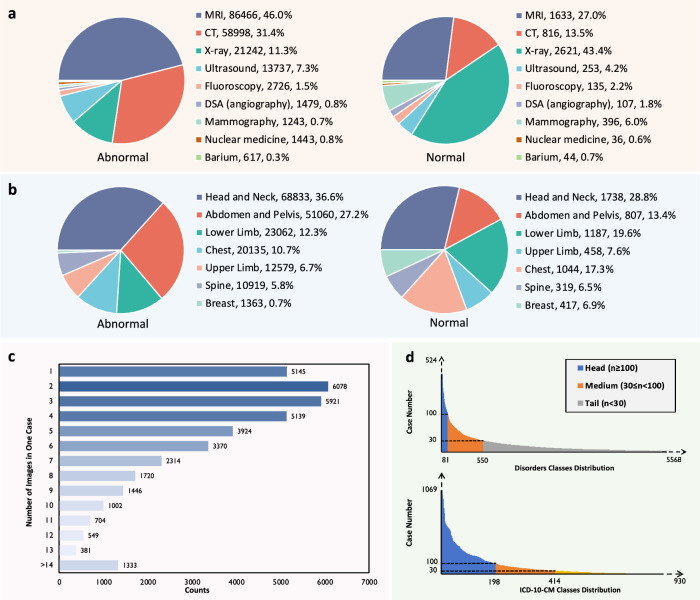
For Modality coverage, RP3D-DiagDS comprises images from 9 modalities, namely, computed tomography (CT), magnetic resonance imaging (MRI), X-ray, Ultrasound, Fluoroscopy, Nuclear medicine, Mammography, DSA (angiography), and Barium Enema. Each case may include images from multiple modalities, to ensure the precise and comprehensive diagnosis of disorders. Overall, approximately 19.4% of the cases comprise images from two modalities, while around 2.9% involve images from three to five modalities. The remaining cases are associated with image scans from a single modality. The distribution of modalities among all abnormal and normal samples are illustrated in Fig. 1a.
Fig. 1. The data distribution analysis on RP3D-DiagDS.
a The distribution of imaging modalities of abnormal (left) and normal (right) cases in RP3D-DiagDS. Each label is annotated with the class name, number of cases, and the corresponding proportion. b The distribution of imaging anatomies of abnormal (left) and normal (right) cases in RP3D-DiagDS. c Case distribution on image numbers. In the bar plot, We show the distribution for the number of images in one case. In RP3D-DiagDS, each case may include multiple images from patient history scans, different modalities, and different angles or conditions. d Case distribution on classes. We demonstrate the long-tailed distributions for disorder and ICD-10-CM classes. We also categorize these classes into three categories: “head class'', “body class” and “tail class” based on the number of cases. Notably, to better show the main part of the case distributions, we clip the axes, indicated by the dotted axes lines. Source data are provided as a Source Data file.
Beyond the comprehensive modality coverage, RP3D-DiagDS also comprises images from various anatomical regions, including the head and neck, spine, chest, breast, abdomen and pelvis, upper limb, and lower limb, providing comprehensive coverage of the entire human body. The statistics are shown in Fig. 1b.
In terms of disorder coverage, for both disorder and disease classification, each case can correspond to multiple disorders, resulting in RP3D-DiagDS a multi-label classification dataset. As shown in Fig. 1d, the distributions exhibit extremely long-tailed patterns, rendering such 2D & 3D image classification problems as a long-tailed multi-label classification task. Totally there are 5568 disorders mapped into 930 ICD-10-CM classes. We define the “head class” category with case counts greater than 100, the “body class” category with case counts between 30 and 100, and the “tail class” category with case counts less than 30.
Ablation studies
We first carry out a series of ablation studies to explore the optimal model architecture and parameter configurations, including the fusion strategy, visual encoder architecture, the depth of 3D scans, and augmentation. Note that, considering the original RP3D-DiagDS is too large to carry out extensive empirical experiments, we construct two subsets, each comprising around 1/4 classes of the entire dataset for ablation studies. Specifically, one subset keeps the top 200 classes, which can result more robust performance comparison, as the head classes have more positive cases. While the other subset contain samples from 200 random classes, that can also reflect the results for body and tail classes. We report the results on different subsets as “xx/xx” in Table 1 and more ablation tables in Supplementary Section C, the former representing the performance on the top subset, and the latter representing that on the random subset. Generally, the results reflected by these two subsets are consistent, thus for simplicity, next we mainly report and analyse the results from the subset with top 200 class.
Table 1.
Ablation study on various settings of visual encoder architectures
| Visual Encoder | Architecture | AUC | AP | F1 | MCC | R@0.01 | R@0.05 | R@0.1 | |
|---|---|---|---|---|---|---|---|---|---|
| Normalisation | Shared Enc | ||||||||
| ViT | 2-layer MLP | 6-layer ViT | 79.98/77.24 | 6.01/4.99 | 13.57/7.36 | 14.69/7.95 | 17.78/10.87 | 33.56/24.66 | 47.21/35.02 |
| 4-layer MLP | 6-layer ViT | 80.57/77.74 | 6.13/5.20 | 13.49/8.41 | 14.78/9.53 | 17.68/11.48 | 34.01/25.79 | 47.44/36.12 | |
| 2-layer MLP | 12-layer ViT | 81.69/78.15 | 6.40/5.26 | 14.71/8.89 | 15.30/9.74 | 18.11/11.33 | 34.73/25.97 | 48.84/36.20 | |
| 4-layer MLP | 12-layer ViT | 82.03/78.59 | 6.67/5.37 | 14.94/8.87 | 15.66/9.77 | 18.20/12.09 | 34.99/26.58 | 49.52/36.74 | |
| ResNet | ResNet-18 | ResNet-18 | 86.91/81.16 | 11.00/5.27 | 16.77/9.21 | 18.63/11.48 | 20.42/12.63 | 41.87/29.20 | 59.38/42.04 |
| ResNet-34 | ResNet-18 | 86.99/81.75 | 11.15/5.70 | 17.14/10.06 | 19.21/11.47 | 20.82/13.61 | 44.67/30.73 | 61.13/43.54 | |
| ResNet-18 | ResNet-34 | 87.06/82.09 | 11.27/6.09 | 17.36/10.15 | 19.23/12.00 | 21.48/1.96 | 44.38/31.46 | 61.54/44.13 | |
| ResNet-34 | ResNet-34 | 87.10/82.44 | 11.31/6.32 | 17.66/10.06 | 19.41/12.34 | 21.33/13.88 | 44.19/31.60 | 62.25/44.24 | |
| ResNet-ViT | ResNet-34 | 6-layer ViT | 88.74/84.02 | 11.52/7.07 | 17.86/11.30 | 20.05/14.11 | 21.92/15.10 | 44.63/33.38 | 63.09/47.81 |
| ResNet-50 | 6-layer ViT | 89.53/84.76 | 11.75/7.74 | 19.59/12.51 | 20.61/15.01 | 23.18/15.33 | 51.34/33.92 | 67.39/48.67 | |
| ResNet-34 | 12-layer ViT | 88.93/84.23 | 11.4/7.52 | 18.07/11.86 | 20.09/14.49 | 22.38/15.19 | 45.23/33.65 | 65.04/48.07 | |
| ResNet-50 | 12-layer ViT | 89.56/84.95 | 11.73/7.72 | 19.73/12.36 | 21.16/14.97 | 22.58/15.81 | 51.64/34.35 | 67.92/48.98 | |
“Normalisation” denotes the separated visual encoder part to perform 2D and 3D normalization and “Shared Enc.” denotes the shared encoder part for both 2D and 3D scans. The value preceding ‘/’ represents the results from the subset with top 200 classes, while the value following ‘/’ denotes the results from the subset with 200 random classes.
As simple experiment baselines, we use images with spatial resolution of 512 × 512, 16 as the 3D scan depth, and ResNet18 + ResNet34 as the visual backbone, without augmentation strategy or knowledge enhancement, adopting the basic max pooling fusion strategy introduced in Supplementary Section B. While conducting an ablation study on certain component, we keep other settings unchanged. More results can be found in Supplementary Section C. Herein, we mainly describe the critical results on comparing image encoder architectures.
Specifically, we investigate three backbone architectures variants for realizing unified diagnosis with arbitrary 2D and 3D scans, i.e., ResNet-based, ViT-based and ResNet-ViT-mixing. More details on architecture design can be found in “Method” Section. As shown in Table 1, we make three observations: (i) The performance of ResNet-ViT-mixing based model significantly surpasses that of ViT-based and ResNet-based ones. (ii) Enhancing the feature dimension of the vision encoder results in notable improvements, e.g., ResNet-34 + 6-layerViT vs. ResNet-50 + 6-layerViT. (iii) Increasing the number of shared encoder layers in the ViT-based model does not yield a significant enhancement in performance but incurs substantial computational overhead, e.g., ResNet-50 + 6-layer ViT vs. ResNet-50 + 12-layer ViT.
Based on the comprehensive ablation results shown in Supplementary Section C, we choose the ResNet-ViT-mixing model with augmentation strategy and unifying the 3D scan depth to 32 as the most suitable settings for the following evaluation on long-tailed case-level multi-modal diagnosis. After deciding the basic configurations, we evaluate the diagnosis ability on both our proposed internal RP3D-DiagDS dataset and various external benchmarks. In internal evaluation, we mainly show the effectiveness of our newly proposed modules, considering the problem as a multi-label classification task under a long-tailed distribution. In external evaluation, we systematically investigate the boundary of our final model, serving as a diagnosis foundation model under both fine-tuning and zero-shot settings.
Evaluation on internal benchmark
In this part, our model is trained on the entire Rad3D-DiagDS training set (30880 cases), evaluated on its test split (7721 cases), as detailed in Table 2. The comparison of AUC curves is depicted in Fig. 2a–f. We conduct experiments at two levels-disorder and ICD-10-CM classes-to evaluate the model’s performance, both with and without text knowledge enhancement. These evaluations consider two integration strategies: early fusion and late fusion. For comprehensive definitions and applications of these fusion strategies, please see Supplementary Section B for more details. The model’s effectiveness is assessed using various metrics on the test set, including the AUC curve’s confidence interval (CI), to gauge its stability and consistency.
Table 2.
Classification results on Disorders and ICD-10-CM levels
| Granularity | Category | Methods | Metrics | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FM | KE | AUC | AP | F1 | MCC | R@0.01 | R@0.05 | R@0.1 | ||
| Disorders | Head | ✗ | ✗ | 92.44 | 15.34 | 24.95 | 26.14 | 34.11 | 63.70 | 76.70 |
| ✓ | ✗ | 93.61 | 18.16 | 28.15 | 29.30 | 39.89 | 67.74 | 80.10 | ||
| ✓ | ✓ | 94.41 | 20.27 | 30.21 | 32.27 | 41.93 | 71.09 | 81.15 | ||
| Medium | ✗ | ✗ | 93.74 | 11.12 | 19.88 | 22.67 | 32.62 | 62.29 | 73.63 | |
| ✓ | ✗ | 94.79 | 13.51 | 22.96 | 25.80 | 37.63 | 66.25 | 78.06 | ||
| ✓ | ✓ | 95.14 | 15.95 | 25.82 | 28.84 | 42.49 | 68.73 | 79.39 | ||
| Tail | ✗ | ✗ | 88.10 | 6.04 | 10.35 | 15.06 | 9.52 | 22.48 | 32.94 | |
| ✓ | ✗ | 90.13 | 6.65 | 10.98 | 15.84 | 11.43 | 27.37 | 43.01 | ||
| ✓ | ✓ | 90.96 | 7.75 | 12.68 | 17.49 | 13.13 | 28.88 | 44.08 | ||
| ICD-10-CM | Head | ✗ | ✗ | 89.83 | 12.31 | 20.27 | 21.58 | 23.81 | 52.05 | 68.45 |
| ✓ | ✗ | 90.25 | 13.94 | 21.78 | 22.98 | 25.94 | 55.22 | 69.15 | ||
| ✓ | ✓ | 91.27 | 14.59 | 22.81 | 25.12 | 27.87 | 57.75 | 72.32 | ||
| Medium | ✗ | ✗ | 90.58 | 8.42 | 16.42 | 19.41 | 23.80 | 50.80 | 66.42 | |
| ✓ | ✗ | 91.12 | 9.18 | 18.02 | 20.05 | 27.10 | 53.77 | 67.45 | ||
| ✓ | ✓ | 92.01 | 10.34 | 19.08 | 22.16 | 28.81 | 55.55 | 69.86 | ||
| Tail | ✗ | ✗ | 86.91 | 4.82 | 8.64 | 12.70 | 10.13 | 29.82 | 51.04 | |
| ✓ | ✗ | 87.32 | 5.09 | 9.29 | 13.23 | 11.21 | 30.00 | 49.39 | ||
| ✓ | ✓ | 88.11 | 5.57 | 10.48 | 14.68 | 12.75 | 30.11 | 51.77 | ||
*All indicators are presented as percentages, with the % omitted in the table.
In the table “FM” represents the fusion module and “KE” represents the knowledge enhancement strategy. We report the results on Head/Medium/Tail category sets separately.
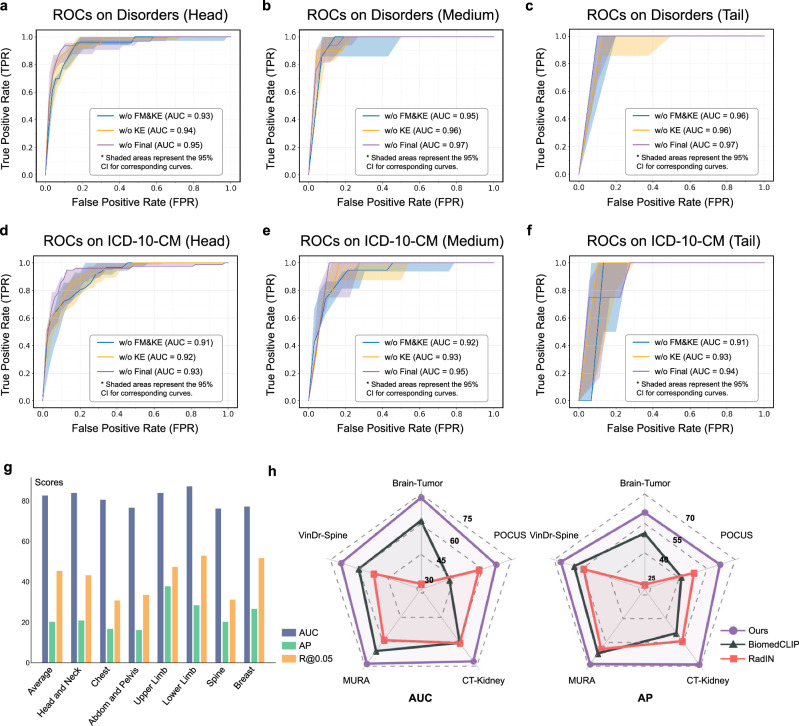
Fig. 2. RP3D-Diag performance evaluation on internal and external datasets.
a–f ROC curves for Disorders and ICD-10-CM, including head, medium, and tail parts of RP3D-DiagDS respectively. The shaded area in each figure represents the 95% Confidence Interval (CI). FM and KE stand for Fusion Module and Knowledge Enhancement, respectively. Additional ROC curves for each class are provided in the supplementary file. g The bar plot illustrates the AUC, AP, and Recall@0.05 of the Normal class across different body regions in the Internal test set of RP3D-DiagDS. h The radar chart compares the AUC and AP performance of RadIN, BiomedCLIP, and our model on external clinical datasets across five distinct anatomical regions. Source data are provided in a separate Source Data file.
To demonstrate the effectiveness of the fusion module, we start with a baseline that adopts max pooling on the predictions of different images from the same case (check Supplementary Section B for more details). Then, we add the fusion module and knowledge enhancement step-by-step, to improve the model’s performance. As shown in Table 2, adding the fusion module can greatly improve the results on Head, Medium, and Tail classes at both disorder and ICD-10-CM levels, showing the critical role of case-level information fusion in diagnosis tasks. These results align well with our expectations, as in clinical practice, the examination of one modality for a diagnosis is often insufficient. A thorough and meticulous diagnostic process typically involves an integrated review of all test results. Each test is weighted differently, depending on how its results correspond with other tests. Our fusion model adeptly mirrors this comprehensive approach, demonstrating its effectiveness in simulating the nuanced process of clinical diagnosis.
Moreover, we experiment with the knowledge-enhanced training strategy, where we first leverage the rich domain knowledge to pre-train a knowledge encoder with natural language and use it to guide the visual representation learning for disease diagnosis. As shown in Table 2, knowledge enhancement further promotes the final diagnosis performance on both disorder and ICD-10-CM classifications, showing that a better text embedding space trained by rich domain knowledge can greatly help visual representation learning.
Beyond the quantitative results, we also analysis on ROC Curves. The solid lines in the Fig. 2a–f represents the median AUC value. This value is derived from a process where, for each category, 1000 random samples are taken and the median AUC value is calculated. This procedure is repeated multiple times (1000), with the final curve representing the median of these values. The ROC curve is illustrated by the solid lines. Accompanying each solid line is a shaded area, which denotes the 95% confidence interval (CI). For each type of implementation, this shaded area is significantly higher than the portion above the solid line. This implies that the AUC value represented by the solid line is generally higher than the class-wise average across different data splits. As shown in the figure, the highest AUC value has reached 0.97, and the lowest value is 0.91, this observed pattern suggests, that there exists a few categories that are more challenging to learn than others.
In clinical scenarios, the proportion of normal cases is significantly higher compared to abnormal cases. Therefore, it is crucial to highlight the evaluation of the model’s ability to distinguish between normal and abnormal cases. As shown in Fig. 2g, our model achieves an AUC of 82.61% for such diagnosis, given the significant disease variations in abnormal cases. Additionally, we also split out six test sub-sets based on the anatomy of cases, including Head and Neck, Chest, Abdomen and Pelvis, Upper Limb, Lower Limb, Spine, and Breast. In each sub-set, all cases are from the same anatomy, thus we can track the performance among a certain anatomy region. As shown in Fig. 2g, the results on each anatomy are generally consistent, indicating that our model robustly demonstrates superior performance across different anatomical regions, without showing notable biases on any specific area.
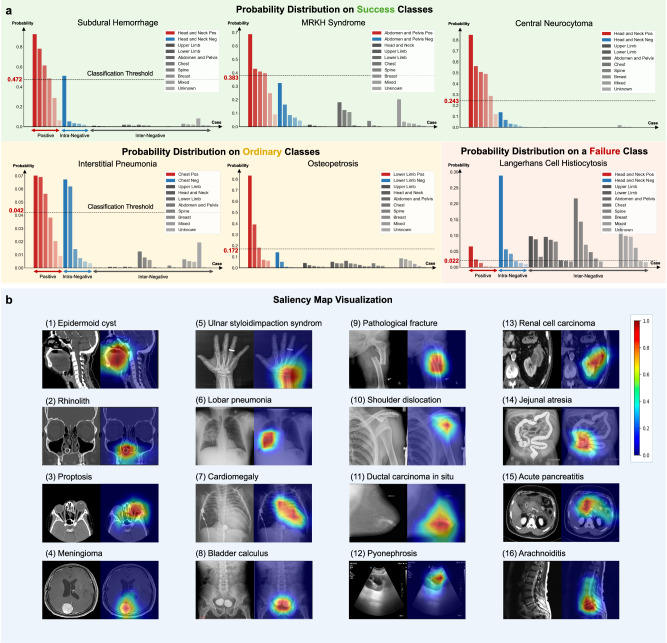
To better understand the model performance, we show prediction probability distributions for some certain classes. To get a deeper and more intuitive understanding of the model’s capabilities, we analyzed its performance across different disorder classes. Given the large number of disorders, we select 48 representative disorders, encompassing various body regions and imaging modalities to analyze their statistical distribution. Here in the main text, we present 6 of these disorders, chosen proportionally based on the statistical patterns of performance across all categories. These 6 cases serve as exemplars shown in Fig. 3. A more comprehensive demonstration of 42 typical disorders is available in Supplementary Section G. All cases are categorized by their related anatomies, with positive cases corresponding to the considered disorder class and negative cases divided into two types: intra-negative cases from the same anatomy, and inter-negative cases from other anatomies. Horizontal dashed lines in the bar plot denote the threshold, with positive and negative cases from the same anatomy colored in red and blue, and negative cases from other anatomies in different shades of gray. Ideally, positive cases should have high scores, while both types of negative cases should have low scores. Therefore, the difference between the actual distribution and the ideal situation can effectively illustrate the model’s performance on the disorder. As shown by the results, for the success classes, the model can distinguish both the intra-negative and inter-negative cases against the positive, while for the ordinary classes, the model may be confused with some intra-negative and positive cases. In the failure class, the model cannot pick out the positive cases. These results demonstrate that our internal evaluation requires the model to perform a two-stage differential diagnosis. The first step is to locate the possible disorders through the shooting anatomy, etc., and then capture the detailed lesion patterns features to make further diagnosis.
Fig. 3. The prediction probability distribution for different classes and saliency map visualization.
a Based on the anatomies of each case, we split them into positive cases, intra-negative cases which are located in the same anatomy as the positive ones, and inter-negative cases which are located in other anatomies. The classification threshold score in the figure denotes the final comparison bar to transform the soft probabilities into binary true/false diagnosis results. The first three probability distribution figures depict the distributions of three relatively successful classes, where the model can clearly distinguish the inter-negative cases and the intra-negative cases are more confusing. We then show two ordinary classes. As shown by the distributions, most errors are caused by the intra-negative cases, and similarly, the inter-negative cases are easily dismissed as well. At last, we show a failure case where the model can hardly distinguish the positive and negative cases regardless of whether they are intra-negative or inter-negative. b Saliency map of the key frames. Red indicates the areas that the model focuses on when inferring the corresponding disorder category. This indicates that RadDiag is capable of accurately identifying the locations of lesions or abnormal regions. Detailed abnormalities and corresponding data source links are provided in Supplementary Section G. Source data are provided as a Source Data file.
At last, we show the saliency map visualization results. Inspired by the proposal of explainable AI (XAI)10, we utilize EigenGradCAM11, a visual explanation method based on class activation maps, to visualize the areas that contribute most to the decision-making process for the target disorder class. As shown in Fig. 3b, we sample several cases from different modalities and anatomies to compute the saliency maps. For instance, as depicted in Fig. 3b(5) for ‘Ulnar styloid impaction syndrome’, our model accurately pinpoints the location of the ulna during inference for this category, without being distracted by other anomalies, such as rings worn on the fingers. In other cases shown in Fig. 3b(6) and b(7), our model correctly localizes the anomalies of ‘Lobar pneumonia’ and ‘Cardiomegaly’, indicating that the diagnosis is conducted by visual cues, rather based on the modality and imaging view. Detailed abnormalities, complete diagnosis labels, and corresponding data source links are provided in Supplementary Section G. These visualizations demonstrate our model’s precision in identifying the relevant anatomical structures for various disorder categories, even amidst other potential distractions or anomalies within the same image. This illustrates its robustness and specificity in medical disorder diagnosis.
Evaluation on external benchmarks
In this part, we further assess the transferring ability of our diagnosis model trained on RP3D-DiagDS. We adopt two settings to demonstrate the transferring ability to External Benchmarks of our model, i.e., zero-shot, and fine-tuning. First, in the zero-shot setting, we evaluate the effectiveness of our model for directly deployed in real clinical scenarios. In detail, as our dataset encompasses a wide range of disorder categories affecting the entire body, it can be tested on most external datasets. More details on how we perform zero-shot transferring for our method and other baselines can be found in the Method section. We have adopted F1, MCC, and ACC for comparison. Second, our model can also serve as a pre-trained model and be fine-tuned on each downstream task to improve the final performance. Specifically, for the dataset with single image input, we simply adopt the pre-trained visual encoder module, i.e., discarding the fusion module. For the dataset with multi-image input, we inherit the entire model. In both cases, the final classification layer will be trained from scratch. In addition to using all available external training data, we also consider using 1%, 10%, and 30% portion data for few-shot learning.
First of all, in zero-shot assessment, we evaluate the transferring ability of our final model on normal/abnormal diagnosis for external dataset, e.g., Brain-Tumor (405 normal, 906 abnormal), POCUS (256 normal, 302 abnormal), CT-Kidney (1037 normal, 1453 abnormal), MURA (1667 normal, 1530 abnormal), and VinDr-Spine (1070 normal, 1007 abnormal), totaling 9634 cases. These datasets are derived from medical centers in different regions, thus is a good assessment of the model’s performance in distinguishing normal from abnormal cases under zero-shot conditions. We directly adopt the “normal” classifier head trained on our RP3D-DiagDS to obtain the prediction probability score and compare it with other foundation models, RadIN12 and BiomedCLIP13, in terms of AUC and AP on the five zero-shot datasets. As shown in Fig. 2h, our model demonstrates significantly better results on all the five considered datasets, i.e., Brain-Tumor, POCUS, CT-Kidney, and MURA, compared to RadIN and BiomedCLIP. These findings show that our model consistently demonstrates superior diagnostic performance on zero-shot normal/abnormal diagnosis, across different anatomies under cross-center real clinical practices, significantly outperforming existing SOTA models.
In addition to the normal/abnormal diagnosis, as shown in Table 3, we also conduct zero-shot evaluation on fine-grained diagnosis tasks introduced by each representative external datasets, spanning six anatomies, including the head and neck, chest, breast, abdomen and pelvis, and spine. These datasets cover five modalities, i.e., CT, MRI, X-ray, Ultrasound, and Mammography, allowing for a comprehensive evaluation of our model’s capabilities. As baselines, we have included RadIN12 and BiomedCLIP13 for comparison in the zero-shot setting. The results demonstrate that our model outperforms the other two models in overall performance, indicating strong transfer learning capabilities. Interestingly, our model’s zero-shot performance can sometimes be even comparable to the results obtained by the model trained from scratch on a subset of the corresponding dataset, demonstrating the superiority of our RadDiag for diagnostic capabilities, which cannot be achieved by former foundation models.
Table 3.
Zero-shot results on 6 external datasets
| Dataset | Anatomy | Modality | Dim | Method | External Data | F1 ↑ | MCC ↑ | ACC ↑ |
|---|---|---|---|---|---|---|---|---|
| Brain-Tumor | Head&Neck | MRI | 2D | RadIN | Zero-shot | 22.29 | 3.23 | 63.63 |
| BiomedCLIP | 56.25 | 32.81 | 57.21 | |||||
| Ours | 61.09 | 47.37 | 73.38 | |||||
| S.T. | n=571(10%) | 61.77 | 46.69 | 72.17 | ||||
| POCUS | Chest | US | Video | RadIN | Zero-shot | 2.22 | 0.17 | 66.67 |
| BiomedCLIP | 50.33 | 11.73 | 45.82 | |||||
| Ours | 65.02 | 47.97 | 79.84 | |||||
| S.T. | n=223(1%) | 64.20 | 48.31 | 80.58 | ||||
| CT-KIDNEY | Abdom&Pelvis | CT | 2D | RadIN | Zero-shot | 38.1 | 17.57 | 64.75 |
| BiomedCLIP | 49.38 | 29.58 | 72.21 | |||||
| Ours | 56.50 | 43.18 | 69.96 | |||||
| S.T. | n=995(10%) | 55.73 | 42.28 | 66.95 | ||||
| MURA | Limb | X-ray | 2D | RadIN | Zero-shot | 31.26 | 4.37 | 52.94 |
| BiomedCLIP | 68.36 | 14.96 | 54.70 | |||||
| Ours | 67.64 | 40.15 | 77.51 | |||||
| S.T. | n=3680(10%) | 66.71 | 41.98 | 78.32 | ||||
| VinDr-Spine | Spine | X-ray | 2D | RadIN | Zero-shot | 4.34 | 0.91 | 87.49 |
| BiomedCLIP | 26.49 | 12.79 | 61.3 | |||||
| Ours | 25.93 | 19.40 | 83.07 | |||||
| S.T. | n=84(1%) | 24.73 | 20.61 | 84.46 | ||||
| TCGA | Chest&Breast | CT&MG | 3D | RadIN | Zero-shot | - | - | - |
| BiomedCLIP | - | - | - | |||||
| Ours | 72.18 | 57.20 | 83.47 | |||||
| S.T. | n=173(30%) | 70.33 | 52.26 | 77.41 |
*All indicators are presented as percentages, with the % omitted in the table.
For our model, we avoid exposing these external data for zero-shot experiments and use the threshold corresponding to the highest F1 score on the evaluation set. For RadIN and BiomedCLIP, we use the threshold corresponding to the highest F1 score on each individual external dataset. We also report the supervised training results with partially down-stream task external data, denoting as “S.T.”, for demonstrating what diagnosis performance our model can achieve zero-shotly. We have evaluated F1, MCC and ACC, and all the results reported in the table are the averaged values across all classes. In the table, “US" represents ultrasound and “MG" represents mammography.
Beyond the zero-shot setting, we also demonstrate the finetuning comparison Results. As shown in Table 4, while finetuning our model on external datasets, we see significant performance improvement on all 22 external datasets with different data portions, compared to models trained from scratch. In most cases, our model can even surpass the specialist SOTAs which are designed carefully for the targeting task, demonstrating that publicly shared medical data on the Internet is a tremendous and valuable resource that can serve as a superior large-scale supervised training dataset for the medical domain.
Table 4.
The AUC score comparison on various external datasets
| Dataset | 1% | 10% | 30% | 100% | SOTA | ||||
|---|---|---|---|---|---|---|---|---|---|
| Scratch | Ours | Scratch | Ours | Scratch | Ours | Scratch | Ours | ||
| VinDr-Mammo | 57.05 | 58.69↑1.64 | 58.22 | 59.55↑1.33 | 62.10 | 63.04↑0.94 | 76.25 | 78.46↑2.21 | 77.50†67 |
| CXR14 | 76.85 | 78.87↑2.02 | 77.93 | 80.66↑2.73 | 78.52 | 81.22↑2.70 | 79.12 | 83.44↑4.32 | 82.50†1 |
| VinDr-Spine | 79.35 | 82.13↑2.78 | 85.02 | 86.43↑1.41 | 86.90 | 87.33↑0.43 | 87.35 | 87.92↑0.57 | 88.90*8 |
| MosMedData | 52.96 | 60.78↑7.82 | 62.11 | 64.66↑2.55 | 65.63 | 70.96↑5.33 | 72.36 | 76.79↑4.43 | 68.47†51 |
| ADNI | 56.47 | 59.19↑2.72 | 61.62 | 65.08↑3.46 | 65.13 | 66.98↑1.85 | 83.44 | 85.61↑2.17 | 79.34†68 |
| NSCLC | 53.19 | 58.00↑4.81 | 55.21 | 59.37↑4.16 | 57.33 | 63.01↑5.68 | 67.25 | 72.54↑5.29 | N/A |
| TCGA | 55.28 | 67.33↑12.05 | 71.16 | 76.91↑5.75 | 78.55 | 85.27↑6.72 | 88.66 | 95.17↑6.51 | N/A |
| ISPY1 | 52.61 | 57.99↑5.38 | 57.32 | 59.89↑2.57 | 58.13 | 62.22↑4.09 | 65.88 | 69.43↑3.55 | N/A |
| ChestX-Det10 | 60.12 | 64.40↑4.28 | 64.55 | 67.61↑3.06 | 71.38 | 75.27↑3.89 | 74.11 | 79.44↑5.33 | N/A |
| CheXpert | 85.14 | 87.55↑2.41 | 87.97 | 89.26↑1.29 | 88.31 | 90.08↑1.77 | 89.52 | 91.27↑1.75 | 93.00*1 |
| COVID-19-Radio | 86.67 | 88.34↑1.67 | 87.47 | 90.38↑2.91 | 90.66 | 92.71↑2.05 | 95.39 | 98.56↑3.17 | N/A |
| IU-Xray | 60.22 | 63.38↑3.16 | 63.74 | 66.39↑2.65 | 67.55 | 70.27↑2.72 | 74.01 | 76.04↑2.03 | N/A |
| LNDb | 55.00 | 57.21↑2.21 | 60.77 | 62.10↑1.33 | 63.55 | 64.31↑0.76 | 68.76 | 70.58↑1.82 | N/A |
| PadChest | 77.10 | 78.36↑1.25 | 81.21 | 82.31↑1.10 | 82.45 | 84.18↑1.73 | 83.66 | 85.87↑2.21 | N/A |
| CC-CCII | 86.55 | 89.30↑2.75 | 94.19 | 96.30↑2.11 | 95.04 | 97.81↑2.77 | 98.27 | 99.46↑1.19 | 97.41†58 |
| RadChest | 59.92 | 61.36↑1.44 | 66.11 | 67.36↑1.25 | 70.09 | 72.18↑2.09 | 74.22 | 76.15↑1.93 | 77.30*69 |
| Brain-Tumor | 75.61 | 77.14↑1.53 | 80.86 | 82.33↑1.47 | 84.57 | 88.77↑4.20 | 91.05 | 93.21↑2.16 | N/A |
| Brain-Tumor-17 | 69.98 | 72.42↑2.44 | 78.78 | 80.31↑1.53 | 87.49 | 91.01↑3.52 | 93.66 | 94.43↑0.77 | N/A |
| POCUS | 78.20 | 79.34↑1.14 | 84.09 | 85.76↑1.67 | 88.42 | 89.66↑1.24 | 94.31 | 95.46↑1.15 | 94.00†70 |
| MURA | 76.28 | 78.19↑1.91 | 81.41 | 84.33↑2.92 | 85.17 | 86.27↑1.10 | 86.25 | 88.31↑2.06 | 92.90*59 |
| KneeMRI | 55.06 | 58.88↑3.82 | 62.88 | 63.12↑0.24 | 67.19 | 68.36↑1.17 | 73.05 | 73.22↑0.17 | N/A |
| CT-Kidney | 76.10 | 79.17↑3.07 | 83.61 | 84.96↑1.35 | 85.99 | 88.22↑2.23 | 91.41 | 93.26↑1.85 | N/A |
*The numbers are borrowed from the referred papers.
†The four SOTAs51,58,68,70 are relatively old. Thus, their network architecture is weaker than the classical 3D ResNet trained from scratch.
-All indicators are presented as percentages, with the % omitted in the table.
For each dataset, we experimented with different training data portions, denoted as 1% to 100% in the table. For example, 30% represents we use 30% of data in the downstream training set for finetuning our model or training from scratch. “SOTA” denotes the best performance of former works (pointed with corresponding reference) on the datasets. We mark the gap between ours and training from scratch on the subscript of uparrows↑ in the table.
Discussion
There have been a number of open-source datasets for disease diagnosis. Notably, large-scale datasets such as NIH ChestX-ray14, MIMIC-CXR15, and CheXpert4 stand out for their comprehensive collection of annotated X-ray images, facilitating extensive research and advancements in automated disease classification. However, it is important to note three key limitations. First, these open-source diagnosis datasets mainly consist of chest X-rays4,6,14–23, thus are 2D images. There are only a few 3D datasets available, with a limited number of volumes5,24–31. Second, a significant portion of these large-scale datasets focuses solely on binary classification of specific diseases18–21. Third, existing large-scale datasets contain a broad range of disease categories in different granularities12,17. For example, in PadChest17, disorders exist at different levels, including infiltrates, interstitial patterns, and reticular interstitial patterns. Such variation in granularity poses a great challenge for representation learning. In summary, these datasets fall short of meeting real-world clinical needs, which often involve complex, multimodal, and multi-image data from a single patient. Therefore, constructing a dataset that mirrors the intricacies of actual clinical scenarios is necessary. In comparison, the large-scale radiology data set we constructed covers a large number of categories of diseases, including various common modalities.
The prevailing paradigm in earlier diagnostic models is a specialized model trained on a limited range of disease categories. These models focus on specific imaging modalities and are targeted towards particular anatomical regions. Specifically, ConvNets are widely used in medical image classification due to their outstanding performance, for example23,32,33, have demonstrated excellent results on identifying a wide range of diseases. Recently, Vision Transformer (ViT) has garnered immense interest in the medical imaging community, numerous innovative approaches1,3,8,34–37 have emerged, leveraging ViTs as a foundation for further advancements in this field.
The other stream of work is generalist medical foundation models38–40. These models represent a paradigm shift in medical AI, aiming to create versatile, comprehensive AI systems capable of handling a wide range of tasks across different medical modalities, by leveraging large-scale, diverse medical data. MedPaLM M38 reaches performance competitive with or exceeding the state-of-the-art (SOTA) on various medical benchmarks, demonstrating the potential of the generalist foundation model in disease diagnosis and beyond. RadFM39 is the first medical foundation model capable of processing 3D multi-image inputs, demonstrating the versatility and adaptability of generalist models in processing complex imaging data. However, the development of GMAI models requires substantial computational power, which is often impractical for exploring sophisticated algorithms in academic labs.
In contrast, leveraging our proposed RP3D-DiagDS dataset, our final model RadDiag is capable of performing universal disease diagnosis and classification tasks within a constrained parameter scope, transcending the limitations of focusing on specific anatomies or disease categories. This capability offers a more versatile and efficient approach to healthcare management, resulting in significant clinical and research implications compared with precious AI models trained on former diagnostic datasets as discussed in Supplementary Section E. Our impacts and contributions can be summarized in the following three aspects:
First of all, our work can support model development for case-Level diagnosis. The RP3D-DiagDS dataset facilitates the creation of AI models capable of case-level diagnosis utilizing multi-modal and multi-scan inputs. This is a critical advancement since patients often undergo multiple radiological examinations across different medical departments during their treatment. Traditional disease classification models, which typically process single-image scans, are inadequate for such complex scenarios. Our dataset, with its case-level labels, promotes the development of comprehensive diagnostic AI systems, thereby addressing a significant gap in clinical practice.
Then, our work can help long-tailed rare disease diagnosis. Unlike existing datasets that primarily cover common diseases, the RP3D-DiagDS encompasses over 5000 disorders, including a significant number of rare diseases. This focus on long-tailed rare disease diagnosis is particularly important for clinical applications, as AI systems can readily assist in diagnosing common diseases but often fall short in identifying rare conditions. Our dataset thus plays a crucial role in enhancing the diagnostic process for rare diseases, which are typically more challenging to diagnose due to their lower prevalence.
Except the model development, we also present the value of online radiology resources. The RP3D-DiagDS dataset, collected from the Internet, serves as a valuable resource for training disease diagnosis model, especially for local medical centers dealing with rare diseases. Given the scarcity of cases for such diseases, our dataset’s ability to support few-shot learning scenarios is of paramount importance. It demonstrates the value that, such publicly shared medical data on the Internet represents a colossal and high-quality resource, holding the potential to pave the way for constructing generalist models aimed at advancing AI4healthcare.
Despite the effectiveness of our proposed dataset and architecture, there remain certain limitations. First, on model design, in the fusion step, we can use more image tokens to represent a scan rather than a pooled single vector. The latter may lead to excessive loss of image information during fusion. The model size can be further increased to investigate the effect of model capacities; Second, new loss functions should be explored to tackle such large-scale, long-tailed disorder/disease classification tasks. Third, on the disorder to ICD-10-CM mapping progress, the annotators are labeled on class-name level, i.e., only disorder names are provided, causing some ambiguous classes unable to find strict corresponding ICD-10-CM codes. Though we mark out this class in our shared data files, if providing more case-level information, the mapping could be more accurate. We treat this as future work.
Methods
In this section, we will detail the following aspects separately: dataset construction, model design, and experiment implementation. Notably, our study is based on data from Radiopaedia. It is a peer-reviewed open-edit radiology resource collection website, aiming to “create the best radiology reference available and to make it available for free, forever and for all”. We have asked various uploaders, along with the founder of Radiopaedia for non-commercial use permission and the relevant ethical regulations are controlled under Radiopaedia privacy-policy.
Dataset Construction
In this part, we present the details of our dataset, RP3D-DiagDS, that follows a similar procedure as RP3D39. Specifically, cases in our dataset are sourced from the Radiopaedia41—a growing peer-reviewed educational radiology resource website, that allows clinicians to upload 3D volumes to better reflect real clinical practice. According to the privacy clarification on Radiopaedia case publishing policy (https://radiopaedia.org/articles/case-publishing-guidelines-2), each case in our study has obtained clear permission from local institutions through either strict de-identification processes following institutional requirements and patient preferences, or through explicit informed consent when necessary. These privacy protection measures were completed during the initial case uploading phase, and all cases were subsequently published under appropriate licensing for non-commercial research use.
It is worth noting that, unlike RP3D39 that contains paired free-form text description and radiology scans for visual-language representation learning, here, we focus on multi-modal, multi-anatomy, and multi-label disease diagnosis (classification) under extremely unbalanced distribution. Overall, the proposed dataset contains 40,936 cases, of 195,010 images from 9 diverse imaging modalities and 7 human anatomy regions, note that, each case may contain images of multiple scans. The data covers 5,568 different disorders, that have been manually mapped into 930 ICD-10-CM7 codes.
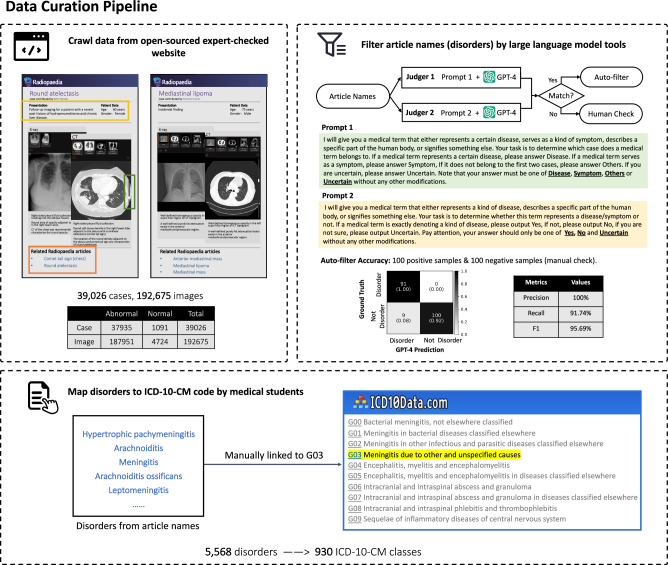
We describe the procedure of our data curation process, shown in Fig. 4. Specifically, we collect three main components from each case on Radiopaedia webpage, namely, “Patient data”, “Radiology images”, and “Articles” (example webpage shown in Fig. 4). “Patient data” includes brief information about this patient, for example, age, gender, etc. “Radiology images” denote a series of radiology examination scans. Notably, when uploading the cases, the physicians have also picked a default key slice to support diagnosis for each scan in “Radiology images”, we also crawl this as a piece of extra information. Figure 1a provides a statistical analysis on the number of images within one single case. “Articles” contains links to related articles named with corresponding disorders, which are treated as diagnosis labels and have been meticulously peer-reviewed by experts in Radiopaedia Editorial Board.
Fig. 4. The data collection pipeline.
First, we crawl data from an open-sourced and expert-checked website, including case descriptions, 2D/3D radiology images, and related Radiopaedia articles. Then, we filter article names by large language model tools (GPT-4) and check manually if mismatch. Finally, we map the article names into ICD-10-CM codes, cross-checked by a 10-year clinician.
To start with, we collect 50,970 cases linked with 10,670 articles from the Radiopaedia website. However, naively adopting the article titles as disorders may lead to ambiguities from three aspects: Firstly, not all articles are related to disorders. Secondly, article titles can be written with different granularity, for example, “pneumonia” and “bacterial pneumonia” can be referred to as different disorders, though they should ideally be arranged in a hierarchical structure. Thirdly, normal cases from Radiopaedia are not balanced in modalities and anatomies. Next, we discuss the 3-stage procedure proposed to alleviate the above-mentioned challenges.
Then, we deploy article filtering to clean the data. Specifically, we leverage the GPT-442 to automatically filter the article list, keeping those referring to disorders. Specifically, taking inspiration from the self-consistency prompts43, we design two different query prompts with similar meanings, as shown in Fig. 4. An article name is labeled as a disorder if GPT-4 consistently gives positive results from both prompts, while for those GPT-4 gives inconsistent results, we manually check them. To measure the quality of filtering, we randomly sample a portion of data for manual checking to control its quality. The confusion metrics are shown in Fig. 4. The 100% precision score indicates our auto-filtering strategy, can strictly ensure the left ones to be disordered. Eventually, 5342 articles can pass the first auto-criterion and 226 pass the second manually checking, resulting in 5,568 disorder classes, as shown in the upper right of Fig. 4. Cases not linked to any articles are excluded, ultimately yielding 38,858 cases.
Afterwards, we map disorders to ICD-10-CM codes. Upon getting disorder classes, they may fall into varying hierarchical granularity levels, here, we hope to map them to internationally recognized standards. For this purpose, we utilize the International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) codes, which is a customized version of the ICD-10 used for coding diagnoses in the U.S. healthcare system. The ICD was originally designed as a healthcare classification system, providing a system of diagnostic codes for classifying diseases, including nuanced classifications of a wide variety of signs, symptoms, abnormal findings, complaints, social circumstances, and external causes of injury or disease. Specifically, we hire ten medical Ph.D. students to manually map the article titles into ICD-10-CM codes. After cross-checking by a ten-year clinician, we have mapped the 5,568 disorders into the corresponding ICD-10-CM codes. We unify various disorders into the first hierarchical level of ICD-10-CM code tree, resulting in 930 ICD-10-CM classes. For example, both “J12.0 Adenoviral pneumonia” and “J12 Viral pneumonia” are mapped to the code “J12 Viral pneumonia, not elsewhere classified”, dismissing the ambiguities caused by diagnosis granularity. Note that, we will provide the ICD-10-CM codes along with the original 5,568 classes, each accompanied by its corresponding definition, thus, the dataset can be employed for training diagnosis or visual-language models.
At last, we supplement some normal cases. Despite there exist normal cases on Radiopaedia, covering most anatomies and modalities, we additionally collect more normal cases from additional external datasets, including Brain-Tumor-17, CheXpert, Vertebrase-Xray, KneeMRI, and VinDr-Mammo, and MISTR, with images available for research under a CC-BY-NC-SA 4.0 license, i.e., simply adding the normal case part introduced in them. The expanded normal cases include 2503 cases.
In total, we get 40,936 cases containing 195,010 images labeled by 5568 disorder classes and 930 ICD-10-CM classes and will continually maintain the dataset, growing the case number.
Model design
Our proposed RadDiag architecture consists of two key components: a visual encoder that processes 2D or 3D input scan and a transformer-based fusion module, merging all information to perform case-level diagnosis.
For visual encoder, we consider two popular variants of the visual encoder, namely, ResNet44 and ViT45, as shown in Fig. 5a. The visual encoding progress can generally be formulated as:
| 2 |
where denotes the input scan, C, H, W refer to the image channel, height, and width of the input scan respectively. D is optional, and only available for 3D input scans. The main challenge for architecture design comes from the requirement to process scans in both 2D and 3D formats. Here, we train separate normalization modules to convert the 2D or 3D inputs into feature maps of the same resolution and further passed into a shared encoding module. We here adopt three visual encoding variants to handle the challenge, i.e., ResNet-based, ViT-based, and ResNet-ViT-mixing.
For ResNet-based modules, considering 3D scans, they are first fed into 3D ResNet, followed by average pooling on depth to aggregate the information of the extra dimension; while for 2D scans, they are fed to the 2D ResNet to perform the same down-sampling ratio on height and width as for 3D. After normalization, both 2D and 3D scans are transformed into feature maps with the same resolution, , where is the intermediate feature dimension and h, w denote the normalized size. Then, the feature maps are passed into a shared ResNet to get the final visual embedding.
For ViT-based modules, regarding 3D scans, we convert the input volume into a series of non-overlapped 3D cubes, and pass them into MLP projection layers, to get vector embeddings; while for 2D scans, the input scan is broken into 2D patches, and projected into vector embeddings with another set of MLP layers. As ViT enables handling sequences of variable tokens, we can now pass the resulting vectors into a shared ViT to get the final visual embedding by confusing the feature embedding from arbitrary patch tokens. For position embedding, we adopt two sets of learnable position embeddings, one for 2D input, and the other for 3D, further indicating the network about the input dimension.
For ResNet-ViT-mixing designs, inspired by former works on pseudo-3D residual networks46. We, herein, also design a new 2D + 1D network architecture mixing both ResNet and ViT structures to encode 3D or 2D scans consistently. Specifically, for 3D scans, we treat them as a sequence of 2D slices, and for 2D scans, we also treat them as a sequence of 2D slices of length 1. We first adopt 2D ResNet to get normalized feature embedding per slice as . We then use transformer architecture to perform aggregation on the feature of slice tokens, and thus is similar to ViT-based method. Thanks to the flexibility of transformer architecture, the model can handle arbitrary number of tokens, obtaining the final visual embedding by integrating features from varying slice feature tokens.
Fig. 5. The overview of our method.
Three parts demonstrate our proposed visual encoders and fusion module, together with the knowledge enhancement strategy respectively. a The three types of vision encoder, i.e., ResNet-based, ViT-based, and ResNet-ViT-mixing. b The architecture of the fusion module. The figure shows the transformer-based fusion module, enabling case-level information fusion. c The knowledge enhancement strategy. We first pre-train a text encoder with extra medical knowledge with contrastive learning, leveraging synonyms, descriptions, and hierarchy. Then we view the text embedding as a natural classifier to guide the diagnosis classification.
Then, to fuse case-level information for case-level diagnosis, we propose to aggregate information from multiple scans with a trainable module. As shown in Fig. 5b, we adopt transformer encoders, specifically, we first initialize a set of learnable modality embeddings, denoted as {p1, …, pM}, where M denotes the total number of possible imaging modalities. Given certain visual embedding (vi) from modality j, we first add the corresponding modality embedding (pj) to it, indicating which radiologic modality it is from. Then, we feed all visual embeddings from one case into the fusion module, and output the fused visual embedding , from the “[CLS]” token, similar to paper45, denoted as:
| 3 |
where vi denotes the visual embedding for each image scan and mi denotes the its corresponding imaging modality, like MRI or CT. Till here, we have computed the case-level visual embedding, which can be passed into a classifier for disease diagnosis. We adopt a knowledge-enhanced training strategy1,8, that has shown to be superior in long-tailed recognition problems.
With knowledge enhancement, we hope to leverage the rich medical domain knowledge to enhance the long-tailed classification. Our key insight is that the long-tailed diseases may fundamentally have some shared symptoms or radiological pathologies with the head classes, which could be encoded in text format. In detail, we first pre-train a text encoder with medical knowledge, termed as knowledge encoder, where names of similar disorders or diseases are projected to similar embeddings, for example, “lung disease” is closer to “pneumonia” in the embedding space than “brain disease”. Then, we freeze the text embeddings, and use them to guide the training of the vision encoder, as shown in Fig. 5c.
To pre-train the knowledge encoder, we leverage several knowledge bases to pre-train a knowledge encoder, including Radiopaedia, ICD10-CM, and UMLS. Specifically, for each disorder term, we collect its definitions, radiologic features from Radiopaedia articles, synonyms, clinical information, and hierarchy structure from ICD10-CM, as well as definitions from UMLS. We aim to train the text encoder with the three considerations, i.e., synonyms, hierarchy, and descriptions. First, if two terms are identified as similar synonyms, we expect them to be close in text embedding space, like “Salmonella enteritis” and “Salmonella gastroenteritis”. Second, in the context of hierarchical relationships, we expect that if a disease is a fine-grained classification of another disease, its embeddings should be closer than those of unrelated diseases. For example, “J93, Pneumothorax and air leak” should exhibit closer embeddings with “J93.0, Spontaneous tension pneumothorax” than with “J96.0, Acute respiratory failure”. Third, for terms associated with descriptions, radiological features, or clinical information, we expect their embeddings to be close to the text embedding space. For example, “Intestinal Neuroendocrine Tumor” and “A well-differentiated, low or intermediate grade tumor with neuroendocrine differentiation that arises from the small or large intestine”.
We start from an off-the-shelf text encoder, namely, MedCPT-Query-Encoder47, and adopt contrastive learning for further finetuning. Given a target medical terminology name encoded by the text encoder, denoted as ftar, its corresponding medical texts, i.e., synonyms, containing terminologies or related descriptions are treated as positive cases. Similarly, we encode them with the text encoder, denoted as f+, and other non-related text embedding is treated as negative cases f−. Note that, we keep positive and negative cases consistent in format, e.g., when the positive case is related to a description sentence, the negative cases are always some non-related description sentences instead of the short name words. The final objective can be formulated as:
| 4 |
where τ refers to the temperature, and N denotes total sampled negative cases. By optimizing the contrastive loss we can further finetune the text encoder, resulting in a knowledge encoder, termed as Φk.
Knowledge-guided Classification. After training the knowledge encoder, we use it to encode the disorder/disease names into text embeddings, for example, denoting the names as {T1, T2, …, Tc}, where Tj is a disorder/disease name like “pneumonia” or “lung tumor”. We embed these free texts with the knowledge encoder as:
| 5 |
where tj denotes the text embeddings that are used for computing dot product with visual embeddings for case-level diagnosis:
| 6 |
where p is the final result. We use classical binary cross entropy (BCE) loss as the final training objective, denoted as:
| 7 |
Evaluation details
In this section, we will describe our evaluation details. First, we build up an internal test-set based on our proposed RP3D-DiagDS, serving for assessing the models’ ability on case-level, multi-modal, multi-scan, long-tailed disease diagnosis. Then we introduce more external datasets for assessing the transferring ability of our final model on other clinical benchmarks. Lastly, we introduce the used evaluaiton metrics, along with the compared baselines.
To start with, for better assessing the model’s ability on case-level diagnosis, we split a test set from our own proposed datasets, RP3D-DiagDS to perform internal evaluation. Specifically, we randomly split our dataset into training (train and validation) and testing sets in a (7:1):2 ratio.
We treat the class with more than (including equal) 100 positive cases as head classes, 30–100 (including 30) cases as medium classes, and less than 30 cases as tail classes. Consequently, from a total of 40,936 cases in our proposed RP3D-DiagDS, the training set comprises 30,880 cases, and the test set includes 7721 cases. Following on the dataset construction procedure, we perform classification tasks on two class sets at different granularities: (i) 5568 disorders + normal; (ii) 930 ICD-10-CM codes + normal. On different sets, we will have different head/medium/tail class sets following our definition. Note that, the normal class is always treated as one of the head classes. Next, we will only discuss the abnormal classes.
At the disorder level, there are 85 head classes, 470 medium classes, and 5014 tail classes. Notably, as the case number of some tailed classes is extremely small, some classes may only appear in training or test sets, but not both. This issue only happens to the tail classes, while our knowledge encoder enables embedding for unseen diseases, ensuring that evaluation is not affected by such challenge. As a result, there are 5260/3391 classes in our training and testing split, respectively.
At the ICD-10-CM level, disorders with similar meanings have been mapped to the same codes, for example, “tuberculosis”, and “primary pulmonary tuberculosis” all correspond to “A15 respiratory tuberculosis”. This merge operation results in more cases in one class and, consequently, more head classes. Specifically, we have 165 head classes, 230 medium classes, and 536 tail classes. Similarly, some tail classes may only be in training or testing split, resulting in 894/754 classes for our final training and testing splits, respectively.
It is important to point out that while we ensure the inclusion of head and medium classes in both the training and test sets, some tail classes are inevitably exclusive from certain divisions, which is treated as our future work to collect more samples of such disorders. We report the results separately for head/medium/tail classes while focusing primarily on the head classes.
In addition to evaluating on our own testset, we also consider zero-shot or finetuning evaluation on other external datasets, demonstrating its transferring abilities on image distribution shift and label space change. In the following sections, we start by introducing the used external datasets, that cover various medical imaging modalities and anatomies. Then we detail the fine-tuning settings. We choose an external evaluation dataset with three principles. Firstly, We hope to cover most radiologic modalities in our external evaluation, e.g., 2D X-ray, 2D CT/MRI slices, and 3D CT/MRI scans demonstrating our model can benefit all of them. Secondly, We hope to cover many human anatomies, e.g., brain, head and neck, chest, spine abdomen, and limb, demonstrating our model can benefit all of them. Finally, We hope to cover two cases, i.e., seen and unseen classes. Seen classes refer to those labels that have appeared in RP3D-DiagDS. Conversely, unseen extra classes denote those not included.
As a result, we pick CXR1414, VinDr-Spine48, VinDr-Mammo49, ADNI50, MosMedData51, NSCLC (Radiogenomics)52, TCGA (Merged), ISPY153, ChestX-Det1054, CheXpert4, COVID-19-Radio23,55, IU-Xray56, LNDb57, PadChest17, CC-CCII58, Brain-Tumor24, Brain-Tumor-1726, POCUS30, MURA59, KneeMRI60, CT-Kidney61 and report AUC scores to compare with others on the external evaluation. For detailed information regarding the dataset, please refer to the supplementary file.
We show the comparison between our model and the existing SOTAs on the external public benchmark. In detail, under the fine-tuning setting, we compare with various specialized state-of-the-arts (SOTAs) for each datasets. Due to the large number of external datasets, we herein do not provide the details of the specific designs per method, while directly indicating the corresponding method by citation in the table, and more details of the designs can be found in the corresponding papers.
In addition to the comparison with various specialized SOTAs, we also compare with other diagnosis foundation models in the zero-shot setting. In detail, we pick out related classes from our class set based on the target disease. It is possible that multiple similar classes in our dataset correspond to a single class in the external dataset due to the definition of class granularity, for example “Tumor” class in external dataset may be linked with “Brain Tumor”, “Chest Tumor” and so on in our class list. We treat the final prediction as positive if any class in the related class list is predicted as positive. Notably, as such union merging is carried out after thresholding the prediction scores, AUC score can not be calculated here. For comparison, we consider two types of baseline models. One is classification models. By training on a wide range of diagnosis data across various diseases, it enables to build a foundation model by simply optimizing the classification loss. Our work falls in this type, and another related work is RadIN12. Similarly to our zero-shot adaptation pipeline, we also adopt RadIN to other datasets zero-shotly, by choosing a most related class from its original class list. The other one is contrastive-learning models. In addition to using classification labels, leveraging vision-language pairs, also enables to build up a foundation model through optimizing contrastive loss62. Inspired by former works2,63, by treating the target disease class name as free-text, these models can also be adapted to zero-shot diagnosis leveraging the cross-modality similarity. We adopt the latest model, BiomedCLIP13 for comparison. It is trained with large-scale image datasets sourced from PubMedCentral papers, and can handle various imaging modalities. However, it is worth emphasizing that both RadIN and BiomedCLIP can only handle 2D input.
At last, we will describe the evaluation metrics in detail. Note that, the following metrics can all be calculated per class. For multi-class cases, we all use macro-average on classes to report the scores by default. For example, “AUC” for multi-class classification denotes the “Macro-averaged AUC on classes”. We report all metrics in percent (%) in tables.
AUC. Area Under Curve64 denotes the area under ROC (receiver operating characteristic) curve. This has been widely used in medical diagnosis, due to its clinical meanings and robustness in unbalanced distribution.
AP. Average Precision (AP) is calculated as the weighted mean of precisions at each threshold. Specifically, for each class, we rank all samples according to the prediction score, then shift the threshold to a series of precision-recall points and draw the precision-recall (PR) curve, AP equals the area under the curve. This score measures whether the unhealthy samples are ranked higher than healthy ones. We report Mean Average Precision (mAP), which is the average of AP of each class.
- F1, MCC and ACC. F1 score is the harmonic mean of the precision and recall. It is widely used in diagnosis tasks. ACC denotes the accuracy score. MCC65 denotes the Matthews Correlation Coefficient metric. It ranges from − 1 to 1 and can be calculated as follow:
All the three metrics need a specific threshold to compute, and we choose one by maximizing F1 on the validation set following former papers1,8,22,34.8 Recall@FPR. We also report the recall scores at different false positive rate (FPR), i.e., sample points from class-wise ROC curves. Specifically, We report the Recall@0.01, Recall@0.05, Recall@0.1 score, denoting the recall scores at 0.01, 0.05, 0.1 FPR respectively.
Implementation
In this part, we will introduce implementation details such as the input and output during training and testing, model hyperparameters, and resource consumption information.
As input to the model, the images are processed into shape of (H, W, D), where H and W represent the image height and width, both set to 256, and D represents the image depth, set to 32. To standardize the processing of both 2D and 3D images, we treat the 3D images as a stack of 2D slices. For images that do not conform to the specified dimensions, we employ bilinear interpolation for the height and width dimensions and uniform resampling for the depth dimension. Finally, we apply feature scaling for normalization.
Based on the results of the ablation study for our model architecture, we select ResNet-50 and a transformer encoder for the visual encoder component. The features output by ResNet-50 have a shape of (2048, 8, 8). After applying average pooling, these features are passed through a transformer encoder to obtain features with a shape of (33, 2048). We choose the features of the first [cls] token for subsequent fusion among different scans. In the transformer encoder used for fusion, the number of heads is set to 4, the number of layers is set to 2, and the output feature dimension is 2048. Finally, through interaction with the knowledge embedding and a fully connected layer, we obtain the final prediction results with a shape of class numbers. For more detailed information, please refer to our Github repository.
The training and testing for this work are conducted using Nvidia A100s (80G). During training, we apply image-level augmentation transformations. Additionally, considering that for each scan we have the manually picked key slice, we will also randomly replace the 3D scan with its 2D key slice. When both early fusion and data augmentation are activated, the GPU memory usage for training is approximately 46G with a batch size of 1. During inference, no data augmentation or random mix-up strategies are activated. Under this setting, GPU memory usage is approximately 7G with early fusion and a batch size of 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Source Data for Supplementary Fig.5,6,7,8,9
Acknowledgements
This work is supported by the National Key R&D Program of China (No. 2022ZD0160702), STCSM (No. 22511106101, No. 18DZ2270700, No. 21DZ1100100), 111 plan (No. BP0719010), and State Key Laboratory of UHD Video and Audio Production and Presentation. Additionally, We sincerely thank all the contributors who uploaded the relevant data to Radiopaedia. We appreciate their willingness to make these valuable cases publicly available, promoting the related research greatly.
Author contributions
All listed authors clearly meet the ICMJE 4 criteria. Specifically, Qiaoyu Zheng, Weike Zhao, Chaoyi Wu, Xiaoman Zhang, Lisong Dai, Hengyu Guan, Yuehua Li, Ya Zhang, Yanfeng Wang, and Weidi Xie all make contributions to the conception or design of the work, and Qiaoyu Zheng, Weike Zhao, Chaoyi Wu further perform acquisition, analysis, or interpretation of data for the work. Lisong Dai and Yuehua Li as experienced clinicians help check the correctness of cases and confirm the medical related knowledge. In writing, Qiaoyu Zheng, Weike Zhao, Chaoyi Wu draft the work and Xiaoman Zhang, Lisong Dai, Hengyu Guan, Yuehua Li, Ya Zhang, Yanfeng Wang, and Weidi Xie review it critically for important intellectual content. All authors approve of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Qiaoyu Zheng, Weike Zhao, Chaoyi Wu contribute equally to the work. Weidi Xie and Yanfeng Wang are the corresponding authors.
Peer review
Peer review information
Nature Communications thanks Marc Dewey, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Due to the Radiopaedia license, we do not directly provide the original dataset. The index data generated in this study have been deposited in the Hugging Face under accession code https://huggingface.co/datasets/QiaoyuZheng/RP3D-DiagDS-Index. The index data are available under the MIT license. The raw data containing the full dataset are available under restricted access due to data licensing reasons. Access can be obtained by contacting Radiopaedia for non-commercial licensing and then reaching out to the article authors. The metadata for the full dataset will then be available at https://huggingface.co/datasets/QiaoyuZheng/RP3D-DiagDS. We also provide the model checkpoint in https://huggingface.co/QiaoyuZheng/RP3D-DiagModel following the apache-2.0 open-source license and all related result numbers can be reproduced based on the provided model checkpoint. Source data for tables and figures is provided with this paper.
Code availability
Our codes can be found in https://github.com/qiaoyu-zheng/RP3D-Diag under the MIT License, cited as66 with all training, evaluation, fine-tuning, and zero-shot evaluation information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qiaoyu Zheng, Weike Zhao, Chaoyi Wu.
Contributor Information
Yanfeng Wang, Email: wangyanfeng622@sjtu.edu.cn.
Weidi Xie, Email: weidi@sjtu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54424-6.
References
- 1.Zhang, X., Wu, C., Zhang, Y., Xie, W. & Wang, Y. Knowledge-enhanced visual-language pre-training on chest radiology images. Nat. Commun.14, 4542 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiu, E. et al. Expert-level detection of pathologies from unannotated chest x-ray images via self-supervised learning. Nat. Biomed. Eng.6, 1399–1406 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai, Y., Gao, Y. & Liu, F. Transmed: transformers advance multi-modal medical image classification, Diagnostics, 11, 1384 (2021). [DOI] [PMC free article] [PubMed]
- 4.Irvin, J. et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison, in Proceedings of the AAAI Conference on Artificial Intelligence.1, 590–597 (2019).
- 5.Bien, N. et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of mrnet, PLoS Medicine, 15, (2018). [DOI] [PMC free article] [PubMed]
- 6.Majkowska, A. et al. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology294, 421–431 (2020). [DOI] [PubMed] [Google Scholar]
- 7.ICD10, https://www.icd10data.com/ICD10CM/Codes (2023).
- 8.Wu, C., Zhang, X., Wang, Y., Zhang, Y. & Xie, W. K-diag: Knowledge-enhanced disease diagnosis in radiographic imaging, Medical Image Computing and Computer Assisted Intervention – MICCAI Workshop (2023).
- 9.Zhang, Y., Jiang, H., Miura, Y., Manning, C. D. & Langlotz, C. P. Contrastive learning of medical visual representations from paired images and text, in Machine Learning for Healthcare Conference. PMLR, 2–25 (2022).
- 10.Joyce, D. W., Kormilitzin, A., Smith, K. A. & Cipriani, A. Explainable artificial intelligence for mental health through transparency and interpretability for understandability, npj Digital Medicine, 6,(2023). [DOI] [PMC free article] [PubMed]
- 11.Muhammad, M. B. & Yeasin, M. Eigen-cam: Class activation map using principal components, in 2020 international joint conference on neural networks (IJCNN). IEEE,1–7(2020).
- 12.Mei, X. et al. Radimagenet: an open radiologic deep learning research dataset for effective transfer learning. Radiology: Artif. Intell.4, e210315 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, S. et al. Biomedclip: a multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs, arXiv preprint arXiv:2303.00915, (2023).
- 14.Wang, X. et al. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2097–2106 (2017).
- 15.Johnson, A. E. et al. Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data6, 317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen, N. H., Pham, H. H., Tran, T. T., Nguyen, T. N. & Nguyen, H. Q., VinDr-PCXR: An open, large-scale chest radiograph dataset for interpretation of common thoracic diseases in children, medRxiv10.1101/2022.03.04.22271937, 2022–03, (2022). [DOI] [PMC free article] [PubMed]
- 17.Bustos, A., Pertusa, A., Salinas, J.-M. & Iglesia-Vayá, Mdela Padchest: A large chest x-ray image dataset with multi-label annotated reports. Med. Image Anal.66, 101797 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Healthcare, J. Object-CXR - Automatic detection of foreign objects on chest X-rays, https://web.archive.org/web/20201127235812/https://jfhealthcare.github.io/object-CXR/ (2020).
- 19.Jaeger, S. et al. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg.4, 475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih, G. et al. Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia. Radiology: Artif. Intell.1, e180041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filice, R. W. et al. Crowdsourcing pneumothorax annotations using machine learning annotations on the nih chest x-ray dataset. J. Digital Imaging33, 490–496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen, H. Q. et al. VinDr-CXR: An open dataset of chest X-rays with radiologist’s annotations. Sci. Data9, 429 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury, M. E. et al. Can AI help in screening viral and covid-19 pneumonia. IEEE Access8, 132,665–132 676 (2020). [Google Scholar]
- 24.Nickparvar, M. Brain tumor mri dataset, https://www.kaggle.com/dsv/2645886 (2021).
- 25.Gupta, S. Brain MRI scans for brain tumor classification, https://www.kaggle.com/datasets/shreyag1103/brain-mri-scans-for-brain-tumor-classification (2023).
- 26.Feltrin, F. Brain tumor MRI images 17 classes, https://www.kaggle.com/datasets/fernando2rad/brain-tumor-mri-images-17-classes (2022).
- 27.Koenig, L. N. et al. Select atrophied regions in Alzheimer's disease (SARA): An improved volumetric model for identifying Alzheimer's disease dementia. NeuroImage: Clin.26, 102248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcus, D. S. et al. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci.19, 1498–1507 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Hssayeni, M. D. et al. Intracranial hemorrhage segmentation using a deep convolutional model. Data5, 14 (2020). [Google Scholar]
- 30.Born, J. et al. POCOVID-Net: automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS), arXiv preprint arXiv:2004.12084, (2020).
- 31.Armato III, S. G. et al. The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Med. Phys.38, 915–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deepak, S. & Ameer, P. Brain tumor classification using deep CNN features via transfer learning. Computers Biol. Med.111, 103345 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Swati, Z. N. K. et al. Brain tumor classification for MR images using transfer learning and fine-tuning. Computerized Med. Imaging Graph.75, 34–46 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Wu, C., Zhang, X., Zhang, Y., Wang, Y. & Xie, W. MedKLIP: Medical Knowledge Enhanced Language-Image Pre-Training, IEEE International Conference on Computer Vision (ICCV), 2023.
- 35.Park, S. et al. Vision transformer for covid-19 cxr diagnosis using chest x-ray feature corpus, arXiv preprint arXiv:2103.07055, (2021).
- 36.Liu, C. & Yin, Q. Automatic diagnosis of covid-19 using a tailored transformer-like network, in Journal of Physics: Conference Series, no. 1. IOP Publishing, 2021, pp. 012175.
- 37.Gao, X., Qian, Y. & Gao, A. COVID-VIT: classification of COVID-19 from CT chest images based on vision transformer models. In 2022 3rd International Conference on Next Generation Computing Applications (NextComp) 1–4 (IEEE, 2022).
- 38.Tu, T. et al. Towards generalist biomedical ai. NEJM AI. 1, AIoa2300138 (2024).
- 39.Wu, C., Zhang, X., Zhang, Y., Wang, Y. & Xie, W. Towards generalist foundation model for radiology by leveraging web-scale 2d&3d medical data, arXiv preprint arXiv:2308.02463, 2023.
- 40.Zhang, X. et al. Pmc-vqa: Visual instruction tuning for medical visual question answering, arXiv preprint arXiv:2305.10415, 2023.
- 41.Radiopaedia, https://radiopaedia.org, Accessed: May 2023.
- 42.OpenAI, GPT-4 Technical Report, arXiv preprint arXiv:2303.08774, 2023.
- 43.Wang, X. et al. Self-consistency improves chain of thought reasoning in language models. In The Eleventh International Conference on Learning Representations (ICLR, 2023).
- 44.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition, 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778, 2015. [Online]. Available: https://api.semanticscholar.org/CorpusID:206594692.
- 45.Dosovitskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. In International Conference on Learning Representations (ICLR, 2020).
- 46.Qiu, Z., Yao, T. & Mei, T. Learning spatio-temporal representation with pseudo-3d residual networks, in Proceedings of the IEEE International Conference on Computer Vision (ICCV), (2017).
- 47.Jin, Q. et al. Medcpt: Contrastive pre-trained transformers with large-scale pubmed search logs for zero-shot biomedical information retrieval. Bioinformatics39, btad651 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen, H. T. et al. VinDr-SpineXR: A deep learning framework for spinal lesions detection and classification from radiographs, in Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part V 24. Springer, 291–301 (2021).
- 49.Nguyen, H. T. et al. VinDr-Mammo: A large-scale benchmark dataset for computer-aided diagnosis in full-field digital mammography. Sci. Data10, 277 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jack, C. R. et al. The Alzheimer’s disease neuroimaging initiative (Adni): Mri methods, Journal of Magnetic Resonance Imaging, 27, 2008. [DOI] [PMC free article] [PubMed]
- 51.Morozov, S. P. et al. Mosmeddata: Chest CT scans with COVID-19 related findings dataset, arXiv preprint arXiv:2005.06465, (2020).
- 52.Bakr, S. et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data5, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hylton, N. M. et al. Neoadjuvant chemotherapy for breast cancer: functional tumor volume by MR imaging predicts recurrence-free survival-results from the acrin 6657/calgb 150007 i-spy 1 trial. Radiology279, 44–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, J., Lian, J. & Yu, Y. Chestx-det10: Chest x-ray dataset on detection of thoracic abnormalities, arXiv preprint arXiv:2006.10550v3, (2020).
- 55.Rahman, T. et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest x-ray images. Computers Biol. Med.132, 104319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pavlopoulos, J., Kougia, V. & Androutsopoulos, I. A survey on biomedical image captioning, in Proceedings of the second workshop on shortcomings in vision and language. 26–36 (2019).
- 57.Pedrosa, J. et al. Lndb: a lung nodule database on computed tomography, arXiv preprint arXiv:1911.08434, (2019).
- 58.Zhang, K. et al. Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell181, 1423–1433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajpurkar, P. et al. Mura: Large dataset for abnormality detection in musculoskeletal radiographs, arXiv preprint arXiv:1712.06957 (2017).
- 60.Štajduhar, I., Mamula, M., Miletić, D. & Uenal, G. Semi-automated detection of anterior cruciate ligament injury from mri. Computer methods Prog. biomedicine140, 151–164 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Islam, M. N. et al. Vision transformer and explainable transfer learning models for auto detection of kidney cyst, stone and tumor from ct-radiography. Sci. Rep.12, 1–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations, in International conference on machine learning. PMLR, 1597–1607 (2020).
- 63.Lin, W. et al. Pmc-clip: Contrastive language-image pre-training using biomedical documents, in Medical Image Computing and Computer Assisted Intervention – MICCAI, (2023).
- 64.Bradley, A. P. The use of the area under the roc curve in the evaluation of machine learning algorithms. Pattern Recognit.30, 1145–1159 (1997). [Google Scholar]
- 65.Chicco, D. & Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over f1 score and accuracy in binary classification evaluation. BMC Genomics21, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng, Q. et al. Large-scale long-tailed disease diagnosis on radioloy images. https://zenodo.org/records/13871377 (2024). [DOI] [PubMed]
- 67.Bhat, S. et al. Aucreshaping: improved sensitivity at high-specificity. Sci. Rep.13, 21097 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korolev, S., Safiullin, A., Belyaev, M. & Dodonova, Y. Residual and plain convolutional neural networks for 3D brain MRI classification, in Proceedings-International Symposium on Biomedical Imaging, 835–838 (2017).
- 69.Draelos, R. L. et al. Machine-learning-based multiple abnormality prediction with large-scale chest computed tomography volumes. Med. image Anal.67, 101857 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Born, J. et al. Accelerating detection of lung pathologies with explainable ultrasound image analysis. Appl. Sci.11, 672 (2021). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Source Data for Supplementary Fig.5,6,7,8,9
Data Availability Statement
Due to the Radiopaedia license, we do not directly provide the original dataset. The index data generated in this study have been deposited in the Hugging Face under accession code https://huggingface.co/datasets/QiaoyuZheng/RP3D-DiagDS-Index. The index data are available under the MIT license. The raw data containing the full dataset are available under restricted access due to data licensing reasons. Access can be obtained by contacting Radiopaedia for non-commercial licensing and then reaching out to the article authors. The metadata for the full dataset will then be available at https://huggingface.co/datasets/QiaoyuZheng/RP3D-DiagDS. We also provide the model checkpoint in https://huggingface.co/QiaoyuZheng/RP3D-DiagModel following the apache-2.0 open-source license and all related result numbers can be reproduced based on the provided model checkpoint. Source data for tables and figures is provided with this paper.
Our codes can be found in https://github.com/qiaoyu-zheng/RP3D-Diag under the MIT License, cited as66 with all training, evaluation, fine-tuning, and zero-shot evaluation information.