Abstract
Introduction
Lung adenocarcinoma (LUAD), a common subtype of NSCLC, has a high mortality rate. Telomere genes are influenced by radiation therapy, affecting treatment response. Additionally, immune cell presence in the tumor microenvironment plays a crucial role in cancer prognosis. However, the role of Radioresistant-Related Telomere Genes (RRTGs) in LUAD prognosis and immune infiltration remains unclear.
Methods
In this research, we utilized diverse bioinformatics techniques to examine our personally tested information along with publicly accessible datasets. We conducted a comprehensive study on the genetic and transcriptional differences, predictive significance, and expression profiles of RRTGs. Afterwards, a RRTGs score was developed to forecast the overall survival (OS) and ascertain its reliable predictive capacity for patients with LUAD. Following this, dependable nomograms were developed to enhance the practicality of RRTGs scoring in a clinical setting. Furthermore, the investigation delved into the associations among RRTGs, infiltration of immune cells, prognosis, and clinical treatments of patients. Gene Set Enrichment Analysis (GSEA) was conducted to explore the potential mechanisms by which RRTGs influence the regulation of LUAD. Then, Western blot, qRT-PCR and Immunohistochemistry were used to detect the expression levels of RRTGs in cell lines and LUAD tumor tissues.
Results
Our research indicates that certain genes related to telomeres have a notable correlation with the prognosis of patients diagnosed with LUAD. The RRTGs score, which includes three key genes (ARRB1, PLK1, and DSG2), was developed to forecast the OS and its dependable predictive capability for individuals diagnosed with LUAD was ascertained. Afterwards, extremely reliable nomograms were developed to improve the practicality of the RRTGs score. Moreover, as illustrated, genetic characteristics can be utilized to assess the infiltration of immune cells in tumors, as well as clinical attributes and prognosis. RRTGs score characterizes tumor mutational burden, immune activity, and notable survival probabilities in addition. Furthermore, GSEA results revealed that RRTGs may influence LUAD by modulating immune-related pathways in high-risk groups and regulating cell cycle and DNA repair processes in low-risk groups. The RRTGs (ARRB1 and PLK1) were upregulated in A549 cells and radiosensitive NSCLC tissues compared to radioresistant A549 cells and NSCLC tissues.
Conclusion
In conclusion, this research emphasizes the significance of RRTGs in the outlook of LUAD. The findings contributed to a better understanding of the link between radiotherapy, telomere-related genes, and prognosis in LUAD, and identified potential therapeutic targets for patients with LUAD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03564-2.
Keywords: Lung adenocarcinoma, Radiotherapy, Telomere genes, Immune Infiltration, Prognostic model
Introduction
On a global level, lung adenocarcinoma (LUAD), which is a type of non-small cell lung cancer (NSCLC), is a dominant and challenging malignancy [1, 2]. Despite significant progress in the early identification and treatment approaches, the outlook for individuals with LUAD remains less than ideal. This underscores the need for persistent research efforts to discover novel biomarkers and treatment targets [3, 4].
Lately, there has been a growing curiosity in exploring the complex relationship between telomeres and the development and advancement of cancer [5]. Telomeres, repetitive DNA sequences situated at chromosomal termini, constitute a paramount safeguard for chromosomal stability and integrity. Maintaining the length of telomeres is crucial for the survival of cells, and disrupting it has been linked to various human illnesses, with cancer being a major area of concern [6, 7]. Disrupted telomeres can engender genomic instability, fostering oncogenic transformation and tumor progression [8].
Radiotherapy plays a crucial role in the treatment of lung cancer by causing DNA harm in cancerous cells, resulting in the halting of cell division and triggering apoptosis [9]. Nevertheless, radioresistance frequently compromises therapeutic efficacy, posing a significant clinical challenge [10]. Interestingly, recent findings highlight a possible link between telomeres and the ability of cancer cells to resist radiation [11]. Telomere dynamics and telomerase activity have been postulated to influence the response to radiation therapy, thus implicating telomeres in modulating the cellular reaction to radiation-induced DNA damage [12, 13].
Moreover, increasing proof highlights the crucial significance of the immune system in the advancement of lung adenocarcinoma and the results for patients [14]. Immune cells that infiltrate, including T cells, B cells, natural killer cells, and macrophages, engage in complex communication with cancer cells and the tumor microenvironment, ultimately impacting tumor growth and the effectiveness of therapy [15, 16]. It is crucial to comprehend the fundamental molecular mechanisms that control immune infiltration in lung adenocarcinoma due to the intricate interaction between cancer cells and immune effectors [17].
Given the possible influence of telomeres on the response to radiotherapy and the biology of tumors, it becomes important to investigate the connection between telomere genes associated with radiotherapy and the infiltration of the immune system in LUAD. Unraveling the impact of telomere-associated genes on the infiltration of immune cells and the tumor microenvironment shows potential in uncovering important knowledge about new treatment approaches, ultimately improving patient outcomes [18, 19].
Using our exclusive sequencing data, combined with knowledge obtained from TCGA and GEO databases, we examined the effects of radiotherapy-associated telomere genes (RRTGs) on the advancement, Tumor Microenvironment (TME), response to immunotherapy, and prognostic results in individuals suffering from LUAD. Furthermore, we classified patients with LUAD into separate categories, allowing us to investigate differences in prognostic significance, molecular traits, responsiveness to anti-cancer medications, and the level of immune cell infiltration. Moreover, we developed a risk classification system based on the assessment of RRTGs, enabling accurate prediction of clinical outcomes and overall survival (OS) measures for patients with LUAD. Our main goal is to improve our comprehension of the complex connection between telomere genes associated with radiotherapy and LUAD, in order to lead the advancement of strong immunotherapeutic strategies focused on LUAD.
Materials and methods
Cell culture
The A549 cells were acquired from the American Type Culture Collection located in Manassas, Virginia, United States. To induce radioresistant cells, A549 cells in the logarithmic growth phase were subjected to fractionated radiation using the TrueBeam linear accelerator from Varian Medical Systems located in Palo Alto, CA, USA. The radiation field size was 10 × 10 cm with a source-to-skin distance of 100 cm. The cells were exposed to a radiation dose of 2 Gy per fraction once daily, with a total of 25 fractions. Following the administration of 25 fractions, the cells were subsequently passaged every 3 days and designated as the A549/X cell line.
Transcriptme RNA sequencing
A549 and A549/X cells were cultured in 6-well dishes for one night, and RNA was extracted using RNAiso Plus following the guidelines provided by the manufacturer. Illumina technology was utilized to conduct high-throughput RNA sequencing (RNA-Seq). RNA quality and integrity were assessed using the Agilent 2100 Bioanalyzer. After enrichment, fragmentation, cDNA synthesis, end repair, and PCR amplification, the Illumina sequencing was performed on the prepared sequencing libraries. Hisat2 was used to align clean reads to the designated reference genome in order to acquire the positional data of reference genes or sequences unique to each sequencing sample. The htseq-count software was used to identify the abundance of every protein-coding gene in each sample by comparing sequences with known reference gene sequences and annotation files. We sequenced three sets of A549 and three sets of A549/X in two runs.
Public data acquisition
Data from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) (TCGA-luad) and Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) databases were collected as public data. To conduct a more thorough examination, we accessed RNA expression data of LUAD from the GEO dataset (GSE72094) and TCGA dataset. We corrected batch effects caused by non-biological technical biases using the “ComBat” algorithm in the SVA package. Single-cell RNA sequencing (scRNA-seq) data of RRTGs in LUAD were analyzed through the TISCH2 website (http://tisch.comp-genomics.org/home/). We utilized the Gene Expression Profiling Interactive Analysis (GEPIA) online database (http://gepia.cancer-pku.cn/index.html) to examine the prognosis of the three model genes and compare their variations between the tumor and normal groups.
Detection of RRTGs with altered expression levels
Expression levels were averaged for repetitive genes in the transcriptome expression matrix acquired from TCGA. Afterwards, a differential analysis was conducted on the matrix of transcriptome expression, using screening criteria of |log2 fold change| > 1 and an adjusted p-value < 0.05. The self-test data underwent the same criteria for differential analysis. Furthermore, genes associated with telomeres were acquired from http://www.cancertelsys.org/telnet/. From the self-test data, the comparison of genes that were expressed differently, along with the comparison of genes expressed differently between tumor and normal groups, and telomere-related genes, resulted in a total of 44 genes that were differentially expressed and related to radiation and telomeres. Differentially expressed genes were analyzed using GO for biological functions and KEGG for pathway involvement. Pathways were considered significantly enriched GO terms and KEGG pathways if they met the criterion of p < 0.05.
Creating and assessing the RRTGs prognostic model
The R package ‘insert’ was used to randomly divide TCGA-LUAD patients into training and internal testing sets in a 1:1 ratio. The RRTGs that showed statistical significance in the univariate Cox regression analysis underwent LASSO regression analysis to precisely calculate the coefficients for each established relationship. The interpretability and predictive capability of the statistical model were enhanced by utilizing the popular LASSO regression technique, which involved the combination of normalization and variable selection. Constructing the risk model for RRTGs involved utilizing the LASSO-Cox regression coefficients of each gene. Risk values for each LUAD patient were calculated using the ‘predict’ function from the survival R package. Based on the median risk value, patients were categorized into low-risk and high-risk groups.
To compare the survival probabilities between the two groups, a Kaplan-Meier analysis was performed using the R packages ‘survival’ and ‘survminer’ with log-rank tests to determine statistical significance (p < 0.05). The R package ‘timeROC’ was used to perform time-dependent receiver operating characteristic (ROC) analysis for survival probabilities at 1-year, 3-year, and 5-year intervals, in order to calculate the values of area under the curve (AUC).
GSEA analysis
To investigate the molecular mechanisms and biological processes associated with the three-gene model, GSEA analysis was conducted between high- and low-risk groups using the KEGG and GO datasets from the molecular signature database (https://www.gsea-msigdb.org/gsea/msigdb). A nominal p < 0.05 and FDR < 0.25 were used as thresholds for statistical significance.
Creating nomograms and conducting an independent prognostic analysis
Obtained were the clinical features of patients with LUAD from the complete TCGA dataset, encompassing factors such as age, grade, and stage. The variables were combined with the score and subjected to both univariate and multivariate Cox regression analyses. In order to obtain individualized forecasts for the survival probabilities of LUAD patients, we created a nomogram utilizing clinical traits and the score derived from the R packages ‘rms’ and ‘regplot’. To assess the accuracy of predicting survival probabilities at 1-year, 3-year, and 5-year intervals, a time-based ROC analysis was employed. The nomogram’s performance was assessed using calibration curves, ROC curves, and decision curve analysis (DCA) curves [20].
RRTGs score and immune infiltration
Established methods were used to determine the immune infiltration status of TCGA database samples in order to investigate the correlation between immune infiltrating cells and risk features. For each LUAD sample, StromalScore, ImmuneScore, and EstimateScore were calculated using the “ESTIMATE” R package based on gene expression patterns of stromal and immune cells, respectively [21]. The CIBERSORT algorithm was utilized to approximate the ratios of 22 immune cell categories from an extensive collection of gene expression data obtained from tumor samples. The TCGA-LUAD RNA-Seq data (normalized using TPM) underwent processing to determine the quantities of immune cells [22]. The differences between high-risk and low-risk groups were analyzed using the Mann-Whitney U test for non-normally distributed data.
Association of the RRTGs score with tumor mutation burden and immune checkpoint
The patient’s response to immunotherapy is linked to the genes Tumor mutation burden (TMB) and immune checkpoint genes (ICGs). To identify variations in mutation status among the score groups of LUAD patients, the TCGA database was utilized to extract mutation annotation format (MAF) using the ‘maftools’ R package. Additionally, the TMB scores were computed for every patient with LUAD in the complete TCGA group.
Assessment of the RRTGs prognostic model in the immune therapy response and its association with the index of tumor stem cells
The TIDE algorithm (http://tide.dfci.harvard.edu) was utilized to assess the response of LUAD patients to immune therapy, aiding in the identification of patients who are better suited for immunotherapy [23]. Moreover, the RNA expression file titled ‘StemnessScores_RNAexp_20170127.2.tsv’ was obtained. The tumor stem cell-like features were determined by analyzing the transcriptome and epigenetic traits of the samples. A correlation analysis was conducted to examine the relationship between RRTGs score and indices of Cancer Stem Cells (CSC).
Correlation of the RRTGs with chemotherapy sensitivity
The TCGA cohort was analyzed using the ‘calcPhenotype’ function from the ‘oncoprecdict’ R package to evaluate the sensitivity scores of frequently prescribed medications in the treatment plans for patients with LUAD. Greater sensitivity to the drug is indicated by a lower estimated drug sensitivity.
Western blot analysis
Total protein was extracted from both A549 and A549/X cell lines using a lysis buffer supplemented with a protease inhibitor cocktail. Protein concentration was quantified using the Bradford assay. Equal amounts of protein from each sample were separated on a 10% SDS-PAGE gel and transferred to PVDF membranes. The membranes were blocked with 5% BSA in TBST for 1 h at room temperature and incubated with primary antibodies against ARRB1 (Abclonal Technology, Wuhan, China), PLK1 (Abclonal Technology, Wuhan, China), and DSG2 (Abclonal Technology, Wuhan, China) overnight at 4 °C. After washing, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Protein bands were detected using ECL substrate and the densitometric analysis was performed using ImageJ software (NIH, Bethesda, MD, USA).
Immunohistochemistry (IHC)
A total of eight patients with recurrent NSCLC after surgical resection, who underwent concurrent chemoradiotherapy at Shanghai Pulmonary Hospital, were included in this study. Tumor response was evaluated based on the Response Evaluation Criteria in Solid Tumors (RECIST, version 1.1). Formalin-fixed, paraffin-embedded sections of NSCLC tumor tissues were deparaffinized in xylene and rehydrated through a graded alcohol series. Antigen retrieval was performed in a citrate buffer (pH 6.0) using a microwave. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide. Non-specific binding was blocked with 5% normal goat serum for 1 h at room temperature. Sections were incubated with primary antibodies against ARRB1 (Abclonal Technology, Wuhan, China), PLK1 (Abclonal Technology, Wuhan, China), and DSG2 (Abclonal Technology, Wuhan, China) overnight at 4 °C. After washing with PBS, the sections were incubated with biotinylated secondary antibody for 30 min at room temperature, followed by treatment with streptavidin-HRP complex. The reaction was visualized with DAB substrate, and the sections were counterstained with hematoxylin, dehydrated, and mounted.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the cell lines using TRIzol, followed by reverse transcription into cDNA with the SYBR Green Master Mix kit. GAPDH served as the endogenous control, and data were analyzed using the comparative Ct method (2−△△Ct).
The primer sequences were as follows:
ARRB1, Forward5’-TGATGACGACATTGTATTTGAGGAC-3’, Reverse5’-AAGAAGACGAGTAAGCATCCGAGT − 3’;
PLK1, Forward5’-AAGAGATCCCGGAGGTCCTA-3’, Reverse5’-TCATTCAGGAAAAGGTTGCC-3’;
DSG2, Forward5’-ATGACGGCTAGGAACACCAC-3’, Reverse5’- GGGTCAGTTTGTGGCTGACT-3’.
And GAPDH Forward5’- TAAAGGGCATCCTGGGCTACACT-3’, Reverse5’- TTACTCCTTGGAGGCCATGTAGG-3’.
Statistical analysis
R software (version 4.1.2) was utilized for all statistical computations. Continuous variables with a normal distribution were analyzed using independent sample t-tests, whereas continuous variables with a non-normal distribution were analyzed using the Mann-Whitney U test. Comparison among three or more groups was conducted using both One-way ANOVA and the Kruskal-Wallis test. The significance of differences was determined by conducting survival analysis using the Kaplan-Meier method and employing the log-rank test. A significance level of less than 0.05 was deemed statistically significant.
Results
Genetic and transcriptomic variations of RRTGs in LUAD
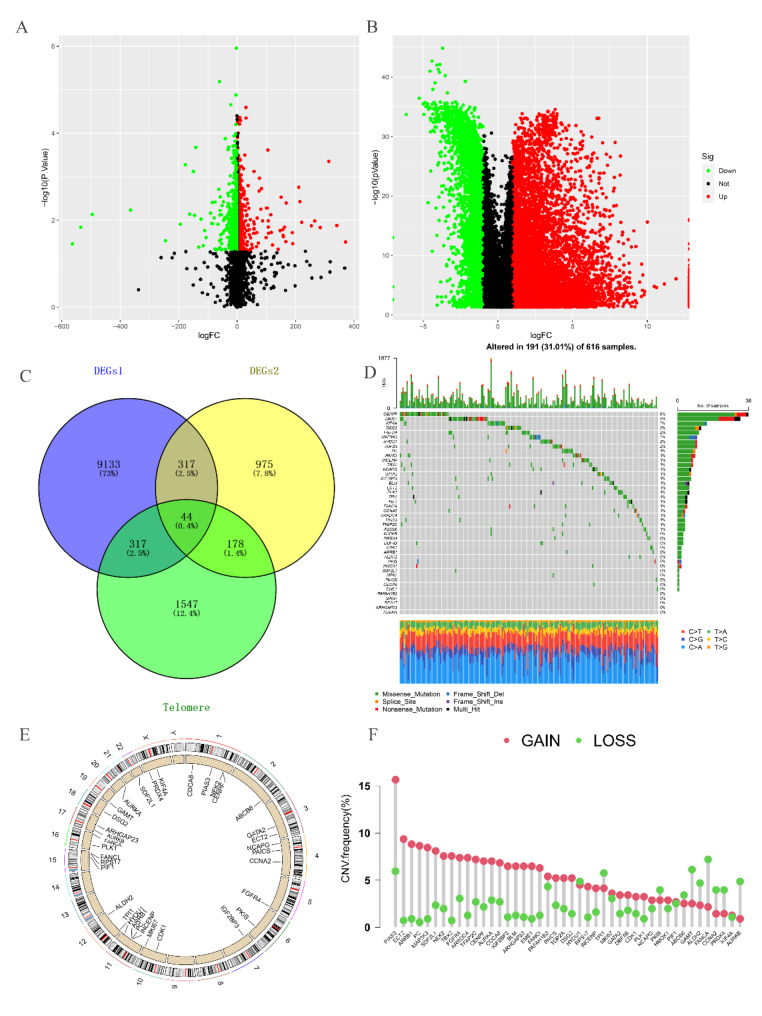
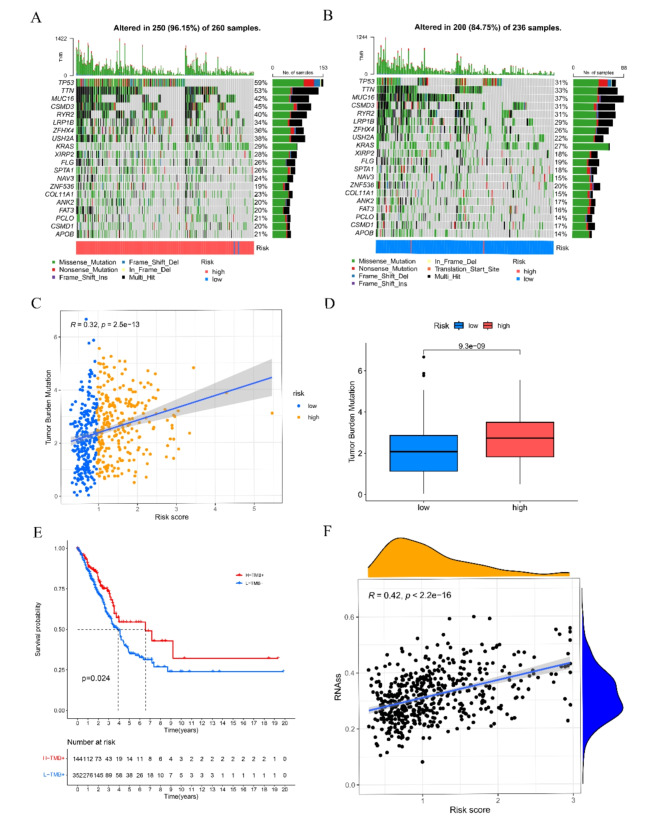
The flowchart of this study is illustrated in Fig. 1. By intersecting the self-test data, the differentially expressed genes between tumor and normal groups in TCGA, and the telomere-related genes (Fig. 2A-C), we were able to identify 44 RRTGs that exhibited differential expression. Figure 2D displays the mutation status of telomere-associated genes in patients with LUAD. Out of the 616 patients with LUAD in the TCGA group, 191 individuals (31.01%) exhibited genetic mutations. The gene mutation rate was highest for CENPF at 6%, with MKI67, DSG2, FGFR4, and MAP3K3 also showing notable mutation frequencies. In addition, we examined the occurrence rate of copy number variations (CNVs) in the 44 genes associated with telomeres in LUAD. In Fig. 2F, PIAS3 exhibited the highest frequency of amplification, whereas GAMT and FANCA experienced extensive losses in CNVs. On 23 chromosomes, Fig. 2E displays the positions of CNV alterations in the 44 genes associated with telomeres.
Fig. 1.
The flowchart of this study
Fig. 2.
Analysis of expression and mutation of RRTGs in LUAD. (A) Volcano map of differentially expressed genes (DEGs) between A549 and A549/X. (B) Volcano map of the differentially expressed genes between tumor and normal groups in TCGA. (C) The Venn diagram of DEGs1, DEGs2, and telomere-related genes. (D) Mutation frequency of 44 RRTGs in LUAD from the TCGA-LUAD dataset. (E) The chromosome distribution of CNV among 44 RRTGs. (F) The frequency of CNV among 44 RRTGs
The GO enrichment analysis identified key biological processes related to cell division and chromosome organization, such as the regulation of nuclear division, G2/M transition of the mitotic cell cycle, mitotic checkpoint signaling, and chromosome segregation. Additionally, activities involving condensed chromosomes, spindle microtubules, and protein kinase regulation were enriched. Meanwhile, KEGG pathway analysis revealed significant enrichment in pathways like progesterone-mediated oocyte maturation, the Fanconi anemia pathway, and the cell cycle pathway, all crucial for DNA repair and cell cycle control. These findings suggest that the 44 RRTGsare closely related to cell cycle regulation and DNA maintenance, which may be linked to radiotherapy resistance (Figure S2A-B).
Developing and assessing the prognostic model for RRTGs
We combined the TCGA and GSE72094 datasets and performed univariate analysis on the 44 differentially expressed telomere genes to identify 32 genes with prognostic significance. Following that, we conducted LASSO and multivariate Cox regression analyses, which led to the discovery of three crucial genes, specifically ARRB1, DSG2, and PLK1, for the development of the prognostic model (Figure S1).
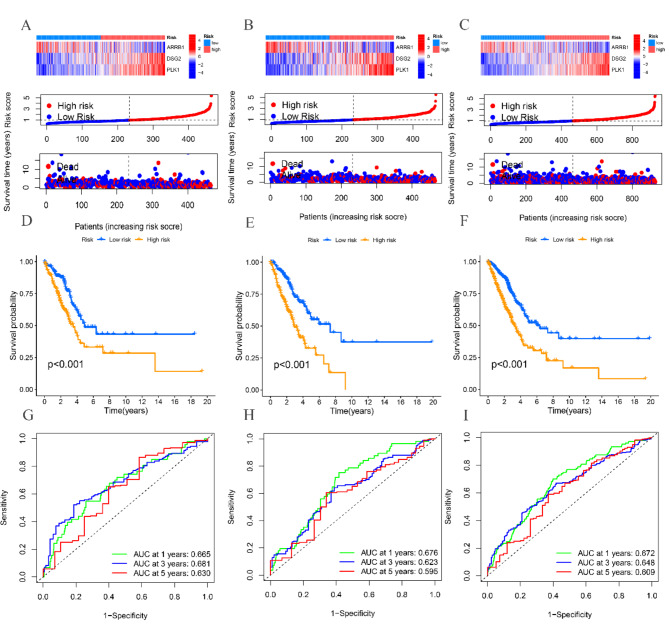
The TCGA cohort patients were split into training and testing cohorts in a 1:1 proportion. Figure 3A-C shows the variation in expression of ARRB1, DSG2, and PLK1 between high-risk and low-risk groups in the TCGA total cohort, training, and testing cohorts, as depicted by the heatmap. Moreover, the risk graph illustrates that individuals with elevated risk scores experienced increased mortality rates and reduced survival durations. According to the Kaplan-Meier analysis, patients in the high-risk group exhibit decreased OS in comparison to patients in the low-risk group across the TCGA total cohort, as well as the training and testing cohorts (P < 0.001) (Fig. 3D-F). Figure 3G-I shows that in the TCGA training group, the AUC values for predicting 1-, 3-, and 5-year OS are 0.676, 0.623, and 0.595, correspondingly. The AUC values for predicting 1-year, 3-year, and 5-year overall survival (OS) in the TCGA testing group are 0.672, 0.648, and 0.609, correspondingly.
Fig. 3.
Construction and validation of the RRTGs risk score model. (A) Heat map and the risk point plot of model gene expression in the high and low-risk groups in the TCGA cohort. (B) Heat map and the risk point plot of model gene expression in the high and low-risk groups in the training set. (C) Heat map and the risk point plot of model gene expression in the high and low-risk groups in the test set. (D) KM survival curves for the high and low-risk groups in the TCGA cohort. (E) KM survival curves for the high and low-risk groups in the training set. (F) KM survival curves for the high and low-risk groups in the test set. (G) ROC curves for the high and low-risk groups in the TCGA cohort. (H) ROC curves for the high and low-risk groups in the training set. (I) ROC curves for the high and low-risk groups in the test set
Creation and verification of the RRTGs Nomogram
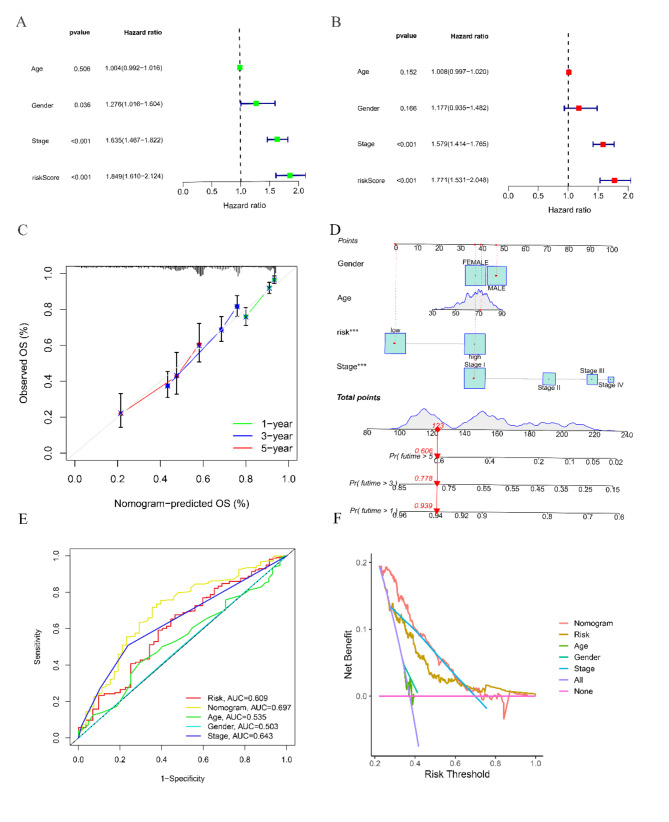
We performed univariate and multivariate Cox regression analyses to assess if the RRTGs score can function as an autonomous prognostic indicator for OS. Significant variations in tumor stage and risk score were observed in the TCGA cohort, as depicted in Fig. 4A and B. Afterwards, we merged the risk score with clinical data of patients, which included age, sex, and tumor stage, in order to create a nomogram within the TCGA group. The nomogram offers a numerical approach to produce individualized prognostic forecasts for patients with LUAD (Fig. 4D). The assessment of the model, utilizing the AUC measurements, calibration curve, and decision curve analysis (DCA), showcased the predictive ability and precision of the nomogram (Fig. 4C, E, F).
Fig. 4.
The nomogram was established in LUAD with RRTGs score. (A) Independent prognosis-univariate Cox Forest plot. (B) Independent prognosis multifactorial Cox Forest plot. (C) 1-, 3-, and 5-year calibration curve. (D) Nomogram for prediction of 1-year, 3-year, and 5-year survival of LUAD patients. (E) ROC curves showed the prognostic performance of the model in the TCGA cohort. (F) DCA curves of the nomogram. (* for P < 0.5, ** for P < 0.01, *** for P < 0.001, and ns for no sense)
Examining the immune microenvironment in groups and GSEA analysis in at high and low risk for RRTGs
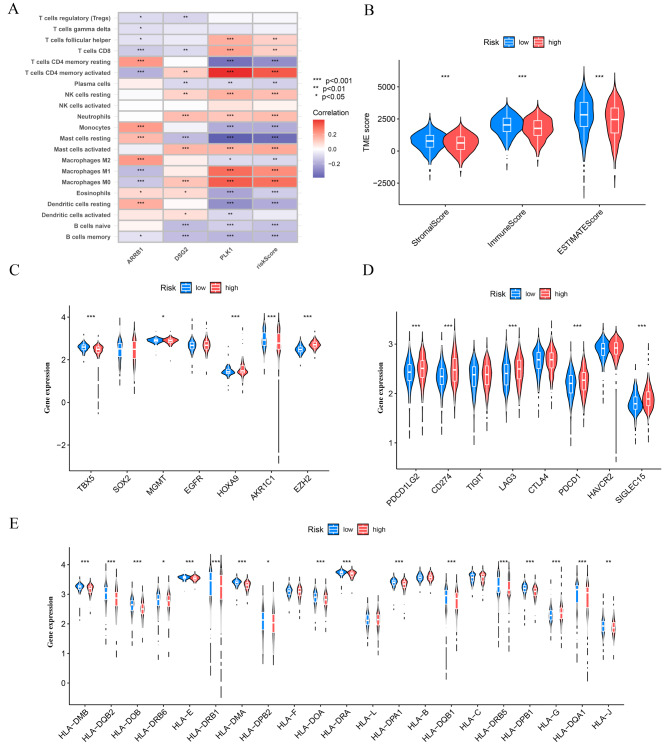
In order to investigate the possible involvement of RRTGs in immune infiltration of LUAD, we employed the ESTIMATE algorithm to compare the tumor TME score and the abundance of immune cells in the high-risk and low-risk groups. Figure 5A demonstrates that ARRB1 displayed a notable positive association with monocytes, M2 macrophages, and quiescent CD4 memory cells, whereas it exhibited a significant negative correlation with activated CD4 memory cells. In addition, ARRB1 exhibited a notable positive association with neutrophils, M0 macrophages, and activated mast cells, while displaying a significant negative association with resting mast cells. Furthermore, PLK1 exhibited noteworthy positive associations with M0 macrophages, M1 macrophages, and activated CD4 memory cells, whereas it demonstrated a substantial negative correlation with resting CD4 memory cells. The results indicate a correlation between these genes and immune cells. In the low-risk group, the TME scores (comprising stromal score, immune score, and ESTIMATE score) exhibited a substantial increase (Fig. 5B). Furthermore, variations were observed in the manifestation of genes associated with radiotherapy and chemotherapy (AKR1C1, EGFR, EZH2, HOXA9, HGMT, SOX2, and TBX5) among the two sets. Furthermore, there were differences in the expression of immune checkpoint genes (PDCD1LG2, CD274, TIGIT, LAG3, CTLA4, PDCD1, HAVCR2, SIGLEC15) and HLA-related genes observed between the high-risk and low-risk groups (Fig. 5C-E).
Fig. 5.
The immune microenvironment was different between the low and high-risk groups. (A) Heat map of immune cell correlation analysis in the high and low-risk groups. (B) Sample tumor purity was assessed by ESTIMATE with the estimated, immune, and stromal scores. (C) Immune checkpoint-related genes in the high and low-risk groups-Violin diagram. (D) Chemoradiotherapy-related genes in the high and low-risk groups-Violin diagram. (E) HLA-related genes in the high and low-risk groups-Violin diagram. *p < 0.05, **p < 0.01, and ***p < 0.001
The GSEA analysis revealed distinct enrichment patterns between the high- and low-risk groups. In the high-risk group, GO terms were significantly enriched in immune-related processes, such as adaptive immune response, antigen processing via MHC class II, complement activation, and MHC class II protein complexes. KEGG pathways in this group were also enriched in immune and metabolism-related pathways, including asthma, cell adhesion molecules, drug metabolism (cytochrome P450), the intestinal immune network for IgA production, and systemic lupus erythematosus. In contrast, the low-risk group showed enrichment in GO terms related to cell division and chromosome dynamics, such as chromosome segregation, mitotic nuclear division, and sister chromatid segregation. KEGG pathways for this group were focused on the cell cycle, DNA replication, oocyte meiosis, p53 signaling, and the spliceosome, indicating an emphasis on cell cycle regulation and genomic stability (Figure S3A-D).
ScRNA-seq data of ARRB1, DSG2, PLK1 in LUAD
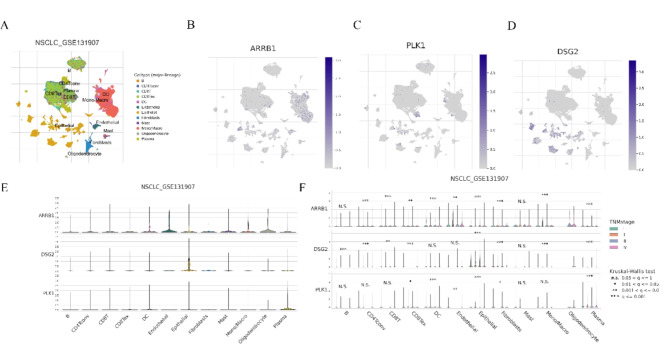
ScRNA-seq data of ARRB1, DSG2, PLK1 in LUAD was analyzed using the online TISCH2 (http://tisch.comp-genomics.org/home/) website to seek their expressions at the single cell level. As displayed in Fig. 6A, the GSE131907 dataset was analyzed. We discovered that ARRB1 is primarily expressed in Endothelial cells, Oligodendrocyte cells, DC cells, and Mono/Macro cells; DSG2 is mainly expressed in Epithelial cells and Fibroblast cells; PLK1 is predominantly expressed in Plasma cells, DC cells, and Epithelial cells (Fig. 6B-E). Moreover, in different stages of LUAD, there are variations in the expression of ARRB1, DSG2, and PLK1 in the aforementioned cells (Fig. 6F).
Fig. 6.
The distribution of the ARRB1, DSG2 and PLK1 in tumor microenvironment in LUAD by single-cell analysis. (A-D) Umap plots of ARRB1, DSG2 and PLK1 in LUAD. (E) Violin plots of ARRB1, DSG2 and PLK1 in tumor microenvironment in LUAD. (F) Violin plots of ARRB1, DSG2 and PLK1 in tumor microenvironment in LUAD in different TMN stage
The relationship between RRTGs associated with radiotherapy and TMB and CSC in LUAD is being examined
Prior research has suggested that an elevated TMB score is linked to improved reaction to immunotherapy. The TP53 mutation rate was considerably greater (59%) in the high-risk group compared to the low-risk group (31%) as shown in Fig. 7A, B. The group at greater risk demonstrated notably elevated TMB scores, and there was a positive correlation between the RRTGs score and TMB scores (Fig. 7C, D). Patients with higher TMB scores had improved overall survival (Fig. 7E) based on the expression level of ICGs, which has been linked to the clinical advantages of checkpoint blockade immunotherapy. Furthermore, there was a noted positive association between the risk score and CSC, as depicted in Fig. 7F.
Fig. 7.
Correlations of the RRTGs risk score with TMB and CSC in LUAD. (A, B) The mutational landscape of LUAD patients in high- and low-risk groups. (C) Correlations between the TMB and the RRTGs score. (D) Difference in the TMB score between high-and low-risk groups. (E) The Kaplan–Meier OS curves among four groups classified by the TMB score and RRTGs score. (F) Correlations between the CSC index and RRTGs risk score
Evaluating the predictive model of RRTGs in the response to immunotherapy for LUAD
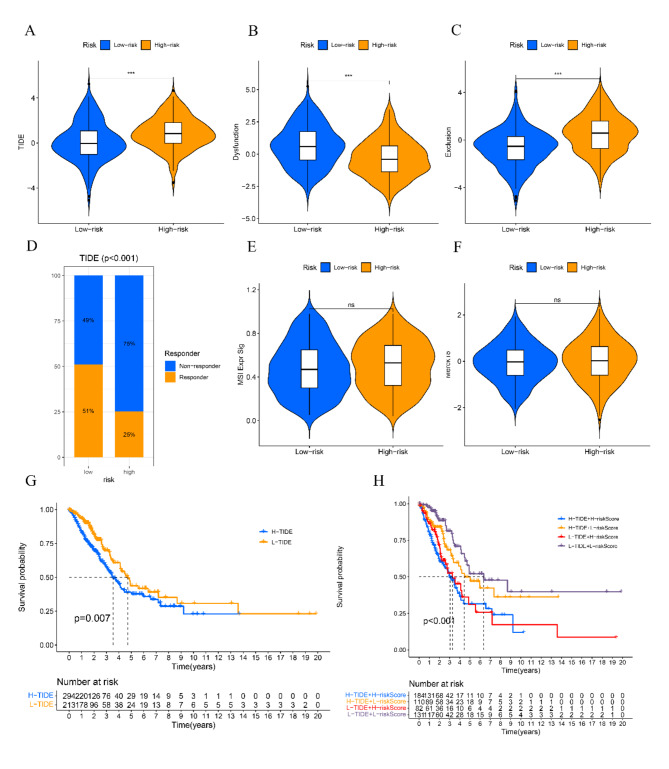
The TIDE algorithm was employed to assess the immunotherapy reaction using transcriptomic information from patients with LUAD. The findings indicated that the TIDE scores exhibited a notable increase in the high-risk cohort as opposed to the low-risk cohort, suggesting that patients belonging to the low-risk group might experience greater advantages from immunotherapy (Fig. 8A). Figure 8B, C show that patients classified as low-risk had elevated levels of functional impairment scores and reduced exclusion scores. Furthermore, individuals who exhibited a positive response to immunotherapy were linked to reduced TIDE scores (as shown in Fig. 8D). No notable disparities were observed in the levels of MSI Expr Sig and Merck18 between the high-risk and low-risk groups, as depicted in Fig. 8E and F. The worst prognosis was linked to the combination of elevated risk and high TIDE scores, as depicted in Fig. 8G, H.
Fig. 8.
Estimation of the RRTGs prognostic model in immunotherapy response in LUAD. (A) Difference in TIDE scores between high- and low-risk groups. (B) Difference in dysfunction scores between high- and low-risk groups. (C) Difference in exclusion scores between high- and low-risk groups. (D) The distribution of immunotherapy response in indicated groups stratified by the telomere-related risk scores based on the TIDE algorithm. (E) Difference in MSI Expr Sig between high- and low-risk groups. (F) Difference in Merck18 between high- and low-risk groups. (G) The Kaplan–Meier OS curves among two groups classified by TIDE score. (H) The Kaplan–Meier OS curves among four groups classified by RRTGs risk score and TIDE score. *p < 0.05, **p < 0.01, and ***p < 0.001.“ns” indicates p ≥ 0.05, not statistically significant
Evaluating patient reactions to different anti-cancer medications using the RRTGs score
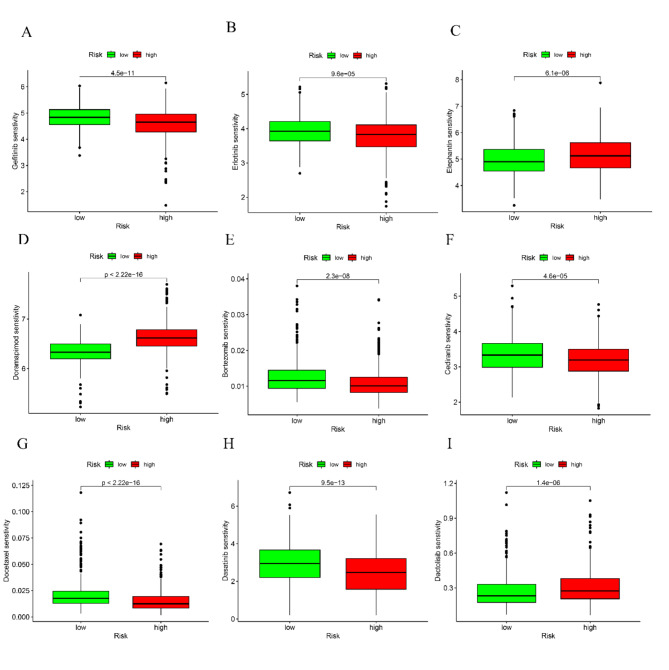
Afterwards, different cancer-fighting medications were chosen to evaluate the responsiveness of individuals in the low-risk and high-risk categories. Patients who obtained high scores exhibited reduced IC50 values for Gefitinib, Erlotinib, Bortezomib, Cediranib, Docetaxel, and Dasatinib, as observed. Conversely, individuals with reduced scores exhibited notably decreased IC50 values for cancer treatment medications, such as Elephantin, Doramapimod, and Dactolisib. The results indicate a connection between the genetic score and the sensitivity of the drug (Fig. 9A-I).
Fig. 9.
Correlations of the RRTGs risk score with drug sensitivity in LUAD. Gefitinib (A), Erlotinib (B), Elephantin (C), Doramapimod (D), Bortezomib (E), Cediranib (F), Docetaxel (G), Dasatinib (H), Dactolisib (I) sensitivity between high- and low-risk groups
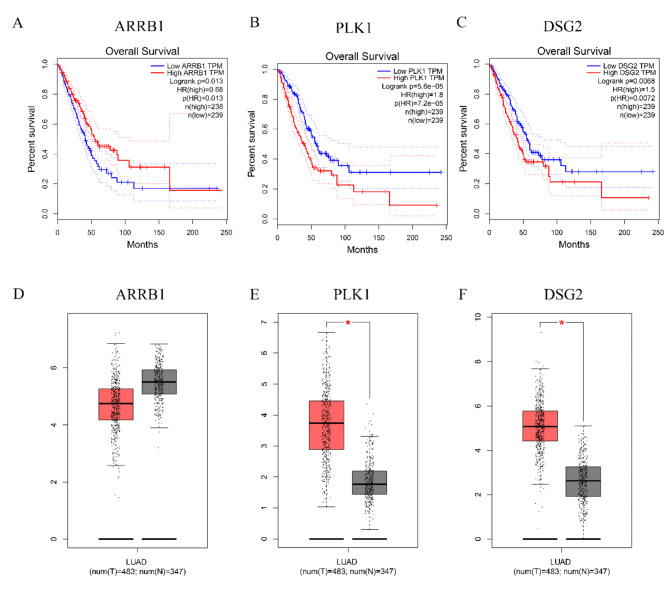
The Kaplan-Meier curves and the gene expression of ARRB1, DSG2, and PLK1 were analyzed in LUAD
A favorable prognosis was linked to reduced levels of DSG2 and PLK1, whereas an unfavorable prognosis was linked to reduced levels of ARRB1 (Fig. 10A-C). DSG2 and PLK1 were highly expressed in tumors, while ARRB1 showed no significant difference in expression between tumor and normal tissues in GEPIA database (Fig. 10D-F).
Fig. 10.
The Kaplan–Meier OS curves and genes expression of ARRB1, DSG2 and PLK1 in LUAD. (A, B, C) The Kaplan–Meier OS curves among two groups classified by ARRB1, DSG2 and PLK1 expression. (D, E, F) ARRB1, DSG2, PLK1 expression between tumor and normal tissues. *p < 0.05
The findings indicate that RRTGs score and the levels of ARRB1, DSG2, and PLK1 expression could have significant implications for the prognosis of LUAD, response to immunotherapy, and sensitivity to drugs. These factors have the potential to serve as valuable biomarkers for clinical decision-making and treatment approaches.
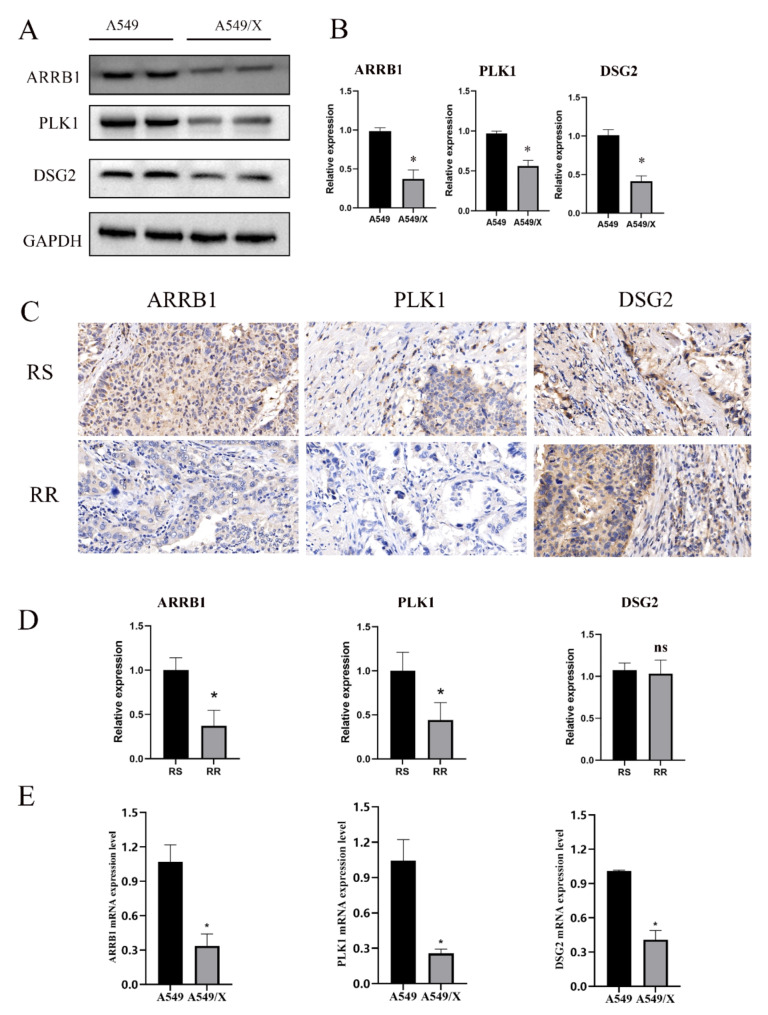
Expression analysis of ARRB1, PLK1, and DSG2 in cell lines and NSCLC tumor tissues
Western blot and qRT-PCR analysis was conducted to assess the expression levels of ARRB1, PLK1, and DSG2 in the A549/X and A549 cell lines. The results demonstrated that both protein and mRNA levels of ARRB1, PLK1, and DSG2 were significantly upregulated in A549 cells compared to A549/X cells (Fig. 11A-B, E). IHC was utilized to determine the protein expression of ARRB1 and PLK1 in NSCLC tumor tissues. ARRB1 and PLK1 were found to be highly expressed in radioresistant NSCLC tissues (n = 4) compared to radiosensitive samples (n = 4). Whereas, DSG2 expression did not show significant variation between the two groups (Fig. 11C-D).
Fig. 11.
Validating characteristic gene expression in cell lines and LUAD tissues. (A, B) Western blot results showing the expression of ARRB1, PLK1 and DSG2 in A549 and A549/X cells. (C, D) IHC results showing the expression of ARRB1, PLK1 and DSG2 in radiosensitive (RS) (n = 4) and radioresistant (RR) (n = 4) LUAD tissues. (E) qRT-PCR results showing the expression of ARRB1, PLK1 and DSG2 in A549 and A549/X cells. *p < 0.05.“ns” indicates p ≥ 0.05, not statistically significant
Discussion
In recent years, several clinical trials with individuals diagnosed with lung adenocarcinoma have verified the significance of radiotherapy in extending lifespan and enhancing overall well-being. The significant discoveries have strengthened the position of radiotherapy in the treatment of LUAD [24, 25]. Radiotherapy’s mechanisms encompass not just its notable tumor-fighting impacts, effectively reducing tumor size, but also its capacity to initiate an immune response against tumors and induce substantial alterations in the tumor microenvironment [26, 27].
Nonetheless, overcoming radiotherapy resistance in lung adenocarcinoma continues to be a substantial challenge in attaining successful therapeutic results [28]. Preliminary studies indicate that focusing on genes associated with telomeres can increase tumor sensitivity to radiotherapy and decrease cancer-induced toxicity [5, 12]. This suggests a close connection between resistance to radiotherapy and telomere-related genes. The GO and KEGG enrichment analyses further support the connection between radiotherapy resistance and telomere-related genes by highlighting key biological processes related to cell division and chromosome organization. Specifically, the regulation of nuclear division, G2/M transition of the mitotic cell cycle, and mitotic checkpoint signaling were identified as critical processes enriched in the 44 RRTGs. These processes are fundamental to cell cycle regulation and ensure proper chromosome segregation and stability. Additionally, the significant enrichment of KEGG pathways like progesterone-mediated oocyte maturation, the Fanconi anemia pathway, and the cell cycle pathway—pathways crucial for DNA repair and cell cycle control—suggests that the RRTGs play a vital role in maintaining genomic stability. Disruptions in these pathways may contribute to radiotherapy resistance, as the cell’s ability to repair radiation-induced DNA damage could be compromised. Thus, targeting these pathways could be a promising therapeutic strategy to overcome resistance and enhance the effectiveness of radiotherapy.
Further analysis led to the identification of three key telomere-associated genes—ARRB1, PLK1, and DSG2—that significantly influence radiotherapy response and prognosis in lung adenocarcinoma. In order to enhance the accuracy of forecasting the life expectancy of individuals diagnosed with lung adenocarcinoma, we employed intricate risk models and nomograms for assessment.
We acquired LUAD samples and relevant information by integrating our sequencing data with the TCGA and GEO databases. Initially, we detected the overlap of genes with altered expression in our dataset, genes with altered expression between tumor and normal groups, and genes associated with telomeres, leading to the identification of 44 genes with altered expression and related to telomeres. Afterwards, we conducted univariate and multivariate Cox regression analyses in order to develop the RRTGs score, which can be used to identify high-risk and low-risk subgroups. Patients who had higher RRTGs scores showed worse clinical outcomes, suggesting that individuals with elevated RRTGs scores experienced a more unfavorable prognosis. Additionally, we combined patient tumor stages and RRTGs scores to construct a nomogram tumor prediction model, which showed good predictive ability.
The interaction of the immune system is a significant factor in the development of cancer and is also a focus for treatment in LUAD. Previous research has suggested that the TME primarily consists of stromal and immune cells. Additionally, the clinical characteristics and prognosis of LUAD are linked to immune and stromal scores, as indicated by prior studies [16, 29]. Applying the ESTI-MATE algorithm for TME score estimation, we observed a notable increase in immune and stromal scores within the low RRTGs score group when compared to the high RRTGs score group. The presence of telomere-related genes in TME implies a potential role in controlling the development and advancement of LUAD. The GSEA analysis further revealed distinct enrichment patterns between high- and low-risk groups, providing additional insight into the biological mechanisms. In the high-risk group, immune-related processes such as adaptive immune response, antigen processing via MHC class II, and complement activation were significantly enriched, as well as pathways related to immune response and metabolism, including asthma and cell adhesion molecules. Conversely, the low-risk group showed enrichment in pathways associated with cell division and chromosome dynamics, such as mitotic nuclear division and chromosome segregation, as well as KEGG pathways involving the cell cycle and DNA replication. These findings highlight the differential involvement of immune activity in high-risk groups and genomic stability processes in low-risk groups, further supporting the potential influence of RRTGs on LUAD prognosis and therapy responses.
Furthermore, researchers have discovered that abnormal immune cells can promote LUAD progression, while immune therapy based on checkpoint inhibitors can improve survival in advanced cancer patients [30, 31]. With RRTGs, an accurate prognostic model representing the TME has been established. Through a thorough examination of RRTGs and considering clinical factors, this aids healthcare professionals in assessing the effectiveness of immunotherapy as a viable treatment choice. Furthermore, notable variations were noted in the clinical prognostic characteristics, genetic mutations, immune infiltration, stromal scores, and drug responsiveness among the two groups of patients. Consequently, the classification of LUAD patients’ prognosis becomes more accurate, offering improved understanding of the genetic foundation of the ailment and suggesting innovative approaches to tumor immunotherapy.
A major constraint of our research is the reliance on genomic datasets that are accessible to the public. While these datasets offer valuable resources for large-scale analyses, they are generated from different studies with varying experimental conditions, platforms, and patient cohorts. As a result, the introduction of batch effects and potential biases may have an impact on the precision and replicability of our findings. The challenge of integrating diverse datasets should not be underestimated, as it requires meticulous preprocessing and normalization to ensure data harmonization. To mitigate these issues, we applied rigorous quality control measures and utilized standardized analytical pipelines [32, 33]. Despite the presence of dataset heterogeneity, it is important to acknowledge that future research could be enhanced by employing more uniform datasets or confirming our discoveries through independent cohorts.
Additionally, the inherent limitations in establishing causal relationships between the identified telomere genes and treatment outcomes or immune infiltration dynamics are posed by the retrospective nature of the analyzed data. Despite employing advanced statistical analyses to control for confounding variables, the possibility of unmeasured or residual confounders cannot be fully ruled out. To understand the functional effects of changed telomere gene expression and their influence on the advancement of lung adenocarcinoma and response to therapy, it is necessary to conduct future studies or experiments using models.
Furthermore, the complexity of telomere biology in lung adenocarcinoma is enhanced by the specific incorporation of three more genes related to telomeres, namely ARRB1, DSG2, and PLK1, in addition to the general restrictions mentioned earlier. ARRB1, a constituent of the arrestin protein group, has been linked to cellular reactions to DNA damage caused by radiation and could impact the results of radiotherapy [34, 35]. DSG2 codes for the enzyme dihydrofolate reductase, which is linked to the regulation of telomeres and the stability of chromosomes, indicating its possible involvement in the advancement of tumors and the response to therapy [36, 37]. However, its involvement in radiotherapy has rarely been reported yet. The dysregulation of PLK1, a kinase that regulates the cell cycle, has been associated with tumor growth and resistance to treatment due to its crucial involvement in telomere maintenance and replication [38–40]. By incorporating these genes into our analysis, we gain a more extensive perspective on the molecular mechanisms implicated in lung adenocarcinoma.
To summarize, we presented a thorough examination of RRTGs in lung adenocarcinoma and developed a novel risk model for the treatment and prognosis of patients with this type of lung cancer. This model includes three genes (ARRB1, PLK1, DSG2) as important components. In addition, we have determined the functions of these genes in lung adenocarcinoma, which impact the immune microenvironment of the tumor, clinical characteristics, approaches to treatment, and future outlook. The results emphasize the possible clinical consequences of RRTGs, indicating that RRTGs could serve as potential treatment targets for individuals diagnosed with LUAD.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
P. L and X. G designed the study. P. L and L. M collected, analyzed and visualized the data. P. L and X. G drafted the initial manuscript. P. L and S. L curated data, H. T and S. L reviewed and edited the article. All authors approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82473378), Shanghai Talents Development Fund Project (2021071), Clinical Research fundation of Shanghai Pulmonary Hospital (SKPY2021006) and the National Natural Science Foundation Cultivation Project of Shanghai Pulmonary Hospital (fkzr2436).
Data availability
Data will be made available on request.
Declarations
Ethical approval
This study was approved by the Ethics Committee at the Shanghai Pulmonary Hospital (approval No. L20-316Z).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Wu G, Yan Y, Cai Y, Peng B, Li J, Huang J, Xu Z, Zhou J. ALKBH1-8 and FTO: potential therapeutic targets and prognostic biomarkers in lung adenocarcinoma Pathogenesis. Front Cell Dev Biol. 2021;9:633927. 10.3389/fcell.2021.633927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen FL, Huang CL, Wu QM, Jiang LL, Chen ST, Chen LY. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J Cell Biochem. 2020;121(3):2525–33. 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Pickett HA. Targeting telomeres: advances in telomere maintenance mechanism-specific cancer therapies. Nat Rev Cancer. 2022;22(9):515–32. 10.1038/s41568-022-00490-1. [DOI] [PubMed] [Google Scholar]
- 6.Daniel M, Peek GW, Tollefsbol TO. Regulation of the human catalytic subunit of telomerase (hTERT). Gene. 2012;498(2):135–46. 10.1016/j.gene.2012.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roake CM, Artandi SE. Regulation of human telomerase in homeostasis and disease. Nat Rev Mol Cell Biol. 2020;21(7):384–97. 10.1038/s41580-020-0234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Dai M, Xing X, Wang X, Qin X, Huang T, Fang Z, Fan Y, Xu D. Genomic, epigenomic, and transcriptomic signatures for telomerase complex components: a pan-cancer analysis. Mol Oncol. 2023;17(1):150–72. 10.1002/1878-0261.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suwa T, Kobayashi M, Nam JM, Harada H. Tumor microenvironment and radioresistance. Exp Mol Med. 2021;53(6):1029–35. 10.1038/s12276-021-00640-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Z, Guan L, Xiang GM, Gao BA. Radiation resistance of the lung adenocarcinoma is related to the AKT-Onzin-POU5F1 axis. Biochem Biophys Res Commun. 2018;499(3):538–43. 10.1016/j.bbrc.2018.03.185. [DOI] [PubMed] [Google Scholar]
- 11.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5(7):577–84. 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 12.Berardinelli F, Coluzzi E, Sgura A, Antoccia A. Targeting telomerase and telomeres to enhance ionizing radiation effects in in vitro and in vivo cancer models. Mutat Res Rev Mutat Res. 2017;773:204–19. 10.1016/j.mrrev.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Eckburg A, Dein J, Berei J, Schrank Z, Puri N. Oligonucleotides and microRNAs targeting telomerase subunits in Cancer Therapy. Cancers (Basel). 2020;12(9). 10.3390/cancers12092337. [DOI] [PMC free article] [PubMed]
- 14.Tian Y, Zhai X, Yan W, Zhu H, Yu J. Clinical outcomes of immune checkpoint blockades and the underlying immune escape mechanisms in squamous and adenocarcinoma NSCLC. Cancer Med. 2021;10(1):3–14. 10.1002/cam4.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, Mcgraw T, Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19(1):9–31. 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen DL, Wang YF, Zhang X, Ding QF, Wang XF, Xue YH, Wang W, Mao YM, Chen C, Chen YB. Characterization of Tumor Microenvironment in Lung Adenocarcinoma Identifies Immune Signatures to Predict Clinical Outcomes and Therapeutic Responses. Frontiers in Oncology, 2021, 11. http://dx.doi.org/ARTN58103010.3389/fonc.2021.581030 [DOI] [PMC free article] [PubMed]
- 17.Gu M, Xu T, Chang P. KRAS/LKB1 and KRAS/TP53 co-mutations create divergent immune signatures in lung adenocarcinomas. Ther Adv Med Oncol. 2021;13:17588359211006950. 10.1177/17588359211006950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebata H, Loo TM, Takahashi A. Telomere Maintenance and the cGAS-STING pathway in Cancer. Cells. 2022;11(12). 10.3390/cells11121958. [DOI] [PMC free article] [PubMed]
- 19.Hu Y, You X, Zhang Z, Zhao H. Telomere-Associated Gene signatures correlate with prognosis, Tumor Microenvironment, and chemosensitivity in breast Cancer. Med Sci Monit. 2023;29:e939921. 10.12659/MSM.939921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu K, Zhao L, Feng S, Zhang S, Zhou Q, Gao X, Guo Y. Colorectal polyp region extraction using saliency detection network with neutrosophic enhancement. Comput Biol Med. 2022;147:105760. 10.1016/j.compbiomed.2022.105760. [DOI] [PubMed] [Google Scholar]
- 21.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Y, Wang Z, He D, Zhu Y, Chen X, Cao K. Identification of novel subtypes based on ssGSEA in immune-related prognostic signature for tongue squamous cell carcinoma. Cancer Med. 2021;10(23):8693–707. 10.1002/cam4.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–8. 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal JP, Pilar A, Mummudi N, Gupta M, Laskar SG, Pathak RS, Tibdewal AR, Kinhikar R, Ghadi Y, Tandon S, Purandare N, Prabhash K, Patil V. Stereotactic body radiation therapy for medically inoperable early-stage lung cancer: Tata Memorial Hospital perspective and practice recommendations. Indian Journal of Cancer, 2020, 57(1): 18–24. http://dx.doi.org/PMID3192923310.4103/ijc.IJC_216_18 [DOI] [PubMed]
- 25.Chi A, Nguyen NP. Rationale for Combing Stereotactic Body Radiation Therapy with Immune Checkpoint Inhibitors in Medically Inoperable Early-Stage Non-Small Cell Lung Cancer. Cancers, 2022, 14(13). http://dx.doi.org/ARTN314410.3390/cancers14133144 [DOI] [PMC free article] [PubMed]
- 26.Krisnawan VE, Stanley JA, Schwarz JK, Denardo DG. Tumor Microenvironment as a Regulator of Radiation Therapy: New Insights into Stromal-Mediated Radioresistance. Cancers, 2020, 12(10). http://dx.doi.org/ARTN291610.3390/cancers12102916 [DOI] [PMC free article] [PubMed]
- 27.Allibhai Z, Taremi M, Bezjak A, Brade A, Hope AJ, Sun A, Cho BCJ. The impact of Tumor size on outcomes after stereotactic body Radiation Therapy for medically inoperable early-stage Non-small Cell Lung Cancer. Int J Radiat Oncol Biol Phys. 2013;87(5):1064–70. 10.1016/j.ijrobp.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Zhou F, Wang YY, Xie HK, Luo SL, Meng L, Su B, Ye Y, Wu KL, Xu YP, Gong XM. Eliminating Radiation Resistance of Non-small Cell Lung Cancer by Dihydroartemisinin through abrogating immunity escaping and promoting Radiation sensitivity by inhibiting PD-L1 Expression. Front Oncol. 2020;595466. 10. http://dx.doi.org/ARTN59546610.3389/fonc.2020.595466 [DOI] [PMC free article] [PubMed]
- 29.Chen ZC, Huang YW, Hu ZY, Zhao MN, Li M, Bi GS, Zheng YS, Liang JQ, Lu T, Jiang W, Xu ST, Zhan C, Xi JJ, Wang Q, Tan LJ. Landscape and dynamics of single tumor and immune cells in early and advanced-stage lung adenocarcinoma. Clinical and Translational Medicine, 2021, 11(3). http://dx.doi.org/ARTNe35010.1002/ctm2.350 [DOI] [PMC free article] [PubMed]
- 30.Sun H, Liu SY, Zhou JY, Xu JT, Zhang HK, Yan HH, Huan JJ, Dai PP, Xu CR, Su J, Guan YF, Yi X, Yu RS, Zhong WZ, Wu YL. Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. Ebiomedicine, 2020, 60. http://dx.doi.org/ARTN10299010.1016/j.ebiom.2020.102990 [DOI] [PMC free article] [PubMed]
- 31.Kang L, Miao MS, Song YG, Fang XY, Zhang J, Zhang YN, Miao JX. Total flavonoids of Taraxacum mongolicum inhibit non-small cell lung cancer by regulating immune function. Journal of Ethnopharmacology, 2021, 281. http://dx.doi.org/ARTN11451410.1016/j.jep.2021.114514 [DOI] [PubMed]
- 32.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 33.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang LG, Wang K, Dong W, Shen HC, Du JJ. Regulation of response to radiotherapy by beta-arrestin 1 in non-small cell lung cancer. J Cancer. 2019;10(17):4085–95. 10.7150/jca.30012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao X, Liu Q, Chen X, Chen S, Yang J, Liu Q, Cheng Y. Screening of tumor grade-related mRNAs and lncRNAs for Esophagus squamous cell Carcinoma. J Clin Lab Anal. 2021;35(6):e23797. 10.1002/jcla.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahlmann NA, Tsoukas RL, Erkens S, Wang HJ, Jonsson F, Aydin M, Naumova EA, Lieber A, Ehrhardt A, Zhang WL. Properties of Adenovirus Vectors with Increased Affinity to DSG2 and the Potential Benefit s of Oncolytic Approaches and Gene Therapy. Viruses-Basel, 2022, 14(8). http://dx.doi.org/ARTN183510.3390/v14081835 [DOI] [PMC free article] [PubMed]
- 37.Jin R, Wang X, Zang R, Liu C, Zheng S, Li H, Sun N, He J. Desmoglein-2 modulates tumor progression and osimertinib drug resistance through the EGFR/Src/PAK1 pathway in lung adenocarcinoma. Cancer Lett. 2020;483:46–58. 10.1016/j.canlet.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Lund-Andersen C, Patzke S, Nahse-Kumpf V, Syljuasen RG. PLK1-inhibition can cause radiosensitization or radioresistance dependent on the treatment schedule. Radiother Oncol. 2014;110(2):355–61. 10.1016/j.radonc.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Zhu DD, Xia J, Liu C, Fang C. Numb/Notch/PLK1 signaling pathway mediated hyperglycemic memory in pancreatic cancer cell radioresistance and the therapeutic effects of metformin. Cellular Signalling, 2022, 93. http://dx.doi.org/ARTN11026810.1016/j.cellsig.2022.110268 [DOI] [PubMed]
- 40.Tatekawa S, Tamari K, Chijimatsu R, Konno M, Motooka D, Mitsufuji S, Akita H, Kobayashi S, Murakumo Y, Doki Y, Eguchi H, Ishii H, Ogawa K. N(6)-methyladenosine methylation-regulated polo-like kinase 1 cell cycle homeostasis as a potential target of radiotherapy in pancreatic adenocarcinoma. Sci Rep. 2022;12(1):11074. 10.1038/s41598-022-15196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.