Abstract
The use of time lapse systems (TLS) in In Vitro Fertilization (IVF) labs to record developing embryos has paved the way for deep-learning based computer vision algorithms to assist embryologists in their morphokinetic evaluation. Today, most of the literature has characterized algorithms that predict pregnancy, ploidy or blastocyst quality, leaving to the side the task of identifying key morphokinetic events. Using a dataset of N = 1909 embryos collected from multiple clinics equipped with EMBRYOSCOPE/EMBRYOSCOPE+ (Vitrolife), GERI (Genea Biomedx) or MIRI (Esco Medical), this study proposes a novel deep-learning architecture to automatically detect 11 kinetic events (from 1-cell to blastocyst). First, a Transformer based video backbone was trained with a custom metric inspired by reverse cross-entropy which enables the model to learn the ordinal structure of the events. Second, embeddings were extracted from the backbone and passed into a Gated Recurrent Unit (GRU) sequence model to account for kinetic dependencies. A weighted average of 66.0%, 67.6% and 66.3% in timing precision, recall and F1-score respectively was reached on a test set of 278 embryos, with a model applicable to multiple TLS.
Keywords: Embryology, Machine learning, Medical Imaging
Subject terms: Machine learning, Embryology
Introduction
With the increasing penetration of time lapse systems in IVF clinics, a growing number of publications have investigated the association between embryo kinetics and embryo viability1–7. This technology has made the monitoring of embryonic development even more important but also more tedious and time consuming for IVF practicians. Furthermore, as shown in Refs.8,9, inter-operator and inter-laboratory agreement on the identification of embryo kinetic events is variable, with a standard deviation ranging from 0.5 h up to 5.4 h. Recent advancements in computer vision and deep learning (DL) could therefore help minimize the need for manual identification while empowering embryologists to make robust and standardized decisions.
Automatic and semi-automatic methods have been developed recently to detect embryo development stages from time lapse systems (TLS) images. Many publications have focused on detecting a single morphokinetic event such as the number of pronuclei10, blastocyst formation11 or direct cleavage12. References13,14 detected several events, but only early ones from 1 cell to 4 cells+. In Ref.15, the suggested method developed on EMBRYOSCOPE videos from a single clinic could only detect cleavage stages as it is counting the number of cells. Reference16 used a multi-task DL network to classify frames, until the fourth cell division only, from EMBRYOSCOPE+ (Vitrolife) videos extracted from a single clinic. In contrast, Ref.17 proposed an automatic annotation system from 1-cell to blastocyst stage, based on gray level coefficient of variation and detection of the zona pellucida thickness. This system was however developed on videos extracted from a single clinic equipped with EMBRYOSCOPE, and was based on classical computer vision algorithms using human-based feature engineering which is at risk of failing to generalize to all embryos. Moreover, their processing time of 20 min per video makes this solution unsuitable for real-time clinic use.
Most publications, even those that have identified more kinetic events, have however characterized their model based on individual frames. This can be problematic because seemingly small inaccuracies at the frame level, can entail missing on the real transition from one event to the other or incoherent kinetic chronology. Recent advances have consisted of applying post processing methods to clean the sequence of predictions at the video-level, to enforce a biologically relevant timeline14,16,18,19. Reference20 built a linear-chain conditional random field on top of a two-stream convolutional neural network to model the monotonic constraints of biological events. However, Refs.16,18–20 still only report accuracy at the frame level which does not capture the biological chronology of the detection in the entire video. References14,16 reported the root mean squared error (RMSE) at the video level, but it is not the most suited metric to capture the ability of the model to precisely detect timing of events. Indeed predictions could constantly oscillate between adjacent events from one frame to the next keeping the RMSE low but making no sense from a biological point of view.
Reference21 therefore suggested a new temporal accuracy metric based on the time difference between biological events predicted transition and actual transition. If the time difference was above a certain threshold the detection was considered inaccurate. Different thresholds were used for each biological event based on the intra-operator standard deviation observed in Ref.9. They reported an average temporal accuracy of 0.66 for the best model. This metric offered the advantage of being more easily interpreted by clinicians, showing how often the model was able to correctly detect events within a certain window. However, performance per event was not reported and since each event uses a specific window of acceptability it is difficult to interpret the reported global temporal accuracy.
In this study, we propose a new model characterized with a more complete set of metrics, reporting not only the timing accuracy but also timing precision, recall and F1-score using a fixed timing window of 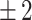 h for all kinetic events. This allows better assessment of the biological coherence of the predicted sequence of events, and therefore the added value of using a DL model to annotate embryo kinetic events in a real clinical setting. Moreover, the DL algorithm described in this study establishes competitive performances on data from three TLS collected across several clinics unlike existing publications that have focused on data from a single clinic and TLS.
h for all kinetic events. This allows better assessment of the biological coherence of the predicted sequence of events, and therefore the added value of using a DL model to annotate embryo kinetic events in a real clinical setting. Moreover, the DL algorithm described in this study establishes competitive performances on data from three TLS collected across several clinics unlike existing publications that have focused on data from a single clinic and TLS.
Materials and methods
Data collection
Data from 3 different Time Lapse Systems (TLS) was used to train the Biological Event Extraction (BEE) model collected either through: (i) an open source dataset21 composed of 702 EmbryoScope videos (of which 24 appeared to be mislabeled and thus were discarded following visual inspection) gathered from a single center between 2011 and 2019 (ii) 4 partner clinics which amounted to N = 1184 retrospective videos, of which 981 from a clinic in France equipped with an EMBRYOSCOPE (Vitrolife), N = 103 from a clinic in France equipped with a GERI (Genea Biomedx) and N = 100 from two clinics in France and Spain equipped with a MIRI (Esco Medical). Regarding non-open source data, for EMBRYOSCOPE a single senior embryologist (5+ years of experience) manually annotated the videos through the EmbryoViewer (without using any of the semi-automatic annotations provided by the software), for GERI and MIRI another senior embryologist (5+ years of experience) manually annotated the videos through an annotation platform which was setup specifically for this study. No automatic labels were used in this study.
Collection of non-opensource retrospective data for this study (N = 1184 videos) was exempted from ethical review and approval, and from the requirement for informed consent, because of the retrospective nature of the analyses, and de-identification of data. We performed and declared our research and development of the algorithms under the reference methodology MR-004 produced by the Commission Nationale de l’Informatique et des Libertés (CNIL), for research not involving the human person in the health field. Due to the retrospective nature of the study, the Commission Nationale de l’Informatique et des Libertés (CNIL) waived the need of obtaining informed consent and approved the current study. All methods were performed in accordance with the relevant guidelines and regulations.
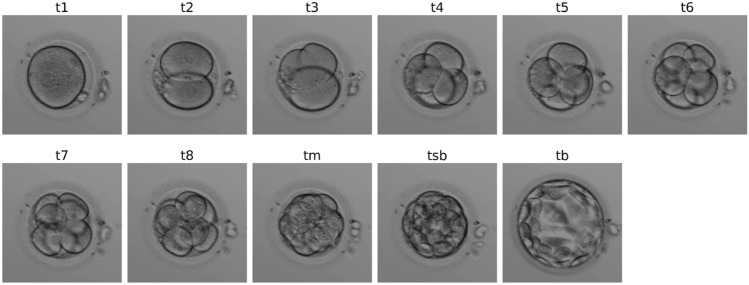
For each video, the assigned embryologist was asked to identify all the frames corresponding to the following 11 biological events, according to the guidelines from Ref.22, in order to reduce as much as possible inter-expert variability: t1, t2, t3, t4, t5, t6, t7, ie 1-cell to 7-cell stages, t8 (8+ cells), tm (morula), tsb (start of blastulation) and tb (full blastocyst). Figure S2 in supplementary material displays the temporal distribution of events per annotator. An example of these stages is shown in Fig. 1. Note that the open-source dataset’s annotations have more biological events, namely tPB2 (polar body detachment), tPNF (pronuclei fading), tEB (extended blastocyst) and tH (hatched blastocyst). Therefore these events were merged into broader categories, i.e. 1-cell (tPB2 & tPNF) and tB (tEB & tH).
Figure 1.
Images of kinetic events from an embryo recorded by a GERI as identified by an embryologist.
The complete dataset thus comprised N = 1909 videos with N = 14,696 corresponding biological events annotated ranging from t1 to t8+, tm, tsb, and tb. Each event was identified with its corresponding start and end frame indices. In total, N = 339,834 frames were annotated. Table 1 presents the distribution of events in the dataset. Videos in the dataset last on average 99.5 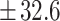 h post insemination (hpi). N = 1305 (68%) and N = 324 (17%) videos were randomly sampled to create the training and validation datasets, respectively. The remaining N = 278 (15%) videos were used as an independent test set, including 41 videos of non developing embryos (defined as embryos which only developed to early cleavage stages, or did not cleave at all throughout 4 days of development) selected from 6 clinics in France. 12% of the test set corresponded to MIRI videos, 17% to GERI, 61% to EMBRYOSCOPE and 10% to EMBRYOSCOPE+.
h post insemination (hpi). N = 1305 (68%) and N = 324 (17%) videos were randomly sampled to create the training and validation datasets, respectively. The remaining N = 278 (15%) videos were used as an independent test set, including 41 videos of non developing embryos (defined as embryos which only developed to early cleavage stages, or did not cleave at all throughout 4 days of development) selected from 6 clinics in France. 12% of the test set corresponded to MIRI videos, 17% to GERI, 61% to EMBRYOSCOPE and 10% to EMBRYOSCOPE+.
Table 1.
Distribution of embryo kinetic events in the complete dataset.
| Biological event | Number of videos (% of total videos) | Number of frames (% of total frames) |
|---|---|---|
| t1 (1-cell) | 1887 (98.8%) | 49,611 (14.6%) |
| t2 (2-cells) | 1828 (95.7%) | 47,338 (13.9%) |
| t3 (3-cells) | 1470 (77%) | 8440 (2.5%) |
| t4 (4-cells) | 1698 (88.9%) | 47,635 (14%) |
| t5 (5-cells) | 1425 (74.6%) | 8071 (2.4%) |
| t6 (6-cells) | 749 (39.2%) | 8387 (2.5%) |
| t7 (7-cells) | 685 (35.8%) | 9505 (2.8%) |
| t8+ (8+-cells) | 1498 (78.5%) | 70,981 (20.9%) |
| tM (morula) | 728 (38.1%) | 25,820 (7.6%) |
| tSB (start blastulation) | 1355 (71.0%) | 29,161 (8.6%) |
| tB (blastocyst) | 1373 (71.9%) | 34,885 (10.3%) |
| Total | 1909 | 339,834 |
All video labels were anonymized and no patient data nor clinical data was collected.
Data preparation
All videos were pre-processed as described in Ref.23, and went through the following steps: temporal normalization, embryo cropping, resizing, removal of the final empty frames and luminosity correction. The goal of this preprocessing was to set all videos to a common interval time (20 min) between each frame, reducing video length by a factor of 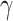 . Other steps involved cropping each frame of the video around the embryo, reducing the video resolution to 192
. Other steps involved cropping each frame of the video around the embryo, reducing the video resolution to 192 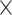 192 and standardizing the luminosity and contrast.
192 and standardizing the luminosity and contrast.
Deep learning model
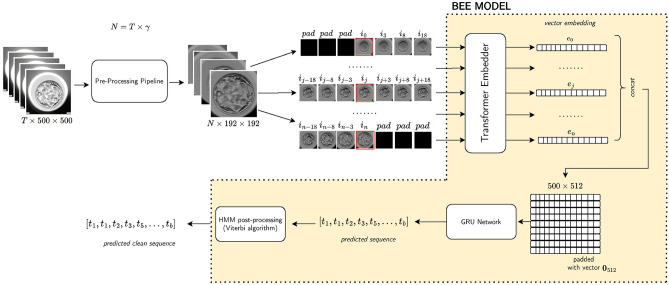
The BEE model was made of 3 main blocks, as illustrated in Fig. 2: (i) a visual encoder to extract a 512-dimensional embedding for each frame in the video, which is based on the Uniformer architecture from Ref.24, (ii) a sequence model which takes as input the frame embeddings of the entire embryo video, using Gated Recurrent Unit (GRU) layers as introduced in Ref.25, (iii) a Hidden Markov Model (HMM) applied on the sequence of scores outputted by the sequence model to smooth the predictions and create a physiologically plausible sequence of events, similarly to Ref.21.
Figure 2.
Biological event extraction (BEE) pipeline: raw videos with T frames are cropped and normalized to 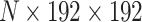 , where
, where 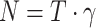 is the number of frames that can span from 80 to 150 hpi. A sub-clip of 7 frames is constructed with the frame of interest as the central frame. These sub-clips are passed to a Transformer model to extract 512-d embeddings. The sequence of N embeddings is fed into a GRU network to output
is the number of frames that can span from 80 to 150 hpi. A sub-clip of 7 frames is constructed with the frame of interest as the central frame. These sub-clips are passed to a Transformer model to extract 512-d embeddings. The sequence of N embeddings is fed into a GRU network to output 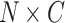 scores, where C is the number of classes. These scores are then converted to predictions using an HMM model.
scores, where C is the number of classes. These scores are then converted to predictions using an HMM model.
The training of the model was done in two stages. First, given a sub-clip of 7 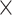 192
192  192 frames from a video, the visual encoder is trained to predict the class of the central frame, being given a context of 3 frames before and 3 frames after, with an increasing stride the further away from the central frame. The second stage consisted of training a sequence model, using as input the embeddings of each frame of the video extracted from the visual encoder. This two-stage design was required as it would have not been possible to fit in GPU memory 300+ sub-clips of 7 frames to process an entire video end to end. Indeed, embryo videos of 5 days of development have 350 frames on average.
192 frames from a video, the visual encoder is trained to predict the class of the central frame, being given a context of 3 frames before and 3 frames after, with an increasing stride the further away from the central frame. The second stage consisted of training a sequence model, using as input the embeddings of each frame of the video extracted from the visual encoder. This two-stage design was required as it would have not been possible to fit in GPU memory 300+ sub-clips of 7 frames to process an entire video end to end. Indeed, embryo videos of 5 days of development have 350 frames on average.
During the training of the visual encoder, data augmentation techniques were applied to prevent overfitting and incite the model to discover patterns that are invariant to certain transformations. Specifically, the augmentations used were random horizontal and vertical flip, random color jitter, random resize and crop, random gaussian blur, random mean pixel gaussian noise. The model was trained for 60 epochs, starting from Kinetics-400 pre-trained weights, with 4 GPUs and a batch size of 32 sub-clips, using the SGD optimizer with a learning rate of 0.005. A custom loss is employed, which mixes cross-entropy (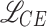 ) and an inspiration from reverse cross-entropy26 to penalize predicted events that are far from the ground truth event. Reverse cross-entropy is defined as
) and an inspiration from reverse cross-entropy26 to penalize predicted events that are far from the ground truth event. Reverse cross-entropy is defined as 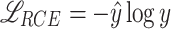 where
where 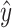 is the predicted score and y is the ground truth. In this implementation (
is the predicted score and y is the ground truth. In this implementation (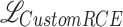 ), designed specifically for this study, y is replaced with
), designed specifically for this study, y is replaced with 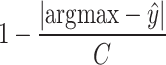 , where
, where 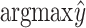 is the predicted class and C is the total number of classes. The final loss is
is the predicted class and C is the total number of classes. The final loss is 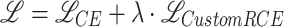 . In our experiments
. In our experiments 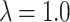 gave best results.
gave best results.
The sequence model was a 2-layer bidirectional Gated Recurrent Unit (GRU) using a 32-dimensional hidden size. GRU25 is a type of Recurrent Neural Network similar to Long Short Term Memory27 but without context vector and output gate which makes it more parameter efficient. In order to train this sequence model, 512-dimensional embeddings are extracted for every frame in the training, validation and test videos using the trained visual encoder, with a maximum sequence length of 500.
To leverage a large amount of data, the visual encoder’s embeddings extracted from the training set were used to train the GRU model. However, to avoid overfitting these training embeddings, random gaussian noise (with standard deviation of 0.05) is applied as well as random cutting of the sequence with a minimum length of 150 for the cut down sequence. The sequence model is trained using Adam optimizer (lr = 0.0004) with cross-entropy loss (ignoring the padding class) with label smoothing and class weights to account for under-represented events. Indeed, some events are much shorter and sometimes not present in annotations (eg t3, t5...), and therefore have much fewer frames with this assigned label. The model is trained with early stopping to keep the best validation weights.
Finally, during inference, an HMM was used to smooth the predictions outputted by the sequence model in order to make the sequence of events predicted more biologically plausible. To this end, a transition matrix is computed based on empirical observations from the dataset, meaning the probability of transitioning from state a to state b is equal to the proportion of such transitions observed in the dataset. The Viterbi algorithm28 is applied to find the most likely sequence of events given the sequence model scores for each frame in the video.
Model evaluation
To assess the model in a clinically relevant setting, its ability to correctly identify the starting time of a biological event in the video was evaluated. A detection was considered correct if the starting time of the event was detected within 2 h of the starting time defined by the embryologist; otherwise it is considered incorrect. This arbitrary threshold was fixed after discussion with embryologists, as it targets a minimum of performance seen as acceptable by the experts. A common window size between all events allows a fair comparison of the metrics between each class. The detection was also considered incorrect if the model detects an event that was not identified by the embryologist. As such, the following metrics were used for each biological event class: 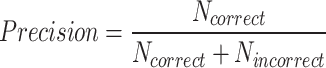 ,
, 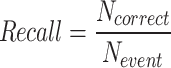 where
where 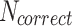 is the number of correct detections (True Positives),
is the number of correct detections (True Positives), 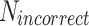 the number of incorrect detections (False Positives) and
the number of incorrect detections (False Positives) and 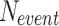 the number of videos where the event was annotated, meaning
the number of videos where the event was annotated, meaning 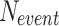 indeed includes events which were not detected by the model (False Negatives). Using these definitions, the F1-score per class can be computed as
indeed includes events which were not detected by the model (False Negatives). Using these definitions, the F1-score per class can be computed as 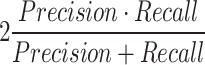 , which is a helpful metric to summarize in a single value the capacity of the model to be precise in its predictions without missing too many actual occurrences of the event in the dataset. These “per class” F1-scores are aggregated as a weighted average based on the number of videos where each event occurs. 1-cell stage metrics are ignored as it is trivial that the starting point of this event should correspond to the beginning of the video. Nonetheless, this class is still useful for the model during the learning process to not mistake other stages with 1-cell.
, which is a helpful metric to summarize in a single value the capacity of the model to be precise in its predictions without missing too many actual occurrences of the event in the dataset. These “per class” F1-scores are aggregated as a weighted average based on the number of videos where each event occurs. 1-cell stage metrics are ignored as it is trivial that the starting point of this event should correspond to the beginning of the video. Nonetheless, this class is still useful for the model during the learning process to not mistake other stages with 1-cell.
As reported in other studies, the average 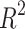 score per event is used to measure the goodness of fit between the detection of event start time versus real event start time. For a particular event class t and video i, let
score per event is used to measure the goodness of fit between the detection of event start time versus real event start time. For a particular event class t and video i, let 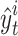 be the detected start time, and
be the detected start time, and 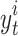 the real start time. The
the real start time. The 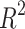 score is defined as
score is defined as 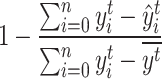 with
with 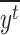 the mean real start time for event t, ie
the mean real start time for event t, ie 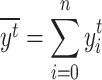 .
.
Moreover, the ability to detect if the embryo reached the blastocyst stage was also evaluated, as a means to help embryologists compute embryonic key performance metrics such as blastulation rates. We considered this stage was reached when tb was identified.
Ablation study
Ablation studies were conducted to understand the impact on performances of each component of the final architecture (see Table 4). First, the visual transformer was evaluated on its capacity to predict the final sequence without the GRU layers on top and without the HMM component. This configuration allows one to measure the capacity of the model to predict the biological events without having access to the full events sequence. The GRU layers were then added and performances were recomputed to evaluate the benefits of leveraging the learned feature of each frame into a sequence. Finally, the HMM is applied on the sequence of scores outputted by the GRU layers and final performances are computed to measure the benefit of this last component.
Table 4.
Ablation: weighted average F1-score on the test set of the different model architectures.
| Model architecture | weighted average F1-score (%) | Absolute added value |
|---|---|---|
| Visual encoder only | 44.0 | N/A |
| Visual encoder + sequence model | 60.7 | +16.7 |
| Visual encoder + sequence model + HMM | 66.3 | +5.6 |
Results
Performances across all embryo kinetic events
The BEE model reached a 66.3% weighted average F1-score on the test set using the 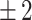 h acceptability window, with F1-scores over 69% on t2, t4, t6, tsb and tb, as shown in Table 2. When using the timing windows defined in21, the weighted average F1-score was 69.6%. Precision and Recall metrics are also reported in this figure. The lower precision of the model on t3 and t5 indicates the model’s tendency to over-detect these events. When removing the 41 non-developing embryos from the test set, the weighted-average F1-score across all biological events increased by +3.6% to reach 68.75% Specifically, t3 and t5 F1-scores increased by +7.3% and +7.7% respectively (detail in Supplementary Table S4). The weighted average F1-score per TLS was 69.5%, 58.9% and 52.3% for EMBRYOSCOPE, GERI and MIRI respectively. Detailed per event results for each TLS can be found in supplementary material Tables S1 to S3. The macro-average F1-score for 2-cell to 8-cell stage, as reported in Ref.15 was 65.1% and 80% when including later events (tm, tsb and tb). The overall
h acceptability window, with F1-scores over 69% on t2, t4, t6, tsb and tb, as shown in Table 2. When using the timing windows defined in21, the weighted average F1-score was 69.6%. Precision and Recall metrics are also reported in this figure. The lower precision of the model on t3 and t5 indicates the model’s tendency to over-detect these events. When removing the 41 non-developing embryos from the test set, the weighted-average F1-score across all biological events increased by +3.6% to reach 68.75% Specifically, t3 and t5 F1-scores increased by +7.3% and +7.7% respectively (detail in Supplementary Table S4). The weighted average F1-score per TLS was 69.5%, 58.9% and 52.3% for EMBRYOSCOPE, GERI and MIRI respectively. Detailed per event results for each TLS can be found in supplementary material Tables S1 to S3. The macro-average F1-score for 2-cell to 8-cell stage, as reported in Ref.15 was 65.1% and 80% when including later events (tm, tsb and tb). The overall 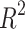 score between ground truth event start and detected event start was 0.95 (see Fig. S1 in supplementary material for detailed
score between ground truth event start and detected event start was 0.95 (see Fig. S1 in supplementary material for detailed 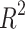 scores per event).
scores per event).
Table 2.
Timing precision, recall and F1-score per event on the test set, using a 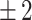 h window.
h window.
| Event | Precision (%) | Recall (%) | F1-score (%) | n samples |
|---|---|---|---|---|
| t2 | 93.3 | 92.2 | 92.8 | 258 |
| t3 | 54.8 | 73.3 | 62.7 | 187 |
| t4 | 84.4 | 64.5 | 73.1 | 234 |
| t5 | 46.6 | 56.9 | 51.3 | 195 |
| t6 | 67.9 | 77.7 | 72.5 | 188 |
| t7 | 56.7 | 69.7 | 62.5 | 165 |
| t8+ | 48.7 | 44.1 | 46.3 | 220 |
| tM | 54.6 | 54.3 | 54.4 | 164 |
| tSB | 72.8 | 68.2 | 70.4 | 157 |
| tB | 67.9 | 70.6 | 69.2 | 153 |
| Weighted average | 66.0 | 67.6 | 66.3 | 278 |
Figure 3 shows the box plot of each event as the time difference between predicted start time and the ground truth. By definition, this time difference can only be computed when the event exists both in ground truth and in detection. Medians of the delta in hours for all biological events are within the acceptance windows, as most of the Q1 and Q2 quartiles. Outliers are represented as white circles in the graph. Visual observation has shown that, for deltas above 10 hours, those are often due to highly fragmented embryos where correct detection of the events is a hard task for the model.
Figure 3.
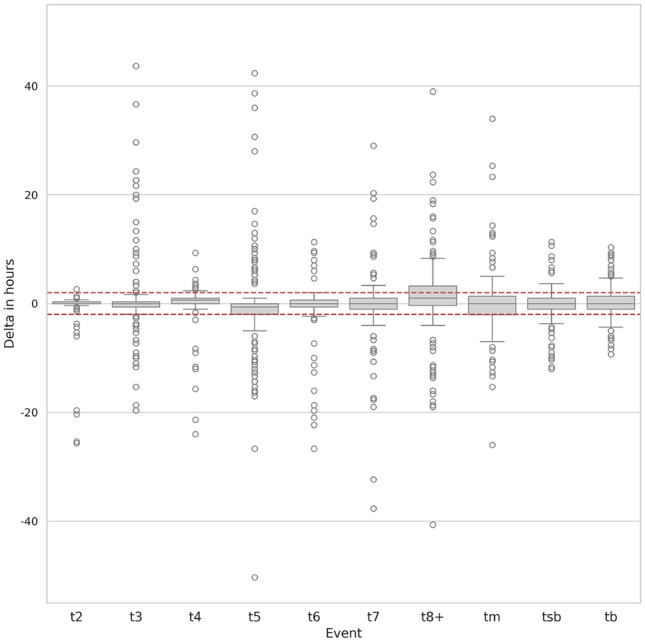
Box plot of the time difference in the test set between detected event start and ground truth. Horizontal red lines represent the 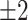 h window.
h window.
Performance of blastulation detection
The BEE model’s ability to correctly detect if an embryo reached the blastocyst stage or not was assessed regardless of the timing outputted by the model. On the test dataset, Table 3 shows that the model achieves 96% average F1-score, Precision and Recall on this task. Using the open-source dataset only and the definition of accuracy for blastocyst detection described in Ref.21 the BEE model achieved 99.5% accuracy.
Table 3.
Performances of the BEE model for blastocyst detection.
| Category | Precision (%) | Recall (%) | F1-score (%) | n samples |
|---|---|---|---|---|
| No blastocyst | 97.5 | 93.5 | 95.5 | 123 |
| Blastocyst | 95.0 | 98.1 | 96.5 | 155 |
| weighted-average | 96.1 | 96.0 | 96.0 | 278 |
Added value of individual components on model’s performances
Table 4 presents the contribution of each component of the BEE model architecture in terms of F1-score. Solely using the visual encoder to detect biological events resulted in 44% weighted average F1-score. When adding the sequence model on top of the visual encoder, the F1-score increased by 16.7 points. Finally, adding the post-processing with the HMM component to smooth the predictions of the sequence model brings an additional 5.6 points improvement.
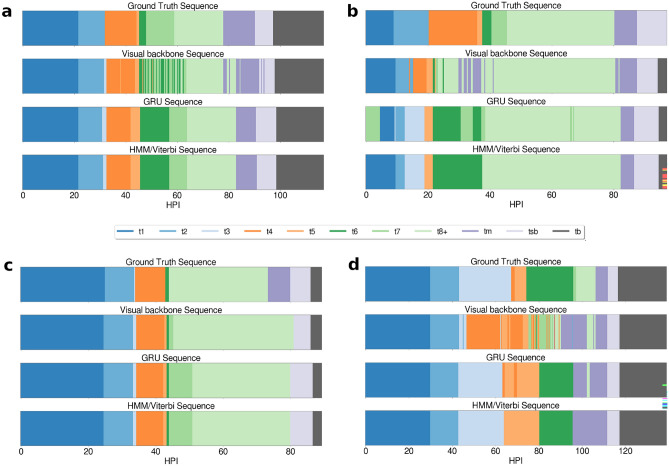
Figure 4 shows examples in the test dataset of 4 sequences of events for the same videos, but at different stages of the full pipeline. (a) and (d) displays sequences of embryos developing until day 5, while (b) and (c) develop until day 4. It is clear from the sequence plot that, during the cleavage stages in embryos (a), (b) and (d), the visual encoder oscillates a lot between frames, switching from one stage to the other. Once the sequence is passed through the GRU layers however, the noise is greatly removed, even though it can still be imperfect as displayed in (b) and (d). Finally, once passed through the HMM component, the sequence is smoothed and oscillations between events are removed.
Figure 4.
Visualization of the sequence of events on 4 videos. The x-axis represents the time in hpi, and each color corresponds to a biological event annotated or detected for this hpi. The first row of color sequence displays the ground truth as annotated by the embryologists. The second row displays the detections of the visual backbone, the third row presents the detections of the sequence model (GRU) using the output feature of the and the fourth row shows the final prediction of the BEE model after smoothing with the HMM component.
In embryo (b), interestingly, the sequence generated after the GRU layers is biologically impossible, as it starts from t7 to go to t1. After visual inspection, this is due to an extensive amount of debris and fragmentation in this specific video. This is later fixed by the HMM component, as visible in the final sequence generated which has events in an order that is biologically correct. However, this does not prevent the model from still making mistakes in the final sequence, with an incorrect blastocyst stage detected.
Embryo (d) shows a detection of t3 that was not annotated in the ground truth. After visual inspection, it was confirmed this was a correct prediction from the model, and instead a human annotation mistake. Small events lasting only a few frames can easily be missed by the annotator. This demonstrates how such DL models can be helpful for the embryologist.
Discussion
This study aims at describing a novel transformer based deep-learning architecture to predict embryo kinetic sequences that are close to the clinical reality with an increased accuracy compared to previous work. This work was based on data from various TLS, showing its capacity to be used in different workflows. Moreover, an new evaluation approach was also introduced, to better assess the clinical relevance of algorithms that help embryologists identify embryo kinetic events. This study’s proposed method extracts embeddings for every frame in the video as input to a sequence model (GRU) which is able to account for temporal dependencies throughout the whole sequence, unlike previous work that processed events either frame by frame or sub-clip by sub-clip. Furthermore, the backbone of this novel architecture was trained using a custom tailored loss inspired by the reverse cross-entropy. Finally, building on previous work that defined the timing accuracy of embryo kinetic event start21, the timing precision, recall and F1-score metrics were defined and reported to better capture the performance of the model at the video level and help embryologists assess the true capacity of a model to detect timings of biological events.
With a 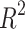 score of 0.95, the BEE model compared favorably with the
score of 0.95, the BEE model compared favorably with the 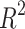 score of 0.92 reported in Ref.17 but below the
score of 0.92 reported in Ref.17 but below the 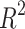 score of 0.99 reported in Ref.19. Compared to Ref.21 which reported a timing accuracy of 65.9% on fivefold cross-validation, the BEE model reached 69.6% on this specific metric and a 99.5% accuracy at detecting blastocyst compared to 99% reported in Ref.21. Reference15 reported a macro-average F1-score of 50.4% for 2-cell to 8-cell stages, while the BEE model achieved 65.1% on the same metric. When including later events (tm, tsb and tb), the macro-average F1-score was 80%, indicating a high frame-level accuracy on later events. This is expected as the blastulation stage is visually easily distinguishable from cleavage stages.
score of 0.99 reported in Ref.19. Compared to Ref.21 which reported a timing accuracy of 65.9% on fivefold cross-validation, the BEE model reached 69.6% on this specific metric and a 99.5% accuracy at detecting blastocyst compared to 99% reported in Ref.21. Reference15 reported a macro-average F1-score of 50.4% for 2-cell to 8-cell stages, while the BEE model achieved 65.1% on the same metric. When including later events (tm, tsb and tb), the macro-average F1-score was 80%, indicating a high frame-level accuracy on later events. This is expected as the blastulation stage is visually easily distinguishable from cleavage stages.
The BEE model robustly detected t2, t4, t6, tsb and tb with timing F1-scores above 69%, with noticeable differences across TLS (Tables S1, S2, S3 in supplementary material). Across all TLS, events such as t3, t5 and t8 were more difficult to correctly detect, either because: (i) these events have short durations and are thus harder to detect; (ii) a single focal plane is analyzed thus making it likelier to miss cells as they accumulate in other focal planes. Future work could try to leverage multiple focal planes to improve performances. It is also worth noting that the test set in this study included 15% of non-developing or degenerated embryos, i.e. embryos with cellular breakdown. When removing these 41 non-developing embryos from the test set, the average F1-score increased by + 3.6%. This suggests that such embryos were not sufficiently represented during the training phase. It could be beneficial in future studies to add more non-developing embryos and embryos with chaotic development in the training and validation datasets.
Ablation studies done in this study demonstrated that each component of this architecture serves a purpose to increase the accuracy of the event detection at the video level. The GRU layers learn biological patterns of events that help to generate plausible sequences, when the visual Transformer alone might be “short-sighted”. Viterbi algorithm, by design, will find the most likely sequence using a transition matrix based on the original data, therefore improving the probability of getting a monotonically increasing sequence of events, which is not a constraint of the deep-learning models. This is clearly illustrated in Fig. 4 where the generated sequences get more accurate as different stages of the pipeline are executed. We acknowledge that no ablation was performed to measure the performance of visual encoder + HMM, without the sequence model.
Finally, some limitations of this study were the lack of cross-validation when training the model and the absence of a clinic hold out set to measure the model’s ability to generalize on new clinical settings. Since the annotations for GERI and MIRI TLS only accounted for 11% of the test dataset, future studies could focus on building a larger and more balanced dataset in terms of TLS type. Similarly, embryos with more diverse qualities could be collected to be even more representative of diverse lab settings, especially for training in order to reinforce the abilities of the model on poor quality embryos. An interesting direction would be to have a panel of embryologists annotating the same set of videos to be able to compare the inter-operator agreement versus their agreement with the BEE detections. This would allow us to evaluate the capacity of the model to be trusted as a consensus opinion between experts, as their annotations may diverge9. Future work could leverage the power of BEE to automatically annotate a larger dataset of embryos and compare their kinetics with multiple clinical endpoints or even train subsequent AI models to use BEE annotations as input.
Supplementary Information
Acknowledgements
The authors want to thank Jessica Vandame and Xavier Pollet-Villard for their help in the annotation process.
Author contributions
GC and AD conceived the study and designed the methodology. AB-C was responsible for project management and supervision of research activity. GC and AD wrote the manuscript and ND helped review it.
Funding
Funding for the study was provided by ImVitro, with grant funding received in part from BPIFrance Aide au Developpement DeepTech (DOS0152872/00).
Data availability
The open-source dataset of 702 videos of embryos is available here: https://zenodo.org/records/7912264. The datasets generated and/or analyzed during the current study are available in the supplementary material.
Declarations
Competing interests
A.B.-C. is a co-owner of, and holds stocks in, ImVitro SAS. A.B.-C. holds a patent for “Devices and processes for machine learning prediction of in vitro fertilization” (EP20305914.2). G.C., A.D., N.D. are or have been employees of ImVitro and have been granted stock options.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guillaume Canat and Antonin Duval.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80565-1.
References
- 1.Pribenszky, C. et al. Pregnancy achieved by transfer of a single blastocyst selected by time-lapse monitoring. Reprod. Biomed. Online21, 533–536. 10.1016/j.rbmo.2010.04.015 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Chamayou, S. et al. The use of morphokinetic parameters to select all embryos with full capacity to implant. J. Assist. Reprod. Genet.30, 703–710. 10.1007/s10815-013-9992-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, J. et al. The human first cell cycle: Impact on implantation. Reprod. Biomed. Online28, 475–484. 10.1016/j.rbmo.2013.11.014 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Sayed, S. et al. Time-lapse imaging derived morphokinetic variables reveal association with implantation and live birth following in vitro fertilization: A retrospective study using data from transferred human embryos. PLoS ONE15, e0242377. 10.1371/journal.pone.0242377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, N., Goldberg, J. M., Austin, C. & Falcone, T. Are cleavage anomalies, multinucleation, or specific cell cycle kinetics observed with time-lapse imaging predictive of embryo developmental capacity or ploidy? Fertil. Steril.109, 665–674. 10.1016/j.fertnstert.2017.12.025 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Bamford, T. et al. Morphological and morphokinetic associations with aneuploidy: A systematic review and meta-analysis. Hum. Reprod. Update28, 656–686. 10.1093/humupd/dmac022 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Canosa, S. et al. A novel machine-learning framework based on early embryo morphokinetics identifies a feature signature associated with blastocyst development. J. Ovar. Res.17, 63. 10.1186/s13048-024-01376-6 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundvall, L., Ingerslev, H. J., Breth Knudsen, U. & Kirkegaard, K. Inter- and intra-observer variability of time-lapse annotations. Hum. Reprod.28, 3215–3221. 10.1093/humrep/det366 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Granados, L. et al. Inter-laboratory agreement on embryo classification and clinical decision: Conventional morphological assessment vs time lapse. PLoS ONE12, e0183328. 10.1371/journal.pone.0183328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukunaga, N. et al. Development of an automated two pronuclei detection system on time-lapse embryo images using deep learning techniques. Reprod. Med. Biol.19, 286–294. 10.1002/rmb2.12331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao, Q. et al. Development of deep learning algorithms for predicting blastocyst formation and quality by time-lapse monitoring. Commun. Biol.4, 415. 10.1038/s42003-021-01937-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theilgaard Lassen, J., Fly Kragh, M., Rimestad, J., Nygård Johansen, M. & Berntsen, J. Development and validation of deep learning based embryo selection across multiple days of transfer. Sci. Rep.13, 4235. 10.1038/s41598-023-31136-3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raudonis, V., Paulauskaite-Taraseviciene, A., Sutiene, K. & Jonaitis, D. Towards the automation of early-stage human embryo development detection. Biomed. Eng. Online18, 1–20. 10.1186/s12938-019-0738-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, T. et al. Embryo staging with weakly-supervised region selection and dynamically-decoded predictions. In Proceedings of the 4th Machine Learning for Healthcare Conference, vol. 106 of Proceedings of Machine Learning Research (eds Doshi-Velez, F. et al.) 663–679 (PMLR, 2019).
- 15.Sharma, A. et al. Detecting human embryo cleavage stages using yolo v5 object detection algorithm. In Nordic Artificial Intelligence Research and Development 81–93. 10.1007/978-3-031-17030-0_7 (Springer, 2022).
- 16.Liu, Z. et al. Multi-task deep learning with dynamic programming for embryo early development stage classification from time-lapse videos. IEEE Access7, 122153–122163. 10.1109/ACCESS.2019.2937765 (2019). [Google Scholar]
- 17.Feyeux, M. et al. Development of automated annotation software for human embryo morphokinetics. Hum. Reprod.35, 557–564. 10.1093/humrep/deaa001 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Leahy, B. D. et al. Automated measurements of key morphological features of human embryos for ivf. In Medical Image Computing and Computer Assisted Intervention-MICCAI: 23rd International Conference, Lima, Peru, October 4–8, 2020. Proceedings, Part V, vol. 23. 10.1007/978-3-030-59722-1_3 (Springer, 2020) [DOI] [PMC free article] [PubMed]
- 19.Zabari, N. et al. Delineating the heterogeneity of embryo preimplantation development using automated and accurate morphokinetic annotation. J. Assist. Reprod. Genet.40, 1391–1406. 10.1007/s10815-023-02806-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukyanenko, S. et al. Developmental stage classification of embryos using two-stream neural network with linear-chain conditional random field. In Medical Image Computing and Computer Assisted Intervention-MICCAI: 24th International Conference, Strasbourg, France, September 27–October 1, 2021. Proceedings, Part VIII, vol. 24. 10.1007/978-3-030-87237-3_35 (Springer, 2021) [DOI] [PMC free article] [PubMed]
- 21.Gomez, T. et al. A time-lapse embryo dataset for morphokinetic parameter prediction. Data Brief.42, 108258. 10.1016/j.dib.2022.108258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciray, H. N. et al. Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Hum. Reprod.29, 2650–2660. 10.1093/humrep/deu278 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Duval, A. et al. A hybrid artificial intelligence model leverages multi-centric clinical data to improve fetal heart rate pregnancy prediction across time-lapse systems. Hum. Reprod.38, 596–608. 10.1093/humrep/dead023 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, K. et al. Uniformer: Unified transformer for efficient spatiotemporal representation learning. 10.48550/arXiv.2201.04676 (2022).
- 25.Cho, K. Learning phrase representations using rnn encoder-decoder for statistical machine translation. 10.48550/arXiv.1406.1078 (2014).
- 26.Pang, T., Du, C., Dong, Y. & Zhu, J. Towards robust detection of adversarial examples. Adv. Neural Inf. Process. Syst.31, 1 (2018). [Google Scholar]
- 27.Hochreiter, S. Long short-term memory. Neural Computation MIT-Press (1997). [DOI] [PubMed]
- 28.Viterbi, A. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory13, 260–269. 10.1109/TIT.1967.1054010 (1967). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The open-source dataset of 702 videos of embryos is available here: https://zenodo.org/records/7912264. The datasets generated and/or analyzed during the current study are available in the supplementary material.