Abstract
Objectives
Total tumor volume (TTV) is associated with overall and recurrence-free survival in patients with colorectal cancer liver metastases (CRLM). However, the labor-intensive nature of such manual assessments has hampered the clinical adoption of TTV as an imaging biomarker. This study aimed to develop and externally evaluate a CRLM auto-segmentation model on CT scans, to facilitate the clinical adoption of TTV.
Methods
We developed an auto-segmentation model to segment CRLM using 783 contrast-enhanced portal venous phase CTs (CT-PVP) of 373 patients. We used a self-learning setup whereby we first trained a teacher model on 99 manually segmented CT-PVPs from three radiologists. The teacher model was then used to segment CRLM in the remaining 663 CT-PVPs for training the student model. We used the DICE score and the intraclass correlation coefficient (ICC) to compare the student model’s segmentations and the TTV obtained from these segmentations to those obtained from the merged segmentations. We evaluated the student model in an external test set of 50 CT-PVPs from 35 patients from the Oslo University Hospital and an internal test set of 21 CT-PVPs from 10 patients from the Amsterdam University Medical Centers.
Results
The model reached a mean DICE score of 0.85 (IQR: 0.05) and 0.83 (IQR: 0.10) on the internal and external test sets, respectively. The ICC between the segmented volumes from the student model and from the merged segmentations was 0.97 on both test sets.
Conclusion
The developed colorectal cancer liver metastases auto-segmentation model achieved a high DICE score and near-perfect agreement for assessing TTV.
Critical relevance statement
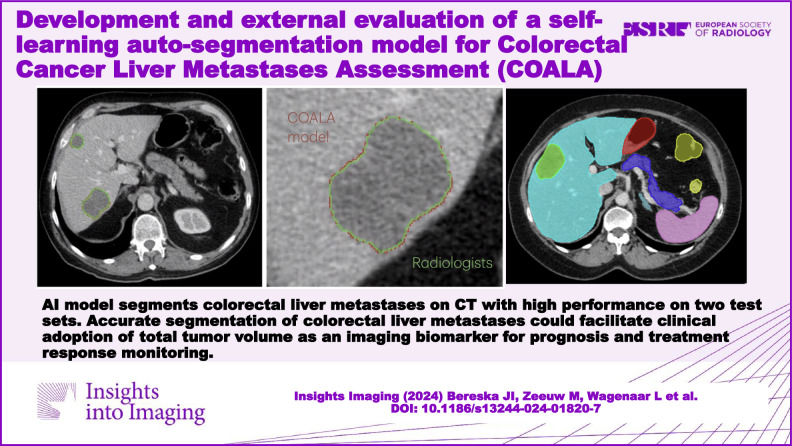
AI model segments colorectal liver metastases on CT with high performance on two test sets. Accurate segmentation of colorectal liver metastases could facilitate the clinical adoption of total tumor volume as an imaging biomarker for prognosis and treatment response monitoring.
Key Points
Developed colorectal liver metastases segmentation model to facilitate total tumor volume assessment.
Model achieved high performance on internal and external test sets.
Model can improve prognostic stratification and treatment planning for colorectal liver metastases.
Graphical Abstract
Keywords: Colorectal neoplasms, Liver, Biomarkers, Tumor, Artificial intelligence
Introduction
Total tumor volume (TTV) at baseline and TTV response to systemic therapy are prognostic for overall and recurrence-free survival in patients with colorectal cancer liver metastases (CRLM) [1–5]. Currently, the evaluation of response to systemic therapy of CRLM is performed using the Response Evaluation Criteria in Solid Tumors (RECIST1.1) [6]. However, the correlation between RECIST1.1 and survival remains indeterminate [7]. Using TTV might lead to more clinically relevant assessments when evaluating the response to systemic therapy of CRLM. Assessing TTV involves manual segmentation of numerous CRLMs, which is a time-consuming task requiring considerable expertise. Moreover, manual segmentation is subjective, leading to inter-observer variability. Thus, despite its potential prognostic value, TTV assessment has not been adopted in clinical practice. Artificial intelligence (AI) CRLM segmentation models may aid clinicians in automatically assessing TTV, facilitating practical application in routine patient care.
Automatic segmentation of the liver and primary liver tumors has been extensively studied in recent years, with various deep learning architectures such as convolutional neural networks, UNet and UNet variants, and generative adversarial networks being employed to segment primary liver tumors like hepatocellular carcinoma with promising results [8–17]. However, this work focuses on CRLM, which presents unique challenges compared to primary liver tumors due to their heterogeneous appearance and less well-defined borders. Although some work has been done on automatic segmentation of CRLM, it is limited compared to the body of research on primary liver tumors. For instance, Vorontsov et al proposed a semi-automatic segmentation method for CRLM, improving segmentation speed compared to manual segmentations but lacking volumetric accuracy [17]. Similarly, Wesdorp et al introduced an automatic segmentation model for CRLM; however, this model fell short in an external test cohort [16]. This lack of segmentation accuracy underlines the imperative for developing more precise models capable of clinical-grade CRLM segmentation to facilitate automated TTV assessments.
To address current limitations in spatial accuracy of automated CRLM segmentation, we developed a self-learning-based segmentation model for COlorectal CAncer Liver metastasis Assessment (COALA) using a large patient cohort. The COALA model leverages the teacher-student dynamic, with a teacher model trained on a smaller segmented dataset guiding a student model learning from a larger unsegmented dataset. By using averaged ground-truth segmentations consolidated from multiple radiologists, we aim to minimize observer-dependent variations and improve the feasibility of employing TTV assessments in clinical practice.
Materials and methods
This study retrospectively included data from two medical centers. The Medical Ethics Review Committee of the Amsterdam UMC, the Regional Ethical Committee of Norway, and the Data Protection Officer of Oslo University Hospital approved this study protocol. All patients were managed per institutional practices. All patients signed a written informed consent form permitting the use of their data for studies.
Datasets
We utilized two datasets for this retrospective study: the INTERNAL and EXTERNAL datasets. The INTERNAL dataset included 783 portal venous phase CT scans (CT-PVPs) from 373 patients registered in the CAIRO5 trial (NCT02162563), a multicenter randomized controlled trial conducted by the Dutch Colorectal Cancer Group between November 2014 and January 2022 in 47 hospitals [18]. The CAIRO5 trial evaluated the optimal systemic induction therapy for patients with initially unresectable liver-only CRLM. The patients were randomized between systemic therapy combinations depending on the primary tumor site and genetic mutation status. These treatment regimens included doublet or triplet chemotherapy in combination with targeted therapy.
The EXTERNAL dataset included 50 CT-PVPs from 35 patients enrolled in the Oslo-COMET trial (NCT01516710), a single-center, randomized superiority trial conducted at the Oslo University Hospital between February 2012 and January 2016. The patients were randomly assigned to undergo laparoscopic or open parenchyma-sparing liver resection [19].
Both datasets consisted of CT-PVPs at baseline before systemic therapy and at follow-up after systemic therapy. We collected information on age, sex, systemic induction therapies, and the number of CRLMs (Table 1). The CT acquisition and reconstruction parameters are detailed in Table S1 in the Supplemental Digital Content.
Table 1.
Patient demographics for the INTERNAL and EXTERNAL datasets
| Characteristics | INTERNAL dataset N = 373 |
EXTERNAL dataset N = 35 |
|---|---|---|
| Sex, n (%), female, male | 136 (36%), 237 (64%) | 19 (54%), 16 (46%) |
| Average age at diagnosis, years (dev) | 62 (10.2) | 64 (9) |
| Average number of CRLM at diagnosis, n (dev) | 12 (15.6) | 1 (1) |
| Average largest CRLM diameter mm (dev) | 44 (32) | 19.5 (11.1) |
| Pre-NAT scans, n (%) | 373 (48%) | 31 (62%) |
| Post-NAT scans, n (%) | 410 (52%) | 19 (38%) |
CRLM colorectal cancer liver metastasis, n number, dev standard deviation, NAT neoadjuvant therapy
Data preparation
For the INTERNAL dataset, two research team members (M.Z., N.W.) semi-automatically segmented a selection of 120 CT-PVPs from 55 patients with 1113 CRLM using the Tumor Tracking Modality feature of IntelliSpace Portal 9.0® (Philips). Initially, IntelliSpace Portal automatically generated a region of interest based on differences in density. The two research team members manually refined these outlines slice-by-slice for precise segmentation. Three specialist abdominal radiologists (J.H.v.W.: 18 years’ experience, J.v.d.B.: 10 years’ experience, I.N.: 2 years’ experience) independently reviewed and adjusted these segmentations as necessary using the IntelliSpace Portal.
For the EXTERNAL dataset, two members of the research team independently performed initial segmentations of 50 CT-PVPs from 35 patients with 72 CRLM using 3DSlicer (5.4.0). Three specialist abdominal radiologists (G.K.:12 years’ experience, T.S.: 15 years’ experience, F.K.K.: 10 years’ experience) each subsequently independently reviewed and, if needed, corrected all these segmentations using 3DSlicer (5.4.0) and MedSeg (1.0).
The corrected segmentations from the three radiologists in the INTERNAL and EXTERNAL datasets were merged into one single segmentation through the Simultaneous Truth and Performance Level Estimation algorithm (STAPLE) algorithm, henceforth referred to as the merged segmentations [20].
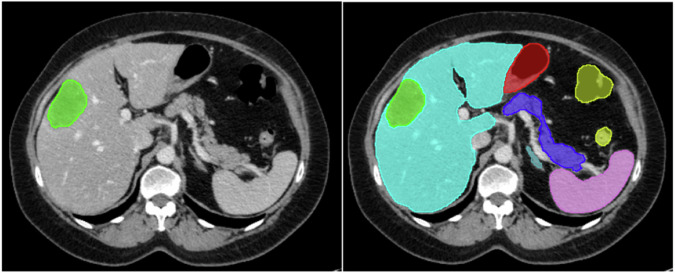
Including surrounding abdominal structures has been shown to increase segmentation model performance; therefore, next to the CRLM segmentations, we included segmentations of twelve pertinent surrounding anatomical structures: the duodenum, pancreas, both adrenal glands, spleen, gallbladder, both kidneys, colon, stomach, small bowel, and liver [21]. These additional segmentations were generated automatically using the anatomical segmentation model, TotalSegmentator, and serve as contextual information for the model, helping it to identify relevant areas of the scan and improve CRLM segmentation accuracy [22]. Figure 1 depicts an example of a segmented CT-PVP from the INTERNAL dataset.
Fig. 1.
Example of a manually segmented portal venous phase axial computed tomography scan performed by a trio of radiologists and combined using the STAPLE algorithm. Green = CRLM, turquoise = liver, pink = spleen, dark blue = pancreas, light blue = adrenal glands, red = stomach, yellow = colon
Model implementation
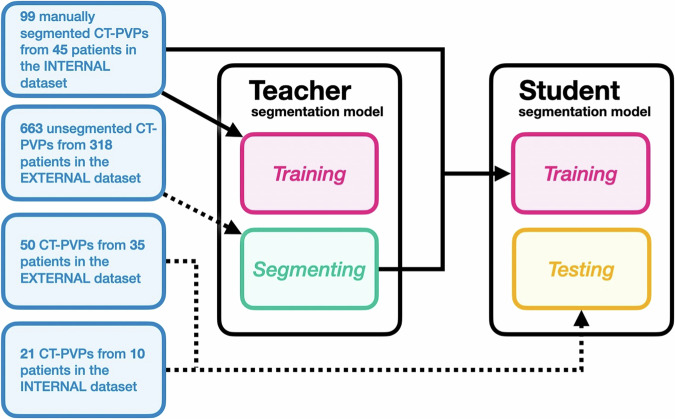
We followed a self-learning approach to train the COALA segmentation model, which is demonstrated schematically in Fig. 2. Self-learning commences with a teacher segmentation model trained on a small set of manually segmented training data. This teacher segmentation model is then used to generate segmentations for the entire unsegmented training dataset. These teacher-generated segmentations are subsequently used to train a student segmentation model. The student segmentation model, through leveraging the additional training data, can exceed the performance of the initial teacher segmentation model. This approach can facilitate a reduction in manual segmentations and an increase in the robustness and generalizability of the segmentation model [23].
Fig. 2.
The proposed learning framework for CRLM and abdominal organ segmentation on contrast-enhanced CT scans. CT-PVP, portal venous phase computed tomography scan
We initially trained a teacher segmentation model using a subset of 99 CT-PVPs from the previously manually segmented 120 CT-PVPs from the INTERNAL dataset. Using this teacher segmentation model, we obtained automatic segmentations of the remaining INTERNAL dataset, comprising 663 CT scans from 318 patients. The resulting automatic and initial 99 segmentations were used to train the student segmentation model. The student model served as the final COALA segmentation model.
We selected a nnUNet network setup that included a two-stage 3D U-Net cascade for both the student and teacher segmentation models [24]. The cascade comprised an initial U-Net trained on down-sampled images to generate low-resolution segmentations, which served as an auxiliary input for training the subsequent full-resolution U-Net. We used 5-fold cross-validation with an 80:20 training-validation split, 1000 steps per fold, and an initial learning rate of 0.05 to train both the low-resolution and full-resolution U-Nets. All models were trained on an NVIDIA A100 GPU, taking roughly one day per fold.
Performance assessment
We assessed the performance of the trained COALA model using the 50 CT-PVPs from the EXTERNAL dataset and a subset of the INTERNAL dataset containing 21 CT-PVPs. To evaluate the spatial accuracy of our model’s CRLM segmentations, we compared the model’s segmentations to the merged segmentations using DICE scores. To evaluate our model’s TTV assessment, we derived the TTV in voxels from the model’s and merged segmentations. We examined the agreement between the model’s and the merged segmentation’s TTV by calculating a two-way mixed-effect intraclass correlation coefficient (ICC), categorizing the results as poor (ICC < 0.4), fair (ICC 0.4–0.59), good (ICC 0.6–0.74), or excellent (ICC 0.75–1.0). Finally, we used Welch’s t-test to compare the model’s performance on pre-NAT and post-NAT scans. A p-value less than 0.05 denoted statistical significance.
Results
Patient characteristics
The INTERNAL dataset contained 783 CT-PVPs from 373 patients, depicting 14,152 CRLM, and the EXTERNAL dataset contained 50 CT-PVPs from 35 patients depicting 72 CRLM. In the INTERNAL dataset, the majority of patients were male (64%), in the EXTERNAL dataset less than half of patients was male (46%). The median number of CRLM at diagnosis (12 versus 1) and the median largest diameter (42 mm versus 19.5 mm) were higher in the INTERNAL dataset. Imaging data consisted of CT-PVPs at baseline before systemic therapy (INTERNAL: 373 (48%), EXTERNAL: 410 (52%)) and at follow-up after systemic therapy (INTERNAL: 31 (62%), EXTERNAL: 19 (38%)). See Table 1.
Segmentation model
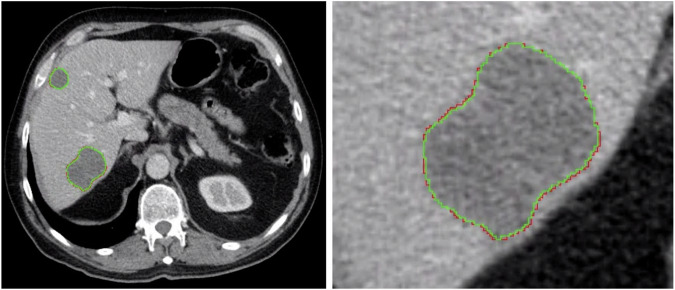
The COALA model achieved a mean DICE score of 0.83 (IQR: 0.10) on the EXTERNAL dataset, with a mean DICE score of 0.84 (0.10) and 0.82 (IQR: 0.05) on pre- and post-NAT scans, respectively. On the withheld subset of the INTERNAL dataset, the COALA model achieved a mean DICE score of 0.85 (IQR: 0.05) with a mean DICE score of 0.87 (IQR: 0.02) and 0.85 (IQR: 0.05) on pre- and post-NAT scans, respectively. A Welch’s t-test revealed no significant difference between the model’s performance on pre- or post-NAT scans on either the EXTERNAL of the INTERNAL dataset (p = 0.64 and p = 0.22). A visual comparison between the segmentation results garnered by the model and the ground-truth merged segmentation is depicted in Fig. 3.
Fig. 3.
Comparison between the COALA model’s segmentation and the merged segmentation within a portal venous phase axial computed tomography scan. Red = automatic segmentation performed by our model, green = merged manual segmentation performed by three radiologists and merged using the STAPLE algorithm
Total tumor volume analysis
The agreement of the TTV derived from the COALA model’s with the volumes from the merged segmentations reached an ICC of 0.97 on both the EXTERNAL dataset INTERNAL datasets. The median, largest, and smallest TTV in voxels obtained from the model’s and the merged segmentations are denoted in Table 2.
Table 2.
TTV derived from the COALA model’s and the merged segmentations in voxels on the INTERNAL and EXTERNAL datasets
| TTV in voxels | EXTERNAL COALA model | EXTERNAL merged | INTERNAL COALA model | INTERNAL merged |
|---|---|---|---|---|
| Median (IQR) | 2,024 (2,573) | 2,315 (3,011) | 59,760 (135,198) | 62,772 (127,396) |
| Largest | 11,240 | 12,546 | 656,040 | 684,647 |
| Smallest | 189 | 122 | 721 | 2601 |
TTV total tumor volume, IQR interquartile range
Discussion
This study presents the development and external evaluation of a fully automatic CRLM segmentation and TTV assessment model COALA. By employing a self-learning training setup with a diverse dataset and consolidating CRLM segmentations from three radiologists into a unified ground truth, we reduced the required manual training samples, enhanced the model’s robustness and generalizability, and mitigated observer-dependent variations. The COALA model showed no significant difference in CRLM segmentation DICE scores and displayed near-perfect agreement for TTV assessment in the external evaluation cohort from the Oslo University Hospital. Collectively, these findings suggest that the proposed COALA model has the potential to provide reliable and consistent TTV assessments in routine clinical practice.
While automatic segmentation of primary liver tumors has been extensively studied using various deep learning architectures [8–17], the segmentation of colorectal liver metastases (CRLM) presents unique challenges due to their heterogeneous appearance and less well-defined borders. Vorontsov et al made significant contributions in applying deep learning to TTV assessment for CRLM [17]. Their methodology, based on fully convolutional networks, did offer improvements in segmentation speed but was compromised by a lack of segmentation and volumetric accuracy. Specifically, the DICE score achieved by their automated and even user-corrected CRLM segmentation model was substantially lower than the DICE score achieved by our COALA model (with 0.68 compared to 0.85). Similarly, Wesdorp et al introduced an automatic segmentation model for CRLM, but it fell short in an external test cohort [16]. These limitations underscore the need for more precise models capable of clinical-grade CRLM segmentation. By utilizing a larger and more diverse training dataset of 833 scans, compared to 115 in the previous study, we sought to enhance the model’s ability to generalize to new, unseen data from various patient populations. Furthermore, we strengthened the reliability of our ground truth by incorporating annotations from three experienced radiologists, reducing the risk of individual bias or errors. Lastly, our study included an external evaluation of the COALA model using data from Oslo University Hospital, demonstrating its applicability and effectiveness across different medical centers.
There are several limitations of our study that should be acknowledged. First, the retrospective nature of our study limits the prediction of the model’s efficacy in prospective clinical settings. Second, the merged segmentations were created by radiologists correcting an existing pre-segmentation, likely resulting in higher inter-rater DICE scores and ICC compared to from-scratch segmentations. Third, the external test cohort differed from the training cohort, specifically in the number of CRLMs per patient. While the model’s good performance despite this discrepancy can be considered a strength, it also poses questions about how representative the training data is for a wide range of clinical scenarios. Finally, we did not evaluate our model on a publicly available benchmark dataset, as existing ones, such as the Liver Tumor Segmentation (LiTS) dataset, mainly comprise primary liver tumors, not CRLM [25]. To address this, we make our internal test set publicly available along with our model for future benchmarking. Future studies should incorporate data from global centers and include more clinically representative test cohorts. Automating manual radiological evaluations, such as response evaluation currently done in clinical practice using RECIST1.1 criteria, presents a promising application.
In conclusion, our study introduces the first fully automatic CRLM segmentation model COALA, which aligns with inter-observer agreement for segmentation and displays near-perfect agreement for TTV assessment. The model’s robustness is highlighted by its external evaluation of data annotated by three radiologists, offering a substantial mitigation of observer-dependent variations. These advancements provide a promising foundation for reliable and consistent TTV measurements, crucial for the effective management of patients with colorectal cancer liver metastases.
Supplementary information
Acknowledgements
Use of Large Language Models: The following manuscript has undergone evaluation, critique, and improvement using ChatGPT-4 and Claude, AI language models developed by OpenAI and Anthropic (last access: June 11th, 2024).
Abbreviations
- AI
Artificial intelligence
- COALA
COlorectal CAncer Liver metastasis Assessment
- CRLM
Colorectal cancer liver metastasis
- CT-PVP
Portal venous phase CT scan
- ICC
Intraclass correlation coefficient
- RECIST
Response Evaluation Criteria in Solid Tumors
- STAPLE
Simultaneous Truth and Performance Level Estimation
- TTV
Total tumor volume
Author contributions
Jacqueline I. Bereska, MSc: conceptualization; data curation; formal analysis; investigation; methodology; project administration; writing—original draft. Michiel Zeeuw, MD: conceptualization; data curation; investigation; methodology; project administration; writing—original draft. Luuk Wagenaar, MSc: conceptualization; data curation; investigation; methodology; project administration. Håvard Bjørke Jenssen, MD: data curation; resources; writing—review & editing. Nina J. Wesdorp, MD: data curation; writing—review & editing. Delanie van der Meulen, MSc: data curation; writing—review & editing; Leonard F. Bereska, MSc: supervision; writing—original draft; writing—review & editing. Efstratios Gavves, PhD: supervision; writing—review & editing. Boris V. Janssen, BSc: writing—review & editing. Marc G. Besselink, MD PhD: supervision; conceptualization; methodology; writing—review & editing; funding acquisition. Henk A. Marquering, PhD: supervision; conceptualization; methodology; writing—review & editing. Jan-Hein T.M. van Waesberghe, MD PhD: data curation. Davit L. Aghayan, MD: data curation; writing—review & editing. Egidijus Pelanis, MD PhD: data curation; writing—review & editing. Janneke van den Bergh, MD: data curation; writing—review & editing. Irene I.M. Nota, MD: data curation; writing—review & editing. Shira Moos, MD PhD: data curation; writing—review & editing. Gunter Kemmerich, MD: data curation; writing—review & editing. Trygve Syversveen, MD PhD: data curation; writing—review & editing. Finn Kristian Kolrud, MD: data curation; writing—review & editing. Joost Huiskens, MD PhD: conceptualization; funding acquisition; methodology; resources; supervision; writing—review & editing. Rutger-Jan Swijnenburg, MD PhD: funding acquisition; resources; supervision; writing—review & editing. Cornelis J.A. Punt, MD PhD: conceptualization; funding acquisition; resources; supervision; writing—review & editing. Jaap Stoker, MD PhD: conceptualization; funding acquisition; methodology; resources; supervision; writing—review & editing. Bjørn Edwin, MD PhD: data curation; writing—review & editing. Åsmund A. Fretland, MD PhD: data curation; conceptualization; resources; writing—review & editing. Geert Kazemier, MD PhD: conceptualization; funding acquisition; methodology; resources; supervision; writing—review & editing. Inez M. Verpalen, MD PhD: conceptualization; methodology; resources; supervision; writing—review & editing.
Funding
This study has received funding from the Dutch Cancer Society (KWF Kankerbestrijding), project number 14002/2021-2, and an unrestricted grant from the Cancer Center Amsterdam Foundation. The funders had no role in the design and conduct of the study, data collection, management, analysis, interpretation, manuscript preparation, review, approval, or decision to submit the manuscript for publication.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material. The documented code, fully trained COALA model, and test set will be made available on GitHub upon publication.
Declarations
Ethics approval and consent to participate
The Medical Ethics Review Committee of the Amsterdam UMC, the Regional Ethical Committee of Norway, and the Data Protection Officer of Oslo University Hospital approved this study protocol. All patients were managed per institutional practices. All patients signed a written informed consent form permitting the use of their data for studies.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lists of authors and their affiliations appear at the end of the paper.
Jacqueline I. Bereska and Michiel Zeeuw contributed equally to this work.
Geert Kazemier and Inez M. Verpalen jointly supervised to this work.
Contributor Information
Jacqueline I. Bereska, Email: j.i.bereska@amsterdamUMC.nl
Inez M. Verpalen, Email: i.m.verpalen@amsterdamUMC.nl
for the Pancreatobiliary and Hepatic Artificial Intelligence Research (PHAIR) consortium:
Giovanni Marchegiani, Domenico Bassi, Riccardo Boetto, Mattia Ballo, Riccardo Carandina, Filippo Crimi, Matteo Fassan, Arantza Farina, Caroline Verbeke, Knut Jørgen Labori, Åsmund Fretland, Mirko D’Onofrio, Giulia Zamboni, Riccardo di Robertis, Claudio Luchini, Alberto Balduzzi, Giuseppe Malleo, Roberto Salvia, Christopher Wolfgang, Ammar Javed, Katie Colborn, Marco Del Chiaro, Jeffrey Kaplan, Toshimasa Clark, Thomas Stoop, Ioana Lupescu, Cristian Mugur Grasu, Cristian Anghel, Mihai Dan Pomohaci, Philipp Mayer, Benedict Kinny-Köster, Martin Loos, and Christoph Michalski
the Dutch Colorectal Cancer Group Liver Expert Panel:
Martinus J. van Amerongen, Marinde J. G. Bond, Thiery Chapelle, Ronald M. van Dam, Marc R. W. Engelbrecht, Michael F. Gerhards, Dirk J. Grunhagen, Thomas M. van Gulik, John J. Hermans, Koert P. de Jong, Joost M. Klaase, Niels F. M. Kok, Wouter K. G. Leclercq, Mike S. L. Liem, Krijn P. van Lienden, I. Quintus Molenaar, Gijs A. Patijn, Arjen M. Rijken, Theo M. Ruers, Cornelis Verhoef, and Johannes H. W. de Wilt
Supplementary information
The online version contains supplementary material available at 10.1186/s13244-024-01820-7.
References
- 1.Shimura Y, Komatsu S, Nagatani Y et al (2023) The usefulness of total tumor volume as a prognostic factor and in selecting the optimal treatment strategy of chemotherapeutic intervention in patients with colorectal liver metastases. Ann Surg Oncol 30:6603–6610. 10.1245/s10434-023-13746-3 [DOI] [PubMed] [Google Scholar]
- 2.He J, Li W, Zhou J et al (2023) Evaluation of total tumor volume reduction ratio in initially unresectable colorectal liver metastases after first-line systemic treatment. Eur J Radiol 165:110950. 10.1016/j.ejrad.2023.110950 [DOI] [PubMed] [Google Scholar]
- 3.Tai K, Komatsu S, Sofue K et al (2020) Total tumour volume as a prognostic factor in patients with resectable colorectal cancer liver metastases. BJS Open 4:456–466. 10.1002/bjs5.50280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wesdorp NJ, Bolhuis K, Roor J et al (2021) Total tumor volume response versus RECIST upon systemic treatment in patients with initially unresectable colorectal liver metastases. HPB 23:S834. 10.1016/j.hpb.2021.08.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michiel Zeeuw J, Wesdorp NJ, Ali M et al (2024) Prognostic value of total tumor volume in patients with colorectal liver metastases: A secondary analysis of the randomized CAIRO5 trial with external cohort validation. Eur J Cancer 207:114185. 10.1016/j.ejca.2024.114185 [DOI] [PubMed] [Google Scholar]
- 6.Eisenhauer EA, Therasse P, Bogaerts J et al (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45:228–247. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 7.Bogaerts J, Ford R, Sargent D et al (2009) Individual patient data analysis to assess modifications to the RECIST criteria. Eur J Cancer 45:248–260. 10.1016/j.ejca.2008.10.027 [DOI] [PubMed] [Google Scholar]
- 8.Christ PF, Ettlinger F, Grün F et al (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. 10.48550/arXiv.1702.05970
- 9.Han X (2017) Automatic liver lesion segmentation using a deep convolutional neural network method. Med Phys 44:1408–1419. 10.1002/mp.12155 [DOI] [PubMed] [Google Scholar]
- 10.Jin Q, Meng Z, Sun C, Cui H, Su R (2020) RA-UNet: a hybrid deep attention-aware network to extract liver and tumor in CT Scans. Front Bioeng Biotechnol 8. 10.3389/fbioe.2020.605132 [DOI] [PMC free article] [PubMed]
- 11.Li S, Tso GKF (2024) Bottleneck supervised U-Net for pixel-wise liver and tumor segmentation. Available: http://arxiv.org/abs/1810.10331
- 12.Long J, Shelhamer E, Darrell T (2024) Fully convolutional networks for semantic segmentation. Presented at the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, 3431–3440. Accessed: Jun. 11, 2024. [Online]. Available: https://openaccess.thecvf.com/content_cvpr_2015/html/Long_Fully_Convolutional_Networks_2015_CVPR_paper.html
- 13.Jiang H, Shi T, Bai Z, Huang L (2019) AHCNet: an application of attention mechanism and hybrid connection for liver tumor segmentation in CT volumes. IEEE Access 7:24898–24909. 10.1109/ACCESS.2019.2899608 [Google Scholar]
- 14.Li W, Jia F, Hu Q (2015) Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J Comput Commun 3:11. 10.4236/jcc.2015.311023
- 15.Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng P-A (2016) 3D deeply supervised network for automatic liver segmentation from CT volumes. Accessed: Jun. 11, 2024. [Online]. Available: http://arxiv.org/abs/1607.00582
- 16.Wesdorp NJ, Zeeuw JM, Postma SCJ et al (2023) Deep learning models for automatic tumor segmentation and total tumor volume assessment in patients with colorectal liver metastases. Eur Radiol Exp 7:75. 10.1186/s41747-023-00383-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vorontsov E, Cerny M, Régnier P et al (2019) Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiol Artif Intell Mar. Accessed: Sep. 29, 2023. [Online]. Available: https://pubs.rsna.org/doi/10.1148/ryai.2019180014 [DOI] [PMC free article] [PubMed]
- 18.Huiskens J, van Gulik TM, van Lienden KP et al (2015) Treatment strategies in colorectal cancer patients with initially unresectable liver-only metastases, a study protocol of the randomised phase 3 CAIRO5 study of the Dutch Colorectal Cancer Group (DCCG). BMC Cancer 15:365. 10.1186/s12885-015-1323-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fretland ÅA, Dagenborg VJ, Bjørnelv GMW et al (2018) Laparoscopic versus open resection for colorectal liver metastases: the OSLO-COMET randomized controlled trial. Ann Surg 267:199–207. 10.1097/SLA.0000000000002353 [DOI] [PubMed] [Google Scholar]
- 20.Warfield SK, Zou KH, Wells WM (2004) Simultaneous Truth and Performance Level Estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23:903–921. 10.1109/TMI.2004.828354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alves N, Schuurmans M, Litjens G, Bosma JS, Hermans J, Huisman H (2022) Fully automatic deep learning framework for pancreatic ductal adenocarcinoma detection on computed tomography. Cancers (Basel) 14:376. 10.3390/cancers14020376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasserthal J, Breit H-C, Meyer MT et al (2023) TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. Radiol Artif Intell 5:e230024. 10.1148/ryai.230024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Zhang Z, Wu C et al (2021) Improving semantic segmentation via efficient self-training. IEEE Trans Pattern Anal Mach Intell 1–1. 10.1109/TPAMI.2021.3138337 [DOI] [PubMed]
- 24.Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:2. 10.1038/s41592-020-01008-z [DOI] [PubMed]
- 25.Bilic P, Christ P, Li HB et al (2023) The Liver Tumor Segmentation Benchmark (LiTS). Med Image Anal 84:102680. 10.1016/j.media.2022.102680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material. The documented code, fully trained COALA model, and test set will be made available on GitHub upon publication.