Abstract
Jalili syndrome, an autosomal recessive disorder causing cone-rod dystrophy and amelogenesis imperfecta, is a rare genetic disorder impacting visual and dental development. Missense variants (c.1474G > T and c.1475G > A) previously identified in patients with Jalili syndrome have been linked to functional impairment of CNNM4, however, the biological consequences of these pathogenic variants remain largely unexplored. In this study, we investigated the functional implications of these CNNM4 missense variants, which correspond to p.(Gly492Cys) and p.(Gly492Asp) substitutions within the CBS domain of the CNNM4 protein. Our findings demonstrated that these variants exhibit significantly reduced protein stability and increased mRNA decay rates compared with wild type. Despite exhibiting normal Mg2+ localization, the mutant proteins demonstrated significantly reduced Mg²⁺ extrusion activity. This suggests that the pathogenic mechanism underlying Jalili syndrome associated with these variants likely involves decreased mRNA and/or protein stability, rather than mislocalization. Our study provides valuable insights into the interplay between genetic variations, molecular stability, and functional consequences in the context of CNNM4-related disorders, highlighting the importance of CNNM4-mediated Mg²⁺ transport in Jalili syndrome. Further investigation into the mechanisms regulating CNNM4 expression and protein stability may reveal potential therapeutic avenues.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80720-8.
Subject terms: Biochemistry, Cell biology, Genetics, Molecular biology, Diseases
Introduction
The cyclin and cystathionine β-synthase (CBS) domain divalent metal cation transporter mediator 4 (CNNM4; MIM#607805), also known as an ancient conserved domain protein 4 (ACDP4), encodes a protein playing a vital role in the transport of metal ions. CNNM4 variants are responsible for Jalili syndrome (JS; OMIM#217080), a rare autosomal recessive disorder characterized by the combined presence of cone-rod dystrophy (CRD) and amelogenesis imperfecta (AI)1–26. Individuals with JS experience significant challenges due to this genetic condition. Vision impairment, often starting in childhood or early adulthood, is a common symptom as retinal degeneration progresses, leading to cone-rod dystrophy. Additionally, amelogenesis imperfecta is present, resulting in enamel defects, increased dental sensitivity, and aesthetic concerns. These dental issues, combined with vision impairment, markedly impact patients’ quality of life, affecting function and self-esteem. Despite these severe clinical complications, the understanding of the pathogenic mechanism related to CNNM4 variants has not been fully elucidated.
CNNM4 comprises 7 exons that encode the protein containing four domains: (1) the extracellular domain, (2) the transmembrane domain, also known as the domain of unknown function 21 or DUF21, (3) the CBS domain, and (4) the cyclic nucleotide-binding homology (CNBH) domain27–29. The intracellular CBS and CNBH domains are involved in protein dimerization and Mg2+-ATP binding. Functional studies of the essential residues in the DUF21, CBS, CNBH domains revealed their impact on Mg2+-ATP binding, protein homodimerization, and/or cellular localization30–35.
Cnnm4 is highly expressed in various murine tissues, including the outer plexiform, inner plexiform, and ganglion cell layers of the neural retina, corneal keratocytes, mature ameloblasts, and the basolateral plasma membrane of intestinal epithelial cells1,8,35. This expression pattern suggests its involvement in the functions of the eyes, enamel, and intestine. The homozygous Cnnm4-knockout mice exhibited low serum and urine magnesium levels and a high fecal magnesium level, compared with those in wild-type mice, indicating that Cnnm4 plays an important role in magnesium absorption35. In addition to its function as a magnesium transporter, Cnnm4 has an essential role in sperm Ca2+ homeostasis during capacitation. The Cnnm4-knockout mice exhibited near infertility attributed to abnormal sperm motility, underscoring the importance of Cnnm4 in male fertility36.
Our previous research identified a missense variant in CNNM4, c.1475G > A, p.(Gly492Asp), among the Lua ethnic group of Northern Thailand20. The affected individuals, in addition to the typical JS features such as CRD and AI, displayed novel clinical features including advanced dental age, crossbite, and delayed tooth development. In this study, to deepen our comprehension of the pathogenic mechanism underlying JS, we conducted functional studies of the variants, p.(Gly492Asp) and p.(Gly492Cys), both entailing substitutions at the Gly492 position, as well as the well-documented variant, p.(Thr495Ile) situated within the Mg2+-ATP binding pocket19,21,32. The aim of this study was to assess the impact of these pathogenic variants on magnesium transport function, thereby advancing our understanding of the mechanisms contributing to JS pathogenesis.
Results
Summary of reported CNNM4 variants
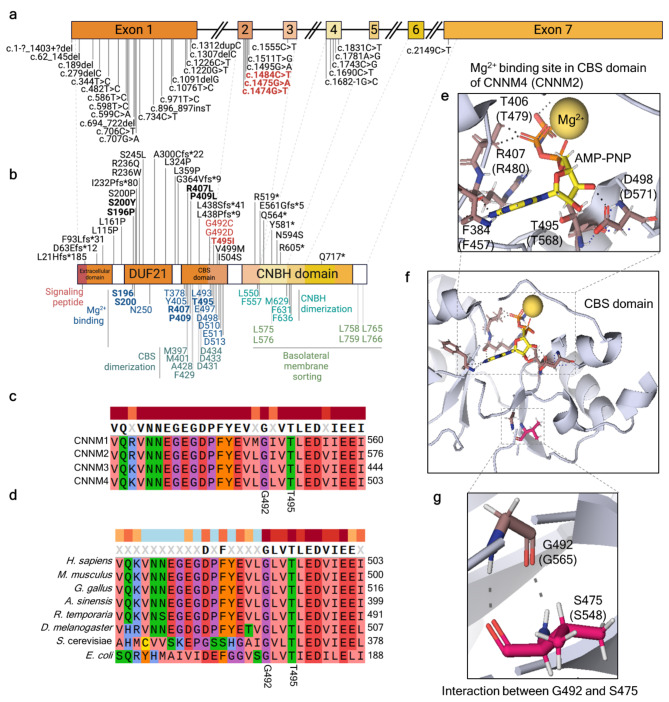
Across 26 studies, 33 CNNM4 variants have been reported as associated with JS. These variants comprise 18 missense, 9 frameshift, 1 deletion, and 5 nonsense variants (Table S1 and Fig. 1a). Exon 1 contains the largest proportion of the observed variants (n = 21), followed by exons 2 and 4 (n = 5 each), exon 3 (n = 1), and exon 7 (n = 1). Three variants are located within the extracellular domain, nine within the DUF21 domain, nine within the cystathionine β-synthase (CBS) domain, and seven within the cyclic nucleotide-binding homology (CNBH) domain (Fig. 1b). Among these pathogenic variants identified in JS patients, five variants were functionally and biochemically characterized in previous reports9,14,21,31,32 (Fig. 1a and Table S1-S2). Briefly, the critical residues for Mg2+-ATP binding in the DUF21 and CBS domains are Ser196, Ser200, Arg407, Pro409, and Thr495. Variants in these residues, except for Pro409, abolish in vitro Mg2+ efflux activity without impacting its localization. Moreover, ATP binding is completely impaired in the Arg407, Pro409, and Thr495 mutant CNNM4 proteins. The JS phenotypes resulting from pathogenic variants in these five residues are attributed to defects in Mg2+-ATP binding, while the effects of other pathogenic variants on JS phenotypes remain unclear.
Fig. 1.
In silico functional prediction of p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile) variant in CNNM4 protein. (a) CNNM4, located on chromosome 2q11.2, consists of seven exons and six introns (created with BioRender.com). All reported pathogenic variants associated with Jalili syndrome (JS) are indicated, with the bold text highlighting the variants selected for functional characterization in this study. (b) Four key domains of the CNNM4 protein are illustrated from the N-terminal to C-terminal: the extracellular domain, domain of unknown function 21 (DUF21), cystathionine β-synthase (CBS) domain, and cyclic nucleotide-binding homology (CNBH) domain. The signal peptide is located at the N-terminal. The top panel displays amino acid alterations corresponding to reported JS variants, while the lower panel shows the functionally characterized residues based on 3D structural determination. Pathogenic variants from JS patients are indicated in bold text, with those included in this functional study highlighted in bold red text (created with BioRender.com). (c) Amino acid alignment within the CNNM protein family. (d) Amino acid sequence alignment of CNNM4 across various organisms. (e, f) Structure of the Mg2+−ATP binding site within the CBS domain. (g) Interaction between Gly492 and Ser475 in the CBS domain of CNNM4 is depicted, with corresponding residues from CNNM2 shown in parentheses. References are provided in Supplementary Tables S1-2.
CNNM4 variants analysis: p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile)
Three CNNM4 variants identified in JS patients in previous reports were two variants at the Gly492 position: the c.1475G > A, p.(Gly492Asp)20 and the c.1474G > T, p.(Gly492Cys)19 and one variant at the Thr495 position: c.1484 C > T, p.(Thr495Ile)14. The p.(Thr495Ile) CNNM4 protein impairs Mg2+ export activity, ATP binding, and protein homodimerization32.
Conservation analysis demonstrated that residues Gly492 and Thr495 are highly conserved within the CNNM protein family and CNNM4 across diverse organisms. (Fig. 1c-d), indicating the crucial role of these amino acids in the CBS domain. The 3D structure analysis of CNNM2 protein with Mg2+-AMP-PNP (used as a substitute for CNNM4 due to the absence of 3D structure with Mg2+-ATP analogue) identified residues Phe457, Thr479, Arg480, Thr568, and Asp571 of CNNM2 protein, corresponding to Phe384, Thr406, Arg407, Thr495, and Asp498, respectively, in CNNM4 protein, as an analogue interacting with Mg2+-ATP32 (Fig. 1e). It was observed that Gly492, positioned distant from the Mg2+-ATP binding site, and its carboxyl and amino groups solely interacted with the backbone of Ser475 within the flexible loop of the CBS domain (Fig. 1e).
Functional analyses of p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile)
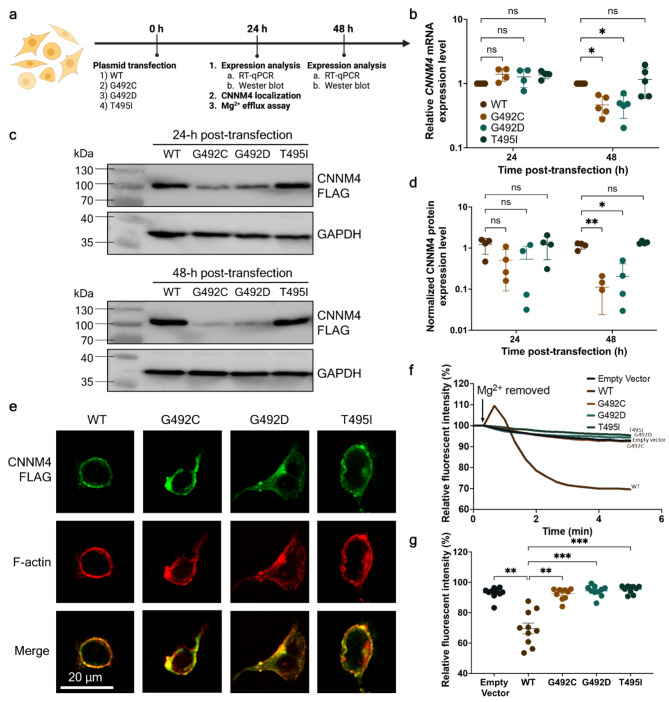
The plasmid constructs pCMV-FLAG-CNNM4-WT, -Gly492Cys, -Gly492Asp, and -Thr495Ile were ectopically overexpressed in a HEK293 cell line (Fig. 2a)32,35. The expression profiles of CNNM4 mRNA and CNNM4 protein were measured by RT-qPCR and Western blot, respectively, at 24- and 48-hours post-transfection.
Fig. 2.
Functional characterization of wild-type p.(Gly492), p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile) CNNM4 protein. (a) Experimental workflow for the functional study of overexpressed wild-type and mutant CNNM4 mRNA and CNNM4 protein. Time points for these experiments were set at 0-, 24-, and 48-hours post-plasmid transfection (created with BioRender.com). (b) Relative mRNA levels of wild-type and mutant CNNM4 at 24- and 48-hours post-transfection. Wild-type mRNA levels at both time points were arbitrarily set to 1, with results derived from four independent experiments (n = 4). (c) Western blot analysis of wild-type and mutant CNNM4 protein at 24- and 48-hours post-transfection. GAPDH was used as a loading control. For (b) and (c), individual data points are presented with means and standard errors. Two-way ANOVA was performed to assess the statistical differences between wild-type and mutant mRNA and protein levels (*p < 0.05, **p < 0.005). (d) Expression levels of wild-type and mutant proteins at 24- and 48-hours post-transfection, with FLAG-tagged CNNM4 protein band intensities normalized to GAPDH. The results are from four independent experiments (n = 4). (e) Localization of overexpressed CNNM4-FLAG in the HEK293 cell line, visualized using immunofluorescence. The green signal indicates the localization of wild-type and mutant FLAG-tagged CNNM4 proteins, while the red signal indicates F-actin location. Merged signals were examined for co-localization (scale bar = 20 μm). (f) Mg2+ extrusion assay: Line plot depicting the average percentage of relative fluorescent intensity over time (minutes) (n = 10), with the initial intensity set to 100%. (g) Remaining fluorescent intensity at 5 min relative to the initial intensity. Individual data points are shown with means and standard errors. The non-parametric Kruskal-Wallis test was conducted to evaluate the statistical differences between the relative fluorescent intensities of wild-type and mutant proteins (n = 10, **p < 0.005, ***p < 0.001).
The relative CNNM4 mRNA levels of WT and all mutants were not significantly different at 24-hours post-transfection. At 48 h post-transfection, the relative mRNA levels of the CNNM4 variants c.1474G > T and c.1475G > A were significantly reduced, showing a 2.1-fold decrease compared with WT mRNA levels. (Fig. 2b, Supplementary Table S3). No significant difference was observed between the WT and c.1484 C > T mRNA levels.
Western blot analysis revealed that the levels of FLAG-tagged p.(Gly492Cys) and p.(Gly492Asp) were lower than those of WT and p.(Thr495Ile) at 24- and 48-hours post-transfection (Fig. 2C, Supplementary Figure S1). The quantitative data of signal intensities did not show any statistical differences among the FLAG-tagged CNNM4 proteins at 24-hours post-transfection (Supplementary Table S4). However, at 48 h, the mean levels of FLAG-tagged p.(Gly492Cys) and p.(Gly492Asp) proteins were significantly decreased compared with that of WT by 10.5-fold and 5.8-fold, respectively. These findings show that both missense variants at p.(Gly492) result in reduced CNNM4 mRNA and protein.
To examine cellular localization, the mutant and WT plasmids were transfected into a HEK293 cell line, and immunofluorescent staining was performed at 24-hours post-transfection (Fig. 2a). Fluorescent signals corresponding to the FLAG-tagged WT and all mutants were detected on the plasma membrane of transfected HEK293 cells (Fig. 2e, Supplementary Figure S2). These signals co-localized with F-actin fluorescent signals at the plasma membrane, indicating that both wild-type and mutant proteins were localized to the plasma membrane.
To assess Mg2+ export activity, Magnesium Green, a fluorescent indicator for Mg2+, was incubated with the transfected HEK293 cells and the Mg2+ efflux assay was conducted 24-hours post-transfection (Fig. 2a). The results showed a rapid decrease in relative fluorescent signals within WT-transfected cells upon Mg2+ removal (Fig. 2f, Supplementary Table S5). Conversely, cells transfected with an empty vector exhibited only a slight decrease in relative fluorescent intensity, as did cells transfected with p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile) mutants. Following Mg2+ removal for 5 min, the remaining fluorescent signal in WT plasmid-transfected cells was at 69.61 ± 11.26%, which was significantly lower than that in cells transfected with the empty vector (92.98 ± 3.80%) and all mutants (p.(Gly492Cys) = 92.46 ± 3.77%, p.(Gly492Asp) = 94.37 ± 3.64%, and p.(Thr495Ile) = 95.40 ± 2.51%) (Fig. 2g, Supplementary Table S6). These findings indicate the impairment of magnesium extrusion activity in cells expressing the p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile) CNNM4 proteins.
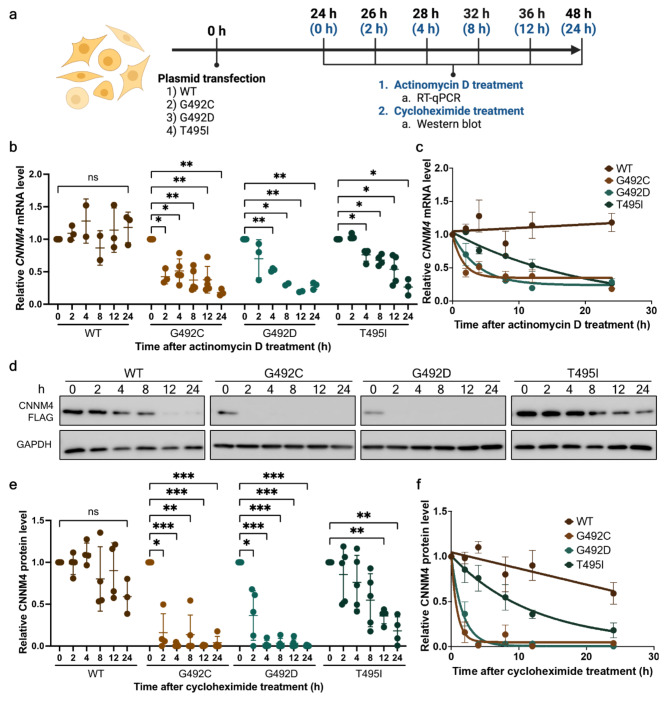
Reduction of CNNM4 mRNA levels and half-life of CNNM4 protein variants
As demonstrated earlier, the levels of c.1474G > T, p.(Gly492Cys) and c.1475G > A, c.1484 C > T, p.(Gly492Asp) mRNA and protein were significantly lower compared with WT and p.(Thr495Ile) protein at 48-hours after transfection (Fig. 2b-d). To validate the mRNA and protein instability of the mutants, the mRNA and protein decay assays were conducted at 24- to 48-hours post-transfection (Fig. 3a). At 24-hours post-transfection, the cells were treated with actinomycin D or cycloheximide to inhibit global transcription or translation, respectively. Cells were collected before treatment (0 h) and after treatment for 2, 4, 8, 12 and 24 h. The CNNM4 mRNA and protein were measured using RT-qPCR and Western blot, respectively. The results demonstrated that the WT CNNM4 mRNA levels were not significantly changed after actinomycin D treatment over 24 h (Fig. 3b), whereas c.1474G > T and c.1475G > A CNNM4 levels decreased significantly to 51.2 ± 18.5% and 52.9 ± 2.4% after actinomycin D treatment for 4 h and were 18 ± 5.0% and 29 ± 5.4%, respectively, after 24 h (Fig. 3b-c, Supplementary Table S7). Relative c.1484 C > T mRNA levels decreased significantly to 76 ± 9.6% and 27 ± 12.5% at 4 and 24 h, respectively, after treatment. To estimate the half-life of the WT and all mutant proteins, the relative mRNA levels were fitted to a one phase decay model (Fig. 3c). Within the 24-hour experimental period, the half-life of WT CNNM4 mRNA could not be estimated, while the half-life of c.1474G > T and c.1475G > A mRNA was estimated at 1.1 h (R2 = 0.71) and 2.6 h (R2 = 0.88), respectively. Additionally, the estimated half-life of c.1484 C > T was at 14 h (R2 = 0.82). These findings indicate that the c.1474G > T, c.1475G > A, and c.1484 C > T lead to reduced CNNM4 mRNA stability.
Fig. 3.
Determination of the wild-type and mutant mRNA and protein turnover rates. (a) Experimental workflow for determining the decay rates of wild-type and mutant CNNM4 mRNA and CNNM4 protein (created with BioRender.com). Black time points represent the duration post-transfection (0–48 h), while blue time points indicate the intervals following treatment with actinomycin D or cycloheximide. (b) Relative levels of wild-type and mutant CNNM4 mRNA after actinomycin D treatment at 0, 2, 4, 8, 12, and 24 h. mRNA levels at 0 h were arbitrarily set to 1. Results at 24 and 48 h were derived from four and five independent experiments, respectively. (c) Data from (b) were fitted to a one-phase decay model to determine the mRNA half-lives. Means and standard deviations are indicated along the fitted decay lines. (d) Western blot analysis results for wild-type and mutant CNNM4 proteins at 24- and 48-hours post-transfection, with GAPDH serving as a loading control. (e) Quantification of wild-type and mutant CNNM4 protein expression levels following cycloheximide treatment at 0, 2, 4, 8, 12, and 24 h. FLAG-tagged CNNM4 protein band intensities were normalized to GAPDH. The results are from four independent experiments (n = 4). (f) Data from (e) were fitted to a one-phase decay model to calculate protein half-lives. Means and standard deviations are shown along the fitted decay trends. For (b) and (e), individual data points are plotted with means and standard deviations. Two-way ANOVA was conducted to assess the statistical differences between wild-type and mutant mRNAs (*p < 0.05, **p < 0.005, ***p < 0.001).
Corresponding to the pattern of mRNA levels, FLAG-tagged WT CNNM4 protein levels remained relatively stable over a 24-hour period (Fig. 3d-e, Supplementary Table S8). In contrast, the relative protein levels of p.(Gly492Cys) and p.(Gly492Asp) significantly decreased to 15.9 ± 22.8% and 36.4 ± 29.6%, respectively, after treatment with cycloheximide for 2 hours (Supplementary Table S8, with p.(Gly492Cys) and p.(Gly492Asp) proteins undetectable after 4 h of treatment (Fig. 3d). For p.(Thr495Ile), its protein level was significantly reduced to 36.0 ± 9.3% after 12 h of treatment. The estimated half-life of p.(Gly492Cys), p.(Gly492Asp) and p.(Thr495Ile) CNNM4 proteins was 0.61 h (R2 = 0.89), 1.18 h (R2 = 0.90), and 12.11 h (R2 = 0.61), respectively, whereas the WT CNNM4 protein data was insufficient to fit the equation (Fig. 3f). These protein decay assays indicate the mutant CNNM4 proteins had reduced half-lives. Consistently, all three mutants exhibited reduced mRNA stability and protein half-life, with p.(Gly492Cys) and p.(Gly492Asp) exhibiting a faster decay rate compared with p.(Thr495Ile).
Discussion
A previous study reported that two Saudi Arabian children affected with JS had the c.1474G > T (p.Gly492Cys) variant in CNNM419. Our previous study also identified three JS patients harboring a homozygous CNNM4 variant c.1475G > A (p.Gly492Asp)20. In the current study, we assessed the functional impact of these missense variants using bioinformatic analysis and various biomolecular techniques.
Amino acid sequence alignment among CNNM family members and CNNM4 proteins from various organisms has revealed high conservation of Gly492, Thr495, and nearby amino acids. Our analyses of the crystal structure using the Mg2+-bound cytosolic fragment of CNNM2 (PDB: 6N7E) showed that Gly565 in CNNM2, equivalent to Gly492 in CNNM4, was located far from Mg2+-ATP. Thus, the role of Gly492 in interaction with Mg2+-ATP could not be concluded from the 3D structure. The backbone of Gly492 only interacts with the backbone of Ser475 in the flexible loop of the CBS domain. Substituting glycine with either cysteine or aspartate might disrupt this interaction. Previous studies proposed that Gly433 of CNNM3, corresponding to Gly492 of CNNM4, might be involved in stabilizing the loop and CBS domain structure20. Functional analysis of the p.(Gly433Asp) variant in CNNM3 showed disrupted interaction between CNNM3 and the protein tyrosine phosphatase PRL-2, resulting in impaired intracellular magnesium regulation. Based on this evidence, it is possible that CNNM4 Gly492 functions in stabilizing the CBS domain structure and Mg2+ efflux activity.
The relative expression levels of WT and mutant CNNM4 mRNAs showed no significant difference at 24-hours post-transfection. However, c.1474G > T and c.1475G > A, but not c.1484 C > T, significantly reduced CNNM4 mRNA levels compared with WT, suggesting that the nucleotide substitutions c.1474G > T and c.1475G > A might affect mRNA half-life at 48-hours post-transfection. The CNNM4 mRNA decay assay using actinomycin D treatment confirmed these reductions, and the half-life of c.1484 C > T mRNA was also significantly decreased. Kato et al. identified that a pathogenic missense variant at the end of exon 5 of LMNA, encoded lamin A/C, in severe dilated cardiomyopathy37. They noted that the LMNA complementary DNA in patients’ lymphocytes and cardiac tissues was undetectable compared with controls. Functional analysis also showed that this missense variant in LMNA affected the splicing process resulting in mRNA degradation. However, the mechanism by which missense variants lead to a reduction in CNNM4 mRNA half-life remains unclear and requires further investigation.
Similarly, WT and mutant CNNM4 protein levels were comparable at 24-hours post-transfection. However, lower mRNA levels led to reduced p.(Gly492Cys) and p.(Gly492Asp) CNNM4 protein levels at 48-hours post-transfection. The CNNM4 protein decay assay using cycloheximide treatment revealed significantly reduced half-lives for the p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile) proteins compared with WT. This reduced mRNA and protein stability may contribute to the CRD and AI phenotypes observed in JS patients. Further investigation is needed to elucidate the specific mechanisms underlying CNNM4 protein degradation.
In our study, the CNNM4 mutants colocalized with F-actin at the plasma membrane, similar to WT. Another group has also shown that the p.(Gly492Asp) expressed in HEK293 cells was localized in the plasma membrane, mainly in the intracellular membrane structure38. The localization of mutant p.(Thr495Ile) observed in our study was similar to a localization experiment in Chen et al.32. Previous studies revealed that CNNM4 was localized to/in the basolateral membrane of mouse intestinal epithelia and an MDCK cell line30,35. Although we noted that these mutant CNNM4 proteins were localized in the plasma membrane in our experiment, it was unclear whether they were in the apical or basolateral membrane.
The Mg2+ efflux assay results demonstrated that the WT CNNM4 protein contributed to the extrusion of intracellular Mg2+. In contrast, intracellular Mg2+ accumulated in cells expressing p.(Gly492Cys), p.(Gly492Asp) and p.(Thr495Ile) CNNM4 indicating loss-of-function variants. The Thr495 residue stabilizes the side chain of Asp498 and facilitates the binding of Mg2+-ATP. Substitution of p.(Thr495Ile) led to impaired Mg2+ binding and extrusion activity32. Taken together, our study demonstrates for the first time that the p.(Gly492) and p.(Thr495) variants lead to a reduction in mRNA and protein half-life, resulting in impaired Mg2+ efflux activity.
This study provides valuable insights into Jalili syndrome by examining how specific missense variants in CNNM4, p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile), affect mRNA and protein stability as well as Mg2+ extrusion activity. The findings highlight that increased decay rates and functional loss in Mg2+ efflux, without localization defects, may underlie the pathogenic mechanisms of the syndrome, offering a focused direction for future research on protein stability and function.
This study has several limitations. Lacking a 3D structure of CNNM4 bound to a Mg²⁺-ATP analogue, we could not definitively determine how the Gly492 residue affects Mg²⁺ extrusion activity. In addition, the mechanism underlying the reduced mRNA half-life of missense variants needs further investigation. Furthermore, although the mutant CNNM4 proteins localize to the plasma membrane, their specific apical or basolateral membrane localization, crucial for their functional roles, requires clarification. Finally, further research is needed to fully understand the broader biological effects of Gly492 variants on CNNM4 activity and their contributions to Jalili syndrome.
Conclusion
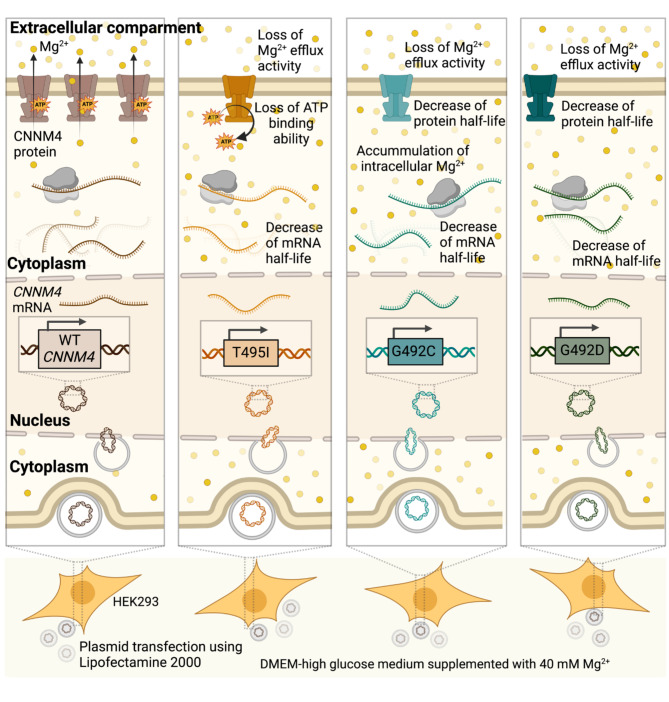
This study demonstrated that the missense variants c.1474G > T, c.1475G > A, and c.1484 C > T in CNNM4, corresponding to p.(Gly492Cys), p.(Gly492Asp) and p.(Thr495Ile), respectively, in the CBS domain of CNNM4 led to significantly increased mRNA and protein decay rates compared with the wild type (Fig. 4). These mutant CNNM4 proteins exhibited a loss of Mg2+ extrusion activity. Importantly, no detectable localization defects were observed for the p.(Gly492Cys), p.(Gly492Asp) and p.(Thr495Ile) mutants. These findings suggest that the rapid degradation rates and loss of Mg2+ efflux activity associated with these variants may serve as potential pathogenic mechanisms underlying the phenotypes of Jalili syndrome.
Fig. 4.
Summary of findings on CNNM4 mutants and their impact on protein function and stability. This graphic summarizes the key findings of this study, illustrating that the missense variants c.1474G > T, c.1475G > A, and c.1484 C > T in CNNM4, which correspond to p.(Gly492Cys), p.(Gly492Asp), and p.(Thr495Ile), respectively, in the CBS domain of the CNNM4 protein, result in significantly increased mRNA and protein decay rates compared with the wild type. Notably, these mutant CNNM4 proteins demonstrate a loss of Mg2+ extrusion activity, with no detectable localization defects.
Materials and methods
Structural determination and amino acid alignment
The amino acid sequence of human CNNM4 (NP_064569.3) was aligned with human CNNM1, CNNM2 and CNNM3 (NP_065081.2, NP_060119.3, NP_060093.3, respectively) and CNNM4 of Mus musculus (NP_291048), Gallus gallus (XP_040507492), Alligator sinensis (XP_025068613), Rana temporaria (XP_040201886), unextended protein (Uex) of Drosophila melanogaster (NP_001104391), Mam3p of Saccharomyces cerevisiae (NP_014581) and CorC of Escherichia coli (CAD6019356) using Clustal MUSCLE in SnapGene software.
The Mg2+-ATP binding site of CNNM4 was obtained from partial alignment of the CBS domain of CNNM4 (residues 366–511) (PDB: 6RS2)39 with the corresponding Mg2+- bound cytosolic fragment of CNNM2 (residues 438–585 with Mg2+ and adenylyl-imidodiphosphate (AMP-PNP) (PDB: 6N7E)32.
Plasmid construction
The human wild-type CNNM4 cDNA was inserted into a mammalian expression vector, pCMV-Tag4A (Agilent Technologies) as previously described with minor modification32,35. Briefly, total RNA was isolated from a HEK293 cell line using RiboEx (GeneAll). The first stand cDNA was synthesized using GoScript Reverse Transcription System with Oligo(dT) primer (Promega). The full length CNNM4 cDNA was amplified using Q5 High-Fidelity DNA polymerase (New England BioLabs) and the primers are shown in Supplementary Table S7. The full length CNNM4 cDNA was cut with NotI-HF and SalI-HF (New England BioLabs) and inserted into pCMV-Tag4A. The pathogenic variants of the CNNM4 plasmid constructs comprising c.1474G > T, p.(Gly492Cys), c.1475G > A, p.(Gly492Asp) and c.1484 C > T, p.(Thr495Ile) were generated by site-directed mutagenesis with the specific primers listed in Supplementary Table S7. The nucleotide sequences of the CNNM4 plasmid constructs were verified by Sanger sequencing at Macrogen Inc. (Korea).
Cell culture
Human embryonic kidney 293 (HEK293) cells obtained from the American Type Culture Collection (ATCC, VA, USA) were used in this study. HEK293 cells are a well-established homologous expression system and have been widely utilized for in vitro functional studies of wild-type and mutant human CNNM430,32,35,38. The cells were cultured in complete medium (DMEM high glucose with 10% (v/v) fetal bovine serum (FBS) (Gibco), 1%(v/v) antibiotic/antimycotic (Gibco) and 1% (v/v) GlutaMAX (Gibco)) and maintained in an incubator at 37ºC, 5% (v/v) CO2. The cells were subcultured after reaching 70–80% confluence. Mycoplasma in culture was detected by PCR using specific primers (Supplementary Table S7). Plasmocin treatment (Invivogen) at 25 µg/ml was supplemented in complete medium to eliminate contaminated mycoplasma.
Plasmid transfection
HEK293 cells (5 × 105) were seeded into 6-well plates containing complete medium and incubated overnight in an incubator at 37ºC, 5% (v/v) CO2. The complete medium was replaced with antibiotic-free DMEM supplemented with 40 mM Mg2+. The empty vector and the CNNM4 constructs (1 µg) were transfected into HEK293 cells using Lipofectamine 2000 following the manufacturer’s instructions and incubated for 24- and 48-hours before cell harvesting. CNNM4 mRNA and protein expression were measured using RT-qPCR and Western blot analysis, respectively.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR)
RNA was isolated using RiboEX (GeneAll) according to the manufacturer’s instructions. The RNA concentration was measured by NanoDrop One (Thermo Scientific). The first strand DNA was synthesized using 2 µg total RNA in iScript Reverse Transcription Supermix (BIO-RAD). The reaction was incubated at 25ºC for 5 min, followed by incubating at 46ºC for 20 min before heat inactivation at 95ºC for 1 min. The cDNA was stored at -80ºC until used. qPCR was performed in a 12 µl-reaction mixture containing 1 µl cDNA and 11 µl master mix containing 1xFastStrat Universal Master Mix, and 300 nM forward and reverse primers (Supplementary Table S9). For qRT-PCR using SYBR, the thermal profiles consisted of an incubation at 95ºC for 10 min followed 40 amplification cycles, each comprising denaturation at 95ºC for 30 s, annealing at 60ºC for 30 s and extension at 72ºC for 30 s in a CFX Connect Real-time PCR Detection System (BIO-RAD). Gene expression was normalized with that of ATCB1 and shown as the relative gene expression. The differential cycle threshold (∆∆CT) method was used to compare fold change.
Western blot analysis
Crude protein lysates were extracted by suspending the cell pellet in radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% (w/v) SDS, 0.25% (w/v) sodium deoxycholate, 1% (V/V) NP-40 and 1x protease inhibitor cocktail (Thermo Scientific)) on ice for 10 min before centrifugation at 12,000xg, 4ºC for 10 min. The supernatant was transferred into a new tube and kept at -80ºC until analysis. The Pierce BCA protein assay kit (Thermo Scientific) was used to determine protein concentration.
Protein samples (15 µg) were mixed with 6x loading dye (0.1 M Tris-Cl pH 6.8, 4% (w/v) SDS, 0.6% (w/v) bromophenol blue, 60% (v/v) glycerol and 0.1 M β-mercaptoethanol and heated at 95ºC for 5 min. The samples were briefly centrifuged before loading in 10% (v/v) SDS-PAGE. Electrophoresis was performed under reducing condition using 1x Tris-glycine (25 mM Tris-HCl pH 8.3, 193 mM glycine, 0.1% (w/v) SDS) as a running buffer at 100 V for 120 min. The proteins were transferred from the gel to a membrane using a Trans-blot Semi-dry transfer cell (Bio-Rad) at constant 18 V for 45 min. The membrane was blocked with 5% (w/v) skim milk or 3% (w/v) BSA in 1xTBST buffer (50 mM Tris-Cl pH 7.6, 150 mM NaCl) and 0.1% (v/v) Tween 20), depending on the primary antibody. The anti-FLAG clone M2 (Sigma-Aldrich, Cat#F1804) or anti-GAPDH (Abcam) primary antibody was incubated with the membrane at 4ºC overnight. The blots were incubated with a secondary antibody for 1 h at room temperature before detecting using the Pierce ECL Western Blotting substrate (Thermo Scientific) by Amersham Imager 600 (GE Healthcare).
Immunofluorescent staining
HEK293 cells (3 × 104) were plated onto cover glasses in 24-well plates containing complete medium and incubated in an incubator at 37ºC, 5% (v/v) CO2 overnight. The medium was replaced with antibiotic-free Opti-MEM reduced serum medium (Gibco) supplemented with 40 mM Mg2+. The cells were transfected using Lipofectamine 2000 as described in a previous section. At 24-hours post-transfection, immunofluorescent staining was performed at room temperature. The transfected cells were washed twice with phosphate buffered saline (PBS) and fixed with 2% (v/v) paraformaldehyde for 15 min. The cells were washed three times with PBS followed by incubation with 0.2% (v/v) TritonX-100 in PBS for 10 min. Nonspecific binding was blocked by incubation with 3% (v/v) FBS and 10% (w/v) bovine serum albumin (BSA) in PBS for 1 h. The anti-FLAG primary antibody (clone M2, Sigma-Aldrich) was diluted with blocking buffer to 1:500 and incubated at 4ºC overnight. The cells were washed three times with PBS before incubation with a secondary antibody tagged with Alexa-488 (BioLegend, 1:1,000 in blocking buffer) for 30 min. The cells were washed three times with PBS and counter-stained with rhodamine-phalloidin (Invitrogen) for 1 h. The cover glasses were mounted with ProLong Gold Antifade Mountant (Invitrogen) and placed on glass slides. The fluorescent signals were visualized using Z-stack optical sectioning by Zeiss Apotome 3 (Zeiss). The fluorescent noise was removed by ZEN 3.8 software.
Mg2+ efflux assay
The Mg2+ extrusion assay was performed according to previous studies32,35. Briefly, HEK293 cells were plated and transfected in 40 mM Mg2+ as described above. At 24-hours post-transfection, the cells were incubated with 2 µM Magnesium Green-AM (Invitrogen) Mg2+-loading buffer (78.1 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 40 mM MgCl2, 5.5 mM glucose, and 5.5 mM HEPES-KOH, pH 7.4) for 30 min at 37ºC. The cells were washed once with Mg2+-loading buffer and observed under a confocal microscope (ZEISS). The fluorescent signal was captured every 20 s under control of the ZEN software (ZEISS). The buffer was replaced with loading buffer without magnesium (138.1 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 5.5 mM glucose, 5.5 mM HEPES-KOH, pH 7.4). The data is presented as line plots of the relative mean fluorescent intensity of ten randomly selected cells.
CNNM4 mRNA and protein decay assay
HEK293 cells were plated and transfected as described above. The transfected cells were treated with 5 µg/ml actinomycin and 50 µg/ml cycloheximide at 24-hours post-transfection. After chemical treatment for 2, 4, 8, 12 and 24 h, the cells were harvested for measuring the CNNM4 mRNA and protein levels. The transfected cells without chemical treatment were used to measure the initial amount of mRNA and protein. CNNM4 mRNA and protein was analyzed as previously described40,41. Briefly, the Ct of wild-type and mutant CNNM4 mRNA levels at different time points were normalized to the level before actinomycin D treatment. The relative mRNA expression was fitted to one phase decay equation using Prism 9 to estimate the mRNA half-life. The analysis of the wild-type and mutant CNNM4 proteins was the same as the analysis of mRNA decay, however, the wild-type and mutant CNNM4 protein levels were normalized to the GAPDH protein level.
Statistical analysis
Information on the statistical analysis is described in the relevant sections of the figure legends and supplementary materials. A normality test was conducted to determine the data distribution. For a small sample study, a non-parametric statistical analysis was applied. The statistical analyses were performed using GraphPad Prism version 9.0.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This project was funded by the Health Systems Research Institute, National Research Council of Thailand (N42A650229), Faculty of Dentistry (DRF68_013), and Thailand Science Research and Innovation Fund Chulalongkorn University. KR was supported by the Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University. We also thank Dr. Kevin Tompkins for revising the language in the manuscript.
Author contributions
KR conceived and designed the study under supervision of TP and VS. KR, MR and KS performed the experiments and collected data. KR and MR analyzed the data and conducted statistical analysis. KR, MR, TP and VS contributed to the interpretation of the results. KR and TP wrote the initial draft of the manuscript. All authors read and approved the final version of the manuscript.
Data availability
All the data produced throughout this research are incorporated in this published research article and its supplementary information. All amino acid sequences for alignment analysis in Fig. 1c-d were obtained from UniProt database (Accession number: Q9NRU3, Q9H8M5, Q8NE01, Q6P4Q7, Q69ZF7, A0A8V0ZES4, A0A3Q0HDP4, A0A2G9R6E9, A0A0B7P9G0, Q12296 and P0AE78). The deposited 3D-structure data from the Protein Data Bank (PDB) (Accession number: 6RS2 and 6N7E) were used for visualizing the Mg2+-ATP binding site in Fig. 1e-g. CNNM4 cDNA (NM_020184.4) was cloned and mutated as reported in ClinVar database (Accession numbers: VCV001342702.1 and VCV001502683.4).
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Parry, D. A. et al. Mutations in CNNM4 cause Jalili syndrome, consisting of autosomal-recessive cone-rod dystrophy and amelogenesis imperfecta. Am. J. Hum. Genet.84, 266–273. 10.1016/j.ajhg.2009.01.009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppieters, F. et al. Identity-by-descent-guided mutation analysis and exome sequencing in consanguineous families reveals unusual clinical and molecular findings in retinal dystrophy. Genet. Med.16, 671–680. 10.1038/gim.2014.24 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Prasov, L. et al. Expanding the genotypic spectrum of Jalili syndrome: Novel CNNM4 variants and uniparental isodisomy in a north American patient cohort. Am. J. Med. Genet. A. 182, 493–497. 10.1002/ajmg.a.61484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, J. H., Park, S. H., Yim, J. S., Kim, M. S. & Kim, S. Y. The first Korean child of Jalili syndrome with a novel missense mutation in cation transport mediator 4 (CNNM4): A case report. Korean J. Ophthalmol.37, 195–197. 10.3341/kjo.2022.0144 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyde, R. A., Kratunova, E., Park, J. C. & McAnany, J. J. Cone pathway dysfunction in Jalili syndrome due to a novel familial variant of CNNM4 revealed by pupillometry and electrophysiologic investigations. Ophthalmic Genet.43, 268–276. 10.1080/13816810.2021.2002916 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, H., Huang, Y., Li, J. & Xie, M. Novel homozygous nonsynonymous variant of CNNM4 gene in a Chinese family with Jalili syndrome. Mol. Genet. Genomic Med.10, e1860. 10.1002/mgg3.1860 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiessling, F., Mitter, D., Langmann, T. & Müller, D. T. H. Novel deletion in the CNNM4 gene in siblings with Jalili syndrome. Int. J. Ophthalmol. Clin. Res.3, 046 (2016). [Google Scholar]
- 8.Polok, B. et al. Mutations in CNNM4 cause recessive cone-rod dystrophy with amelogenesis imperfecta. Am. J. Hum. Genet.84, 259–265. 10.1016/j.ajhg.2009.01.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirji, N. et al. Jalili Syndrome: Cross-sectional and longitudinal features of seven patients with cone-rod dystrophy and amelogenesis imperfecta. Am. J. Ophthalmol.188, 123–130. 10.1016/j.ajo.2018.01.029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, H. et al. Comprehensive molecular diagnosis of a large Chinese leber congenital amaurosis cohort. Invest. Ophthalmol. Vis. Sci.56, 3642–3655. 10.1167/iovs.14-15972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maia, C. M. F. et al. Report of two unrelated families with Jalili syndrome and a novel nonsense heterozygous mutation in CNNM4 gene. Eur. J. Med. Genet.61, 384–387. 10.1016/j.ejmg.2018.02.003 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Wawrocka, A. et al. Co-occurrence of Jalili syndrome and muscular overgrowth. Am. J. Med. Genet. A. 173, 2280–2283. 10.1002/ajmg.a.38318 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Rahimi-Aliabadi, S. et al. A novel mutation and variable phenotypic expression in a large consanguineous pedigree with Jalili syndrome. Eye30, 1424–1432. 10.1038/eye.2016.137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parveen, A. et al. A novel pathogenic missense variant in CNNM4 underlying Jalili syndrome: Insights from molecular dynamics simulations. Mol. Genet. Genomic Med.7, e902. 10.1002/mgg3.902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaelides, M., Bloch-Zupan, A., Holder, G. E., Hunt, D. M. & Moore, A. T. An autosomal recessive cone-rod dystrophy associated with amelogenesis imperfecta. J. Med. Genet.41, 468–473. 10.1136/jmg.2003.015792 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zobor, D. et al. Cone-rod dystrophy associated with amelogenesis imperfecta in a child with neurofibromatosis type 1. Ophthalmic Genet.33, 34–38. 10.3109/13816810.2011.592178 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Luder, H. U., Gerth-Kahlert, C., Ostertag-Benzinger, S. & Schorderet, D. F. Dental phenotype in Jalili syndrome due to a c.1312 dupC homozygous mutation in the CNNM4 gene. PLoS One. 8, e78529. 10.1371/journal.pone.0078529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerth-Kahlert, C. et al. Intra-familial phenotype variability in patients with Jalili syndrome. Eye (Lond). 29, 712–716. 10.1038/eye.2014.314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez Torres, L., Schorderet, D., Valmaggia, C. & Todorova, M. A novel mutation in CNNM 4 (G492C) associated with Jalili syndrome. Acta Ophthalmol.93 (2015).
- 20.Rattanapornsompong, K. et al. Novel CNNM4 variant and clinical features of Jalili syndrome. Clin. Genet.103, 256–257. 10.1111/cge.14258 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Abu-Safieh, L. et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res.23, 236–247. 10.1101/gr.144105.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad, M. K. et al. A targeted next-generation sequencing assay for the molecular diagnosis of genetic disorders with orodental involvement. J. Med. Genet.53, 98–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doucette, L. et al. Molecular genetics of achromatopsia in Newfoundland reveal genetic heterogeneity, founder effects and the first cases of Jalili syndrome in North America. Ophthalmic Genet.34, 119–129. 10.3109/13816810.2013.763993 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Jaouad, I. C. et al. Novel splice site mutation in CNNM4 gene in a family with Jalili syndrome. Eur. J. Med. Genet.60, 239–244 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Topçu, V. et al. A new familial case of Jalili syndrome caused by a novel mutation in CNNM4. Ophthalmic Genet.38, 161–166. 10.3109/13816810.2016.1164192 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Li, S. et al. Identification of a mutation in CNNM4 by whole exome sequencing in an amish family and functional link between CNNM4 and IQCB1. Mol. Genet. Genomics. 293, 699–710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimenez-Mascarell, P. et al. Current structural knowledge on the CNNM family of magnesium transport mediators. Int. J. Mol. Sci.2010.3390/ijms20051135 (2019). [DOI] [PMC free article] [PubMed]
- 28.Funato, Y. & Miki, H. The emerging roles and therapeutic potential of cyclin M/CorC family of mg(2+) transporters. J. Pharmacol. Sci.148, 14–18. 10.1016/j.jphs.2021.09.004 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Daneshmandpour, Y., Darvish, H., Pashazadeh, F. & Emamalizadeh, B. Features, genetics and their correlation in Jalili syndrome: A systematic review. J. Med. Genet.56, 358–369. 10.1136/jmedgenet-2018-105716 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Hirata, Y., Funato, Y. & Miki, H. Basolateral sorting of the mg(2)(+) transporter CNNM4 requires interaction with AP-1A and AP-1B. Biochem. Biophys. Res. Commun.455, 184–189. 10.1016/j.bbrc.2014.10.138 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Huang, Y. et al. Structural basis for the mg(2+) recognition and regulation of the CorC mg(2+) transporter. Sci. Adv.710.1126/sciadv.abe6140 (2021). [DOI] [PMC free article] [PubMed]
- 32.Chen, Y. S. et al. Mg(2+)-ATP sensing in CNNM, a putative magnesium transporter. Structure 28, 324–335.e324 (2020). 10.1016/j.str.2019.11.016 [DOI] [PubMed]
- 33.Hirata, Y., Funato, Y., Takano, Y. & Miki, H. Mg2+-dependent interactions of ATP with the cystathionine-beta-synthase (CBS) domains of a magnesium transporter. J. Biol. Chem.289, 14731–14739. 10.1074/jbc.M114.551176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, Y. S. et al. The cyclic nucleotide-binding homology domain of the integral membrane protein CNNM mediates dimerization and is required for mg(2+) efflux activity. J. Biol. Chem.293, 19998–20007. 10.1074/jbc.RA118.005672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamazaki, D. et al. Basolateral Mg2+ extrusion via CNNM4 mediates transcellular Mg2+ transport across epithelia: A mouse model. PLoS Genet.9, e1003983. 10.1371/journal.pgen.1003983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki, D. et al. The Mg2+ transporter CNNM4 regulates sperm Ca2+ homeostasis and is essential for reproduction. J. Cell. Sci.129, 1940–1949. 10.1242/jcs.182220 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Kato, K. et al. LMNA missense mutation causes nonsense-mediated mRNA decay and severe dilated cardiomyopathy. Circ. Genom Precis Med.13, 435–443. 10.1161/circgen.119.002853 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Gulerez, I. et al. Phosphocysteine in the PRL-CNNM pathway mediates magnesium homeostasis. EMBO Rep.17, 1890–1900. 10.15252/embr.201643393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gimenez-Mascarell, P. et al. Structural insights into the intracellular region of the human magnesium transport mediator CNNM4. Int. J. Mol. Sci.2010.3390/ijms20246279 (2019). [DOI] [PMC free article] [PubMed]
- 40.Ratnadiwakara, M. & Änkö, M. L. mRNA stability assay using transcription inhibition by actinomycin D in mouse pluripotent stem cells. Bio Protoc.8, e3072. 10.21769/BioProtoc.3072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra, R. & Banerjea, A. C. SARS-CoV-2 spike targets USP33-IRF9 axis via exosomal miR-148a to activate human microglia. Front. Immunol.12, 656700. 10.3389/fimmu.2021.656700 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data produced throughout this research are incorporated in this published research article and its supplementary information. All amino acid sequences for alignment analysis in Fig. 1c-d were obtained from UniProt database (Accession number: Q9NRU3, Q9H8M5, Q8NE01, Q6P4Q7, Q69ZF7, A0A8V0ZES4, A0A3Q0HDP4, A0A2G9R6E9, A0A0B7P9G0, Q12296 and P0AE78). The deposited 3D-structure data from the Protein Data Bank (PDB) (Accession number: 6RS2 and 6N7E) were used for visualizing the Mg2+-ATP binding site in Fig. 1e-g. CNNM4 cDNA (NM_020184.4) was cloned and mutated as reported in ClinVar database (Accession numbers: VCV001342702.1 and VCV001502683.4).