Abstract
Parkinson’s Disease (PD) is a debilitating neurodegenerative disorder, characterized by motor and cognitive impairments, that affects >1% of the population over the age of 60. The pathogenesis of PD is complex and remains largely unknown. Due to the cellular heterogeneity of the human brain and changes in cell type composition with disease progression, this complexity cannot be fully captured with bulk tissue studies. To address this, we generated single-nucleus RNA sequencing and whole-genome sequencing data from 100 postmortem cases and controls, carefully selected to represent the entire spectrum of PD neuropathological severity and diverse clinical symptoms. The single nucleus data were generated from five brain regions, capturing the subcortical and cortical spread of PD pathology. Rigorous preprocessing and quality control were applied to ensure data reliability. Committed to collaborative research and open science, this dataset is available on the AMP PD Knowledge Platform, offering researchers a valuable tool to explore the molecular bases of PD and accelerate advances in understanding and treating the disease.
Subject terms: Parkinson's disease, Gene expression, Computational biology and bioinformatics
Background & Summary
Parkinson’s disease (PD) is a complex neurodegenerative disorder that significantly diminishes the quality of life of affected individuals by impairing motor skills and often impacting cognitive function. Neuropathological characteristics include the buildup of α-synuclein protein within neurons, leading to the formation of Lewy bodies and Lewy neurites1,2, alongside the degeneration of dopamine-producing neurons3. This condition manifests through symptoms such as tremors, stiffness, and memory loss, which progressively worsen, affecting daily activities and overall well-being4. The complex pathophysiological mechanisms that erode cognitive abilities in Parkinson's are still not fully understood. This knowledge gap highlights the need to examine changes in gene expression, which could reveal the underlying mechanisms of PD progression and enhance the potential for early diagnosis and the development of more effective treatments.
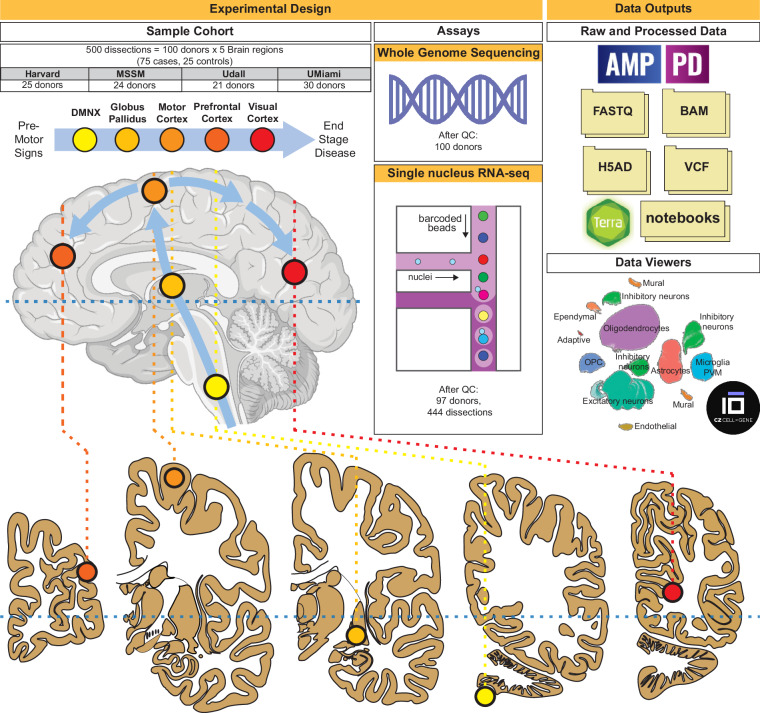
Alterations in gene expression and cell type composition are common disruptions in neurodegenerative disorders, including PD5–10. Traditional approaches11 such as bulk or cell-sorted tissue analyses have not been able to fully capture the complex molecular changes in PD, mainly due to the confounding effects of changes in cell composition12. This problem is worsened by the narrow focus of previous research, which has tended to concentrate on small sample sizes and specific brain areas, notably the substantia nigra, where there is a substantial decrease in dopaminergic neurons3. Moreover, the typical use of case-control studies, primarily involving patients in advanced stages of PD, fails to effectively track the gradual changes in gene expression that occur as the disease progresses. In our study, we have adopted several strategies to overcome these limitations: First, we utilized a single-nucleus RNA-seq assay (snRNA-seq) for data generation, enabling cell-specific analysis. Secondly, in collaboration with four different brain banks, we gathered extensive clinical and demographic records for over 600 PD cases. From this collection, we selected age- and sex- balanced specimens from 75 PD patients at varying disease stages, based on neuropathological evaluations and staging, and included 25 unaffected controls. To ensure coverage of all disease stages, we utilized Braak PD staging13, which quantifies regional disease progression and accumulation of Lewy bodies, primarily composed of α-synuclein. This approach enabled the inclusion of donors with early-stage PD pathology. Furthermore, we expanded the scope of transcriptomic profiling to include five brain regions beyond the extensively studied substantia nigra, tracking the progression of abnormal immunostaining α-synuclein patterns as defined by Braak PD stages13 (Fig. 1). This included early affected (DMNX: dorsal motor nucleus of the Xth nerve; GPI: globus pallidus interna) to late-affected regions (PMC: primary motor cortex, DLPFC: dorsolateral prefrontal cortex), as well as a largely unaffected region (PVC: primary visual cortex). The corresponding Brodmann areas (BA) for these regions are as follows: PMC = BA4, DLPFC = BA9 and PVC = BA17.
Fig. 1.
Schematic overview of dataset collection and main study deliverables. Harvard: NIH NeuroBiobank at the Harvard Brain Tissue Resource Center; MSSM: NIH NeuroBioBank at the Mount Sinai Brain Bank; Udall: University of Miami Udall Center of Excellence for Parkinson's Disease Research; UMiami: NIH NeuroBioBank at the University of Miami and University of Miami Brain Endowment Bank.
For whole genome sequencing (WGS), we utilized genomic DNA extracted from the PVC (Fig. 1). The data described in this study represent the largest PD-oriented single nucleus data collection to date and is available at the AMP PD Knowledge Platform.
Methods
Cohort data collection
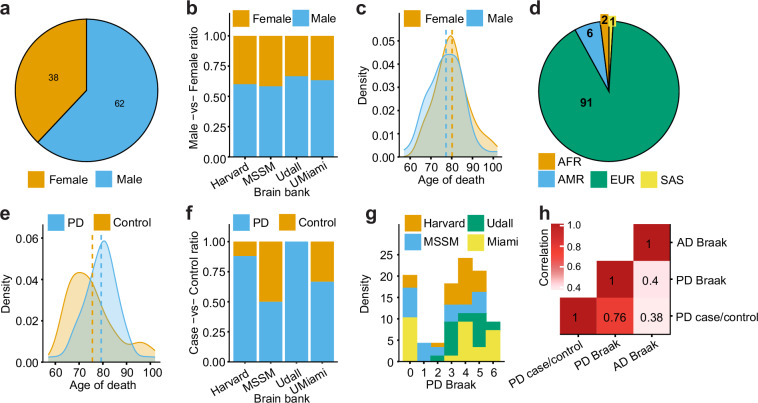
The cohort consists of genetic (WGS) and transcriptomic (snRNA-seq) assays collected using a cohort of 100 donors sourced from the following brain banks: NIH NeuroBioBank at the Mount Sinai School of Medicine, NIH NeuroBioBank at the Harvard Brain Tissue Resource Center, NIH NeuroBioBank at the University of Miami, the University of Miami Brain Endowment Bank and the University of Miami Udall Center of Excellence for Parkinson's Disease Research (Fig. 1 and Table 1). All data were obtained from biobanks with appropriate informed consent from all participants. Detailed cognitive, neuropathological, and demographic information was gathered for all donors, who were mainly of European descent with a male-to-female ratio of 3:2 (Fig. 2a–d). The study included 75 donors across a spectrum of PD severity, based on the Braak PD staging (Braak et al.13), which tracks the spread of Lewy body pathology (Fig. 2e–g), and 25 donors without the disease as control subjects (Table 1). Additionally, data were collected on Alzheimer’s disease (AD) Braak staging14, which evaluates tau neurofibrillary tangle accumulation, and the Hoehn and Yahr scale15, assessing functional disability in PD. The importance of examining both neuropathological (Braak PD staging) and detailed clinical characteristics of PD is evident from only a limited correlation observed between those phenotypes (Fig. 2h). Furthermore, the availability of both Braak AD and Braak PD stages provides an opportunity to explore the transcriptomics basis of symptomatic, clinical16, and, to a lesser extent, genetic overlaps17,18 between Tau and Lewy body accumulation. However, it is important to note that all donors were either unaffected controls or clinically diagnosed exclusively with PD but no other neurological or major neuropsychiatric diseases, including AD.
Table 1.
Summary of clinical and demographics data stratified by source brain bank.
| Brain Bank | # | Disease Status | Sex | Age | Data Available | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Male | Female | <70 | 70–84 | >85 | Clinical | WGS | snRNAseq | ||
| UDa | 21 | 21 | 0 | 14 | 7 | 1 | 13 | 7 | 21 | 21 | 21 |
| HAb | 25 | 22 | 3 | 15 | 10 | 6 | 15 | 4 | 25 | 25 | 25 |
| UMc NIH NeuroBioBank | 17 | 12 | 5 | 10 | 7 | 3 | 9 | 5 | 17 | 17 | 15 |
| UMc Brain Endowment Bank | 13 | 8 | 5 | 9 | 4 | 3 | 7 | 3 | 13 | 13 | 12 |
| MSd | 24 | 12 | 12 | 14 | 10 | 3 | 19 | 2 | 24 | 24 | 24 |
| Total | 100 | 75 | 25 | 62 | 38 | 16 | 63 | 21 | 100 | 100 | 97 |
aUdall Center of Excellence for Parkinson’s Disease Research (UD).
bNIH NeuroBioBank at Harvard Brain Tissue Resource Center (HA).
cUniversity of Miami (UM).
dNIH NeuroBioBank at the Mount Sinai Brain Bank (MS).
Fig. 2.
Study cohort characteristics. (a–c) Numbers and distributions of male and female donors across brain regions and age. (d) Distribution of donors by the most similar ancestry superpopulation predicted by QDA (AFR: African, AMR: Admixed American, EUR: European, SAS: South Asian). (e–g) Distribution of donors by age, source brain bank and Braak PD staging. (h) Spearman correlation coefficients among PD-related phenotypes. Harvard: NIH NeuroBiobank at the Harvard Brain Tissue Resource Center; MSSM: NIH NeuroBioBank at the Mount Sinai Brain Bank; Udall: University of Miami Udall Center of Excellence for Parkinson's Disease Research; UMiami: NIH NeuroBioBank at the University of Miami and University of Miami Brain Endowment Bank.
snRNA-seq data generation
Nuclei isolation and snRNA-seq library preparation
All buffers were supplemented with RNAse inhibitors (Takara). Six samples, each from a different individual, were processed in parallel. Twenty-five mg of frozen postmortem human brain tissue from each specimen was homogenized in cold lysis buffer (0.32 M Sucrose, 5 mM CaCl2, 3 mM Magnesium acetate, 0.1 mM, EDTA, 10 mM Tris-HCl, pH8, 1 mM DTT, 0.1% Triton X-100) and filtered through a 40 µm cell strainer. The flow-through was underlaid with sucrose solution (1.8 M Sucrose, 3 mM Magnesium acetate, 1 mM DTT, 10 mM Tris-HCl, pH8) and centrifuged at 107,000 g for 1 hour at 4°C. Pellets were resuspended in PBS and quantified (Countess II, Life Technologies). 2 million nuclei from each sample were then pelleted at 500 g for 5 minutes at 4˚C and re-suspended in 100 µl staining buffer (2% BSA, 0.02% Tween-20, 10 mM Tris, 146 mM NaCl, 1 mM CaCl2 and 21 mM MgCl2). Each sample was incubated with 1 µg of a distinct TotalSeq-A nuclear hashing antibody (Biolegend) for 30 min at 4°C. Prior to Fluorescence-Activated Nuclei Sorting (FANS), volumes were brought up to 250 µl with staining buffer and 7-AAD (Invitrogen) added to facilitate the detection of nuclei. 7-AAD positive nuclei were sorted into tubes pre-coated with 5% BSA using a FACSAria flow cytometer (BD Biosciences).
Following FANS, nuclei were washed in staining buffer before being re-suspended in 22 µl PBS and quantified. Nuclei concentrations were normalized and equal amounts from each sample were pooled together. Two aliquots of 60,000 pooled nuclei (i.e. 10,000 per sample) were processed in parallel using 3’ v3.1 reagents (10x Genomics). At the cDNA amplification step, reactions were supplemented with a hash-tag oligo (HTO) cDNA “additive” primer (GTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T; *Phosphorothioate bond). Following cDNA amplification, supernatants from the 0.6x SPRI selection step were retained for HTO library15 generation. Otherwise, cDNA libraries were prepared according to the manufacturer’s instructions (10x Genomics). HTO libraries were prepared as described previously19. All libraries were sequenced at the New York Genome Center (NYGC) using the Novaseq 6000 platform (Illumina).
Computational processing
Alignment of sequencing reads from each multiplexed sample batch was conducted using the STARsolo (v.2.7.9a)20,21 algorithm against the hg38 reference genome. To assign the cells from each sequencing pool to their respective donors, we used a genotype-based demultiplexing strategy followed by a genotype concordance check. Initially, cellSNP-lite (v.1.2.0)22 collected allele data from polymorphic loci overlapping snRNA-seq reads from genes expressed in at least 10 cells. These polymorphic loci were required to display a minor allele frequency of at least 0.1 and meet the UMI threshold of 20. Subsequently, the vireo (v.0.5.8)23 segregated cells into clusters that corresponded to the six distinct donors in each batch. Identity verification for each cell cluster was performed through a genotype concordance analysis using the QTLtools-mbv (v.1.3)24, comparing cell clusters against WGS. To ensure the accuracy of this procedure, we filtered out cells not meeting baseline quality control (QC) metrics, i.e. minimum number of expressed genes (n ≥ 1,000) and maximum fraction of mitochondrial reads (less than 5%). Despite most pools containing the expected donors, genotype concordance data was crucial for identifying and correcting sporadic instances of sample mislabeling or swapping.
Following alignment and donor assignment, a stringent, three-tiered QC protocol was employed to eliminate ambient RNA and ensure only viable cells were retained for subsequent analyses. Initially, a rigorous cell-level QC was implemented, which built on preliminary checks from the demultiplexing phase. Cells falling outside the defined ranges for UMI counts (1,500 to 110,000), gene expression (1,100 to 12,500 genes), and mitochondrial content (below 2%) were excluded. This stage also included assessment for potential ambient RNA contamination, particularly from non-messenger RNAs such as rRNA, sRNA, pseudogenes, and the lncRNA MALAT1. Additionally, cell doublets were identified and removed using the Scrublet (v.0.2.3)25. The second QC stage focused on gene expression, removing genes not consistently expressed in at least 0.05% of nuclei. The final QC step targeted the sample level, excluding samples represented by fewer than 50 cells to minimize noise in downstream analysis.
Cell clustering
All 2,232,626 nuclei resulting from the previous QC steps were unified into a single dataset. Normalization and clustering were performed using the SCANPY (v.1.9.3)26 and Pegasus package27. Briefly, counts for all nuclei were scaled by the total library size and logarithmically transformed. Subsequently, 6,000 highly variable genes were identified based on dispersion and mean (excluding sex chromosomes and mitochondria-related genes), followed by regression of the technical influence of the total number of counts, percentage of mitotic counts and cell cycle difference using pg.regress_out() function in Pegasus. Furthermore, the data were also corrected for batch effects coming from different brain banks using the Harmony28 approach via the pg.run_harmony() function. Principal Component Analysis (PCA) was carried out on the variable genes, followed by Uniform Manifold Approximation and Projection (UMAP)29 dimensionality reduction on the top 30 principal components (PCs). We confirmed that more than 30 PCs capture 100% of the data variance. The top 50 PCs were utilized to construct a k-nearest-neighbors cell–cell graph with k=100 neighbors. The Leiden algorithm was then applied to identify cell clusters. These analyses were performed using the functions pg.pca(), pg.elbowplot(), pg.neighbours() in Pegasus, and leiden clustering using sc.tl.leiden() in SCANPY. Differential gene expression analysis for each cluster was conducted using the variance-adjusted t-test implemented in the sc.tl.rank_genes_groups() function in SCANPY. The top 300 ranking genes for each cluster were extracted and tested for overlap with previously reported markers30–32. Subsequently, during iterative sub-clustering, additional potentially dubious clusters representing low-quality or doublet cells were identified based on extreme separation from the rest of the sub-cluster population from the same cell type. Among these, clusters characterized by a distinctly high number of total counts or/and mixed expression of markers from different cell types were detected as potential doublets and excluded from downstream analyses, resulting in a total of 2,096,155 nuclei retained. Furthermore, cellular identities at the class level of taxonomy were confirmed by examining cosine similarity correlations which compared to pseudo bulk-level transcriptome of detected Leiden clusters with reference datasets30–32.
Whole-genome sequencing
Library preparation
DNA was extracted from tissue samples using the QIAmp DNA kit (Qiagen, kit number 51306), according to the manufacturer’s instructions. Once DNA was extracted, samples were quantified using the Qubit Fluorometer (Life Technologies) and PicoGreen (Thermo Fisher), and sample quality was evaluated by checking Fragment Analyzer (Advanced Analytical) traces. WGS libraries were prepared using the Truseq DNA PCR-free Library Preparation Kit (Illumina, kit number 20015965, lot numbers 20698565 and 20706057) in accordance with the manufacturer’s instructions. Briefly, 1 µg of DNA was sheared using a Covaris LE220 sonicator (adaptive focused acoustics). DNA fragments underwent bead-based size selection and were subsequently end-repaired, adenylated, and ligated to IDT for Illumina TruSeq DNA UD Indexes (kit 20040870, lot number 20704419). Final libraries were quantified using the Qubit Fluorometer (Life Technologies) or Spectromax M2 (Molecular Devices) and library size determined using a Fragment Analyzer (Advanced Analytical) or Agilent 2100 BioAnalyzer. Libraries were sequenced on an Illumina Novaseq 6000 sequencer using 2x150bp cycles and S4 reagent kit v1.5 (catalog number 20028312).
Computational processing
Sequencing reads were aligned using BWA-mem33 to the hg38 reference genome. WGS variant calling was performed according to the Genome Analysis Toolkit (GATK, v.3.9.0) best practice recommendations34. Briefly, sample-level nucleotide variants (SNVs) and insertions/deletions (indels) were called using the GATK’s HaplotypeCaller and GenotypeGVCFs tools. To refine and annotate variants, Variant Quality Score Recalibration (VQSR) was conducted within the GATK framework. Sample-level QC followed established described pipelines35–37 involving an assessment of relatedness, DNA contamination (by VerifyBamID, v.1.1.3)38, sample-level missingness (exclusion when > 0.05), and overall coverage (exclusion when < 25x). Furthermore, outlier samples were checked against various metrics, including the number of called SNVs and indels, insert size length, alignment mapping quality score, CRAM file size, transition/transversion (Ti/Tv) ratio, the ratio of novel variants to all variants, and the mapped reads to paired reads ratio as previously described35–37. Variant-level filtering eliminated variants with missingness > 0.10 and high heterozygosity levels (InbreedingCoeff < −0.8). Individual genotype calls with depth < 10 or genotype quality < 20 were set as missing. Analyses were restricted to biallelic variants only.
Ancestry estimation
Based on the success of Mahalanobis distance techniques in ancestry assignment39,40, we leveraged quadratic discriminant analysis (QDA) to assign ancestry using scikit-learn (v.1.14.4)41. For each sample, we identified the most similar genetic ancestry group among the 1000 Genomes Project’s five superpopulations42. First, unimputed genotypes were merged with GRCh38 v2a 1000 Genomes Project data42 using BCFtools 1.9). PCs of the merged genotypes were computed using PLINK (v.2.0) PCA after variant-level filtering, i.e. retaining SNVs with minor allele frequency ≥ 0.01, Hardy-Weinberg equilibrium p-value ≥ 10−10 and variant-level missingness ≤ 0.01, followed by linkage disequilibrium pruning (window size = 1,000 kb, step size = 10, R2 = 0.2). Forward selection was used to select PC1 to PC6 to train the QDA models with regularization parameter 5−7.
Data Records
All data described herein are available for use by the research community and have been deposited in the AMP PD Knowledge Platform43 (https://app.terra.bio/#workspaces/amp-pd-public/AMP-PD-In-Terra; select “AMP PD Release 4”). The released dataset encompasses files for snRNA-seq and WGS. For snRNA-seq, the data includes raw multiplexed sequencing files (FASTQ), sample-level and combined gene expression profiles (h5ad), and sample-level metadata. For WGS, the dataset comprises sample-level aligned sequencing data (CRAM), genomic variants (gVCF), and a set of QC files produced by Picard (v.2.22.3) and GATK (v.3.9.0), including insert size metrics, duplication metrics, GC bias metrics, alignment summary metrics, and WGS-specific metrics. The dataset webpage includes several Terra notebooks in Python and R for data analysis in a cloud environment. Access to this data is governed by a Data Use Agreement that permits its use for approved research and educational purposes by registered and compliant users. Single-cell data can be further inspected at CELLxGENE (RRID:SCR_021059) portal (https://cellxgene.cziscience.com/collections/d5d0df8f-4eee-49d8-a221-a288f50a1590).
Technical Validation
snRNA-seq data quality control
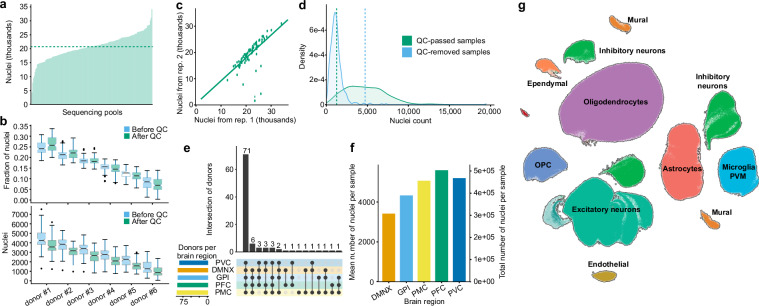
After completing the QC process and excluding all low-quality samples, our snRNA-seq dataset included a total of 2,096,155 nuclei distributed across 161 pools. Each pool comprised six samples, and each pool was sequenced in replicate across two different flowcells. The average yield per pool was 20,640 nuclei (Fig. 3a) and these were expected to be evenly distributed across the samples. Despite this expectation, substantial variability in cell counts was observed, largely due to differences in the quality and condition of the samples within the same pool, which influenced cell viability and capture efficiency44. Typically, the largest sample captured about 25% of the nuclei (µ=3,680 nuclei), while the smallest captured about 7% (µ=1,043 nuclei) (Fig. 3b). Such variability is, however, not unusual and has been observed in other studies23,44,45. Despite these variations in per-sample nuclei count, the comparison of total number of nuclei between replicates showed almost perfect correlation (Spearman’s ρ=0.79), suggesting data robustness (Fig. 3c). In contrast, samples that were removed at the QC step contained approximately 70% fewer nuclei compared to those that passed QC (1,407 vs 4,721 cells, Fig. 3d). Donor representation across brain regions also varied: 71 donors were represented in each of the five brain regions and 18 were missing samples from only one region (Fig. 3e). Conversely, only one donor had samples from a single brain region and 3 donors did not generate any snRNA-seq data. Notably, regions affected early in Parkinson's disease exhibited significantly fewer cells than later affected, or putatively unaffected, regions (Fig. 3f). This suggests regional variations in cellular vulnerability and pathological progression, underscoring the importance of targeted studies to elucidate region-specific disease mechanisms in PD. Cell taxonomy identified nine major cell type clusters that are known to be present in the investigated brain regions (Fig. 3g).
Fig. 3.
Analysis of snRNA-seq dataset. (a) Distribution of the number of nuclei across sequencing pools. Horizontal dashed line denotes a mean value. (b) Distribution of nuclei to replicates within pools, ordered by cell count. Each replicate is depicted using two boxplots representing the nuclei distribution before and after QC. The center line (black) indicates the median, the box shows the interquartile range, and the whiskers indicate the highest/lowest values within 1.5× the interquartile range. (c) Comparison of QC-passed nuclei counts between pairs of replicates from the same pools shows high consistency (Spearman’s ρ=0.79). (d) Distribution of nuclei counts in samples that passed or failed QC (vertical line indicates the mean values). (e) Sample counts and intersections among brain regions. (f) nuclei distribution across five brain regions. The left y-axis shows the average number of nuclei per sample for each region, while the right y-axis indicates the total number of nuclei detected in all samples from each region. (g) UMAP visualization of single nuclei defined by RNA-seq data shows eight major cell type clusters that are expected to be presented in the investigated brain regions.
Whole-genome sequencing quality control
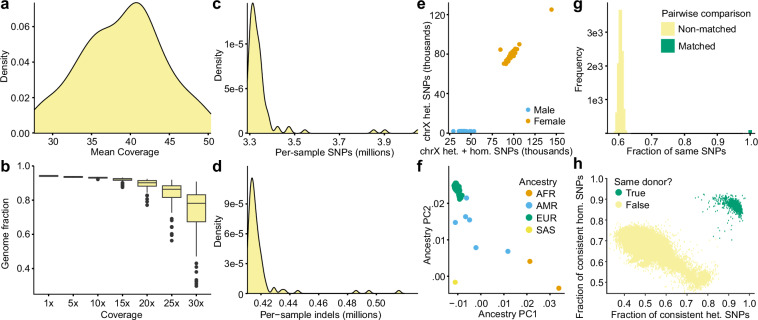
The mean mapped coverage across all samples was 39x (±5x; Fig. 4a), with 94.2% (±0.3%) of the genome achieving at least 1x coverage and 93.1% (±0.3%) reaching at least 10x coverage (Fig. 4b). On average, each sample contained approximately 3.35 million SNPs (±0.15 million) and 418,813 indels (±15,274; Fig. 4c,d). Additionally, our analysis clearly distinguished male from female samples and demonstrated high concordance between inferred and self-reported ancestry across all donors (Fig. 4e,f). Pairwise genotype comparison between WGS samples as well as genotype comparison between WGS and snRNA-seq samples confirmed a clear separation between pairs from the same donors compared to those from different donors (Fig. 4g,h).
Fig. 4.
Quality control of WGS data. (a) Distribution of mean coverage indicating the average number of high-quality sequencing reads per base after applying all QC steps. (b) The fraction of the genome sequenced at different depths. The center line (black) indicates the median, the box shows the interquartile range, and the whiskers indicate the highest/lowest values within 1.5× the interquartile range. (c,d) Number of per-sample SNPs and indels. (e) Sex check based on comparison of the counts of heterozygous and homozygous alleles. (f) The first two PCs of genetic ancestry. (g) Pairwise comparison of the SNPs among all putatively matched and non-matched combinations of WGS samples. (h) Pairwise comparison of the genetic similarities calculated by QTLtools-mbv between WGS samples and genotypes called from snRNA-seq data.
Usage Notes
We would like to highlight the availability of additional clinical metadata beyond Braak PD staging and the binary clinical definition of PD case/control status. Specifically, we provide Braak AD staging data for 83% of the donors. Additionally, other metrics, including longitudinal data and detailed clinical features of PD, are available. These metrics use the Movement Disorder Society – Unified Parkinson’s Disease Rating Scale46, Mini-mental state examination47, Modified Schwab & England scale48, Epworth Sleepiness Scale49, and are primarily available for donors from UD (Table 2). The UD cohort consists of 21 donors, all diagnosed with Parkinson's disease and having a Braak PD stage of at least 2. Thus, the UD cohort does not cover the entire spectrum of disease progression, and users utilizing only data from UD should consider this limitation in their analyses.
Table 2.
Summary of availability of clinical metadata stratified by source brain bank.
| Table Name | Description | Brain Bank | Ref [PMID] | |||
|---|---|---|---|---|---|---|
| UD | HA | UM | MS | |||
| PD Medical History | initiation and use of PD medication, changes of diagnosis over time, indication of surgeries | X | X | X | X | |
| Family History PD | indication of PD diagnosis for father, mother and/or other relatives | X | X | |||
| Smoking and alcohol history | indication and quantification of severity of smoking and alcohol consumption | X | X | X | ||
| Epworth Sleepiness Scale | general level of daytime sleepiness | X | 1798888 | |||
| LBD_Cohort_Clinical_Data | clinical symptoms (e.g. visual hallucionations, loss of memory, mood disorder) | X | X | X | X | |
| LBD_Cohort_Path_Data | neuropathological assessments (CERADa, AD Braak, PD Braak) | X | X | X | X | |
| MDS UPDRSb Part I | non-motor symptoms covering aspects like mood, cognition and sleep | X | 12815652 | |||
| MDS UPDRSb Part II | motor symptoms impacting daily activities | X | X | 12815652 | ||
| MDS UPDRSb Part IV | motor complications related to PD disease treatment | X | 12815652 | |||
| MMSEc | screening for cognitive impairment and monitoring fo changes in mental status over time | X | X | 1202204 | ||
| Modified Schwab & England (ADLd) | assessment of the capabilities of people with impaired mobility | X | ||||
| UPDRSb | other UPDRS metrics not involved in Part I-IV | X | 12815652 | |||
aCERAD: Consortium to Establish a Registry for Alzheimer’s Disease.
bMDS UPDRS: Movement Disorder Society – Unified Parkinson’s Disease Rating Scale.
cMMSE: Mini Mental State Examination.
dADL: Activities of Daily Living.
Complete description of all metrics at https://amp-pd.org/harmonized-clinical-assessments.
Our study involves tissue samples from four brain banks, each contributing different proportions of cases and controls, males and females, and varying age-at-death distributions. To account for unwanted biological and technical variation, we recommend adjusting for relevant covariates from the metadata. The choice of covariates should align with the specific research question; for instance, sex should not be adjusted if studying sex-specific differences. In most of our studies, we typically account for demographic and technical factors such as sex, age, brain bank, RNA integrity number (RIN), and postmortem interval (PMI).
Access to the AMP PD Knowledge Platform data can be obtained by following the registration process outlined at http://amp-pd.org/register-for-amp-pd. This process includes the submission of a registration form, obtaining approval, and compliance with the AMP PD Data Use Agreement.
Acknowledgements
Human tissue was obtained from the NIH NeuroBioBank at the following sites: the University of Miami, the Mount Sinai Brain Bank and the Harvard Brain Tissue Resource Center. Human tissue was also obtained from the University of Miami Brain Endowment Bank the University of Miami Udall Center of Excellence for Parkinson's Disease Research. We thank members of the Roussos laboratory for thoughtful advice and critique. This study was supported by grants from the National Institute of Neurological Disorders and Stroke (NINDS), the National Institutes of Health (NIH) grant U01NS125580 (to P.R., V.H. and W.K.S) and the Accelerating Medicine Partnership® (AMP®) Parkinson’s Disease (AMP PD) program (https://www.amp-pd.org). The AMP® PD program is a public-private partnership managed by the Foundation for the National Institutes of Health and funded by the National Institute of Neurological Disorders and Stroke (NINDS) in partnership with the Aligning Science Across Parkinson's (ASAP) initiative; Celgene Corporation, a subsidiary of Bristol-Myers Squibb Company; GlaxoSmithKline plc (GSK); The Michael J. Fox Foundation for Parkinson's Research; Pfizer Inc.; AbbVie Inc.; Sanofi US Services Inc.; and Verily Life Sciences. This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences. Research reported in this publication was also supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD026880 and S10OD030463. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
P.R. conceived and designed the study. A.B., D.A.D., R.T.V., S.P.G., J.M.V., L.S., M.C., S.B., V.H. and W.K.S., contributed to the selection of donors, obtained informed consents, reviewed clinical examinations and/or pathological analysis and dissected tissue. J.F.F., C.C., E.H., M.A. and J.K. processed tissue and generated snRNA-seq and/or WGS libraries. P.N.M., T.C., D.M., D.B. and J.B. conducted bioinformatics data analysis. D.L. and G.V. supervised bioinformatics data analysis. D.V. and B.L. processed the data into the analysis-ready formats at the AMP PD Knowledge Platform. P.N.M., J.F.F., T.C., J.B. and P.R. wrote and edited the manuscript with input from all co-authors.
Code availability
The source code used to analyze the metadata and create figures for this manuscript can be found on GitHub at this location: https://github.com/DiseaseNeuroGenomics/AMP-PD_SciData. Additionally, jupyter notebooks for working with the dataset at Terra platform are available at this location: https://app.terra.bio/#workspaces/amp-pd-release-v4/Getting%20Started%20Tier%202%20-%20Clinical%20and%20Omics%20Access.
Competing interests
Panos Roussos is an Editorial Board Member for Scientific Data.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Prashant N. M., John F. Fullard.
These authors jointly supervised this work: Jaroslav Bendl, Panos Roussos.
Contributor Information
Jaroslav Bendl, Email: jaroslav.bendl@mssm.edu.
Panos Roussos, Email: panagiotis.roussos@mssm.edu.
References
- 1.Goedert, M., Spillantini, M. G., Del Tredici, K. & Braak, H. 100 years of Lewy pathology. Nat. Rev. Neurol.9, 13–24 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Shahmoradian, S. H. et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci.22, 1099–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Iarkov, A., Barreto, G. E., Grizzell, J. A. & Echeverria, V. Strategies for the Treatment of Parkinson’s Disease: Beyond Dopamine. Front. Aging Neurosci.12, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascherio, A. & Schwarzschild, M. A. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol.15, 1257–1272 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kamath, T. et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci.25, 588–595 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryois, J. et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet.52, 482–493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, Q. et al. Molecular profiling of human substantia nigra identifies diverse neuron types associated with vulnerability in Parkinson’s disease. Sci Adv10, eadi8287 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatila, Z. K. et al. RNA- and ATAC-sequencing Reveals a Unique CD83+ Microglial Population Focally Depleted in Parkinson’s Disease. bioRxiv10.1101/2023.05.17.540842 (2023).
- 9.Toker, L. et al. Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson’s disease brain. Mol. Neurodegener.16, 31 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, Y. J. K. et al. Identifying consensus disease pathways in Parkinson’s disease using an integrative systems biology approach. PLoS One6, e16917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendl, J. et al. The three-dimensional landscape of cortical chromatin accessibility in Alzheimer’s disease. Nat. Neurosci.25, 1366–1378 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClymont, S. A. et al. Parkinson-Associated SNCA Enhancer Variants Revealed by Open Chromatin in Mouse Dopamine Neurons. Am. J. Hum. Genet.103, 874–892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging24, 197–211 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Braak, H., Alafuzoff, I., Arzberger, T., Kretzschmar, H. & Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol.112, 389–404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabey, J. M. & Korczyn, A. D. The Hoehn and Yahr Rating Scale for Parkinson’s Disease. in Instrumental Methods and Scoring in Extrapyramidal Disorders 7–17 (Springer Berlin Heidelberg, 1995).
- 16.Jellinger, K. A. Neuropathological aspects of Alzheimer disease, Parkinson disease and frontotemporal dementia. Neurodegener. Dis.5, 118–121 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Wightman, D. P. et al. The genetic overlap between Alzheimer’s disease, amyotrophic lateral sclerosis, Lewy body dementia, and Parkinson's disease. Neurobiol. Aging127, 99–112 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Wainberg, M., Andrews, S. J. & Tripathy, S. J. Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson's disease, and amyotrophic lateral sclerosis. Alzheimers. Res. Ther.15, 113 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol.19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminow, B., Yunusov, D. & Dobin, A. STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. bioRxiv 2021.05.05.442755, 10.1101/2021.05.05.442755 (2021).
- 22.Huang, X. & Huang, Y. Cellsnp-lite: an efficient tool for genotyping single cells. Bioinformatics10.1093/bioinformatics/btab358 (2021). [DOI] [PubMed]
- 23.Huang, Y., McCarthy, D. J. & Stegle, O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol.20, 273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fort, A. et al. MBV: a method to solve sample mislabeling and detect technical bias in large combined genotype and sequencing assay datasets. Bioinformatics33, 1895–1897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, B. et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods17, 793–798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Lake, B. B. et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol.36, 70–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science382, eadd7046 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Mathys, H. et al. Single-cell atlas reveals correlates of high cognitive function, dementia, and resilience to Alzheimer’s disease pathology. Cell186, 4365–4385.e27 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN] (2013).
- 34.Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178, 10.1101/201178 (2018).
- 35.Adelson, R. P. et al. Empirical design of a variant quality control pipeline for whole genome sequencing data using replicate discordance. Sci. Rep.9, 16156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naj, A. C. et al. Quality control and integration of genotypes from two calling pipelines for whole genome sequence data in the Alzheimer’s disease sequencing project. Genomics111, 808–818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jun, G. et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet.91, 839–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson, R. E. et al. The utility of empirically assigning ancestry groups in cross-population genetic studies of addiction. Am. J. Addict.26, 494–501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conomos, M. P. et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet.98, 165–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 42.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nm, P. et al. AMP-PD release 4: snRNA-seq and WGS dataset. AMP-PD Knowledge Portal 10.5281/ZENODO.13830696 (2023).
- 44.Brown, D. V. et al. A risk-reward examination of sample multiplexing reagents for single cell RNA-Seq. Genomics116, 110793 (2024). [DOI] [PubMed] [Google Scholar]
- 45.Cardiello, J. F. et al. Evaluation of genetic demultiplexing of single-cell sequencing data from model species. Life Sci Alliance6 (2023). [DOI] [PMC free article] [PubMed]
- 46.Movement Disorder Society Task Force on Rating Scales for Parkinson’s Disease. The Unified Parkinson’s Disease Rating Scale (UPDRS): status and recommendations. Mov. Disord.18, 738–750 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Folstein, M. F., Folstein, S. E. & McHugh, P. R. ‘Mini-mental state’. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res.12, 189–198 (1975). [DOI] [PubMed] [Google Scholar]
- 48.Schwab, R. S. Projection technique for evaluating surgery in Parkinson’s disease. in Third symposium on Parkinson’s disease 152–157 (E&S Livingstone, 1969).
- 49.Johns, M. W. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep14, 540–545 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source code used to analyze the metadata and create figures for this manuscript can be found on GitHub at this location: https://github.com/DiseaseNeuroGenomics/AMP-PD_SciData. Additionally, jupyter notebooks for working with the dataset at Terra platform are available at this location: https://app.terra.bio/#workspaces/amp-pd-release-v4/Getting%20Started%20Tier%202%20-%20Clinical%20and%20Omics%20Access.