Abstract
Musk, secreted by adult male forest musk deer, is a kind of precious Chinese traditional medicine for treating cardiovascular, cerebrovascular and neurogenic diseases. However, a lack of knowledge on musk biosynthetic mechanism and limited musk deer population have seriously hindered the development of the musk industry. Fortunately, given that muskrat musk has similar constituents and pharmacological action with deer musk, muskrat is an ideal model animal for exploring musk biosynthetic mechanism. To explore the biosynthetic mechanism of muskrat musk, in the current study, transcriptomic analysis in the liver, kidney and musk glands of male muskrats between musk secreting and non-musk secreting stages was conducted. The findings indicated that the role of muskrat liver on musk biosynthesis was altering sugar, lipid and amino acid metabolism as well as producing basic resources to support musk glands. Moreover, Tigar, Slc11a2, Gpt, Hmgcr, Slc27a4, and Elovl1 were identified as candidate genes for musk biosynthesis via a remotely controlled process. Expression of the Tigar, Slc11a2, and Gpt genes in the liver are downregulated to support the production of musk in muskrat musk gland. And the Hmgcr, Slc27a4, and Elovl1 genes in the musk gland participate in muskrat musk synthesis by influencing lipid metabolism in the musk secreting period. This study provided novel insights into the musk biosynthetic pathway in muskrat by transcriptomic analysis and preliminarily suggested the remote control of metabolism from the liver to musk gland during musk biosynthesis, which was useful to further understanding the musk biosynthetic process and improve musk production in the future.
Keywords: Muskrats, Musk biosynthetic mechanism, Transcriptomic analysis, Remote control
Subject terms: Gene expression, Gene regulation, RNA sequencing
Introduction
Musk, mainly secreted by adult male forest musk deer (Moschus berezovskii), is a kind of precious Chinese traditional medicine with great economic and medicinal value, including curative effects on cardiovascular, cerebrovascular, and neurogenic diseases 1–3. Sadly, the forest musk deer is near extinction because of illegal poaching and trading. In addition, the biosynthesis mechanism of musk remains largely unclear, which limits the effective utilization and development of the musk industry 4,5. Fortunately, another kind of musk, secreted by muskrat (Ondatra zibethicus), has similar pharmacological effects to deer musk, which indicates that muskrat is an ideal model animal for the exploration of musk biosynthetic mechanism 2,6,7. Previous studies have indicated that the musk biosynthetic mechanism is related to biosynthetic processes of long chain fatty acid skeletons and steroids, including 13-tetradecenal, 8-cyclohexadecen-1-one, cholesterol and trans-dehydroandrosterone 8,9. In terms of molecular structure, long-chain fatty acids and steroids are very similar to muscone, trans-dehydroandrosterone and so on 9,10. Therefore, pertinent genes involved in pro-chemical or musk synthesis have been considered to participate in the formation of musk and the substrates necessary for musk synthesis 7,11,12.
The metabolic processes of three major chemicals, namely, glucose, lipids, and amino acids, showed great potential for being associated with musk biosynthesis because a large proportion of musk chemicals or pro-chemicals are resemblance with the secondary products of these three major chemicals 6,10–12. Moreover, the results indicated that metabolisms, including that of glucose, lipid, and amino acids, were closely associated with the biosynthesis of long-chain fatty acids and steroids. In addition, the metabolisms were also involved in ketone synthesis, allosteric reactions and other oxidation–reduction reactions, such as carbon source generation, carbon chain hydrogenation and carboxyl activation.
In glycometabolism, the pivotal genes encoding rate-limiting enzymes involved in gluconeogenesis and glycolysis, such as hexokinase, phosphofructokinase, pyruvate kinase, glucose-6-phosphatase, fructose diphosphatase, and pyruvate carboxylase, have been considered as candidates 13,14. In fatty acid, sterol and amino acid metabolism, genes expressing fatty acid synthase, transaminase, thiolase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and Fe2+ transporters have been indicated as key candidate genes 15–17. Therefore, expressional information of genes associated with musk production occurs in breeding season is very important for understanding the musk biosynthesis mechanism. Furthermore, considering the significance of liver and kidney in providing the requisite substrates 18,19, we monitored the difference in gene expression in the liver, kidney, and musk gland tissues of muskrats between the musk secreting and non-musk secreting periods by mRNA transcriptomic sequencing and examined the transcript expression levels of the above-mentioned genes to reveal the musk biosynthesis mechanism in muskrat and provide a theoretical basis for enhancing musk production in the near future.
Materials and methods
Ethics statement
The experimental protocol and procedures for animal testing were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (permit number: S20191017). All operations were carried out according to the European Commission (1997), and all efforts were made to minimize suffering. And this study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Animals and sample collection
A total of eight healthy, one-year-old, half-blooded, male muskrats weighing 1.3-1.4 kg from the Chongqing Institute of Medicinal Plant Cultivation (Chongqing, China) were selected as experimental animals. All the animals were randomly divided into two groups, including a control group (n=4, non-musk secreting period) and an experimental group (n=4, musk secreting period). Every animal was randomly housed in a separated 1.2-square-metre enclosure including two layers of down pool and an upper rest area where fodder had been thrown. The concentrate-to-forage ratio of the feed was 1:3. The concentrates included 37% alfalfa meal, 45% corn, 15% full-fat soybean, 0.4% NaCl, 1.5% CaCO3, 0.8% CaHPO4, 0.15% multi-vitamins, and 0.15% multi-minerals. Feeding with chicory leaves and ryegrass was performed at 08:30 every morning, and feeding with concentrated fodder was performed at 16:30 every afternoon. And muskrat could drink water freely. No unnecessary noise was allowed, and regular disinfection was conducted on the farm.
The experimental and control individuals were anaesthetized by Telazol injection (4.5 mg/kg) and euthanized by cervical dislocation in June of the musk secreting period and October of the non-musk secreting period, respectively. Pieces of musk gland, liver, and kidney tissues were collected with a small aseptic scalpel and tweezers, stored in a sterile EP tube, and then immediately frozen in liquid nitrogen to reduce RNA degradation.
RNA sequencing and data analysis
Total RNA from each sample was extracted with TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The quantity and integrity of RNA were examined using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA) and 1% agarose gel electrophoresis. The mRNA was purified from total RNA using poly-T oligo-attached magnetic beads, and high-throughput sequencing was performed using the Illumina 2500 sequencing platform with PE150. A total of 24 samples, which contained 4 biological replicates for musk gland, liver and kidney for musk secreting and non-musk secreting period, respectively, were used for subsequent transcriptome analysis.
The sequencing quality was controlled by removing reads containing adapters, reads containing N bases and low-quality reads from the raw data. All downstream analyses were based on clean high-quality data. The Trinity method 20 was used to assemble transcripts and acquire unigenes. Then, unigene terms were annotated based on the NCBI nonredundant protein sequence (Nr), NCBI nonredundant nucleotide sequence (Nt), Protein family (Pfam), Clusters of Orthologous Groups of proteins (KOG/COG), Swiss-Prot (a manually annotated and reviewed protein sequence database), KEGG Ortholog (KO), and Gene Ontology (GO). Differentially expressed gene (DEG; with |log2-fold change| > 1 and P < 0.05), volcano map, heatmap, KEGG enrichment plot, and correlation matrix analyses and principal component analysis (PCA) of the candidate genes were performed by using R software version 4.0.2 (https://www.r-project.org/) with the ggplot2 (https://cran.r-project. org/web/packages/ggplot2/), pheatmap (https://cran.r-project.org/web/packages/pheatmap/), scatterplot3d (https://cran.r-project.org/web/packages/scatterplot3d/), and performance Analytics (https://cran.r-project.org/web/packages/PerformanceAnalytics/) packages. KOBAS Version 3.0 (http://kobas.cbi.pku.edu.cn/genelist/) was utilized for KEGG enrichment analysis when the DEGs had a P < 0.05. Then, candidate genes were screened out from the DEGs by inspecting related metabolic pathway genes involved in sugar, lipid and amino acid metabolism and above 1000 annotation scores from the Nr and Nt databases. In this study, Student’s T test, sampling test, Pearson analysis and PCA were performed by R 4.1.2 and KOBAS 3.0.
Validation of the gene expression by qPCR
We selected 6 candidate genes to confirm the expression profiles of RNA-seq by Real-Time PCR (qPCR). The expression levels of the selected genes were normalized to beta-actin. Primers for the candidate genes were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table 1). Total RNA was converted to cDNA using the EasyScript One-Step gDNA Removel and cDNA Synthesis SuperMix (Transgen Biotech, China).
Table 1.
The primer sequences used for qPCR.
| Genes | Accession ID | Forward primer | Reverse primer | Product length |
|---|---|---|---|---|
| Tigar | XM_055395513.1 | CAGTCAGCCCTAACACAGGG | AAGTCAAGAGGCACTCGCTC | 192bp |
| Slc11a2 | NM_001399169.1 | CTCCTGGGATATGGAGTCGC | CAGAAGCACCATCGTCTGGA | 135bp |
| Gpt | NM_031039.2 | GGTGTTGTCTTACCTCTCGCA | GTGAGGCCATGACTGCTCAA | 110bp |
| Hmgcr | NM_013134.2 | CCTCCATTGAGATCCGGAGG | AAGTGTCACCGTTCCCACAA | 123bp |
| Slc27a4 | NM_001100706.1 | GTACAGACCCTACAAGGGCG | CCCACCAGAGATGCTCCAAG | 182bp |
| Elovl1 | NM_001044275.1 | GGCTGGTCAAAGGTGAGTCC | AGGCCCAAGTGATAGGACGA | 170bp |
| β-actin | NM_031144.3 | GCGCAAGTACTCTGTGTGGA | AGGGTGTAAAACGCAGCTCAG | 159bp |
Results
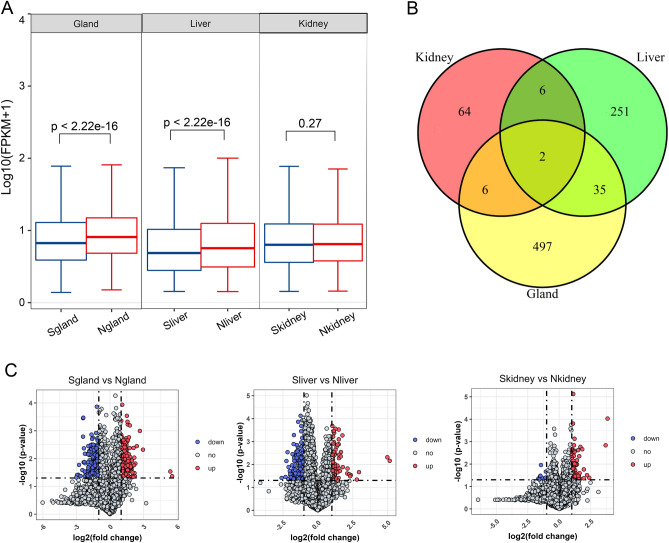
A total of 13,611 genes were detected in all samples (Supplementary Table 1), and all the normalized FPKM values of each gene are shown in the boxplot in Fig. 1A. Compared with musk secreting period, the genes expressed in musk gland and liver of non-musk secreting period were significant higher. In total, 540, 294 and 78 DEGs were detected in the musk gland, liver, and kidney, respectively, discovered by comparing musk secreting and non-musk secreting periods with |log2-fold change| > 1 and Padj < 0.05. Moreover, as illustrated in the volcano map (Fig. 1B), the DEGs included 199 upregulated genes and 341 downregulated genes in the musk gland; 78 upregulated genes and 216 downregulated genes in the liver; and 68 upregulated genes and 10 downregulated genes in the kidney in the musk secreting period compared with the non-musk secreting period, respectively. Moreover, many DEGs were enriched in the muskrat musk gland and liver, but there were fewer DEGs in the kidney tissue in the musk secreting period versus the non-musk secreting period. The set theory results are displayed in Fig. 1C, which showed that there were two DEGs (HBA and HBB) present in all three tissues of muskrats. Hemoglobin subunit alpha and beta are the main constitutes of red blood cells, which is one of the important proteins that are capable of transporting O2 and CO2 and participating in Oxidation-Reduction Reactions. The liver and musk gland both had 35 common DEGs, but every other pair contained only 6 common DEGs. For displaying entire expression information of all the detected genes of three tissues for these two groups.
Fig. 1.
The expression profile of genes in the musk gland, liver and kidney of muskrat. (A) Boxplot of all expressed genes in muskrat tissues. (B) Venn diagrams of DEGs in the musk gland, liver and kidney of muskrat. (C) Volcano maps of DEGs in the musk gland (left), liver (middle) and kidney (right) of muskrat based on the comparisons between the musk secreting and non-musk secreting periods. “S” represents samples of musk secreting period, and “N” represents samples of non-musk secreting period.
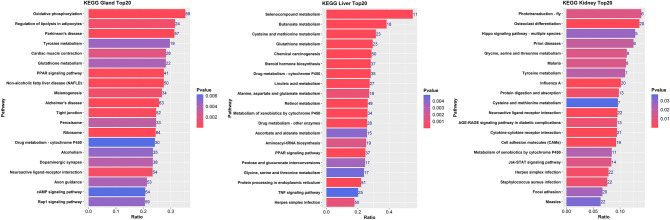
The detailed results of KEGG enrichment analysis of the DEGs by KOBAS 3.0 are shown in Fig. 2; these results helped to identify signalling pathways related to muskrat musk biosynthesis, but only DEGs with P < 0.05 between the musk secreting and non-musk secreting periods were selected. In particular, the KEGG enrichment analysis results revealed that pertinent signalling pathways closely related to glucose, lipid, and amino acid metabolism, such as oxidative phosphorylation (ko00190), regulation of lipolysis in adipocytes (ko04923), amino metabolism (ko00350, ko00480, ko00260, ko00350, ko00270), the PPAR signalling pathway (ko03320), linoleic acid metabolism (ko00591), butanoate metabolism (ko00650), etc., in musk gland, liver, and kidney tissues were influenced in the musk production periods.
Fig. 2.
The top 20 enriched pathways in the musk gland, liver, and kidney.
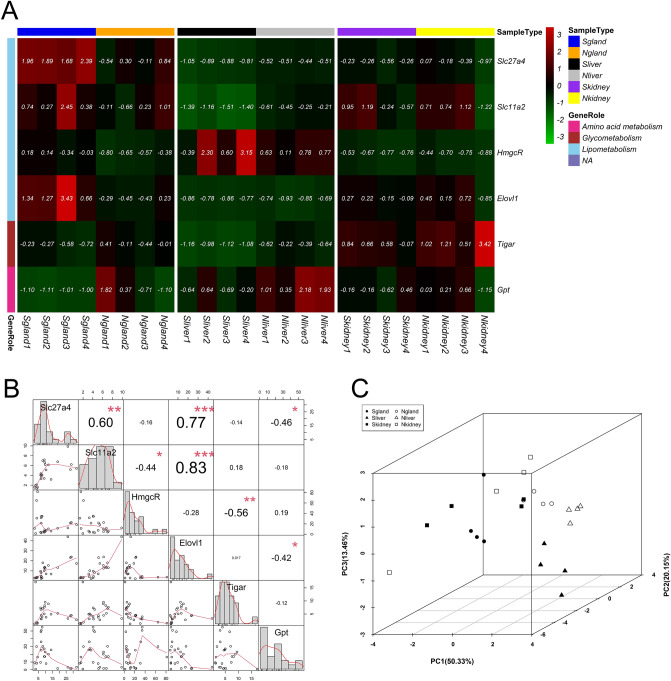
To further screen candidate genes associated with musk production in muskrat, we selected 6 candidate DEGs, namely, glutamic-pyruvic transaminase (Gpt), solute carrier family 27 member 4 (Slc27a4), solute carrier family 11 member 2 (Slc11a2), elongation of very long-chain fatty acids protein (Elovl1), 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), and TP53-induced glycolysis regulatory phosphatase (Tigar). Among which, Slc27a4, Slc11a2, Hmgcr, and Elovl1 were involved in lipometabolism; Tigar was enriched in glycometabolism; and Gpt participated in amino acid metabolism (Fig. 3A). Meanwhile, the expressional correlation between candidate genes was dissected by constructing a relevant correlation matrix, as in Fig. 3B. The correlation between gene pairs were shown in the upper right triangular matrix, and expression scatter plots of genes were illustrated in the lower left triangular matrix. The matrix frame of the symmetric line presents the expression statistics distribution of candidate genes. Moreover, the upregulated electron carriers, such as nicotinamide adenine dinucleotide dehydrogenases (Ndufa9, Ndufa1, Ndufa2, Ndufb10 and Ndufb7), were also selected as judgement indexes (Supplementary Figure 1). In liver tissue, the Ndufb10 and Ndufb7 genes were upregulated, and the Slc11a, Tigar, and Gpt genes were downregulated, during the musk secreting period. Upregulation of Slc27a4, Elovl1, Hmgcr, Ndufa9, Ndufa1, and Ndufa2 was detected in the musk gland during the musk secreting period. However, the six candidate genes and five electron carriers showed no significant difference between different stages in kidney tissue.
Fig. 3.
Identification of potential candidate genes associated with musk synthesis and secretion. (A) The expression heatmap of 6 candidate genes in all samples. (B) The correlation matrix (upper right) and expression levels (lower left) of 6 candidate genes. “*” represents p < 0.05, “**” represents p < 0.01, and “***” represents p < 0.001. (C) PCA of 6 candidate genes and physiological stages. “S” represents samples of musk secreting period, and “N” represents samples of non-musk secreting period.
To further explore the relationships between candidate genes at different periods, PCA was performed to verify the effects of candidate genes on the identification of sample types. As shown in Fig. 3C, the first three principle components explained 83.93% of the variance. Musk gland and liver samples were distinguished in the musk secreting and non-musk secreting periods using expression data for candidate genes, but the plot for kidney samples showed a random distribution.
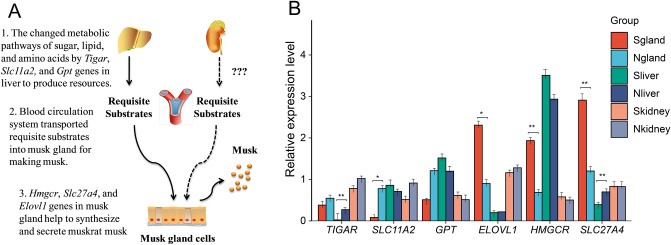
Figure 4A shows the simple mode of musk biosynthesis in organs and glands of muskrats, impacting musk production in the musk secreting period, based on the results of transcriptomic sequencing analysis. The three steps are as follows: first, the metabolic pathways of sugar, lipid, and amino acids are changed by the Tigar, Slc11a2, and Gpt genes in the liver for resource production. Second, the blood circulation system transports the requisite substrates into the musk gland for musk production. Third, the Hmgcr, Slc27a4, and Elovl1 genes in musk glands help in the synthesis and secretion of muskrat musk.
Fig. 4.
Musk biosynthesis mechanism (A) and candidate genes validation (B) in muskrats.
The candidate genes that were associated with musk production in muskrat were selected to confirm the sequencing data. Tigar, Slc11a2 and Gpt were related to the synthesis of requisite substrates in liver; Hmgcr, Slc27a4, and Elovl1 were associated to musk production in musk gland. The relative expression of genes detected by qPCR were compared between musk secreting and non-musk secreting periods. The results illustrated that Tigar, Slc11a2 and Slc27a4 over-expressed in liver, gland and liver of non-musk secreting period, respectively. While Elovl1, Hmgcr and Slc27a4 significant highly expressed in liver of musk-secreting period (Fig. 4B).
Discussion
Musk has high value in medicinal and fragrance products in the market, but the mechanism of musk biosynthesis remains largely unknown, and forest musk deer, which secrete musk, have become very rare, which limits the effective development and utilization of musk products. To overcome this obstacle, musk secreted by muskrats has been selected as a suitable substitute for musk produced by forest musk deer. In addition, it has potential applications in scientific research to comprehensively understand the mechanism of musk biosynthesis and improve musk production in the future. As we known, the liver and kidney are important organs in mammals and provide basic chemical resources by using blood circulation for biosynthesizing final products in exocrine glands 21,22. Therefore, in the current study, we screened DEGs in the liver, kidney, and musk gland tissues of muskrats by comparing individuals between the musk secreting and non-musk secreting periods using mRNA transcriptomic sequencing to explore the musk biosynthesis pathways. There were 540, 294 and 78 DEGs in musk gland, liver, and kidney tissues, respectively, which indicated that the largest change in metabolic activity occurred in the musk gland, followed by the liver and kidney. Intriguingly, HBB and HBA were found to be significantly higher expressed among three tissues in musk secreting period, which indicated the increasing hemoglobin concentration facilitated oxygen carrying capacity in the blood and enhanced oxygen delivery to other tissues. Meanwhile, oxidation-reduction reactions catalyzed by them were beneficial to the formation of acetyl-CoA and promoted its participation in various metabolic pathways to synthesize fatty acids, cholesterol, ketone bodies, etc. It hinted the intricate multi-organ synthesis mechanisms during musk production as well. According to previous evidence, on the one hand, the liver in mammals is a major organ associated with carbohydrate and lipid metabolism, including in the production of cholesterol, an important chemical analogue of musk, so the liver may generate musk pro-chemicals such as cholesterol, free fatty acids, and pyruvic acid, which are imported into musk gland cells by the blood circulation system for further processing 15,21. On the other hand, the kidney is also an important organ for amino acid reabsorption and metabolism, providing amino acids or carbon skeletons such as tyrosine and pyruvic acid as resources to support musk biosynthesis 17,18.
Combined with the KEGG enrichment analysis of DEGs in the liver and other evidence 7,11,12, these results implied the importance of the liver for musk biosynthesis; signalling pathways related to lipid and amino acid metabolism, such as steroid hormone biosynthesis and butanoate, linoleic acid, and amino acid metabolism, as well as the PPAR signalling pathway were found in liver tissue. Notably, oxidative phosphorylation, regulation of lipolysis in adipocytes, the PPAR signalling pathway, and tyrosine metabolism were found in the musk gland, similar to the results of other studies 11,23, which suggested that similar metabolic processes in the liver tissue and musk gland could support musk synthesis via the generation of basic carbon resources. In the kidney, DEGs were found to be enriched in tyrosine, glycine, serine and threonine metabolism, which further verified the hypothesis that the kidney provides amino acids or carbon skeletons for the musk biosynthesis process according to the roles of the kidney 18.
It was indicated in this study that a similar change in the metabolic pathway occurred in the liver and musk gland during musk biosynthesis in muskrat. As the liver and musk gland participate in metabolite exchange via blood circulation to ensure musk production, for instance, pyruvic acid 24 made by the liver is transported into musk glands by blood circulation as an ingredient to assist musk biosynthesis. Therefore, the set theory results again suggested the importance of the liver for musk production by the musk gland and provided more evidence in support of the hypothesis that the liver is a key organ for the generation of basic resources for sustaining musk production in the musk gland.
To identify key genes related to musk biosynthesis, we screened out some candidate genes from all the DEGs by identifying potential protein enzymes associated with musk components and the metabolic processes of carbohydrates, lipids, and amino acids, including glycolysis, gluconeogenesis, fatty acid synthesis, sterol synthesis. Result indicated that the main chemicals presented in musk, were long-chain carbon skeletons, steroids, and amino acids, such as muscone, 13-tetradecenal, 8-cyclohexadecen-1-one, cholesterol, trans-dehydroandrosterone, and tyrosine 9,10. Therefore, glutamic-pyruvic transaminase (Gpt) 25, solute carrier family 27 member 4 (Slc27a4) 26, solute carrier family 11 member 2 (Slc11a2) 27, elongation of very long-chain fatty acids protein (Elovl1) 28, 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr) 29, and TP53-induced glycolysis regulatory phosphatase (Tigar) 30 were selected as candidate genes involved in musk biosynthesis. In addition to upregulated nicotinamide adenine dinucleotide dehydrogenases (Ndufa9, Ndufa1, Ndufa2, Ndufb10 and Ndufb7) 31, as electron carriers, have been reported to display metabolic activity during musk production. Carriers of the respiratory chain, including Ndufa9, Ndufa1, Ndufa2, Ndufb10 and Ndufb7, were significantly upregulated in liver and musk gland tissues, which indicated the high abundance of oxidation–reduction reactions for musk biosynthesis in the liver and musk gland, such as carbon chain hydrogenation, carboxyl activation, and alcohol oxidation 31 improving the formation of carbon chain rings and ketones for the synthesis of muscone (3-Methylcyclopentadecanone).
Plentiful steroid substances have been detected in musk by gas chromatography–mass spectrometry, and the Hmgcr gene encodes a key rate-limiting enzyme that is upregulated in musk glands during the musk secreting period to generate steroid substances 11,23,29. Moreover, steroid substances are not only major components of musk but also critical ingredients for the production of dehydroandrosterone, which is also the main component in musk. Therefore, analysis of the expression of the Hmgcr gene in muskrat musk glands explained some aspects of the mechanism of musk biosynthesis 11,23. Upregulating of Hmgcr gene in musk glands improved the level of steroid substances in musk and influenced the physical properties and chemical composition of musk. This is an exciting direction of inquiry and it is very meaningful to explore in detail the relationship between the Hmgcr gene and musk quality, but further evidence is needed. In the musk gland, the Slc27a4 gene 26, encoding a transporter of long-chain fatty acids, was upregulated during musk secretion, which implied that effective transport of long-chain fatty acids could result in the secretion of musk from the muskrat musk gland. Interestingly, the Elovl1 gene 28, which plays critical roles in the lipid metabolism process for the elongation of long-chain fatty acids as well as sphingomyelin synthesis, was upregulated in musk glands during the musk secreting period, which indicated the importance of the Elovl1 gene in musk biosynthesis processes, such as the production of carbon skeletons or musk pro-chemicals, and helped identify novel research targets to improve musk yield in the future.
Conclusions
In summary, this paper preliminarily revealed the mechanisms of musk biosynthesis in muskrats, suggesting the significance of the liver in sugar, lipid, and amino acid biosynthesis. Lipid metabolism in musk glands has also been indicated as a key pathway for musk biosynthesis, which is quite meaningful and will be useful for clearly elucidating the process of musk biosynthesis and will contribute to improving musk production for use in medicinal and fragrance products in the near future. In addition, candidate genes, including the Tigar, Slc11a2, and Gpt genes in the liver and the Hmgcr, Slc27a4, and Elovl1 genes in the musk gland, were identified and shown to be closely related to musk biosynthesis.
Supplementary Information
Author contributions
Z.X. contributed to designing and conceptualizing the idea, writing the original draft, and editing the manuscript. Y.C. performed the experiments, organized the figures and tables, and wrote this paper. D.Z. and X.S revised the paper and analysed the data. T.Z. and C.Z. conducted animal feeding and musk sampling. X.F., L.Y. and G.Z. were responsible for the bioinformation analysis. H.J. conceived, designed and conducted this experiment. All the authors read and approved the final manuscript.
Funding
This research was funded by Chongqing Talent project (cstc2021ycjh-bgzxm0201), Fundamental Research Funds of Chongqing (2022JK017; 2023jbky-003) and the Fundamental Research Funds of China West Normal University (21E037).
Data availability
The datasets generated during the current study are available at https://www.ncbi.nlm.nih.gov/sra/?term = PRJNA751138 (accession no. PRJNA751138).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhongxian Xu and Yinglian Chen.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80080-3.
References
- 1.Wang, J. et al. Pharmacological effects and mechanisms of muscone. J. Ethnopharmacol.262, 113120 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Lee, D., Kim, Y. S., Song, J. & Kim, H. Neuroprotective effects of musk of muskrat on transient focal cerebral ischemia in rats. Evid.-based Complement. Altern. Med.: eCAM2019, 9817949 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, X. et al. Beneficial effects of muscone on cardiac remodeling in a mouse model of myocardial infarction. Int. J. Mol. Med.34(1), 103–111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh, P. B. et al. Projected distribution and climate refugia of endangered Kashmir musk deer Moschus cupreus in greater Himalaya, South Asia. Sci. Rep.10(1), 1511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang, F. et al. Musk deer (Moschus spp.) face redistribution to higher elevations and latitudes under climate change in China. Sci. Total Environ.704, 135335 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Sun, F. F. et al. GC-MS analysis of the differences in the chemical components of muskrat and musk. Nat. Prod. Res. Dev.33, 1643–1648 (2021). [Google Scholar]
- 7.Shi, X. et al. Correlation analysis between muskrat (Ondatra zibethicus) musk and traditional musk. Animals (Basel).13(10), 1678 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, D. et al. The musk chemical composition and microbiota of Chinese forest musk deer males. Sci. Rep.6, 18975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, T. et al. Study of compositions of musks in different types secreted by forest musk deer (Moschus berezovskii). PloS One16(3), e0245677 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie, W. et al. Seasonal expressions of SF-1, StAR and P450scc in the scent glands of the muskrats (Ondatra zibethicus). J. Steroid Biochem. Mol. Biol.204, 105766 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Zhang, M. et al. Regulatory roles of peroxisomal metabolic pathways involved in musk secretion in muskrats. J. Memb. Biol.252(1), 61–75 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Stoolman, J. S. & Chandel, N. S. Glucose metabolism linked to antiviral responses. Cell178(1), 10–11 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Liu, X. et al. Acetate production from glucose and coupling to mitochondrial metabolism in mammals. Cell175(2), 502-513.e13. 10.1016/j.cell.2018.08.040 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez Campos, S., Alza, N. P. & Salvador, G. A. Lipid metabolism alterations in the neuronal response to A53T alpha-synuclein and Fe-induced injury. Arch. Biochem. Biophys.655, 43–54 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Muret, K. et al. Long noncoding RNAs in lipid metabolism: literature review and conservation analysis across species. BMC Genomics20(1), 882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair, K. S. Amino acid and protein metabolism in chronic renal failure. J. Renal Nutr. Off. J. Council Renal Nutr. Nat. Kidney Found.15(1), 28–33 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Li, X., Zheng, S. & Wu, G. Amino acid metabolism in the kidneys: Nutritional and physiological significance. Adv. Exp. Med. Biol.1265, 71–95 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Kühnlein, R. P. Lipid droplet-based storage fat metabolism in Drosophila. J. Lipid Res.53(8), 1430–1436. 10.1194/jlr.R024299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols8(8), 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulley, J. F., Hargreaves, A. D., Hegarty, M. J., Heller, R. S. & Swain, M. T. Transcriptomic analysis of the lesser spotted catshark (Scyliorhinus canicula) pancreas, liver and brain reveals molecular level conservation of vertebrate pancreas function. BMC Genomics15, 1074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McManaman, J. L., Reyland, M. E. & Thrower, E. C. Secretion and fluid transport mechanisms in the mammary gland: comparisons with the exocrine pancreas and the salivary gland. J. Mammary Gland Biol. Neoplasia11(3–4), 249–268 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Han, W. et al. Seasonal expression of P450c17 and 5alpha-reductase-2 in the scented gland of male muskrats (Ondatra zibethicus). General Comp. Endocrinol.254, 60–67 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Wespi, P., Steinhauser, J., Kwiatkowski, G. & Kozerke, S. Overestimation of cardiac lactate production caused by liver metabolism of hyperpolarized [1-(13) C]pyruvate. Magnetic Reson. Med.80(5), 1882–1890 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Lin, J. D. et al. Serum glutamic-oxaloacetic transaminase (GOT) and glutamic-pyruvic transaminase (GPT) levels in children and adolescents with intellectual disabilities. Res. Dev. Disab.31(1), 172–177 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Xu, Z. Y., Xiong, Y. Z., Lei, M. G., Li, F. E. & Zuo, B. Genetic polymorphisms and preliminary association analysis with production traits of the porcine SLC27A4 gene. Mol. Biol. Rep.36(6), 1427–1432 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Wang, H. et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology66(2), 449–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sassa, T., Tadaki, M., Kiyonari, H. & Kihara, A. Very long-chain tear film lipids produced by fatty acid elongase ELOVL1 prevent dry eye disease in mice. FASEB J.Off. Publ. Fed. American Soc. Exp. Biol.32(6), 2966–2978 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Li, X. et al. Epigenetic regulation of key enzymes CYP7a1 and HMGCR affect hepatic cholesterol metabolism in different breeds of piglets. Front. Vet. Sci.7, 231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy, N. Metabolism: a TIGAR tale. Nat. Rev. Cancer13(8), 522 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Breitenbach, M. et al. The defense and signaling role of NADPH oxidases in eukaryotic cells : Review. Wiener medizinische Wochenschrift168(11–12), 286–299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chini, C. C. S., Zeidler, J. D., Kashyap, S., Warner, G. & Chini, E. N. Evolving concepts in NAD(+) metabolism. Cell Metab.33(6), 1076–1087. 10.1016/j.cmet.2021.04.003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available at https://www.ncbi.nlm.nih.gov/sra/?term = PRJNA751138 (accession no. PRJNA751138).