Abstract
LC3-associated phagocytosis (LAP) is critical in host defense against invading pathogens, but the molecular mechanism for LAP activation is still unclear. Here, we find programmed cell death 6 (PDCD6) as a negative regulator of LAP. PDCD6 deficiency in mice and macrophages induces enhanced bactericidal activity and LAP formation. In parallel, lactate dehydrogenase A (LDHA) activity and lactate production is induced in macrophages challenged with bacteria, Zymosan or Pam3CSK4, while genetic ablation or pharmacological inhibition of LDHA reduces lactate levels and impairs bactericidal activity in vivo and in vitro. Mechanistically, PDCD6 interacts with LDHA to downregulate lactate metabolism, leading to reduced RUBCN lactylation at lysine33 (K33). By contrast, PDCD6-deficiency increases RUBCN lactylation, thereby promotes RUBCN interaction with VPS34, LAP formation, and protective responses. Our results thus suggest a PDCD6-LDHA-lactate-RUBCN axis of innate immunity regulation that may both contribute to protection from infectious diseases and serve as targets for therapeutic development.
Subject terms: Infection, Bacterial host response, Autophagy, Post-translational modifications
The molecular mechanism of LAP activation against invading pathogens is not fully understood. Here the authors demonstrate that programmed cell death 6 (PDCD6) negatively regulates LAP formation and the antibacterial response by interacting with lactate dehydrogenase A (LDHA) to downregulate lactate metabolism and RUBCN lactylation-mediated signaling.
Introduction
Macrophages serve as the body’s first line of defense against invading pathogens, recognizing pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRR), thereby allowing rapid pathogen elimination1. Various mechanisms contribute to the clearance of intracellular bacteria, including autophagy, pyroptosis, proinflammatory cytokine and chemokine production, and LC3-associated phagocytosis (LAP). LAP is a noncanonical autophagy pathway linking PRR activation to phagosome formation recently identified as being crucial in the defense against pathogens, including bacteria and fungi2,3. This autophagic immune response involves the assembly of a UVRAG-containing class III PI3K complex (Beclin 1, VPS34), leading to sustained PtdIns(3)P production, NADPH oxidase 2 (NOX2) stabilization, reactive oxygen species (ROS) release, and lipidated LC3 (LC3-II) conjugation on the phagosome. Rubicon, a key regulator of LAP, binds to the UVRAG-containing complex and inhibits canonical autophagy by blocking the formation of the PI3K/ATG14L complex. Despite these insights, knowledge of the upstream signaling pathways involved in LC3+ phagosome (LAPosome) formation and the role of NOX activation in this process remains limited4.
Programmed cell death protein 6 (PDCD6, also known as apoptosis-linked gene-2 [ALG-2]) is a 191-amino-acid protein with a molecular mass of 21.7 kDa. It features an N-terminal AGP structure and an EF-hand domain comprising 5 EF hands and 8 helices5. The EF-hand domain contains 168 amino acid residues with 5 “helix-ring-helix” calcium ion binding domains. Fluorescence spectroscopic analysis of PDCD6 indicated that it undergoes conformational changes and hydrophobic pocket exposure in response to increased calcium ion levels, suggesting that it functions as a calcium ion receptor6. PDCD6 was first identified as a proapoptotic protein in T-cell hybridomas7. However, it has since been found to be involved in the regulation of cell death, cell division, and signal transduction8,9, as well as play a critical role in vesicle transport from the endoplasmic reticulum (ER) to the Golgi by localizing at ER exit sites (ERES) and interacting with SEC31A to stabilize the protein. In vitro, PDCD6 was shown to inhibit coat protein complex II (COPII) vesicle fusion, thereby affecting transport dynamics10, as well as disrupting COPII vesicle budding and trafficking11,12. PDCD6 also acts as a regulator of cytosolic DNA-induced immune responses by modulating STING transport13. Its expression is elevated in virus-infected organisms, suggesting that it plays a role in antiviral immunity, and also has potential as a diagnostic marker for COVID-1914. However, PDCD6 deletion mice show normal development and immune function15, highlighting the need for further studies to clarify its function in antibacterial immunity.
Immunometabolic changes influence host–pathogen interactions. Macrophages shift to aerobic glycolysis upon bacterial detection16, and this metabolic reprogramming promotes pathogen clearance by modulating immune responses17,18. However, bacteria can exploit these metabolic shifts to enhance their survival. For instance, Listeria monocytogenes can modulate mitochondrial Ca²+ signaling, thus altering metabolism and promoting phagosomal escape19.
In this study, we found that PDCD6 downregulates antibacterial activity and serves as a negative regulator of LAP. PDCD6 deficiency in both macrophages and mice enhanced defenses against intracellular bacteria by enhancing LAP. PDCD6 interacts with LDHA, reducing lactate metabolism and, subsequently, RUBCN lactylation at lysine 33 (K33), a modification that is essential for RUBCN-mediated LAP activation. Our findings highlight a key role for PDCD6 in linking lactate metabolism to LAP during host defenses against bacterial infection.
Results
PDCD6 deficiency boosts host defenses against intracellular bacterial infection
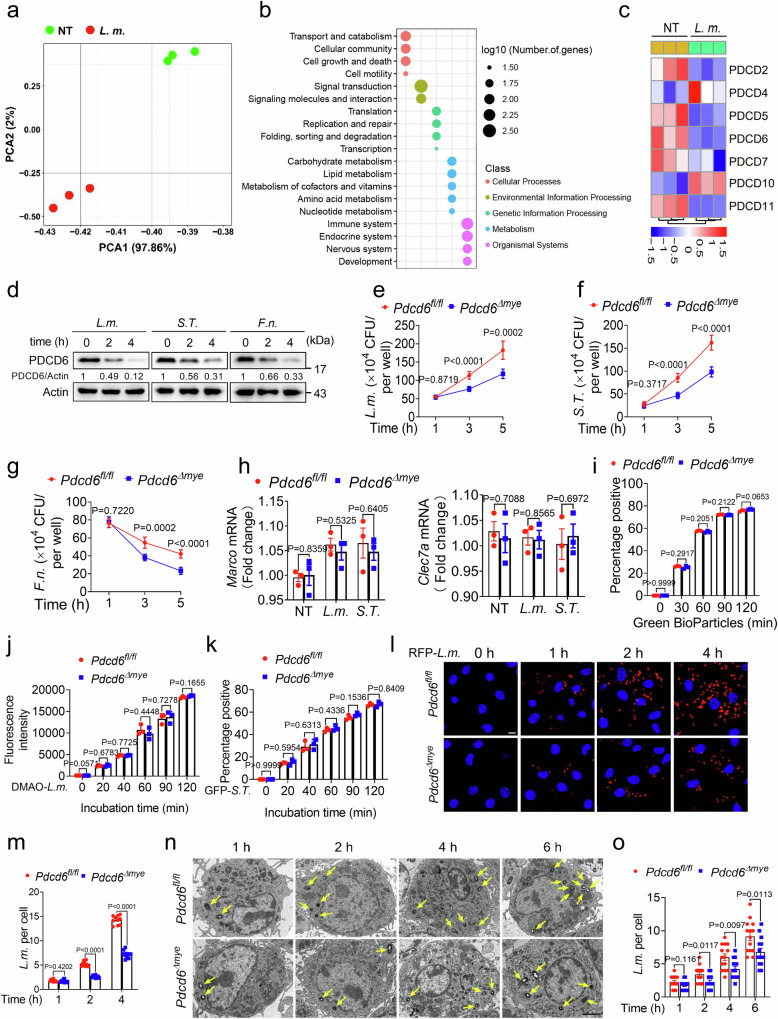
To investigate RNA expression changes during bacterial infection, mouse bone marrow-derived macrophages (BMDMs) were challenged with Listeria monocytogenes (L. monocytogenes) (DP-L5538), and then subjected to RNA sequencing. Principal component analysis (PCA) showed that there was a significant alteration in the RNA profile of the cells following L. monocytogenes challenge (Fig. 1a). Pathway enrichment analysis also identified changes in key pathways, including those involved in cellular processes, metabolism, and immune response (Fig. 1b). Given the critical role of cell death-related genes in host defenses against pathogens20,21, we subsequently focused on PDCD genes. After 2 h of infection, the expression levels of Pdcd2, Pdcd5, Pdcd6, and Pdcd11 were downregulated, while that of Pdcd10 remained unchanged (Fig. 1c, Supplementary Table 1). Notably, Pdcd6 exhibited the highest baseline expression level (Supplementary Table 1), indicative of its potential importance in infection and innate immunity13. We further confirmed that the protein expression of PDCD6 was significantly decreased following challenge with L. monocytogenes (DP-L5538), Salmonella Typhimurium (S. Typhimurium), or Francisella novicida (F. novicida) (Fig. 1d). These findings demonstrated that intracellular bacterial infection significantly decreased PDCD6 expression in macrophages.
Fig. 1. PDCD6 deficiency facilitates bactericidal activity.
a, b RNA sequencing-based total RNA profiling of mouse bone marrow-derived macrophages (BMDMs) treated with L. monocytogenes (DP-L5538) (multiplicity of infection [MOI]: 5) for 0 or 2 h was assessed by principal component analysis (a) and pathway enrichment analysis (b). c Heatmap of programmed cell death (PDCD) family genes in BMDMs challenged with L. monocytogenes for 0 or 2 h. d Relative protein expression of PDCD6 in BMDMs stimulated or not with L. monocytogenes (DP-L5538) (MOI: 5), S. Typhimurium (MOI: 5), or F. novicida (MOI: 20). Numbers indicate the values of the densitometric analysis of PDCD6 signals relative to actin. e–g Gentamicin protection assays in Pdcd6fl/fl and Pdcd6Δmye BMDMs infected with the indicated bacteria for 1, 3, and 5 h. Five minutes after the addition of gentamicin-containing medium, the incubation time was considered as 0 h. h Relative mRNA expression of Marco and Clec7a in Pdcd6fl/fl and Pdcd6Δmye BMDMs. i–k Flow cytometric analysis of Green BioParticles (i), DMAO-labeled L. monocytogenes (j), and GFP-S. Typhimurium (k) in Pdcd6fl/fl and Pdcd6Δmye BMDMs for the indicated periods. l, m Fluorescence microscopic images of RFP (l) and the quantification of fluorescent signals (m) to determine the bacteria-killing ability of Pdcd6fl/fl and Pdcd6Δmye BMDMs. Scale bar, 5 µm. n, o Transmission electron micrographs of Pdcd6fl/fl and Pdcd6Δmye BMDMs challenged with L. monocytogenes for a set period (n) and the quantification of intracellular L. monocytogenes (o). Scale bar, 2 µm. The averages of n = 6 (e–g), n = 3 (h–k), n = 10 (m), and n = 14 (o) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (e–k, m, o) was determined using the t test (and nonparametric tests). The data in (d–o) are representative of three independent experiments.
To explore the role of PDCD6 in antibacterial defenses, a mouse line with myeloid-specific Pdcd6 deletion (Pdcd6Δmye) was generated by crossing Pdcd6fl/fl mice with Lyz2 Cre mice (Supplementary Fig. 1a). Pdcd6 knockout was confirmed via genotyping and immunoblotting (Supplementary Fig. 1b, c). BMDMs from Pdcd6Δmye mice displayed significantly reduced intracellular bacterial growth compared with that in Pdcd6fl/fl BMDMs in a gentamicin protection assay (Fig. 1e–g). However, no differences in the expression levels of the phagocytic receptors MARCO and CLEC7A (Fig. 1h) or the phagocytosis of bioparticles, L. monocytogenes, or S. Typhimurium were observed between BMDMs derived from Pdcd6fl/fl and Pdcd6Δmye mice (Fig. 1i–k). The gating strategies for cell sorting were optimized for the detection of green fluorescence signals (Supplementary Fig. 1d). Fluorescence microscopy and transmission electron microscopy (TEM) further confirmed that bacterial growth was decreased in Pdcd6Δmye BMDMs (Fig. 1l–o), with similar results being obtained in Pdcd6Δmye peritoneal macrophages (Supplementary Fig. 1e–h). To determine whether the attenuated intracellular bacterial growth observed in Pdcd6Δmye BMDMs was due to enhanced bacterial killing, we analyzed the intracellular growth of the L. monocytogenes Δhly strain. Bacteria of this strain lack the hly gene, which encodes listeriolysin O (LLO), a membrane pore-forming protein implicated in bacterial escape, and remain confined within the phagosome. We found that the number of live bacteria was notably reduced in Pdcd6Δmye BMDMs compared with that in Pdcd6fl/fl BMDMs, suggesting that phagosomal bacterial killing was enhanced in the former (Supplementary Fig. 1i).
Because L. monocytogenes (DP-L5538) shows a similar phenotype (Fig. 1e and Supplementary Fig. 1e) to L. monocytogenes (10403S) (Supplementary Fig. 1f), we used DP-L5538 in this study. The knockdown of PDCD6 using siRNA also significantly reduced intracellular bacterial growth in THP-1 cells (Supplementary Fig. 1j, k). However, no differences in cell viability or calcium mobilization were observed between Pdcd6fl/fl and Pdcd6Δmye BMDMs, with or without bacterial stimulation (Supplementary Fig. 1l–n). This indicated that PDCD6 downregulation restricts intracellular bacterial growth in both human and mouse cells.
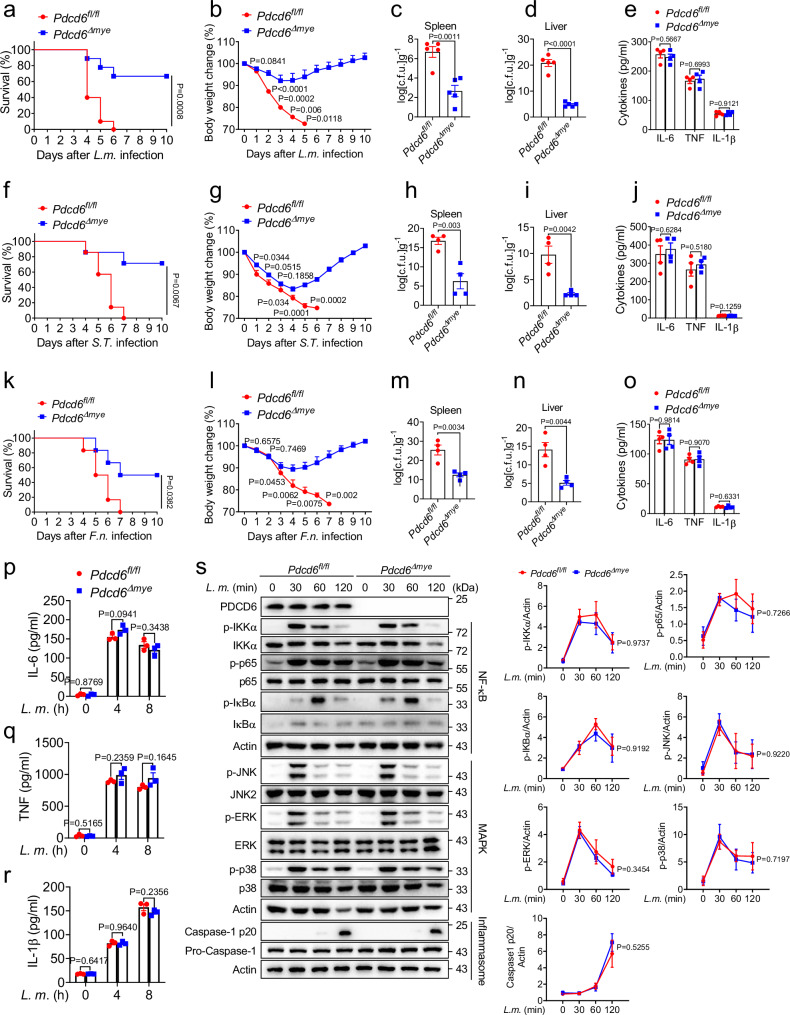
To clarify the role of PDCD6 in bacterial infection, Pdcd6fl/fl and Pdcd6Δmye mice were intraperitoneally infected with L. monocytogenes. Pdcd6Δmye mice showed reduced mortality (Fig. 2a), less body weight loss (Fig. 2b), and lower bacterial burdens in the spleen and liver (Fig. 2c, d) relative to Pdcd6fl/fl mice. Similar effects were observed with S. Typhimurium and F. novicida infections (Fig. 2f–i, k–n). Additionally, mice of the two groups generated comparable levels of IL-6, TNF, and IL-1β in response to infection (Fig. 2e, j, o).
Fig. 2. PDCD6 regulates intracellular bacterial infection in vivo.
a, b Survival (a) and body-weight changes (b) in Pdcd6fl/fl and Pdcd6Δmye mice (n = 10/group, 5 males and 5 females) after treatment with L. monocytogenes (DP-L5538) (0.1 × 106 CFUs). c–e Bacterial loads in the spleen (c) and liver (d), and IL-6, TNF, and IL-1β levels (e) in serum from Pdcd6fl/fl and Pdcd6Δmye mice challenged with L. monocytogenes for 24 h (n = 5/group, 3 males and 2 females). f, g Survival (f) and body-weight changes (g) in Pdcd6fl/fl and Pdcd6Δmye mice (n = 7/group, 3 males and 4 females) after treatment with S. Typhimurium (0.1 × 106 CFUs). h–j Bacterial loads in the spleen (h) and liver (i), and IL-6, TNF, and IL-1β levels (j) in serum from Pdcd6fl/fl and Pdcd6Δmye mice challenged with S. Typhimurium for 24 h (n = 4/group, 2 males and 2 females). k, l Survival (k) and body-weight changes (l) in Pdcd6fl/fl and Pdcd6Δmye mice (n = 6/group, 3 males and 3 females) after the intraperitoneal injection of F. novicida (1 × 106 CFUs). m–o Bacterial loads in the spleen (m) and liver (n), and IL-6, TNF, and IL-1β levels (o) in serum from Pdcd6fl/fl and Pdcd6Δmye mice (n = 4/group, 2 males and 2 females) challenged with F. novicida for 24 h. p–r Cytokine concentrations in the supernatants of mouse Pdcd6fl/fl and Pdcd6Δmye bone marrow-derived macrophages (BMDMs) stimulated or not with L. monocytogenes for the indicated periods. s NF-κB-, MAPK-, and inflammasome activation-related signaling molecules in Pdcd6fl/fl and Pdcd6Δmye BMDMs infected or not with L. monocytogenes (MOI: 5) for the indicated periods and their densitometric analysis. The averages of n = 10 (a, b), n = 5 (c–e), n = 7 (f, g), n = 4 (h–j, m–o), n = 6 (k, l), and n = 3 (p–s) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (a–s) was determined using t tests (and nonparametric tests). The data presented in (a–s) are representative of three independent experiments.
NRAMP1 (SlC11A1/Ity/Lsh/Bcg) plays a vital part in the innate immune response to Salmonella infection22–24 and Nramp1 mutations in C57BL/6 mice affect infection outcomes in a manner that is dependent on factors such as bacterial strain and dose25–27. Here, we found that a high dose (5 × 105 CFU/mouse, administered intraperitoneally) of S. Typhimurium (ATCC 14028) caused rapid mortality and weight loss in both Pdcd6fl/fl and Pdcd6Δmye mice, reflecting the Nramp1 mutation phenotype (Supplementary Fig. 2a, b)25,26. However, at a lower dose (1 × 105 CFU/mouse), Pdcd6Δmye mice showed prolonged survival and more robust antibacterial responses (Fig. 2f–i), supporting that PDCD6 deficiency enhances the resilience of mice to bacterial infection.
Macrophages trigger the inflammatory response following PAMP recognition by pattern recognition receptors28,29. Accordingly, we next examined whether PDCD6 deficiency affects cytokine production during bacterial infection. Pdcd6-deficient BMDMs produced similar levels of IL-6, TNF, and IL-1β (Fig. 2p–r; Supplementary Fig. 2c–i) and showed no differences in NF-κB or MAPK signaling or inflammasome activation (Fig. 2s; Supplementary Fig. 2j) when stimulated with L. monocytogenes or S. Typhimurium. Pdcd6Δmye BMDMs also exhibited comparable IL-1β production in response to ATP and nigericin (Supplementary Fig. 2k, l), indicating that PDCD6 is dispensable for NLRP3 inflammasome activation. These findings imply that Pdcd6 deficiency enhances defenses against intracellular bacteria without altering cytokine production.
PDCD6 deficiency promotes LAP
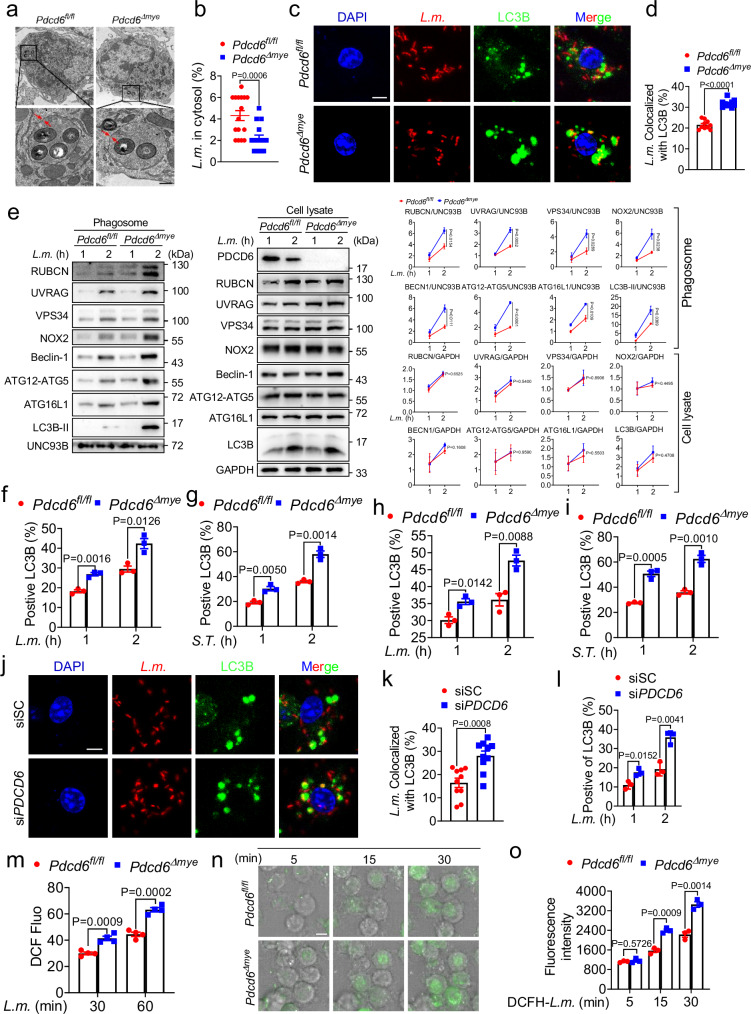
LAP represents a key defense against pathogens such as bacteria and fungi2,3,30. Here, we found that intracellular L. monocytogenes were mostly retained within single-membrane phagosomes, and fewer bacteria escaped in Pdcd6Δmye BMDMs than in Pdcd6fl/fl BMDMs after 2 h of infection (Fig. 3a, b). Additionally, there was significantly more widespread colocalization between L. monocytogenes and LC3B (microtubule-associated protein 1 light chain 3 beta, MAP1LC3B) puncta in Pdcd6Δmye BMDMs than in Pdcd6fl/fl BMDMs (Fig. 3c, d). Immunoblotting confirmed that LAP-associated molecule accumulation was greater in Pdcd6Δmye phagosomes than in the controls (Fig. 3e), while flow cytometry indicated that the LC3B positivity ratio during LAP was significantly higher in both Pdcd6Δmye BMDMs (Fig. 3f, g) and peritoneal macrophages (Fig. 3h, i) than in Pdcd6fl/fl cells following challenge with L. monocytogenes or S. Typhimurium. This indicated that Pdcd6 deletion enhanced LAPosome formation. The gating strategies employed for cell sorting were specifically designed to detect LC3B levels (Supplementary Fig. 3a).
Fig. 3. PDCD6 deficiency promotes LAP.
a, b Transmission electron micrographs of bacteria-containing single-membrane phagosomes (a) and quantification of cytosolic bacteria in bone marrow-derived macrophages (BMDMs) derived from Pdcd6fl/fl and Pdcd6Δmye mice (50 cells per group) (b) infected with L. monocytogenes. Scale bar, 0.5 µm. c, d Confocal microscopic images (c) and quantification (d) of the colocalization between L. monocytogenes (red) and LC3B puncta (green) in Pdcd6fl/fl and Pdcd6Δmye BMDMs (30 cells per group) challenged with RFP-L. monocytogenes. Scale bar, 2 µm. e Immunoblotting of LAP-associated molecules in phagosomes (left) and whole-cell lysates (right) isolated from Pdcd6fl/fl and Pdcd6Δmye BMDMs after bacterial challenge. f–i Flow cytometric analysis of the LC3B positivity ratio in Pdcd6fl/fl and Pdcd6Δmye BMDMs (f, g) or peritoneal macrophages (h, i) infected with L. monocytogenes (MOI: 5) (f, h) or S. Typhimurium (MOI: 5) (g, i) for the indicated periods. j, k Confocal microscopic images (j) and quantification (k) of the colocalization between L. monocytogenes (red) and LC3B puncta (green) in siCtrl and siPDCD6 THP-1 cells. Scale bar, 2 µm. l Flow cytometric analysis of the LC3B positivity ratio in siCtrl and siPDCD6 THP-1 cells infected with L. monocytogenes (MOI: 5) for the indicated periods. m–o Determination of reactive oxygen species (ROS) levels using DCFH-DA in the indicated BMDMs stimulated with L. monocytogenes. (m) Time-lapse confocal microscopic images (n) and quantification (o) of ROS fluorescence in phagosomes containing DCFH-SE-labeled L. monocytogenes (green). n Time (minutes) is indicated at the top of each panel. Scale bar, 5 µm. The averages of n = 16 (b), n = 10 (d, k), n = 3 (f–i, l, o), and n = 4 (m) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (b, d, f–i, k–m, o) was determined using the t test (and nonparametric tests). The data presented in (a, c, e–j, l–o) are representative of three independent experiments.
As LAP promotes bacterial killing by increasing phagosome–lysosome fusion31, we investigated whether PDCD6 was involved in this process. We found that bacteria/LAMP1 colocalization was greater in Pdcd6Δmye BMDMs than in Pdcd6fl/fl BMDMs (Supplementary Fig. 3b, c). Moreover, Förster resonance energy transfer (FRET) analysis confirmed that Pdcd6 deficiency enhanced phagosome–lysosome fusion (Supplementary Fig. 3d).
We further assessed the role of PDCD6 in human antibacterial immunity by knocking down PDCD6 in THP-1 cells via siRNA transfection. Compared with that seen in the control condition (scrambled siRNA treatment), the knockdown of PDCD6 led to increased colocalization of RFP-L. monocytogenes with LC3B (Fig. 3j, k), accompanied by an increase in the LC3B positivity ratio during LAP (Fig. 3l, Supplementary Fig. 3e). LAP requires NOX2-mediated ROS production31,32. In line with this, we observed that Pdcd6Δmye BMDMs exhibited higher total and phagosomal ROS levels following bacterial challenge compared with those in Pdcd6fl/fl BMDMs (Fig. 3m–o, Supplementary Movie 1 and 2). Combined, the above results indicated that Pdcd6 deficiency enhances LAP and phagosome–lysosome fusion.
LAP is required for the PDCD6 deficiency-mediated enhancement of bacterial clearance
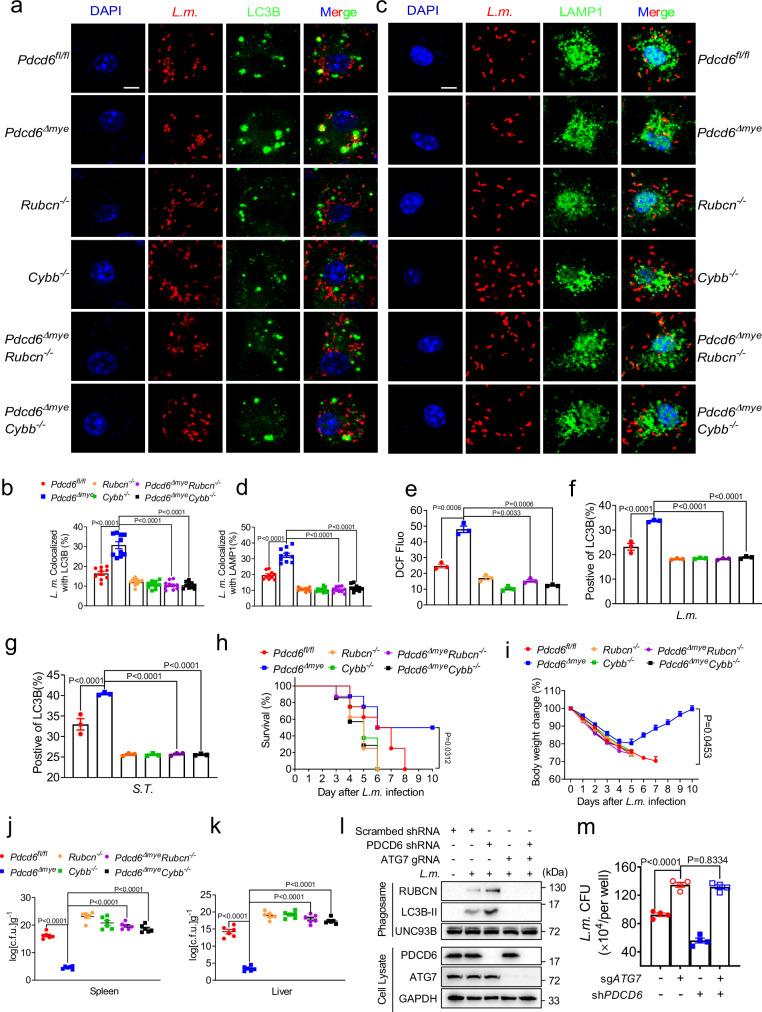
Given that we found that Pdcd6 deficiency promoted LAPosome formation and maturation, we sought to determine whether LAP plays a role in the Pdcd6 deficiency-mediated enhancement of bacterial clearance. For this, we generated Pdcd6ΔmyeRubcn−/− and Pdcd6ΔmyeCybb−/− mouse lines by crossing Pdcd6Δmye mice with mice lacking the Rubcn and Cybb genes, respectively, both of which are important regulators of LAPosome formation. BMDMs derived from both the Pdcd6ΔmyeRubcn−/− and Pdcd6ΔmyeCybb−/− lines showed reduced bacteria-LC3B colocalization (Fig. 4a, b), phagosome–lysosome fusion (Fig. 4c, d), and ROS production (Fig. 4e, Supplementary Fig. 4a). The LC3B positivity ratio during LAP was also diminished (Fig. 4f, g), leading to defective bacterial clearance in vitro (Supplementary Fig. 4b, c). Mice with these deletions exhibited higher mortality rates and bacterial loads after L. monocytogenes challenge (Fig. 4h–k). Furthermore, the knockdown of PDCD6 in THP-1 cells lacking the ATG7 gene, which plays an essential role in LAPosome formation, disrupted LAP and bacterial killing (Fig. 4l, m). These data demonstrated that LAP is essential for the efficient clearance of intracellular bacteria in Pdcd6-deficient macrophages and mice.
Fig. 4. PDCD6 deficiency-mediated enhancement of bacterial clearance requires LAP.
a–d Confocal microscopic images (a, c) and quantification (b, d) of the colocalization between L. monocytogenes (red) and LC3B (green) (a, b) or between L. monocytogenes (red) and LAMP1 (green) (c, d) in Pdcd6fl/fl, Pdcd6Δmye, Rubcn−/−, Pdcd6ΔmyeRubcn−/−, Cybb−/−, and Pdcd6ΔmyeCybb−/− mouse bone marrow-derived macrophages (BMDMs) challenged with RFP-L. monocytogenes. Scale bar, 2 µm. e DCFH-DA-based determination of reactive oxygen species (ROS) levels in the indicated BMDMs stimulated with L. monocytogenes. f, g Flow cytometric analysis of LC3B fluorescence in Pdcd6fl/fl, Pdcd6Δmye, Rubcn−/−, Pdcd6ΔmyeRubcn−/−, Cybb−/−, and Pdcd6ΔmyeCybb−/− BMDMs infected with L. monocytogenes (MOI: 5) (f) or S.Typhimurium (MOI: 5) (g) for 2 h. h, i Survival (h) (one-way ANOVA with multiple comparisons; Pdcd6Δmye vs. Rubcn−/−, Pdcd6ΔmyeRubcn−/−, Cybb−/−, and Pdcd6ΔmyeCybb−/−, P = 0.0312) and body-weight changes (i) (one-way ANOVA with multiple comparisons; Pdcd6Δmye vs. Rubcn−/−, Pdcd6ΔmyeRubcn−/−, Cybb−/−, and Pdcd6ΔmyeCybb−/−, P = 0.0453) in Pdcd6fl/fl, Pdcd6Δmye, Rubcn−/−, Pdcd6ΔmyeRubcn−/−, Cybb−/−, and Pdcd6ΔmyeCybb−/− mice (n = 9 per group, 5 males and 4 females) intraperitoneally injected with L. monocytogenes. j, k Bacterial loads in the spleen (j) and liver (k) of Pdcd6fl/fl, Pdcd6Δmye, Rubcn−/−, Pdcd6ΔmyeRubcn−/−, Cybb−/−, and Pdcd6ΔmyeCybb−/− mice challenged with L. monocytogenes for 24 h (n = 6 per group, 3 males and 3 females). l, m Immunoblotting of LAP-associated molecules in isolated phagosomes or total cell lysates (l) and gentamicin protection assays (m) in wild-type, PDCD6-knockdown (KD), ATG7-knockout (KO), or PDCD6/ATG7 double-KO THP-1 cells following L. monocytogenes challenge. The averages of n = 10 (b, d), n = 3 (e–g), n = 9 (h, i), n = 6 (j, k), and n = 4 (m) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (b, d–k, m) was determined using one-way ANOVA with multiple comparisons. The data presented in (a, c, e–m) are representative of three independent experiments.
To assess whether PDCD6 influences bacterial clearance via autophagy, we examined L. monocytogenes/p62 colocalization in Pdcd6fl/fl and Pdcd6Δmye BMDMs. No significant differences were found between the two groups (Supplementary Fig. 4d, e), suggesting that PDCD6 does not impact autophagic clearance. Because NOX2 reduces phagosome acidification33, we tested whether the LAP-mediated augmentation of antibacterial effects in Pdcdc6-deficient cells was dependent on NOX2 or lysosomal acidification. Pretreatment with N-acetylcysteine (NAC; a ROS scavenger) or NH4Cl (raises vesicular pH)34,35 increased bacterial growth in both Pdcd6fl/fl and Pdcd6Δmye BMDMs (Supplementary Fig. 4f). NOX2-generated ROS plays a significant role in facilitating LC3B recruitment to phagosomes31,32,36 as well as in raising phagosomal pH to activate the V-ATPase/ATG16L1 axis for LC3B conjugation to phagosomal membranes37. Accordingly, NAC treatment likely impaired LAP formation, leading to decreased L. monocytogene clearance. This inhibitory effect parallels the disruption observed when lysosomal function is impaired, providing compelling evidence that enhanced LAPosome formation represents the primary mechanism underlying the heightened bactericidal activity observed in Pdcd6-deficient cells.
Elevated lactate levels augment host defenses against intracellular bacteria
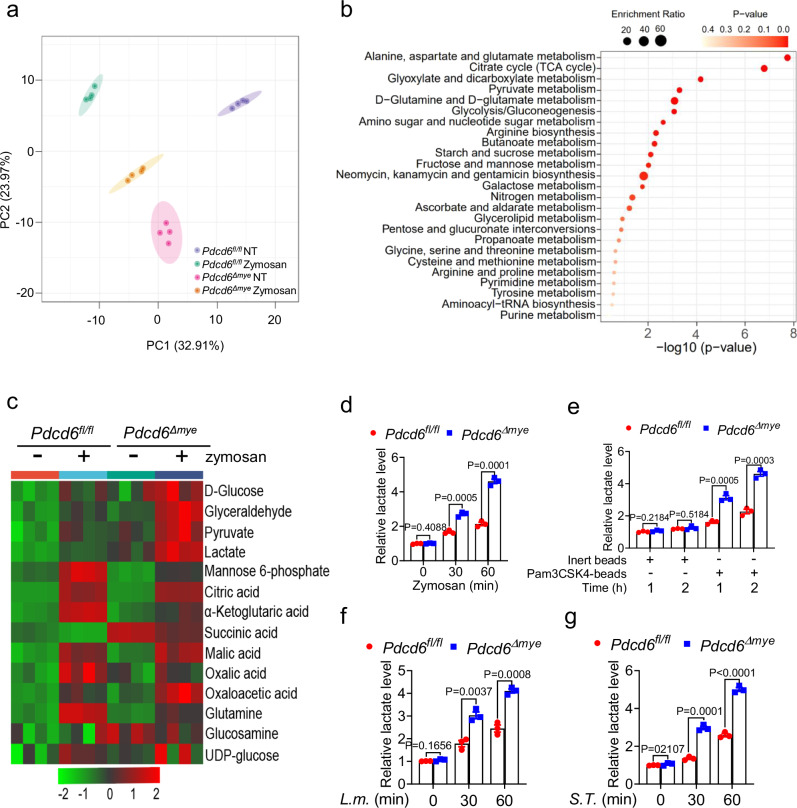
Microbial infection triggers metabolic reprogramming in macrophages, leading to the accumulation of metabolites that support pathogen clearance16. However, the interplay between LAP and metabolism remains largely unexplored. To identify changes in the metabolic profile that occur during LAP, metabolomic assays were conducted on Pdcd6fl/fl and Pdcd6Δmye BMDMs following zymosan stimulation. PCA revealed the presence of distinct metabolic profiles between Pdcd6fl/fl and Pdcd6Δmye BMDMs (Fig. 5a). Furthermore, pathway enrichment analysis highlighted significant alterations in key pathways, such as those involved in sugar, nucleotide, amino acid, and glycerolipid metabolism (Fig. 5b). Notably, compared with control cells, glycolysis and pyruvate metabolism were upregulated in Pdcd6Δmye BMDMs, as evidenced by the significant increase in lactate levels in the latter (Fig. 5c–g, Supplementary Table 2). Similar trends were detected in PDCD6-knockdown THP-1 cells (Supplementary Fig. 5a–d), suggesting that PDCD6 suppresses lactate accumulation.
Fig. 5. PDCD6 deficiency elevates lactate metabolism.
a, b LC–MS/MS-based total metabolite profiling using principle component analysis (a) and pathway enrichment analysis (b) in Pdcd6fl/fl and Pdcd6Δmye bone marrow-derived macrophages (BMDMs) treated with zymosan (10 μg/mL) for 2 h. c Heatmap of glucose metabolites in Pdcd6fl/fl and Pdcd6Δmye BMDMs treated or not with zymosan. d–g Lactate levels in Pdcd6fl/fl and Pdcd6Δmye BMDMs treated with zymosan (10 μg/mL) (d), Pam3CSK4-coated beads (10 μg/mL) (e), L. monocytogenes (MOI: 5) (f), or S. Typhimurium (MOI: 5) (g) for the indicated periods. The averages of n = 3 (d–g) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (d–g) was determined using the t test (and nonparametric tests). The data presented in (d–g) are representative of three independent experiments.
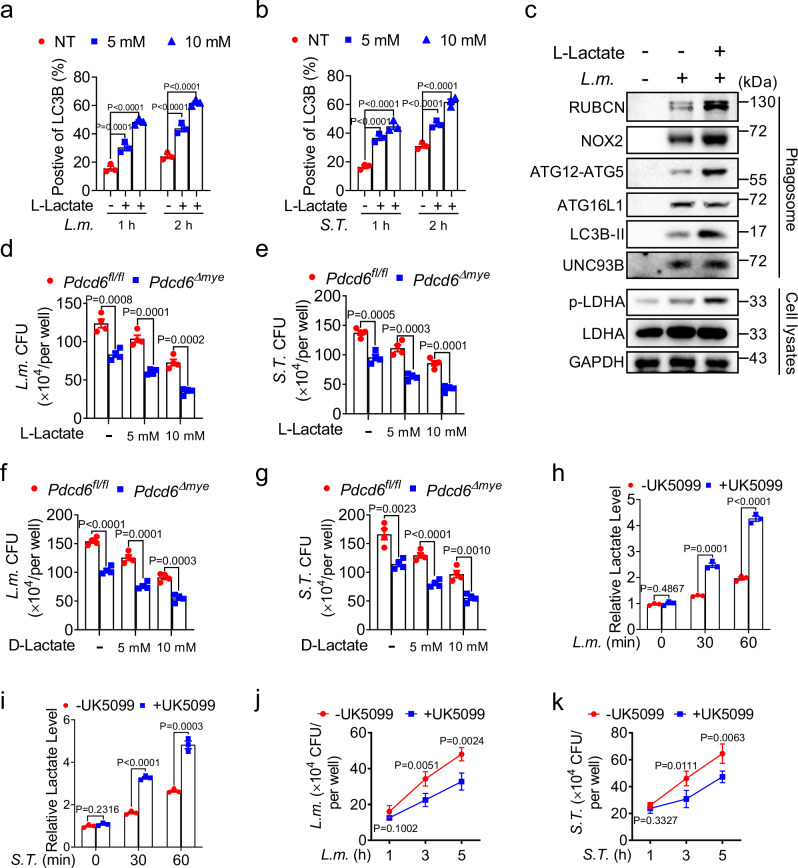
To investigate the role of lactate in LAP, flow cytometric analysis of LC3B and phagosome immunoblotting assays were conducted after L. monocytogenes challenge. Sodium lactate treatment enhanced LAP in a dose- and time-dependent manner (Fig. 6a–c) while pretreatment with either L- or D-sodium lactate increased the antibacterial activity of Pdcd6Δmye BMDMs (Fig. 6d–g). Similarly, treatment with UK-5099, a pyruvate carrier inhibitor, raised lactate levels (Fig. 6h, i) and boosted bactericidal effects (Fig. 6j, k). These results suggested that augmented lactate levels in Pdcd6Δmye BMDMs promote intracellular bacteria clearance.
Fig. 6. Lactate enhances host defenses against bacterial infection.
a, b Flow cytometric analysis of LC3B fluorescence in wild-type (WT) bone marrow-derived macrophages (BMDMs) pretreated or not with sodium L-lactate (10 or 20 mM) and then stimulated with L. monocytogenes (a) or S. Typhimurium (b) for the indicated periods. c Immunoblotting of LAP-related molecules in phagosomes and whole-cell lysates isolated from BMDMs pretreated or not with sodium L-lactate and then stimulated with L. monocytogenes. d–g Gentamicin protection assays in Pdcd6fl/fl and Pdcd6Δmye BMDMs treated or not with sodium L-lactate (d, e) or sodium D-lactate (f, g) and then challenged with L. monocytogenes (d, f) or S. Typhimurium (e, g). h, i Lactate levels in BMDMs pretreated or not with UK5099 (10 μM) for 16 h, and then challenged with L. monocytogenes (h) or S. Typhimurium (i) for the indicated periods. j, k Gentamicin protection assays in BMDMs pretreated with UK5099 and then further stimulated with L. monocytogenes (j) or S. Typhimurium (k) for the indicated durations. The averages of n = 3 (a, b, h, i) and n = 4 (d–g, j, k) biologically independent samples are shown. Data are shown as the mean ± SEM. a, b Statistical significance was determined using one-way ANOVA with multiple comparisons; in (d–k), the t test (and nonparametric tests) was employed. The data presented in (a–k) are representative of three independent experiments.
LDHA is required for the PDCD6 deficiency-mediated enhancement of bactericidal effects
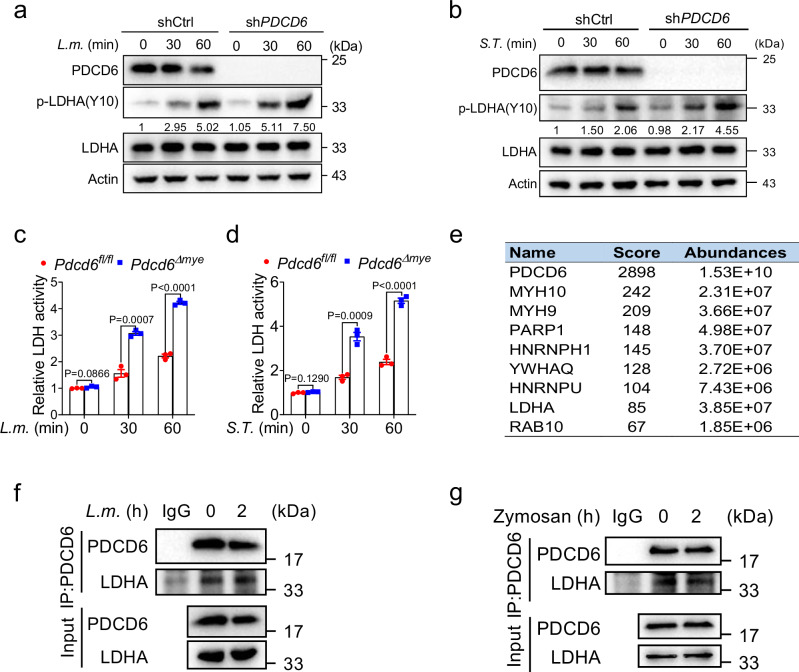
Lactate dehydrogenase A (LDHA) catalyzes the conversion of pyruvate to lactate (Supplementary Fig. 6a). The increase in lactate levels observed in Pdcd6-deficient cells suggested that PDCD6 suppresses LDHA activity. As Y10 phosphorylation is known to enhance LDHA activity38, we assessed LDHA expression and phosphorylation levels in wild-type and PDCD6-knockdown THP-1 cells after L. monocytogenes challenge. While LDHA expression was comparable between the two cell lines, Y10 phosphorylation was significantly higher in PDCD6-knockdown cells (Fig. 7a). Similar results were recorded with S. Typhimurium and zymosan challenge (Fig. 7b, Supplementary Fig. 6b). LDH activity assays confirmed that compared with the respective controls, LDH activity was increased in Pdcd6Δmye BMDMs (Fig. 7c, d) and PDCD6-knockdown cells (Supplementary Fig. 6c), whereas the overexpression of PDCD6 induced the opposite effect (Supplementary Fig. 6d). These findings suggested that PDCD6 downregulates LDHA activity.
Fig. 7. PDCD6 interacts with LDHA.
a, b Immunoblotting of p-LDHA in wild-type and PDCD6-knockdown (KD) THP-1 cells treated with L. monocytogenes (a) or S. Typhimurium (b) for the indicated periods. Numbers indicate the densitometric values of p-LDHA signals relative to actin. c, d LDH activity in Pdcd6fl/fl and Pdcd6Δmye bone marrow-derived macrophages (BMDMs) stimulated with L. monocytogenes (c) or S. Typhimurium (d) for the indicated times. e PDCD6 binding partners with high scores were identified through mass spectrometry. f, g Total PDCD6 was immunoprecipitated from BMDMs challenged or not with L. monocytogenes (f) or zymosan (g) for 2 h, followed by immunoblotting with anti-LDHA antibody. The averages of n = 3 (c, d) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (c, d) was determined using the t test (and nonparametric tests). The data presented in (a–d, f, g) are representative of three independent experiments.
To investigate how PDCD6 influences LDHA activity, a mass spectrometry-based proteomic analysis was performed on the PDCD6 interactome in HEK293T cells using Flag-tagged PDCD6. The results showed that LDHA was a binding partner of PDCD6 (Fig. 7e, Supplementary Data 1), an effect that was confirmed through immunoprecipitation assays (Supplementary Fig. 6e–g). Additionally, endogenous PDCD6 was noted to interact with LDHA following L. monocytogenes or zymosan challenge (Fig. 7f, g). Overall, these findings suggested that LDHA is a bona fide partner of PDCD6.
FGFR1 interacts with LDHA and mediates its phosphorylation at the Y10 site38. There were no significant changes in FGFR1 expression in Pdcd6fl/fl or Pdcd6Δmye BMDMs following exposure to L. monocytogenes or S. Typhimurium, indicating that PDCD6 does not regulate FGFR1 expression (Supplementary Fig. 7a). Immunoprecipitation assays showed that increasing PDCD6 levels weakened the FGFR1–LDHA interaction, resulting in reduced LDHA Y10 phosphorylation (Supplementary Fig. 7b). Notably, no direct interaction was observed between PDCD6 and FGFR1 (Supplementary Fig. 7c). Finally, L. monocytogenes challenge resulted in a significant increase in the endogenous interaction between LDHA and FGFR1, which was more pronounced in Pdcd6Δmye BMDMs. These results further confirmed that PDCD6 disrupted the FGFR1–LDHA interaction, diminishing the FGFR1-mediated activation of LDHA without altering FGFR1 expression in input samples (Supplementary Fig. 7d). These findings suggested that PDCD6 inhibits LDHA activity by disrupting FGFR1–LDHA interaction, leading to reduced FGFR1-mediated phosphorylation and activation of LDHA.
LDHA is mainly localized in the cytoplasm for glycolysis but has also been detected in mitochondria and nuclei39,40. To assess the effect of PDCD6 on LDHA localization, we analyzed LDHA distribution in Pdcd6fl/fl and Pdcd6Δmye BMDMs with or without L. monocytogenes challenge. Mitochondria were labeled with Mitotracker Red, and nuclei were marked with DAPI (Supplementary Fig. 7e). No significant difference in LDHA localization was detected between the two groups of cells, indicating that PDCD6 does not affect LDHA cellular localization. We next examined LDHA stability using cycloheximide (CHX) and found that LDHA stability was similar between Pdcd6fl/fl and Pdcd6Δmye BMDMs (Supplementary Fig. 7f). Analogous results were obtained with GFP-PDCD6-overexpressing THP-1 cells treated with CHX and the corresponding controls (Supplementary Fig. 7g). These data further suggested that PDCD6–LDHA interaction does not affect LDHA protein stability.
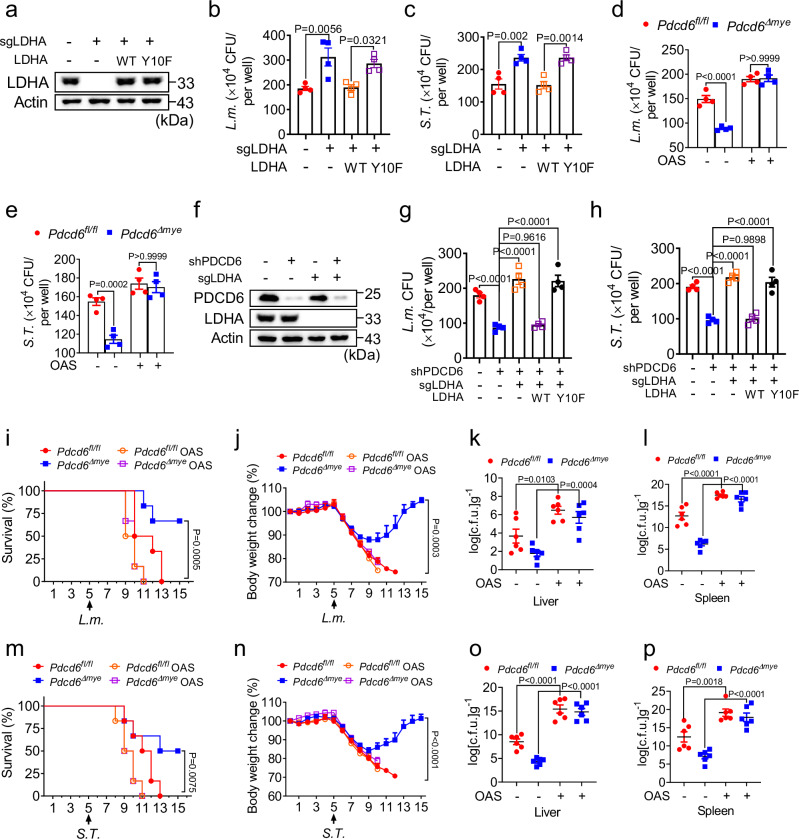
LDHA degradation is regulated by both the proteasome and lysosome in a post-translational modification-dependent manner. LDHA interacts with the E3 ligase FBW7, leading to LDHA ubiquitination and proteasomal degradation41. The CPT1A-mediated succinylation of LDHA at K222 reduces its lysosomal degradation42, while LDHA acetylation at K5 facilitates its lysosomal targeting via the HSC70 chaperone43. Here, we investigated these modifications in LDHA under elevated PDCD6 expression and found that ubiquitination, succinylation (Ksn), or acetylation (Ac-K) levels were not affected. This suggested that PDCD6 does not influence LDHA degradation pathways (Supplementary Fig. 7h). Lactate accumulation due to elevated LDHA activity in PDCD6-deficient cells may contribute to host defenses by enhancing LAP. To explore the role of LDHA in antibacterial immunity, we employed both genetic and pharmacologic approaches. LDHA knockout in THP-1 cells resulted in increased intracellular bacterial growth after L. monocytogenes or S. Typhimurium infection, similar to the effect seen in cells expressing the LDHA Y10F mutant, while the reintroduction of LDHA restored the normal antibacterial response (Fig. 8a–c). Treatment with sodium oxamate (OAS), an LDHA inhibitor44, also reduced lactate levels (Supplementary Fig. 8a), LAP-associated molecule recruitment (Supplementary Fig. 8b), and bactericidal activity (Fig. 8d–e). These data suggested that LDHA enzymatic activity positively regulates antibacterial immunity.
Fig. 8. LDHA is required for the PDCD6 deficiency-mediated enhancement of antibacterial effects.
a–c Immunoblotting and gentamicin protection assays in LDHA-knockout (KO) THP-1 cells transduced with empty vector, Flag-tagged wild-type (WT) LDHA, or LDHA Y10F (a) and then challenged with L. monocytogenes (b) or S. Typhimurium (c). d, e Gentamicin protection assays in Pdcd6fl/fl and Pdcd6Δmye bone marrow-derived macrophages (BMDMs) pretreated or not with oxamic acid sodium salt (OAS) (10 μM) for 24 h, and then challenged with L. monocytogenes (d) or S. Typhimurium (e). f–h Immunoblotting and gentamicin protection assays in PDCD6-knockdown (KD)/LDHA-knockout (KO) THP-1 cells (f) transduced with empty vector, Flag-tagged WT LDHA, or LDHA Y10F following stimulation with L. monocytogenes (g) or S. Typhimurium (h). i–p Survival (i, m) (one-way ANOVA with multiple comparisons; Pdcd6Δmye vs. Pdcd6fl/fl, Pdcd6fl/fl OAS, and Pdcd6Δmye OAS; P = 0.0005 [(i, P = 0.0075 [(m]), body-weight changes (j, n) (one-way ANOVA with multiple comparisons; Pdcd6Δmye vs. Pdcd6fl/fl, Pdcd6fl/fl OAS, and Pdcd6Δmye OAS; P = 0.0003 [(j], P < 0.0001 [(n]), and bacterial loads in the liver (k, o) and spleen (l, p) of Pdcd6fl/fl and Pdcd6Δmye mice pretreated with OAS for 5 days, and then treated with L. monocytogenes (n = 6 per group, 3 males and 3 females) (i–l) or S. Typhimurium (n = 6 per group, 3 males and 3 females) (m–p) for the indicated periods. The averages of n = 4 (b–e, g, h) and n = 6 (i–p) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance was determined using one-way ANOVA with multiple comparisons in (g–p) and the t test (and nonparametric tests) in (b–e). The data presented in (a–p) are representative of three independent experiments.
We further evaluated whether LDHA is essential for the enhanced bactericidal effect seen in PDCD6-deficient cells. The reconstitution of wild-type LDHA, but not the LDHA Y10F mutant, in THP-1 PDCD6-knockdown/LDHA-knockout cells restored the enhancement of bactericidal activity (Fig. 8f–h). To assess the role of LDHA enzymatic activity in vivo, OAS was administered to Pdcd6fl/fl and Pdcd6Δmye mice, resulting in the loss of the survival benefit and bacterial control in Pdcd6Δmye mice challenged with L. monocytogenes or S. Typhimurium (Fig. 8i–p).
To determine whether the LDHA Y10F mutation inhibits the recruitment of the PI3K complex and NOX2 to phagosomes, a phagosome isolation assay was performed in LDHA-knockout THP-1 cells reconstituted with wild-type LDHA or LDHA Y10F after L. monocytogenes challenge. Immunoblotting showed that wild-type LDHA, but not LDHA Y10F restored VPS34 and NOX2 levels (Supplementary Fig. 8c). Gentamicin protection assays confirmed that LDHA Y10F failed to restore the augmentation of the antibacterial effects seen with wild-type LDHA (Supplementary Fig. 8d). Together, these results demonstrated that LDHA is required for the PDCD6 deficiency-mediated enhancement of bactericidal effects.
The lactylation of RUBCN enhances RUBCN–VPS34 interaction
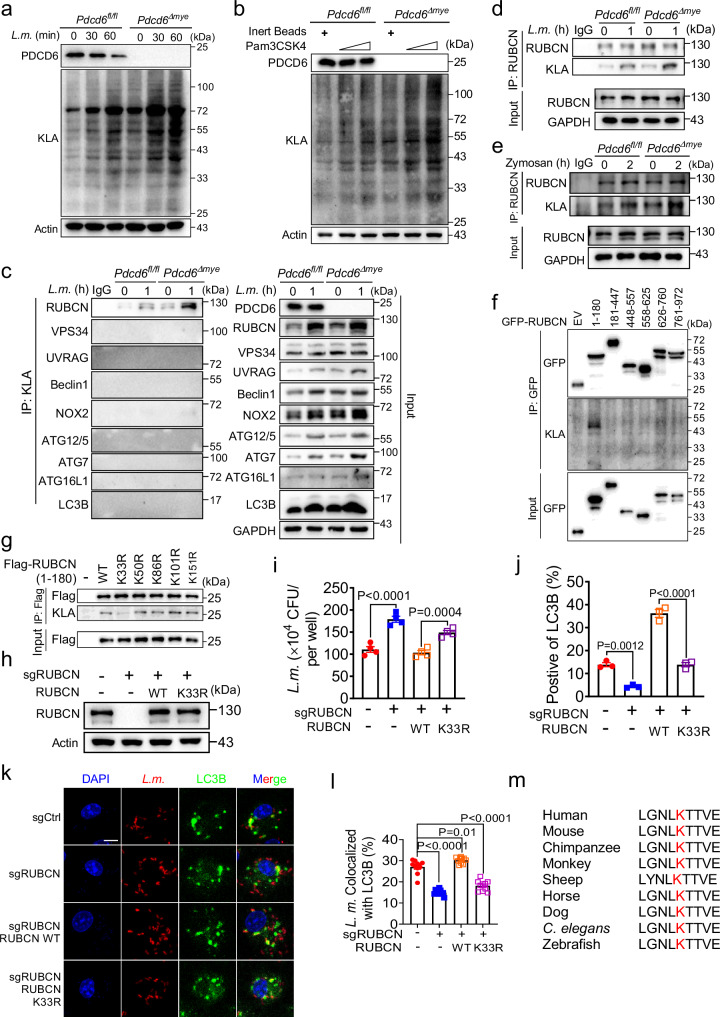
Lactyl-CoA, a downstream metabolite of lactate, donates lactyl groups for lysine lactylation (Supplementary Fig. 9a), a recently identified post-translational modification involved in the regulation of inflammation and immune responses45,46. Here, we evaluated total protein lactylation levels following bacterial infection and found that they were significantly higher in Pdcd6Δmye BMDMs than in control cells after challenge with L. monocytogenes or Pam3CSK4-coated beads (Fig. 9a, b). That elevated lactate levels enhanced LAP (Fig. 6c) while the inhibition of LDHA activity by OAS exerted the opposite effect (Supplementary Fig. 8b) suggested that upregulated protein lactylation may improve LAP during bacterial infection. Immunoprecipitation assays showed that Rubicon, but not other LAP-associated molecules, was lactylated, with higher levels being detected in Pdcd6Δmye BMDMs (Fig. 9c–e).
Fig. 9. Rubicon lactylation promotes LAP activation.
a, b Immunoblotting of total lactylation in Pdcd6fl/fl and Pdcd6Δmye BMDMs treated or not with L. monocytogenes (a) or Pam3CSK4 (b) for the indicated periods. c–e Total Kla (c) and Rubicon (d, e) were immunoprecipitated from Pdcd6fl/fl, and Pdcd6Δmye BMDMs challenged or not with L. monocytogenes (MOI: 5) (c, d) or zymosan (e), followed by immunoblotting with the indicated antibodies. f, g Lactylation mapping in RUBCN truncation mutants (f) or RUBCN K-to-M mutants (aa 1–180) (g) by immunoprecipitation using HEK293T cells. h Reconstitution of THP-1 cells with RUBCN WT or RUBCN K33R mutant. i–l Gentamicin protection assays (i), flow cytometric analysis of LC3B fluorescence (j), and colocalization analysis between LC3B and RFP-L. monocytogenes (k, l) in THP-1 cells with the reconstitution of either RUBCN WT or the RUBCN K33R mutant. Scale bar, 2 µm. m Cross-species sequence alignment of RUBCN highlighted the conservation of the K33 lactylation site. The averages of n = 4 (i), n = 3 (j), and n = 10 (l) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance in (i, j, l) was determined using the t test (and nonparametric tests). The data presented in (a–k) are representative of three independent experiments.
To identify lactylation sites on Rubicon, we transfected plasmids harboring truncated Rubicon into HEK293T cells and performed an immunoprecipitation assay with an anti-Kla antibody. Rubicon was found to be lactylated on the fragment containing amino acids (aa) 1–180 (Fig. 9f). Lysine-to-arginine mutations in this region greatly reduced RUBCN K33R lactylation levels (Fig. 9g). To confirm the importance of K33 in LAPosome formation and antibacterial activity, RUBCN-knockout THP-1 cells were reconstituted with wild-type RUBCN or RUBCN harboring the K33R mutation. Unlike that observed with wild-type RUBCN, reconstitution with RUBCN K33R did not result in bacterial growth inhibition, LAPosome formation, or RUBCN colocalization with LC3B (Fig. 9h–l, Supplementary Fig. 9b, c), and also failed to restore VPS34 and NOX2 levels (Supplementary Fig. 9d) and antibacterial effects in RUBCN-knockout THP-1 cells (Supplementary Fig. 9e). Cross-species alignment indicated that the K33 site was highly conserved, highlighting its biological importance (Fig. 9m).
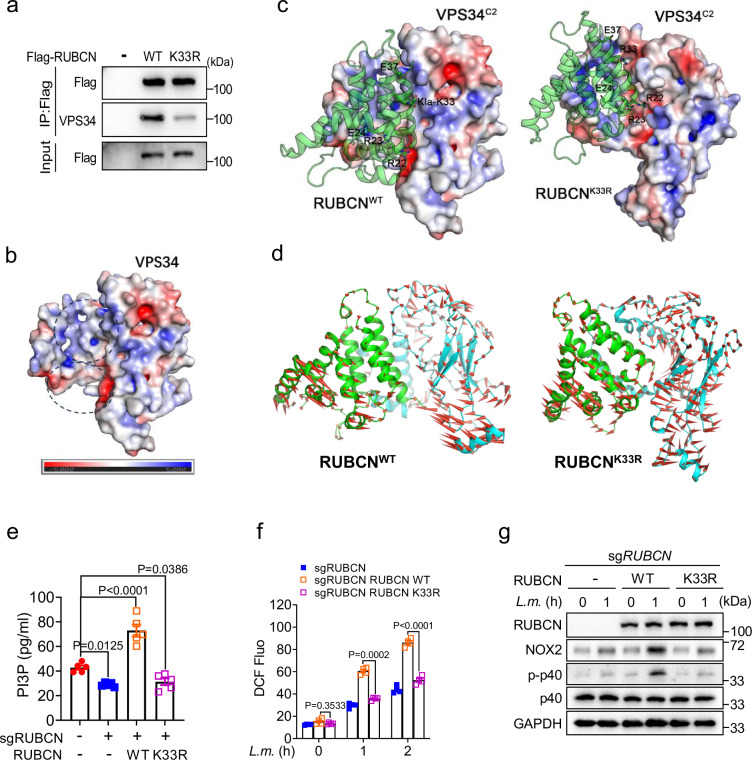
The K33 residue in RUBCN, located close to the RUN domain (aa 49–180) required for VPS34 binding47, was examined for its influence on RUBCN–VPS34 interaction. Immunoprecipitation analysis revealed that the RUBCN K33R mutation resulted in a significantly weakened interaction between RUBCN and VPS34 compared with that seen with wild-type RUBCN (Fig. 10a). To explore the effect of RUBCN lactylation on the interaction between these two proteins, we undertook a molecular simulation analysis. The RUBCN structure was predicted using AlphaFold248. The resulting model, which included the N-terminus and the RUN domain (RUBCNRUN, aa: 17–177), was docked with the PI3K-type C2 domain of VPS34 (VPS34C2, aa: 5–245, cryo-EM structure 7BL1), which is responsible for Rab5A binding49. Analysis of the molecular dynamics (MD) simulation trajectories of lactylated wild-type RUBCN or non-lactylated RUBCNK33R complexed with VPS34C2 demonstrated that the interaction between RUBCNK33R and VPS34C2 was significantly weaker than that between RUBCNWT and VPS34C2, as evidenced by the calculated binding free energy value (Supplementary Table 3). Further analysis indicated that the underlying cause of the difference was related to the surface charge properties surrounding the K33 lactylation site. The area involved in the binding of RUBCNRUN to VPS34C2 exhibited distinct surface charge properties in different parts, with the upper half possessing a strong positive charge and the lower part a strong negative charge (Fig. 10b). Lactylation at K33 altered the surface charge distribution (Supplementary Fig. 9f) of RUBCNRUN, particularly the area around the K33 site, resulting in a shift from a positive to a negative charge. This area is located close to the positively charged portion of the VPS34C2 binding area, and the change resulted in surface charge complementarity in the RUBCNWT system, involving not only the area around K33 but also the other residues on the interface (Fig. 10c). PCA is typically used to describe the overall dynamics of systems in MD simulations. The related dynamics states were exactly the opposite in the RUBCNWT and RUBCNK33R systems. In the former, a stable interaction caused the two proteins to show a strong coupling trend, which also confirmed the trends observed in RoG and interface area curves (Fig. 10d). Thus, these data indicated that Rubicon lactylation on K33 promotes Rubicon–VPS34 interaction. We explored alternatives to lysine lactylation using natural amino acids, but identifying suitable mimics proved challenging due to the unique structural and physical properties of this modification. Mutating RUBCN K33 to arginine (R), glutamic acid (E), aspartic acid (D), glutamine (Q), asparagine (N), or threonine (T) led to a significant reduction in lactylation (Supplementary Fig. 9g). Additionally, although R33, E33, and D33 showed some similarities to K33, the charge differences posed limitations. Similarly, neutral polar residues such as Q33, N33, and T33 also shared some traits with K33 but failed to fully mimic lysine lactylation.
Fig. 10. The lactylation of RUBCN enhances its interaction with VPS34.
a RUBCN–VPS34 interaction was assessed in HEK293T cells transfected with either wild-type (WT) RUBCN or its K33R variant. b Computed structure model of VPS34. The black dashed circles indicate the upper and lower parts of VPS34. The color bar represents the surface charge distribution of VPS34, with red denoting a negative charge and blue a positive charge. c The molecular dynamics (MD) simulation results for the RUBCNWT and RUBCNK33R systems. In the last frame of the MD simulation, the sticks represent key charged residues on the interface, with E denoting a negatively charged sidechain and R a positively charged sidechain. d Principal component analysis (PCA) for RUBCN-WT and RUBCN-K33R was performed based on the latter half (250–500 ns) of the simulation. The arrows indicate the direction of motion of RUBCN (green) and VPS34 (cyan) during the trajectories. e–g PtdIns(3)P contents (e), reactive oxygen species (ROS) production (f), and NOX complex activity (g) in THP-1 cells with reconstitution of WT RUBCN or its K33R mutant and treated or not with L. monocytogenes for the indicated periods. The averages of n = 5 (e) and n = 4 (f) biologically independent samples are shown. Data are shown as the mean ± SEM. Statistical significance was determined using one-way ANOVA with multiple comparisons in (e) and two-way ANOVA with multiple comparisons in (f). The data presented in (a, e–g) are representative of three independent experiments.
Rubicon acts as a negative regulator of the UVRAG/VPS34 complex, affecting autophagosome–lysosome fusion50. Our study showed that increased Rubicon lactylation enhanced its interaction with VPS34, suggesting that PDCD6 deficiency may impair autophagosome maturation. To test this, we assessed p62 degradation in Pdcd6fl/fl and Pdcd6Δmye BMDMs during starvation-induced autophagy. The results revealed that p62 degradation was impaired in Pdcd6Δmye BMDMs (Supplementary Fig. 10a). No differences in the activation of the AMPK or AKT/mTOR pathways, key markers of autophagy induction51, were observed between the two groups of cells following L. monocytogenes challenge (Supplementary Fig. 10b). Treatment with LY294002, a PI3K inhibitor, and chloroquine (CQ), an autophagy inhibitor, indicated that PDCD6 deficiency mainly led to impaired autophagosome maturation, as evidenced by an increase in p62 levels in Pdcd6Δmye BMDMs following CQ treatment (Supplementary Fig. 10c, d). Together, these results indicated that PDCD6 deficiency leads to a decrease in autophagosome maturation rather than autophagy induction.
Rubicon enhances VPS34 activity and maintains PI3P on LAPosomes while also stabilizing the NOX2 complex for ROS production, both of which are essential for LAP progression19. To determine whether RUBCN lactylation influences these processes, RUBCN-knockout THP-1 cells were reconstituted with RUBCN K33R or wild-type RUBCN, and the effects were compared. Compared with that seen with wild-type RUBCN, cells reconstituted with RUBCN K33R displayed significantly reduced PI3P levels (Fig. 10e), diminished ROS production (Fig. 10f), and decreased p40phox phosphorylation, indicative of a reduction in NOX complex activity (Fig. 10g). These findings suggested that RUBCN lactylation at K33 is crucial for regulating LAP-mediated antibacterial responses (Supplementary Fig. 10e).
Discussion
Various forms of cell death, such as apoptosis, pyroptosis, and necroptosis, contribute to innate immunity and host defenses against microbial infections20,52,53. Proteins such as caspase-1, caspase-3, caspase-8, FADD, RIPK1, and RIPK3 regulate immune responses by activating signaling pathways, promoting cytokine production, and modulating inflammasome activation54–56. Our study revealed that PDCD6, a novel member of the programmed cell death family, plays a key role in defending against intracellular bacterial infections. Notably, we found that PDCD6 deficiency enhances host defenses by promoting LAP, thereby providing new insights into host–pathogen interactions.
L. monocytogenes, S. Typhimurium, and F. novicida are important bacterial pathogens responsible for severe infectious diseases. In this study, we leveraged these pathogens to examine how PDCD6 regulates antibacterial defenses via LAP. While differences exist in how these pathogens affect cellular signaling and metabolism, each is recognized through distinct PAMPs, and they may share regulatory mechanisms involving calcium signaling. Infections with L. monocytogenes and F. novicida increase intracellular calcium levels, thereby enhancing pyruvate dehydrogenase (PDH) activity and acetyl-CoA production. This leads to the acetylation of RUBCN, which modulates its interaction with the NOX complex and influences LAP activation19. Similarly, S. Typhimurium infection can cause a substantial elevation in intracellular calcium ion concentrations57, hinting at a possible parallel mechanism that may affect PDH activity, acetyl-CoA concentrations, and RUBCN acetylation, ultimately impacting LAP activation.
Studies have shown that PDCD6 is involved in vesicular trafficking by interacting with SEC31, thereby supporting ER-to-Golgi transport via COPII vesicles58, as well as with TRPML1, which promotes retrograde endosome/lysosome transport42. COPII vesicles mediate anterograde ER-Golgi trafficking, transporting their cargos to the Golgi after budding from ER exit sites (ERESs)59, while retrograde transport moves cargos from endosomes to the trans-Golgi network (TGN). In LAP, however, phagocytosed cargos are delivered to lysosomes, which is a spatially distinct process. COPII vesicles consist of SAR1, SEC23, SEC24, SEC13, and SEC3159, while the retromer complex, involved in endosome-to-Golgi transport, includes VPS35, VPS29, and VPS2660. None of these proteins participate in LAP, and there is no evidence linking COPII vesicles or retrograde transport to LAP, highlighting that there is no clear connection between PDCD6-mediated trafficking and LAP.
A recent study revealed that PDCD6 suppresses antiviral innate immunity13. However, how PDCD6 regulates innate immunity during bacterial infections remains poorly understood. In our study, we found that intracellular bacterial infections led to the downregulation of PDCD6 expression. Myeloid-specific PDCD6 deletion in macrophages impaired bactericidal activity without affecting phagocytosis, cell death, or calcium mobilization. In vivo, Pdcd6 knockout mice demonstrated significantly enhanced antibacterial responses relative to their wild-type counterparts, indicating that PDCD6 negatively regulates antibacterial activity.
Following the recognition of microbial products, innate immune receptors trigger a range of host inflammatory responses, which are key to controlling infections2,61. No significant differences in autophagy induction or maturation were found between PDCD6-deficient and control cells under either classical pro-autophagic stimuli or bacterial infections. However, PDCD6-deficient cells showed a marked increase in LAPosome formation and maturation. The enhancement of bactericidal effects observed in Pdcd6-deficient macrophages or mice was abolished with Pdcd6/Rubcn or Pdcd6/Cybb double knockout, suggesting that LAP plays an essential role in PDCD6-mediated antibacterial activity.
Metabolic reprogramming during microbial infection influences immune functions such as cell migration, phagocytosis, and cytokine production, all of which are crucial for host defenses19,62. Our study confirmed that L. monocytogenes infection upregulates glucose metabolism, with acetyl-CoA playing a role in bacterial clearance via protein acetylation19. Although the link between metabolism and LAP activation is unclear, our analysis of zymosan-stimulated macrophages showed that lactate and glucose metabolites were significantly increased in these cells, particularly PDCD6-deficient BMDMs. Moreover, our findings indicated that there is a direct correlation between elevated lactate levels and LAP.
Lactate, produced via the activity of LDHA during glycolysis, plays diverse roles in regulating cellular processes. Our results showed that PDCD6 binds to LDHA and inhibits its activation, leading to reduced lactate production. Lactate interacts with proteins and modulates their activity. For instance, lactate can inhibit MAVS, and can influence a range of biological processes through its role in lactylation44–46. Here, we identified RUBCN as a lactylated protein, where lactylation at K33 enhances its interaction with VPS34, leading to increased PtdIns(3)P production (Fig. 10e), ROS generation (Fig. 10f), and NOX activation (Fig. 10g), all of which are essential for LAP induction. LAP represents a major defense mechanism against pathogens such as Legionella dumoffii, S. Typhimurium, and Candida albicans2,3,30. Modulating LAP by targeting PDCD6 or lactate metabolism may augment immune responses, thereby offering novel therapeutic strategies for infectious diseases.
Methods
Mice
Pdcd6 floxed mice (strain S-CKO-04201) in the C57BL6/J background were purchased from Cyagen Biosciences and Lysozyme M-Cre mice (004781) were obtained from Jackson Laboratories. Pdcd6Δmye mice were generated by crossing Pdcd6fl/fl mice with Lysozyme M-Cre mice. All mice were housed at 22 °C under standard specific-pathogen-free (SPF) conditions, with 60% ± 10% humidity and a 12-h/12-h light/dark cycle. They were fed a standard diet and had ad libitum access to sterilized water. Both male and female mice, aged 6 to 8 weeks, were used for experiments. Pdcd6fl/fl mice were genotyped using PCR amplification of tail genomic DNA using the following primers: Forward: 5′-CTCTCTTCTACAATAGCACAGGGTTC-3′, reverse: 5′-CTCTTTTCACTGATGCCGTTTTCC-3′. The PCR conditions were 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s. Pdcd6ΔmyeCybb−/− and Pdcd6ΔmyeRubcn−/− double knockout mice were generated by crossing Pdcd6Δmye mice with Cybb−/− and Rubcn−/− mice, respectively. All animal-related experiments adhered to National Institute of Health guidelines and were approved by the Ethics Committee of Shandong Normal University. For euthanasia, mice were placed in a chamber, and CO2 was introduced at a rate of 30% to 70% of the chamber volume per minute. Death was subsequently confirmed by cervical dislocation. After confirmation, the carcass was placed in a bag and stored in a freezer. Mice were euthanized if their weight loss equaled or exceeded 20%.
Cell culture and stimulation
HEK293T (ATCC CRL-11268) and L929 (ATCC CCL-1) cells were purchased from ATCC and maintained under a humidified atmosphere with 5% CO2 at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). THP-1 cells (SCSP-567) were purchased from ATCC and maintained in RPMI 1640 medium supplemented with 10% FBS. BMDMs were generated from Pdcd6fl/fl, Pdcd6Δmye, Cybb−/−, Rubcn−/−, Pdcd6ΔmyeCybb−/−, and Pdcd6ΔmyeRubcn−/− mice in the presence of L929 conditional medium. Peritoneal macrophages were isolated by peritoneal lavage with 10 mL of sterile DPBS containing 2% FBS.
Gentamicin protection assay
THP-1 cells or BMDMs were seeded in a 96-well plate at a density of 0.2 × 106 cells per well and infected with L. monocytogenes, S. Typhimurium, or F. novicida at a multiplicity of infection (MOI) of 5, 5, and 20, respectively, for 1 h. The medium was then replaced with gentamicin-containing medium (100 μg/mL). After an additional 5 min (considered the 1-h time point), and after 1, 2, 4, and 6 h (considered the 2-, 3-, 5-, and 7-h time points), the cells were washed three times with PBS and lysed in 0.1% Triton X-100 in PBS (100 μL). Tenfold serial dilutions of the lysates were prepared in PBS and 10 μL was dropped onto an LB agar plate without antibiotics. After 36 h of incubation at 37 °C, colonies were counted. This experiment was repeated three times, with triplicate wells used each time.
Bacteria
L. monocytogenes-RFP (DP-L5538)63 was generously provided by Cai Zhan (Shandong University, China). L. monocytogenes (10403S)19 was kindly donated by Shijun Zheng (China Agricultural University, China). F. novicida (strain U112)64 was a kind gift from Xiaopeng Qi (Shandong University, China). S. Typhimurium (ATCC 14028)65 was kindly supplied by Bingqing Li (Shandong First Medical University, China). L. monocytogenes was cultured in Brain Heart Infused (BHI) medium to an OD600 of 0.5, while S. Typhimurium and F. novicida were grown on LB plates, and bacteria collected from the respective plates were suspended in PBS to an OD600 of 1 for use in subsequent experiments.
Quantitative real-time PCR
Total RNA was extracted from in vitro-cultured cells using Trizol reagent (R401-01, Vazyme) and reverse-transcribed to cDNA using HiScript II Q RT SuperMix for qPCR (R223-01, Vazyme). qPCR was performed using SYBR Green PCR Master Mix (11201ES08, Yeasen Biotechnology) in a LightCycler 480 II System (Roche). The ΔΔCt method was employed to determine the fold difference in mRNA expression levels between treatment groups, and β-actin served as the internal control. The primers used for qPCR are listed in the Supplementary Table 4.
Immunoblotting
Cultured cells were lysed in RIPA buffer, and the extracted protein was separated by SDS–PAGE (SDS–PAGE Gel Preparation Kit (20325ES62, Yeasen Biotechnology)) according to the manufacturer’s protocol, transferred to nitrocellulose membranes (Bio-Rad), and sequentially incubated with primary antibodies and HRP-conjugated secondary antibodies. Signals were detected using Enhanced Chemiluminescence Reagent (36222ES76, Yeasen Biotechnology). The dilutions and catalog numbers of the antibodies are indicated in Supplementary Data 2. Images were acquired with a Chemiluminescence Imaging System (Tanon).
ELISA
Cytokines in the supernatant of in vitro cultured cells, peritoneal lavage fluid, or serum obtained from animal experiments were measured using ELISA kits for mouse IL-6 (DY406), IL-1β (DY406), TNF (DY406) (all from R&D Systems), and PI3P (HB575-Mu, Hengyuan Biotechnology). Measurements were performed following the manufacturer’s instructions.
Phagocytosis assay
The phagocytosis assay was performed as previously described66. Briefly, L. monocytogenes was fluorescently labeled with DMAO (C2030S, Beyotime Biotechnology), and either DMAO-labeled L. monocytogenes or GFP-S. Typhimurium was used to infect Pdcd6fl/fl and Pdcd6Δmye BMDMs in a 12-well plate for 20, 40, 60, 90, or 120 min at 37 °C. Non-ingested bacteria were removed by treatment with gentamicin (50 μg/mL) for 1 h, and trypan blue was applied to quench surface fluorescence. Cells were then fixed in 4% paraformaldehyde and a minimum of 10,000 cells per sample were subjected to flow cytometry (BD LSR Fortessa) to assess phagocytosis.
The phagocytosis assay involving pHrodo Green E. coli BioParticles Conjugate (P35366, Invitrogen) was performed following the manufacturer’s protocol. Briefly, BMDMs were challenged or not with the BioParticles (100 µg/mL) for 30, 60, 90, and 120 min in complete DMEM, and then analyzed by flow cytometry.
Plasmids and cloning
The plasmids for expressing PDCD6, RUBCN, LDHA, and VPS34 were purchased from WZ Biosciences and the plasmid for expressing FGFR1 was purchased from Miaoling Biology. To generate RUBCN truncation and mutant constructs, the ClonExpress II One Step Cloning Kit (C112-01, Vazyme) and the Mut Express MultiS Fast Mutagenesis Kit (C215-01, Vazyme) were used, respectively, according to the manufacturer’s instructions. The primers used are listed in the Supplementary Table 5. The complete nucleotide sequences of all RUBCN mutants were confirmed by sequencing.
Cell transfection
HEK293T and THP-1 cells were transfected for 30 h with a combination of the indicated expression plasmids using Liposomal Transfection Reagent (40802ES03, Yeasen Biotechnology), followed by immunoprecipitation and immunoblotting assays. siRNAs were synthesized by GenePharma (Shanghai, China) and were transfected into cells using INTERFERin siRNA transfection reagent (101000028, PolyPlus, France) as instructed by the manufacturer. The sequences of the siRNA targeting human PDCD6 were #1: 5′-gtagctgtatcgttctaat and #2: 5′-gcagttagatgctgttctt.
Laser scanning confocal microscopy
BMDMs were cultured in an 8-well-chamber slide and infected with RFP-expressing L. monocytogenes at a MOI of 5 for 1 h. Gentamicin was then applied to kill extracellular bacteria and the cells were fixed and permeabilized. LC3B (M152-3, MBL International’s), LAMP1 (AF7353, Beyotime), LysoTracker Green (40738ES50, Yeasen Biotechnology), and DAPI (H-1200, Vector Laboratories) were used to visualize different compartments. Images were acquired with a laser scanning confocal fluorescence microscope (Leica TCS SP8 MP).
Transmission electron microscopic analysis
Pdcd6fl/fl and Pdcd6Δmye BMDMs were infected with L. monocytogenes at a MOI of 5 in 6-well plates. After 1 or 2 h of infection, the cells were washed with cold PBS, collected by centrifugation, and fixed in a solution containing 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer for 24 h at 4 °C. The samples were then post-fixed using 1% osmium tetroxide, dehydrated through a graded alcohol series, and embedded in Epon 812 resin (Electron Microscopy Sciences). Subsequently, the samples were trimmed, sectioned, and stained with 2% uranyl acetate and lead citrate. Images were captured with a Hitachi HT7800 transmission electron microscope and bacterial counting was done in a double-blind manner.
ROS measurement
BMDMs were seeded in black 96-well plates, stimulated with L. monocytogenes at a MOI of 5 for 2 h, and then exposed to 10 μM DCFH-DA (S0033S, Beyotime) for 20 min. Mean fluorescence intensity was quantified using a Spectra Max M5 microplate reader with excitation and emission wavelengths of 488 and 525 nm, respectively.
Calcium measurement
BMDMs were seeded in black 96-well plates, treated with Fluo-4/AM (273221-67-3, Yeasen Biotechnology) at a concentration of 5 mM in the extracellular medium, and incubated in the dark at 37 °C for 60 min. Subsequently, intracellular calcium concentrations were measured through fluorescence readings using a Spectra Max M5 microplate reader with excitation and emission wavelengths of 494 and 516 nm, respectively.
Phagosome purification
The phagosome isolation assay was performed as previously described19. Briefly, BMDMs were cultured with Pam3CSK4-coated beads and then washed with cold PBS. The cells were suspended in 2 mL of homogenization buffer containing a protease inhibitor cocktail and homogenized on ice using a Dounce homogenizer. Phagosomes were separated from the homogenized mixture using a magnetic column, washed six times with hypotonic buffer, and finally resuspended in 100 μL of SDS sample loading buffer for further analysis.
Detection of LC3-associated phagocytosis using flow cytometry
LC3-associated phagocytosis was detected using antibody staining and flow cytometry as previously described67. Briefly, BMDMs, peritoneal macrophages, or THP-1 cells were seeded in 6-well plates and treated with rapamycin and chloroquine as controls. Following incubation, the cells were exposed to L. monocytogenes or S. Typhimurium stimulation. The cells were subsequently collected, washed, permeabilized, and incubated with anti-LC3B antibody conjugated to Alexa Fluor 488 or Alexa Fluor 594. After additional washing steps, the cells were analyzed using a flow cytometer equipped with lasers for excitation at 488 nm and 561 nm. The mean fluorescence intensity of LC3B was measured specifically in events positive for L. monocytogenes or S. Typhimurium, providing quantitative information on LC3B expression levels in the desired cell populations.
CRISPR/Cas9-mediated knockdown
A short hairpin sequence (5′-caactccgggatgatcgataa) targeting human PDCD6 was cloned into the pPLK-GFP-Puro vector. RUBCN-targeting gRNA (5′-TATACTGTCTATCCCCGAGC) and ATG7-targeting gRNA (5′-CACCGGATACTCGTTCAGCTTCTTC) were cloned into the lentiCRISPR v2 backbone (Addgene; 52961). Lentiviruses were packaged with gene-specific or empty vectors in HEK293T cells with pMDL and VSV-G. THP-1 cells were transduced with the lentiviral vectors in the presence of polybrene, and single-cell colonies were selected using limiting dilution. Cells with PDCD6, RUBCN, LDHA, or ATG7 deletions were used for further assays. To reconstitute RUBCN or LDHA expression in knockout cells, p3xFLAG-CMV-7.1-RUBCN or pENTER-Flag-6×His-LDHA plasmids were used. Synonymous mutations were introduced to prevent Cas9-mediated transgene editing.
Metabolomic analysis
A total of 1 × 107 BMDMs derived from Pdcd6fl/fl or Pdcd6Δmye mice were treated or not with zymosan for 4 h, and then used for metabolomic analysis. To extract metabolites from cells, 1 mL of cold methanol: acetonitrile: H2O (2:2:1, v/v/v) was added to 100 μL of each sample (1 × 107 cells). The resulting mixture was vortexed for 60 s, ultrasonicated twice for 30 min at 4 °C, incubated at −20 °C for 1 h, and centrifuged at 14,000 × g for 20 min at 4 °C. The liquid phase (supernatant) of each sample was transferred to a new tube for HILIC LC–MS/MS analysis. The samples were analyzed using an AB5500 QqQ mass spectrometer (AB Sciex, USA) coupled with a Waters I-class HPLC system (Waters, Ireland). Data acquisition and analysis were performed by Shanghai Applied Protein Technology Co., Ltd.
Lactate measurement
Intracellular lactate levels were quantified after the indicated treatments using an L-lactate assay kit (E-BC-K044-M, Elabscience) according to the manufacturer’s protocol. Following treatment with activators, cells were detached with TrypLE, and the cell pellet was homogenized in 100 μL of lactate assay buffer with a G26 needle and deprotonated using TCA (Abcam, ab204708). The lactic acid content was calculated by measuring the OD at 530 nm.
LDH activity assay
The LDH activity assay was performed as previously described38. Briefly, LDH activity was determined by measuring the rate of NADH oxidation in a reaction mixture containing 20 mM HEPES (pH 7.2), 20 μM NADH, 0.05% bovine serum albumin, and 2 mM pyruvate. Fluorescence was measured using a microplate reader (excitation wavelength: 340 nm, emission wavelength: 460 nm).
Library construction for RNA sequencing and sequencing procedures
Total RNA was isolated using a RNeasy Mini Kit (Qiagen). Paired-end libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina, USA) following the manufacturer’s instructions. Briefly, polyA-containing mRNA was purified using poly-T oligo-attached magnetic beads and fragmented into small pieces with divalent cations at 94 °C for 8 min. The generated RNA fragments were reverse-transcribed into first-strand cDNA using random primers, followed by second-strand cDNA synthesis using DNA Polymerase I and RNase H. The resulting cDNA fragments were end-repaired, followed by the addition of a single ‘A’ base and adapter ligation. The products were then purified and PCR-amplified, yielding the final cDNA library. Purified libraries were quantified using a Qubit 2.0 Fluorometer (Life Technologies, USA). An Agilent 2100 bioanalyzer (Agilent Technologies, USA) was employed to confirm the insert size and calculate the mole concentration. Sample clusters were generated using cBot with the library diluted to 10 pM and these were then sequenced on the Illumina HiSeq XTen platform (Illumina, USA). Library construction and sequencing were performed at Shanghai Biotechnology Corporation.
Bacterial infection of mice
Sex-matched Pdcd6fl/fl, Pdcd6Δmye, Cybb−/−, Rubcn−/−, Pdcd6Δmye Cybb−/−, or Pdcd6ΔmyeRubcn−/− mice, 6–8 weeks old, were infected with L. monocytogenes (0.1 × 106 CFU/mouse), S. Typhimurium (0.1 × 106 CFU/mouse), or F. novicida U112 (1 × 106 CFU/mouse) using a single intraperitoneal injection, as previously described19. Mice were weighed and monitored daily over 14 days. Serum and tissues (spleen and liver) were collected 24 h post-injection for immunological and bacterial burden analyses.
Molecular simulation
Interactions between the N-terminus and RUN domain (aa 1–177) of RUBCN from the Alphafold2 structural model and the cryo-EM structure (PDB ID: 7BL1, chain BBB) of the PI3K-type C2 domain (aa 5–245) of VPS34 were predicted using the macromolecule docking software ZDOCK 3.0.268. The interaction conformation and site of interaction between VPS34 and Rab5A in 7BL1 served as the reference for analyzing the docking results. The structure of the RUBCN/VPS34 complex was used to conduct a 100-ns molecular simulation to relax and refine the structure. Lactylation and single-site mutations in K33 of RUBCN were inserted into the structure chosen from the largest cluster in the equilibration period of the MD simulation for further study. To investigate the effect of lactylation and the K33R mutation on the RUBCN-VPS34 complex, a 500-ns MD simulation was conducted in parallel. All MD simulations were performed in Amber20 using Amber14 force field69. The structure was solvated in a cubic box filled with TIP3P water molecules, with a 1-nm distance between the solute and the edge of the box, and was neutralized by sodium ions. After four steps of energy minimization, the system was gradually heated to 300 K over 100 ps to perform the 10-ns NVT equilibration and the 10-ns NPT equilibration. The MD simulations were performed at 300 K and 1 atm using the LINCS algorithm to restrain the hydrogen positions at their equilibrium distances, which allowed the use of an integration time step of 2 fs. Energies and coordinates were saved every 10 ps for the postproduction analysis of the MD simulations. All MD simulations were performed on a high-performance computer cluster running the Linux operating system.
DCFH2-SE labeling of L. monocytogenes for ROS detection
L. monocytogenes was cultured in Brain Heart Infusion medium to an OD600 of approximately 0.5. After centrifugation and two washes with PBS, the bacteria were resuspended in a 100 mM NaHCO3 solution at pH 8.3 to a concentration of 1 × 109 cells/mL, and labeled with DCFH2-SE. A 1.5-M hydroxylamine solution at pH 8.5 was then added to a final concentration of 100 mM and, after a 45-min incubation and centrifugation at 1100 × g for 5 min, the pellets were washed three times with PBS and left to stand overnight. The labeled L. monocytogenes cells were then washed with PBS and aliquoted (20-μL volumes, 1 × 109 cells/mL). For phagocytosis, the labeled bacteria were opsonized with FBS in a 1:1 ratio, incubated at 37 °C for 1 h, washed twice with PBS, and resuspended in a physiological buffer containing 10 mM HEPES, pH 7.4, 140 mM NaCl, 5 mM KCl, divalent ions (1 mM Mg2+, 2 mM Ca2+), 1 mM glucose, and 1% FBS. BMDMs were then exposed to DCFH2-SE-labeled L. monocytogenes on 25-mm coverslips. To observe real-time dynamics of ROS production within infected BMDMs, time-lapse imaging of ROS generation was performed at an excitation wavelength of 450 nm and an emission wavelength of 520 nm, with readings taken at 1-min intervals over 1 h.
Phagosome–lysosome fusion detection using FRET
Phagosome–lysosome fusion detection was performed using FRET as previously described70. Briefly, BMDMs were seeded in a 96-well plate and incubated for 2 h, after which the medium was replaced with fresh medium containing 100 μg/mL Alexa Fluor 594 hydrazide. After 4 h of incubation, the medium was aspirated, and the cells were washed twice with PBS and cultured overnight in a normal medium. Subsequently, Alexa Fluor 488-labeled zymosan (MOI: 10) in binding buffer (PBS supplemented with 2.7 mM KCl, 0.5 mM MgCl2, 1 mM CaCl2, 0.25% gelatin, and 5 mM glucose) was added to the plates followed by incubation at 37 °C. Alexa Fluor 488 fluorescence (excitation: 450 nm, emission: 520 nm), Alexa Fluor 594 fluorescence (excitation: 584 nm, emission: 612 nm), and the FRET signal (excitation: 485 nm, emission: 612 nm) were measured at 1-min intervals over 3 h using a multi-label microplate reader.
Statistical analysis
The statistical methods and the sample sizes (n) are indicated in the figure legends. Statistical analysis was performed using GraphPad Prism 8.2.1 software. For in vivo studies, where “n” represents the number of mice per group, and in vitro studies, where “n” denotes the number of biological replicates, statistical significance was assessed using t tests, nonparametric tests, or two-way ANOVA with multiple comparisons. Data are shown as the mean ± SEM. Statistical significance was defined as P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Dr. Xiaopeng Qi from Shandong University for F. novicida (strain U112), Dr. Shijun Zheng from China Agricultural University for L. monocytogenes (10403S), and Dr. Qingchao Li for PDCD6 plasmid. We thank members of the Li lab for their discussions. This work was supported by grants from the Natural Science Foundation of China (32170727, 82341078 to T.L., and 32200716 to Y.L.), the Natural Science Foundation of Shandong Province (ZR202103010337 to T.L.), Shandong Province Excellent Youth Science Fund (2022HWYQ-076 to T.L.), the Taishan Scholar Program Special Funding (tsqn202211107 to T.L.), and Natural Science Foundation of Jiangsu Province (no. BK20200253 to S.W.).
Author contributions
T.L., J.Z., and Y.L. designed the experiments, supervised the study, and interpreted the data; L.S., S.W., H.W., T.Z., and M.Z. performed experiments and provided intellectual input; S.W. performed computational modeling of Rubicon-VPS34 interaction and provided intellectual input; X.B. and X.Z. performed biostatistical analyses and provided intellectual input; B.L. and C.Z. contributed intellectual input and generated critical reagents; T.L. and Y.L. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Christian Münz, Nengming Xiao and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. The RNA sequencing data have been deposited in NCBI under the accession code PRJNA1164990, https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=1164990. The FACS data have been deposited in FLOW Repository under the accession codes FR-FCM-Z8B2, http://flowrepository.org/id/RvFr1va8imDwYNAMKipG57vN85gzpUHraKwIckYVJi3wQuMJtpNNht1R6jh4Kkkk, and FR-FCM-Z8B3, http://flowrepository.org/id/RvFr1mn21engVcfD5gojLWGmgP2zqsqhTSE0QSy8REJkDCNOKPhFTcmE13vNSky1. The mass spectrometry proteomics data have been deposited in ProteomeXchange Consortium via the PRIDE partner repository under the accession code PXD056047, Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lulu Sun, Sijin Wu.
Change history
2/11/2025
A Correction to this paper has been published: 10.1038/s41467-025-56837-3
Contributor Information
Jun Zhou, Email: junzhou@sdnu.edu.cn.
Tianliang Li, Email: li.tianliang@outlook.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54377-w.
References
- 1.Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol.11, 373–384 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Mitchell, G. & Isberg, R. R. Innate immunity to intracellular pathogens: balancing microbial elimination and inflammation. Cell Host Microbe22, 166–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta, P., Henault, J., Kolbeck, R. & Sanjuan, M. A. Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr. Opin. Immunol.26, 69–75 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kyrmizi, I. et al. Calcium sequestration by fungal melanin inhibits calcium-calmodulin signalling to prevent LC3-associated phagocytosis. Nat. Microbiol.3, 791–803 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Maki, M., Takahara, T. & Shibata, H. Multifaceted roles of ALG-2 in Ca(2+)-regulated membrane trafficking. Int. J. Mol. Sci.17, 1401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki, H. et al. Structural basis for Ca2+ -dependent formation of ALG-2/Alix peptide complex: Ca2+/EF3-driven arginine switch mechanism. Structure16, 1562–1573 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Vito, P., Lacana, E. & D’Adamio, L. Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science271, 521–525 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Tarabykina, S., Mollerup, J., Winding, P. & Berchtold, M. W. ALG-2, a multifunctional calcium binding protein? Front. Biosci.9, 1817–1832 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Sadoul, R. Do Alix and ALG-2 really control endosomes for better or for worse? Biol. Cell98, 69–77 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Shibata, H., Suzuki, H., Yoshida, H. & Maki, M. ALG-2 directly binds Sec31A and localizes at endoplasmic reticulum exit sites in a Ca2+-dependent manner. Biochem. Biophys. Res. Commun.353, 756–763 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Bentley, M. et al. Vesicular calcium regulates coat retention, fusogenicity, and size of pre-Golgi intermediates. Mol. Biol. Cell21, 1033–1046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.la Cour, J. M., Schindler, A. J., Berchtold, M. W. & Schekman, R. ALG-2 attenuates COPII budding in vitro and stabilizes the Sec23/Sec31A complex. PLoS One8, e75309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, W., Zhang, L., Xu, X. & Liu, X. ALG2 regulates type I interferon responses by inhibiting STING trafficking. J. Cell Sci.134, jcs259060 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Chen, Z. J., Xiao, J. & Chen, H. H. Identification of key genes related to immune cells in patients with COVID-19 via integrated bioinformatics-based analysis. Biochem. Genet.61, 2650–2671 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang, I. K., Hu, R., Lacana, E., D’Adamio, L. & Gu, H. Apoptosis-linked gene 2-deficient mice exhibit normal T-cell development and function. Mol. Cell Biol.22, 4094–4100 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg, G., Riquelme, S., Prince, A. & Avraham, R. Immunometabolic crosstalk during bacterial infection. Nat. Microbiol.7, 497–507 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Ryan, D. G. & O’Neill, L. A. J. Krebs cycle reborn in macrophage immunometabolism. Annu. Rev. Immunol.38, 289–313 (2020). [DOI] [PubMed] [Google Scholar]
- 18.O’Brien, K. L. & Finlay, D. K. Immunometabolism and natural killer cell responses. Nat. Rev. Immunol.19, 282–290 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Li, T. et al. Listeria monocytogenes upregulates mitochondrial calcium signalling to inhibit LC3-associated phagocytosis as a survival strategy. Nat. Microbiol.6, 366–379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen, I., Rayamajhi, M. & Miao, E. A. Programmed cell death as a defence against infection. Nat. Rev. Immunol.17, 151–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng, M., Karki, R., Vogel, P. & Kanneganti, T. D. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell181, 674–687.e613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwell, J. M., Searle, S., Mohamed, H. & White, J. K. Divalent cation transport and susceptibility to infectious and autoimmune disease: continuation of the Ity/Lsh/Bcg/Nramp1/Slc11a1 gene story. Immunol. Lett.85, 197–203 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Vidal, S. M., Malo, D., Vogan, K., Skamene, E. & Gros, P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell73, 469–485 (1993). [DOI] [PubMed] [Google Scholar]
- 24.Benjamin, W. H. Jr, Hall, P., Roberts, S. J. & Briles, D. E. The primary effect of the Ity locus is on the rate of growth of Salmonella typhimurium that are relatively protected from killing. J. Immunol.144, 3143–3151 (1990). [PubMed] [Google Scholar]
- 25.Vidal, S. et al. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J. Exp. Med182, 655–666 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauler, T. J. et al. Salmonella Meningitis Associated with Monocyte Infiltration in Mice. Am. J. Pathol.187, 187–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Govoni, G. et al. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect. Immun.64, 2923–2929 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arango Duque, G. & Descoteaux, A. Macrophage cytokines: involvement in immunity and infectious diseases. Front. Immunol.5, 491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol.16, 35–50 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Huang, J. & Brumell, J. H. Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microbiol.12, 101–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gluschko, A. et al. The beta(2) Integrin Mac-1 Induces Protective LC3-Associated Phagocytosis of Listeria monocytogenes. Cell Host Microbe23, 324–337.e325 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Huang, J. et al. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl Acad. Sci. USA106, 6226–6231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savina, A. et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell126, 205–218 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Germic, N., Frangez, Z., Yousefi, S. & Simon, H. U. Regulation of the innate immune system by autophagy: neutrophils, eosinophils, mast cells, NK cells. Cell Death Differ.26, 703–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coryell, P. R. et al. Autophagy regulates the localization and degradation of p16(INK4a). Aging Cell19, e13171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ligeon, L. A. et al. Oxidation inhibits autophagy protein deconjugation from phagosomes to sustain MHC class II restricted antigen presentation. Nat. Commun.12, 1508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y. et al. Control of infection by LC3-associated phagocytosis, CASM, and detection of raised vacuolar pH by the V-ATPase-ATG16L1 axis. Sci. Adv.8, eabn3298 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin, L. et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene36, 3797–3806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks, G. A., Dubouchaud, H., Brown, M., Sicurello, J. P. & Butz, C. E. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc. Natl. Acad. Sci. USA96, 1129–1134 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Y. et al. Nuclear lactate dehydrogenase A senses ROS to produce alpha-hydroxybutyrate for HPV-induced cervical tumor growth. Nat. Commun.9, 4429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao, S. et al. Fbw7 inhibits the progression of activated B-cell like diffuse large B-cell lymphoma by targeting the positive feedback loop of the LDHA/lactate/miR-223 axis. Front. Oncol.12, 842356 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, X. et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol.18, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, D. et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell23, 464–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, W. et al. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell178, 176–189.e115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature574, 575–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, K. et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ.29, 133–146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, Q. et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J. Biol. Chem.286, 185–191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tremel, S. et al. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun.12, 1564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsunaga, K. et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol.11, 385–396 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol.13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasparakis, M. & Vandenabeele, P. Necroptosis and its role in inflammation. Nature517, 311–320 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Riera Romo, M. Cell death as part of innate immunity: cause or consequence? Immunology163, 399–415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He, S. & Wang, X. RIP kinases as modulators of inflammation and immunity. Nat. Immunol.19, 912–922 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Wallach, D., Kang, T. B., Dillon, C. P. & Green, D. R. Programmed necrosis in inflammation: toward identification of the effector molecules. Science352, aaf2154 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Gurung, P. & Kanneganti, T. D. Novel roles for caspase-8 in IL-1beta and inflammasome regulation. Am. J. Pathol.185, 17–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gewirtz, A. T. et al. Salmonella typhimurium induces epithelial IL-8 expression via Ca(2+)-mediated activation of the NF-kappaB pathway. J. Clin. Invest.105, 79–92 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helm, J. R. et al. Apoptosis-linked gene-2 (ALG-2)/Sec31 interactions regulate endoplasmic reticulum (ER)-to-Golgi transport: a potential effector pathway for luminal calcium. J. Biol. Chem.289, 23609–23628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen, D. & Schekman, R. COPII-mediated vesicle formation at a glance. J. Cell Sci.124, 1–4 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Cullen, P. J. & Steinberg, F. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol.19, 679–696 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Galan, J. E. Salmonella Typhimurium and inflammation: a pathogen-centric affair. Nat. Rev. Microbiol.19, 716–725 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med.213, 15–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Czuczman, M. A. et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature509, 230–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo, Y. et al. HUWE1 mediates inflammasome activation and promotes host defense against bacterial infection. J. Clin. Invest.130, 6301–6316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai, Y. et al. Salmonella manipulates macrophage migration via SteC-mediated myosin light chain activation to penetrate the gut-vascular barrier. EMBO J.43, 1499–1518 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, C. S. et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe11, 264–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez, J. Detection of LC3-associated phagocytosis (LAP). Curr. Protoc. Cell Biol.87, e104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pierce, B. G., Hourai, Y. & Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One6, e24657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Case, D. A. et al. The Amber biomolecular simulation programs. J. Comput. Chem.26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pryor, P. R. Analyzing lysosomes in live cells. Methods Enzymol.505, 145–157 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. The RNA sequencing data have been deposited in NCBI under the accession code PRJNA1164990, https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=1164990. The FACS data have been deposited in FLOW Repository under the accession codes FR-FCM-Z8B2, http://flowrepository.org/id/RvFr1va8imDwYNAMKipG57vN85gzpUHraKwIckYVJi3wQuMJtpNNht1R6jh4Kkkk, and FR-FCM-Z8B3, http://flowrepository.org/id/RvFr1mn21engVcfD5gojLWGmgP2zqsqhTSE0QSy8REJkDCNOKPhFTcmE13vNSky1. The mass spectrometry proteomics data have been deposited in ProteomeXchange Consortium via the PRIDE partner repository under the accession code PXD056047, Source data are provided with this paper.